Abstract
The cytosolic chaperonin CCT (chaperonin containing TCP-1) is an ATP-dependent double-ring protein machine mediating the folding of members of the eukaryotic cytoskeletal protein families. The actins and tubulins are obligate substrates of CCT because they are completely dependent on CCT activity to reach their native states. Genetic and proteomic analysis of the CCT interactome in the yeast Saccharomyces cerevisiae revealed a CCT network of approximately 300 genes and proteins involved in many fundamental biological processes. We classified network members into sets such as substrates, CCT cofactors and CCT-mediated assembly processes. Many members of the 7-bladed propeller family of proteins are commonly found tightly bound to CCT isolated from human and plant cells and yeasts. The anaphase promoting complex (APC/C) cofactor propellers, Cdh1p and Cdc20p, are also obligate substrates since they both require CCT for folding and functional activation. In vitro translation analysis in prokaryotic and eukaryotic cell extracts of a set of yeast propellers demonstrates their highly differential interactions with CCT and GroEL (another chaperonin). Individual propeller proteins have idiosyncratic interaction modes with CCT because they emerged independently with neo-functions many times throughout eukaryotic evolution. We present a toy model in which cytoskeletal protein biogenesis and folding flux through CCT couples cell growth and size control to time dependent cell cycle mechanisms.
This article is part of a discussion meeting issue ‘Allostery and molecular machines’.
Keywords: chaperonin CCT, protein folding, actin, tubulin, 7-bladed WD40 propellers, APC/C
1. Introduction
The cytosolic chaperonin CCT is a 1 MDa multi-subunit protein complex which functions in cytoskeletal protein folding in all eukaryotes [1–3]. CCT stands for chaperonin containing tailless complex polypeptide 1 (TCP-1), and is also named TRiC (TCP-1 Ring Complex). CCT is a Group II chaperonin and like all chaperonins has a double-toroid shape enclosing a central cavity and can, upon binding and hydrolysis of ATP, assist in the folding of non-native proteins or in the assembly of substrates with their respective partners. CCT is constructed from eight protein subunits, encoded by eight homologous genes, positioned in each ring in a precise arrangement [4,5]. Each CCT subunit consists of three domains: the equatorial domain harbouring the ATP binding site, an intermediate domain, and the apical domain involved in substrate binding, as first identified in the X-ray crystal structure of the Group I chaperonin GroEL [6]. The sequence differences between the eight CCT subunits are located mainly in their apical domains [4,7] which provide the substrate binding specificity of each subunit [8]. The selective nature of the CCT machinery and its individual subunits was revealed in cryo-EM maps of CCT-actin [9] and CCT-tubulin complexes [10,11] and through direct biochemical analyses of actin binding sites [12–14]. CCT shows sequential allosteric behaviour in its ATP-binding and hydrolysis regime [15,16] and in its actin-folding regime [4,11,17]. Our long-standing view is that CCT actively folds a very restricted group of non-cytoskeletal proteins which includes many members of the 7-bladed WD40-propeller repeat protein family. The core set of obligate CCT substrates, together with their cofactors and other binding partners, is involved in ancient and fundamental biological processes including transcription, chromatin modification and secretion. Figure 1 shows the core CCT interactome in pictorial form to aid the reader grasp the depth and breadth of the system.
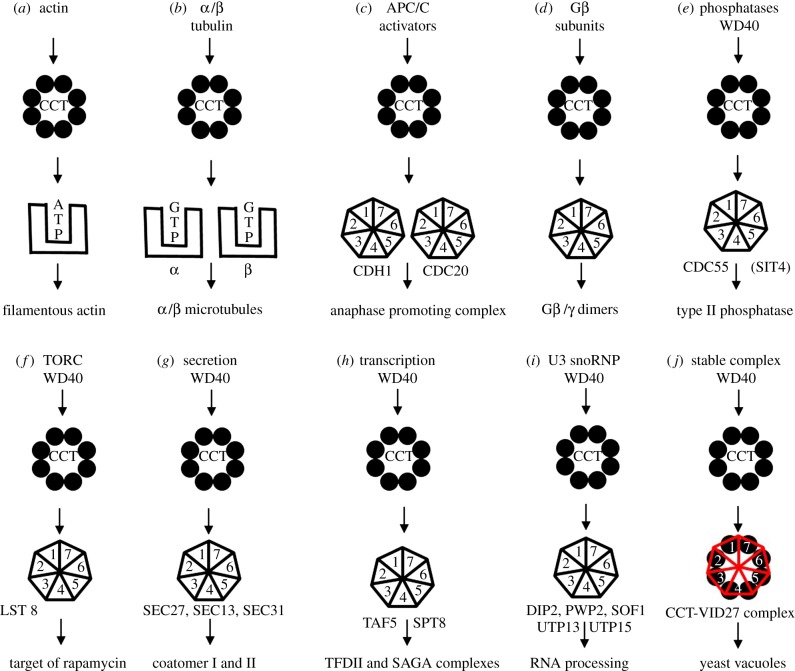
Figure 1.
Cartoons of schemes for CCT action upon its substrates and binding complex formation. (a) Direct folding to native state: actin filaments (F-actin) are composed of a single monomer species (G-actin) and CCT activity yields assembly competent monomers. It is not yet known if ATP loading is concomitant with folding or whether nucleotide-free actin is released into the 2–5 mM ATP pool present in cell cytoplasm and then equilibrates with free ATP. (b) Folding and assembly: tubulin filaments are composed of two monomer species (α-tubulin and β-tubulin) which both bind guanosine triphosphate (GTP). GTP concentration is low in cells (50 µM) and CCT may well play a direct role in GTP loading of β-tubulin. There is strong evidence for post-CCT-dependent folding and assembly step(s), involving several cofactor proteins, which produce filament-assembly competent tubulin monomers. (c) Production of the APC activators: Cdc20p and Cdh1p propeller proteins are CCT-dependent. (d) Assembly of G-protein complexes [18]. (e) Type II phosphatase complex—CCT interaction conserved from yeast to humans. (f) TORC complex: WD40 subunits. (g) Secretory complexes ER and Golgi. (h) Transcription factor and several histone deacetylase complexes interact with CCT [19]. (i) RNA processing. (j) Holding activity of the Saccharomyces cerevisiae Vid27p propeller protein involved in autophagy. (Online version in colour.)
Willison [20] has reviewed the CCT structure and allosteric mechanism and its role in normal biological processes and in disease. New data on the WD40 interactions with CCT in Saccharomyces cerevisiae and mutagenesis of CCT-binding motifs in the Cdh1 co-activator of APC/C are presented and reviewed in this paper. We then calculate the folding capacity of the CCT pool and the variance in CCT complex copy number in growing yeast and propose that access to CCT interaction may be a mechanism through which proteins can detect the instantaneous flux of actin and tubulin synthesis via CCT-binding site availability manifested through their individual effective association rate constants.
