 By establishing a chemical labeling method in combination with liquid chromatography-mass spectrometry analysis, we reported the widespread existence of various modified nucleoside triphosphates in eukaryotes.
By establishing a chemical labeling method in combination with liquid chromatography-mass spectrometry analysis, we reported the widespread existence of various modified nucleoside triphosphates in eukaryotes.
Abstract
DNA and RNA contain diverse chemical modifications that exert important influences in a variety of cellular processes. In addition to enzyme-mediated modifications of DNA and RNA, previous in vitro studies showed that pre-modified nucleoside triphosphates (NTPs) can be incorporated into DNA and RNA during replication and transcription. Herein, we established a chemical labeling method in combination with liquid chromatography-electrospray ionization-mass spectrometry (LC-ESI-MS) analysis for the determination of endogenous NTPs in the mammalian cells and tissues. We synthesized 8-(diazomethyl)quinoline (8-DMQ) that could efficiently react with the phosphate group under mild condition to label NTPs. The developed method allowed sensitive detection of NTPs, with the detection limits improved by 56–137 folds. The results showed that 12 types of endogenous modified NTPs were distinctly determined in the mammalian cells and tissues. In addition, the majority of these modified NTPs exhibited significantly decreased contents in human hepatocellular carcinoma (HCC) tissues compared to tumor-adjacent normal tissues. Taken together, our study revealed the widespread existence of various modified NTPs in eukaryotes.
Introduction
DNA and RNA carry diverse chemical modifications that exert essential and critical influences in a variety of cellular processes.1–3 Abnormal DNA and RNA modifications can cause many human diseases, such as diabetes,4,5 neurological disorders6 and cancers.7–10 To properly carry out the biological functions, the distribution of modifications in DNA and RNA needs to be tightly controlled.1 DNA cytosine methylation (5-methylcytosine, 5-mC) has been considered to be the most important epigenetic mark that plays vital roles in various physiological processes.11 In mammals, DNA cytosine methylation is carried out by the DNA methyltransferase (DNMT) family of enzymes that catalyze the transfer of a methyl group from S-adenosyl-l-methionine to DNA.12 5-mC can also be generated in the RNA of mammals by enzymes such as DNMT2, NSun2 and NSun4.13
In addition to enzymatically mediated methylation of DNA and RNA, in vitro studies showed that pre-methylated nucleoside triphosphates (NTPs) can be incorporated into DNA and RNA during replication and transcription.14 NTPs are generally synthesized by the corresponding nucleosides and phosphokinases and they act as the endogenous source to participate in DNA and RNA synthesis.15 Modified NTPs can be theoretically incorporated into DNA and RNA during replication and transcription. While these kinds of non-enzymatic modifications of DNA and RNA could be random, they may alter the normal distribution of modifications in DNA and RNA and eventually lead to the dysregulation of gene expression. For example, it has been known that inosine-5′-triphosphate (ITP) can be incorporated into RNA by RNA polymerase II.16 The incorporated inosine could affect the structure and function of RNA and lead to aberrant translation. And the oxidation of nucleotide pools by reactive oxygen species could result in various cellular dysfunctions.17 The representative example of oxidation of NTPs is the formation of 8-oxo-2′-deoxyguanosine-5′-triphosphate (8-oxo-dGTP), which can be incorporated into DNA and can eventually induce mutations, cellular senescence, neurological diseases and cancers.17–19
To control the quality of nucleotide pools, living organisms possess a number of NTP pyrophosphatases which hydrolyze non-canonical NTPs to their corresponding monophosphates. For example, inosine triphosphate pyrophosphatase (ITPase) can hydrolyze deaminated purine nucleoside triphosphates, such as ITP and dITP, to nucleoside monophosphate.20,21 These enzymes are termed “sanitizing” or “house-cleaning” enzymes.22,23 House-cleaning NTP pyrophosphatases targeting non-canonical NTPs belong to several structural superfamilies, including Nudix hydrolases, dUTPase, ITPase, all-α NTP pyrophosphatases, and cytidine deaminases.23,24 It has been established that 8-oxo-dGTP is frequently incorporated into DNA by DNA polymerases in mammals,25 indicating that the “house-cleaning” enzymes may not fully remove the endogenous modified NTPs. In addition, these “house-cleaning” enzymes that generally have high affinity for their natural substrates may not be able to efficiently hydrolyze other types of modified NTPs.23 In this respect, it is possible that certain modified NTPs may escape the degradation by these enzymes.
Sensitive detection methods are necessary to explore the potential existence of various modified NTPs in cells since these NTPs could be present in low abundance in vivo. Some methods were developed to detect NTPs, such as thin-layer chromatography detection,26 capillary electrophoresis (CE) with UV detection,27 high-performance liquid chromatography (HPLC) and reversed-phase ion pair liquid chromatography with UV28 and mass spectrometry29 detection. However, these methods lack enough sensitivity for the detection of low-abundant NTPs, especially for modified NTPs. In addition, NTPs are generally unstable and may dephosphorylate spontaneously.30 Owing to the low abundance and the low ionization efficiency of NTPs during mass spectrometry detection, determination of endogenous modified NTPs is still a challenging task.
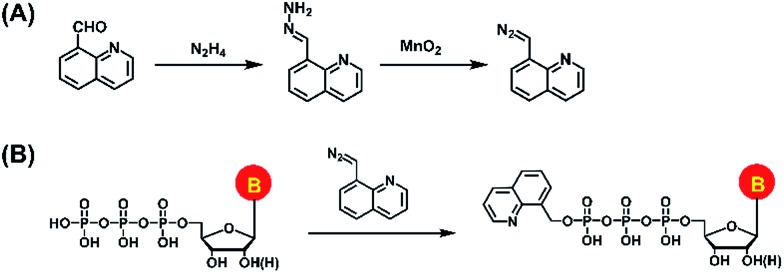
In the current study, we established a chemical labeling method coupled with liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) analysis for sensitive and simultaneous determination of endogenous modified NTPs. We designed and synthesized 8-(diazomethyl)quinoline (8-DMQ) (Fig. 1A) as the labeling reagent to selectively and efficiently react with the phosphate group of NTPs under mild condition (Fig. 1B). Using the developed method, we were able to distinctly determine 12 types of modified NTPs in addition to 8 kinds of canonical NTPs in mammalian cells and tissues, which provides evidence for the existence of various endogenous modified NTPs. In addition, we observed that the contents of the majority of these modified NTPs were significantly decreased in hepatocellular carcinoma (HCC) tissues compared to the tumor-adjacent normal tissues.
