Abstract
The orphan nuclear receptor Nurr1 (also called nuclear receptor-4A2) regulates inflammatory gene expression in glial cells, as well as genes associated with homeostatic and trophic function in dopaminergic neurons. Despite these known functions of Nurr1, an endogenous ligand has not been discovered. We postulated that the activation of Nurr1 would suppress the activation of glia and thereby protect against loss of dopamine (DA) neurons after subacute lesioning with 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP). Our previous studies have shown that a synthetic Nurr1 ligand, 1,1-bis(3′-indolyl)-1-(p-chlorophenyl)methane (C-DIM12), suppresses inflammatory gene expression in primary astrocytes and induces a dopaminergic phenotype in neurons. Pharmacokinetic analysis of C-DIM12 in mice by liquid chromatography-mass spectrometry demonstrated that approximately three times more compound concentrated in the brain than in plasma. Mice treated with four doses of MPTP + probenecid over 14 days were monitored for neurobehavioral function, loss of dopaminergic neurons, and glial activation. C-DIM12 protected against the loss of DA neurons in the substantia nigra pars compacta and DA terminals in the striatum, maintained a ramified phenotype in microglia, and suppressed activation of astrocytes. In vitro reporter assays demonstrated that C-DIM12 was an effective activator of Nurr1 transcription in neuronal cell lines. Computational modeling of C-DIM12 binding to the three-dimensional structure of human Nurr1 identified a high-affinity binding interaction with Nurr1 at the coactivator domain. Taken together, these data suggest that C-DIM12 is an activator of Nurr1 that suppresses glial activation and neuronal loss in vivo after treatment with MPTP, and that this receptor could be an efficacious target for disease modification in individuals with Parkinson’s disease and related disorders.
Introduction
Gene expression necessary for the synthesis and regulation of dopamine (DA) in neurons of the substantia nigra pars compacta (SNpc) is controlled by the orphan nuclear receptor related 1 [Nurr1 (also called NR4A2)] protein (Zetterström et al., 1996; Sakurada et al., 1999). Nurr1 is highly expressed in the ventral midbrain and is downregulated in Parkinson’s disease (PD) patients (Kadkhodaei et al., 2013; Montarolo et al., 2016). The homeostatic function of Nurr1 in DA neurons is mediated by nuclear binding to the nerve growth factor binding recognition element (NBRE) as monomers, homodimers, or heterodimers with the coactivator protein retinoic acid receptor (Saijo et al., 2009; García-Yagüe et al., 2013). NBRE sequences are recognized in the upstream promoter regions of DA genes such as tyrosine hydroxylase (TH), vesicular monoamine transporter 2 (VMAT2), DA transporter (DAT), and aromatic amino acid decarboxylase (Smits et al., 2003). Thus, Nurr1 activity is crucial for DA neuronal differentiation during development to maintenance throughout adulthood (Sakurada et al., 1999; Jankovic et al., 2005).
Homozygous Nurr1 (−/−) mice do not survive past postnatal day 1, and Nurr1 (+/−) mice are more susceptible to 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)–induced neurotoxicity (Saucedo-Cardenas et al., 1998; Le et al., 1999). Inhibition of mitochondrial complex 1 by the active metabolite of MPTP, MPP+, causes the loss of DA neurons in the nigrostriatal system and is accompanied by significant glial activation and neuroinflammation that is thought to exacerbate neuronal injury (Wu et al., 2003a; Glass et al., 2010). Nurr1 is now recognized as a critical regulator of inflammatory gene expression in glial cells, where it acts as a tonic regulator of NF-κB–regulated inflammatory genes by stabilizing nuclear corepressor proteins at p65/p50 cis-acting promoter elements, thereby limiting the expression of inflammatory genes (Saijo et al., 2009).
Despite the known transcriptional regulatory functions of Nurr1 (NR4A2), an endogenous ligand for this receptor is yet to be identified. Therefore, like orphan nuclear receptor related 77 (Nur77) [nuclear receptor-4A1 (NR4A1)] and Nor1 [neuron-derived orphan receptor 1 (nuclear receptor-4A3)], Nurr1 is classified as an orphan nuclear receptor (Safe et al., 2016). However, multiple studies have demonstrated modulation of Nurr1 transcriptional activity using synthetic compounds (Ordentlich et al., 2003; Li et al., 2012; Smith et al., 2015). Our previous studies using several phenyl-substituted 3,3′-diindolylmethane (C-DIM) compounds demonstrated that selected analogs are structure-dependent activators of NR4A orphan nuclear receptors (Inamoto et al., 2008; Safe et al., 2008). One compound from this series with high specific activity toward Nurr1, 1,1-bis(3′-indolyl)-1-(p-chlorophenyl)methane (C-DIM12), activates Nurr1 and induces antineoplastic effects in cancer cells (Li et al., 2012). We also reported that C-DIM12 inhibits the expression of NF-κB–regulated genes in glial cells and induces a dopaminergic phenotype in neuronal cell lines (De Miranda et al., 2015; Hammond et al., 2015). RNA interference knockdown of Nurr1 in both glia and neurons ablated the effects of C-DIM12 in each cell type, demonstrating that in neural cells C-DIM12 acts through a Nurr1. In vivo, C-DIM12 displayed favorable pharmacokinetics and neuroprotective efficacy with oral dosing, including high bioavailability and distribution to the central nervous system, as well as protection against the loss of DA neurons in the SNpc in the MPTP/probenecid (MPTPp) model of PD (De Miranda et al., 2013, 2014). These studies demonstrate that C-DIM12 inhibits neuroinflammatory activation of microglia and astrocytes and protects against the loss of DA neurons. However, it remains to be determined whether lower doses of C-DIM12 will have similar pharmacodynamic efficacy during concurrent lesioning with MPTPp and whether they will provide a similar degree of protection against an ongoing neurotoxic insult mimicking the complex I deficits in PD.
Based on the demonstrated neuroprotective and antineuroinflammatory effects of C-DIM12, we postulated that this Nurr1 activator would protect against the loss of dopaminergic neurons during lesioning with MPTPp, despite the neurotoxic stress of mitochondrial complex 1 inhibition. To test this hypothesis, C57BL/6 mice were dosed with MPTPp twice weekly for 2 weeks and were concurrently given C-DIM12 (25 mg/kg daily, by mouth) once daily throughout the treatment period. Neurobehavioral analysis was conducted during the study to detect motor deficits consistent with deprecations in nigrostriatal DA. Brain tissue [SN and striatum (ST)] was collected to determine the pharmacokinetic parameters of C-DIM12 at this dose, as well as stereological determination of DA neuron numbers, glial activation, and quantitative polymerase chain reaction (qPCR) array measurement of gene expression for inflammatory and cell death pathways. In addition, we conducted transactivation reporter studies in neural cell lines, as well as in silico modeling to identify putative binding sites for C-DIM12 within the ligand binding domain of human Nurr1. These findings demonstrate that C-DIM12 is an activating ligand of Nurr1 that induces an anti-inflammatory phenotype in glial cells and preserves dopaminergic soma even during concurrent lesioning with MPTPp.
Materials and Methods
Chemicals and Reagents
C-DIM12 was synthesized and obtained by the laboratory of Dr. Stephen Safe at Texas A&M University (College Station, TX). Working concentrations of C-DIM12 were diluted in corn oil (C.O.) and sonicated in hot water bath until solubilized. MPTP (Sigma-Aldrich, St. Louis, MO) was solubilized at a final working concentration in saline (0.9% NaCl2). Probenecid (Sigma-Aldrich) was prepared in 5% sodium bicarbonate/Milli-Q water (MilliporeSigma, Burlington, MA) to a final working concentration (pH 7.5). All additional reagents were obtained from Sigma-Aldrich, unless stated otherwise.
Animals and Treatment Regimen
Inbred C57BL/6 male mice (∼24 weeks of age; 25–30 g in weight) were acquired by Charles River Laboratories (Wilmington, MA) and housed on 12-hour light/dark cycles in a temperature-controlled room (maintained at 22–24°C) with access to standard chow and water ad libitum. Mice were administered C-DIM12 (25 mg/kg) or C.O. (vehicle control) by oral gavage. Mice were dosed with MPTPp twice weekly for 2 weeks, with each dose delivered 4 days apart. On the day of dosing, probenecid was delivered in the morning by intraperitoneal injection (100 mg/kg), and then MPTP (20 mg/kg) or saline (0.9% NaCl) was administered 4 hours later by subcutaneous injection, per our previously published protocol (De Miranda et al., 2014). C-DIM12 or C.O. was administered daily by intragastric gavage (14 doses total) throughout the treatment period. At the conclusion of the study, mice were anesthetized under deep isoflurane anesthesia and transcardially perfused with 0.1 M phosphate-buffered saline (PBS)-cacodylate/heparin (10 U/ml) and 3% paraformaldehyde/PBS. After perfusion, brains were dissected and stored in paraformaldehyde at 4°C overnight, and then stored in sodium-cacodylate-PBS (pH 7.2) containing 15%–30% sucrose at 4°C until processed for cryosectioning. For neurochemical sample collection, animals were also administered deep isoflurane before rapid removal of ST and ventral midbrain for flash freezing in liquid nitrogen. Brain samples were then transferred to −80°C storage until processed for RNA, protein, and high-performance liquid chromatography (HPLC) analysis.
Real-Time Video Gait Analysis and Open Field Behavioral Testing.
Changes in gait were determined by analysis of unrestricted movement along a fixed trackway using a video-based system constructed in our laboratory. Briefly, mice were allowed to walk along a 1-m-long glass trackway with fixed sides and top that was illuminated with green light-emitting diode lights such that paw placement results in total internal reflection of light downward toward a high-speed video camera for digital recording. Animals were backlit from above with low-power red light-emitting diode lights to distinguish the silhouetted form of the mouse (black) from paw prints (green) for digital analysis. Incentive to traverse the trackway was provided by placement of the home cage of the animal at the end of the trackway, thereby permitting reliable detection of unrestricted gait and motion. Mice were habituated to the trackway once daily for 2 days prior to the onset of the treatment period, and a baseline of gait was recorded on the first day of the study prior to dosing with MPTPp. Video recording of the mouse gait was filmed using a GoPro Hero3+ camera (GoPro, San Mateo, CA) at 60 frames/s, 1080 dots per inch, and analysis code was written in Matlab (MathWorks, Natick, MA) in conjunction with the Department of Electrical and Computer Engineering at Colorado State University (Fort Collins, CO). Open field activity parameters were acquired with the Versamax System (Omnitech Electronics, Inc., Columbus, OH). Mice were allowed to acclimate to open-field activity testing (OFT) behavioral chambers for 2 days prior to study for measurement of x-, y-, and z-planes of activity for a monitoring period of 5 minutes under white noise and ambient light. OFT activity was then monitored for animals on days 0, 7, and 14 of the study. Parameters were analyzed using Fusion software (Omnitech Electronics, Inc.) for the detection of the total margin and center time activity. All values were normalized for the difference from baseline at day 0.
Three-Dimensional Design–Based Stereology of Neuronal Cell Bodies and Assessment of Striatal Terminal Density.
