Abstract
Novel methylindoles were identified as endobiotic and xenobiotic ligands of the human aryl hydrocarbon receptor (AhR). We examined the effects of 22 methylated and methoxylated indoles on the transcriptional activity of AhRs. Employing reporter gene assays in AZ-AHR transgenic cells, we determined full agonist, partial agonist, or antagonist activities of tested compounds, having substantially variable EC50, IC50, and relative efficacies. The most effective agonists (EMAX relative to 5 nM dioxin) of the AhR were 4-Me-indole (134%), 6-Me-indole (91%), and 7-MeO-indole (80%), respectively. The most effective antagonists of the AhR included 3-Me-indole (IC50; 19 μM), 2,3-diMe-indole (IC50; 11 μM), and 2,3,7-triMe-indole (IC50; 12 μM). Reverse transcription polymerase chain reaction analyses of CYP1A1 mRNA in LS180 cells confirmed the data from gene reporter assays. The compound leads, 4-Me-indole and 7-MeO-indole, induced substantial nuclear translocation of the AhR and enriched binding of the AhR to the CYP1A1 promoter, as observed using fluorescent immunohistochemistry and chromatin immunoprecipitation assays, respectively. Molecular modeling and docking studies suggest the agonists and antagonists likely share the same binding pocket but have unique binding modes that code for their affinity. Binding pocket analysis further revealed that 4-methylindole and 7-methoxyindole can simultaneously bind to the pocket and produce synergistic interactions. Together, these data show a dependence on subtle and specific chemical indole structures as AhR modulators and furthermore underscore the importance of complete evaluation of indole compounds as nuclear receptor ligands.
Introduction
Aryl hydrocarbon receptor (AhR), is a ligand-activated transcriptional factor that regulates xenobiotic metabolizing enzymes, but also plays a role in many physiologic functions, including regulation of the cell cycle and proliferation, apoptosis, DNA repair, immune response, circadian rhythm, tumor promotion, the expression of lipid metabolism genes, etc. (Kolluri et al., 2017). AhR is activated by a number of exogenous and endogenous compounds (Denison and Nagy, 2003; Stejskalova et al., 2011). The examples of exogenous AhR ligands are polyaromatic hydrocarbons, polyhalogenated hydrocarbons, halogenated dioxins and furans, plant polyphenolics, alkaloids, and many synthetic compounds including pesticides, drugs, kinase inhibitors, etc. Examples of endogenous AhR ligands are eicosanoids and kynurenic acid. A variety of compounds containing indole in their structure was described as ligands (mainly endogenous and/or foodborne) of human AhR, including agonists and antagonists. Examples include, but are not limited to, indirubin and indigo (Adachi et al., 2001); heme metabolites including bilirubin, biliverdin, and hemin (Sinal and Bend, 1997); diindolylmethane (Chen et al., 1998) and other di-indole derivatives (Chowdhury et al., 2009); indole-3-carbinol (Chen et al., 1996); indoxyl-3-sulfate (Schroeder et al., 2010; Rothhammer et al., 2016); tryptophan and its metabolites tryptamine and indole-3-acetic acid (Heath-Pagliuso et al., 1998); and UV photoproducts of tryptophan (Helferich and Denison, 1991) including 6-formylindolo[3,2-b]carbazole (Bergander et al., 2003), indole-3-aldehyde (Rothhammer et al., 2016), marine brominated indoles (DeGroot et al., 2015), and indole itself as a human AhR-specific ligand (Hubbard et al., 2015).
There are few studies of methylindoles, and no reports are available on methoxyindoles. Rasmussen et al. (2016) demonstrated that skatole (3-Me-indole), which is a tryptophan metabolite produced by intestinal microbiome, is a partial agonist of human AhR and an inducer of CYP1A1, CYP1A2, and CYP1B1 in human cell lines and hepatocytes at physiologically detected concentrations (Li et al., 2009). These authors also showed that a metabolite of skatole, indole-3-carbinol (∼10 μM, which is physiologically relevant) (Anderton et al., 2004), is an activator of human AhR and an inducer of CYP1A1 mRNA. Induction of CYP1A1 by 3-Me-indole was also described in primary normal human bronchial epithelial cells at physiologically relevant concentrations (Weems and Yost, 2010). ethoxy-resorufine-O-deethylase activity was induced by 3-Me-indole (but also by xenobiotic 2-Me-indole) in Zebra fish (Brown et al., 2015) and Atlantic killifish embryos (Brown et al., 2016). Activation of human AhR in gene reporter assays was observed for 3-Me-indole, but not for 2-Me-indole and 1-Me-indole; however, experiments were performed only up to 10 μM concentrations of methylindoles (Hubbard et al., 2015).
Besides 3-methylindole, the metabolism of tested methoxyindoles and methylindoles in man is mostly unknown. Hepatic metabolites of skatole, identified using human liver microsomes, are mainly hydroxylated and oxidized products (Lanza and Yost, 2001) and their conjugates with glutathione (Yan et al., 2007). Also, pulmonary metabolites of skatole have been described (Ruangyuttikarn et al., 1991). However, it has been demonstrated that indole-3-carbinol, a metabolite of skatole, has AhR biologic activity (Chen et al., 1996), thus the potential for other indolic metabolites as potential AhR ligands is very probable. Indeed, as the indole chemistry is optimized, several indoles could be developed as potential lead or drug-like AhR ligands. In this light, given that there is little known about whether other methyl or methoxy indoles activate AhR and/or may therefore have biologic effects, we undertook a very focused study on 22 methylated and methoxylated indoles on transcriptional activity of AhRs, by the means of reporter gene assays in AZ-AHR transgenic cells, reverse transcription polymerase chain reaction (PCR) analyses of CYP1A1 mRNA, chromatin immunoprecipitation (ChIP), and fluorescent immunohistochemistry. These in vitro results were then compared with the predicted binding modes of all 22 ligands that were docked to a modeled three-dimensional structure of human AhR. It is important to note that with all of these indoles, on a systems level, AhR agonists such as the classic indole molecule could in a context-dependent manner act as antagonists (Jin et al., 2014). Thus, studies were also conducted in an antagonist mode to characterize the effect of potential agonist on strong (dioxin) mediated AhR activation.
Materials and Methods
Chemicals.
4-Me-indole (97% purity) was purchased from Energy Chemical (Shanghai, China); 1,3-diMe-indole (95% purity) was from Shanghai SINKH Pharmaceuticals Tech. Co., Ltd. (Shanghai, China); 2,3,7-triMe-indole (97% purity; checked by gas chromatography) was purchased from Georganics Ltd. (Bratislava, Slovak Republic); and 7-MeO-4-Me-indole (95% purity) was purchased from 1 ClickChemistry Inc. (Kendall Park, NJ). All other indole-derived test compounds (where the purity ranged from 97% to 99% as assessed by gas chromatography at the supplier), dimethylsulfoxide (DMSO), Triton X-100, bovine serum albumin, and hygromycin B were purchased from Sigma-Aldrich (Prague, Czech Republic). 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) was purchased from Ultra Scientific (North Kingstown, RI); luciferase lysis buffer was purchased from Promega (Madison, CA); and 4′,6-diamino-2-phenylindole was purchased from Serva (Heidelberg, Germany). All other chemicals were of the highest quality commercially available.
Cell Lines.
Human Caucasian colon adenocarcinoma cells LS180 (European Collection of Cell Cultures No. 87021202; Salisbury, UK) were purchased from the European Collection of Cell Cultures and used in passage numbers 5–12. Stably transfected gene reporter cell line AZ-AHR has been described elsewhere (Novotna et al., 2011). The same AZ-AHR clone was used in passages for all experiments conducted. Cells were maintained at 37°C and 5% CO2 in a humidified incubator.
Cytotoxicity Assay.
Cells were incubated for 24 hours with tested compounds, vehicle (untreated; 0.1% v/v DMSO) and Triton X-100 (1%, v/v), using multiwell culture plates of 96 wells. MTT (3-[4,5-dimethylthiazole-2-yl]-2,5-diphenyltetrazolium bromide) test was performed and absorbance was measured spectrophotometrically at 540 nm on Infinite M200 (Schoeller Instruments, Prague, Czech Republic). The data were expressed as the percentage of cell viability, where 100% and 0% represent the treatments with vehicle and Triton X-100, respectively.
Reporter Gene Assay.
The stably transfected gene reporter cell line AZ-AhR was used for the evaluation of transcriptional activity of AhRs. Cells were seeded in 96-well culture plates and incubated for 24 hours with tested compounds in the presence or absence of TCDD (5 nM). Thereafter, the cells were lysed and luciferase activity was measured on a Tecan Infinite M200 Pro Plate Reader (Schoeller Instruments). The IC50 and EC50 values were calculated using GraphPad Prism 6 software (GraphPad Software, San Diego, CA).
Isolation of RNA and Quantitative Reverse Transcription PCR.
The total RNA was isolated by TRI Reagent (Sigma-Aldrich, Prague, Czech Republic) and cDNA was synthesized from 1000 ng of total RNA using M-MuLV Reverse Transcriptase (New England Biolabs, Ipswich, MA) at 42°C for 60 minutes in the presence of random hexamers (New England Biolabs). The levels of CYP1A1 and GAPDH mRNAs were determined using the Light Cycler 480 II apparatus (Roche Diagnostic Corporation, Prague, Czech Republic), as described elsewhere (Vrzal et al., 2013). Measurements were carried out in triplicate. Gene expression was normalized to GAPDH as a housekeeping gene. The data were processed by the delta-delta method.
Intracellular Distribution of AhRs: Immunohistochemistry.
LS180 cells were seeded on chamber slides (ibidi GmbH, Planegg, Germany) and cultured for 2 days. Then, cells were treated for 90 minutes with 4-methylindole or 7-methoxyindole at 1, 10, and 100 μM concentrations; 5 nM TCDD and 0.1% DMSO were used as the positive and negative controls, respectively. After the treatment, cells were washed by phosphate-buffered saline (PBS) and fixed with 4% formaldehyde in PBS for 30 minutes. The permeabilization using 0.1% Triton X-100 for 5 minutes was followed by blocking with 3% bovine serum albumin in PBS for 45 minutes at room temperature. Then, cells were incubated with Alexa Fluor 488 labeled primary antibody against AhR (Santa Cruz Biotechnology, Dallas, TX) diluted 1:500 in 0.5% bovine serum albumin at 4°C overnight. The next day, nuclei were stained with 4′,6-diamino-2-phenylindole, and cells were enclosed by VectaShield Antifade Mounting Medium (Vector Laboratories Inc., Peterborough, UK). AhR translocation into the nucleus was visualized and evaluated using the IX73 fluorescence microscope (Olympus, Tokyo, Japan). The entire staining protocol was performed in two independent experiments with all tested compounds in duplication. The AhR translocation was evaluated visually depending on the distinct signal intensity of the AhR antibody in the nucleus and cytosol. For percentage calculation, approximately 100 cells from at least four randomly selected fields of view in each replicate were used.