Fig. 1. (A) Synthesis route of the labeling reagent of 8-DMQ. (B) Chemical labeling reaction of NTPs by 8-DMQ.
Experimental section
Chemicals and reagents
All NTPs were purchased from TriLink BioTechnologies (San Diego, CA) and Sigma-Aldrich (Beijing, China) (Table S1 and Fig. S1 in ESI†). The stable isotopic ATP and dATP (13C10,15N5-ATP and 13C10,15N5-dATP) were purchased from Sigma-Aldrich (Beijing, China). Quinoline-8-carbaldehyde, activated manganese dioxide (MnO2), l-methionine-(methyl-D3) (D3-Met) and phosphodiesterase I were purchased from Sigma-Aldrich (Beijing, China). S1 nuclease and alkaline phosphatase were from Takara Biotechnology Co., Ltd (Dalian, China). The bicinchoninic acid (BCA) protein assay kit was purchased from Beyotime (Shanghai, China).
Biological and clinical samples
Human 293T cells, Jurkat-T cells and HeLa cells were obtained from the China Center for Type Culture Collection and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA), 100 U mL–1 penicillin, and 100 μg mL–1 streptomycin. Cells were maintained in a humidified atmosphere with 5% CO2 at 37 °C. As for the D3-Met labeling, 30 mg of D3-Met was added into the l-methionine-free DMEM-KO medium supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA), 100 U mL–1 penicillin, and 100 μg mL–1 streptomycin.
A total of 10 pairs of hepatocellular carcinoma (HCC) tissues and matched tumor-adjacent normal tissues were collected from Hubei Cancer Hospital. An approval was granted by the Hubei Cancer Hospital Ethics Committee and the study met the requirements of the declaration of Helsinki. All the experiments were performed in accordance with Hubei Cancer Hospital Ethics Committee's guidelines and regulations.
Extraction of endogenous NTPs from cultured cells and tissue samples
Human 293T cells, Jurkat-T cells and HeLa cells, at a density of approximately 1.0 × 106 cells per mL (10 mL), were collected by centrifugation at 380g under 4 °C for 5 min and then washed twice with ice-cold phosphate buffered saline to remove the fetal bovine serum.
For the extraction of endogenous NTPs from cells, the samples were rapidly quenched with pre-chilled 80% aqueous methanol (0 °C, 2 mL) and incubated at –20 °C for 30 min according to a previous report.31 For the extraction of endogenous NTPs from tissues, the tissue samples were extracted by homogenization in pre-chilled 80% aqueous methanol (0 °C, 1 mL). Then the extracts of cells and tissues were centrifuged at 14 000 g for 15 min at 4 °C to remove precipitated proteins. Supernatants were collected and dried under nitrogen gas and then stored at –80 °C. The protein concentrations were used to normalize the contents of NTPs. The protein concentrations of all the cells and tissue samples were measured using the BCA protein assay kit.
Synthesis of 8-(diazomethyl)quinoline (8-DMQ)
8-(Diazomethyl)quinoline (8-DMQ) was synthesized from quinoline-8-carbaldehyde according to a previous report with some modifications.32 The synthesis route is shown in Fig. 1A. Briefly, 40 mg of quinoline-8-carbaldehyde in DMSO (985 μL) was incubated with hydrazine hydrate (15 μL) at 60 °C for 2 h. When the solution mixture cooled down to room temperature, anhydrous magnesium sulfate (10 mg) was added to remove the water. The supernatant was collected and activated MnO2 (400 mg) was added. The suspension was stirred vigorously for 4 h at 25 °C. Finally, the precipitate was spun down and the supernatant containing 8-DMQ was collected. Because the diazo reagent of 8-DMQ was sensitive to protic solvents, it was stored in 50 mM DMSO solution at –20 °C. The synthesized 8-DMQ was examined by high-resolution mass spectrometry and nuclear magnetic resonance (NMR) analysis.
Chemical labeling
Eight canonical NTPs (dATP, dGTP, dCTP, TTP, ATP, GTP, CTP, and UTP) were used to optimize the chemical labeling conditions. To achieve the best labeling efficiency, we optimized the reaction time and temperature as well as the amounts of labeling reagents. All the reactions were performed in a mixture containing 50 mM borate buffer (pH 6.9, 160 μL) and DMSO (40 μL) with 50 pmol of each NTP.
Analysis of 8-DMQ-labeled NTPs by LC-ESI-MS/MS
The 8-DMQ-labeled NTPs (8-MQ-NTPs) were analyzed on an LC-ESI-MS/MS system consisting of a Shimadzu MS-8040 triple quadrupole mass spectrometer (Tokyo, Japan) with an electrospray ionization (ESI) source (Turbo Ionspray) and a Shimadzu LC-20AD HPLC system (Tokyo, Japan). An Inertsil ODS-3 column (250 mm × 2.1 mm i.d., 3.5 μm, GL Science, Shanghai, China) was used for the separation. The column temperature was set at 35 °C. Ammonium bicarbonate in water (2 mM, pH 6.8, solvent A) and methanol (solvent B) were employed as mobile phases. A gradient of 0–2.5 min 5% B, 2.5–4.5 min 5% to 12% B, 4.5–12.5 min 12% to 85% B and 12.5–20.5 min 85% was used, and the flow rate was set at 0.2 mL min–1.
The LC-ESI-MS/MS analysis was performed under multiple reaction monitoring (MRM) in the positive-ion mode. The MRM parameters of the analytes were optimized to achieve maximal detection sensitivity (Table S2 in ESI†). The optimal ESI source conditions were as follows: DL temperature, 250 °C; heat block temperature, 400 °C; nebulizing gas flow rate, 3 L min–1; and drying gas flow rate, 15 L min–1.
High-resolution mass spectrometry analysis
The 8-MQ-NTPs were examined by an LTQ-Orbitrap Elite high-resolution mass spectrometer (Thermo-Fisher Scientific, Waltham, MA, USA) equipped with an electrospray ionization (ESI) source and a Dionex Ultimate 3000 UPLC system (Thermo-Fisher Scientific, Waltham, MA, USA). The LC separation conditions were the same as those for the Shimadzu MS-8040 triple quadrupole mass spectrometer system. Full MS scans were acquired in the positive-ion mode at a resolution of 60 000. The molecular masses of the 8-MQ-NTPs were listed as the precursor ion for further MS/MS analysis. Collision induced dissociation (CID) with the collision energy of 35 eV was used. The fragments were acquired with a mass range of m/z 100–700 at a resolution of 60 000. The source and ion transfer parameters applied were as follows: heater temperature, 300 °C; capillary temperature, 350 °C; sheath gas flow, 35 arbitrary units; auxiliary gas flow, 15 arbitrary units; spray voltage, 3.5 kV; capillary voltage, 35 V; and the S-lens RF level, 60%. The data analysis was carried out using Xcalibur v3.0.63 (Thermo-Fisher Scientific, Waltham, MA, USA).
