Abstract
X-rays are widely applied in the medical field for the diagnosis and treatment of diseases. Among the uses of X-rays in diagnosis, computed tomography (CT) has been established as one of the most informative diagnostic radiology examinations. Moreover, recent advances in CT scan technology have made this examination much easier and more informative and increased its application, especially in Japan. However, the radiation dose of CT scans is higher than that of simple X-ray examinations. Therefore, the health risk of a CT scan has been discussed in various studies, but is still controversial. Consequently, the biological and cytogenetic effects of CT scans are being analyzed. Here, we summarize the recent findings concerning the biological and cytogenetic effects of ionizing radiation from a CT scan, by focusing on DNA damage and chromosome aberrations.
Keywords: CT scan, medical radiation, DNA damage, chromosome aberration, biological dosimetry
INTRODUCTION
Applications of radiation in medicine, such as for the diagnosis of diseases or injuries, started just after the discovery of X-rays in 1895. One of the most common usages of radiation in current medical diagnosis is computed tomography (CT) using X-rays, since CT scan images provide more detailed information than that provided by simple X-ray examinations. In addition to facilitating the diagnosis of various diseases, including cancer and vascular diseases, CT scans are useful in the follow-up care of cancer patients (for estimating the effectiveness of therapy, by examining the tumor size and detecting metastasis). Recent advances in CT scanning technology now allow the acquisition of appropriate images within a short scanning time and enable the examination of children without anesthesia [1]. Due to these benefits and technological advancements, CT scans are gaining popularity around the world.
However, the use of X-rays is a double-edged sword. On the one hand, doctors can make an accurate diagnosis of diseases with the information provided by diagnostic imaging, and improve the prognosis of cancer patients through radiation therapy. On the other hand, medical radiation has become a major source of radiation exposure for the general population. Since the increased risk of leukemia and solid cancer after high-dose irradiation has been reported in various studies, including the epidemiological studies of A-bomb survivors in Hiroshima and Nagasaki, the effects of medical radiation have become a public health issue [2–4]. Several reports concerning the cancer risk after medical radiation imaging, such as CT scans, have been published. Brenner et al. first reported the quantitative estimates of radiation risks associated with pediatric CT [5]. In 2012 and 2013, large cohort studies of the cancer risks in people exposed to CT scans in childhood provided evidence of the association between cancer risk and CT scans [6–8]. In contrast, other studies failed to observe the association of childhood cancer risk with medical diagnostic radiation [9, 10]. Therefore, it is difficult to draw a conclusion about the health effects of medical diagnostic imaging radiation from these cohort studies [11].
Ionizing radiation induces various types of DNA damage, including DNA double-strand breaks (DSBs). The DNA damage is normally repaired by the corresponding DNA repair system. However, if mistakes are made in the repair of the damaged DNA, then the genetic information may be changed, leading to health effects such as cancer and vascular events. Therefore, accurately quantifying the amounts of DNA damage, especially DSBs, induced by ionizing radiation is important in studying the effects of medical radiation exposure. Various proteins related to the DNA damage response or DNA repair, including the phosphorylated form of histone H2AX (γ-H2AX), form ‘radiation-induced foci’ at sites with DNA damage. Therefore, the analysis of γ-H2AX foci has been applied to examining the radiation effects of CT scans.
Several methods have been developed for estimating the radiation dose in the field of radiation emergency medicine. Among the biological dosimetry procedures applied in radiation emergency medicine, conventional chromosome analysis using Giemsa-stained samples to detect dicentric and ring chromosomes has been established as the gold standard [12]. However, the analysis of Giemsa-stained chromosomes is time-consuming and requires well-trained technicians. Therefore, other methods, such as the fluorescence in situ hybridization (FISH) technique and the micronuclei (MN) assay, have been developed to overcome the weak points of Giemsa analysis and to facilitate dose estimation.
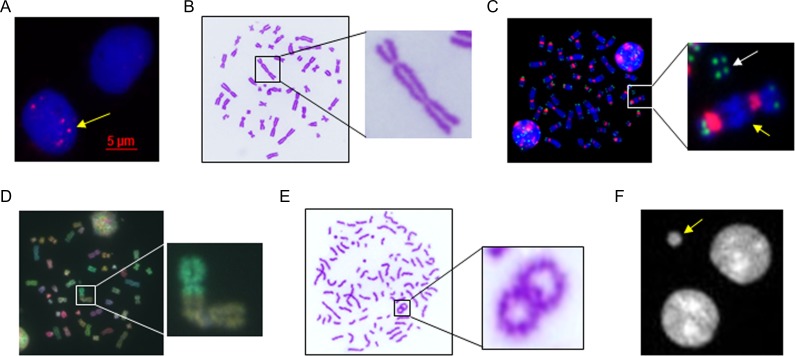
The biological effects of low-dose medical radiation exposure, including that from CT scans, have been investigated by using these molecular biological and cytogenetic methods, normally applied in radiation emergency medicine. Here, we summarize the reports related to the effects of CT scans, one of the major radiation sources in medicine, to discuss the biological effects of medical radiation exposure on human chromosomal DNA (Fig. 1).
Fig. 1.
Various methods used for the estimation of the biological effects of medical radiation. (A) γ-H2AX foci (yellow arrow) in PBLs after irradiation; (B) dicentric chromosome in a metaphase lymphocyte after Giemsa staining; (C) dicentric chromosome (yellow arrow) and acentric fragment (white arrow) in metaphase chromosomes after PNA-FISH using telomere/centromere probes; (D) chromosome aberration in metaphase chromosomes after multi-color FISH using chromosome painting probes; (E) PCC-ring chromosome in PBLs after irradiation; (F) MN (yellow arrow) in binucleated cells after DAPI staining.
