Abstract
DNA barcoding is emerging as a useful tool not only for species identification but also for studying evolutionary and ecological processes. Although plant DNA barcodes do not always provide species-level resolution, the generation of large DNA barcode data sets can provide insights into the mechanisms underlying the generation of species diversity. Here, we study evolutionary processes in taxonomically complex British Euphrasia (Orobanchaceae), a group with multiple ploidy levels, frequent self-fertilization, young species divergence and widespread hybridization. We use a phylogenetic approach to investigate the colonization history of British Euphrasia, followed by a DNA barcoding survey and population genetic analyses to reveal the causes of shared sequence variation. Phylogenetic analysis shows Euphrasia have colonized Britain from mainland Europe on multiple occasions. DNA barcoding reveals that no British Euphrasia species has a consistent diagnostic sequence profile, and instead, plastid haplotypes are either widespread across species, or are population specific. The partitioning of nuclear genetic variation suggests differences in ploidy act as a barrier to gene exchange, while the divergence between diploid and tetraploid ITS sequences supports the polyploids being allotetraploid in origin. Overall, these results show that even when lacking species-level resolution, analyses of DNA barcoding data can reveal evolutionary patterns in taxonomically complex genera.
Keywords: British flora, DNA barcoding, Euphrasia, Orobanchaceae, phylogeny, polyploidy, taxonomic complexity
The sequencing of short standardized pieces of DNA (DNA barcodes) is commonly used to tell species apart. This approach is thought to be of limited value in complex plant groups where species have evolved recently. We use DNA barcoding in taxonomically complex British Euphrasia, and while we find no species with a diagnostic sequence profile, we do see evolutionary patterns, such as divergence between two different species groups with different chromosome numbers. This shows DNA barcoding can be useful for studying evolutionary patterns in complex plant groups.
Introduction
DNA barcoding is a valuable tool for discriminating among species, and these data often give insights into identity that are overlooked based on morphology alone (Hebert and Gregory 2005). DNA barcoding relies on sequencing a small set of gene regions (such as the core plant DNA barcode rbcL + matK, often supplemented with ITS and other regions), and using these data for species identification (CBOL Plant Working Group 2009). Successful applications of DNA barcoding include species discovery, reconstructing historical vegetation types from frozen sediments, surveying environmental variation and many other research topics (reviewed in Hollingsworth et al. 2016). However, there are numerous reports of taxon groups where the standard DNA barcode sequences do not provide exact plant species identification, and where DNA barcode sequences are shared among related species (Spooner 2009; Percy et al. 2014; Zarrei et al. 2015; Yan et al. 2015a, b). Even in these cases, however, the generation of large data sets of DNA sequences from multiple individuals of multiple species can shed light onto evolutionary relationships and patterns of divergence, without a need for the barcode markers to track species boundaries.
Postglacial species radiations of taxonomically complex groups in Northern Europe are a case where we may not expect a clear cut-off between intraspecific variation and interspecific divergence and thus DNA barcoding may provide limited discriminatory power. Such postglacial groups include the Arabidopsis arenosa complex (Schmickl et al. 2012), Cerastium (Brysting et al. 2007), Epipactis (Squirrell et al. 2002) and Galium (Kolář et al. 2015). Despite this complexity, DNA barcoding may still be valuable if used to identify evolutionary and ecological processes that result in shared sequence variation. For example, many postglacial taxa are characterized by a combination of: (i) recent postglacial speciation, (ii) extensive hybridization, (iii) frequent self-fertilization, (iv) divergence involving polyploidy. Our expectation is that factors (i) + (ii) will cause DNA barcode sequences to be shared among geographically proximate taxa, while (iii) will cause barcodes to be population rather than species specific (Hollingsworth et al. 2011; Naciri et al. 2012). Factor (iv), polyploidy, will manifest as shared variation between recent polyploids and their parental progenitors, or deep allelic divergence in older polyploid groups, where ploidy acts as a reproductive isolating barrier and allows congeneric taxa to accumulate genetic differences.
One example of a taxonomically challenging group showing postglacial divergence is British Euphrasia species (Ennos et al. 2005). This group of 19 taxa is renowned for their difficult species identification, and at present only a handful of experts can identify these species in the field. Morphological species identification is difficult due to their small stature, combined with species being defined by a complex suite of overlapping characters (Yeo 1978). They are also generalist hemiparasites and thus phenotypes are plastic and may depend upon host quality (Svensson and Carlsson 2004). DNA barcode-based identification could partly resolve these identification issues, and lead to a greater understanding of species diversity and distributions in this under-recorded group. This is particularly important as a number of Euphrasia species are critically rare and of conservation concern, while others are ecological specialists that are useful indicators of habitat type (French et al. 2008). More generally, DNA barcoding data could reveal the processes structuring genetic diversity and those that are responsible for recent speciation.
Previous broad-scale surveys of Euphrasia using amplified fragment length polymorphisms (AFLPs) and microsatellites have shown a significant proportion of genetic variation is partitioned between two ploidy groups (diploids and tetraploids), and by species, despite extensive hybridization (French et al. 2008). Here, we follow-on from this population genetic study by using DNA sequence data to investigate the processes underlying the regional assembly of British Euphrasia diversity. Our first aim is to understand whether British endemic Euphrasia are a product of speciation within a single clade, or if speciation has occurred within multiple groups of genetically diverse European relatives. We address this question of regional assembly by placing British Euphrasia species in the context of a global Euphrasia phylogeny. In the light of our phylogenetic analysis, our second aim is to deploy DNA barcoding across a large British sample set to characterize patterns of genetic diversity and the potential factors underlying shared sequence variation. The combination of our two data sets also sheds light on the role that polyploidy plays in shaping genetic diversity in Euphrasia, with our phylogenetic analysis revealing whether polyploidization has occurred recently in British taxa (or occurred before colonizing the UK), while our DNA barcoding shows whether ploidy differences are a barrier to gene exchange. Overall, these results are used to improve our understanding of the evolution of a complex regional plant assembly, and to test the efficacy of DNA barcoding for studying species-level variation in a taxonomically complex group.