Statistical analysis
The statistical data were processed with SPSS 19.0 software (IBM SPSS Inc, USA). The paired t-test was performed to evaluate the content differences of NTPs between HCC tissues and tumor-adjacent normal tissues. All p values were two-sided, and generally, p values <0.05 were considered to have statistical significance.
Results and discussion
Strategy for the determination of modified NTPs by chemical labeling coupled with LC-ESI-MS/MS analysis
Systematic exploration of endogenous modified NTPs in mammalian cells is still a challenging task, possibly due to their low in vivo abundance as well as the poor ionization efficiencies during mass spectrometry analysis. In addition, the molecular structures as well as the physical and chemical properties of NTPs are similar, which can result in poor separation during chromatographic separation and therefore restrict their identifications.
Introduction of an easily ionizable group to targeted analytes could enhance the ionization efficiency in mass spectrometry analysis.33,34 Our group recently established chemical labeling methods for the sensitive detection of endogenous low-abundant compounds by LC-MS analysis.35–38 Along this line, here we synthesized 8-DMQ that harbors a diazo group to efficiently react with the phosphate group of NTPs under mild condition (Fig. 1). The quinoline group of 8-DMQ could increase the ionization efficiency during mass spectrometry analysis, and the hydrophobic phenyl group introduced from 8-DMQ enhances the retention of NTPs on reversed-phase LC that can improve the chromatographic separation. As a result, the detection sensitivities of these 8-DMQ-labeled NTPs dramatically increased during LC-ESI-MS/MS analysis. Using the developed method, we were able to readily determine the endogenous modified NTPs in mammalian cells and tissues.
Chemical labeling
We first synthesized the labeling reagent of 8-DMQ and the synthesis route is shown in Fig. 1A. The high-resolution mass spectrometry and NMR analysis demonstrated that 8-DMQ was successfully synthesized (Fig. S2 and S3 in ESI†). The LC chromatograms showed that the synthesis reaction was complete, and the unpurified 8-DMQ did not contain the starting material of quinoline-8-carbaldehyde and 8-(hydrazonomethyl)quinolone (Fig. S4 in ESI†). Because 8-DMQ is not very stable, the synthesized 8-DMQ was directly used for the labeling of NTPs.
We examined the 8-DMQ-labeled NTPs (8-MQ-NTPs) using 34 NTP standards (Table S1 and Fig. S1 in the ESI†) by high-resolution mass spectrometry analysis. The results showed that the measured precursor ions and product ions of 8-MQ-NTPs were identical to the theoretical values (Fig. S5 in ESI†), suggesting that all the NTPs successfully reacted with 8-DMQ and formed the desired derivatives of 8-MQ-NTPs. Moreover, only one phosphate group of NTPs could be labeled (Fig. S6 in ESI†). We also observed that the stabilities of the NTPs increased upon 8-DMQ labeling (Fig. S7 in ESI†), indicating that the 8-DMQ labeling may delay the degradation of NTPs. The improved stabilities of NTP derivatives (8-MQ-NTPs) benefit the accurate quantification of NTPs.
To achieve the best chemical labeling efficiencies of NTPs by 8-DMQ, we optimized the labeling reaction conditions. We first optimized the molar ratios of 8-DMQ/NTPs ranging from 10/1 to 200/1. The results showed that the peak areas of 8-MQ-NTPs reached a plateau when the molar ratio of 8-DMQ/NTPs was 50/1 (Fig. S8A in ESI†). Therefore, the molar ratio of 50/1 (8-DMQ/NTPs) was chosen. Next, the reaction temperature was optimized ranging from 25 °C to 50 °C. The results showed that the labeling could achieve better reaction efficiencies under 25 °C (Fig. S8B in ESI†). So, we chose 25 °C as the reaction temperature. Finally, the reaction time was investigated ranging from 10 min to 4 h. The results showed that 8-DMQ could react with NTPs within 10 min and the peak areas of 8-MQ-NTPs decreased after 1 h (Fig. S8C in ESI†). Therefore, 10 min was used for the following experiments.
Taken together, the optimized labeling conditions by 8-DMQ were 25 °C for 10 min with a 50/1 molar ratio of 8-DMQ/NTPs. Under optimized labeling conditions, the labeling efficiencies of nucleotides were over 99% (Fig. S9 in ESI†).