QUANTIFICATION OF MEDICAL IRRADIATION-INDUCED DNA DAMAGE BY THE γ-H2AX FOCI ASSAY
γ-H2AX focus formation after induction of DNA damage
The histone variant H2AX becomes phosphorylated at Ser139 by PIKK family kinases, such as ataxia telangiectasia mutated (ATM), ATR or DNA-PK, after the induction of DNA damage, and is referred to as γ-H2AX. γ-H2AX accumulates at a site containing a DSBs to form a focus within several minutes after ionizing irradiation exposure (Fig. 1A) [13–15]. The number of γ-H2AX foci in cultured cells reaches a peak within 60 min after irradiation in vitro and shows a linear correlation with the exposed dose, even within the low-dose range of <100 mGy [16, 17]. Given their easy sampling and circulation throughout the body, human peripheral blood lymphocytes (PBLs) are most commonly used to assess the DNA damage induced by CT scans. Several studies have reported the increase in γ-H2AX foci in PBLs after CT scans in vitro and in vivo (Table 1). The CT dose index (CTDI), the dose length product (DLP) and the size-specific dose estimates (SSDEs) are the parameters used for estimating the dose absorbed during a CT scan. The γ-H2AX foci assay revealed that the DNA damage induced by a CT scan correlated with the CTDI, DLP and SSDE [18–20]. These findings suggest that CT scans induce DNA damage in PBLs in a dose-dependent manner.
Table 1.
Overview of published reports about γ-H2AX foci formation after CT scans
| Publication | Type of study | Number of patients | Radiation effect of CT | Dose-enhancing effect of contrast material (CM) | Foci count | Toxicity of CM itself |
|---|---|---|---|---|---|---|
| Löbrich et al., 2005; [18] | In vitro & in vivo | Adult patients: n = 23 | Yes | Manual | ||
| Rothkamm et al., 2007; [55] | in vitro & in vivo | Adult patients: n = 13 | Yes | Manual | ||
| Jost et al., 2009; [28] | In vitro | Yes | No (within diagnostic dose range) | Automatic | ||
| Grudzenski et al., 2009; [25] | In vitro & in vivo | Adult patients; contrast-enhanced CT: n = 18; unenhanced CT: n = 19 | Yes | Yes (in vitro: 33 mg/ml blood; in vivo: 27–49 g/patients) | Manual | No |
| Kuefner et al., 2010; [56] | In vivo | Adult patients: n = 36 | Yes | Manual | ||
| Kuefner et al., 2010; [33] | In vivo | Adult patients: n = 34 | Yes | Manual | ||
| Pathe et al., 2011; [21] | In vitro & in vivo | Contrast-enhanced CT: n = 15; unenhanced CT: n = 15 | Yes | Yes (37–44.4 g/patient) | Manual | No |
| Geisel et al., 2012; [57] | In vivo | Adult patients: n = 56 | Yes | Automatic | ||
| Beels et al., 2012; [19] | In vitro & in vivo | Adult patients: n = 69 (contrast-enhanced CT) | Yes | No (in vitro) | Manual | No |
| May et al., 2012; [58] | In vivo | Adult patients: n = 33 | Yes (18F-Fdg PET/CT) | Manual | ||
| Brand et al., 2012; [59] | In vivo | Adult patients: n = 66 | Yes | Manual | ||
| Kuefner et al., 2013; [23] | In vitro & in vivo | Adult patients: n = 7 | Yes | Manual | ||
| Halm et al., 2014; [34] | In vivo | Pediatric patients: n = 3 | Yes | Manual | ||
| Piechowiak et al., 2015; [26] | In vivo | Contrast-enhanced CT: n = 179; unenhanced CT: n = 66 | Yes | Yes (18.651 g/patient) | Manual | |
| Nguyen et al., 2015; [60] | In vivo | Adult patients: n = 67 (cardiac CT angiography) | Yes | Flow cytometry & manual | ||
| Vandevoorde et al., 2015; [17] | In vitro & in vivo | Pediatric patients: n = 51 | Yes | Manual | ||
| Fukumoto et al., 2017; [20] | In vitro & in vivo | Adult patients: n = 45 | Yes | Automatic | ||
| Wang et al., 2017; [27] | In vivo | Adult patients; contrast-enhanced CT: n = 48; unenhanced CT: n = 22 | Yes | Yes (33.3 g/patient) | Manual | Yes |
Following the formation of γ-H2AX foci, the DNA repair pathway is activated and DNA repair proteins accumulate at sites with damaged DNA. It is commonly accepted that the disappearance of γ-H2AX foci suggests that the DNA damage has been repaired. After the peak of the γ-H2AX focus formation, ~1 h after the induction of DNA damage, the number of foci decreases and returns to almost the basal level at 24 h after radiation exposure [18, 21]. Therefore, the rapid kinetics of γ-H2AX foci formation may lead to underestimation of the amount of DNA damage induced by radiation. To avoid this underestimation, the phosphatase inhibitor calyculin A has been employed to prevent the disappearance of γ-H2AX foci. Calyculin A itself does not induce DNA damage, and a treatment with 1 nM calyculin A reportedly inhibited the decrease in γ-H2AX foci in irradiated cells [22, 23]. When compared with patients’ blood treated with calyculin A, the number of CT-induced γ-H2AX foci in blood without treatment was lower by ~35% [23]. Hence, calyculin A would be useful for enhancing the accurate detection of CT-induced DNA damage by disturbing the time-dependent disappearance of γ-H2AX foci.