Chromatin Immunoprecipitation.
HepG2 cells (3.5 millions) were seeded in a 60-mm dish in Dulbecco’s modified Eagle’s medium (D6546; Sigma Aldrich), and the following day they were incubated with DMSO, TCDD (5 and 50 nM), 4-methylindole (200 µM), 7-methoxyindole (200 µM), or the mixture of 4-methylindole and 7-methoxyindole (100 µM each) for 90 minutes at 37°C. Then, the procedure followed the manufacturer’s recommendations as described in the manual for the SimpleChIP Plus Enzymatic Chromatin IP Kit (Magnetic Beads; Cell Signaling Technology, Leiden, The Netherlands) with minor modifications. Briefly, DNA-protein complexes were crosslinked by the addition of 108 µl 37% formaldehyde to 4 ml of media for 9 minutes at room temperature. Thereafter, 400 µl of 10 × glycine was applied for 5 minutes at room temperature. The cells were rinsed twice with ice-cold PBS and collected in 1 ml of ice-cold PBS (supplemented with protease inhibitor cocktail). Cells were centrifuged at 2000 rpm for 5 minutes at 4°C (Eppendorf Centrifuge 5415R; Eppendorf, Stevenage, UK) and the pellets were resuspended in 1 ml of cold buffer A and incubated on ice for 10 minutes, reverting the tubes every 3 minutes. Then, the lysates were centrifuged (3000 rpm for 5 minutes at 4°C), and the pellets were resuspended in 1 ml of ice-cold buffer B, followed by centrifugation (3000 rpm for 5 minutes at 4°C). The resulting pellets were resuspended in 100 µl of ice-cold buffer B supplemented with 0.66 µl of nuclease and incubated for 20 minutes at 37°C with shaking (1000 rpm) in a TS-100C Thermo-Shaker (with SC-24NC block; BioSan, Riga, Latvia) to digest DNA into fragments of sizes between 150 and 900 base pairs. Digestion was stopped by adding 10 µl of 0.5 M EDTA and the reaction mixture was centrifuged at 13,000 rpm for 1 minute at 4°C. The pellets were resuspended in 100 µl of ChIP buffer, incubated on ice for 10 minutes, and then sonicated five times for 30 seconds in a sonicator (Sonorex Digitec DT31; Bandelin, Berlin, Germany) with a 30-second rest on ice between sonication cycles. Cellular debris was removed by centrifugation (10,000 rpm for 10 minutes at 4°C) and DNA concentration was determined in the supernatants using a NanoDrop Lite spectrophotometer (Thermo Scientific, Prague, Czech Republic). An aliquot of 25 µg of DNA was resuspended in a total volume of 500 µl of ChIP buffer. Immunoprecipitation was achieved by the addition of 5 µl of anti-AhR antibody (D5S6H; Cell Signaling Technology) to samples or 1 µl of normal rabbit IgG as the negative control to the DMSO-treated sample with rotation at 4°C overnight.
The next day, 30 µl of Protein G Magnetic Beads was added and the samples were incubated for 2 hours at 4°C with rotation. Beads were briefly pelleted in Magnetic Separation Rack 14654 (Cell Signaling Technology) and washed three times by 1 ml of low-salt buffer for 5 minutes at 4°C with rotation. Then, 1 ml of high-salt buffer was applied for 5 minutes at 4°C with rotation, and after this step the pellets were resuspended in 150 µl of ChIP elution buffer and chromatin was eluted for 30 minutes at 65°C with 1200 rpm shaking in the TS-100C Thermo-Shaker (BioSan). After separation in the magnetic rack, eluted chromatin was incubated in ChIP elution buffer with 6 µl 5 M NaCl and 2 µl of proteinase K for 15 minutes at 65°C in the Thermo-Shaker (1200 rpm). Consequently, 750 µl of DNA binding buffer was added to each sample and centrifuged at 13,200 rpm for 30 seconds in DNA spin columns. After application of 750 µl of DNA wash buffer and repeated centrifugation at 13,200 rpm for 30 seconds in DNA spin columns, 50 µl of DNA elution buffer was applied into DNA spin columns and eluted purified DNA was used for quantitative PCR in LightCycler 480 (Roche, Prague, Czech Republic). Then, 2 μl of DNA was used in the PCR reaction together with nuclease-free water, SYBR-Green (LightCycler 480 SYBR Green I Master; Roche), and 5 µM CYP1A1 promotor primers (5′-AGCTAGGCCATGCCAAAT-3′ and 5′-AAGGGTCTAGGTCTGCGTGT-3′). The PCR program was as follows: enzyme activation at 95°C for 10 minutes and denaturation with annealing and elongation for 40 cycles at 95°C for 15 seconds and at 60°C for 60 seconds. Then, the fold enrichment method was applied and the results were expressed as fold enrichment next to the DMSO-treated sample. The PCR product was electrophoresed on a 2% agarose gel after PCR with elongation for 29 cycles only.
Molecular Modeling and Docking.
The crystal structure of a fragment of the AhR in complex with the AhR nuclear translocator has been solved (Protein Data Bank ID 5NJ8 [https://www.rcsb.org/structure/5NJ8]) (Schulte et al., 2017); however, the fragment does not include the ligand binding domain. Hence, an attempt was made to model residues 34–400 of human AhR using homology modeling techniques. A template search using PSI-BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome) revealed that the crystal structure of neuronal PAS1 (Protein Data Bank ID 5SY5 [https://www.rcsb.org/structure/5SY5]) (Wu et al., 2016) had large homology with 29% identity and >40% similarity in the region between 34 and 400 amino acids of the AhR (Supplemental Fig. 1B). Hence, the nPAS1 protein was used as a template to model residues 34–400 of the AhR using the program MODELER version 9.19 (https://salilab.org/modeller/9.19/release.html) (Sali and Blundell, 1993). The resulting model was energy minimized and subjected to 10 nanoseconds of constrained molecular dynamics simulation to obtain a stable, low-energy AhR model that was subsequently used for docking studies.
All 22 ligands and a known AhR ligand TCDD were modeled using the builder module of the molecular operating environment program, MOE version 2016.0801 (Chemical Computing Group ULC, Montreal, QC, Canada). The ligands were all minimized and the partial charges were set to the AMBER force field as adapted in MOE. Recent studies using site-directed mutagenesis techniques on mouse AhR have identified the putative binding pocket for TCDD in mouse AhR (Motto et al., 2011; Soshilov and Denison, 2014). According to these studies, residues His285, Phe289, Phe318, Gln377, Thr283, Leu347, Phe345, Met342, and Ser359 of mouse AhR were found to be involved in promoting strong binding interactions with the AhR ligands. We used these studies to derive the binding pocket information to guide our molecular docking studies. Accordingly the pocket lined by residues His291, Phe295, Phe324, Gln383, Thr289, Leu353, Phe351, Met348, and Ser364 of human AhR was used to dock all of the ligands. Molecular docking was performed using GOLD version 5.2 (Cambridge Crystallographic Data Centre, Cambridge, UK) (Jones et al., 1995) with 20 iterations for each ligand to explore the conformational space. The protein-ligand complexes were ranked using GOLDSCORE and the best ranking conformation for each ligand was scored and analyzed visually using the MOE ligand interactions module.
Statistics.
Student’s t test, one-way analysis of variance, and Dunnett’s test, as well as the EC50 and IC50 values, were calculated using GraphPad Prism version 6.0 for Windows (GraphPad Software, La Jolla, CA). The log[agonist/inhibitor]-variable slope four-parameter fit was used to determine the EC50/IC50 values. The least-squares fitting method was used, and for the EC50 calculations the data were normalized such that the bottom value (the lowest one) was fixed as zero and highest value was fixed as 100%. No weighting was applied because, in general, the Y values did not increase in distribution in proportion to the X values (i.e., the constant coefficient of variation). Each replicate Y value was treated as an individual data point and 95% confidence interval values were imputed and selected using an asymmetrical (likelihood) confidence interval. A maximum of 1000 iterations was used to fit the curve and goodness of fit was selected using the R-squared, sum of squares, and Sy/x values. The D’Agostino-Pearson omnibus normality test was used and strict convergence criteria were selected. The R-squared value was checked in all of the calculations and did not drop below 0.8.
Results
Effects of the Tested Indoles on the Transcriptional Activity of AhRs in AZ-AHR Cells by Reporter Gene Assay.
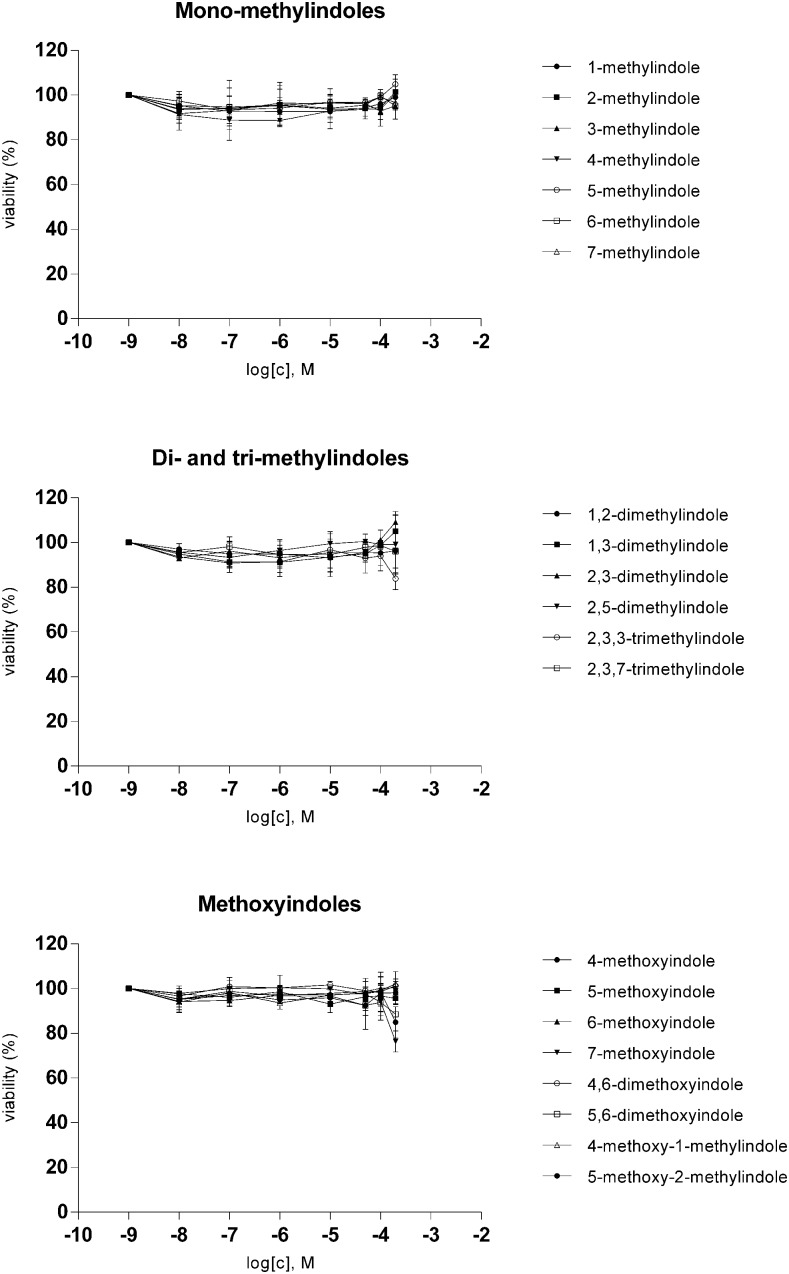
We used stably transfected human hepatoma AZ-AHR cells to study the effects of the tested indoles on the transcriptional activity of AhRs. Prior to the reporter gene assays, we performed a cytotoxicity assay in AZ-AHR, using the conventional MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) test. For this purpose, AZ-AHR cells were incubated for 24 hours with tested compounds in concentrations ranging from 1 nM to 200 μM. All tested indoles were not cytotoxic in AZ-AHR cells, causing less than 20% drop in cell viability (Fig. 1).