Improvements of detection sensitivity and LC separation upon chemical labeling
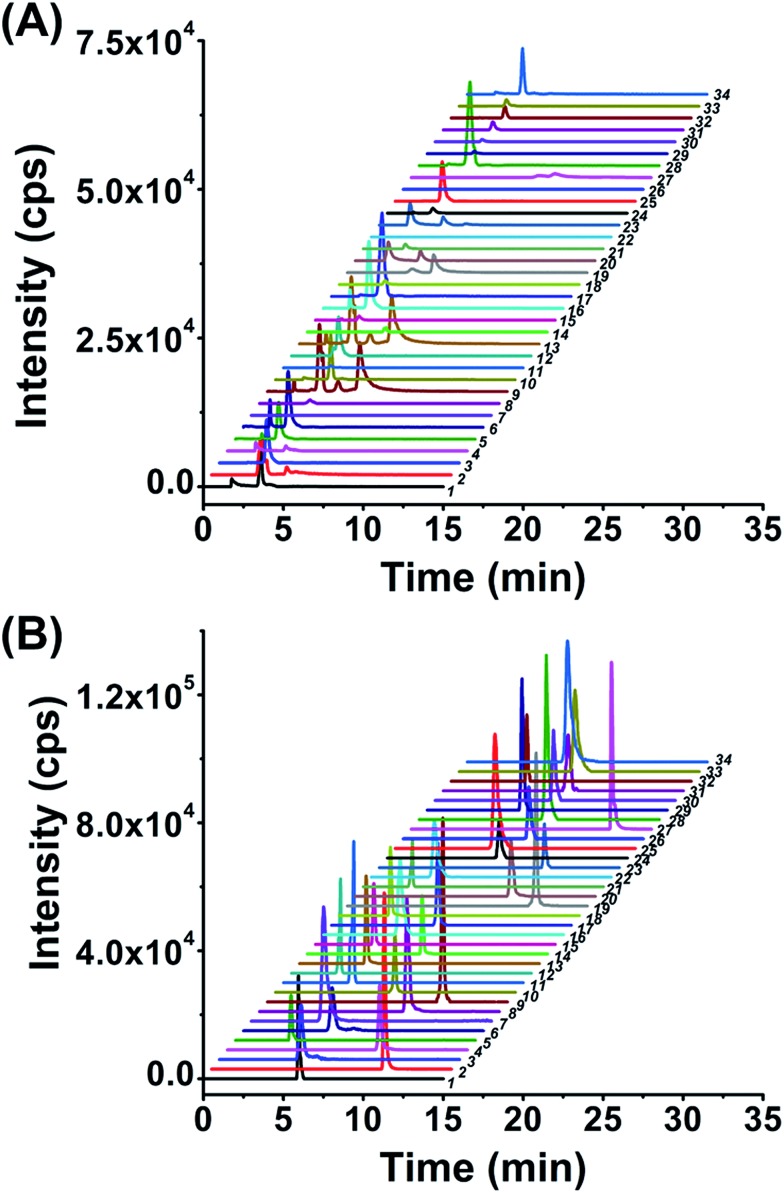
After optimization of the labeling reaction conditions, we then established 8-DMQ labeling in combination with LC-ESI-MS/MS analysis for the determination of modified NTPs. We used 34 kinds of 8-MQ-NTPs to optimize the chromatographic separation and MS parameters. The extracted ion chromatograms of 34 native NTPs and their corresponding labeled products of 8-MQ-NTPs are shown in Fig. 2A and B, respectively. The results suggested that the retention of native NTPs was relatively weak and the separation of NTPs was poor. The peaks typically tailed on the reversed-phase chromatographic column even under optimized separation conditions (Fig. 2A). However, after 8-DMQ labeling, the retention and separation resolution effectively improved (Fig. 2B), which can be attributed to the increased hydrophobicity of these derivatives.
Fig. 2. Extracted ion chromatograms of the 34 NTPs without labeling (A) or with 8-DMQ labeling (B) followed by LC-ESI-MS/MS analysis. 1, ATP; 2, dATP; 3, GTP; 4, dGTP; 5, CTP; 6, dCTP; 7, UTP; 8, TTP; 9, N6-meATP; 10, 7-meGTP; 11, ITP; 12, XTP; 13, N1-meATP; 14, 2-S-UTP; 15, 5-hmCTP; 16, 5-meCTP; 17, 5-medCTP; 18, 5-hmdCTP; 19, 2′-O-meATP; 20, 2′-O-me-GTP; 21, 5-cadCTP; 22, YTP; 23, 2′-O-me-ITP; 24, 5-meUTP; 25, 2′-O-meCTP; 26, 5-caseUTP; 27, N6-medATP; 28, N4-medCTP; 29, 2′-O-meYTP; 30, 2′-O-meUTP; 31, 5-fodCTP; 32, 5-foCTP; 33, O6-meGTP; and 34, N1-meGTP.
Compared to the native forms of NTPs, 8-DMQ labeling could also dramatically increase their detection sensitivities. We used 8 canonical NTPs to evaluate the enhancement of detection sensitivities of NTPs upon 8-DMQ labeling. The limits of detection (LODs) defined as the amounts of the analytes at a signal-to-noise ratio (S/N) of 3 were employed to evaluate the improved detection limits of NTPs by 8-DMQ labeling. The measured LODs of NTPs with and without chemical labeling are listed in Table 1. We found that the detection sensitivities of NTPs improved by 56 to 137 folds upon 8-DMQ labeling (Table 1). The introduction of methylquinoline group to NTPs could increase the ionization efficiency during MS analysis. In addition, the increased hydrophobicity of these 8-DMQ-labeled NTPs resulted in longer retention times by the elution with a higher ratio of organic solvent. Thus, the derivatives could be ionized more effectively in the ESI source. Compared to other methods, our method exhibited the best detection sensitivities towards NTPs (Table S3 in ESI†). Taken together, 8-DMQ labeling in combination with LC-ESI-MS/MS analysis can efficiently increase the detection performance of NTPs.
Table 1. The limits of detection (LODs) of 8 canonical NTPs with and without 8-DMQ labeling by LC-ESI-MS/MS analysis under the optimized detection conditions.
| Analytes | Unlabeled (LODs, fmol) | After labelling (LODs, fmol) | Detection limit improved (folds) |
| ATP | 33.6 | 0.6 | 56 |
| GTP | 46.2 | 0.6 | 77 |
| CTP | 104.0 | 1.3 | 80 |
| UTP | 411.0 | 3.0 | 137 |
| dATP | 17.2 | 0.4 | 43 |
| dGTP | 124.3 | 1.1 | 113 |
| dCTP | 58.2 | 0.6 | 97 |
| TTP | 44.5 | 0.5 | 89 |
Determination of endogenous modified NTPs in mammalian cells
With the established method, we explored the possible existence of endogenous modified NTPs in cultured human cells, including 293T cells, Jurkat-T cells and HeLa cells.