2. The CCT interactome
All eight CCT genes are essential in yeast [19] and even minor perturbations of any of their ATP-binding site motifs [21] and ATP-hydrolysis kinetics [22] result in loss of viability and severe growth defects which include abnormally large cell sizes and defective cell shapes. Although the genetic and physical CCT interactome comprises approximately 300 members in yeast [19] most of the components are not actively folded by CCT but are cofactors and ancillary genetic effectors. The reason for the existence of such a large and complex network is that CCT has become coupled to many fundamental biological processes because it co-evolved with, and facilitated, the novel biophysical properties of the emerging cytoskeleton in the primitive eukaryotic cell [1,8,23]. Recent independent computational analysis [24] and global experiments in yeast concur with the view that CCT is highly selective towards the cytoskeleton in its network specificity [25].
We produced the comprehensive yeast CCT ‘interactome’ by employing mass spectrometric proteomics experiments using CCT-3CBP and CCT-6CBP tagged complexes and a Synthetic Genetic Array (SGA) screen for synthetic lethal interactions with a temperature-sensitive cct1-2 allele [19]. Using the combined results of these integrative approaches, a large number of novel CCT physical and genetic interactors were identified, defining several functional interaction networks of CCT. Integral to the proteomics experiment was the focus on ATP-dependent release from CCT, which was taken as a potential indicator of interacting proteins that are folding substrates. Before subjecting CCT to liquid chromatography-electrospray ionisation mass spectrometry (LC-ESI-MS), the chaperonin, while bound to calmodulin-resin through the affinity tag in the CCT3 or the CCT6 subunit, was washed with either a buffer containing ATP or a plain buffer. The rationale behind this was that CCT, which is still active when bound to calmodulin-resin, will fold and release its substrates only in the presence of ATP, and the released substrates that are subsequently washed away will not pass through the mass spectrometer. We identified many ATP-elutable CCT interacting proteins, of which the most abundant matches included known CCT interacting proteins, namely the folding substrates actin and tubulin, and their respective folding regulators Plp2p and Plp1p [26,27]. Dekker et al. [19] only identified one WD40 protein in the physical interaction set but subsequent higher resolution LC-ESI-MS analysis in our group, using more advanced mass spectrometers, routinely recovers the following avid CCT-binding WD40 proteins: Vid27p, Sec31p, Spt8p, Sec13p and Cdc55p. It is difficult to estimate the coverage of the MS analysis in our work but the dataset is certainly incomplete for many technical reasons such as variable biochemical stringency of CCT interactions and low cellular abundance of some partner proteins such as the APC/C activators, Cdh1p and Cdc20p, which are dependent on CCT for their folding and maturation in vivo [28] and in vitro [29]. Arava et al. [30] calculated protein synthesis rates from 5701 mRNAs in yeast; Actp is synthesized at a rate of 2.2 proteins per second whereas Cdh1p and Cdc20p are synthesized 100x and 366x more slowly. It also is unsurprising that we did not detect CCT-bound Cdh1p and Cdc20p because SILAC mass spectrometry fails to recover these two proteins from over 4000 proteins identified [31]. In the early global protein interaction screen in yeast the 16 CCT-binding WD40 proteins identified were all over-expressed as high copy number baits supporting the suggestion that many of the WD40 interactors are indeed low copy number proteins [2,32]. The core evolutionarily conserved WD40 proteins are components of the Type II phosphatase system (Cdc55p) and TOR complex (Lst8), RNA polymerase II and chromatin modification (Taf5p), RNA processing (U3 snoRNP), secretion (Coatomer I and II) and cell cycle control through protein degradation control (Cdh1p and Cdc20p) as defined in Dekker et al. [19] and sketched in figure 1.
3. 7-bladed WD40-repeat protein family
The WD40 repeat proteins contain multiple, tandem copies of an approximately 40 amino acid repeat which forms a blade composed of four anti-parallel β-sheets (figure 2). The WD repeat begins with a glycine-histidine (GH) dipeptide and ends with the tryptophan-aspartic acid (WD) dipeptide, hence the name [33,34]. Individual WD repeats show great sequence variation at every residue position and even the GH and WD dipeptides are not invariant [34] as observed for Cdh1p in the alignment in figure 4. In addition, the number of tandem repeats differs and variable-sized insertions are found both within and between them, which makes the bioinformatic identification and characterization of WD-repeat proteins very difficult. For example, Ski8p, a protein regulating mRNA degradation, had been predicted to contain five WD repeats in its primary sequence but crystallographic analysis revealed seven WD repeats within the sequence and the two extra repeats are so divergent from the predicted WD-consensus sequence that they cannot be detected from the protein's primary sequence [35]. Although there are examples of WD proteins having eight blades, such as Cdc4p [36] and Sif2p [37], the most common arrangement is for seven tandem WD repeats to form the circularized β-propeller structure as in Tup1p (figure 2). A newly constructed database of WD40-repeat proteins (WDSPdb) lists 83 yeast proteins containing more than six repeats [38]. We have constructed a 30-member sub-family of homologous 7-bladed WD40 proteins in S. cerevisiae using sequential Psi-BLAST homology searching, Phyre analysis and hand alignment (table 1). The members of this family in S. cerevisiae participate in many different cellular functions but the common property of the WD40 propeller domains is that they mediate protein–protein interactions, often as components of large protein complexes. The β-sheets of the propeller structure provide a stable platform for protein–protein interaction via the loops that connect them. Structure/function analysis of Doa1p shows that its propeller binds ubiquitin as do those from Cop1p, Sec27p and Taf5p [39] and the APC/C cofactors. No enzyme activity has yet been attributed to any propeller domain.
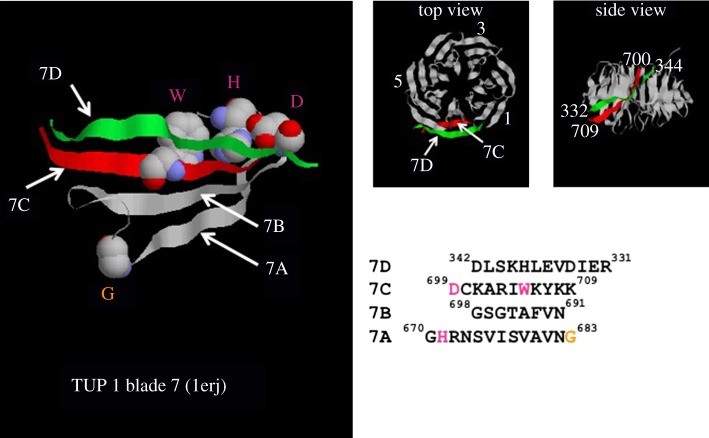
Figure 2.