Fig. 1.
Cytotoxicity of tested indoles in AZ-AHR cells. AZ-AHR cells were incubated for 24 hours with vehicle (DMSO; 0.1% v/v), TCDD (5 nM), and tested compounds in concentrations ranging from 1 nM to 200 μM. MTT test was performed and absorbance was measured at 540 nm. Experiments were carried out in two consecutive passages of AZ-AHR cells and the representative experiment is shown. The data are mean ± S.D. from measurements performed in triplicate and are expressed as a percentage of viability of control cells.
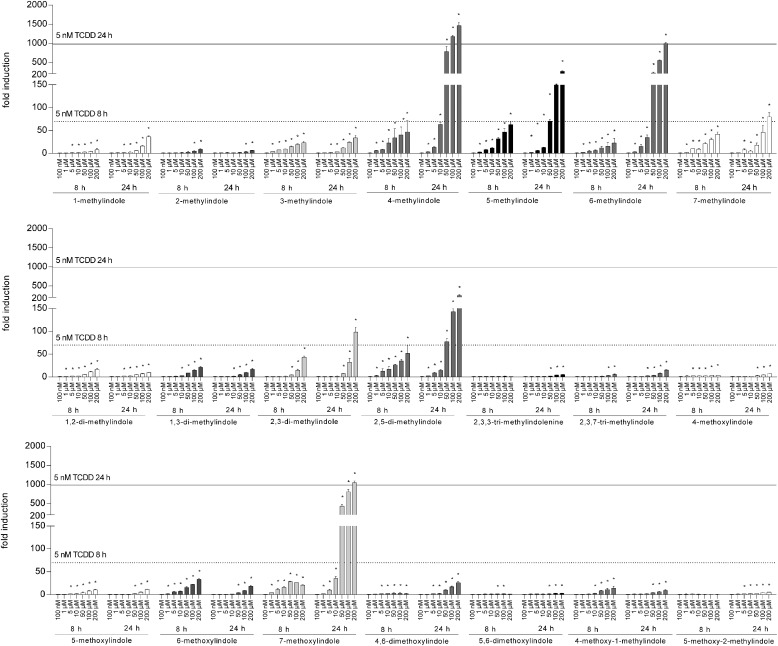
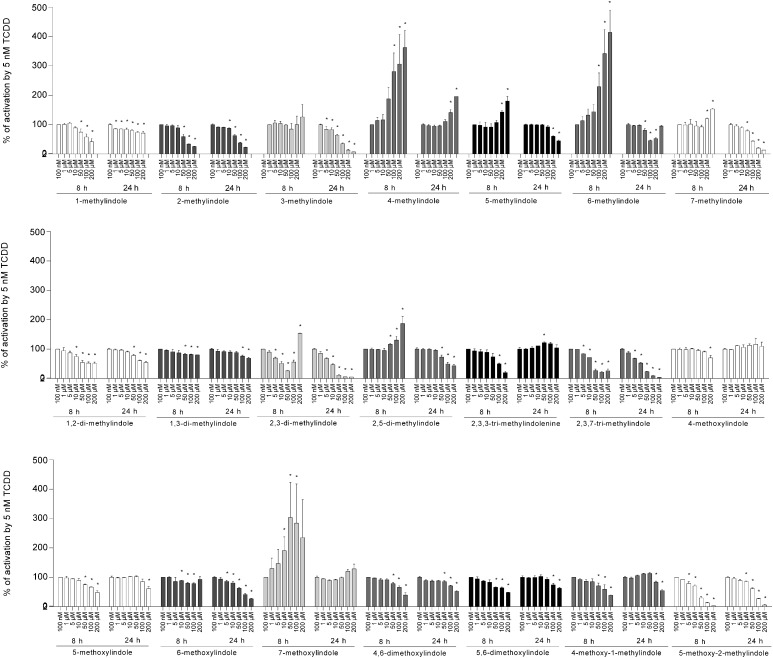
Reporter gene assays were carried out in the absence (agonist mode) or the presence (antagonist mode) of TCDD, a prototypical AhR ligand. Induction of luciferase activity by 5 nM TCDD in four consecutive cell passages varied from 579- to 1438-fold. All tested compounds displayed full agonist, partial agonist, or antagonist activities; however, their EC50, IC50, and relative efficacies varied substantially. The most efficacious agonists of AhRs were 4-Me-indole, 6-Me-indole, and 7-MeO-indole, displaying relative efficacies of 134%, 91%, and 80%, respectively, compared with 5 nM TCDD (Fig. 2). The strongest antagonist activities were achieved by 3-Me-indole (IC50; 19 μM), 2,3-diMe-indole (IC50; 11 μM), and 2,3,7-triMe-indole (IC50; 12 μM) (Fig. 3). Cells were incubated with tested indoles for 8 and 24 hours to avoid putative chemical instability and/or transient effects of the compounds in the cell culture. Consistent with the findings of Rasmussen et al. (2016), who studied 3-Me-indole, we observed that indole derivatives have a higher magnitude of AhR activation compared with TCDD over short periods of time (8 hours); however, the opposite is true for incubations over a long period of time (24 hours). Accordingly, the relative efficacies of the majority of tested indoles were higher at 8 hours compared with 24 hours of treatment (Table 1). Interestingly, the exception that proves the rule was the time course by the strongest AhR agonists, i.e., 4-Me-indole, 6-Me-indole, and 7-MeO-indole. Half-maximal effective concentrations (potency; EC50) were calculated only for about one-half of the tested compounds because the plateau was not reached by many compounds even at 200 μM concentration. Regardless of the treatment time, the potencies of the tested indoles ranged from 4 × 10−6 to 8 × 10−5 M; therefore, the relative potencies compared with TCDD were very low. A comprehensive overview of the pharmacological parameters of the tested indoles at AhR is presented in Table 1.
Fig. 2.
Transcriptional activity of AhR by tested indoles. AZ-AHR cells were incubated for 8 and 24 hours with vehicle (DMSO; 0.1% v/v), TCDD (5 nM), and tested compounds in concentrations ranging from 100 nM to 200 μM. Cells were lysed and luciferase activity was measured. Experiments were performed in four consecutive passages of AZ-AHR cells and the representative experiment is shown. Data are expressed as a fold induction of luciferase activity over control cells and are the mean ± S.D. from measurements performed in quadruplicate. Inserted horizontal lines refer to activation attained by 5 nM TCDD. *P < 0.05 indicates a value different from control cells.
Fig. 3.
Effects of tested indoles on TCDD-inducible transcriptional activity of AhR. AZ-AHR cells were incubated for 8 and 24 hours with vehicle (DMSO; 0.1% v/v) and tested compounds in concentrations ranging from 100 nM to 200 μM. Incubations were carried out in the presence of TCDD (5 nM). Cells were lysed and luciferase activity was measured. Experiments were performed in four consecutive passages of AZ-AHR cells and the representative experiment is shown. Data are expressed as a percentage of the activation attained by 5 nM TCDD and are the mean ± S.D. from measurements performed in quadruplicate. *P < 0.05 indicates a value different from TCDD-treated cells.
TABLE 1.
AhR activity: gene reporter assay and AZ-AHR cells (n = 2)
PubChem Bioassay AID2796 (https://pubchem.ncbi.nlm.nih.gov/bioassay/2796): “Luminescence-based primary cell-based high throughput screening assay to identify activators of the Aryl Hydrocarbon Receptor (AHR)”; relative efficacy = 100 × fold induction (200 μM compound)/fold induction (10 nM TCDD); relative potency = 100 × EC50 TCCD/EC50 compound.