Enhancement of DNA damage induction by contrast media
Contrast media are used to enhance the contrast of human internal organs, structures or fluids within the body in CT imaging. Contrast media reportedly lack intrinsic genotoxicity detectable by the γ-H2AX assay [24, 25]. However, increases in CT-induced DNA damage by contrast media have been reported by several groups [16, 21, 24, 26]. In those studies, the number of radiation-induced γ-H2AX foci was associated with the concentration of the contrast media. A significant increase in radiation-induced DSBs caused by contrast media was observed when the blood concentration of iodine was >17.5 mg/ml, in an in vitro study [24]. Wang et al. reported a slight but significant increase in γ-H2AX foci in PBLs from patients after the injection of contrast media but before the CT scan [27]. However, several studies have also shown that the administration of contrast media within the pharmacologically appropriate concentration range and a radiation dose corresponding to that of a CT scan failed to enhance the induction of DNA damage [17, 19, 28].
Although further studies are required in order to clarify the enhancement of DNA damage by and the organ toxicity of contrast medium, the enhanced radiation effect associated with contrast medium may be due to the secondary electrons generated during a contrast-enhanced CT scan [29, 30]. Through dose estimation from the change in attenuation measured in Hounsfield units, some studies have revealed an increase in the radiation dose in several organs after contrast-enhanced CT as compared with un-enhanced CT [31, 32]. Therefore, the enhancement of radiation-induced DNA damage by contrast media is still controversial.
The advantages and disadvantages of the γ-H2AX foci assay
Among the various methods for detecting radiation-induced DNA damage, the γ-H2AX foci assay is one of the fastest methods, and can be completed within several hours of the collection of blood samples. Furthermore, the γ-H2AX foci assay is a sensitive method that can detect DNA damage induced by X-ray irradiation at ~1 mGy [33, 34]. Therefore, the γ-H2AX foci assay is an ideal method for analyzing the DNA damage induced by medical diagnostic radiation exposure, such as from a CT scan.
Manual quantification of the γ-H2AX foci, however, is laborious and time-consuming work. Therefore, image analysis software, such as Image J and FociCounter, is used for the quantification of γ-H2AX foci [13, 35]. Several microscope systems allow automatic image capture and quantification of foci, including the MetaCyte (MetaSystems GmbH, Germany) and In Cell Analyzer (GE Healthcare) systems [20, 36]. Although it is possible to count the numbers of γ-H2AX foci automatically using these software and microscope systems, the parameter values used to detect γ-H2AX foci, such as focus size or intensity, must be determined and implemented subjectively by operators. This could lead to inter- or intralaboratory variations, even for the analysis of the same samples [37]. Besides this problem, differences in the quality or sensitivity of the antibodies used for the detection of γ-H2AX foci, could also affect the quantification of γ-H2AX foci. Therefore, a standardized method for detecting γ-H2AX foci, including how to prepare the internal control, should be established for the practical use of the γ-H2AX foci assay in the evaluation of DNA damage induced by medical radiation exposure.
CHROMOSOME ABERRATIONS INDUCED BY MEDICAL IRRADIATION
Misjoining of the wrong DNA ends of DSBs leads to structural abnormalities of chromosomes. Among the chromosome aberrations, dicentric and ring chromosomes are unstable abnormalities, since cells carrying these abnormalities are defective in chromosome segregation, leading to cell death. Therefore, the scoring of dicentric chromosomes in lymphocytes arrested at the first mitosis after stimulation with phytohemagglutinin (PHA) has been established as the gold standard for the estimation of radiation dose after a recent exposure. In contrast, reciprocal translocations, which can be transferred to daughter cells, are stable abnormalities persisting for many years after radiation exposure. Scoring of translocations is useful for chronic radiation exposure estimations or retrospective studies of past radiation exposure. Although the dose response of the increase in abnormal chromosomes after low-dose irradiation (<100 mGy) is still unclear, there are several reports concerning chromosome analysis for patients receiving a CT scan, in which the sensitivity of the analysis was increased by analyzing >1000 cells per sample.
Conventional Giemsa staining analysis
Conventional Giemsa staining analysis is the most common method applied for the biological dosimetry of ionizing radiation, because of the short, easy staining procedures and low cost (Fig. 1B). Using conventional Giemsa staining analysis, the increase in dicentric chromosomes after in vitro exposure of PBLs to radiation from a CT scan was observed, and the relationship between the chromosome abnormalities and the tube voltage/phantom size of the CT scanner was suggested [38]. For accurate evaluation of the radiation dose, only chromosome abnormalities in the first mitosis after the PHA stimulation of PBLs should be counted. For this purpose, the fluorescence plus Giemsa (FPG) method is used to identify the lymphocytes arrested at the first mitosis. Using the FPG staining method, a significant increase in dicentric chromosomes was observed in the lymphocytes from 10 pediatric patients <10 years old after a CT scan, while the increase in the older group of children from 10 to 15 years old was not significant [39]. This finding suggests that children <10 years old may be more sensitive to the low-dose radiation from a CT scan, as compared with older children. In contrast, a significant increase in chromosome aberrations in PBLs from adult patients after a CT scan has also been reported [40, 41]. These different results regarding the increase in chromosome abnormalities after a CT scan could be due to the radiation dose/conditions of the CT scan (Table 2). Another possibility is the interlaboratory variability in the scoring of dicentric chromosomes using Giemsa staining [42, 43]. The identification of abnormal chromosomes by Giemsa analyses is difficult and requires skilled technicians. Therefore, the IAEA recommends that laboratories involved in radiation dosimetry establish their own dose–response curves [12]. About 10 dicentric chromosomes in 100 cells are reportedly formed after 1 Gy γ-ray irradiation of human blood in vitro at a dose rate of ~110 mGy/min [44]. Therefore, the scoring of >1000 cells is recommended for the estimation of radiation doses of <100 mGy, because of the low numbers of abnormal chromosomes. It would not be practical to apply the Giemsa-staining method to analyzing many samples manually for the evaluation of the effects of low-dose irradiation, including that from CT scans.