The Velcro cap. Structural model of the propeller of TUP1 (PDB: 1erj). TUP1 is a 713-amino acid residue protein with a single 7-bladed propeller. The left-hand panel shows a ribbon diagram with the W, D and H residues of the WD40 repeat highlighted in pink which interact to form a structural element connecting strands 7A and 7C across which strand 7D binds like a ‘Velcro’ cap [33]. The two right-hand panels show top and side views of the propeller, a conical frustum. The amino acid sequences of the interaction region between the β-sheets are shown below, with the four sequences aligned to reflect the registration of the blades in three-dimensional space (KR Willison 2011, unpublished analysis).
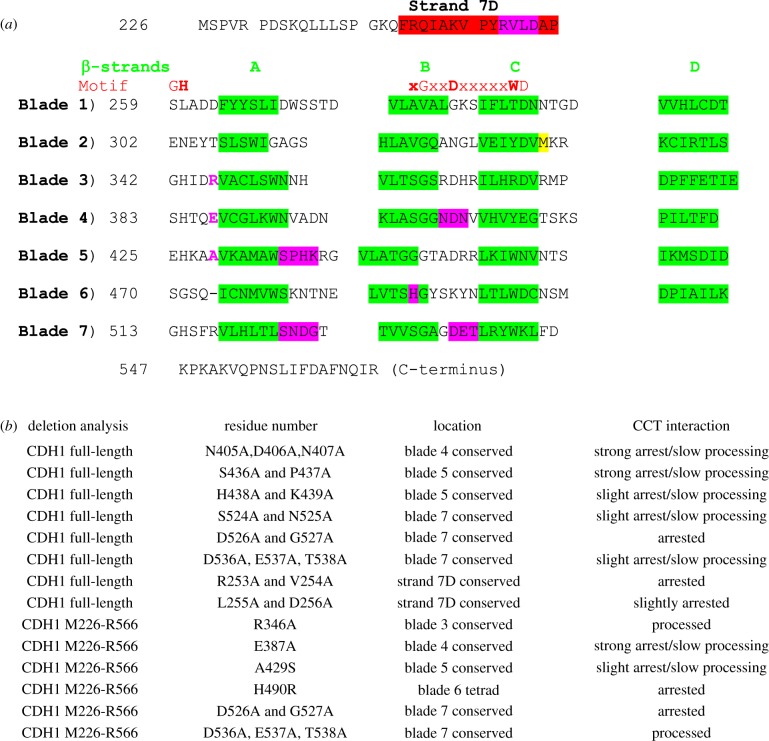
Figure 4.
Mapping CCT-binding residues in the Cdh1p propeller. Mutants in the Cdh1p propeller (amino acid residues 226–566) were screened for strength of binding and degree of processing by CCT using in vitro translation/native gel analysis. Six mutations were tested in the context of the propeller domain only. (a) The β-strands in the blades are highlighted in green. Motif: the red bold letters highlight the residues (GH-G-D-WD) forming the hydrogen-bonding network (‘structural tetrad’) which stabilizes the blades. The M332A mutation, coloured yellow, is included to prevent internal translation initiation at this methionine which reduces the accuracy of quantitation in these assays [27]; the equivalent residue in human CDH1 is alanine. The point mutants which arrest the processing are highlighted in magenta. (b) List of 14 mutants and their CCT interaction behaviour. Strong arrest indicates that greater than 15% of the counts become associated with CCT; slow processing means that the appearance of the folded Cdh1 (either full-length or the propeller domain) is retarded compared to wild-type control.
Table 1.
List of the core 7-bladed WD40-repeat-containing proteins in yeast. This set was constructed using sequential Psi-BLAST homology searching, Phyre analysis and hand alignment—full alignment file is available on request. Gene name, protein complex, length in amino acids and haploid versus diploid protein expression ratios are from Saccharomyces cerevisiae. The UNIPROT entry list shows the genes from Encephalitis cuniculi parasite which is a eukaryote with a minimal (approx. 1990 genes) and compacted genome; all the WD40 proteins are detected by homology except TUP1.
| number | gene name | protein complex | UNIPROT | length aa | haploid versus diploid |
|---|---|---|---|---|---|
| 1 | COP1 | coatomer I | Q8SSJ2 | 1201 | 1 |
| 2 | SEC27 | coatomer I | Q8SRA6 | 889 | 0.96 |
| 3 | SEC13 | coatomer II | Q8SQT8 | 297 | 1.05 |
| 4 | SEC31 | coatomer II | Q8SRF6 | 1273 | 0.97 |
| 5 | ASC1 | ribosome | Q8SRB0 | 319 | 0.93 |
| 6 | RSA4 | ribosome | Q8SW59 | 515 | 1.01 |
| 7 | TAF5 | TFIID | Q8SQS4 | 798 | 1.12 |
| 8 | ELP2 | Pol II elongation | Q8SSL8 | 788 | 1.02 |
| 9 | HIR1 | histone repressor | Q8SSG4 | 875 | 1.13 |
| 10 | HAT2 | acetyltransferase | Q8SRK1 | 401 | 1.03 |
| 11 | SWD2 | Set1p complex | Q8SVQ1 | 329 | 1.15 |
| 12 | DOA1 | ubiquitin | Q8SQK6 | 715 | 0.98 |
| 13 | CAC2 | CAF1-p60 | Q8SRA5 | 468 | 1.12 |
| 14 | TUP1 | transcription | ND | 713 | 0.91 |
| 15 | CDC55 | PP2A | Q8SRX4 | 526 | 1.08 |
| 16 | CDC20 | APC/C | Q8SS21 | 610 | not detected |
| 17 | CDH1 | APC/C | Q8SVZ6 | 566 | not detected |
| 18 | PRP46 | RNA splicing | Q8STZ5 | 451 | 1.04 |
| 19 | PFS2 | PolyA factor | Q8SW96 | 465 | 1.03 |
| 20 | GLE2 | nuclear pore (NPC) | Q8SRM6 | 365 | 0.98 |
| 21 | DIP2 | U3 snoRNP (SSU) | Q8SW87 | 943 | 0.98 |
| 22 | PWP2 | U3 snoRNP (SSU) | Q8SSO7 | 923 | 1.02 |
| 23 | SOF1 | U3 snoRNP (SSU) | Q8SSI4 | 489 | 1.02 |
| 24 | UTP13 | U3 snoRNP (SSU) | Q8SVM7 | 817 | 0.99 |
| 25 | UTP15 | U3 snoRNP (SSU) | Q8SU33 | 513 | 1.01 |
| 26 | TIF34 | initiation factor 3 | Q8SR77 | 347 | 0.98 |
| 27 | RRB1 | ribosome biogenesis | Q8SRW1 | 512 | 0.95 |
| 28 | CIA1 | Fe/S protein assembly | Q8STN6 | 330 | 1.03 |
| 29 | YL149 | hypothetical | Q8STM2 | 730 | 1.12 |
| 30 | VID27 | vacuole | Q8STM0 | 782 | 1.32 |
Early studies on folding of WD40-repeat proteins from the group of Neer lead to the ‘Velcro’ model of folding [40] so named because it was noticed that the seventh blade of the propeller structure is composed of three contiguous β-strands located at the C-terminus of the sequence but the fourth is encoded at the N-terminus of the repeats. The two segments associate together in three-dimensional space and interact to form the Velcro cap and thus close the propeller fold. Figure 2 shows structural details of the Tup1p Velcro cap. Bringing these four strands together in space to complete folding must be under careful kinetic control and it is likely that different WD40 folds have evolved with different folding speeds and therefore different chaperone dependencies due to kinetic partitioning. Kinetic partitioning of proteins due to alterations of local folding rates in their folding pathways is likely to be responsible for GroE-dependent versus GroE-independent folding behaviour and accounts for the fact that single point mutations can convert MetK from Ureaplasma urealyticum, a bacterium with no GroE chaperonin system, into an obligate substrate of E. coli GroEL/ES [41].