Methods
Specimen sampling
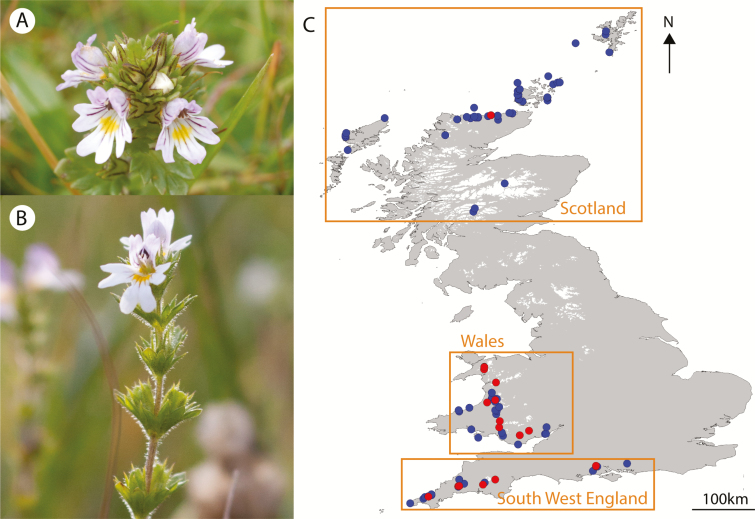
The 19 currently recognized British Euphrasia species are all annual, selfing or mixed-mating small herbaceous plants, which occur in a range of habitats including coastal turf, chalk downland, mountain ridges and heather moorland (French et al. 2004). The species can be divided into two groups, glabrous or short eglandular hairy tetraploids (15 species, Fig. 1A), or long glandular hairy diploids (4 species, Fig. 1B). Our sampling includes representatives of all British species (Fig. 1C; Supporting Information—Table S1). Samples were collected in South West England and Wales to allow us to include mixed populations of diploids and tetraploids, early generation diploid × tetraploid hybrids, and two diploid hybrid species hypothesized to be derived from diploid × tetraploid crosses (E. vigursii, parentage: E. rostkoviana × E. micrantha; E. rivularis, parentage: E. anglica × E. micrantha; Yeo 1956). Samples from Scotland allows us to sample complex tetraploid taxa and tetraploid hybrids, plus scarcer Scottish diploids. Our sampling scheme investigated range-wide variation by targeting many taxa and populations, with a focus on collecting multiple species in areas of sympatry. We chose not to include detailed intrapopulation sampling because prior work has shown low intrapopulation diversity, with populations frequently fixed for a given allele (French et al. 2008). All samples collected prior to 2012 were identified by former Euphrasia referee Alan Silverside, while recent samples were identified by current referee Chris Metherell.
Figure 1.
Euphrasia samples used in this study. (A) Tetraploid British Euphrasia (here E. arctica) have glabrous leaves sometimes with sparse short eglandular hairs or bristles. (B) Diploid British Euphrasia have long glandular hairs. (C) Collection sites of Euphrasia DNA samples. Diploids are shown in red, tetraploids in blue. Orange boxes correspond to the three broad sampling areas. Photo credits: Alex Twyford, Max Brown.
For our molecular phylogenetic analysis aimed at understanding the colonization history of British Euphrasia, we expanded the sampling in the phylogeny of the genus by Gussarova et al. (2008) to include a detailed sample of British taxa. The previous analysis included 41 taxa for the nuclear ribosomal internal transcribed spacer (ITS), and 50 taxa for plastid DNA (Gussarova et al. 2008). We sequenced samples to match the previous data matrix, which included: the trnL intron (Taberlet et al. 1991), intergenic spacers atpB-rbcL (Hodges and Arnold 1994) and trnL-trnF (Taberlet et al. 1991), and ITS (White et al. 1990).
For our population-level DNA barcoding study, we analysed a total of 133 individuals, with 106 samples representing 19 species, as well as 27 samples from 14 putative hybrid taxa. We sequenced samples for the core plant DNA barcoding loci, matK and rbcL (CBOL Plant Working Group 2009), as well as partial sequences of ITS (ITS2), which has been suggested to be incorporated into the core DNA barcode (China Plant BOL Group 2011). We also followed the recommendation of Hollingsworth et al. (2011), to add a non-coding gene to our set of plastid loci to help resolve recent haplotype divergence. We used rpl32-trnLUAG, which has been informative in prior population studies of Euphrasia (Stone 2013).
DNA extraction, PCR amplification and sequencing
DNA was extracted from silica-dried tissue using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol, but with an extended incubation of 1 h at 65 °C. These DNA samples were added to existing DNA extractions of 68 individuals from French et al. (2008).
We performed PCRs in 10 μL reactions, with DNA amplification and PCR conditions for each primer given in Supporting Information—Table S2. We visualized PCR products on a 1 % agarose gel, with 5 μL of PCR product purified for sequencing with ExoSAP-IT (USB Corporation, Cleveland, OH, USA) using standard protocols. Sequencing was performed in 10 μL reactions containing 1.5 μL 5× BigDye buffer (Life Technologies, Carlsbad, CA, USA), 0.88 μL BigDye enhancing buffer BD × 64 (MCLAB, San Francisco, CA, USA), 0.125 μL BigDye v3.1 (Life Technologies), 0.32 μM primer and 1 μM of purified PCR product. We sequenced PCR products on the ABI 3730 DNA Analyser (Applied Biosystems, Foster City, CA, USA) at Edinburgh Genomics. In addition to these newly generated sequences, a subset of sequences were generated as part of the effort to DNA barcode the UK Flora, and followed a different set of protocols, detailed in de Vere et al. (2012).
We assembled, manually edited and aligned sequences using Geneious v. 8 (Biomatters, Auckland, New Zealand). We scored indels as unordered binary characters and appended them to the matrices. We used gap coding as implemented in Gapcoder (Young and Healy 2003), with indels treated as point mutations and equally weighted with other mutations.
Phylogenetic analysis of global Euphrasia
We used Bayesian phylogenetic analyses in MrBayes v. 3.1.2 (Huelsenbeck and Ronquist 2001) to infer species relationships and broad-scale patterns of colonization. Our analyses used a sequence matrix that included our newly sampled British taxa in addition to previous global Euphrasia samples from Gussarova et al. (2008). We selected the best fitting model of nucleotide substitution using the Akaike Information Criterion (AIC) with an empirical correction for small sample sizes implemented in MrAIC (Nylander 2004). Using GTR + G as the best model for the plastid data set and SYM + G for the ITS data set we ran two sets of four Markov Chain Monte Carlo (MCMC) runs for 5000000 generations. Indels were included as a separate partition with a restriction site (binary) model. We sampled every 1000th generation and discarded the first 25 % as burn-in. We confirmed chain convergence by observing the average standard deviation of split frequencies and by plotting parameter values in Tracer v. 1.6 (Rambaut and Drummond 2013). The alignment and trees are deposited in TreeBase under accession number 22492 (https://treebase.org).