Table 2.
Overview of published reports about chromosome aberrations induced by CT scans
| Publication | Type of study | Number of patients | Type of chromosome aberrations | Staining method | Exposed dose | Number of lymphocytes analyzed | Radiation effect of CT |
|---|---|---|---|---|---|---|---|
| M’kacher et al., 2003; [45] | In vitro & in vivo | Adult patients (n = 10) | Translocations, dicentrics, rings, insertions and acentric fragments of painted chromosomes in metaphase and chromosome fragments in interphase (PCC) | FISH (chromosome DNA probes 1, 3, 4) | In vivo: 0.057 Gy; in vitro: 0.05, 0.1, 0.2 Gy | Significant increase in chromosomal fragments (PCC) but not chromosome aberrations in metaphase | |
| Stephan et al., 2007; [39] | In vivo | Pediatric patients (n = 10) | Dicentric (dic) & excess acentrics (ace) | Fluorescence plus Giemsa (FPG) staining | Mean dose to blood: 12.9 mGy | >20 000 cells (average: 1000 cells/subject) | Yes (only in patients <10 years old) |
| Jost et al., 2009; [28] | In vitro | Dicentric | Fluorescence plus Giemsa (FPG) staining | 0.025, 0.05, 0.1, 1 Gy | >1000 metaphase/subject (0, 0.025, 0.05, 0.1 Gy); >200 metaphase for subjects after 1 Gy IR | Yes | |
| Golfier et al., 2009; [61] | In vitro | Dicentric | Fluorescence plus Giemsa (FPG) staining | 0.025, 0.05, 0.1, 1 Gy | >1000 metaphase/subject (0, 0.025, 0.05, 0.1 Gy); >200 metaphase for subjects after 1 Gy IR | Yes | |
| Abe et al., 2015; [41] | In vivo | Adult patients (n = 10) | Dicentric | Giemsa staining & PNA-FISH | 5.78–60.27 mSv: mean 24.24 mSv | 2000 metaphase/subject | Yes |
| Kanagaraj et al., 2015 [40] | In vivo | Adult patients (contrast scan: n = 14; brain plain scan: n = 13) | Chromosome aberrations (dicentric, chromosome/chromatid break) & MN | Giemsa staining | Eye: 2~520 mGy; forehead: 0.84–210 mGy; thyroid: 1.79~ 185 | 250–300 metaphase/subject (chromosome aberrations); 1000 binucleated cells (MN) | Yes |
| Abe et al., 2016; [62] | In vivo | Adult patients (n = 12) | Translocation | FISH (chromosome painting DNA probes 1, 2, 4) | 5.78–60.27 mSv: mean 24.24 mSv | >5000 metaphase/subject | No significant increase in translocation after CT scan |
Chromosome analyses using FISH
The FISH technique can score abnormal chromosomes sensitively, easily and accurately, by marking specific DNA regions or chromosomes with different fluorescent dyes (Fig. 1C and D). Therefore, to overcome the difficulty in identifying the chromosomal aberrations in Giemsa-stained lymphocytes, the FISH technique has been applied for karyotyping. The probes used in the FISH analysis, however, are very expensive, especially those for differentially painting 24 chromosomes. This has led to widespread application of the analysis using three chromosome painting probes to estimate the numbers of chromosome translocations in biological dosimetry. FISH analysis using three chromosome painting probes failed to show an increase in translocations or chromosome aberrations in metaphase lymphocytes after a CT scan [41, 45]. This could be due to confounding factors, such as smoking and aging, which are also associated with chromosome translocations [46]. Chromosome translocations are stable abnormalities that can persist for many years, and a lifelong accumulation of chromosome translocations in hematopoietic stem cells may also make it difficult to detect the slight effects of low-dose irradiation in lymphocytes. The other possibility is the low sensitivity of the analysis, due to the detection of chromosome translocations involving only three chromosomes. To increase the accuracy of the translocation assay, FISH analyses using probes differentially painting 24 chromosomes (22 + X + Y) should be performed.
PNA-FISH, using telomere and centromere PNA probes, can identify dicentric and ring chromosomes in PBLs after high-dose irradiation more easily and accurately than a Giemsa analysis [47]. Moreover, the PNA probes for centromeres and telomeres are not expensive. A telomere/centromere PNA-FISH analysis revealed an increase in dicentric chromosomes in lymphocytes from 10 patients after a CT scan [41]. Therefore, at present, the PNA-FISH analysis of dicentric and ring chromosomes could detect a slight increase in abnormal chromosomes after low-dose irradiation, such as from a CT scan. Further study is required to establish the usefulness of the PNA-FISH analysis in this point.