| Compound | CID | AID2796 | Relative Efficacy |
EC50 (Exp1/Exp2) |
Relative Potency |
IC50 (Exp1/Exp2) |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| 8 hours | 24 hours | 8 hours | 24 hours | 8 hours | 24 hours | 8 hours | 24 hours | |||
| % | % | % | % | |||||||
| 1-Methylindole | 11781 | Not tested | 6.8–11.1 | 2.7–3.0 | N.C. | N.C. | N.C. | N.C. | 103.7 (CI 77.7–138.6 μM)/191.1 μM (CI 133.2–274.1 μM) | >200 μM |
| 2-Methylindole | 7224 | Not tested | 8.7–11.3 | <2 | N.C. | N.C. | N.C. | N.C. | 73.8 (CI 60.6–88.9 μM)/56.3 μM (CI 46.7–68.0 μM) | 68.0 (CI 52.1–88.6 μM)/62.9μM (CI 49.2–80.4 μM) |
| 3-Methylindole | 6736 | Inactive | 24.3–27.6 | 2.5–6.7 | 12.4 (CI 7.5–20.3 μM)/19.8 μM (CI 11.4–34.3 μM) | 66.5 (CI 57.3–77.1 μM)/84.9 μM (CI 63.9–112.7 μM) | 0.0029 | 0.0193 | Potentiation | 20.1 (CI 13.2–30.7 μM)/18.3 μM (CI 15.5–21.6 μM) |
| 4-Methylindole | 85262 | Not tested | 31.9–76.0 | 110.1–156.9 | 16.3 (CI 10.2–26.1 μM)/12.7 μM (CI 7.2–22.2 μM) | 47.3 (CI 42.2–52.8 μM)/47.7 μM (CI 36.8–61.8 μM) | 0.0163 | 0.0065 | Potentiation | Potentiation |
| 5-Methylindole | 1197B | Not tested | 62.0–80.3 | 19.9–21.6 | 43.6 (CI 31.1–61.0 μM)/38.0 μM (CI 23.7–60.8 μM) | N.C. | 0.0054 | N.C. | Potentiation | 161.5 (CI 126.8–205.7 μM)/153.1 μM (CI 121.4–193.2 μM) |
| 6-methylindole | 137928 | Not tested | 16.6–35.5 | 74.6–106.4 | 39.6 (CI 22.9–68.3 μM)/28.5 μM (CI 14.2–57.4 μM) | N.C. | 0.0065 | N.C. | Potentiation | Potentiation |
| 7-Methylindole | 70275 | Not tested | 45.5–49.1 | 6.0–7.1 | 36.9 (CI 19.2–71.0 μM)/36.0 μM (CI 20.6–62.8 μM) | N.C. | 0.0060 | N.C. | Potentiation | 35.0 (CI 29.5–41.5 μM)/41.0 μM (CI 29.7–56.8 μM) |
| 1,2-di-methylindole | 13408 | Not tested | 18.6–19.5 | <2 | 69.0 (CI 54.2–87.6 μM)/72.3 μM (CI 59.9–87.2 μM) | 50.9 (CI 32.7–79.2 μM)/70.8 μM (CI 46.1–108.9 μM) | 0.0031 | 0.0051 | >200 μM | 227.7 (CI 167.5–309.5 μM)/159.3 μM (CI 117.6–215.7 μM) |
| 1,3-Di-methylindole | 70130 | Not tested | 22.9–24.2 | <2 | 58.6 (CI 46.6–73.8 μM)/63.9 μM (CI 52.3–78.0 μM) | N.C. | 0.0036 | N.C. | >200 μM | >200 μM |
| 2,3-Di-methylindole | 7053 | Not tested | 48.7–49.1 | 7.4–12.8 | N.C. | N.C. | N.C. | N.C. | 14.6 (CI 11.7–18.2 μM)/10.2 μM (CI 6.9–15.2 μM) | 8.9 (CI 7.3–10.9 μM)/12.1 μM (CI 5.4–27.0 μM) |
| 2,5-Di-methylindole | 70965 | Not tested | 47.1–68.8 | 18.6–29.9 | 11.0 (CI 6.3–19.0 μM)/55.1 μM (CI 29.8–101.9 μM) | N.C. | 0.0066 | N.C. | Potentiation | >200 μM |
| 2,3,3-Tri-methylindolenine | 15427 | Not tested | <2 | <2 | N.C. | 70.6 (CI 64.3–77.5 μM)/42.2 μM (CI 28.3–65.2 μM) | N.C. | 0.0055 | 106.1 (CI 96.4–116.8 μM)/70.2 μM (CI 41.3–119.3 μM) | >200 μM |
| 2,3,7-Tri-methylindole | 96869 | Not tested | 6.6–7.0 | <2 | N.C. | N.C. | N.C. | N.C. | 23.4 (CI 14.7–37.2 μM)/29.5μM (CI 16.8–51.8 μM) | 11.0 (CI 9.2–13.2 μM)/13.l μM (CI 10.5–16.2 μM) |
| 4-Methoxyindole | 138363 | Not tested | 4.0–4.5 | <2 | N.C. | N.C. | N.C. | N.C. | >200 μM | >200 μM |
| 5-Methoxyindole | 13872 | Not tested | 10.4–13.0 | <2 | N.C. | N.C. | N.C. | N.C. | >200 μM | >200 μM |
| 6-Methoxyindole | 76659 | Inactive | 37.3–38.6 | <2 | N.C. | N.C. | N.C. | N.C. | >200 μM | 67.8 (CI 51.4–89.6 μM)/76.7 μM (CI 67.7–86.9 μM) |
| 7-Methoxyindole | 76660 | Not tested | 23.1–24.7 | 78.8–81.3 | 8.8 (CI 5.8–13.4 μM)/6.2 μM (CI 4.6–8.3 μM) | 57.9 (CI 53.0–63.3 μM)/49.5 μM (CI 47.9–51.2 μM) | 0.0293 | 0.0058 | Potentiation | Potentiation |
| 4,6-Di-methoxyindole | 828760 | Inactive | 2.3–3.3 | 2.0 ± 3.5 | 2.3 (CI 1.6–3.3 μM)/5.9 μM (CI 4.1–8.5 μM) | 68.5 (CI 55.5–84.5 μM)/49.7 μM (CI 38.6–64.3 μM) | 0.0537 | 0.0052 | 132.1 (CI 87.1–200.4 μM)/180.5 μM (CI 144.8–225.0 μM) | >200 μM |
| 5,6-Di-methoxyindole | 84431 | Not tested | <2 | <2 | N.C. | 22.6 (CI 7.4–68.9 μM)/28.2 μM (CI 18.1–43.9 μM) | N.C. | 0.0122 | 178.1 (CI 125.4–252.9 μM)/192.1 μM (CI 117.2–314.8 μM) | >200 μM |
| 4-Methoxy-l-Me-indole | 595238 | Not tested | 13.3–18.5 | <2 | 26.5 (CI 19.2–36.8 μM)/46.7 μM (CI 36.1–60.4 μM) | 59.3 (CI 44.3–79.5 μM)/74.5 μM (CI 56.3–98.6 μM) | 0.0059 | 0.0046 | 107.0 (CI 84.6–135.2 μM)/174.1 μM (CI 42.4–714.1 μM) | 210.4 μM (CI 145.8– 303.6 μM)/162.0 μM (CI 141.7–185.3 μM) |
| 5-Methoxy-2-Me-indole | 70642 | Not tested | <2 | <2 | N.C. | 15.3 (CI 10.4–22.5 μM)/6.0 μM (CI 3.3–11.0 μM) | N.C. | 0.0292 | 22.9 (CI 17.8–29.4 μM)/17.1 μM (CI 13.3–21.9 μM) | 55.8 (CI 39.0–79.7 μM)/37.6 μM (CI.28.9–48.9 μM) |
| TCDD | — | 1000 | 100 | 2.1 (CI 1.6–2.8 nM)/2.3 nM(CI 2.1–2.5 nM) | 3.1 (CI 2.2–4.4 nM)/3.1 nM(CI 2.7–3.6 nM) | 100 | 100 | — | ||
CI, confidence interval; CID, compound identification (PubChem); Exp1, experiment 1; Exp2, experiment 2; N.C., not calculated.
Effects of the Tested Indoles on the Expression of CYP1A1 mRNA in LS180 Cells.
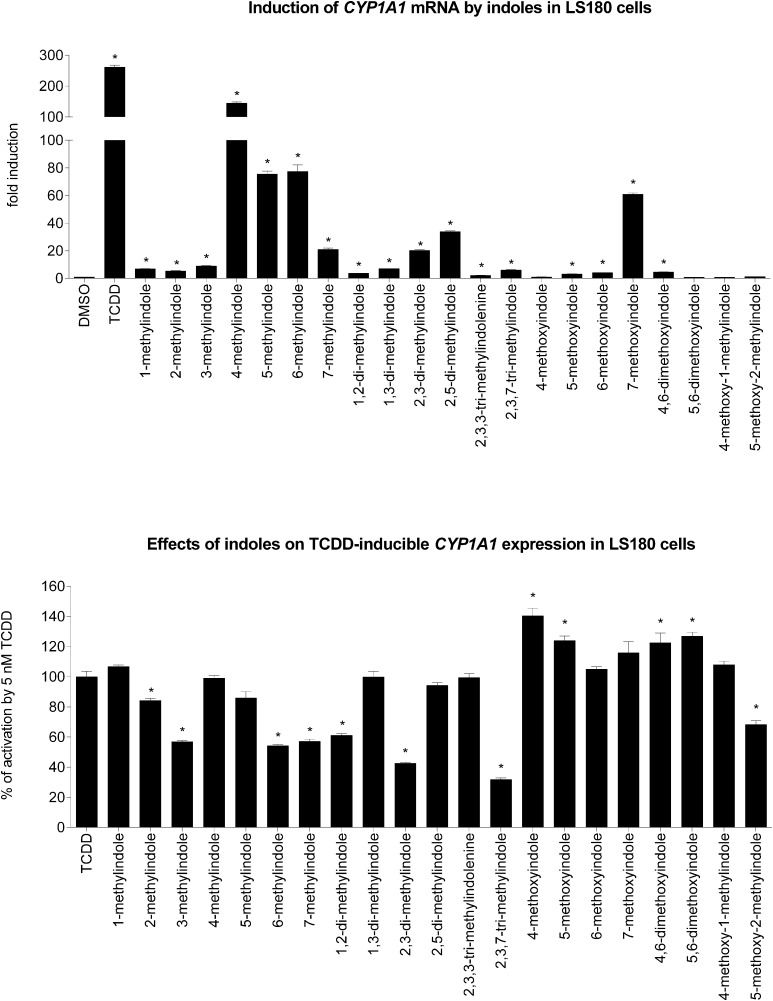
We tested the effects of the tested indoles on basal and TCDD-inducible expression of CYP1A1, which is a typical AhR target gene. Human Caucasian colon adenocarcinoma LS180 cells were incubated for 24 hours with the tested indoles (200 μM) in the presence or absence of 5 nM TCDD, and the levels of CYP1A1 mRNA were measured by quantitative reverse transcription PCR. The level of CYP1A1 mRNA was increased (250-fold) in cells incubated with TCDD. Most of the tested indoles induced CYP1A1 mRNA, and the strongest inducers of CYP1A1 mRNA were 4-Me-indole (140-fold), 5-Me-indole (75-fold), 6-Me-indole (80-fold), and 7-MeO-indole (60-fold) (Fig. 4, upper panel). Strong inhibition of TCDD-inducible expression of CYP1A1 mRNA was caused by 2-Me-indole, 3-Me-indole, 6-Me-indole, 7-Me-indole, 2,3-diMe-indole, 2,3,7-triMe-indole, and 5-MeO-2-Me-indole (Fig. 4, lower panel). Collectively, the data on CYP1A1 mRNA expression in LS180 cells were consistent with those from gene reporter assays.
Fig. 4.
Effects of tested indoles on the expression of CYP1A1 mRNA in LS180 cells. Cells were incubated for 24 hours with vehicle (DMSO; 0.1% v/v) and tested indoles (200 μM) in the presence or absence of TCDD (5 nM). Incubations were carried out in two consecutive passages of LS180 cells and the representative experiment is shown. The level of CYP1A1 mRNA was determined by reverse transcription PCR and the data were normalized to the GAPDH mRNA level. Data are mean ± S.D. from experiments performed in triplicate. *P < 0.05 indicates a value different from control cells. (Upper graph) Incubations were carried out in the absence of TCDD. Data are expressed as a fold induction of CYP1A1 mRNA over control cells. (Lower graph) Incubations were carried out in the presence of TCDD (5 nM). Data are expressed as a percentage of the activation attained by 5 nM TCDD.