DNA barcoding survey of British taxa
We examined patterns of sequence variation using a range of population genetic methods. We investigated the amount of sequence diversity across species using descriptive statistics, and then tested the cohesiveness of taxa using analysis of molecular variance (AMOVA) and related methods. Analyses were performed separately on ITS2 and a concatenated matrix of either all sampled plastid loci, or just the core DNA barcode loci. For plastid data, haplotypes were determined from nucleotide substitutions and indels of the aligned sequences. Basic population genetic statistics were performed in Arlequin Version 3.0 (Excoffier and Lischer 2010), and this included the number of haplotypes, as well as hierarchical AMOVA in groups according to: (i) ploidy level (diploid vs. tetraploid); (ii) geographic regions (Wales, England, Scotland); (iii) species. Analysis of molecular variance was performed on all taxa, and repeated for ploidy levels and geographic regions on a data set only including confirmed species (i.e. excluding hybrids). Sequence diversity and divergence statistics were estimated with DnaSP (Librado and Rozas 2009), which included: average nucleotide diversity across taxa (π), Watterson’s θ (per site), Tajima’s D and divergence between ploidy levels (DXY).
Genetic divergence among sampling localities was explored with spatial analysis of molecular variance (SAMOVA; Dupanloup et al. 2002), implemented in SPADS v.1.0 (Dellicour and Mardulyn 2014). Spatial analysis of molecular variance maximizes the proportion of genetic variance due to differences among populations (FCT) for a given number of genetic clusters (K-value). We considered the best grouping to have the highest FCT value after 100 repetitions. This analysis investigated interspecific differentiation, thus only used species samples, excluding hybrids.
The relationships between haplotypes were inferred by constructing median-joining networks (Bandelt et al. 1999) with the program NETWORK v.4.6.1.1 (available at http://www.fluxus-engineering.com/), treating gaps as single evolutionary events.
Results
Global phylogenetic analysis
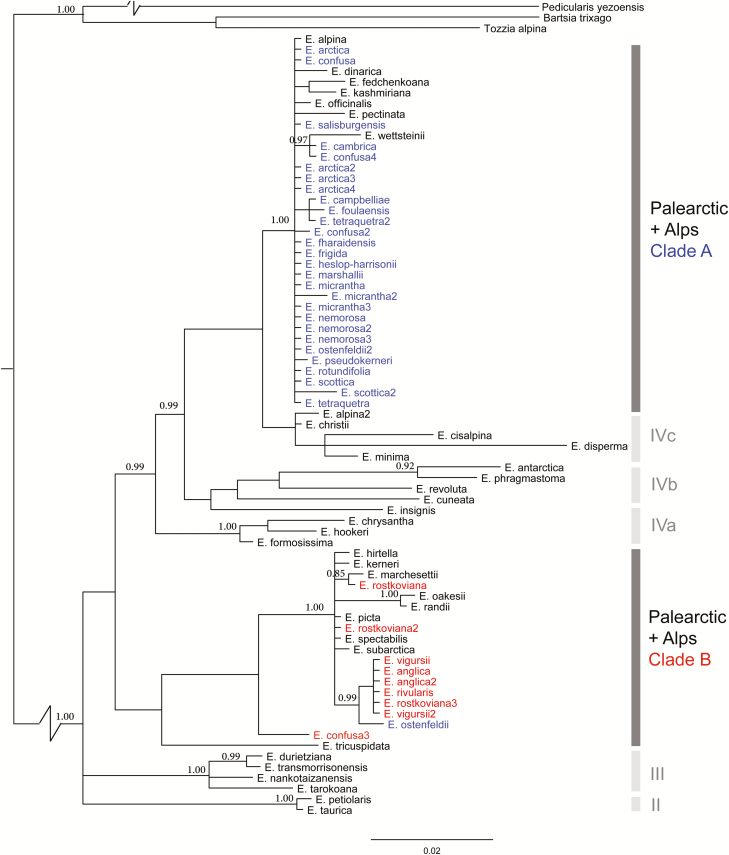
The final ITS alignment was 638 bp in length, for a total of 76 Euphrasia samples, including those from Gussarova et al. (2008). Our broad-scale global Euphrasia phylogenetic analysis performed using MrBayes gave meaningful clusters of species, though the tree topology was generally poorly supported with many polytomies (Fig. 2). British taxa predominantly belonged to two clusters: a tetraploid clade of Holarctic taxa from Sect. Euphrasia (posterior probability support, pp = 1.00, Clade A, Fig. 2), and a well-supported geographically restricted Palearctic diploid lineage (pp = 1.0, Clade B, Fig. 2). The tetraploid clade included a mix of British and European taxa, and is sister to a mixed clade of alpine diploid species and tetraploid E. minima (Clade IVc, Fig. 2). The diploid clade includes British diploids E. anglica, E. rivularis, E. rostkoviana, E. vigursii and European relatives (diploids or taxa without chromosome counts). The only non-diploid in the clade is one individual of tetraploid British E. ostenfeldii, which appears to be correctly identified and thus may have captured the diploid ITS variant through hybridization. Overall, terminal branches of the tree are short, indicative of limited divergence. The only exception was the long branch of E. disperma from New Zealand, a result seen in previous Bayesian analyses (cf. Gussarova et al. 2008, Fig. 2) but not in parsimony analyses, where it clusters together with the other southern hemisphere species on a shorter branch (Gussarova et al. 2008).
Figure 2.
Majority rule consensus phylogeny of Euphrasia inferred from the ITS region using MrBayes. Posterior probabilities >0.85 are indicated. Individuals are coloured by ploidy and geography: British diploids (red), British tetraploids (blue), other geographic areas (black). Clade A and Clade B correspond to the main study groups, with additional clades corresponding to Gussarova et al. (2008) also marked: II northern tetraploids; III Taiwan; IVa South American/Tasmanian; IVb complex (South American, New Zealand, Japan); IVc Alpine European.