When I first made the CCT-WD40 connection it was obvious that the CCT ring had the required shape and dimensions to facilitate the binding of the propeller [2]. Recent cryo-electron microscopy structures have shown that the human Gβ1 propeller is near native and bound to the CCT6, CCT3, CCT1, CCT4 side of the ring of insect CCT [42]. If higher resolution structures can demonstrate that the role of CCT in WD40 propeller folding turns out to be the one of permitting the two ends of the repeat to interact effectively to form blade 7 it would be analogous to our mechanistic model for the role of CCT in actin folding in which the manipulation of the C-terminus of the actin intermediate is the second critical reaction step preceding release of folded actin [4,17,20,43].
4. In vitro interactions between CCT and WD40-repeat proteins
In order to probe different WD40-repeat proteins for their CCT interaction behaviour 15 yeast proteins (table 2) from the core 30-member 7-bladed WD40 set (table 1) and the 8-bladed Cdc4p were tested. They were tested for CCT and GroEL binding using standard in vitro translation time course systems [13,47]. The results are a clear demonstration of multiple modes of interaction with CCT (table 2 and figure 3). Some proteins do not bind CCT or GroEL whereas others bind both chaperonins strongly (sample assay data in figure 3). Although 14 of these 16 proteins showed some degree of binding to CCT, only five demonstrated very strong interaction with it. These strong CCT interactors are Cdc55p, Vid27p, Taf5p, Cdh1p and Cdc20p, each of which has been identified in other genetic and physical studies (table 1 and figure 1). It is striking that these proteins accumulate on CCT over time unlike actin which has a demonstrable precursor-product relationship in the rabbit reticulocyte lysate system because it is being actively processed and folded by CCT [13]. This reinforces a common role for CCT in the assembly, rather than folding, of these WD40 proteins with elements of their respective protein complexes, as is clearly the case for the Gβ-Gγ heterodimer assembly process [42]. The stalling of the folding of WD40 propellers due to the blade-7 completion problem permits CCT to intervene in the process and provide a holding platform until binding partners arrive.
Table 2.
Screening WD40-repeat proteins for CCT interaction. Fifteen yeast proteins predicted to contain 7-blade WD40 repeats, by 3DPSSM structure-based sequence alignment [44], and CDC4, an 8-bladed WD40 [36], non-CCT interacting protein functional in cct1-2 mutant cells at 37°C [28] were analysed for interaction with CCT (column 3) and GroEL (column 4) by quantitative in vitro translation and native gel analysis. The extent of binding to CCT or GroEL was determined by calculating the fraction bound to chaperonin compared to total protein synthesized (weak ≤ 5%, medium = 5–15%, strong ≥ 15%). Results from independent studies are shown in column 5 and referenced in column 6. nd, not done; nt, not previously studied.
| protein | protein complex/function | CCT binding this study | GroEL binding this study | CCT binding published work | ref. |
|---|---|---|---|---|---|
| ASC1 | ribosome subunit/signalling | weak | weak | nt | |
| CAC2 | CAF1-p60/chromatin | medium | yes | nt | |
| CDC4 | Skip1/ubiquitin | weak | yes | no | [28] |
| CDC20 | APC/ubiquitin E3 | strong | yes | yes + in vivo | [32,28] |
| CDC55 | PP2A regB/phosphatase | strong | nd | yes + in vivo | [32,29] |
| CDH1 | APC/ubiquitin E3 | strong | yes | yes + in vivo | [32,28,29] |
| DOA1 | ubiquitin | weak | no | nt | |
| ELP2 | Pol II elongation/transcription | negative | no | nt | |
| HAT2 | histoneAT Type B/acetylation | weak | no | nt | |
| SEC13 | coatomer II/secretion | weak | no | nt | |
| SEC27 | coatomer I/secretion | medium | nd | yes | [32] |
| TAF5 | TFIID/transcription | strong | yes | yes | [32,29] |
| TIF34 | initiation factor 3/translation | negative | no | nt | |
| TUP1 | Tup1-Ssn6/transcription repression | medium | nd | nt | |
| UTP13 | U3 snoRNA (SSU)/translation | medium | yes | nt | |
| VID27 | vacuole/protein degradation | strong | nd | yes | [45,46] |
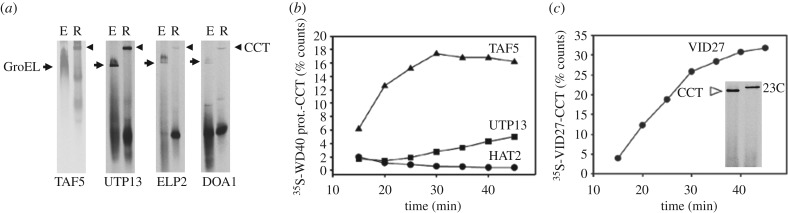
Figure 3.
Screening WD40-repeat proteins for CCT interaction. (a) 6% native PAGE analysis of TAF5, UTP13, ELP2 and DOA1 after in vitro translation in E. coli lysate (E) or rabbit reticulocyte lysate (R) in vitro transcription/translation system to detect GroEL (filled arrowhead) or CCT interaction (open arrowhead) respectively. (b) Graphs of time courses of CCT-bound protein by 6% native PAGE analysis of rabbit reticulocyte in vitro translation assays of TAF5, UTP13 and HAT2. (c) 6% native PAGE analysis of time course of rabbit reticulocyte in vitro translation assay of VID27. Inset image of sample at the end of the time course showing interaction of CCT with Vid27p which can be shifted with the anti-TCP1 antibody 23C, demonstrating specificity of CCT binding. Vid27p is the most avid CCT binding protein in yeast [19,25,46]. Vid27 is a little studied, non-essential, WD40-repeat protein most closely related to BUB3 and possibly involved in vacuolar protein degradation and is found in yeasts and plants but not in higher eukaryotes.