Effects of the Lead Indoles (4-Me and 7-MeO) and the Coupled 7-MeO-4-Me-Indole on the AhR-CYP1A1 Pathway.
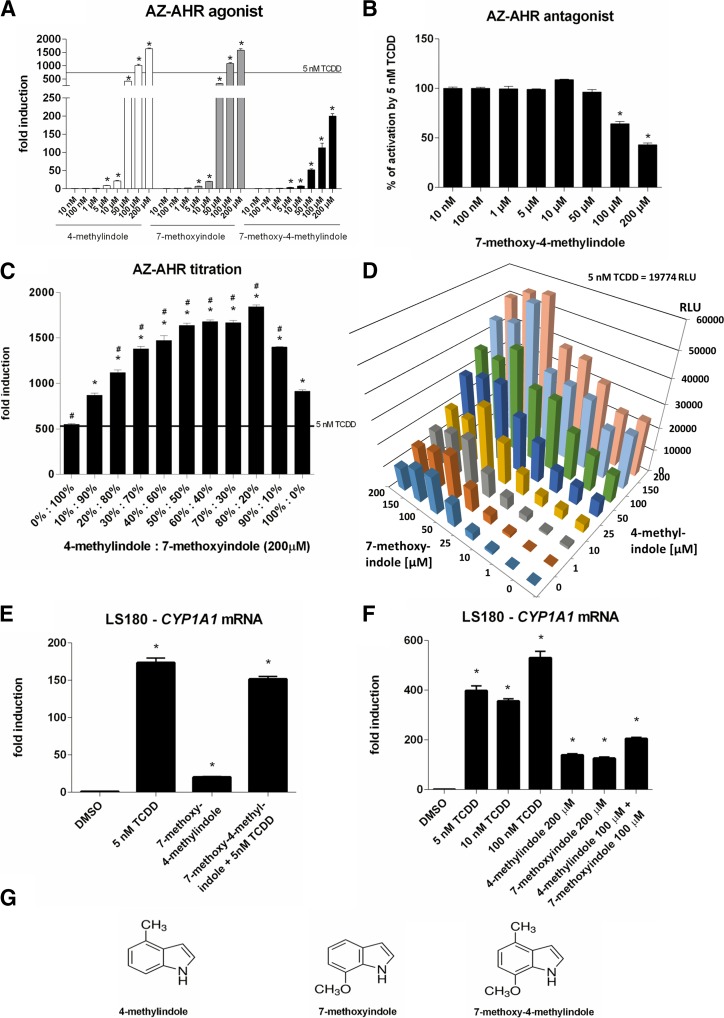
For further investigations, we selected highly efficacious activators of AhRs, 4-Me-indole and 7-MeO-indole as the lead indoles, representing methylindoles and methoxyindoles, respectively. Taking into consideration that methyl substitution at position 4 and methoxy substitution at position 7 results in massive activation of the AhR, we also investigated the coupled compound 7-MeO-4-Me-indole, which comprises both AhR-active substitutions. Surprisingly, while 7-MeO-4-Me-indole dose dependently activated the AhR, as revealed by the gene reporter assay, its relative efficacy (fold induction) was approximately 10% of that for 4-Me-indole and 7-MeO-indole (Fig. 5A). In addition, whereas 4-Me-indole and 7-MeO-indole slightly augmented TCDD-inducible AhR activity, 7-MeO-4-Me-indole displayed a dose-dependent antagonist effect on AhR (Fig. 5B). Accordingly, 7-MeO-4-Me-indole was a substantially weaker inducer of CYP1A1 mRNA in LS180 cells compared with 4-Me-indole and 7-MeO-indole. On the other hand, all three compounds had negligible effect on TCDD-inducible CYP1A1 mRNA in LS180 cells (Figs. 4; Fig. 5, E and F). Despite our expectations, combining highly efficacious 4-Me and 7-MeO substitutions in coupled derivative 7-MeO-4-Me-indole did not result in a superefficacious activator of AhRs. To resolve the roles of 4-Me and 7-MeO at AhRs, individually and in combination, we performed titration experiments. In a binary titration assay, we incubated AZ-AHR cells for 24 hours with mixtures of 4-Me-indole and 7-MeO indole (200 μM final concentration), applying their ratios from 0%:100% to 100%:0% with a step of 10%. We obtained an asymmetric inverse U-shaped profile revealing a synergistic effect between 4-Me-indole and 7-MeO indole. While 4-Me-indole and 7-MeO indole activated AhR approximately 900- and 500-fold, respectively, their mixtures 50%:50%, 60%:40%, and 70%:30% caused approximately 1700-fold activation of AhRs (Fig. 5C). We also performed a large-scale titration assay, in which we incubated AZ-AHR cells for 24 hours with mixtures of 4-Me-indole and 7-MeO-indole; each compound in concentrations of 1, 10, 25, 50, 100, 150, and 200 μM. Hence, 64 different combinations of 4-Me-indole/7-MeO-indole were tested, with the highest total concentration of indoles being 400 μM. The results showed that for a wide array of concentration ratios the combined incubations of AZ-AHR cells with 4-Me-indole and 7-MeO-indole yielded enhanced activation of AhRs compared with the incubations of individual compounds (Fig. 5D). Consistently, combined incubation of LS180 cells with 4-Me-indole (100 μM) and 7-MeO-indole (100 μM) resulted in stronger induction of CYP1A1 mRNA compared with incubations of individual compounds at 200 μM (Fig. 5F). Overall, combining 4-Me and 7-MeO substituents in a coupled molecule did not increase the efficacy of the compound, but combining 4-Me-indole and 7-MeO-indole in the mixture resulted in synergistic (cooperative) activation of AhRs. A speculative explanation for such behavior could be the binding of more than one indole-derived molecule to the AhR protein.
Fig. 5.
Effects of lead indoles (4-Me, 7-MeO) and coupled 7-MeO-4-Me-indole on AhR-CYP1A1 pathway. (A and B) Reporter gene assay: dose response. AZ-AHR cells were incubated for 24 hours with vehicle (DMSO; 0.1% v/v) and tested compounds (10 nM–200 μM) in the absence (A) or presence (B) of TCDD (5 nM). Experiments were carried out in two consecutive passages of AZ-AHR cells and the representative experiment is shown. Data are mean ± S.D. from measurements performed in quadruplicate. Inserted horizontal line refers to activation attained by 5 nM TCDD. *P < 0.05 indicates a value different from control cells. (C) Reporter gene assay: binary titration. AZ-AHR cells were incubated for 24 hours with mixtures of 4-Me-indole and 7-MeO-indole in final concentrations of 200 μM. Different ratios of 4-Me:7-MeO were applied, ranging from 0%:100% to 100%:0%, applying a step of 10%. Experiments were carried out in two consecutive passages of AZ-AHR cells and the representative experiment is shown. Data are mean ± S.D. from measurements performed in quadruplicate. Inserted horizontal line refers to an activation attained by 5 nM TCDD. *P < 0.05 indicates a value different from cells incubated with 7-MeO-indole; #P < 0.05 indicates a value different from cells incubated with 4-Me-indole. (D) Reporter gene assay: large-scale three-dimensional titration. AZ-AHR cells were incubated for 24 hours with mixtures of 4-Me-indole and 7-MeO-indole, each compound in concentrations of 1, 10, 25, 50, 100, 150, and 200 μM. In total, 64 different combinations were tested, with the highest total concentration of indoles being 400 μM. Experiments were carried out in two consecutive passages of AZ-AHR cells and the representative experiment is shown. Measurements were performed in quadruplicate. Inserted text refers to an activation attained by 5 nM TCDD. (E and F) Reverse transcription (RT)-PCR: CYP1A1 mRNA. LS180 cells were incubated for 24 hours with vehicle (DMSO; 0.1% v/v), TCDD (5, 10, and 100 nM), 4-Me-indole, 7-MeO-indole, and 7-MeO-4-Me-indole or the combinations. Incubations were carried out in two consecutive passages of LS180 cells and the representative experiment is shown. The level of CYP1A1 mRNA was determined by RT-PCR and the data were normalized to the GAPDH mRNA level. Data are mean ± S.D. from experiments performed in triplicate. *P < 0.05 indicates a value different from control cells. (G) Chemical structures of tested compounds.
Nuclear Translocation of AhRs by the Lead Indoles (4-Me and 7-MeO).
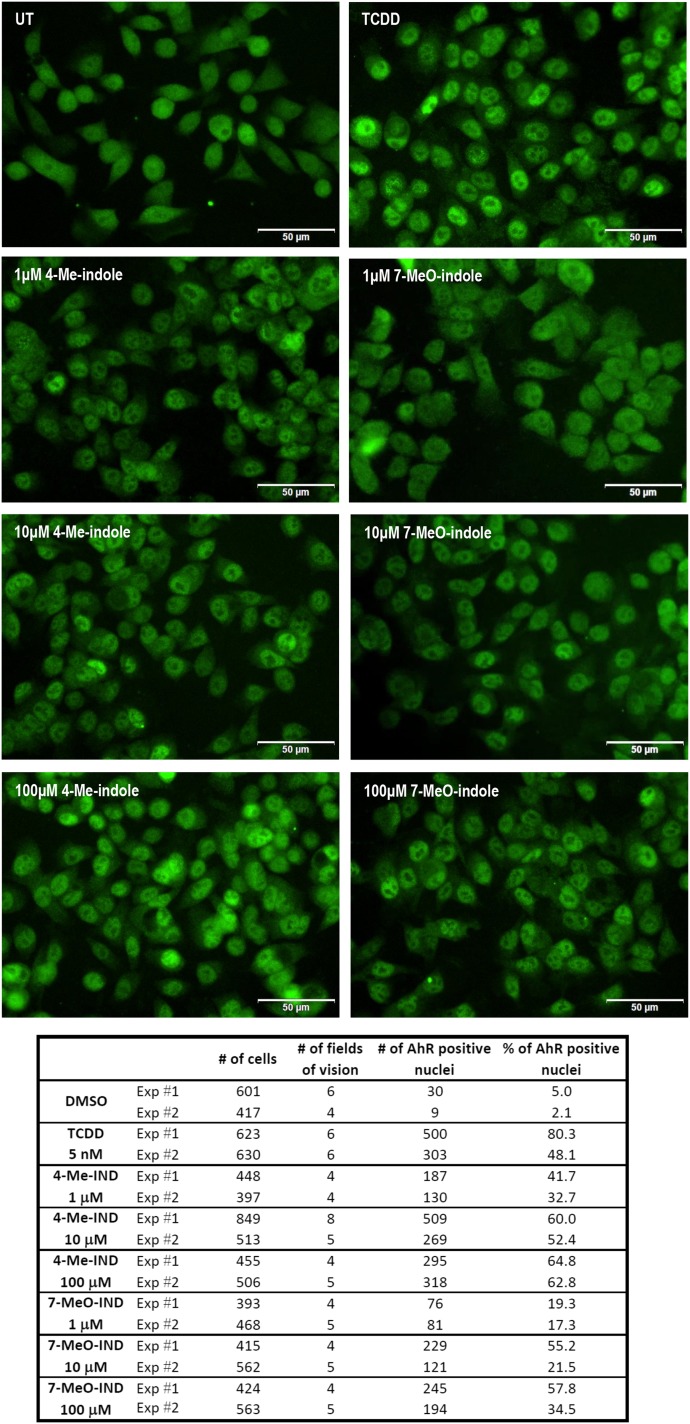
An essential event in the process of AhR activation and function is the translocation of ligand-bound AhR protein from cytosol to the cell nucleus. Therefore, we investigated the effects of the lead indoles on the nuclear translocation of AhRs by immunohistochemistry, using fluorescently labeled AhR antibody. We incubated LS180 cells for 90 minutes with TCDD (5 nM), DMSO (0.1% V/V), 4-Me-indole, or 7-MeO-indole at 1, 10, and 100 μM concentrations. In cells incubated with vehicle (negative control), only a negligible part of the cells (∼4%) displayed AhRs in the nuclei. Addition of TCDD to the cells caused massive nuclear translocation of AhRs, resulting in approximately 64% of AhR-positive nuclei. Both 4-methylindole and 7-methoxyindole strongly induced nuclear translocation of AhRs in all of the applied concentrations. The effects were dose dependent, and the percentage of AhR-positive nuclei ranged between 17% and 65% (Fig. 6). Hence, the capability of the lead indoles to induce nuclear translocation of AhRs was comparable with that of TCDD, which was consistent with the data from reporter gene assays and CYP1A1 mRNA expression.