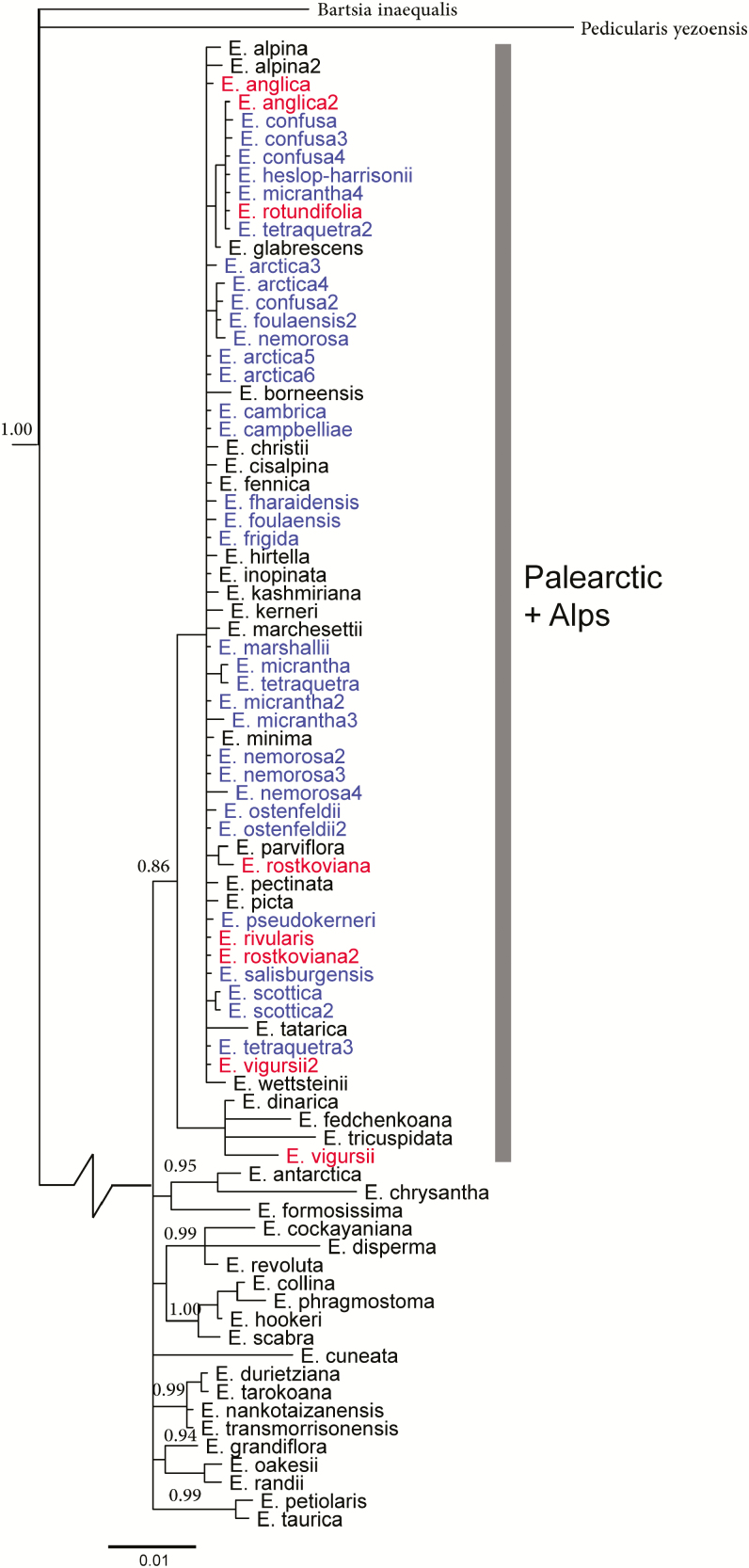
The final concatenated plastid alignment was 1692 bp in length, for a total of 82 Euphrasia samples, including those from Gussarova et al. (2008). This alignment included the trnL intron (517 bp, 73 variable sites), trnL-trnF (420 bp, 85 variable sites) and atpB-rbcL (754 bp, 89 variable sites). The plastid tree (Fig. 3) recovered the geographic clades reported in Gussarova et al. (2008). All diploid and tetraploid British samples possessed plastid haplotypes from the broad Palearctic clade, which also includes E. borneensis (Borneo) and E. fedtschenkoana (Tian Shan). This clade received moderate support in our analysis (pp = 0.85). While partially informative of broad-scale relationships, most terminal branches were extremely short, and gave no information on interspecific relationships.
Figure 3.
Majority rule consensus phylogeny of Euphrasia inferred from a concatenation of plastid trnL intron, trnL-trnF and atpB-rbcL using MrBayes. Posterior probabilities >0.85 are indicated, and British diploids (red) and British tetraploids (blue) are coloured.
UK DNA barcoding
ITS diversity.
The final ITS2 alignment contained 130 individuals representative of all 19 British taxa, and was 380 bp in length. Only two samples (of E. scottica) presented double peaks, and were excluded from analyses. Overall diversity across taxa was modest, with a nucleotide diversity (π) of 2.3 %, and θ (per site) of 0.01781. There were 33 nucleotide substitutions and one indel, from which we called 23 alleles.
The ITS2 data revealed strong partitioning by ploidy. Of the 23 alleles, three (H1, H20 and H21) were restricted to diploids, and 19 to tetraploids, with only one allele (H2) shared across ploidy levels (Table 1). H2 was not only shared across ploidy levels but was also the most widespread variant, found in 67 samples across 34 populations. This included geographically distinct species such as the Scottish endemic E. marshallii and the predominantly English and Welsh E. anglica, and ecologically contrasting taxa such as the dry heathland specialist E. micrantha and the (currently unpublished) obligate coastal ‘E. fharaidensis’. Overall, 86 % of taxa had one of six widespread alleles. There were also a large number of rare variants, with over two-thirds restricted to a single population (17 alleles: H4, H7–H10, H12–H21 and H23; Table 1). The remaining alleles found in multiple populations (H1, H2, H3, H5, H6, H11) showed no clear pattern of geography, with three found in all geographic regions (England, Scotland, Wales) and the remaining three shared between two geographic regions. Similarly, patterns of shared sequence variation do not follow species boundaries. Of the eight species with multiple populations (excluding hybrids), none of them had a diagnostic ITS2 sequence. Despite variants being shared across taxa, there was no evidence for this being due to non-neutral processes, as the value of Tajima’s D (−0.17) was not significantly different from zero.
Table 1.
The distribution of ITS2 variation between species and geographic regions for British Euphrasia. Allele numbers correspond to the network in Fig. 4. Population-specific variants are aggregated under one column. n = number of samples. Regions refer to: W, Wales; S, Scotland; SW, South West England.