Assays for chromosomal aberrations require a lot of time, usually several days from the blood sample collection to the completion of the analysis, as compared with the γ-H2AX foci assay. Lymphocytes must be cultured in medium containing PHA for 48 h, for cell-cycle progression to obtain metaphase cells. The time and cost required for the staining and analyses differ quite a lot between the Giemsa and FISH staining methods. Giemsa staining is the fastest and least expensive, but the identification and scoring of chromosome aberrations is time-consuming work. FISH using chromosome painting probes is the most expensive and requires long-term hybridization of the probe with the target DNA, usually more than 12 h. As compared with Giemsa staining, FISH using chromosome painting probes is faster, easier and more accurate for chromosome analysis. PNA-FISH overcomes the disadvantages of Giemsa staining and FISH using chromosome painting probes, due to the short hybridization time, low cost and easily identified chromosome aberrations.
Due to the low numbers of chromosome aberrations induced by low-dose radiation, including that from CT scans, the analysis of more than 1000 cells is recommended to increase the accuracy. More than 3 h are required for the analysis of 1000 metaphase chromosomes, even in the PNA-FISH analysis of dicentric and ring chromosomes. Computer-assisted microscopy, such as the Metafer microscope platform (MetaSystems, Germany), can be used to search for all of the metaphase cells on slides and obtain high-resolution images automatically. Similar to the γ-H2AX foci assay, the scoring of abnormal chromosomes can also be processed by software, such as DCScore and Ikaros. Since the detection efficiency of dicentric chromosomes by these software programs is low compared with that of human observers, dose–response curves for automatic or semi-automatic analyses are required for dose estimation [48, 49]. Automatic or semi-automatic chromosome aberration scoring has been suggested as a useful method for the rapid triage of potentially exposed individuals in a large-scale radiation accident [48, 49]. Therefore, the development of software to automatically and accurately identify abnormal chromosomes is awaited for the detection of the effects of low-dose medical irradiation.
Premature chromosome condensation
The fusion of interphase cells with mitotic cells or drug treatment can induce the condensation of chromosomes, in a phenomenon referred to as premature chromosome condensation (PCC) (Fig. 1E). The combination of PCC with the chromosome staining technique is applied to the detecting of chromosome fragments or ring chromosomes in interphase lymphocytes after ionizing radiation exposure. Since few ring chromosomes are induced by low-dose irradiation, the PCC ring assay is suited for the estimation of the radiation doses of samples after high-dose radiation, usually >10 Gy [12]. For chromosome analyses, PCC fusion using mitotic cells is the fastest method of evaluating the effects of ionizing radiation, since the chromosome aberrations can be scored within several hours after blood sampling. PCC associated with the FISH technique using painting probes seems to be more sensitive, compared with the FISH analysis of metaphase lymphocytes using three chromosome painting probes. The numbers of translocations in metaphase lymphocytes showed no significant changes after a CT scan, using the samples from the same patients, but this could be due to the increase in chromosomal fragments in interphase lymphocytes immediately after a CT scan [45].
The PCC assay requires skilled technicians, as is the case for chromosome analysis of Giemsa-stained samples. Some reports have noted the usefulness of software in the PCC analysis of samples after high-dose irradiation [50, 51]. The application of such software for studies of the effects of low-dose irradiation, such as that from a CT scan, is awaited.
MN assay
MN are considered to result from a lagging acentric chromosome, or chromosome aberrations not included in the daughter nuclei, due to failed attachment to the spindle during the segregation of chromosomes in anaphase. The cell cycle progression of PHA-stimulated lymphocytes can be blocked at the stage of cytokinesis after the first mitosis by using cytochalasin B, for the efficient detection of MN. Therefore, the cytokinesis-blocked MN (CBMN) assay has been established as a useful and sensitive method for the quantification of radiation effects on chromosomal DNA (Fig. 1F). A significant increase in MN has been reported in lymphocytes from patients undergoing plain or contrast CT scans by the CBMN assay [40]. Computer-assisted microscopy and software are also used in the MN assay [52, 53]. Lyulko et al. reported the automatic generation of dose–response curves of MN by using the RABiT image analysis software, and the numbers of MN scored correlated well with the manual scoring results [54]. The automatic MN assay, however, has not been applied for evaluation of the radiation effects of CT scans.
SUMMARY
Adverse health effects from low-dose irradiation by a CT scan, especially possible associations with cancer risk, have been suggested, but such effects remain controversial. However, the induction of DNA damage by a CT scan examination has been clearly demonstrated by the γ-H2AX foci assay. Several reports have also suggested the induction of chromosome aberrations after a CT scan, using conventional chromosome analysis methods, but the technical difficulty of chromosome analysis has prevented the large-scale study required for evaluation of the effects of low-dose irradiation by diagnostic exposure. Recent advances in microscopy and image analysis will overcome this technical difficulty of chromosome analysis, by enabling high-throughput analysis of the minor increase in chromosome aberrations after low-dose radiation exposure. Moreover, recent advancements in biomedical science, such as the development of next-generation sequencers, will also make it possible to perform high-throughput analyses of genome information. Therefore, the combined applications of these new technologies will enable a breakthrough in understanding the biological and cytogenetic effects of low-dose irradiation, including CT scan examinations, in the near future. The development of these new technologies will also facilitate the establishment of precision medicine, by providing information about an individual’s genome instability.