Stereological determination for the number of neurons in the SNpc was performed using a three-dimensional (3D) design–based stereology method, as we previously reported (De Miranda et al., 2014). The entire SNpc was serially sectioned from rostral to caudal, which was demarcated by the subthalamic nucleus to the retrorubral field, respectively. Every fifth free-floating section (10 total) was selected from each animal and immunostained with antibodies against TH (Abcam, Cambridge, UK) and microtubule-associated protein 2 (MAP2) (Abcam). Stereological counting of both cell markers was performed using Slidebook software (version 5.0; Intelligent Imaging Innovations, Denver, CO). The SNpc boundary was marked on a 10× objective montage image of the entire section. The optical fractionator method was employed with a randomized 40× objective z-stack image-sampling setup frame size (100 × 100 μm), frame spacing (200 × 200 μm), dissector height (30 μm), and upper guard distance (2 μm). Each z-stack image was blindly quantitated for TH+ and MAP2+ cells to calculate a total estimate of neuronal cell bodies within the SNpc of each animal. For measurement of TH+ striatal terminals, two ST sections per animal with similar anatomic landmarks were selected and immunostained for TH. The 10× objective montage images were generated for a masked outline of the caudate putamen. Each masked region was normalized for background subtraction and quantified for mean intensity fluorescence of TH.
Immunofluorescence Staining
All brain tissue processed for immunofluorescence (IF) was frozen with optimal cutting temperature compound on a microtome stage and sectioned/collected for ST (25 μm in thickness) and SN (40 μm in thickness) regions. Tissue sections were stored in cryoprotectant (30% sucrose, 30% ethylene glycol, and 0.5 M phosphate buffer, pH 7.2) at 20°C until selected for immunostaining. IF staining was conducted as previously described by Miller et al. (2011), and all antibody dilutions were 1:500, unless stated otherwise (Miller et al., 2011). For stereology, SN sections were immunostained with anti-TH (cat. no. AB152; Merck Millipore, Burlington, MA) and anti-MAP2 (cat. no. AB5392; Abcam). For gliosis, SN and ST sections were immunostained with either anti–ionized calcium-binding adapter molecule 1 (IBA-1) (1:250; cat. no. 016-20001; Wako Chemicals USA, Inc., Richmond, VA) or anti–glial fibrillary acidic protein (GFAP) (cat. no. Z0334; Agilent Technologies, Santa Clara, CA) and anti-TH (cat. no. AB76442, Abcam). SN tissue was also immunostained with anti-TH and anti-Nurr1 (1:200, SC991; Santa Cruz Biotechnology, Dallas, TX) for mean intensity measurements of Nurr1. All secondary antibodies used for IF were Alexa Fluor 488, 555, and 647 (Thermo Fisher Scientific, Carlsbad, CA).
Measurement of Gliosis and IF Imaging
Quantitation of IBA-1+ and GFAP+ cell bodies was conducted on two sections of the SNpc and ST regions per animal. The optical fractionator method that was employed was adjusted with counting frame size (150 × 150 μm) and frame spacing (for SN, 250 × 250 μm; for ST, 550 × 550 μm). Blind quantitation of cell bodies for the relative number of glial cells per region was conducted on 40× z-stack images of IBA+ cells from the SNpc. The same IBA-1+ cell images were also converted to a maximum projection after a binary transformation and then were rendered to a skeletonized image in ImageJ, as previously described by Morrison and Filosa (2013), Schneider et al. (2012), and Morrison and Filosa (2013). Images for quantitative measurements were all acquired with an Axiovert 200 M inverted fluorescent microscope (Carl Zeiss Microscopy GmbH, Jena, Germany) equipped with a Hamamatsu ORCA-ER-cooled charge coupled device camera using a 10× and 40× air objective (Hamamatsu Photonics, Hamamatsu City, Japan). High magnification representative images were generated with a Plan-Apochromat 100× oil objective lens. High-magnification images of 3D IBA-1+ cells were acquired with a 63× objective lens on a Zeiss LSM 510 laser-scanning confocal microscope, using Zen software (Oberkochen, Germany). The z-stack images of microglia were surfaced rendered in Imaris software (Bitplane, Zurich, Switzerland) for a voxel view of morphology. Low-magnification montage images of tissue sections were acquired with an air 10× objective lens using a Hamamatsu Photonics ORCA-Flash4.0 digital complementary metal-oxide semiconductor camera, a ProScan III stage controller (Prior, Rockland, MA), and CellSens Dimension software (version 1.12; Olympus, Center Valley, PA).
Western Blotting
Striatal tissue was homogenized and lysed in radioimmunoprecipitation assay buffer with protease inhibitor for Western blot analysis. Protein concentration was determined using a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, Inc., Rockford, IL). Thirty micrograms of protein was loaded into a polyacrylamide 12% separating and 4% stacking gel. Anti-DAT (1:500, Ab1591P; Merck Millipore); anti-VMAT2 (1:750; gift from the laboratory of Dr. Gary Miller, Emory University, Atlanta, GA); and anti-TH (1:1000; cat. no. AB152), anti-Nurr1 (1:100; cat. no. SC991), anti-GFAP (1:1000; cat. no. Z0334), and anti–β actin (1:2000, cat. no. A1978; Sigma-Aldrich) were diluted in 5% milk/Tris-buffered saline with tween (0.1%) blocking buffer. All Western blots were imaged on a Bio-Rad Laboratories (Hercules, CA) ChemiDoc MP Imaging System, and raw TIFF files were analyzed for mean optical band density with ImageJ analysis software (Schneider et al., 2012).
Pharmacokinetic Analysis of C-DIM12 and Determination of Catecholamine Content
For pharmacokinetic analysis of C-DIM12, male C57BL/6 mice (27–30 g) were administered C-DIM12 (25 mg/kg) dissolved in C.O. by intragastric gavage. After administration, mice were euthanized at 0, 0.5, 1, 2, 4, 8, and 24 hours (N = 4/time point) by decapitation under isoflurane anesthesia. Midbrain tissue samples were rapidly dissected using a 1 mm brain block. Plasma samples were collected from trunk blood and centrifuged at 1500 rpm for 5 minutes. Frozen brain and plasma samples were then processed for liquid chromatography-mass spectrometry by the Pharmacology Core at Colorado State University, as reported in previous studies from our group (De Miranda et al., 2013). Striatal samples from each experimental group were measured for DA and 3,4-dihydroxyphenylacetic acid (DOPAC) by HPLC coupled with electrochemical detection. The Neurochemistry Core Laboratory at the Vanderbilt University Center for Molecular Neuroscience Research group (Nashville, TN) processed all tissue samples from each experimental group with coded labeling for unbiased analysis.
Computational Modeling
Small-molecule docking studies were conducted using Accelrys Discovery Studio 4.5 (Accelrys Inc., San Diego, CA), and the crystal structure coordinates for the human Nurr1 (NR4A2) ligand binding domain (Protein Data Bank identification, 1OVL) (Wang et al., 2003) were downloaded from the Protein Data Bank (http://www.rcsb.org/pdb). The protein was prepared and subjected to energy minimization using the conjugate gradient minimization protocol with a CHARMm (Chemistry at Harvard Macromolecular Mechanics) force field (Brooks et al., 2009) and the generalized born implicit solvent model with simple switching (Feig et al., 2004) that converged to a root mean squared gradient of <0.01 kcal/mol. The Flexible Docking protocol (Koska et al., 2008), which allows flexibility in both the protein and the ligand during the docking calculations, was used to predict the binding of C-DIM12 in the regions of both the coactivator and ligand binding pockets of NR4A2. Predicted binding poses were energy minimized in situ using the CDOCKER protocol (Wu et al., 2003b) prior to final ranking of docked poses via consensus scoring combining the Jain (1996), proteolipid protein 2 (Parrill and Rami Reddy, 1999), and Ludi3 (Böhm, 1994) scoring functions. Predicted binding energies were calculated using the distance-dependent dielectric model.
Transfections and Luciferase Assays
Neuron-like, PC12 cells were plated on 12-well plates at 2.5 × 105 per well in Dulbecco’s modified Eagle’s medium/F12 supplemented with 2.5% charcoal-stripped fetal bovine serum and were allowed to attach and settle for overnight. Luciferase (Luc) plasmids (UASx5-Luc, NBREx3-Luc, and NurREx3-Luc) and corresponding expression plasmids (GAL4-NR4A2 or FLAG-NR4A2) were cotransfected in cells at a 10:1 ratio (i.e., 1000 ng Luc and 100 ng NR4A2 per well). Lipofectamine 2000 Reagent (Thermo Fisher Scientific) was used for transfection according to the manufacturer protocol, and all plasmids have previously been described (Li et al., 2012). After 6 hours of transfection, cells were treated with medium (as described above) containing either solvent (dimethylsulfoxide) or the indicated concentration of C-DIM12 for 18 hours. Cells were then lysed using a freeze-thaw protocol, and cell extracts were used for Luc assays. Luc activity values were normalized against corresponding protein concentrations determined in a Bradford assay. Luc assays were run in triplicate for each determination, and results are expressed as the mean ± S.D.
RNA Isolation and Reverse-Transcription-qPCR Array Analysis
Midbrain tissue samples from each experimental group were homogenized and lysed using Qiashredder columns along with on-column and in solution DNase treatment (Qiagen, Hilden, Germany). Samples were purified for RNA using an RNeasy kit, eluted with RNase-free water and concentrations were determined using a Nanodrop One Spectrophotometer (Thermo Scientific, Waltham, MA). mRNA was reverse transcribed to cDNA with iScript (Bio-Rad Laboratories) reverse transcriptase enzyme for a total of 250 ng, and qPCRs were prepared in SYBRgreen mastermix (Bio-Rad Laboratories). Samples from each experimental group were amplified using RT2 Profiler PCR arrays (Qiagen) for NF-κB signaling pathway target genes (PAMM-025ZG-4) and PD-associated genes (cat. no. PAMM-124ZG-4) for a total analysis of 168 genes. Both sets of 384-well pathway array plates were run according the manufacturer protocol on a LightCycler 480 Real-Time PCR System (Roche Molecular Diagnostics, Branford, CT). Gene expression fold change was analyzed using the SABiosciences (Frederick, MD) software.
Statistical Analysis
Data were presented as the mean ± S.E.M., unless noted otherwise. All experimental values from each mean were analyzed with a Grubb’s test (α = 0.05) for the exclusion of significant outliers. Differences among three experimental groups were analyzed with a one-way analysis of variance followed by a Tukey post hoc multiple comparisons test. Two group comparisons for densitometry analysis was conducted with an unpaired student’s t test followed by Welch’s correction. A two-way analysis of variance was performed when incorporating “day” as an experimental variable for behavioral tests. Significance was identified as ++P < 0.01, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. All statistical analyses were conducted using Prism (version 6.0; GraphPad Software, San Diego, CA).
Results
C-DIM12 Is Highly Concentrated in Brain Tissue.