| Taxa name | Region | n | Widespread haplotypes | Population-specific haplotypes | |||||
|---|---|---|---|---|---|---|---|---|---|
| H1 | H2 | H3 | H5 | H6 | H11 | ||||
| E. anglica | SW | 4 | 4 | 0 | |||||
| E. anglica | W | 3 | 2 | 1 | 0 | ||||
| E. arctica | S | 3 | 1 | 2 | 0 | ||||
| E. arctica | SW | 2 | 2 | 0 | |||||
| E. arctica | W | 2 | 1 | 1 | 0 | ||||
| E. arctica × confusa | S | 1 | 1 | ||||||
| E. arctica × foulaensis | S | 1 | 1 | 0 | |||||
| E. arctica × micrantha | S | 3 | 1 | 1 | 1 | 0 | |||
| E. arctica × nemorosa | S | 2 | 1 | 1 | 0 | ||||
| E. arctica × rostkoviana | S | 3 | 1 | 2 | 0 | ||||
| E. cambrica | W | 3 | 1 | 1 | 1 | ||||
| E. campbelliae | S | 3 | 3 | 0 | |||||
| E. confusa | S | 4 | 4 | 0 | |||||
| E. confusa | SW | 3 | 1 | 1 | 1 | ||||
| E. confusa | W | 3 | 2 | 1 | |||||
| E. confusa × micrantha | S | 2 | 1 | 1 | |||||
| “E. fharaidensis” | S | 2 | 1 | 1 | 0 | ||||
| E. foulaensis | S | 6 | 2 | 2 | 2 | 0 | |||
| E. foulaensis × marshllii | S | 2 | 1 | 1 | 0 | ||||
| E. foulaensis × nemorosa | S | 1 | 1 | 0 | |||||
| E. foulaensis × ostenfeldii | S | 1 | 1 | 0 | |||||
| E. frigida | S | 6 | 5 | 1 | 0 | ||||
| E. heslop-harrisonii | S | 6 | 5 | 1 | |||||
| E. marshllii | S | 3 | 3 | 0 | |||||
| E. marshallii × micrantha | S | 2 | 2 | 0 | |||||
| E. micrantha | S | 5 | 4 | 1 | |||||
| E. micrantha | SW | 3 | 2 | 1 | |||||
| E. micrantha | W | 3 | 2 | 1 | |||||
| E. micrantha × nemorosa | SW | 1 | 1 | 0 | |||||
| E. micrantha × scottica | W | 4 | 4 | 0 | |||||
| E. nemorosa | S | 3 | 3 | 0 | |||||
| E. nemorosa | SW | 2 | 1 | 1 | |||||
| E. nemorosa | W | 3 | 2 | 1 | |||||
| E. nemorosa × tetraquetra | SW | 1 | 1 | 0 | |||||
| E. ostenfeldii | S | 5 | 3 | 2 | |||||
| E. ostenfeldii | W | 1 | 1 | 0 | |||||
| E. pseudokerneri | W | 3 | 2 | 1 | 0 | ||||
| E. rivularis | W | 3 | 2 | 1 | |||||
| E. rostkoviana | W | 3 | 2 | 1 | |||||
| E. rotundifolia | S | 1 | 1 | 0 | |||||
| E. scottica | S | 3 | 3 | 0 | |||||
| E. scottica | W | 3 | 3 | ||||||
| E. tetraquetra | SW | 3 | 3 | 0 | |||||
| E. tetraquetra | W | 3 | 1 | 2 | 0 | ||||
| E. tetraquetra × vigursii | SW | 2 | 1 | 1 | 0 | ||||
| E. vigursii | SW | 4 | 4 | 0 | |||||
| Total | 130 | 15 | 67 | 12 | 4 | 11 | 3 | 18 | |
The putative hybrid species, E. vigursii and E. rivularis, possessed allele H1, which is common to other diploid taxa, or population-specific variants (H20, H21), but no tetraploid-specific allelic variation. The two sampled diploid–tetraploid hybrids (E. arctica × rostkoviana, E. tetraquetra × vigursii) possessed the full range of alleles: H2, which is common across ploidy levels, tetraploid-specific H3 and diploid-specific H1. Most (9/12) tetraploid hybrid populations had alleles shared with their putative parents, while the other populations had unique alleles.
The highest FCT value in the SAMOVA was when K = 2 [see Supporting Information—Table S3], and this corresponded to the diploid–tetraploid divide described above. At K = 3, SAMOVA distinguished clusters corresponding to the two ploidy groups, and a third group of hybrid species derived from inter-ploidy mating. Analysis of molecular variance also supported the strong division by ploidy, with 88.2 % of variation attributed to ploidy differences (Table 2; P < 0.001). A high proportion of variation was also partitioned by taxa (63.2 %), and regions (25 %), though this may be inflated by limited sampling within species.
Table 2.
Hierarchical AMOVA of British Euphrasia populations. Analyses performed between (A) taxa, (B) three geographic locations (Wales, South West England, Scotland), (C) diploids and tetraploids. Number in parentheses is the result only including species (excluding hybrids). d.f. = degrees of freedom. **P < 0.001; *P < 0.05.
| Source of variation | ITS | Plastid DNA | ||
|---|---|---|---|---|
| d.f. | % Total variance | d.f. | % Total variance | |
| (A) Taxa | ||||
| Between taxa | 33 (19) | 63.17** (65.62**) | 33 (19) | 15.87** (26.48**) |
| Within taxa | 96 (77) | 36.83 (34.38) | 96 (68) | 84.13 (73.52) |
| (B) Location | ||||
| Between regions | 2 (2) | 25.52** (25.92**) | 2 (2) | 5.40** (4.91*) |
| Within regions | 127 (94) | 74.48 (74.08) | 127 (85) | 94.60 (95.09) |
| (C) Ploidy | ||||
| Between ploidy groups | 1 (1) | 88.24** (84.39**) | 1 (1) | 11.05* (18.76*) |
| Within diploids and tetraploids | 123 (95) | 11.76 (15.61) | 123 (86) | 88.95 (81.24) |
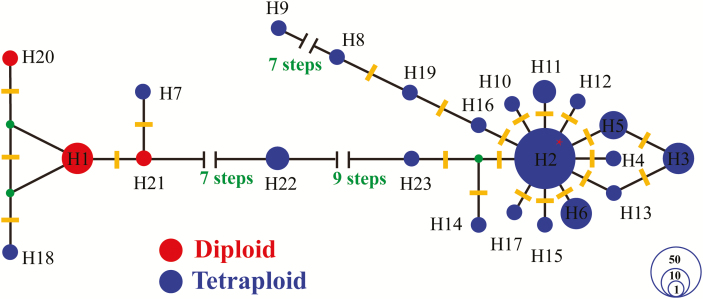
Summary statistics and a network analysis revealed substantial divergence between diploid and tetraploid variants. An average of 18.3 site differences were found between unique diploid and tetraploid alleles, with divergence measured as DXY = 0.051 (5.1 %). The phylogenetic network revealed broad clusters that largely corresponded to diploids and tetraploids, separated by many mutations (Fig. 4). The diploid cluster centres round allele H1, found in 15 individuals from five diploid species and one diploid–tetraploid hybrid. The only allele from this part of the network present in tetraploids is H18, found in a single sample of E. ostenfeldii. Within the predominantly tetraploid cluster, widespread variant H2 is at the centre (found across tetraploids and a single diploid individual), surrounded by other widespread tetraploid variants (H3, eight populations, 12 samples; H6, seven populations, 11 samples), and singleton variants.
Figure 4.
Median-joining network of ITS2 sequences in British Euphrasia. Numbers correspond to the ITS2 copies in Table 1. Alleles are coloured by ploidy, with diploids in red and tetraploids in blue. Hypothetical (unsampled) alleles are represented by filled green circles. H2 is the only allele shared across ploidy groups, found in 66 tetraploids and 1 diploid (indicate by the red asterisk). Yellow bar represents variation steps. The circle size represents the approximate numbers of individuals, and the scale is provided in the lower right corner.
Plastid haplotype diversity.