Fig. 6.
Nuclear translocation of AhR. LS180 cells were seeded on chamber slides and cultured for 2 days. Then, the cells were incubated for 90 minutes with DMSO (0.1% v/v), TCDD (5 nM), 4-Me-indole (1, 10, and 100 μM), and 7-MeO-indole (1, 10, and 100 μM). Microscopic specimens were prepared according to the common protocol, using Alexa Fluor 488 labeled primary antibody against AhR, 4′,6-diamino-2-phenylindole, and VectaShield Antifade Mounting Medium. AhR was visualized and evaluated using a fluorescence microscope. Percentage of cells with nuclear AhR was calculated by visual comparison of antibody signal intensity in the nucleus and cytosol, when at least four random locations per sample with approximately 100 cells were evaluated. Experiments were performed in two consecutive cell passages, with all tested compounds in duplicate. The representative images are shown together with an inset containing the total and AhR-positive counts of cells.
Binding of the AhR to the CYP1A1 Promoter by the Lead Indoles (4-Me and 7-MeO).
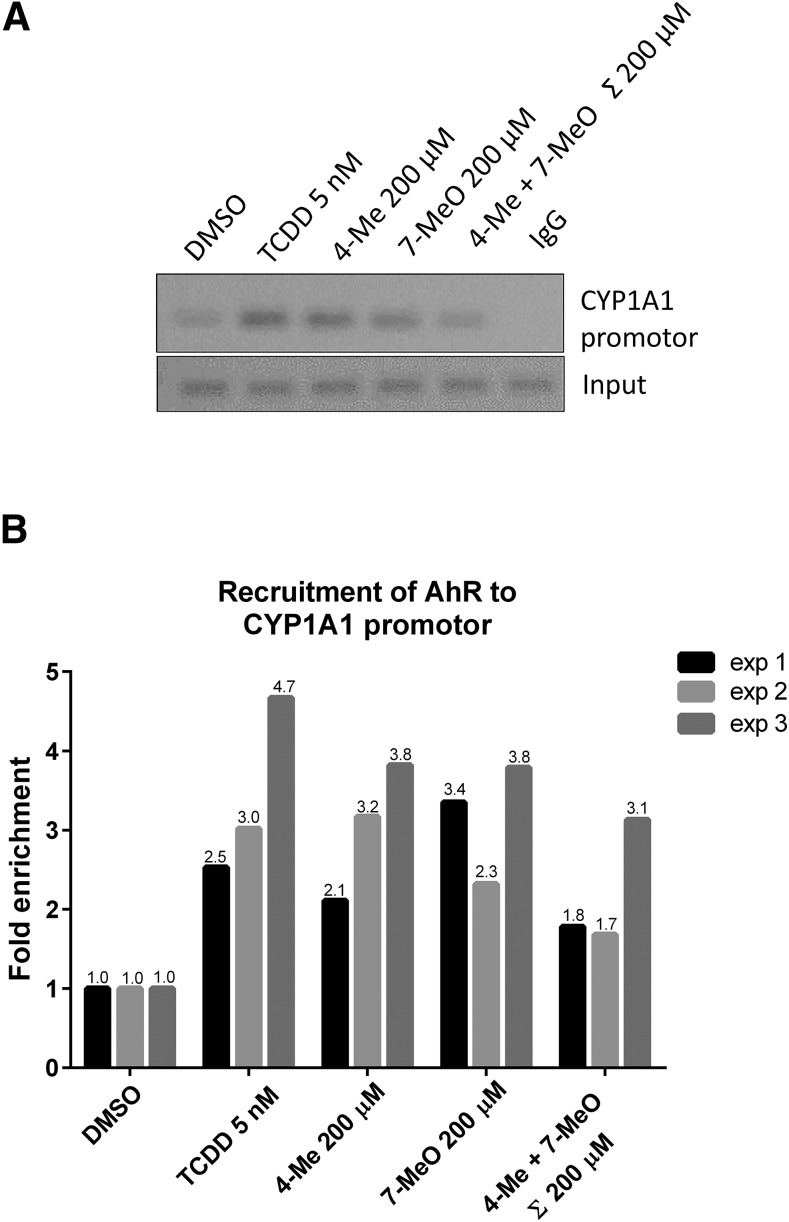
The capability of the AhR to bind to a promoter of its target genes was assessed by ChIP assay. For this purpose, we incubated HepG2 cells with vehicle, TCDD (5 and 50 nM), 4-Me-indole (200 µM), 7-MeO-indole (200 µM), or the mixture of 4-Me-indole and 7-MeO-indole (100 µM each) for 90 minutes. In cells incubated with TCDD, the enrichment of CYP1A1 promoter with the AhR ranged from 2.5- to 4.7-fold. The lead indoles, including 4-Me-indole and 7-MeO-indole, caused enrichment of CYP1A1 with the AhR to a similar extend as did TCDD. Unlike in the AhR gene reporter assays and CYP1A1 mRNA induction, in the current AhR-ChIP assay a combination of 4-Me-indole and 7-MeO-indole did not attain synergistic effects (but rather the opposite) compared with individual compounds (Fig. 7). Collectively, the lead methyl- and methoxy-indoles elicit nuclear translocation of AhRs, which consequently binds the CYP1A1 promoter and triggers the expression of CYP1A1.
Fig. 7.
Chromatin immunoprecipitation: ChIP (AhR–CYP1A1). HepG2 cells were treated as described in Materials and Methods and subjected to ChIP analysis using anti-AhR antibody. (A) Representative DNA fragments amplified by PCR analyzed on a 2% agarose gel are from the third experiment. (B) Association of AhR to CYP1A1 promotor was quantified by real-time PCR. Results shown represent three independent experiments and are expressed as fold enrichment to vehicle (DMSO) control.
Molecular Modeling and Docking.
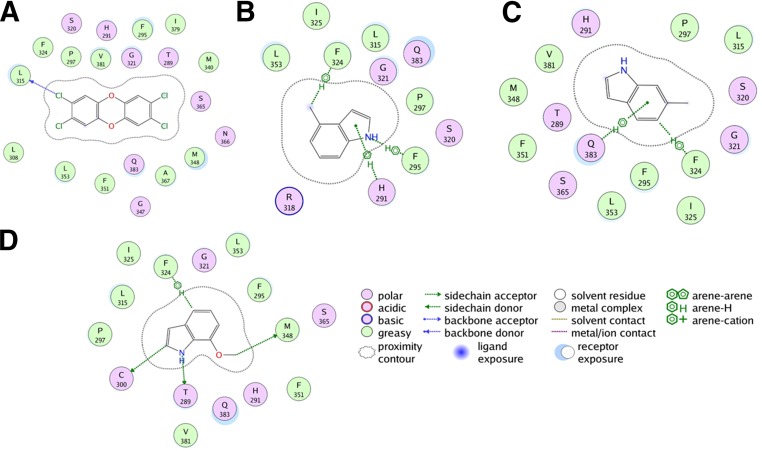
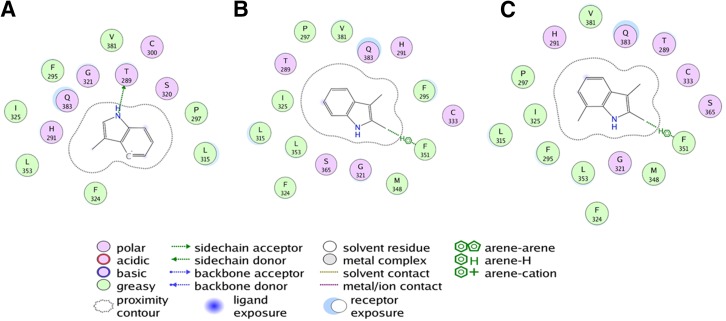
The AhR model (34–400) was superimposed onto the AhR crystal structure (112–272) (Supplemental Fig. 1A), where the root-mean-square deviation was 1.98 Å. TCDD was docked to the binding site formed by residues His291, Ser365, Val381, Gln383, Met340, Ala367, Thr289, and Phe295 of human AhR. TCDD docked with a high score of 60.14 to this site with conserved interactions for several hydrophobic residues including Ala367, Leu315, and the aromatic cluster formed by Phe295, Phe324, and Phe351 (Fig. 8A). In addition, TCDD’s binding to AhRs was also coordinated by electrostatic interactions with Thr289, Gln383, His291, and Ser365 (Fig. 8A). Previous modeling and site-directed mutagenesis have validated the role of several of these residues including Ala367, which when mutated had a substantial effect on TCDD-mediated transcriptional activity (Motto et al., 2011; Soshilov and Denison, 2014). This docking site was also previously confirmed by Hubbard et al. (2015). For the binding of methylindole compounds to AhRs, all 22 compounds were docked to the binding site. Due to the small volume, all molecules docked to the binding pocket without any steric hindrance. However, there were substantial differences in the binding mode of agonists and antagonists. Among the key agonists, 4-Me-indole, 6-Me-indole, and 7-MeO-indole had higher docking scores of 56.22, 57.26, and 58.42, respectively. The binding mode of these agonists showed several favorable interactions with the receptor residues lining the binding pocket. Key interactions included the hydrogen bond with Thr289 and aromatic interactions with Phe324 and His291, and arene-H interactions with Gln383 in addition to several hydrophobic and hydrophilic interactions (Fig. 8, B–D), which likely contribute to their high affinity. A close observation of the binding pocket revealed that both 4-Me-indole and 7-MeO-indole can simultaneously bind to the AhR ligand binding domain with high affinity, suggesting they may have a synergistic effect on AhR. Antagonists such as 3-Me-indole (Fig. 9A), 2,3-diMe-indole (Fig. 9B), and 2,3,7-triMe-indole (Fig. 9C) bind with good docking scores of 48.58, 40.87, and 45.97, respectively. However, their binding mode suggests that all of these molecules do not have all of the conserved interactions that favor agonist binding. Instead, 3-Me-indole has a hydrogen bond interaction with T289 (Fig. 9A), while the di- and tri- substituted indoles have aromatic interactions with Phe351 and low-range interactions with several hydrophobic and hydrophilic residues. Thus, it is likely that both agonists and antagonists bind at the same site, suggesting a competitive mode of inhibition. These results are in close agreement with the in vitro results described previously.