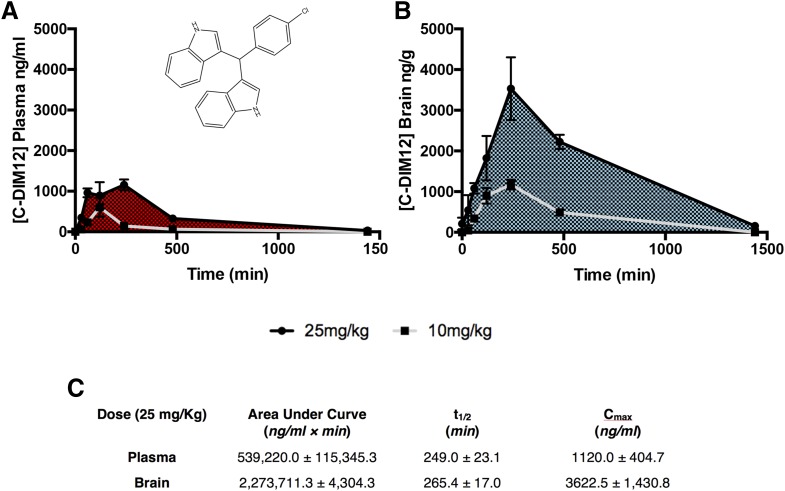
We previously conducted neuroprotection studies with several C-DIM compounds known to activate NR4A1 or NR4A2, using daily oral dosing at 50 mg/kg (De Miranda et al., 2014). To determine whether a lower dose would have similar neuroprotective efficacy, we first performed a pharmacokinetic study with C-DIM12 (chemical structure in Fig. 1A) at 25 mg/kg by administering this dose by mouth and sampling plasma and brain concentrations at 0, 0.5, 1, 2, 4, 8, and 24 hours Plasma and midbrain samples were analyzed by LC-MS analysis per our previously published methods (De Miranda et al., 2013). As shown in Fig. 1, C-DIM12 reached a Cmax at 4 hours of 1120.0 ± 404.7 ng/ml and 3622.5 ± 1430.8 ng/g in plasma and brain, respectively. The half-life was 249 ± 23 minutes in plasma and 264 ± 17 minutes in brain, approximately 15 minutes longer in brain tissue compared with plasma (Fig. 1C). Timing for the Cmax and total drug clearance is consistent with the previous dosage of 10 mg/kg (Fig. 1, A and B) (N = 4 animals per time point).
Fig. 1.
Pharmacokinetic analysis depicts a high concentration of C-DIM12 in brain tissue. Plasma (A) and brain (B) samples were collected from mice at 24, 8, 4, 2, 1, 0.5, and 0 hours after oral gavage of C-DIM12 (25 mg/kg, black line; 10 mg/kg, gray line) (De Miranda et al., 2013) and were analyzed for C-DIM12 concentration via LC-MS. (C) Chart of AUC, half-life (t1/2), and Cmax depict a higher partitioning of C-DIM12 to brain tissue (n = 4 animals/time point).
Daily Administration of C-DIM12 Protects against Loss of Dopaminergic Neurons during Concurrent Neurotoxic Challenge with MPTPp.
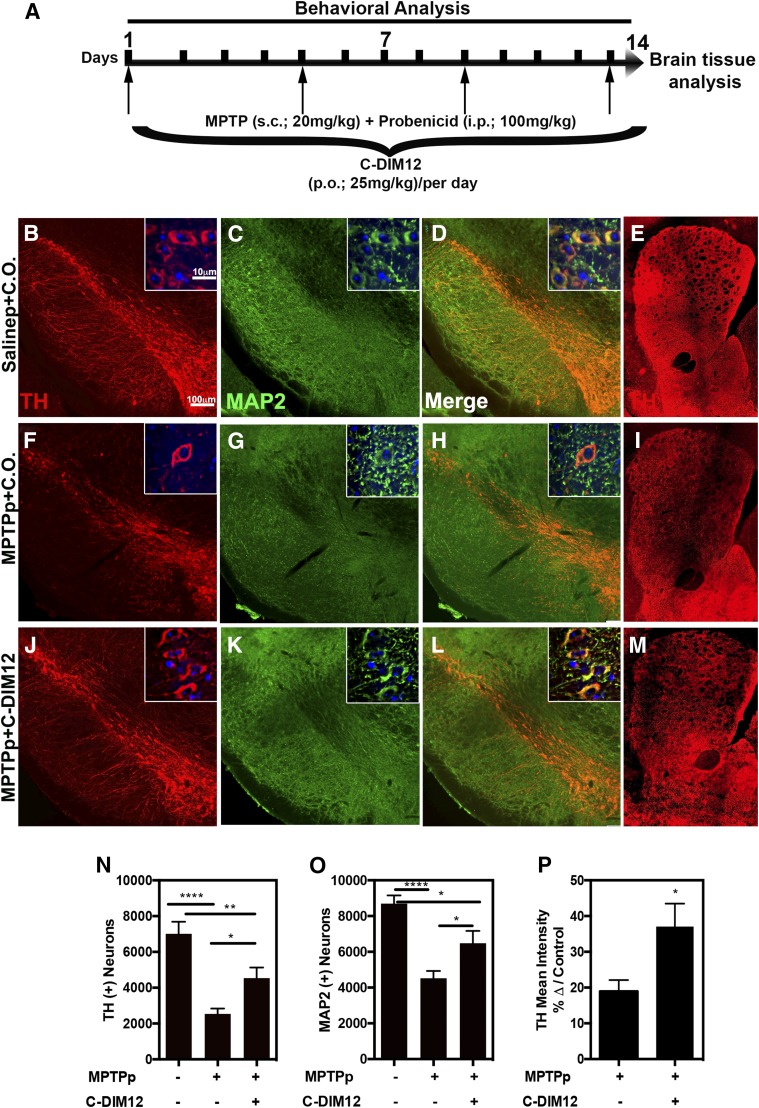
Brain tissue was collected after 14 days of concurrent dosing with MPTPp and the presence or absence of C-DIM12 (25 mg/kg), as depicted in the dosing schematic in Fig. 2A. IF images were generated for the number of TH+ cell bodies (red) in the SNpc on tissue from saline + C.O. (Fig. 2B), MPTPp + C.O. (Fig. 2F), and MPTPp + C-DIM12 (Fig. 2J) experimental groups, as depicted in representative images. The total number of neurons was determined by staining with the general neuronal marker MAP2 (green) (Fig. 2, C, G, and K). Based on unbiased 3D design–based stereological quantitation of the number of TH+ neurons in the SNpc, mice treated with MPTPp + C.O. had a 63.75% ± 4.27% loss of TH+ neurons, whereas mice treated with MPTPp + C-DIM12 had only a 35.28% ± 8.46% loss of TH+ neurons (Fig. 2N). Similarly, the loss of MAP2+ cells was 48.00% ± 4.74% in the MPTP + C.O. treatment group, in contrast to only a 25.50% ± 7.92% loss of total neurons in mice lesioned with MPTPp in the presence of C-DIM12 compared with control (Fig. 2O) (*P < 0.05, **P < 0.01, ****P < 0.0001; N = 10 animals/group). TH+ fiber innervations of the ST were also immunostained as depicted in representative images of Salinep + C.O. (Fig. 2E), MPTPp + C.O. (Fig. 2I), and MPTPp + C-DIM12 (Fig. 2M). The density of TH+ fibers was measured by the mean pixel intensity of the caudate putamen region and was compared to the saline control (set at 100%). Mice treated with MPTPp + C.O. had a 81.05% ± 1.05% loss of TH+ terminals in the ST compared with only a 62.92% ± 6.40% loss of TH+ fibers in mice treated with MPTPp + C-DIM12 (Fig. 2P) (*P < 0.05; n = 12 animals per group). Thus, treatment with C-DIM12 reduced the loss of TH+ fibers in the ST but did not completely protect against MPTPp-induced damage to innervating dopaminergic fibers projecting from the SNpc.
Fig. 2.
C-DIM12 protects DA neuron bodies/terminals from MPTP-induced degradation. (A) Concurrent dosing schematic with C-DIM12 and MPTPp treatments: 10× and 100× objective images of SN/ST tissue immunostained for TH (red) and MAP2 (green) treated with Salinep + C.O. (B–E), MPTPp + C.O. (F–I), and MPTPp + C-DIM12 (J–M). Stereological counts for TH+ (N) and MAP2+ (O) cell bodies of the SNpc demonstrate significantly more neurons in the MPTP + C-DIM12–treated group vs. the MPTP + C.O. group. (P) TH immunoreactivity intensity levels also measured significantly more in the C-DIM12–treated group (*P < 0.05; ** P < 0.01; ****P < 0.0001; N = 10–12 animals/group).
Assessment of Neurobehavioral Deficits Associated with PD and Neurochemical Analysis of the Nigrostriatal System.