Initial sequencing of rbcL in 48 samples revealed no polymorphism and was excluded from subsequent analyses. The final matK alignment was 844 bp with one indel, and revealed 17 haplotypes. Our supplemental locus, the rpl32-trnL region, was 630 bp with nine indels. The final concatenated alignment of variable plastid loci (matK + rpl32-trnL) was 1474 bp for 130 samples, with 2.7 % segregating sites (40 sites) across 38 haplotypes. Nucleotide diversity was exceptionally low with π = 0.3 %, and θ (per site) was similarly low at 0.00381. Tajima’s D was not significantly different from zero (−0.27).
Similar to ITS, most plastid haplotypes were individual or population specific (63 %, 24/38 haplotypes found in one population only), with only four haplotypes being widespread (H4: 15 populations, 24 samples; H5: 13 populations, 18 samples; H2: 11 populations, 16 samples; H1, 10 populations, 14 samples; Supporting Information—Table S4). However, unlike ITS, plastid haplotypes revealed complex patterns unrelated to ploidy. Most widespread haplotypes were shared across ploidy levels. An AMOVA found a moderate degree of genetic diversity was partitioned by ploidy (18.7 %) and species (26.5 %), with these values being reduced when hybrids were included [see Supporting Information—Table S2]. Despite geography explaining little of the variation across the total data set (4.9 %), or being evident in the SAMOVA [see Supporting Information—Table S5], localized haplotype sharing was apparent in Scottish tetraploids. For example, haplotype H7 is shared across Scottish populations of E. arctica (and its hybrids), E. foulaensis and E. micrantha (and its hybrids), while H10 is also shared across three species in Scotland. Results from just the barcoding locus matK were consistent with the concatenated alignment of variable plastid loci [see Supporting Information—Tables S6 and S7].
Discussion
In this study, we used a broad-scale phylogenetic analysis and a regional DNA barcoding survey to investigate the evolution of taxonomically complex Euphrasia in Britain. Our phylogeny revealed deep divergence of ITS sequences between two clades of British Euphrasia, supporting the regional assembly of species diversity from a diverse pool of continental taxa. Our DNA barcoding survey revealed that British Euphrasia species do not possess diagnostic ITS and plastid sequence profiles, and instead variants are either widespread across taxa, or are individual or population specific. However, fixed sequence differences between sympatric diploids and tetraploids suggest ploidy is an important reproductive barrier partitioning genetic variation. Overall, our results shed light on the maintenance of genetic variation in one of the most renowned taxonomically challenging plant groups.
Colonization history and polyploidy in British Euphrasia
Characterizing the factors driving species richness requires an understanding of the relative importance of local and regional processes. Our phylogenetic analysis allows us to estimate the number of times that Euphrasia has colonized Britain from mainland Europe, after which local speciation events give rise to endemic taxa. British taxa are found in two clades of the Euphrasia phylogeny, with each clade nested within diverse European relatives. The dated molecular phylogeny of Gussarova et al. (2008) shows divergence of the crown group including the two clades of British taxa occurred at around 8 million years ago (Ma), with diversity within each clade accumulating after that date but before c. 1 Ma (A. D. Twyford et al., unpubl. res.). As such, clade divergence, and much of the speciation present within British Euphrasia, long pre-dates recent glacial divergence and the origin of young British endemic taxa. Overall, this suggests that British Euphrasia diversity has been assembled from a diverse pool of European and amphi-Atlantic taxa. Evidence for recurrent rounds of colonization from continental Europe have also been seen in phylogeographic studies of various other taxa, in particular tree species such as Fagus sylvatica (Rendell and Ennos 2003) and Quercus petraea (Cottrell et al. 2002).
It is notable from our DNA barcoding survey that ITS sequence variation is strongly partitioned by ploidy, despite the fact that diploids and tetraploids often grow in sympatry and are known to hybridize (Stace et al. 2015). This supports previous population surveys with AFLPs (French et al. 2008) where strong reproductive barriers are inferred between ploidy groups. The extent of divergence between diploid and tetraploid ITS sequences adds further weight to the British tetraploids being allopolyploids. Other lines of support for an allotetraploid origin come from the high number of tetraploid-specific AFLP bands (French et al. 2008), fixed microsatellite heterozygosity indicative of disomic inheritance (Stone 2013), and tetraploid genome assemblies of double the size of the diploids (A. D. Twyford and R. W. Ness, unpubl. data). While it is difficult to decipher the parentage of British tetraploids from our data, our phylogenetic analysis places these tetraploid British taxa in a clade composed exclusively of tetraploids (Clade IVd, Gusarova et al. 2008), and thus the polyploid event likely pre-dates dispersal to the UK.
DNA barcoding in taxonomically complex genera
Taxonomically complex Euphrasia have many characteristics that would make a DNA-based identification system desirable. In particular, molecular identification tools could be used to confirm species identities and subsequently revise the distribution of these under-recorded taxa. More generally, genetic data could be used to investigate which British species are genetically cohesive ‘good’ taxa. Our data show, however, that barcode sequences do not correlate with species boundaries as defined by morphology. Instead, variants are often individual or population specific (for both ITS and plastid DNA), and where widespread are either shared across species within a ploidy level (for ITS) or across ploidy levels and species (plastid DNA). Patterns of shared sequences are often surprising, including between geographically disparate populations 100+ km apart, and between ecologically specialized taxa that seldom co-occur in the wild.
One possibility is that the species are not discrete genetic entities, and that the current taxonomy reflects a blend of discrete lineages, polytopic taxa and morphotypes determined by a small number of genes. Alternatively, the species may represent meaningful biological entities, but with boundaries permeable to gene flow. In either case, the lack of species-specific or morphotype-specific barcodes may be attributed to one of many evolutionary factors, including recent speciation, hybridization and self-fertilization. Recent speciation and lack of sequence variation no doubt affect the resolution of our phylogeny, and in part affect our DNA barcoding survey to look at patterns of shared haplotypic variation. While elevated plastid diversity is a hallmark of some parasitic taxa, this is not the case for facultative hemiparasites like Euphrasia, which generally show similar patterns of mutation to autotrophic taxa (Wicke et al. 2016). The opposing forces of self-fertilization, which results in population-level differentiation, and hybridization, which may cause transpecific polymorphism, will leave a complex signal of genetic variation. The large number of reported hybrids and hybrid species based on morphology (Preston and Pearman 2015; Stace et al. 2015), as well as the prevalence of hybridization in genetic data (Liebst 2008; Stone 2013), point to hybridization being a key factor shaping genetic diversity in Euphrasia. Future genomic surveys will pinpoint the scale at which genetic variation is partitioned, and estimate the proportion of loci introgressing across species barrier in models that explicitly account for incomplete lineage sorting (Twyford and Ennos 2012).