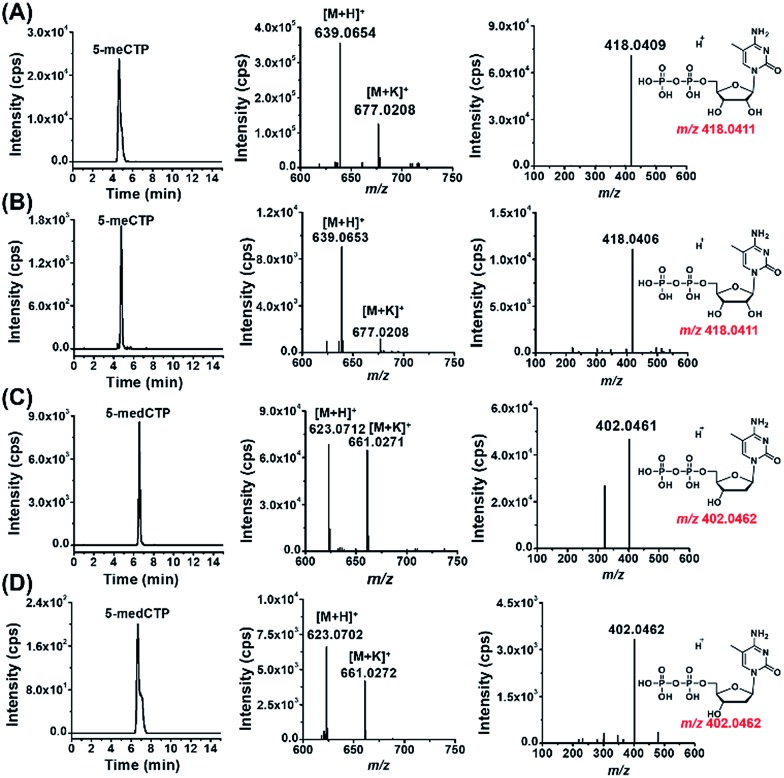
The results showed that the extracted ion chromatograms, high-resolution MS spectra and product ion spectra of 8-DMQ-labeled 5-methylcytidine-5′-triphosphate (5-meCTP) (Fig. 3A) and 5-methyl-2′-deoxycytidine-5′-triphosphate (5-medCTP) (Fig. 3C) from 293T cells were identical to the 8-DMQ-labeled 5-meCTP and 5-medCTP standards (Fig. 3B and D), clearly demonstrating the existence of 5-meCTP and 5-medCTP in 293T cells. Similarly, the extracted ion chromatograms, high-resolution MS spectra and product ion spectra of the other 18 kinds of NTPs (10 kinds of modified NTPs and 8 kinds of canonical NTPs) for the cultured 293T cells are identical to the NTP standards (Fig. S10 in ESI†). In addition to the 293T cells, these 20 kinds of NTPs could also be distinctly detected in Jurkat-T cells and HeLa cells. These results demonstrated the universal existence of 12 types of modified NTPs in mammals.
Fig. 3. Determination of 5-meCTP and 5-medCTP in 293T cells. The extracted ion chromatograms (left panel), precursor ion spectra (middle panel) and product ion spectra (right panel) of (A) 5-meCTP standard, (B) 5-meCTP detected in 293T cells, (C) 5-medCTP standard, and (D) 5-medCTP detected in 293T cells.
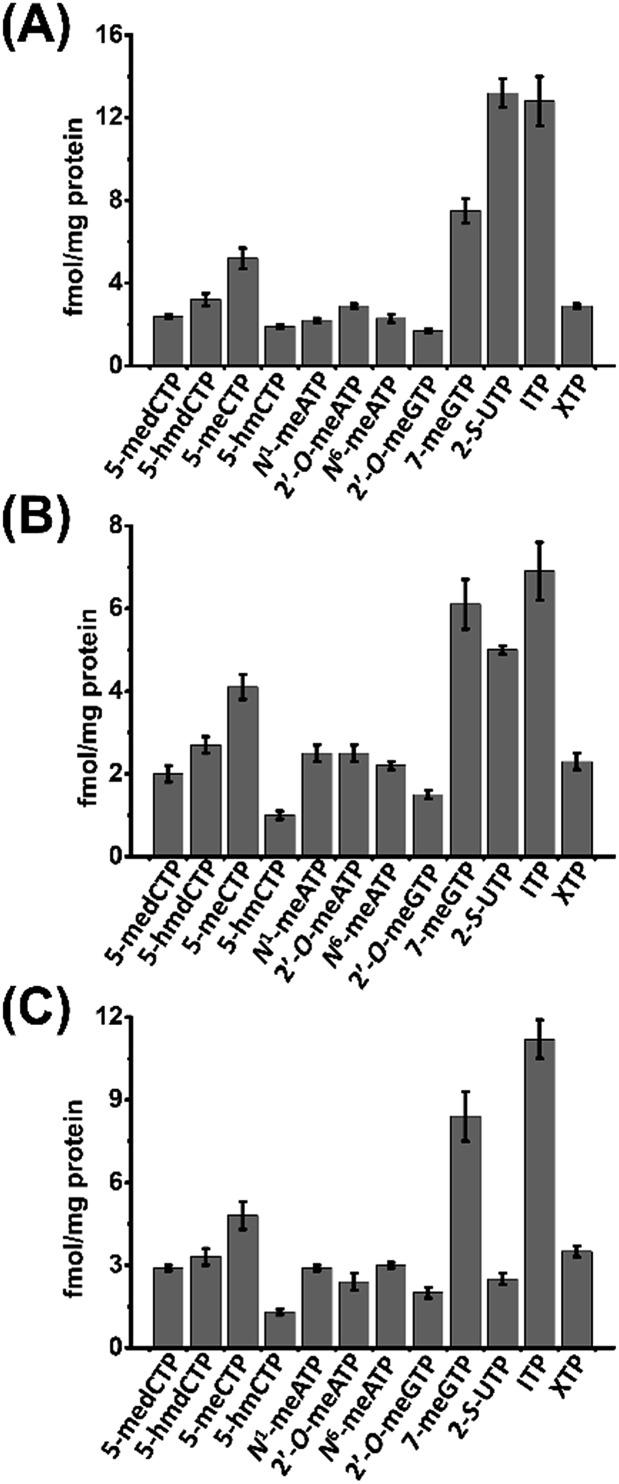
We then next measured the contents of these modified NTPs. To this end, we constructed the calibration curves and validated the established method. The detailed method validation can be found in the ESI.† The quantification results showed that the levels of 12 modified NTPs ranged from 1.0 to 13.2 fmol mg–1 protein in cultured cells (Fig. 4 and Table S4 in ESI†). A previous study reported that certain genes can be silenced by the incorporation of 5-medCTP into DNA in Chinese hamster ovary (CHO) cells treated with 5-medCTP.39 In addition to 5-medCTP, previous studies also demonstrated that many other modified NTPs can be incorporated into DNA and RNA during in vitro replication and transcription.14,16,40–42 In this respect, these detectable endogenous modified NTPs can be theoretically incorporated into DNA and RNA, which may compromise the regulatory roles of modifications.
Fig. 4. Contents of the measured modified NTPs in cultured cells. (A) 293T cells, (B) HeLa cells, and (C) Jurkat-T cells.
Methyl groups of modified NTPs originate from SAM
As a means to study the origin of the methyl groups these modified NTPs, we used stable isotope tracing monitored by mass spectrometry (Fig. S11 in ESI†). S-Adenosyl-l-methionine (SAM) made from ATP and l-methionine by methionine adenosyltransferase is a universal methylating agent and provides methyl groups to DNA and RNA.43 Therefore, human 293T cells were cultured in the methionine-free DMEM-KO medium with the added stable isotope labeled methionine, D3-Met, to metabolically label the DNA and RNA with methyl-CD3 group.