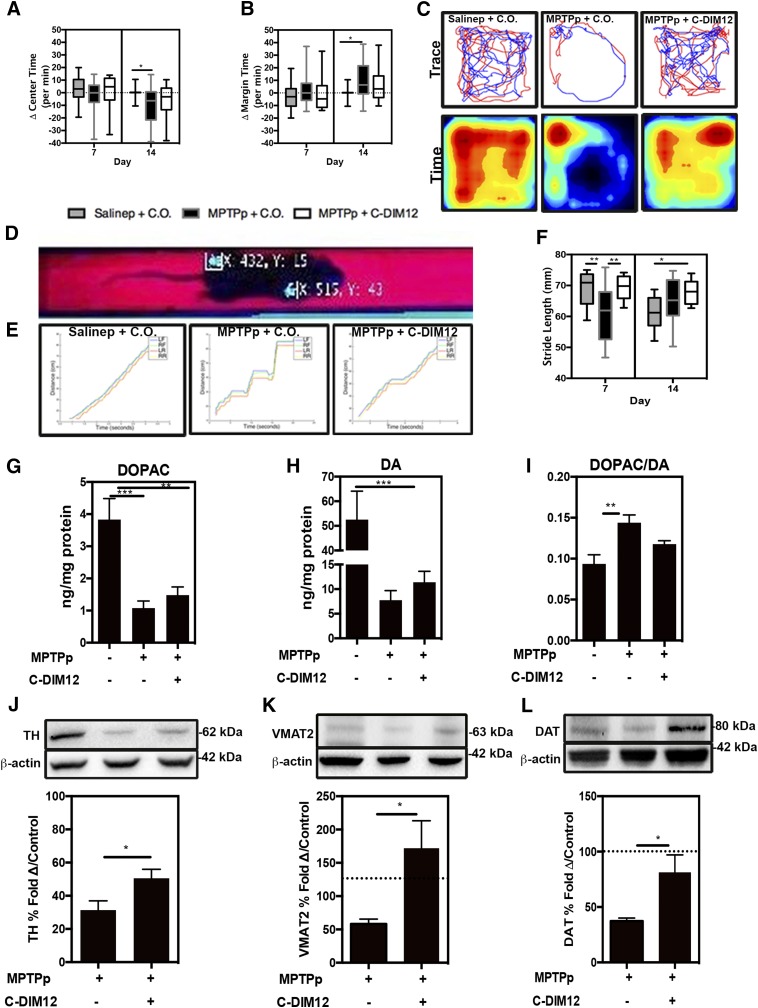
Neurobehavioral function was assessed by OFT on days 0, 7, and 14 during the 2-week study. Mice treated with MPTPp + C.O. spent significantly less time in the center of the chamber (Fig. 3A, dotted line represents baseline subtracted from day 0) compared with controls, whereas the MPTPp + C-DIM12 treatment group was not significantly different from control or the MPTPp + C.O. group at day 14 (Fig. 3A). The quantitation of time spent along the margins of the chamber (a measure of anxiety) indicated that MPTP + C.O.–treated mice spent significantly more time in the margin compared with saline-treated controls. Mice treated with MPTPp + C-DIM12 were not significantly different from controls or the MPTPp + C.O. group at day 14 (Fig. 3B) (*P < 0.05; N = 19–25 animals per group). Individual time traces and pseudocolored heat maps of activity across area over time (red, most time; blue, least time) plots from day 14 of OFT in each treatment group are represented in Fig. 3C, demonstrating the trend toward decreased center time and increased margin time in MPTPp-treated mice that was prevented by concurrent treatment with C-DIM12. To assess locomotor function, we used a real-time video analysis system to determine stride length along a fixed track to measure alterations in gait related to changes in striatal DA. Representative images of illuminated paw print coordinates generated from video analysis are depicted in Fig. 3D. The overall rate of movement along the trackway was analyzed independently for left front, right front, left rear, and right rear paws to identify changes in gait for each treatment group (Fig. 3E). An increased number of stop times and a correspondingly slower overall traverse rate along the trackway was noted in mice treated with MPTPp + C.O., which were largely restored to control levels at day 7 in mice treated with MPTPp + C-DIM12. Hind limb stride length analysis at day 7 also indicated a significantly shorter stride in the MPTPp + C.O. group and a longer stride length in the MPTPp + C-DIM12 group (*P < 0.05, **P < 0.01; N = 6–12 animals per group) (Fig. 3F). The MPTPp + C.O. and MPTPp + C-DIM12 groups were not significantly different at day 14, and the stride length of Salinep + C.O.–treated animals was decreased potentially due to either stress or habituation to the trackway by the conclusion of the treatment period. HPLC was performed on ST tissue for the measurement of DA neurotransmitter content and its metabolite DOPAC. Significantly lower DOPAC and DA levels were detected in MPTP + C.O.– and MPTP + C-DIM12–treated animals compared with saline-treated controls (Fig. 3, G and H). Mice treated with MPTPp + C-DIM12 showed a trend toward protection against the loss of DOPAC and DA compared with the MPTPp + C.O. group, but this was not statistically significant. The DOPAC/DA ratio was also higher in MPTPp + C.O.–treated animals compared with control mice, and the MPTPp + C-DIM12 group showed an intermediate effect that was not different from either control or the MPTPp + C.O. group (**P < 0.01, ***P < 0.001; N = 9 to 10 animals per group) (Fig. 3I). Proteins associated with the production and release of DA in ST tissue was also measured by Western blot. Levels of TH were significantly depleted in both MPTPp + C.O.– and MPTPp + C-DIM12–treated animals compared with saline-treated controls (set at 100%), but C-DIM12 treatment mitigated the loss of TH compared with the MPTP-only group (Fig. 3J). Correspondingly, the amount of VMAT2 (*P < 0.05) (Fig. 3K) and DAT (*P < 0.05) (Fig. 3L) was depleted in MPTPp + C.O.–treated groups and higher in C-DIM12–treated animals (N = 6–8 animals per group).
Fig. 3.
Neurobehavioral deficits associated with PD are ameliorated and DA proteins are preserved with C-DIM12 treatment. OFT was conducted on mice at days 0, 7, and 14 during the study. The change from day 0 per minute of OFT center time (A) and margin time (B) was quantitated at days 7 and 14. (C) Trace and pseudo-colored time plots represent OFT mouse movement at day 14. (D) Representative image shows a real-time video gait analysis system used to detect paw coordinates for quantitative measurement. (E) Gait images depict distance vs. time graphs in animals in each group at day 7. (F) Graph displays stride length measured at days 7 and 14. HPLC was used for the measurement of DA metabolite, DOPAC (G), DA (H), and the DOPAC/DA ratio (I) from each experimental group. Western blot analysis of striatal protein depicts levels of TH (J), VMAT2 (K), and DAT (L) (*P < 0.05; N = 10-12 animals/group for behavior, N = 6–8 animals/group for Western Blot).
C-DIM12 Suppresses Microglial Activation and Preserves a Ramified Morphologic Phenotype in the Substantia Nigra.
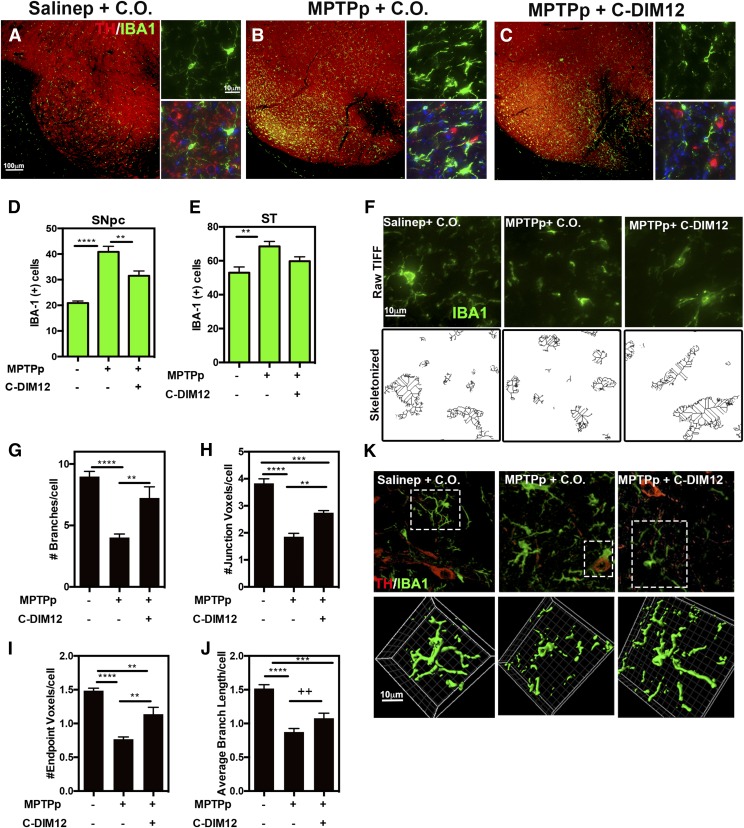
The relative number of microglia in the SN and ST were determined by immunostaining for IBA-1 (green), with counterstaining for TH (red) to demarcate each region, as depicted in the representative images in Fig. 4, A–C. An increased number of activated microglia compared with controls was evident in MPTPp-treated SNpc tissue (Fig. 4, B and C). To quantitate these observations, stereological assessment of IBA-1+ cells was performed on SNpc and ST. In the SNpc, an increased number of IBA-1+ cells was counted in both MPTP-receiving mice, characterized by intense staining for IBA-1 and a change in morphology from a ramified phenotype to a more amoeboid shape. C-DIM12 treatment significantly suppressed the number of IBA-1+ cells in the SNpc compared with MPTP + C.O. treatment; however, in the ST the number of IBA-1+ cells was not significantly lower than in the MPTPp + C.O. group but also was not significantly different from the control group (P = 0.1162 for MPTPp + C.O. vs. MPTPp + C-DIM12; **P < 0.01; ****P < 0.0001; N = 10 animals/group for SN and N = 12 animals/group for ST) (Fig. 4, D and E). For analysis of a microglia morphologic phenotype within the SNpc, the same 40× images used for IBA-1 counts were also rendered for skeletonization as depicted in the representative images in Fig. 4F. Skeletonized images were quantitated for the number of branches per cell (Fig. 4G), junction voxels per cell (Fig. 4H), endpoint voxels per cell (Fig. 4I), and the average branch length per cell (Fig. 4J). C-DIM12 increased the number and complexity of each morphologic parameter, except for the average branch length per cell, which showed only a trend toward an increase relative to the MPTPp-treated group (++P < 0.1), whereas all other parameters in the MPTPp + C-DIM12 group were increased compared with the control group (**P < 0.01,***P < 0.001 ****P < 0.0001, respectively; N = 6 animals/group). The morphology of IBA-1+ cells within the SNpc was also characterized by high-resolution confocal microscopy to visualize 3D morphology (Fig. 4K; Supplemental Videos 1–3). Multiple optical z-planes were acquired and rendered in 3D using Imaris software (Bitplane) to produce volumetric surface renderings of microglia from each treatment group. Microglia in the SNpc of MPTPp-treated mice displayed a loss of ramified cytoplasmic processes and a generally amoeboid shape, whereas mice treated with MPTPp + C-DIM12 had a cellular morphology more consistent with microglia from the control group. Amoeboid-shaped microglia were also seen phagocytosing TH+ neurons in the SNpc in the MPTPp + C.O. group (Supplemental Video 4).
Fig. 4.
C-DIM12 suppresses microglia proliferation and maintains a ramified morphologic phenotype; 10× and 100× objective representative images of TH (red) and microglia marker IBA-1 (green) in the SN are depicted for animals treated with Salinep + C.O. (A), MPTPp + C.O. (B), and MPTPp + C-DIM12 (C). Gliosis counts of the SNpc (D) and ST (E) tissue were conducted for IBA+ cells. To visualize microglia morphology changes of IBA+ cells in the SNpc (F), multiple 40× objective images were also transformed in ImageJ for skeletonization and quantitation of microglia branches (G), junctions (H), endpoints (I), and average branch lengths (J) (++P < 0.1; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; N = 12 animals/group for counts, n = 6 animals/group for skeletonization). (K) The 64× objective, 3D images of microglia in SNpc were surfaced rendered on the channel for IBA-1 in each experimental group (boxed cells from top images with IBA-1/TH were selected; see Supplemental Videos 1–4).
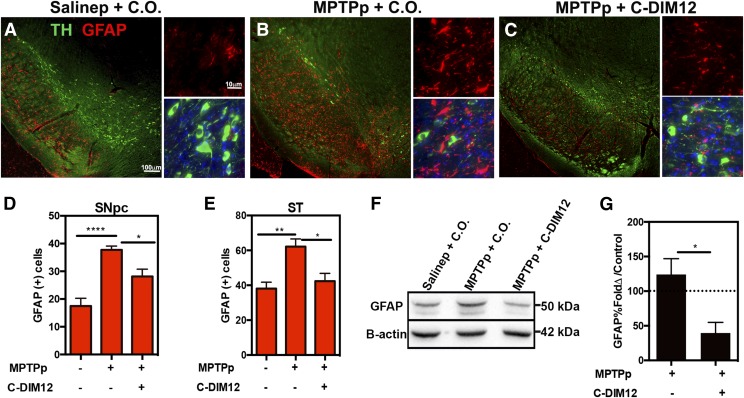
C-DIM12 Suppresses Astrocyte Activation during Progressive DA Neuronal Loss.