5. CCT and co-activators of the anaphase promoting complex (APC/C)
Cdh1p and Cdc20p are co-activators of the anaphase promoting complex/cyclosome (APC/C), a ubiquitin ligase that has a prominent role in regulating cell cycle progression. The APC/C controls mitosis by regulating entry into anaphase, progression through anaphase, exit of mitosis and G1 phase, by degrading sequentially different cell cycle proteins. It is a 1.5 MDa protein complex and contains 13 core subunits, and three related adaptors, two of which are Cdh1p and Ccd20p [48]. The adaptor proteins regulate the APC/C's activity by binding to the complex to deliver target substrates for ubiquitination. The interaction of Cdh1p and Cdc20p with APC/C allows APC/C's ubiquitinase activity. The WD40-repeat domains of Cdh1 and Cdc20p have been shown to be critical in substrate recognition [49,50].
CCT is the only known folding pathway for the production of functional and biologically active Cdh1p and Cdc20p in vivo [28] and in vitro [29]. Camasses et al. [28] mapped the CCT-interaction sites of Cdc20p to the propeller blades 3, 4 and 5. Some new data which maps CCT-binding sites in Cdh1p have been obtained. It was first shown that the Cdc20p and Cdh1p propellers are interchangeable for CCT interaction by swapping them over and finding that the hybrid proteins are processed normally by rabbit CCT. Consequently, mutations in evolutionarily conserved residues were screened for CCT interaction by in vitro translation time courses. Figure 4 shows the results. There are consequential CCT-interaction sites in blades 4 (N405A, D406A, N407A) and 5 (S436A, P437A). Disrupting the histidine (H490) of the tetrad motif in blade 6 arrests processing. Mutations in the ‘Velcro’ interaction strand were discovered in blade 7D, R253A and V254A, which cause complete arrest of CCT processing of the full-length protein. This strongly suggests the important activity of CCT towards propellers is to facilitate the joining of the two ends of the protein domain together.
6. Connecting cytoskeletal protein biogenesis and folding flux to cellular networks
Proteins interact with the CCT system for reasons that include folding, for the actins and tubulins, and just-in-time assembly processes [51]. Trying to locate a newly synthesized protein about to leave one of the 200 000 ribosomes without a high-abundance and/or high-affinity detection system is a difficult task due to the law of mass action. For a protein such as Cdh1p, with a rate of synthesis 100 times less than actin, when considering the bulk cell synthesis rate not the translation rate, the ability to interact with one of the 3000 CCT complexes is effectively a ‘concentrating’ process that moves a nascent Cdh1 protein from one of 200 000 to one of 3000 points in cytoplasmic space and is readily accomplished because of the very high affinity that the Cdh1 folding intermediate has for CCT [29]. As is the case with actin, whose native state has no affinity for CCT, the Cdh1 protein can fold or assemble with other partners on CCT and then leave, never to return, because its native state or assembled state has relinquished its affinity for CCT. It is likely that there are many variations on this theme controlled by kinetic partitioning and other time-dependent processes such as chemical modifications. A further detection enhancement will occur if a binding partner also has independent binding affinity for separate CCT subunits because the combined affinities become multiplicative through addition of logarithmic association rates constants. This effect is observed for the yeast CCT-ACT1-PLP2 system where the PLP2 cofactor increases the rate of native actin production 30-fold [27].
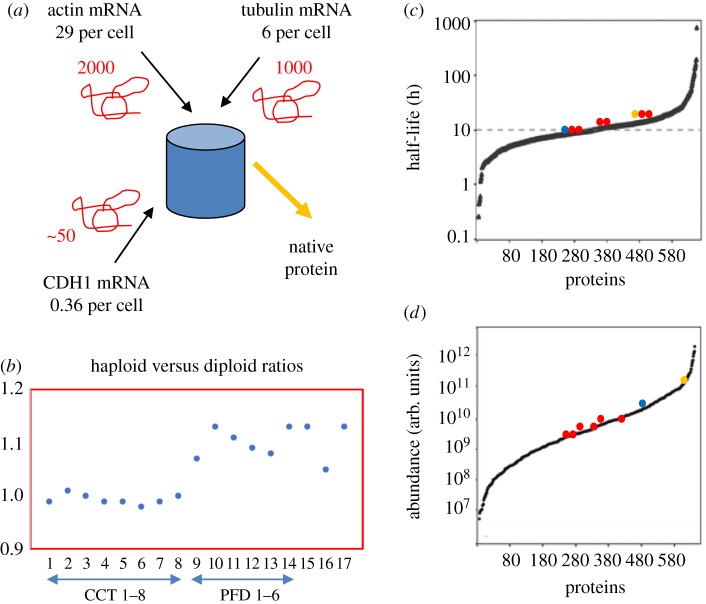
We suggest that the availability of unoccupied and therefore unemployed CCT complexes will be a function of the flux through the CCT system of its two most abundant co-dependent partners, actin and tubulin. The number of available CCT binding sites can be used as a metric for any binding partner to register the instantaneous values of dActin/dt + dTubulin/dt through the copy number of the empty CCT complexes, physically manifested as the effective association rate constant. In S. cerevisiae the number of available CCT complexes will be in the low thousands since CCT is present at between 3000 (biochemical analysis; [47]) and 6000 (mass spectrometry; [52]) copies per yeast cell. Using mass spectrometry analysis of pure CCT–substrate complexes [19] and calculations our laboratory estimates that 50–60% of CCT is occupied with actin and tubulin folding (figure 5a). This flux-counting mechanism is probably the reason for actin and CCT subunit genes being among the most haplo-insufficient genes for growth in yeast YPD medium [54], as are the exosome and U3 snoRNP subunits which are components of the CCT core (figure 1h,j). Haplo-insufficiency is loss of fitness due to absence of one gene copy in a diploid. When gene copy number is increased in aneuploid yeast strains [55] many components of protein complexes show dosage compensation, including all the 137 haplo-insufficient ribosomal subunits. However, CCT subunit levels are not attenuated and continue to maintain the tightest distribution of all the 84 protein complexes analysed. A further genetic test examines epistatic interactions between CCT and other genes. Epistasis is a measurement of the fitness of pairs of double mutants compared to wild-type and single mutants and has been measured for 61 gene pairs in S. cerevisiae [56]. CCT2 is strongly positively epistatic with ACT1, NUS1, RPC10 and RP55. A direct way to measure the variance in CCT subunit levels is to use the protein signals obtained by comparing haploid versus diploid yeast in datasets from mass spectrometric SILAC experiments [31]. Figure 5b shows a plot of haploid versus diploid protein abundance ratios for CCT and prefoldin (PFD) complexes, and actin and tubulin and PLP2. The standard deviation of the eight CCT subunits (mean = 0.994) is 2.8 times less than the six prefoldin subunits (mean = 1.017) and 6.5 times less than the 27-member WD40 protein set (values in table 1—excluding Vid27p).