The sharing of DNA barcode sequences among related Euphrasia species parallels a number of other studies where DNA barcoding has failed to provide species-level information. Extensive plastid haplotype sharing has been reported between two species of Rhinanthus (Vrancken et al. 2012), a related genus in the Orobanchaceae. Another notable example is in willows, where hybridization and selective sweeps have caused a single haplotype to spread across highly divergent taxa and between geographic regions (Percy et al. 2014; Twyford, 2014). Poor species discrimination from DNA barcoding is also seen in the rapid radiation of Chinese Primula (Yan et al. 2015a) and Rhododendron (Yan et al. 2015b), which is likely a product of hybridization and recent species divergence.
Future DNA barcoding systems that target large quantities of nuclear sequence variation are extremely promosing for species discrimination in taxonomically complex groups (Coissac et al. 2016; Hollingsworth et al. 2016). These data have the joint benefit of providing many nucleotide characters from unlinked loci, while also moving away from genomic regions that have atypical inheritance and patterns of evolution (i.e. plastids). Analyses of many nuclear genes (or entire genomes) would be particularly valuable for Euphrasia, where it may be possible to identify adaptive variants maintained in the face of hybridization. These adaptive genes may underlie differences between species or ecotypes (Twyford and Friedman 2015), and these loci could then potentially be used for future species identification.
Conclusions
This study highlights how DNA barcoding data may fail to distinguish between species in taxonomically complex groups such as Euphrasia. No species in our study possessed a consistent diagnostic sequence profile. Widespread haplotype sharing among species, in conjunction with high levels of intraspecific variation, makes Euphrasia a particular challenge for DNA barcoding. However, our results are able to help us understand the maintenance of genetic diversity and the evolutionary importance of polyploidy.
Accession Numbers
All sequences obtained from fresh plant specimens have been deposited in the GenBank database under accession numbers MH202267–MH202656.
Sources of Funding
This study formed part of an international exchange programme for X.W., funded by the Chinese Scholarship Council (CSC). P.M.H. and M.R. acknowledge funding from the Scottish Government’s Rural and Environment Science and Analytical Services Division (RESAS). Research by G.G. is supported by the Research Council of Norway: Projects N248799 and N257642; and Norwegian Taxonomy Initiative: Project N70184215. Research by A.D.T. is supported by a Natural Environment Research Council (NERC) Fellowship NE/L011336/1.
Contributions by the Authors
A.D.T and P.M.H. conceived and designed the study. A.D.T., G.G., N.d.V. and C.M. provided samples. C.M. identified the specimens. X.W. and M.R. performed the sequencing. X.W., G.G., M.R. and A.D.T analysed the data. A.D.T. and X.W. wrote the manuscript, with comments from all other authors.
Conflict of Interest
None declared.
Supporting Information
The following additional information is available in the online version of this article—
Table S1. Voucher information and population details of British Euphrasia samples used for UK DNA barcoding. The cpDNA regions include matK and rpl32-trnL, with rbcL generated for a subset of taxa but excluded from the final analysis (see main text). The columns headed ‘cpDNA’ and ‘ITS 2’ indicate whether the sample was successfully sequenced (1) or not (0). Taxa with an asterisk were also included in the global phylogenetic analysis.
Table S2. PCR conditions and primer sequences for regions sequenced in this study. Loci used for DNA barcoding were matK, rbcL and ITS2, supplemented with rpl32-trnL. Regions used for phylogenetic analysis were atpB-rbcL, trnL intron, trnL-trnF and ITS.
Table S3. Spatial analysis of molecular variation (SAMOVA) of ITS sequence data across British Euphrasia populations.
Table S4. Plastid haplotype frequencies across British Euphrasia species.
Table S5. Spatial analysis of molecular variation (SAMOVA) of plastid sequence data across British Euphrasia populations.
Table S6. The distribution of matK haplotypes for populations of British Euphrasia. The cpDNA (n) column indicates the sample sizes.
Table S7. Hierarchical analysis of molecular variance (AMOVA) for matK sequenced in British Euphrasia populations.
Supplementary Material
Acknowledgements
We thank L. Feng and D. Zeng for assistance with data analysis, and two anonymous reviewers for their helpful comments on the manuscript. Some analyses were run using high performance computing resources provided by The Lifeportal at UiO https://lifeportal.uio.no/ and CIPRES Science Gateway V. 3.3 http://www.phylo.org/.
Literature Cited
- Bandelt HJ, Forster P, Röhl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16:37–48. [DOI] [PubMed] [Google Scholar]
- Brysting AK, Oxelman B, Huber KT, Moulton V, Brochmann C, Renner S. 2007. Untangling complex histories of genome mergings in high polyploids. Systematic Biology 56:467–476. [DOI] [PubMed] [Google Scholar]
- CBOL Plant Working Group 2009. A DNA barcode for land plants. Proceedings of the National Academy of Sciences 106:12794–12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- China Plant BOL Group 2011. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proceedings of the National Academy of Sciences 108:19641–19646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coissac E, Hollingsworth PM, Lavergne S, Taberlet P. 2016. From barcodes to genomes: extending the concept of DNA barcoding. Molecular Ecology 25:1423–1428. [DOI] [PubMed] [Google Scholar]
- Cottrell JE, Munro RC, Tabbener HE, Gillies ACM, Forrest GI, Deans JD, Lowe AJ. 2002. Distribution of chloroplast DNA variation in British oaks (Quercus robur and Q. petraea): the influence of postglacial colonisation and human management. Forest Ecology and Management 156:181–195. [Google Scholar]
- Dellicour S, Mardulyn P. 2014. Spads 1.0: a toolbox to perform spatial analyses on DNA sequence data sets. Molecular Ecology Resources 14:647–651. [DOI] [PubMed] [Google Scholar]
- Dupanloup I, Schneider S, Excoffier L. 2002. A simulated annealing approach to define the genetic structure of populations. Molecular Ecology 11:2571–2581. [DOI] [PubMed] [Google Scholar]
- Ennos RA, French GC, Hollingsworth PM. 2005. Conserving taxonomic complexity. Trends in Ecology & Evolution 20:164–168. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HE. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources 10:564–567. [DOI] [PubMed] [Google Scholar]
- French GC, Ennos RA, Silverside AJ, Hollingsworth PM. 2004. The relationship between flower size, inbreeding coefficient and inferred selfing rate in British Euphrasia species. Heredity 94:44–51. [DOI] [PubMed] [Google Scholar]