The relative activation of astrocytes after MPTPp treatment was measured by quantitation of the intermediate filament, GFAP, in cells in the SN and ST. Total levels of GFAP in the ST were also analyzed by Western immunoblotting. SN tissue was immunostained for GFAP (red) and TH (green) to delineate the pars compacta region for stereological counts, as depicted in representative high- and low-magnification images (Fig. 5, A–C). A basal level of GFAP+-expressing cells within the SN pars reticulate was evident in saline-treated control animals (Fig. 5A) and astrocyte proliferation noticeably increased within the SNpc upon treatment with MPTPp (Fig. 5B). Based on stereological counts of GFAP+-expressing cell bodies, MPTPp + C.O.–treated animals exhibited significantly more astrocytes within the SNpc compared with saline control. Mice that received MPTPp + C-DIM12 had significantly fewer GFAP+ cells within the SNpc compared with animals treated only with MPTP (Fig. 5, C and D). Similarly, C-DIM12 suppressed the number of GFAP+ cells within the ST, which was comparable to that in saline-treated controls (N = 9 animals/group for SN; N = 6 animals/per group for ST; *P < 0.05, **P < 0.01, ****P < 0.0001) (Fig. 5, D and E). GFAP expression levels within the ST were also confirmed by Western blot analysis, as depicted in Fig. 4F. Based on optical density measurements of GFAP protein, compared with control (saline set at 100%), C-DIM12 treatment significantly decreased the total levels of striatal GFAP (*P < 0.05; N = 4 animals/group) (Fig. 4G).
Fig. 5.
Astrocyte activation is suppressed by C-DIM12. Objective images, 10× and 100×, of SN tissue sections isolated from animals treated with Salinep + C.O. (A), MPTPp + C.O. (B), and MPTPp + C-DIM12 (C) depict TH+ neurons (green) and GFAP+ astrocytes (red). Stereological counts of GFAP+ cells in the SNpc (D) and ST (E) demonstrate significantly less astrocyte proliferation in the C-DIM12 group. Western blot of striatal protein has significantly less GFAP with C-DIM12 treatment (F) as quantitated in (G) (*P < 0.05; **P < 0.01; ****P < 0.0001; N = 6 animals/per group).
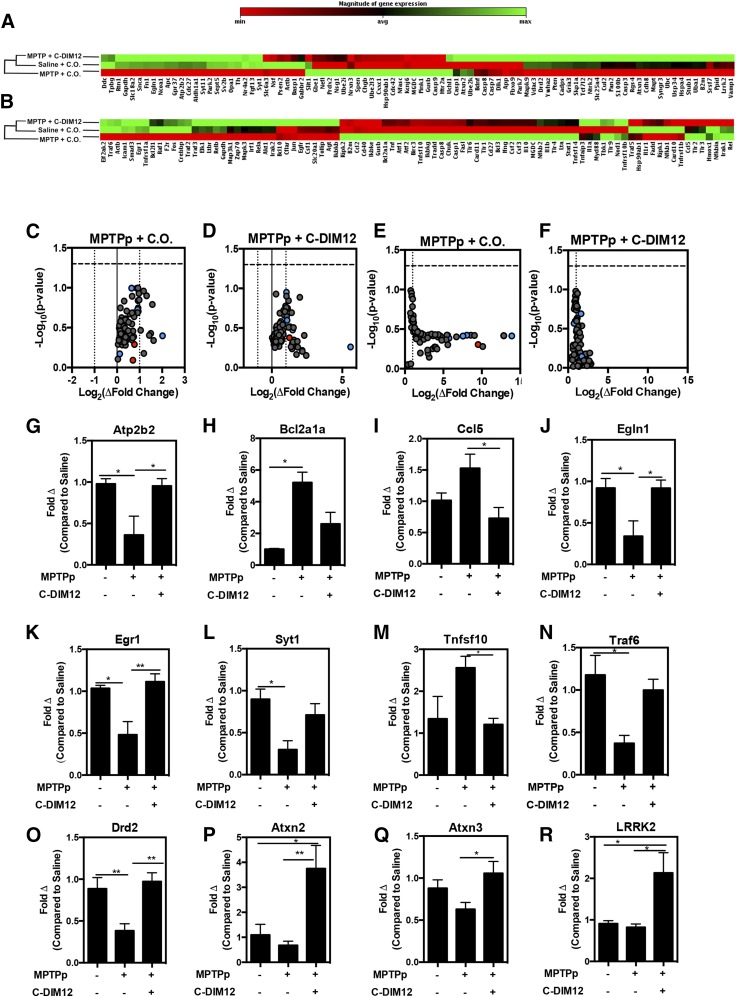
C-DIM12 Reverses MPTP-Induced Changes in Gene Expression Related to PD and NF-κB Signaling.
To analyze the patterns of gene expression in each experimental group, we isolated mRNA from midbrain tissue for real-time reverse-transcription PCR analysis of 168 genes in PD-associated and NF-κB–regulated gene arrays. Heat maps for PD-associated (Fig. 6A) and NF-κB (Fig. 6B) gene arrays depict ontology dendrograms indicating that transcript expression in the MPTPp + C-DIM12 group clustered with the saline (control) group, whereas mRNA levels in the MPTPp + C.O. group segregated independently, based on clustergram analysis. Volcano plots for NF-κB–regulated (Fig. 6, C and D) and PD-associated genes (Fig. 6, E and F) identified a number of genes upregulated in the MPTPp + C.O. group relative to the control group (Fig. 6, C and D) that were downregulated in the MPTPp + C-DIM12 group (Fig. 6, D and F). Several unique transcripts identified in volcano plots that were significantly altered by MPTPp but were not different from controls in the MPTPp + C-DIM12 group included the plasma membrane Ca2+-transporting ATPase (Atp2b2) (Fig. 6G), cell death regulator B cell leukemia/lymphoma 2 related protein Bcl2a1a (Fig. 6H), motif chemokine ligand Ccl5 (RANTES) (Fig. 6I), hypoxia-inducible factor prolyl hydroxylase 2 (Egln10) (Fig. 6J), neural plasticity transcription factor early growth factor 1 (Egr1) (Fig. 6K), synaptotagmin 1 (Syt1) (Fig. 6L), tumor necrosis factor (ligand) superfamily, member 10 (Tnfs10) (Fig. 6M), and tumor necrosis factor receptor–associated factor 6 (Traf6) (Fig. 6N) (*P < 0.05, **P < 0.01; N = 4 animals/group). Additional neuronal PD-associated genes, DrD2 (Fig. 6O), Atxn2 (Fig. 6P), Atxn3 (Fig. 6Q), and Lrrk2 (Fig. 6R), were also found to be induced by C-DIM12 (N = 9–10).
Fig. 6.
PD-associated/NF-κB–regulated gene expression is preserved in MPTPp + C-DIM12–treated mice. (A) Ontology dendrogram heat maps of 84 PD-associated genes from SNpc RNA depicts a cluster grouping with MPTP + C-DIM12–treated and saline-treated groups. Notice that NR4A2 (Nurr1) is lower in the MPTP + C.O.–treated group. (B) Heat map of 84 NF-κB–regulated genes analyzed from SNpc RNA. Map depicts a cluster grouping to saline control levels in MPTP + C-DIM12–treated tissue (red = increase; green = decrease). Volcano plots from PD (C and D) and NF-κB (E and F) arrays show fold changes for MPTP + C.O. and MPTPp + C-DIM12 groups compared with saline controls (red = minimum expression; green = maximum expression). Quantitative expression data for Atp2b2 (G) Bcl2a1a (H), Ccl5 (I), Egln1 (J), Egr1 (K), Syt1 (L) Tnfsf10 (M), and Traf6 (N) depict significant gene fold changes with experimental groups (*P < 0.05; **P < 0.01; N = 4 to 5 mice/group). Additional neuronal genes DrD2 (O), Atxn2 (P), Atxn3 (Q), and LRRK2 (R) were also examined for C-DIM12–induced fold changes (*P < 0.05; **P < 0.01; N = 9–10 mice/group).
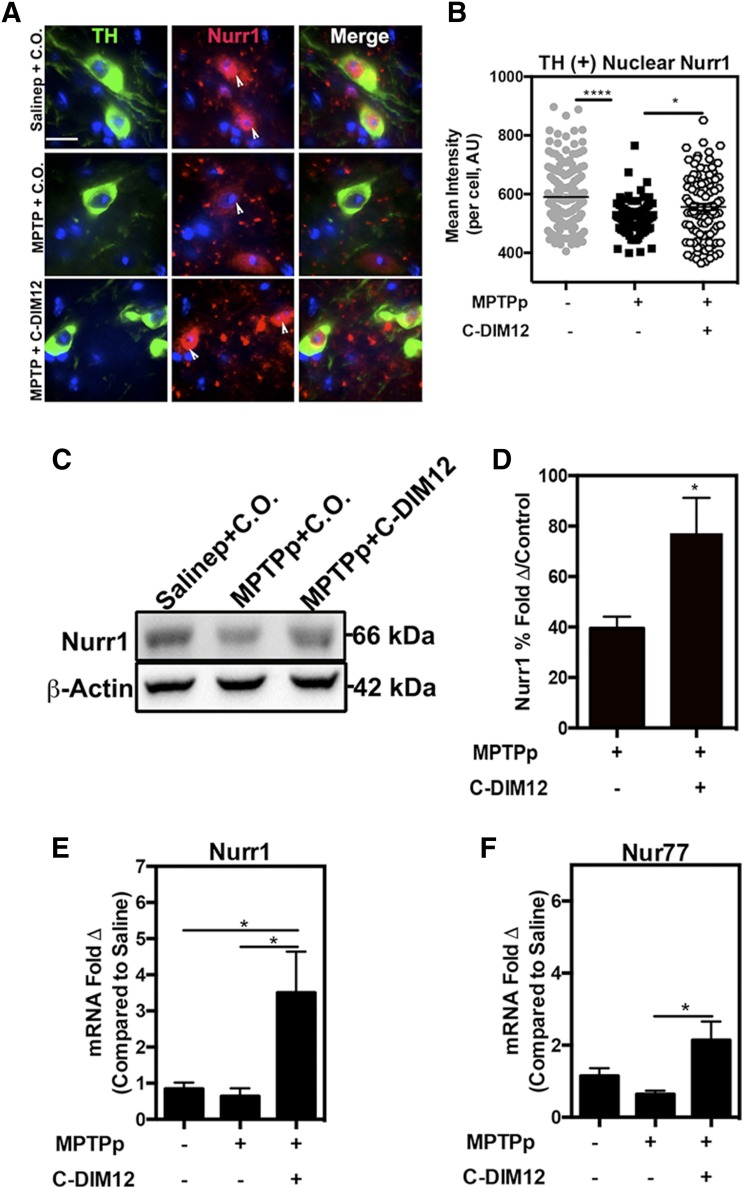
C-DIM12 Maintains Nuclear Nurr1 Localization in DA Neurons and Prevents MPTPp-Induced Nurr1 Protein Expression Changes in SNpc and ST.