Figure 5.
Yeast CCT capacity. Number estimates for the components of this yeast model are from the following sources. (a) Calculated mRNA copy numbers per cell based upon 15 000 transcripts per cell growing at 30°C - ACT1: 28.97, CDH1: 0.36, TUB2: 5.94, TCP1: 1.81, CCT2: 1.56, CCT3: 1.62, CCT4: 2.16, CCT5: 2.24, CCT6: 2.61, CCT7: 2.23, CCT8: 1.7, CDH1: 0.36 [30]. Red squiggles represent estimate of steady state number of nascent chains bound to CCT. (b) Haploid versus diploid protein ratios for CCT subunits (1–8), prefoldin subunits (9–14), ACT1 (15), TUB2 (16), PLP2 (17) taken from de Godoy et al. [31]. (c,d) Half-live distributions (hours) and steady-state abundance distributions (arbitrary units) of 641 proteins [53] are plotted according to increasing value on the x-axis. Half-lives and adundances of CCT subunits (red dots), ACT1 (yellow dot) and TUB2 (blue dot) are highlighted on each distribution. (Online version in colour.)
The tighter the number distributions of the main substrates, actin and tubulin, and the recipient CCT complex pool the more accurate and less noisy and more sensitive the flux measurement system becomes. In this respect it is also significant that actin, tubulin and CCT subunits sit in the centre of the Gaussian mean of the protein abundance distribution in yeast (figure 5d) and protein half-life distribution (figure 5c) because they will have median concentrations throughout the cytoplasm and median half-lives which are well in excess of the 3-hour cell cycle time for yeast growing in YPD medium at 30°C. In conclusion, according to this model, CCT is not only a physical gatekeeper between folding space and native space but also a device which can help transmit information about the state of the cell to the many elements of the essential protein complexes which bind it.
7. Discussion
All our experiments concerning the CCT substrate spectrum in mammalian cells and yeast teach us that CCT has a highly restricted set of substrates. Other laboratories disagree with us and originally put the figure at 9–15% of all cytosolic proteins [57]. The most recent study suggests 7% of cytosolic proteins are substrates based upon binding of in vitro denatured, pulse-chase-labelled total cell extracts to chaperonins [58]. Our laboratory has performed pulse chase analysis of newly translated polypeptides in mammalian cells in vivo which never recovers more than 1% of the total protein counts bound to CCT [59,60]. The labelled proteins that are bound are dominated by actin and tubulin signals. It is becoming even more difficult to sustain the view that CCT is a general cytosolic chaperone in the light of global gene and protein network analysis of the CCT interactome in yeast [19,25] which points to the central involvement of CCT in cytoskeletal function and several essential cellular networks involved in chromatin remodelling, secretory pathways, phosphatase activity and cell cycle regulation via control of the folding of the APC/C regulators. It is remarkable how, in each network, always one or more CCT-dependent WD40-repeat proteins are involved (figure 1).
Acknowledgements
Elizabeth McCormack provided technical assistance.
Data accessibility
This article has no additional data.
Competing interests
We declare we have no competing interests.
Funding
This work was funded by Cancer Research UK and an EPSRC Platform Grant.
References
- 1.Willison KR. 1999. Composition and function of the eukaryotic cytosolic chaperonin containing TCP-1. In Molecular chaperones and folding catalysts. Regulation, cellular function and mechanisms (ed. Bukau B.), pp. 555–571. Amsterdam, The Netherlands: Harwood Academic Publishers. [Google Scholar]
- 2.Valpuesta JM, Martin-Benito J, Gomez-Puertas P, Carrascosa JL, Willison KR. 2002. Structure and function of a protein folding machine: the eukaryotic cytosolic chaperonin CCT. FEBS Lett. 529, 11–16. ( 10.1016/S0014-5793(02)03180-0) [DOI] [PubMed] [Google Scholar]
- 3.Valpuesta JM, Carrascosa JL, Willison KR. 2005. Structure and function of the cytosolic chaperonin CCT. In Protein folding handbook (eds Buchner J, Kiefhaber T), pp. 725–755. Weinheim, Germany: Wiley-VCH Verlag GmbH. [Google Scholar]
- 4.Dekker C, Roe SM, McCormack EA, Beuron F, Pearl LH, Willison KR. 2011. The crystal structure of yeast CCT reveals intrinsic asymmetry of eukaryotic cytosolic chaperonins. EMBO J. 30, 3078–3090. ( 10.1038/emboj.2011.208) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalisman N, Adams CM, Levitt M. 2012. Subunit order of eukaryotic TRiC/CCT chaperonin by cross-linking, mass spectrometry, and combinatorial homology modeling. Proc. Natl Acad. Sci. USA 109, 2884–2889. ( 10.1073/pnas.1119472109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braig K, Otwinowski Z, Hegde R, Boisvert DC, Joachimiak A, Horwich AL, Sigler PB. 1994. The crystal-structure of the bacterial chaperonin GroEl at 2.8 Å. Nature 371, 578–586. ( 10.1038/371578a0) [DOI] [PubMed] [Google Scholar]
- 7.Pappenberger G, Wilsher JA, Roe SM, Counsell DJ, Willison KR, Pearl LH. 2002. Crystal structure of the CCTγ apical domain: implications for substrate binding to the eukaryotic cytosolic chaperonin. J. Mol. Biol. 318, 1367–1379. ( 10.1016/S0022-2836(02)00190-0) [DOI] [PubMed] [Google Scholar]
- 8.Kubota H, Hynes G, Carne A, Ashworth A, Willison K. 1994. Identification of six Tcp-1-related genes encoding divergent subunits of the TCP-1-containing chaperonin. Curr. Biol. 4, 89–99. ( 10.1016/S0960-9822(94)00024-2) [DOI] [PubMed] [Google Scholar]
- 9.Llorca O, McCormack EA, Hynes G, Grantham J, Cordell J, Carrascosa JL, Willison KR, Fernandez JJ, Valpuesta JM. 1999. Eukaryotic type II chaperonin CCT interacts with actin through specific subunits. Nature 402, 693–696. ( 10.1038/45294) [DOI] [PubMed] [Google Scholar]