Fig. 8.
Schematic representation of AhR ligand binding domain docked with agonists TCDD (A), 4-Me-indole (B), 6-Me-indole (C), and 7-MeO-indole (D). TCDD binds favorably to the pocket with several hydrophobic residues facilitating the interactions with the TCDD core. All indole-substituted agonists bind with a similar binding mode and have conserved arene or cation-pi interactions with the aromatic ring of indole and hydrogen bond interaction with the amine group. In addition, many aromatic and hydrophobic residues facilitate the binding of these molecules. All residues that lie within a 5 Å radius from the center of the binding pocket are listed and colored according to their amphiphilicity profile. The schematic legend details the nature of the interactions of agonists with the residues in the binding pocket. The images were generated using the ligand interactions module in the MOE program.
Fig. 9.
Schematic representation of AhR ligand binding domain docked with antagonists 3-Me-indole (A), 2,3-diMe-indole (B), and 2,3,7-triMe-indole (C). All antagonists bind to the same pocket as the agonists and share a conserved hydrogen bond interaction between the amine group and threonine 289 or arene interactions with aromatic residues in the pocket. All residues that lie within a 5 Å radius from the center of the binding pocket are listed and colored according to their amphiphilicity profile. The schematic legend details the nature of the interactions of antagonists with the residues in the binding pocket. The images were generated using the ligand interactions module in the MOE program.
Discussion
A plethora of compounds, containing indole in their structure, were described as ligands of AhRs. These compounds comprise three main spheres: 1) products of intermediary metabolism of tryptophan (e.g., tryptamine, indole-3-acetic acid, and kynurenine) and heme (e.g., bilirubin and biliverdin), having rather systemic effects; 2) photoproducts of tryptophan, e.g., 6-formylindolo[3,2-b]carbazole and others (Pilditch et al., 2010 acting mainly in skin, but also inducing hepatic AhR-dependent genes; and 3) foodborne compounds and their metabolites, produced by hepatic and intestinal metabolism but also by intestinal microbiota, playing roles in immunity and homeostasis (e.g., indole-3-carbinol, indoxyl-3-sulfate, indole-3-acetate, and indole-3-aldehyde). Skatole, which is an intestinal microbiota-produced metabolite of tryptophan, was recently described as a partial agonist of AhRs (Rasmussen et al., 2016). Another methylated indole, xenobiotic 2-Me-indole, was described as an activator of AhRs in aquatic species (Brown et al., 2015, 2016). In addition, the activation of the human AhR in gene reporter assays was observed for 3-Me-indole, but not for 2-Me-indole and 1-Me-indole; however, experiments were performed only up to 10 μM concentrations of methylindoles (Hubbard et al., 2015). To add to the complexity of the interactions, it is important to note that with all of these indoles, on a systems level, AhR agonists like the classic indole molecule could, in a context-dependent manner, act as antagonists (Jin et al., 2014). Thus, studies were also conducted in the antagonist mode to characterize the effect of potential agonists on strong (dioxin) mediated AhR activation.
Interestingly, besides 1-Me-indole, 2-Me-indole, and 3-Me-indole, other methylated indoles were not studied for their capability to activate or inhibit AhRs, despite their chemical simplicity and structural similarity. On the topic of biologic relevance or physiologic relevance of the indoles studied, only skatole is made endogenously in humans and rodents. The others, while being xenobiotics, have biologic effects in other species, and when making them to test against AhRs could have biologic relevance to humans. One example is 5-MeO-2-Me-indole, which is an environmental pollutant that mimics signaling by peptide hormones in some jellyfish during strobilation (Fuchs et al., 2014). In addition, the relevance of studying other exogenous or xenobiotic indoles with respect to AhRs is that it completes our knowledge of which steric positions of the methyl group hinder or enhance AhR activity. This information is seminal to developing skatole-like analogs; specifically those placing certain R groups on certain carbons on the indole ring, which could assist in further obtaining potent and drug-like molecules. Structure/function in this regard would be crucial to know when developing skatole-like mimics that enhance AhR activity (e.g., for use as anti-inflammatory agents, etc.). The importance of evaluating the structure/function of xenobiotic indoles with respect to AhRs, is that it will help future drug-like discovery efforts with respect to the stereochemistry of methylation and methoxylation of simple indoles and these compounds. In general, these compounds are nontoxic and are likely contenders for future drug-like AhR activity. Thus, the importance of studying these chemical entities lies in their ability to focus efforts on very simple indoles that would be potent yet nontoxic molecules for in vivo modulation of AhRs.
In the current paper, we studied the effects of monomethyl- (seven isomers), dimethyl- (four isomers), trimethyl- (two isomers), monomethoxy- (four isomers), dimethoxy- (two isomers), and monomethyl-monomethoxy indoles (two isomers) on AhR-CYP1A1 signaling. Using one hybrid gene reporter assay (AZ-AHR cells) and measurement of CYP1A1 mRNA levels, we identified highly efficacious agonists of AhR involving 4-Me-indole, 6-Me-indole, and 7-MeO-indole, displaying relative efficacies of 134%, 91%, and 80%, respectively, compared with 5 nM dioxin. On the other hand, the tested compounds have much lower potencies (higher EC50 values) at AhRs compared with dioxin. However, their physico-chemical and structural properties are suitable for drug-able substances. In particular, in accordance with Lipinski’s rule of five, and related rules, the tested compounds have molecular masses ranging from 131 to 177 Da, which is much less than 500 or 300 Da, determined as the limit values by Lipinski’s rule of five or modified rule of three, respectively. Similarly, octanol-water partition coefficient log P for tested compounds is in the range from 2.12 to 3.31, which is again much lower than log P < 5 required by Lipinski’s rule of five. Therefore, the tested methylindoles and methoxyindoles are suitable for lead discovery in future drug-like modifications (Teague et al., 1999). Substantial antagonist activities (IC50; ∼15 μM) were achieved by 3-Me-indole, 2,3-diMe-indole, and 2,3,7-triMe-indole. Rasmussen et al. (2016) observed time-dependent activation of the AhR by 3-Me-indole, peaking at 8 hours after incubation; i.e., relative efficacy of 3-methylindole was higher after 8 hours compared with 24 hours after incubation. For this reason, and also to register possible metabolic degradation of the indoles, we tested all derivatives at two incubation times, i.e., 8 and 24 hours. We confirmed the observations of Rasmussen et al. (2016) for weak activators (e.g., 2-Me-indole and 3-Me-indole) in lower concentrations. On the other hand, strong activators of AhRs such as 4-Me-indole, 5-Me-indole, 6-Me-indole, and 7-MeO-indole caused progressive increase of luciferase activity expressed as both absolute and relative efficacy.
Taking into account that methyl substitution at position 4 and methoxy substitution at position 7 result in massive activation of the AhR, we anticipated that coupled compound 7-MeO-4-Me-indole, should act as a superactivator of the AhR. Surprisingly, 7-MeO-4-Me-indole displayed approximately 10 times lower relative efficacy compared with that of 4-Me-indole and 7-MeO-indole. In addition, while 4-Me-indole and 7-MeO-indole augmented TCDD-inducible AhR activity, 7-MeO-4-Me-indole displayed an antagonist effect on AhR. Incubation of AZ-AHR cells with mixtures of 4-Me-indole and 7-MeO-indole clearly showed enhanced activation of the AhR compared with the incubations of individual compounds in a wide array of concentration ratios. Consistently, synergistic induction of CYP1A1 mRNA by a mixture of 4-Me-indole and 7-MeO-indole was observed. Collectively, whereas combining 4-Me and 7-MeO substituents in coupled molecule 7-MeO-4-Me-indole resulted in drastic decrease of efficacy, combining 4-Me-indole and 7-MeO indole in a mixture caused synergistic activation of the AhR. A speculative explanation for such behavior could be the binding of more than one indole-derived molecule to the AhR protein, which in the future could be verified using the AhR ligand binding domain and/or protein mutants. This is consistent with the finding by Hubbard et al. (2015), who performed homology docking and found that two molecules of indole or 3-Me-indole can effectively mimic the binding of indirubin in the human AhR.
Indeed, molecular modeling revealed that the binding mode of strong AhR agonists 4-Me-indole and 7-MeO-indole shows several favorable interactions with the AhR residues lining the binding pocket. This pocket was previously validated for the binding of TCDD and other indole molecules using modeling and site-directed mutagenesis studies (Motto et al., 2011; Soshilov and Denison, 2014). In particular, agonist binding is dominated by strong arene interactions with Gln383, aromatic interactions with Phe324 and His291, and hydrogen bond with Thr289. Agonist binding is also coordinated by several hydrophobic and hydrophilic interactions, likely contributing to the high affinity of 4-Me-indole and 7-MeO-indole. Given the small size of 4-Me-indole and 7-MeO-indole, we observed that both of these compounds can simultaneously bind to the AhR ligand binding domain, and hence may have a synergistic effect on the AhR.
The binding mode of antagonists such as 3-Me-indole, 2,3-diMe-indole, and 2,3,7-triMe-indole suggests that all of these molecules also bind to the same pocket as agonists, suggesting competing binding properties; however, antagonists bind with lower binding scores. These results are in close agreement with our in vitro results. The antagonist interactions are mediated by a hydrogen bond interaction with T289 in the case of 3-Me-indole and arene interactions with Phe351.
Collectively, this is a systematic study that shows critical indole carbons and modifications on those carbons that dictate AhR activity. We have also shown that some indoles, which are active as AhR agonists, could in a context-dependent manner (i.e., the presence of dioxin) act as antagonists. This could be studied further in the future using indole as the activator rather than dioxin. Modeling provides a theoretical binding mode based on well-conceived and clear experimental data. Future studies would involve proof of these binding models using mutants of the AhR at the genetic and protein level. In short, we have provided a very discrete structure-function analysis of methylated and methoxylated indoles (Jin et al., 2014) that allows for future application of these data toward a systems biology study of indoles and host health.