To visualize the subcellular localization of NR4A2/Nurr1 shuttling in DA neurons after treatment with MPTPp, we immunostained SNpc tissue for TH (green) and Nurr1 (red) protein expression, as depicted in the representative high-magnification images in Fig. 7A (white arrows depict subcellular localization). Nurr1 in TH+ neurons was quantified for mean fluorescence intensity and indicated higher levels of Nurr1 in DA neurons within the SNpc in mice treated with MPTPp + C-DIM12 compared with DA neurons in mice treated with MPTPp + C.O. (*P < 0.05, ****P < 0.0001; N = 4 animals/per group) (Fig. 7B). Additionally, total Nurr1 protein from ST tissue was analyzed by Western blot and demonstrates a 39.5% ± 4.6% depletion of Nurr1 with MPTP + C.O. treatment that was prevented by treatment with C-DIM12 (to 77.2% ± 14.40% compared with control levels) (Fig. 7, C and D). qPCR analysis of mRNA isolated from SN tissue demonstrates increased expression of Nurr1 and NR4A family members and Nur77 mRNA after C-DIM12 treatment (*P < 0.05 compared with MPTPp + C.O.; N = 8 animals/group) (Fig. 7, E and F).
Fig. 7.
Subcellular localization and expression of Nurr1 are modified by C-DIM12 treatment in vivo. (A) The 100× objective images of TH+ neurons (green) show that Nurr1 (red) is sequestered to the nucleus with C-DIM12 treatment, and white arrows depict nuclear localization. (B) Mean intensity of TH+ nuclear Nurr1 is significantly higher in the C-DIM12 group compared with the MPTP + C.O. group (*P < 0.05; ****P < 0.0001; N = 4 animals/group). Western blot of Nurr1 protein isolated from ST tissue shows that C-DIM12 prevents MPTPp-induced protein changes (C), as illustrated in the quantitative measurement of mean optical density (D) (control set to 100%; *P < 0.05; N = 6–8 animals/group). qPCR data of mRNA isolated from SNpc for Nurr1 (E) and Nur77 (NR4A1) (F) expression show C-DIM12 induces higher levels of NR4A2 (*P = 0.05; N = 8 animals/group).
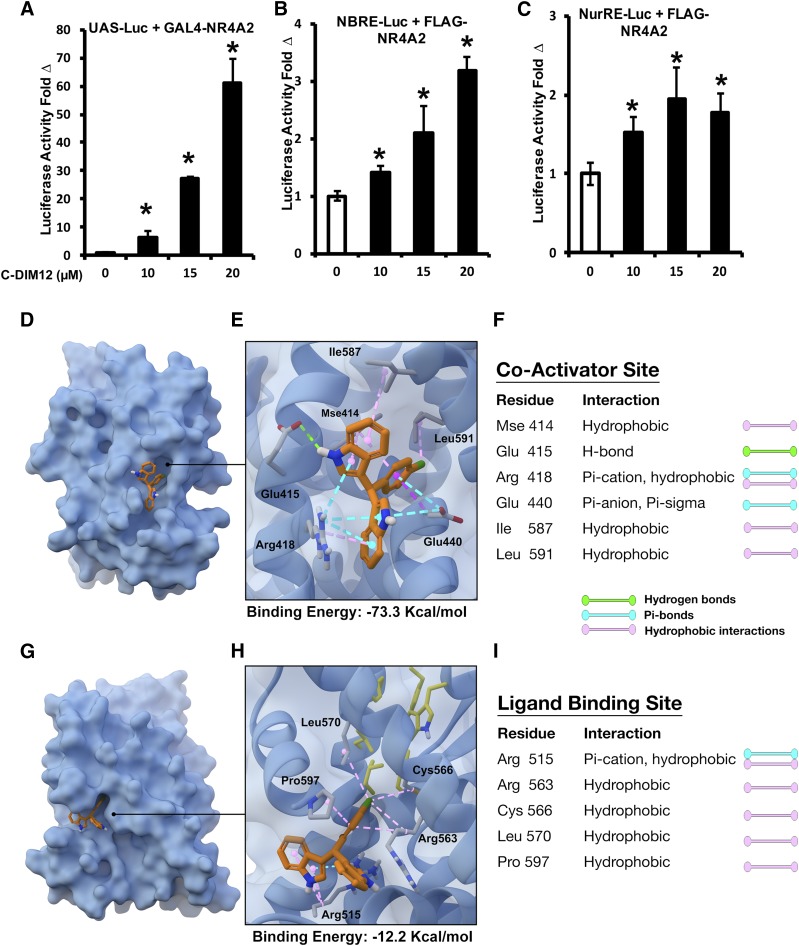
C-DIM12 Activates Nurr1-Dependent Transactivation and Modeling Interactions of C-DIM12 with Nurr1.
C-DIM12 also induced NR4A-dependent transactivation in PC12 cells transfected with a GAL4-NR4A2 chimera (full-length human NR4A2 fused to the yeast GAL4 DNA binding domain) and a Luc reporter gene construct containing five GAL4 response elements (UAS-Luc) (Fig. 8A). In addition, PC12 cells were transfected with NBRE-Luc and NurRE-Luc constructs containing binding sites for NR4A2 monomer and homodimer, respectively (Li et al., 2012) (Fig. 8, B and C). C-DIM12 activated transactivation in PC12 cells in all three assay systems, with the highest responses observed for the GAL4-NR4A2/UAS-Luc assay (Fig. 8A). To examine the potential for direct binding of C-DIM12 to Nurr1, we performed computation-based small-molecule docking studies to predict its possible binding orientation in either the coactivator binding site or the ligand binding site. The modeling results in Fig. 8, D–F indicated that C-DIM12 was predicted to bind with high affinity to the coactivator binding site (binding energy, −73.3 kcal/mol), with the chlorobenzene ring buried in the hydrophobic region of the pocket, which includes Mse414, Ile587, and Leu591, and the indole moieties participating in hydrogen bond interactions with Glu415, π-anion and π-σ interactions with Glu440 and π-cation interactions with Arg418. The only substantive predicted interaction at the ligand binding site involved a π-cation interaction between one of the indole moieties and Arg515 (binding energy, −12.2 kcal/mol), with the balance consisting of comparatively weak hydrophobic interactions with Arg563, Cys563, and Leu570 (Fig. 8, G–I).
Fig. 8.
C-DIM12–dependent transactivation of Nurr1 reporter constructs in neuronal cells and putative binding sites within the ligand binding domain of human NR4A2. Luc expression of PC12 cells cotransfected with UAS-Luc + GAL4-Nurr1 (A), NBRE-Luc + FLAG-Nurr1 (B), and NurRE-Luc + FLAG-Nurr1 (C) in the presence of 0–20 µM C-DIM12 (*P < 0.05; N = 3/experiment). In silico modeling of the Nurr1 ligand binding domain displays putative binding sites for C-DIM12 at the coactivator interface with a calculated binding energy of −73.3 kcal/mol (D–F) and at the ligand binding pocket with a calculated binding energy of −12.2 kcal/mol (G–I).
Discussion
Efficacy of small-molecule therapeutics for neurodegenerative disease is dependent on the capacity to penetrate the blood-brain barrier. We previously demonstrated that selected C-DIM compounds have excellent structure-dependent bioavailability, with C-DIM12 having the greatest area under the curve (AUC) in the brain compared with other p-phenyl–substituted analogs when administered orally with a 50 mg/kg dose daily (De Miranda et al., 2013). To assess brain/plasma distribution at the lower dose of 25 mg/kg used in the current study, we administered C-DIM12 to mice orally over a 24-hour period and observed an ∼3.5 times greater Cmax at 4 hours compared with our previous pharmacokinetic study that examined plasma and brain distributions at 10 mg/kg. At a dose of 25 mg/kg by mouth, the AUC in the brain was ∼4.2 times higher in the brain than in plasma, representing a brain/plasma AUC ratio indicative of highly favorable penetrance of the central nervous system, as indicated by the data in Fig. 1. This dose-dependent increase in brain levels of C-DIM12 confirms the utility of this analog for reaching the molecular target in the brain as a pharmacological modulator of neuroinflammation.
At the time of diagnosis, an individual with PD has already lost approximately 60% of dopaminergic neurons in the SNpc and 70% of striatal DA (Marsden, 1982; Cooper et al., 2009). The subacute MPTPp mouse model used in this study conferred similar lesioning with an approximately 63% loss of SNpc TH+ neurons, a 52% loss of SNpc MAP2+ neurons, and an 81% loss of DA terminals in the ST (Fig. 2, N–P). Based on stereological analysis of neuronal numbers after 2 weeks of exposure to MPTPp, concurrent treatment with C-DIM12 ameliorated neuronal loss to only 35% and 63%, respectively, for DA cell bodies and terminals compared with controls. However, C-DIM12 provided less protection against the loss of striatal DA, which is consistent with the use of MPTP as a potent toxicant damaging neuronal mitochondria in nerve terminals in projecting dopaminergic fibers in the ST (Giovanni et al., 1994). The protective effect of C-DIM12 was consistently observed in MAP2+ neurons in the SNpc in addition to TH+ cells, demonstrating that neuroprotection was not confined to dopaminergic neurons only, but rather to a general protective mechanism consistent with the anti-inflammatory activity of C-DIM12.
Subacute administration of MPTPp in C57BL/6 mice causes cognitive and locomotor dysfunction resembling that seen in PD (Goldberg et al., 2011; Wang et al., 2012). Generally, anxiety-like thigmotaxis behavior of mice is accompanied by reduced exploratory and spontaneous activity (Simon et al., 1994; George et al., 2008), consistent with the trends we observed in open field activity assays (Fig. 3, A–C). We also noted decreased hind limb stride length in MPTPp-treated mice that was prevented by cotreatment with C-DIM12 (Fig. 4, D and F), indicating protection against deprecations in striatal DA. However, this effect was not detected at day 14 (Fig. 4 D–F), possibly due to the characteristically severe loss of DA within the nigrostriatal system caused by MPTPp treatment that was partially prevented by C-DIM12 (Fig. 3, G–I). Yet, C-DIM12 preserved protein expression of TH within the ST, as well as the synaptic and vesicular DATs DAT and VMAT2, respectively (Fig. 3, J–L). The transporter proteins were induced at higher levels upon C-DIM12 treatment compared with TH, suggesting an imbalance of DA transport within the ST or due to vulnerability of TH to MPP+-induced oxidation.
Animal studies suggest that microglial activation could be an etiologic factor in pathogenesis as well as a sequela of neurodegeneration due to MPTP exposure (McGeer and McGeer, 2008; Ramsey and Tansey, 2014). Irrespective of the events, microglial activation exacerbates DA neuronal loss; therefore, suppressing inflammatory activation of microglia could be a viable therapeutic strategy for slowing the progression of PD. Previous studies with the Nurr1 agonist SA00025 showed that the suppression of microglia activation and neuroinflammatory cytokine production was neuroprotective in the 6-hydroxydopamine rat model of PD (Smith et al., 2015). Similarly, we demonstrated that C-DIM12 blocked neuroinflammatory gene expression in LPS-treated BV-2 microglial cells (De Miranda et al., 2015) and suppressed the relative number of IBA-1+ cells in the SNpc and ST after treatment with MPTPp in vivo (De Miranda et al., 2014). In the current study, microglial numbers were significantly reduced within the SNpc, but only slightly reduced in the ST, by concurrent treatment with C-DIM12 (Fig. 4, A–E). Microglia within the SNpc were further examined to discriminate ramified, resting morphology from an amoeboid activated–like cell type using 3D IBA+ morphometric analysis (Fig. 4, F–K; Supplemental Videos 1–3). Based on the morphometry of skeletonized images, MPTPp + C.O. treatment reduced ramification and increased the number of phagocytic microglia surrounding DA neurons (Fig. 4K; Supplementary Video 4). C-DIM12 preserved a ramified morphology in SNpc microglia (Fig. 4, G–K), indicating that the inhibition of inflammatory activation directly correlated with the preservation of dopaminergic soma.