- 10.Llorca O, Martín-Benito J, Ritco-Vonsovici M, Grantham J, Hynes GM, Willison KR, Carrascosa JL, Valpuesta JM. 2000. Eukaryotic chaperonin CCT stabilizes actin and tubulin folding intermediates in open quasi-native conformations. EMBO J. 19, 5971–5979. ( 10.1093/emboj/19.22.5971) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Llorca O, Martın-Benito J, Grantham J, Ritco-Vonsovici M, Willison KR, Carrascosa JL, Valpuesta JM. 2001. The ‘sequential allosteric ring’ mechanism in the eukaryotic chaperonin-assisted folding of actin and tubulin. EMBO J. 20, 4065–4075. ( 10.1093/emboj/20.15.4065) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hynes GM, Willison KR. 2000. Individual subunits of the eukaryotic cytosolic chaperonin mediate interactions with binding sites located on subdomains of beta-actin. J. Biol. Chem. 275, 18 985–18 994. ( 10.1074/jbc.M910297199) [DOI] [PubMed] [Google Scholar]
- 13.McCormack EA, Llorca O, Carrascosa JL, Valpuesta JM, Willison KR. 2001. Point mutations in a hinge linking the small and large domains of β-actin result in trapped folding intermediates bound to cytosolic chaperonin CCT. J. Struct. Biol. 135, 198–204. ( 10.1006/jsbi.2001.4385) [DOI] [PubMed] [Google Scholar]
- 14.Neirynck K, Waterschoot D, Vandekerckhove J, Ampe C, Rommelaere H. 2006. Actin interacts with CCT via discrete binding sites: a binding transition-release model for CCT-mediated actin folding. J. Mol. Biol. 355, 124–138. ( 10.1016/j.jmb.2005.10.051) [DOI] [PubMed] [Google Scholar]
- 15.Horovitz A, Willison KR. 2005. Allosteric regulation of chaperonins. Curr. Opin Struct. Biol. 15, 646–651. ( 10.1016/j.sbi.2005.10.001) [DOI] [PubMed] [Google Scholar]
- 16.Gruber R, Levitt M, Horovitz A. 2017. Sequential allosteric mechanism of ATP hydrolysis by the CCT/TRiC chaperone is revealed through Arrhenius analysis. Proc. Natl Acad. Sci. USA 14, 5189–5194. ( 10.1073/pnas.1617746114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stuart SF, Leatherbarrow RJ, Willison KR. 2011. A two-step mechanism for the folding of actin by the yeast cytosolic chaperonin. J. Biol. Chem. 286, 178–184. ( 10.1074/jbc.M110.166256) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willardson BM, Howlett AC. 2007. Function of transducin-like proteins in G protein signalling and chaperone-assisted protein folding. Cell. Sign. 19, 2417–2427. ( 10.1016/j.cellsig.2007.06.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dekker C, et al. 2008. The interaction network of the chaperonin CCT. EMBO J. 27, 1827–1839. ( 10.1038/emboj.2008.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willison KR. Submitted The structure and evolution of eukaryotic chaperonin containing TCP-1 focussing on its mechanism for folding actin into a protein spring. Biochem. J. [DOI] [PubMed] [Google Scholar]
- 21.Amit M, Weisberg SJ, Nadler-Holly M, McCormack EA, Feldmesser E, Kaganovich D, Willison KR, Horovitz A. 2010. Equivalent mutations in the eight subunits of the chaperonin CCT produce dramatically different cellular and gene expression phenotypes. J. Mol. Biol. 401, 532–543. ( 10.1016/j.jmb.2010.06.037) [DOI] [PubMed] [Google Scholar]
- 22.Shimon L, Hynes GM, McCormack EA, Willison KR, Horovitz A. 2008. ATP-induced allostery in the eukaryotic chaperonin CCT is abolished by the mutation G345D in CCT4 that renders yeast temperature-sensitive for growth. J. Mol. Biol. 377, 469–477. ( 10.1016/j.jmb.2008.01.011) [DOI] [PubMed] [Google Scholar]
- 23.Dekker C, Willison KR, Taylor WR. 2011. On the evolutionary origin of chaperonins. Proteins 79, 1172–1192. ( 10.1002/prot.22952) [DOI] [PubMed] [Google Scholar]
- 24.Aswathy N, Pullepu D, Kabir MA. 2016. The interactome of CCT complex—a computational analysis. Comput. Biol. Chem. 64, 396–402. ( 10.1016/j.compbiolchem.2016.09.002) [DOI] [PubMed] [Google Scholar]
- 25.Rizzolo K, et al. 2017. Features of the chaperone cellular network revealed through systematic interaction mapping. Cell Rep. 20, 2735–2748. ( 10.1016/j.celrep.2017.08.074) [DOI] [PubMed] [Google Scholar]
- 26.Stirling PC, Srayko M, Takhar KS, Pozniakovsky A, Hyman AA, Leroux MR. 2007. Functional interaction between phosducin-like protein 2 and cytosolic chaperonin is essential for cytoskeletal protein function and cell cycle progression. Mol. Biol.Cell 18, 336–2345. ( 10.1091/mbc.E07-01-0069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCormack EA, Altschuler GM, Dekker C, Filmore H, Willison KR. 2009. Yeast phosducin-like protein 2 acts as a stimulatory co-factor for the folding of actin by the chaperonin CCT via a ternary complex. J. Mol. Biol. 391, 192–206. ( 10.1016/j.jmb.2009.06.003) [DOI] [PubMed] [Google Scholar]
- 28.Camasses A, Bogdanova A, Shevchenko A, Zachariae W. 2003. The CCT chaperonin promotes activation of the anaphase-promoting complex through the generation of functional Cdc20. Mol. Cell 12, 87–100. ( 10.1016/S1097-2765(03)00244-2) [DOI] [PubMed] [Google Scholar]
- 29.Passmore L, McCormack EA, Au SWN, Paul A, Willison KR, Harper JW, Barford D. 2003. Doc1 mediates the activity of the anaphase-promoting complex by contributing to substrate recognition. EMBO J. 22, 786–796. ( 10.1093/emboj/cdg084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arava Y, Wang Y, Storey JD, Liu CL, Brown PO, Herschlag D. 2003. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 100, 3889–3894. ( 10.1073/pnas.0635171100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Godoy LMF, Olsen JV, Cox J, Nielsen ML, Hubner NC, Frohlich F, Walther TC, Mann M. 2008. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251–1254. ( 10.1038/nature07341) [DOI] [PubMed] [Google Scholar]
- 32.Ho Y, et al. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180–183. ( 10.1038/415180a) [DOI] [PubMed] [Google Scholar]
- 33.Neer EJ, Schmidt CJ, Nambudripad R, Smith TF. 1994. The ancient regulatory-protein family of WD-repeat proteins. Nature 371, 297–300. ( 10.1038/371297a0) [DOI] [PubMed] [Google Scholar]
- 34.Smith TF, Gaitatzes C, Saxena K, Neer EJ. 1999. The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24, 181–185. ( 10.1016/S0968-0004(99)01384-5) [DOI] [PubMed] [Google Scholar]