ACKNOWLEDGEMENTS
We thank Shinya Matsuura and Kurumi Fujioka from Hiroshima University for providing the photos of MN and multi-color FISH.
CONFLICT OF INTEREST
The authors declare that there are no competing interests.
FUNDING
This study was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI (Grant numbers JP16H01312, JP15H02821, JP26430114) and the Platform Project for Supporting in Drug Discovery and Life Science Research (Platform for Dynamic Approaches to Living System) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) and the Japan Agency for Medical Research and Development (AMED). Part of this study was also supported by a grant from the Center of World Intelligence Project for Nuclear S&T and Human Resource Development by MEXT, entitled ‘Development of a high-throughput radiation dose estimation system by using PNA-FISH method’. This work was partially supported by the International Atomic Energy Agency (IAEA CRP E3.50.08) and the Program of the Network-type Joint Usage/Research Center for Radiation Disaster Medical Science of Hiroshima University, Nagasaki University and Fukushima Medical University.
REFERENCES
- 1. Brenner DJ, Hall EJ. Computed tomography—an increasing source of radiation exposure. N Engl J Med 2007;357:2277–84. [DOI] [PubMed] [Google Scholar]
- 2. Preston DL, Shimizu Y, Pierce DA et al. Studies of mortality of atomic bomb survivors. Report 13: solid cancer and noncancer disease mortality: 1950–1997. Radiat Res 2003;160:381–407. [DOI] [PubMed] [Google Scholar]
- 3. Preston DL, Ron E, Tokuoka S et al. Solid cancer incidence in atomic bomb survivors: 1958–1998. Radiat Res 2007;168:1–64. [DOI] [PubMed] [Google Scholar]
- 4. Preston DL, Pierce DA, Shimizu Y et al. Effect of recent changes in atomic bomb survivor dosimetry on cancer mortality risk estimates. Radiat Res 2004;162:377–89. [DOI] [PubMed] [Google Scholar]
- 5. Brenner D, Elliston C, Hall E et al. Estimated risks of radiation-induced fatal cancer from pediatric CT. Am J Roentgenol 2001;176:289–96. [DOI] [PubMed] [Google Scholar]
- 6. Pearce MS, Salotti JA, Little MP et al. Radiation exposure from CT scans in childhood and subsequent risk of leukaemia and brain tumours: a retrospective cohort study. Lancet 2012;380:499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mathews JD, Forsythe AV, Brady Z et al. Cancer risk in 680,000 people exposed to computed tomography scans in childhood or adolescence: data linkage study of 11 million Australians. BMJ 2013;346:f2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Gonzalez AB, Salotti JA, McHugh K et al. Relationship between paediatric CT scans and subsequent risk of leukaemia and brain tumours: assessment of the impact of underlying conditions. Br J Cancer 2016;114:388–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hammer GP, Seidenbusch MC, Schneider K et al. A cohort study of childhood cancer incidence after postnatal diagnostic X-ray exposure. Radiat Res 2009;171:504–12. [DOI] [PubMed] [Google Scholar]
- 10. Hammer GP, Seidenbusch MC, Regulla DF et al. Childhood cancer risk from conventional radiographic examinations for selected referral criteria: results from a large cohort study. Am J Roentgenol 2011;197:217–23. [DOI] [PubMed] [Google Scholar]
- 11. Costello JE, Cecava ND, Tucker JE et al. CT radiation dose: current controversies and dose reduction strategies. Am J Roentgenol 2013;201:1283–90. [DOI] [PubMed] [Google Scholar]
- 12. IAEA Ctogenetic Dosimetry: Applications in Preparedness for and Response to Radiation Emergencies Vienna: International Atomic Energy Agency, 2011. [Google Scholar]
- 13. Jucha A, Wegierek-Ciuk A, Koza Z et al. FociCounter: a freely available PC programme for quantitative and qualitative analysis of gamma-H2AX foci. Mutat Res 2010;696:16–20. [DOI] [PubMed] [Google Scholar]
- 14. Franken NA, ten Cate R, Krawczyk PM et al. Comparison of RBE values of high-LET alpha-particles for the induction of DNA-DSBs, chromosome aberrations and cell reproductive death. Radiat Oncol 2011;6:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Horn S, Barnard S, Rothkamm K. Gamma-H2AX-based dose estimation for whole and partial body radiation exposure. PLoS One 2011;6:e25113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grudzenski S, Raths A, Conrad S et al. Inducible response required for repair of low-dose radiation damage in human fibroblasts. Proc Natl Acad Sci U S A 2010;107:14205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vandevoorde C, Gomolka M, Roessler U et al. EPI-CT: in vitro assessment of the applicability of the gamma-H2AX-foci assay as cellular biomarker for exposure in a multicentre study of children in diagnostic radiology. Int J Radiat Biol 2015;91:653–63. [DOI] [PubMed] [Google Scholar]