Increased astrocyte proliferation and hypertrophy surrounding DA neurons occurs at approximately 3–4 days after MPTP exposure in mice (Breidert et al., 2002; Hirsch and Hunot, 2010). Although a certain level of astrogliosis may be neurotropic (Liddelow et al., 2017), severe activation leads to neuronal death (Carbone et al., 2009). In astrocyte cultures, by neurotoxin-induced nitric oxide synthase 2, C-DIM compounds are potently anti-inflammatory and neuroprotective (Tjalkens et al., 2008; Carbone et al., 2009). Based on the gliosis quantitation of GFAP+ cells in both the SNpc and ST, concurrent C-DIM12 treatment suppressed gliosis in both regions (Fig. 5, A–E). Interestingly, there was a greater reduction of striatal GFAP+ astrocytes than of striatal IBA-1+ microglia (Fig. 4E; Fig. 5, E–G). This cell-specific effect of C-DIM12 could be due to an established deficit of striatal IBA-1+ cells by the end of the MPTPp treatment regimen or to astrocyte activation persisting as a more chronically activated state in PD animal models (Hirsch and Hunot, 2009). Overall, C-DIM12 appears to modulate both glial cell types in MPTPp-induced models, suggesting the regulatory role of inflammatory gene expression of C-DIM12 to be a promising neuroprotective mechanism.
Neuroinflammatory gene expression mediated by NF-κB is upregulated in PD postmortem brain tissue (Hirsch and Hunot, 2009). It is also essential for glial cross-talk signaling and has been demonstrated to be affected by multiple Nurr1 agonists in in vivo models (De Miranda et al., 2014; Smith et al., 2015; Kirkley et al., 2017). Based on heat map analysis of 168 genes from both reverse-transcription qPCR arrays, C-DIM12 gene pattern expression was clustered similarly to saline treatment control levels compared with MPTP + C.O. treatment levels (Fig. 6, A and B). NF-κB–related genes Traf-6, Tnfsf10, CCl5, Bcl2a1a, and Egr1 were found to be dysregulated by MPTP and preserved in expression with C-DIM12. Genes differentially regulated by C-DIM12 under the PD array were calcium homeostasis genes Atp2B2 and Syt1, as well as the redox sensing transcription factor Egnl1 (Fig. 6, G–N). C-DIM12 also induced neuronal genes DrD2, Atxn2, Atxn3, and, interestingly, Lrrk2 (Fig. 6R). The autosomal-dominant LRRK2 mutation is the most common PD-causing genetic variant, perhaps due to normal LRRK2 function being necessary for synaptogenesis and DA synaptic function (Parisiadou et al., 2014; Hui et al., 2018). These data indicate that C-DIM12 transcriptional activation may modulate death signaling regulated by NF-κB–dependent gene expression and DA synaptic health, although whether these effects are from neurons, glia, or both remains to be validated in future studies.
Previous studies show that C-DIM12 induced NR4A2-dependent transactivation in pancreatic and bladder cancer cells (Inamoto et al., 2008; Li et al., 2012) and that this response was consistently observed in neuronal-like PC12 cells (Fig. 8, A–C). The results of these transactivation studies demonstrate that C-DIM12 directly activates a Gal4-NR4A2 chimera, indicating that ligand-induced transactivation is NR4A2 dependent. This is confirmed in transactivation assays where ligand-induced transactivation is not only NR4A2 dependent but also is dependent on their interactions with cis-acting promoter elements that contain binding sites for an NR4A2 monomer (NBRE) or dimer Nurr1 response element (NuRE). These results support the modeling data, and future studies will use mutational analysis and investigate the key amino acid residues required for ligand-mediated activation of NR4A2. C-DIM analogs have strong binding affinity to the ligand binding pockets of the closely related NR4A family member Nur77 (Lee et al., 2014). X-ray crystal studies of the Nurr1 structure revealed that the classic ligand binding pocket is blocked by several bulky hydrophobic amino acid residues, signifying that an alternative coactivator domain may allow ligand binding for transcriptional activation (Wang et al., 2003; Volakakis et al., 2006). Our computational modeling results predicted substantial greater binding affinity for C-DIM12 at the coactivator site (−73.3 kcal/mol) than to the ligand binding site (−12.2 kcal/mol) (Fig. 8, E and H). Therefore, C-DIM12 interactions at the coactivator binding site likely modulates transcriptional activity of Nurr1.
Nuclear import and export sequences also mediate Nurr1 shuttling from the nucleus to the cytosol during oxidative stress (García-Yagüe et al., 2013). We consistently demonstrated that C-DIM12 maintains nuclear localization of Nurr1 in DA neurons, which was lost in DA neurons of the SNpc after lesioning with MPTPp in the absence of C-DIM12 (Fig. 7, A and B). We previously reported similar effects on nuclear localization of Nurr1 in both DA neurons in vivo (De Miranda et al., 2014) and in primary microglia (De Miranda et al., 2015), suggesting a common mechanism of interaction with this receptor in multiple neural cell types. Total levels of Nurr1 protein from ST were depleted by approximately 60% with MPTP + C.O. treatment and significantly preserved to ∼80% of control levels in C-DIM12–treated animals (Fig. 7, C and D). Last, qPCR analysis of Nurr1 mRNA expression from total SN RNA demonstrated that C-DIM12 induced Nurr1 3.5-fold more than both control and MPTP + C.O. (Fig. 7E). There was also a >2-fold induction of NR4A/Nur77 (Fig. 7F), which is consistent to previous studies that show C-DIM12 binding to Nur77, albeit to a lesser extent than C-DIM12 binding to Nurr1 (Lee et al., 2014).
In conclusion, we demonstrated that C-DIM12 crosses the blood-brain barrier, suppresses glial activation, protects against DA neuronal cell body loss, preserves DA terminals, improves neurobehavioral function, and decreases NF-κB–regulated neuroinflammatory and neuronal gene expression in the MPTPp mouse model of PD by modulating the transcriptional activity of Nurr1. Given these findings, we conclude that C-DIM12 is a functional modulator of Nurr1 with distinct effects on neurons and glia that could represent a disease-modifying treatment strategy for PD.
Acknowledgments
The authors thank Dr. Gary Miller from Emory University for the gift and use of the anti-VMAT2 polyclonal antibody. The authors also thank Dr. Kelly Kirkley and Dr. Briana De Miranda for expert advice in the field, and Collin Bantle for the technical assistance.
Abbreviations
- AUC
area under the curve
- C-DIM
phenyl-substituted 3,3′-diindolylmethane
- C-DIM12
1,1-bis(3′-indolyl)-1-(p-chlorophenyl)methane
- C.O.
corn oil
- DA
dopamine
- DAT
dopamine transporter
- DOPAC
3,4-dihydroxyphenylacetic acid
- GFAP
glial fibrillary acidic protein
- HPLC
high-performance liquid chromatography
- IBA-1
ionized calcium-binding adapter molecule 1
- IF
immunofluorescence
- LC-MS
liquid chromatography-mass spectrometry
- Luc
luciferase
- MAP2
mitogen-associated protein 2
- MPTP
1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine
- MPTPp
1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine + probenecid
- NBRE
nerve growth factor binding recognition element
- NF-κB
nuclear factor-κB
- NR4A1
nuclear receptor-4A1
- NR4A2
nuclear receptor-4A2
- Nur77
orphan nuclear receptor related 77
- Nurr1
orphan nuclear receptor related 1
- OFT
open-field activity testing
- PBS
phosphate-buffered saline
- PD
Parkinson’s disease
- qPCR
quantitative polymerase chain reaction
- SNpc
substantia nigra pars compacta
- ST
striatum
- TH
tyrosine hydroxylase
- 3D
three-dimensional
- VMAT2
vesticular monoamine transporter 2
Authorship Contributions
Participated in research design: Hammond, Popichak, Tjalkens.
Conducted experiments: Hammond, Popichak, Li, Hunt, Richman.
Contributed new reagents or analytic tools: Damale, Chong, Backos.
Performed data analysis: Hammond, Popichak, Li, Backos.
Wrote or contributed to the writing of the manuscript: Hammond, Safe, Tjalkens.
Footnotes
This work was supported by National Institutes of Health Grants ES021656 (R.B.T.), NS096841 (S.L.H.), and ES025713 (S.S.); as well as a grant from the Consolidated Anti-Aging Foundation (R.B.T.). No potential conflicts of interest relevant to this article are reported.
 This article has supplemental material available at jpet.aspetjournals.org.
This article has supplemental material available at jpet.aspetjournals.org.