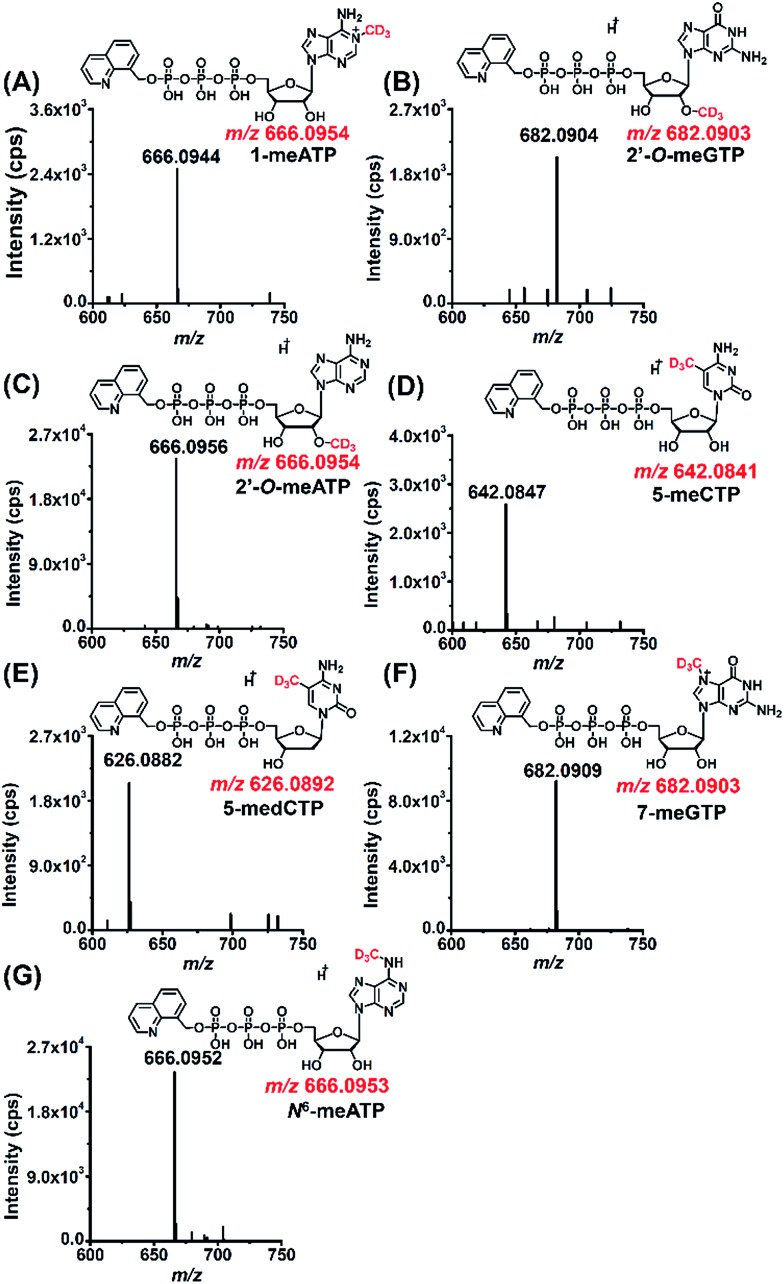
The stable isotope labeling efficiency were calculated to be between 78% and 97% (Fig. S12 in ESI†), suggesting good stable isotope labeling of DNA and RNA (detailed evaluation of the stable isotope labeling efficiency can be found in the ESI†). We further determined the modified NTPs in the D3-Met treated cells. The results showed that all the 7 methylated NTPs (CD3-1-meATP, CD3-2′-O-meGTP, CD3-2′-O-meATP, CD3-5-meCTP, CD3-5-medCTP, CD3-7-meGTP, and CD3-N6-meATP) carry a CD3 group (Fig. 5), indicating that the methyl group in these modified NTPs derive from methionine.
Fig. 5. MS spectra of modified NTPs carrying a CD3 group detected in 293T cells upon D3-Met metabolomic isotope labeling. (A) CD3-1-meATP, (B) CD3-2′-O-meGTP, (C) CD3-2′-O-meATP, (D) CD3-5-meCTP, (E) CD3-5-medCTP, (F) CD3-7-meGTP, and (G) CD3-N6-meATP.
In addition, we analyzed the modified nucleoside monophosphates (NMPs) and nucleoside diphosphates (NDPs) from 293T cells. The results showed that most of the modified NMPs and NDPs related to the modified NTPs were detectable (Fig. S13 in ESI†). As for the several modified NMPs and NDPs that were not detected, it might be that the contents of these modified NMPs and NDPs are lower than the detection limits of the current analytical method. The results together with the stable isotope tracing analysis suggested that these modified NTPs may originate from the degradation of nucleic acids.
Content change of modified NTPs in HCC tissues
The study of physiological consequence of the existence of endogenous modified NTPs will promote our understanding of their biological functions. Here we explored whether these modified NTPs display different contents in cancer tissues. Hepatocellular carcinoma (HCC) is a common human cancer and one of the leading causes of cancer deaths worldwide.44 With the developed analytical method, we investigated the correlation of the endogenous modified NTPs with human HCC.