- 18. Lobrich M, Rief N, Kuhne M et al. In vivo formation and repair of DNA double-strand breaks after computed tomography examinations. Proc Natl Acad Sci U S A 2005;102:8984–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Beels L, Bacher K, Smeets P et al. Dose-length product of scanners correlates with DNA damage in patients undergoing contrast CT. Eur J Radiol 2012;81:1495–9. [DOI] [PubMed] [Google Scholar]
- 20. Fukumoto W, Ishida M, Sakai C et al. DNA damage in lymphocytes induced by cardiac CT and comparison with physical exposure parameters. Eur Radiol 2017;27:1660–6. [DOI] [PubMed] [Google Scholar]
- 21. Pathe C, Eble K, Schmitz-Beuting D et al. The presence of iodinated contrast agents amplifies DNA radiation damage in computed tomography. Contrast Media Mol Imaging 2011;6:507–13. [DOI] [PubMed] [Google Scholar]
- 22. Antonelli F, Belli M, Cuttone G et al. Induction and repair of DNA double-strand breaks in human cells: dephosphorylation of histone H2AX and its inhibition by calyculin A. Radiat Res 2005;164:514–7. [DOI] [PubMed] [Google Scholar]
- 23. Kuefner MA, Brand M, Engert C. The effect of calyculin A on the dephosphorylation of the histone γ-H2AX after formation of X-ray–induced DNA double-strand breaks in human blood lymphocytes. Int J Radiat Biol 2013;89:424–32. [DOI] [PubMed] [Google Scholar]
- 24. Gould R, McFadden SL, Horn S et al. Assessment of DNA double-strand breaks induced by intravascular iodinated contrast media following in vitro irradiation and in vivo, during paediatric cardiac catheterization. Contrast Media Mol Imaging 2016;11:122–9. [DOI] [PubMed] [Google Scholar]
- 25. Grudzenski S, Kuefner MA, Heckmann MB et al. Contrast medium-enhanced radiation damage caused by CT examinations. Radiology 2009;253:706–14. [DOI] [PubMed] [Google Scholar]
- 26. Piechowiak EI, Peter JF, Kleb B et al. Intravenous iodinated contrast agents amplify DNA radiation damage at CT. Radiology 2015;275:692–7. [DOI] [PubMed] [Google Scholar]
- 27. Wang L, Li Q, Wang XM et al. Enhanced radiation damage caused by iodinated contrast agents during CT examination. Eur J Radiol 2017;92:72–7. [DOI] [PubMed] [Google Scholar]
- 28. Jost G, Golfier S, Pietsch H et al. The influence of x-ray contrast agents in computed tomography on the induction of dicentrics and gamma-H2AX foci in lymphocytes of human blood samples. Phys Med Biol 2009;54:6029–39. [DOI] [PubMed] [Google Scholar]
- 29. Hill MA. The variation in biological effectiveness of X-rays and gamma rays with energy. Radiat Prot Dosimetry 2004;112:471–81. [DOI] [PubMed] [Google Scholar]
- 30. Kraft G, Krämer M, Scholz M. LET, track structure and models. Radiat Environ Biophys 1992;31:161–80. [DOI] [PubMed] [Google Scholar]
- 31. Amato E, Lizio D, Settineri N et al. A method to evaluate the dose increase in CT with iodinated contrast medium. Med Phys 2010;37:4249–56. [DOI] [PubMed] [Google Scholar]
- 32. Amato E, Salamone I, Naso S et al. Can contrast media increase organ doses in CT examinations? A clinical study. Am J Roentgenol 2013;200:1288–93. [DOI] [PubMed] [Google Scholar]
- 33. Kuefner MA, Hinkmann FM, Alibek S et al. Reduction of X-ray induced DNA double-strand breaks in blood lymphocytes during coronary CT angiography using high-pitch spiral data acquisition with prospective ECG-triggering. Invest Radiol 2010;45:182–7. [DOI] [PubMed] [Google Scholar]
- 34. Halm BM, Franke AA, Lai JF et al. γ-H2AX foci are increased in lymphocytes in vivo in young children 1 h after very low-dose X-irradiation: a pilot study. Pediatr Radiol 2014;44:1310–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cai Z, Vallis KA, Reilly RM. Computational analysis of the number, area and density of γ -H2AX foci in breast cancer cells exposed to 111In-DTPA-hEGF or gamma-rays using Image-J software. Int J Radiat Biol 2009;85:262–71. [DOI] [PubMed] [Google Scholar]
- 36. Moquet J, Barnard S, Staynova A et al. The second gamma-H2AX assay inter-comparison exercise carried out in the framework of the European biodosimetry network (RENEB). Int J Radiat Biol 2017;93:58–64. [DOI] [PubMed] [Google Scholar]
- 37. Barnard S, Ainsbury EA, Al-hafidh J et al. The first gamma-H2AX biodosimetry intercomparison exercise of the developing European biodosimetry network RENEB. Radiat Prot Dosimetry 2015;164:265–70. [DOI] [PubMed] [Google Scholar]
- 38. Jost G, Lengsfeld P, Voth M et al. The influence of tube voltage and phantom size in computed tomography on the dose–response relationship of dicentrics in human blood samples. Phys Med Biol 2010;55:3237–48. [DOI] [PubMed] [Google Scholar]
- 39. Stephan G, Schneider K, Panzer W et al. Enhanced yield of chromosome aberrations after CT examinations in paediatric patients. Int J Radiat Biol 2007;83:281–7. [DOI] [PubMed] [Google Scholar]
- 40. Kanagaraj K, Abdul Syed Basheerudeen S, Tamizh Selvan G et al. Assessment of dose and DNA damages in individuals exposed to low dose and low dose rate ionizing radiations during computed tomography imaging. Mutat Res Genet Toxicol Environ Mutagen 2015;789–790:1–6. [DOI] [PubMed] [Google Scholar]