- 35.Madrona AY, Wilson DK. 2004. The structure of Ski8p, a protein regulating mRNA degradation: implications for WD protein structure. Protein Sci. 13, 1557–1565. ( 10.1110/ps.04704704) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. 2003. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell 112, 243–256. ( 10.1016/S0092-8674(03)00034-5) [DOI] [PubMed] [Google Scholar]
- 37.Cerna D, Wilson DK. 2005. The structure of Sif2p, a WD repeat protein functioning in the SET3 corepressor complex. J. Mol. Biol. 351, 923–935. ( 10.1016/j.jmb.2005.06.025) [DOI] [PubMed] [Google Scholar]
- 38.Wang Y, Hu XJ, Wu XH, Ye ZQ, Wu YD. 2015. WDSPdb* a database for WD40-repeat proteins. Nucl. Acids Res. 43, 339–344. (doiorg/10.1093/nan/gkv1023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pashkova N, Gakhar L, Winistorfer SC, Yu L, Ramaswamy S, Piper RC. 2010. WD40 repeat propellers define a ubiquitin-binding domain that regulates turnover of F Box proteins. Mol. Cell 40, 433–443. ( 10.1016/j.molcel.2010.10.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garcia-Higuera I, Gaitatzes C, Smith TF, Neer EJ. 1998. Folding a WD repeat propeller. Role of highly conserved aspartic acid residues in the G protein beta subunit and Sec. 13. J. Biol. Chem. 273, 9041–9049. [DOI] [PubMed] [Google Scholar]
- 41.Ishimoto T, Fujiwara K, Niwa T, Taguchi H. 2014. Conversion of a chaperonin GroEL-independent protein into an obligate substrate. J. Biol. Chem. 289, 32 073–32 080. ( 10.1074/jbc.M114.610444) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plimpton RL, et al. 2015. Structures of the Gβ-CCT and PhLP1–Gβ-CCT complexes reveal a mechanism for G-protein β-subunit folding and Gβγ dimer assembly. Proc. Natl Acad. Sci. USA 112, 2413–2418. ( 10.1073/pnas.1419595112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olshina MA, Baumann A, Willison KR, Baum J. 2016. Plasmodium actin is incompletely folded by heterologous protein-folding machinery and likely requires the native Plasmodium chaperonin complex to enter a mature functional state. FASEB J. 30, 405–416. ( 10.1096/fj.15-276618) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kelley LA, Sternberg MJE. 2009. Protein structure prediction on the web: a case study using the Phyre server. Nat. Protoc. 4, 363–371. ( 10.1038/nprot.2009.2) [DOI] [PubMed] [Google Scholar]
- 45.Gavin AC, et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147. ( 10.1038/415141a) [DOI] [PubMed] [Google Scholar]
- 46.Aloy P, et al. 2004. Structure-based assembly of protein complexes in yeast. Science 303, 2026–2029. ( 10.1126/science.1092645) [DOI] [PubMed] [Google Scholar]
- 47.Pappenberger G, McCormack EA, Willison KR. 2006. Quantitative actin folding reactions using yeast CCT purified via an internal tag in the CCT3/γ subunit. J. Mol. Biol. 360, 484–496. ( 10.1016/j.jmb.2006.05.003) [DOI] [PubMed] [Google Scholar]
- 48.Yu H. 2007. Cdc20: a WD40 activator for a cell cycle degradation machine. Mol. Cell 27, 3 ( 10.1016/j.molcel.2007.06.009) [DOI] [PubMed] [Google Scholar]
- 49.Kraft C, Vodermaier HC, Maurer-Stroh S, Eisenhaber F, Peters J-M. 2005. The WD40 propeller domain of Cdh1 functions as a destruction box receptor for APC/C substrates. Mol. Cell 18, 543–553. ( 10.1016/j.molcel.2005.04.023) [DOI] [PubMed] [Google Scholar]
- 50.Burton JL, Tsakraklides V, Solomon MJ. 2005. Assembly of an APC-Cdh1-substrate complex is stimulated by engagement of a destruction box. Mol. Cell 18, 533–542. ( 10.1016/j.molcel.2005.04.022) [DOI] [PubMed] [Google Scholar]
- 51.Dekker C. 2010. On the role of the chaperonin CCT in the just-in-time assembly process of APC/CCdc20. FEBS Lett. 584, 477–481. ( 10.1016/j.febslet.2009.11.088) [DOI] [PubMed] [Google Scholar]
- 52.Brownridge P, Lawless C, Payapilly AB, Lanthaler K, Holman SW, Harman VM, Grant CM, Beynon RJ, Hubbard SJ. 2013. Quantitative analysis of chaperone network throughput in budding yeast. Proteomics 13, 1276–1291. ( 10.1002/pmic.201200412) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Helbig AO, Daran-Lapujade P, van Maris AJA, de Hulster EAF, de Ridder D, Pronk JT, Heck AJR, Slijper M. 2011. The diversity of protein turnover and abundance under nitrogen-limited steady-state conditions in Saccharomyces cerevisiae. Mol. Biol. Syst. 7, 3316–3326. ( 10.1039/c1mb05250k) [DOI] [PubMed] [Google Scholar]
- 54.Deutschbauer AM, Jaramillo DF, Proctor DF, Kumm J, Hillenmeyer ME, Davis RW, Nislow C, Giaever G. 2005. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics 169, 1915–1925. ( 10.1534/genetics.104.036871) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dephoure N, Hwang S, O'Sullivan C, Dodgson SE, Gygi SP, Amon A, Torres EM. 2014. Quantitative proteomic analysis reveals posttranslational responses to aneuploidy in yeast. eLife 3, e03023 ( 10.7554/eLife.03023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He X, Qian W, Wang Z, Lil Y, Zhang J. 2010. Prevalent positive epistasis in Escherichia coli and Saccharomyces cerevisiae metabolic networks. Nat. Genet. 42, 272–276. ( 10.1038/ng.524) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thulasiraman V, Yang C, Frydman J. 1999. In vivo newly translated polypeptides are sequestered in a protected folding environment. EMBO J. 18, 85–95. ( 10.1093/emboj/18.1.85) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yam AY, Xia Y, Lin HT, Burlingame A, Gerstein M, Frydman J. 2008. Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat. Struct. Mol. Biol. 15, 1255–1262. ( 10.1038/nsmb.1515) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sternlicht H, Farr GW, Sternlicht ML, Driscoll JK, Willison KR, Yaffe MB. 1993. The t-complex polypeptide 1 complex is a chaperonin for tubulin and actin in vivo. Proc. Natl Acad. Sci. USA 90, 9422–9426. ( 10.1073/pnas.90.20.9422) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grantham J, Brackley KI, Willison KR. 2006. Substantial CCT activity is required for cell cycle progression and cytoskeletal organization in mammalian cells. Exp. Cell Res. 312, 2309–2324. ( 10.1016/j.yexcr.2006.03.028) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.