Abbreviations
- AhR
aryl hydrocarbon receptor
- ChIP
chromatin immunoprecipitation
- DMSO
dimethylsulfoxide
- MTT
3-[4,5-dimethylthiazole-2-yl]-2,5-diphenyltetrazolium bromide
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- TCDD
2,3,7,8-tetrachlorodibenzo-p-dioxin
Authorship Contributions
Participated in research design: Mani, Dvorak.
Conducted experiments: Stepankova, Bartonkova, Jiskrova, Vrzal, Kortagere.
Contributed new reagents and analytic tools: Dvorak.
Performed data analysis: Stepankova, Bartonkova, Jiskrova, Vrzal, Kortagere, Dvorak.
Wrote or contributed to the writing of the manuscript: Dvorak, Kortagere, Mani.
Footnotes
Financial support from the Czech Science Foundation [Grant P303/12/G163]; Palacky University in Olomouc [Grants PrF-2017-004 and PrF-2018-005]; Operational Programme Research, Development and Education, European Regional Development Fund, Ministry of Education, Youth and Sports of the Czech Republic [Grant CZ.02.1.01/0.0/0.0/16_019/0000754]; Institute for Clinical and Translational Research Pilot Award [AECOM]; Broad Medical Research Program at Crohn’s & Colitis Foundation of America [Grant 362520]; Department of Defence Partnering PI [Grants PR160167, R43DK105694, and P30DK041296]; and National Institutes of Health [Grants CA127231 and CA161879], National Institutes of Health Diabetes Research Center [Grant P30 DK020541], and National Institutes of Health Cancer Center [Grant P30 CA013330] is acknowledged.
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Adachi J, Mori Y, Matsui S, Takigami H, Fujino J, Kitagawa H, Miller CA, III, Kato T, Saeki K, Matsuda T. (2001) Indirubin and indigo are potent aryl hydrocarbon receptor ligands present in human urine. J Biol Chem 276:31475–31478. [DOI] [PubMed] [Google Scholar]
- Anderton MJ, Manson MM, Verschoyle RD, Gescher A, Lamb JH, Farmer PB, Steward WP, Williams ML. (2004) Pharmacokinetics and tissue disposition of indole-3-carbinol and its acid condensation products after oral administration to mice. Clin Cancer Res 10:5233–5241. [DOI] [PubMed] [Google Scholar]
- Bergander L, Wahlström N, Alsberg T, Bergman J, Rannug A, Rannug U. (2003) Characterization of in vitro metabolites of the aryl hydrocarbon receptor ligand 6-formylindolo[3,2-b]carbazole by liquid chromatography-mass spectrometry and NMR. Drug Metab Dispos 31:233–241. [DOI] [PubMed] [Google Scholar]
- Brown DR, Clark BW, Garner LV, Di Giulio RT. (2015) Zebrafish cardiotoxicity: the effects of CYP1A inhibition and AHR2 knockdown following exposure to weak aryl hydrocarbon receptor agonists. Environ Sci Pollut Res Int 22:8329–8338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DR, Clark BW, Garner LV, Di Giulio RT. (2016) Embryonic cardiotoxicity of weak aryl hydrocarbon receptor agonists and CYP1A inhibitor fluoranthene in the Atlantic killifish (Fundulus heteroclitus). Comp Biochem Physiol C Toxicol Pharmacol 188:45–51. [DOI] [PubMed] [Google Scholar]
- Chen I, McDougal A, Wang F, Safe S. (1998) Aryl hydrocarbon receptor-mediated antiestrogenic and antitumorigenic activity of diindolylmethane. Carcinogenesis 19:1631–1639. [DOI] [PubMed] [Google Scholar]
- Chen I, Safe S, Bjeldanes L. (1996) Indole-3-carbinol and diindolylmethane as aryl hydrocarbon (Ah) receptor agonists and antagonists in T47D human breast cancer cells. Biochem Pharmacol 51:1069–1076. [DOI] [PubMed] [Google Scholar]
- Chowdhury G, Dostalek M, Hsu EL, Nguyen LP, Stec DF, Bradfield CA, Guengerich FP. (2009) Structural identification of diindole agonists of the aryl hydrocarbon receptor derived from degradation of indole-3-pyruvic acid. Chem Res Toxicol 22:1905–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGroot DE, Franks DG, Higa T, Tanaka J, Hahn ME, Denison MS. (2015) Naturally occurring marine brominated indoles are aryl hydrocarbon receptor ligands/agonists. Chem Res Toxicol 28:1176–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison MS, Nagy SR. (2003) Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol 43:309–334. [DOI] [PubMed] [Google Scholar]
- Fuchs B, Wang W, Graspeuntner S, Li Y, Insua S, Herbst EM, Dirksen P, Böhm AM, Hemmrich G, Sommer F, et al. (2014) Regulation of polyp-to-jellyfish transition in Aurelia aurita. Curr Biol 24:263–273. [DOI] [PubMed] [Google Scholar]
- Heath-Pagliuso S, Rogers WJ, Tullis K, Seidel SD, Cenijn PH, Brouwer A, Denison MS. (1998) Activation of the Ah receptor by tryptophan and tryptophan metabolites. Biochemistry 37:11508–11515. [DOI] [PubMed] [Google Scholar]
- Helferich WG, Denison MS. (1991) Ultraviolet photoproducts of tryptophan can act as dioxin agonists. Mol Pharmacol 40:674–678. [PubMed] [Google Scholar]
- Hubbard TD, Murray IA, Bisson WH, Lahoti TS, Gowda K, Amin SG, Patterson AD, Perdew GH. (2015) Adaptation of the human aryl hydrocarbon receptor to sense microbiota-derived indoles. Sci Rep 5:12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin UH, Lee SO, Sridharan G, Lee K, Davidson LA, Jayaraman A, Chapkin RS, Alaniz R, Safe S. (2014) Microbiome-derived tryptophan metabolites and their aryl hydrocarbon receptor-dependent agonist and antagonist activities. Mol Pharmacol 85:777–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G, Willett P, Glen RC. (1995) Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. J Mol Biol 245:43–53. [DOI] [PubMed] [Google Scholar]
- Kolluri SK, Jin UH, Safe S. (2017) Role of the aryl hydrocarbon receptor in carcinogenesis and potential as an anti-cancer drug target. Arch Toxicol 91:2497–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanza DL, Yost GS. (2001) Selective dehydrogenation/oxygenation of 3-methylindole by cytochrome P450 enzymes. Drug Metab Dispos 29:950–953. [PubMed] [Google Scholar]
- Li CY, Wu C, Liu JX, Wang YZ, Wang JK. (2009) Spatial variation of intestinal skatole production and microbial community in Jinhua and Landrace pigs. J Sci Food Agric 89:639–644. [Google Scholar]
- Motto I, Bordogna A, Soshilov AA, Denison MS, Bonati L. (2011) New aryl hydrocarbon receptor homology model targeted to improve docking reliability. J Chem Inf Model 51:2868–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotna A, Pavek P, Dvorak Z. (2011) Novel stably transfected gene reporter human hepatoma cell line for assessment of aryl hydrocarbon receptor transcriptional activity: construction and characterization. Environ Sci Technol 45:10133–10139. [DOI] [PubMed] [Google Scholar]
- Pilditch RL, Lizardi I, Nelson BC, Johnson MM. (2010) inventors, Bare Conductive, assignee. Skin-Safe Conductive Ink and Method for Application on the Body. U.S. patent 20120020033A1. 2010 Feb 4. [Google Scholar]
- Rasmussen MK, Balaguer P, Ekstrand B, Daujat-Chavanieu M, Gerbal-Chaloin S. (2016) Skatole (3-methylindole) is a partial aryl hydrocarbon receptor agonist and induces CYP1A1/2 and CYP1B1 expression in primary human hepatocytes. PLoS One 11:e0154629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothhammer V, Mascanfroni ID, Bunse L, Takenaka MC, Kenison JE, Mayo L, Chao CC, Patel B, Yan R, Blain M, et al. (2016) Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat Med 22:586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruangyuttikarn W, Appleton ML, Yost GS. (1991) Metabolism of 3-methylindole in human tissues. Drug Metab Dispos 19:977–984. [PubMed] [Google Scholar]
- Sali A, Blundell TL. (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815. [DOI] [PubMed] [Google Scholar]
- Schroeder JC, Dinatale BC, Murray IA, Flaveny CA, Liu Q, Laurenzana EM, Lin JM, Strom SC, Omiecinski CJ, Amin S, et al. (2010) The uremic toxin 3-indoxyl sulfate is a potent endogenous agonist for the human aryl hydrocarbon receptor. Biochemistry 49:393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte KW, Green E, Wilz A, Platten M, Daumke O. (2017) Structural basis for aryl hydrocarbon receptor-mediated gene activation. Structure 25:1025–1033.e1023. [DOI] [PubMed] [Google Scholar]
- Sinal CJ, Bend JR. (1997) Aryl hydrocarbon receptor-dependent induction of Cyp1a1 by bilirubin in mouse hepatoma Hepa 1c1c7 cells. Mol Pharmacol 52:590–599. [DOI] [PubMed] [Google Scholar]
- Soshilov AA, Denison MS. (2014) Ligand promiscuity of aryl hydrocarbon receptor agonists and antagonists revealed by site-directed mutagenesis. Mol Cell Biol 34:1707–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stejskalova L, Dvorak Z, Pavek P. (2011) Endogenous and exogenous ligands of aryl hydrocarbon receptor: current state of art. Curr Drug Metab 12:198–212. [DOI] [PubMed] [Google Scholar]
- Teague SJ, Davis AM, Leeson PD, Oprea T. (1999) The design of leadlike combinatorial libraries. Angew Chem Int Ed Engl 38:3743–3748. [DOI] [PubMed] [Google Scholar]
- Vrzal R, Knoppová B, Bachleda P, Dvořák Z. (2013) Effects of oral anorexiant sibutramine on the expression of cytochromes P450s in human hepatocytes and cancer cell lines. J Biochem Mol Toxicol 27:515–521. [DOI] [PubMed] [Google Scholar]
- Weems JM, Yost GS. (2010) 3-Methylindole metabolites induce lung CYP1A1 and CYP2F1 enzymes by AhR and non-AhR mechanisms, respectively. Chem Res Toxicol 23:696–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Su X, Potluri N, Kim Y, Rastinejad F. (2016) NPAS1-ARNT and NPAS3-ARNT crystal structures implicate the bHLH-PAS family as multi-ligand binding transcription factors. eLife 5:e18790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Easterwood LM, Maher N, Torres R, Huebert N, Yost GS. (2007) Metabolism and bioactivation of 3-methylindole by human liver microsomes. Chem Res Toxicol 20:140–148. [DOI] [PubMed] [Google Scholar]