References
- Böhm HJ. (1994) On the use of LUDI to search the fine chemicals directory for ligands of proteins of known three-dimensional structure. J Comput Aided Mol Des 8:623–632. [DOI] [PubMed] [Google Scholar]
- Breidert T, Callebert J, Heneka MT, Landreth G, Launay JM, Hirsch EC. (2002) Protective action of the peroxisome proliferator-activated receptor-gamma agonist pioglitazone in a mouse model of Parkinson’s disease. J Neurochem 82:615–624. [DOI] [PubMed] [Google Scholar]
- Brooks BR, Brooks CL, III, Mackerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, et al. (2009) CHARMM: the biomolecular simulation program. J Comput Chem 30:1545–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone DL, Popichak KA, Moreno JA, Safe S, Tjalkens RB. (2009) Suppression of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-induced nitric-oxide synthase 2 expression in astrocytes by a novel diindolylmethane analog protects striatal neurons against apoptosis. Mol Pharmacol 75:35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper O, Astradsson A, Hallett P, Robertson H, Mendez I, Isacson O. (2009) Lack of functional relevance of isolated cell damage in transplants of Parkinson’s disease patients. J Neurol 256 (Suppl 3):310–316. [DOI] [PubMed] [Google Scholar]
- De Miranda BR, Miller JA, Hansen RJ, Lunghofer PJ, Safe S, Gustafson DL, Colagiovanni D, Tjalkens RB. (2013) Neuroprotective efficacy and pharmacokinetic behavior of novel anti-inflammatory para-phenyl substituted diindolylmethanes in a mouse model of Parkinson’s disease. J Pharmacol Exp Ther 345:125–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Miranda BR, Popichak KA, Hammond SL, Jorgensen BA, Phillips AT, Safe S, Tjalkens RB. (2015) The Nurr1 activator 1,1-Bis(3′-Indolyl)-1-(p-Chlorophenyl)methane blocks inflammatory gene expression in BV-2 microglial cells by inhibiting nuclear factor κB. Mol Pharmacol 87:1021–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Miranda BR, Popichak KA, Hammond SL, Miller JA, Safe S, Tjalkens RB. (2014) Novel para-phenyl substituted diindolylmethanes protect against MPTP neurotoxicity and suppress glial activation in a mouse model of Parkinson’s disease. Toxicol Sci 143:360–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig M, Onufriev A, Lee MS, Im W, Case DA, Brooks CL., III (2004) Performance comparison of generalized born and Poisson methods in the calculation of electrostatic solvation energies for protein structures. J Comput Chem 25:265–284. [DOI] [PubMed] [Google Scholar]
- García-Yagüe ÁJ, Rada P, Rojo AI, Lastres-Becker I, Cuadrado A. (2013) Nuclear import and export signals control the subcellular localization of Nurr1 protein in response to oxidative stress. J Biol Chem 288:5506–5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George S, van den Buuse M, San Mok S, Masters CL, Li Q-X, Culvenor JG. (2008) Alpha-synuclein transgenic mice exhibit reduced anxiety-like behaviour. Exp Neurol 210:788–792. [DOI] [PubMed] [Google Scholar]
- Giovanni A, Sieber BA, Heikkila RE, Sonsalla PK. (1994) Studies on species sensitivity to the dopaminergic neurotoxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine. Part 1: systemic administration. J Pharmacol Exp Ther 270:1000–1007. [PubMed] [Google Scholar]
- Glass CK, Saijo K, Winner B, Marchetto MC, Gage FH. (2010) Mechanisms underlying inflammation in neurodegeneration. Cell 140:918–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg NRS, Hampton T, McCue S, Kale A, Meshul CK. (2011) Profiling changes in gait dynamics resulting from progressive 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-induced nigrostriatal lesioning. J Neurosci Res 89:1698–1706. [DOI] [PubMed] [Google Scholar]
- Hammond SL, Safe S, Tjalkens RB. (2015) A novel synthetic activator of Nurr1 induces dopaminergic gene expression and protects against 6-hydroxydopamine neurotoxicity in vitro. Neurosci Lett 607:83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch EC, Hunot S. (2009) Neuroinflammation in Parkinson’s disease: a target for neuroprotection? Lancet Neurol 8:382–397. [DOI] [PubMed] [Google Scholar]
- Hirsch EC, Hunot S. (2010) Pathophysiological involvement of neuroinflammation in various neurological disorders. J Neural Transm (Vienna) 117:897–898. [DOI] [PubMed] [Google Scholar]
- Hui KY, Fernandez-Hernandez H, Hu J, Schaffner A, Pankratz N, NY Hsu, Chuang LS, Carmi S, Villaverde N, Li X, et al. 2018. Functional variants in the LRRK2 gene confer shared effects on risk for Crohn‘s disease and Parkinson’s disease. Sci Transl Med 10:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamoto T, Papineni S, Chintharlapalli S, Cho SD, Safe S, Kamat AM. (2008) 1,1-Bis(3′-indolyl)-1-(p-chlorophenyl)methane activates the orphan nuclear receptor Nurr1 and inhibits bladder cancer growth. Mol Cancer Ther 7:3825–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain AN. (1996) Scoring noncovalent protein-ligand interactions: a continuous differentiable function tuned to compute binding affinities. J Comput Aided Mol Des 10:427–440. [DOI] [PubMed] [Google Scholar]
- Jankovic J, Chen S, Le WD. (2005) The role of Nurr1 in the development of dopaminergic neurons and Parkinson’s disease. Prog Neurobiol 77:128–138. [DOI] [PubMed] [Google Scholar]
- Kadkhodaei B, Alvarsson A, Schintu N, Ramsköld D, Volakakis N, Joodmardi E, Yoshitake T, Kehr J, Decressac M, Björklund A, et al. (2013) Transcription factor Nurr1 maintains fiber integrity and nuclear-encoded mitochondrial gene expression in dopamine neurons. Proc Natl Acad Sci U S A 110:2360–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkley KS, Popichak KA, Afzali MF, Legare ME, Tjalkens RB. (2017) Microglia amplify inflammatory activation of astrocytes in manganese neurotoxicity. J Neuroinflammation 14:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koska J, Spassov VZ, Maynard AJ, Yan L, Austin N, Flook PK, Venkatachalam CM. (2008) Fully automated molecular mechanics based induced fit protein-ligand docking method. J Chem Inf Model 48:1965–1973. [DOI] [PubMed] [Google Scholar]
- Le W, Conneely OM, He Y, Jankovic J, Appel SH. (1999) Reduced Nurr1 expression increases the vulnerability of mesencephalic dopamine neurons to MPTP-induced injury. J Neurochem 73:2218–2221. [PubMed] [Google Scholar]
- Lee S-O, Li X, Hedrick E, Jin U-H, Tjalkens RB, Backos DS, Li L, Zhang Y, Wu Q, Safe S. (2014) Diindolylmethane analogs bind NR4A1 and are NR4A1 antagonists in colon cancer cells. Mol Endocrinol 28:1729–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Lee S-O, Safe S. (2012) Structure-dependent activation of NR4A2 (Nurr1) by 1,1-bis(3′-indolyl)-1-(aromatic)methane analogs in pancreatic cancer cells. Biochem Pharmacol 83:1445–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddelow SA, Guttenplan KA, Clarke LE, Bennett FC, Bohlen CJ, Schirmer L, Bennett ML, Münch AE, Chung W-S, Peterson TC, et al. (2017) Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541:481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden CD. (1982) [Functions of the basal ganglia]. Rinsho Shinkeigaku 22:1093–1094. [PubMed] [Google Scholar]
- McGeer PL, McGeer EG. (2008) Glial reactions in Parkinson’s disease. Mov Disord 23:474–483. [DOI] [PubMed] [Google Scholar]
- Miller JA, Trout BR, Sullivan KA, Bialecki RA, Roberts RA, Tjalkens RB. (2011) Low-dose 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine causes inflammatory activation of astrocytes in nuclear factor-κB reporter mice prior to loss of dopaminergic neurons. J Neurosci Res 89:406–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montarolo F, Perga S, Martire S, Navone DN, Marchet A, Leotta D, Bertolotto A. (2016) Altered NR4A subfamily gene expression level in peripheral blood of Parkinson’s and Alzheimer’s disease patients. Neurotox Res 30:338–344. [DOI] [PubMed] [Google Scholar]
- Morrison HW, Filosa JA. (2013) A quantitative spatiotemporal analysis of microglia morphology during ischemic stroke and reperfusion. J Neuroinflammation 10:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordentlich P, Yan Y, Zhou S, Heyman RA. (2003) Identification of the antineoplastic agent 6-mercaptopurine as an activator of the orphan nuclear hormone receptor Nurr1. J Biol Chem 278:24791–24799. [DOI] [PubMed] [Google Scholar]
- Parisiadou L, Yu J, Sgobio C, Xie C, Liu G, Sun L, Gu XL, Lin X, Crowley NA, Lovinger DM, et al. (2014) LRRK2 regulates synaptogenesis and dopamine receptor activation through modulation of PKA activity. Nat Neurosci 17:367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrill AL, Rami Reddy M. (1999) Rational Drug Design: Novel Methodology and Practical Applications, American Chemical Society, Washington, DC. [Google Scholar]
- Ramsey CP, Tansey MG. (2014) A survey from 2012 of evidence for the role of neuroinflammation in neurotoxin animal models of Parkinson’s disease and potential molecular targets. Exp Neurol 256:126–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safe S, Jin UH, Morpurgo B, Abudayyeh A, Singh M, Tjalkens RB. (2016) Nuclear receptor 4A (NR4A) family - orphans no more. J Steroid Biochem Mol Biol 157:48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safe S, Papineni S, Chintharlapalli S. (2008) Cancer chemotherapy with indole-3-carbinol, bis(3′-indolyl)methane and synthetic analogs. Cancer Lett 269:326–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo K, Winner B, Carson CT, Collier JG, Boyer L, Rosenfeld MG, Gage FH, Glass CK. (2009) A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell 137:47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurada K, Ohshima-Sakurada M, Palmer TD, Gage FH. (1999) Nurr1, an orphan nuclear receptor, is a transcriptional activator of endogenous tyrosine hydroxylase in neural progenitor cells derived from the adult brain. Development 126:4017–4026. [DOI] [PubMed] [Google Scholar]
- Saucedo-Cardenas O, Quintana-Hau JD, Le WD, Smidt MP, Cox JJ, De Mayo F, Burbach JP, Conneely OM. (1998) Nurr1 is essential for the induction of the dopaminergic phenotype and the survival of ventral mesencephalic late dopaminergic precursor neurons. Proc Natl Acad Sci U S A 95:4013–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon P, Dupuis R, Costentin J. (1994) Thigmotaxis as an index of anxiety in mice. Influence of dopaminergic transmissions. Behav Brain Res 61:59–64. [DOI] [PubMed] [Google Scholar]
- Smith GA, Rocha EM, Rooney T, Barneoud P, McLean JR, Beagan J, Osborn T, Coimbra M, Luo Y, Hallett PJ, et al. (2015) A Nurr1 agonist causes neuroprotection in a Parkinson’s disease lesion model primed with the toll-like receptor 3 dsRNA inflammatory stimulant poly(I:C). PLoS One 10:e0121072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits SM, Ponnio T, Conneely OM, Burbach JPH, Smidt MP. (2003) Involvement of Nurr1 in specifying the neurotransmitter identity of ventral midbrain dopaminergic neurons. Eur J Neurosci 18:1731–1738. [DOI] [PubMed] [Google Scholar]
- Tjalkens RB, Liu X, Mohl B, Wright T, Moreno JA, Carbone DL, Safe S. (2008) The peroxisome proliferator-activated receptor-gamma agonist 1,1-bis(3′-indolyl)-1-(p-trifluoromethylphenyl)methane suppresses manganese-induced production of nitric oxide in astrocytes and inhibits apoptosis in cocultured PC12 cells. J Neurosci Res 86:618–629. [DOI] [PubMed] [Google Scholar]
- Volakakis N, Malewicz M, Kadkhodai B, Perlmann T, Benoit G. (2006) Characterization of the Nurr1 ligand-binding domain co-activator interaction surface. J Mol Endocrinol 37:317–326. [DOI] [PubMed] [Google Scholar]
- Wang XH, Lu G, Hu X, Tsang KS, Kwong WH, Wu FX, Meng HW, Jiang S, Liu SW, Ng HK, et al. (2012) Quantitative assessment of gait and neurochemical correlation in a classical murine model of Parkinson’s disease. BMC Neurosci 13:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Benoit G, Liu J, Prasad S, Aarnisalo P, Liu X, Xu H, Walker NPC, Perlmann T. (2003) Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature 423:555–560. [DOI] [PubMed] [Google Scholar]
- Wu DC, Teismann P, Tieu K, Vila M, Jackson-Lewis V, Ischiropoulos H, Przedborski S. (2003a) NADPH oxidase mediates oxidative stress in the 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine model of Parkinson’s disease. Proc Natl Acad Sci U S A 100:6145–6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Robertson DH, Brooks CL, III, Vieth M. (2003b) Detailed analysis of grid-based molecular docking: a case study of CDOCKER-A CHARMm-based MD docking algorithm. J Comput Chem 24:1549–1562. [DOI] [PubMed] [Google Scholar]
- Zetterström RH, Williams R, Perlmann T, Olson L. (1996) Cellular expression of the immediate early transcription factors Nurr1 and NGFI-B suggests a gene regulatory role in several brain regions including the nigrostriatal dopamine system. Brain Res Mol Brain Res 41:111–120. [DOI] [PubMed] [Google Scholar]