- 41. Abe Y, Miura T, Yoshida MA et al. Increase in dicentric chromosome formation after a single CT scan in adults. Sci Rep 2015;5:13882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lloyd DC, Edwards AA, Prosser JS et al. A collaborative exercise on cytogenetic dosimetry for simulated whole and partial body accidental irradiation. Mutat Res 1987;179:197–208. [DOI] [PubMed] [Google Scholar]
- 43. Lloyd DC, Edwards AA, Leonard A et al. Chromosomal aberrations in human lymphocytes induced in vitro by very low doses of X-rays. Int J Radiat Biol 1992;61:335–43. [DOI] [PubMed] [Google Scholar]
- 44. Barquinero JF, Barrios L, Caballin MR et al. Establishment and validation of a dose–effect curve for gamma-rays by cytogenetic analysis. Mutat Res 1995;326:65–9. [DOI] [PubMed] [Google Scholar]
- 45. M’Kacher R, Violot D, Aubert B et al. Premature chromosome condensation associated with fluorescence in situ hybridisation detects cytogenetic abnormalities after a CT scan: evaluaton of the low-dose effect. Radiat Prot Dosimetry 2003;103:35–40. [DOI] [PubMed] [Google Scholar]
- 46. Ramsey MJ, Moore DH II, Briner JF et al. The effects of age and lifestyle factors on the accumulation of cytogenetic damage as measured by chromosome painting. Mutat Res 1995;338:95–106. [DOI] [PubMed] [Google Scholar]
- 47. Shi L, Fujioka K, Sun J et al. A modified system for analyzing ionizing radiation–induced chromosome abnormalities. Radiat Res 2012;177:533–8. [DOI] [PubMed] [Google Scholar]
- 48. Vaurijoux A, Gruel G, Pouzoulet F et al. Strategy for population triage based on dicentric analysis. Radiat Res 2009;171:541–8. [DOI] [PubMed] [Google Scholar]
- 49. Romm H, Ainsbury E, Barnard S et al. Automatic scoring of dicentric chromosomes as a tool in large scale radiation accidents. Mutat Res 2013;756:174–83. [DOI] [PubMed] [Google Scholar]
- 50. Gonzalez JE, Romero I, Gregoire E et al. Biodosimetry estimation using the ratio of the longest:shortest length in the premature chromosome condensation (PCC) method applying autocapture and automatic image analysis. J Radiat Res 2014;55:862–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. M’Kacher R, El Maalouf E, Terzoudi G et al. Detection and automated scoring of dicentric chromosomes in nonstimulated lymphocyte prematurely condensed chromosomes after telomere and centromere staining. Int J Radiat Oncol Biol Phys 2015;91:640–9. [DOI] [PubMed] [Google Scholar]
- 52. Castelain P, Van Hummelen P, Deleener A et al. Automated detection of cytochalasin-B blocked binucleated lymphocytes for scoring micronuclei. Mutagenesis 1993;8:285–93. [DOI] [PubMed] [Google Scholar]
- 53. Depuydt J, Baeyens A, Barnard S et al. RENEB intercomparison exercises analyzing micronuclei (Cytokinesis-block Micronucleus Assay). Int J Radiat Biol 2017;93:36–47. [DOI] [PubMed] [Google Scholar]
- 54. Lyulko OV, Garty G, Randers-Pehrson G et al. Fast image analysis for the micronucleus assay in a fully automated high-throughput biodosimetry system. Radiat Res 2014;181:146–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rothkamm K, Balroop S, Shekhdar J et al. Leukocyte DNA damage after multi-detector row CT: a quantitative biomarker of low-level radiation exposure. Radiology 2007;242:244–51. [DOI] [PubMed] [Google Scholar]
- 56. Kuefner MA, Grudzenski S, Hamann J et al. Effect of CT scan protocols on x-ray–induced DNA double-strand breaks in blood lymphocytes of patients undergoing coronary CT angiography. Eur Radiol 2010;20:2917–24. [DOI] [PubMed] [Google Scholar]
- 57. Geisel D, Zimmermann E, Rief M et al. DNA double-strand breaks as potential indicators for the biological effects of ionising radiation exposure from cardiac CT and conventional coronary angiography: a randomised, controlled study. Eur Radiol 2012;22:1641–50. [DOI] [PubMed] [Google Scholar]
- 58. May MS, Brand M, Wuest W et al. Induction and repair of DNA double-strand breaks in blood lymphocytes of patients undergoing 18F-FDG PET/CT examinations. Eur J Nucl Med Mol Imaging 2012;39:1712–9. [DOI] [PubMed] [Google Scholar]
- 59. Brand M, Sommer M, Achenbach S et al. X-ray induced DNA double-strand breaks in coronary CT angiography: comparison of sequential, low-pitch helical and high-pitch helical data acquisition. Eur J Radiol 2012;81:e357–62. [DOI] [PubMed] [Google Scholar]
- 60. Nguyen PK, Lee WH, Li YF et al. Assessment of the radiation effects of cardiac CT angiography using protein and genetic biomarkers. JACC Cardiovasc Imaging 2015;8:873–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Golfier S, Jost G, Pietsch H et al. Dicentric chromosomes and γ-H2AX foci formation in lymphocytes of human blood samples exposed to a CT scanner: a direct comparison of dose response relationships. Radiat Prot Dosimetry 2009;134:55–61. [DOI] [PubMed] [Google Scholar]
- 62. Abe Y, Miura T, Yoshida MA et al. Analysis of chromosome translocation frequency after a single CT scan in adults. J Radiat Res 2016;57:220–6. [DOI] [PMC free article] [PubMed] [Google Scholar]