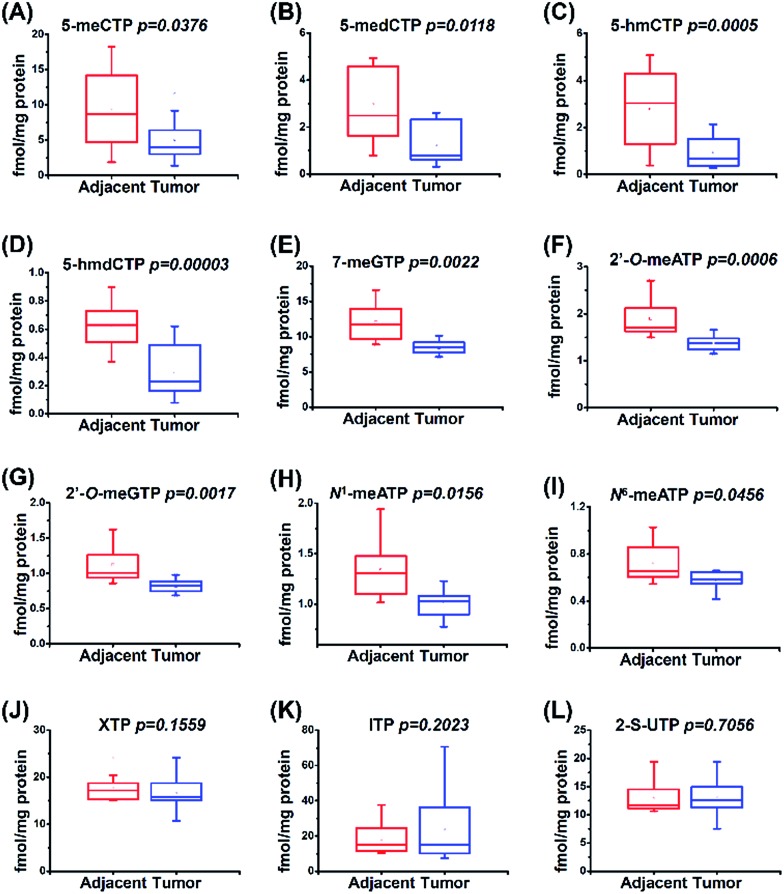
A total of 20 tissue samples derived from 10 HCC patients were analyzed. The statistical results showed that the contents of most of the modified NTPs, including 5-meCTP, 5-medCTP, 5-hmCTP, 5-hmdCTP, 7-meGTP, 2′-O-ATP, 2′-O-GTP, N1-meATP and N6-meATP, exhibited significant decreases in the HCC tissues compared to the tumor-adjacent normal tissues (Fig. 6). 5-mC, 5-mdC, 5-hmC, and 5-hmdC have been demonstrated to exert important epigenetic regulatory roles in gene expressions.1 Previous studies also revealed that 5-hmC and m1A favour mRNA translation and the decreased content of 5-hmC can impair brain development.45,46 In addition, it was reported that m6A modification could affect the stability of mRNA,47 eventually changing the ribonucleoprotein contents of mRNAs and leading to the alteration of synthesized proteins. The modifications occurring in DNA and RNA exhibit important biological roles and therefore need to be tightly controlled to exert their functions. Most of the modified NTPs in the current study haven't been detected before, which may introduce a new source of modifications of DNA and RNA and eventually lead to the physiological dysregulation. The underlying mechanism requires further in-depth investigation. The present study provides the basis for a systematic biochemical analysis of the biological role of modified NTPs in the future.
Fig. 6. Quantification and statistical analysis of the contents of 12 modified NTPs in human HCC tissues and tumor-adjacent normal tissues.
Conclusions
In the current study, we established a method by chemical labeling in combination with LC-ESI-MS/MS analysis for the determination of endogenous modified NTPs in mammals. Using the developed method, 12 types of endogenous modified NTPs were distinctly determined in the mammalian cells and tissues, which provides evidence for the widespread existence of various modified NTPs in eukaryotes. In addition, we observed that 9 out of the 12 kinds of modified NTPs exhibited significant decreases in HCC tissues compared to tumor-adjacent normal tissues. Our study indicates a new potential source for modifications of DNA and RNA, which may contribute to the development and formation of diseases.
Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Conflicts of interest
The authors declare no competing financial interest.
Supplementary Material
Acknowledgments
The work is supported by the National Key R&D Program of China (2017YFC0906800) and the National Natural Science Foundation of China (21522507, 21672166, 21635006, and 21721005).
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c7sc05472f
References
- Chen K., Zhao B. S., He C. Cell Chem. Biol. 2016;23:74–85. doi: 10.1016/j.chembiol.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. A. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Carell T., Brandmayr C., Hienzsch A., Muller M., Pearson D., Reiter V., Thoma I., Thumbs P., Wagner M. Angew. Chem., Int. Ed. 2012;51:7110–7131. doi: 10.1002/anie.201201193. [DOI] [PubMed] [Google Scholar]
- Nilsson E., Jansson P. A., Perfilyev A., Volkov P., Pedersen M., Svensson M. K., Poulsen P., Ribel-Madsen R., Pedersen N. L., Almgren P., Fadista J., Ronn T., Klarlund Pedersen B., Scheele C., Vaag A., Ling C. Diabetes. 2014;63:2962–2976. doi: 10.2337/db13-1459. [DOI] [PubMed] [Google Scholar]
- Shen F., Huang W., Huang J. T., Xiong J., Yang Y., Wu K., Jia G. F., Chen J., Feng Y. Q., Yuan B. F., Liu S. M. J. Clin. Endocrinol. Metab. 2015;100:E148–E154. doi: 10.1210/jc.2014-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urdinguio R. G., Sanchez-Mut J. V., Esteller M. Lancet Neurol. 2009;8:1056–1072. doi: 10.1016/S1474-4422(09)70262-5. [DOI] [PubMed] [Google Scholar]
- Timp W., Feinberg A. P. Nat. Rev. Cancer. 2013;13:497–510. doi: 10.1038/nrc3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. L., Shen F., Huang W., Qi J. H., Wang Y., Feng Y. Q., Liu S. M., Yuan B. F. Clin. Chem. 2013;59:824–832. doi: 10.1373/clinchem.2012.193938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Lan M. D., Qi C. B., Zheng S. J., Wei S. Z., Yuan B. F., Feng Y. Q. Chem. Sci. 2016;7:5495–5502. doi: 10.1039/c6sc01589a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Zheng S. J., Qi C. B., Feng Y. Q., Yuan B. F. Anal. Chem. 2015;87:3445–3452. doi: 10.1021/ac504786r. [DOI] [PubMed] [Google Scholar]
- Egger G., Liang G., Aparicio A., Jones P. A. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- Ko M., An J., Rao A. Curr. Opin. Cell Biol. 2015;37:91–101. doi: 10.1016/j.ceb.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco S., Frye M. Curr. Opin. Cell Biol. 2014;31:1–7. doi: 10.1016/j.ceb.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Luo X., Dong Y. Bioconjugate Chem. 2016;27:849–853. doi: 10.1021/acs.bioconjchem.6b00090. [DOI] [PubMed] [Google Scholar]
- Zauri M., Berridge G., Thezenas M. L., Pugh K. M., Goldin R., Kessler B. M., Kriaucionis S. Nature. 2015;524:114–118. doi: 10.1038/nature14948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M. J., Platas A. A., Hawley D. K. Cell. 1998;93:627–637. doi: 10.1016/s0092-8674(00)81191-5. [DOI] [PubMed] [Google Scholar]