- French G, Hollingsworth P, Silverside A, Ennos R. 2008. Genetics, taxonomy and the conservation of British Euphrasia. Conservation Genetics 9:1547–1562. [Google Scholar]
- Gussarova G, Popp M, Vitek E, Brochmann C. 2008. Molecular phylogeny and biogeography of the bipolar Euphrasia (Orobanchaceae): recent radiations in an old genus. Molecular Phylogenetics and Evolution 48:444–460. [DOI] [PubMed] [Google Scholar]
- Hebert PD, Gregory TR. 2005. The promise of DNA barcoding for taxonomy. Systematic Biology 54:852–859. [DOI] [PubMed] [Google Scholar]
- Hodges SA, Arnold ML. 1994. Columbines: a geographically widespread species flock. Proceedings of the National Academy of Sciences 91:5129–5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth P, De-Zhu L, Van der Bank M, Twyford A. 2016. Telling plant species apart with DNA: from barcodes to genomes. Philosophical Transactions of the Royal Society B 371:20150338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth PM, Graham SW, Little DP. 2011. Choosing and using a plant DNA barcode. PLoS One 6:e19254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17:754–755. [DOI] [PubMed] [Google Scholar]
- Kolář F, Píšová S, Záveská E, Fér T, Weiser M, Ehrendorfer F, Suda J. 2015. The origin of unique diversity in deglaciated areas: traces of Pleistocene processes in north‐European endemics from the Galium pusillum polyploid complex (Rubiaceae). Molecular Ecology 24:1311–1334. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. [DOI] [PubMed] [Google Scholar]
- Liebst B. 2008. Do they really hybridize? A field study in artificially established mixed populations of Euphrasia minima and E. salisburgensis (Orobanchaceae) in the Swiss Alps. Plant Systematics and Evolution 273:179–189. [Google Scholar]
- Naciri Y, Caetano S, Salamin N. 2012. Plant DNA barcodes and the influence of gene flow. Molecular Ecology Resources 12:575–580. [DOI] [PubMed] [Google Scholar]
- Nylander J. 2004. MrModeltest v2. Program distributed by the author. [Google Scholar]
- Percy DM, Argus GW, Cronk QC, Fazekas AJ, Kesanakurti PR, Burgess KS, Husband BC, Newmaster SG, Barrett SC, Graham SW. 2014. Understanding the spectacular failure of DNA barcoding in willows (Salix): does this result from a trans-specific selective sweep?Molecular Ecology 23:4737–4756. [DOI] [PubMed] [Google Scholar]
- Preston CD, Pearman DA. 2015. Plant hybrids in the wild: evidence from biological recording. Biological Journal of the Linnean Society 115:555–572. [Google Scholar]
- Rambaut A, Drummond A, Suchard M. 2013. Tracer v1. 6—MCMC trace analysis package. UK: Institute of Evolutionary Biology, University of Edinburgh. [Google Scholar]
- Rendell S, Ennos RA. 2003. Chloroplast DNA diversity of the dioecious European tree Ilex aquifolium L. (English holly). Molecular Ecology 12:2681–2688. [DOI] [PubMed] [Google Scholar]
- Schmickl R, Paule J, Klein J, Marhold K, Koch MA. 2012. The evolutionary history of the Arabidopsis arenosa complex: diverse tetraploids mask the western carpathian center of species and genetic diversity. PLoS One 7:e42691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spooner DM. 2009. DNA barcoding will frequently fail in complicated groups: an example in wild potatoes. American Journal of Botany 96:1177–1189. [DOI] [PubMed] [Google Scholar]
- Squirrell J, Hollingsworth PM, Bateman RM, Tebbitt MC, Hollingsworth ML. 2002. Taxonomic complexity and breeding system transitions: conservation genetics of the Epipactis leptochila complex (Orchidaceae). Molecular Ecology 11:1957–1964. [DOI] [PubMed] [Google Scholar]
- Stace CA, Preston CD, Pearman DA. 2015. Hybrid flora of the British Isles. Bristol: Botanical Society of Britain and Ireland. [Google Scholar]
- Stone H. 2013. Evolution and conservation of tetraploid Euphrasia L. in Britain. Unpublished PhD Thesis, Edinburgh: The University of Edinburgh. [Google Scholar]
- Svensson BM, Carlsson BÅ. 2004. Significance of time of attachment, host type, and neighbouring hemiparasites in determining fitness in two endangered grassland hemiparasites. Annales Botanici Fennici 41:63–75. [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. 1991. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology 17:1105–1109. [DOI] [PubMed] [Google Scholar]
- Twyford AD. 2014. Testing evolutionary hypotheses for DNA barcoding failure in willows. Molecular Ecology 23:4674–4676. [DOI] [PubMed] [Google Scholar]
- Twyford AD, Ennos RA. 2012. Next-generation hybridization and introgression. Heredity 108:179–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twyford AD, Friedman J. 2015. Adaptive divergence in the monkey flower Mimulus guttatus is maintained by a chromosomal inversion. Evolution 69:1476–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vere N, Rich TC, Ford CR, Trinder SA, Long C, Moore CW, Satterthwaite D, Davies H, Allainguillaume J, Ronca S, Tatarinova T, Garbett H, Walker K, Wilkinson MJ. 2012. DNA barcoding the native flowering plants and conifers of wales. PLoS One 7:e37945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrancken J, Brochmann C, Wesselingh RA. 2012. A European phylogeography of Rhinanthus minor compared to Rhinanthus angustifolius: unexpected splits and signs of hybridization. Ecology and Evolution 2:1531–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S, Müller KF, Quandt D, Bellot S, Schneeweiss GM. 2016. Mechanistic model of evolutionary rate variation en route to a nonphotosynthetic lifestyle in plants. Proceedings of the National Academy of Sciences 113:9045–9050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications 18:315–322. [Google Scholar]
- Yan LJ, Liu J, Möller M, Zhang L, Zhang XM, Li DZ, Gao LM. 2015a. DNA barcoding of Rhododendron (Ericaceae), the largest Chinese plant genus in biodiversity hotspots of the Himalaya–Hengduan Mountains. Molecular Ecology Resources 15:932–944. [DOI] [PubMed] [Google Scholar]
- Yan HF, Liu YJ, Xie XF, Zhang CY, Hu CM, Hao G, Ge XJ. 2015b. DNA barcoding evaluation and its taxonomic implications in the species-rich Genus primula L. in China. PLoS One 10:e0122903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo P. 1956. Hybridization between diploid and tetraploid species of Euphrasia. Watsonia 3:253–269. [Google Scholar]
- Yeo PF. 1978. A taxonomic revision of Euphrasia in Europe. Botanical Journal of the Linnean Society 77:223–334. [Google Scholar]
- Young ND, Healy J. 2003. Gapcoder automates the use of indel characters in phylogenetic analysis. BMC Bioinformatics 4:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrei M, Talent N, Kuzmina M, Lee J, Lund J, Shipley PR, Stefanović S, Dickinson TA. 2015. DNA barcodes from four loci provide poor resolution of taxonomic groups in the genus Crataegus. AoB PLANTS 7:plv045; doi:10.1093/aobpla/plv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.