- Pursell Z. F., McDonald J. T., Mathews C. K., Kunkel T. A. Nucleic Acids Res. 2008;36:2174–2181. doi: 10.1093/nar/gkn062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya H. Mutat. Res. 2010;703:32–36. doi: 10.1016/j.mrgentox.2010.06.003. [DOI] [PubMed] [Google Scholar]
- Russo M. T., Blasi M. F., Chiera F., Fortini P., Degan P., Macpherson P., Furuichi M., Nakabeppu Y., Karran P., Aquilina G., Bignami M. Mol. Cell. Biol. 2004;24:465–474. doi: 10.1128/MCB.24.1.465-474.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakumi K., Abolhassani N., Behmanesh M., Iyama T., Tsuchimoto D., Nakabeppu Y. Mutat. Res. 2010;703:43–50. doi: 10.1016/j.mrgentox.2010.06.009. [DOI] [PubMed] [Google Scholar]
- Hwang K. Y., Chung J. H., Kim S. H., Han Y. S., Cho Y. Nat. Struct. Biol. 1999;6:691–696. doi: 10.1038/10745. [DOI] [PubMed] [Google Scholar]
- Bessman M. J., Frick D. N., O'Handley S. F. J. Biol. Chem. 1996;271:25059–25062. doi: 10.1074/jbc.271.41.25059. [DOI] [PubMed] [Google Scholar]
- Galperin M. Y., Moroz O. V., Wilson K. S., Murzin A. G. Mol. Microbiol. 2006;59:5–19. doi: 10.1111/j.1365-2958.2005.04950.x. [DOI] [PubMed] [Google Scholar]
- Rudd S. G., Valerie N. C. K., Helleday T. DNA Repair. 2016;44:193–204. doi: 10.1016/j.dnarep.2016.05.032. [DOI] [PubMed] [Google Scholar]
- Nakabeppu Y., Oka S., Sheng Z., Tsuchimoto D., Sakumi K. Mutat. Res. 2010;703:51–58. doi: 10.1016/j.mrgentox.2010.06.006. [DOI] [PubMed] [Google Scholar]
- Rajagopal V., Lorsch J. R. Methods Enzymol. 2013;533:325–334. doi: 10.1016/B978-0-12-420067-8.00025-8. [DOI] [PubMed] [Google Scholar]
- Feng H. T., Wong N., Wee S., Lee M. M. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2008;870:131–134. doi: 10.1016/j.jchromb.2008.05.038. [DOI] [PubMed] [Google Scholar]
- Abi Jaoude M., Randon J. Anal. Bioanal. Chem. 2011;400:1241–1249. doi: 10.1007/s00216-011-4681-4. [DOI] [PubMed] [Google Scholar]
- Magdenoska O., Martinussen J., Thykaer J., Nielsen K. F. Anal. Biochem. 2013;440:166–177. doi: 10.1016/j.ab.2013.05.023. [DOI] [PubMed] [Google Scholar]
- Simmonds H. A., Micheli V., Davies P. M., McBride M. B. Clin. Chim. Acta. 1990;192:121–132. doi: 10.1016/0009-8981(90)90076-5. [DOI] [PubMed] [Google Scholar]
- Lu W., Kimball E., Rabinowitz J. D. J. Am. Soc. Mass Spectrom. 2006;17:37–50. doi: 10.1016/j.jasms.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Bourget C., Trevisiol E., Lhomme J., Kotera M., Tora C., Bernal-Mendez E., Laayoun A. Nucleosides, Nucleotides Nucleic Acids. 2003;22:1371–1373. doi: 10.1081/NCN-120022968. [DOI] [PubMed] [Google Scholar]
- Qi B. L., Liu P., Wang Q. Y., Cai W. J., Yuan B. F., Feng Y. Q. TrAC, Trends Anal. Chem. 2014;59:121–132. [Google Scholar]
- Xiong J., Liu X., Cheng Q. Y., Xiao S., Xia L. X., Yuan B. F., Feng Y. Q. ACS Chem. Biol. 2017;12:1636–1643. doi: 10.1021/acschembio.7b00170. [DOI] [PubMed] [Google Scholar]
- Tang Y., Xiong J., Jiang H. P., Zheng S. J., Feng Y. Q., Yuan B. F. Anal. Chem. 2014;86:7764–7772. doi: 10.1021/ac5016886. [DOI] [PubMed] [Google Scholar]
- Liu P., Huang Y. Q., Cai W. J., Yuan B. F., Feng Y. Q. Anal. Chem. 2014;86:9765–9773. doi: 10.1021/ac5023315. [DOI] [PubMed] [Google Scholar]
- Zhang H. Y., Xiong J., Qi B. L., Feng Y. Q., Yuan B. F. Chem. Commun. 2016;52:737–740. doi: 10.1039/c5cc07354e. [DOI] [PubMed] [Google Scholar]
- Cheng Q. Y., Xiong J., Wang F., Yuan B. F., Feng Y. Q. Chin. Chem. Lett. 2018;29:115–118. [Google Scholar]
- Holliday R., Ho T. Somatic Cell Mol. Genet. 1991;17:537–542. doi: 10.1007/BF01233618. [DOI] [PubMed] [Google Scholar]
- Kuchta R. D. Curr. Protoc. Chem Biol. 2010;2:111–124. doi: 10.1002/9780470559277.ch090203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katafuchi A., Nohmi T. Mutat. Res. 2010;703:24–31. doi: 10.1016/j.mrgentox.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Paul N., Yee J. BioTechniques. 2010;48:333–334. [Google Scholar]
- Landgraf B. J., McCarthy E. L., Booker S. J. Annu. Rev. Biochem. 2016;85:485–514. doi: 10.1146/annurev-biochem-060713-035504. [DOI] [PubMed] [Google Scholar]
- Siegel R. L., Miller K. D., Jemal A. Ca-Cancer J. Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- Dominissini D., Nachtergaele S., Moshitch-Moshkovitz S., Peer E., Kol N., Ben-Haim M. S., Dai Q., Di Segni A., Salmon-Divon M., Clark W. C., Zheng G., Pan T., Solomon O., Eyal E., Hershkovitz V., Han D., Dore L. C., Amariglio N., Rechavi G., He C. Nature. 2016;530:441–446. doi: 10.1038/nature16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatte B., Wang F., Ngoc L. V., Collignon E., Bonvin E., Deplus R., Calonne E., Hassabi B., Putmans P., Awe S., Wetzel C., Kreher J., Soin R., Creppe C., Limbach P. A., Gueydan C., Kruys V., Brehm A., Minakhina S., Defrance M., Steward R., Fuks F. Science. 2016;351:282–285. doi: 10.1126/science.aac5253. [DOI] [PubMed] [Google Scholar]
- Wang X., Lu Z., Gomez A., Hon G. C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G., Ren B., Pan T., He C. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.