Abstract
Purpose
Accurate reconstruction of myocardial T1 maps from series of T1-weighted (T1-w) images is compromised by cardiac motions induced from breathing and diaphragmatic drifts. We propose and evaluate a new framework based on Active Shape Models (ASM) to correct for motion in myocardial T1 maps.
Methods
Multiple appearance models were built at different inversion time intervals to model the blood-myocardium contrast and brightness changes during the longitudinal relaxation. Myocardial inner and outer borders were automatically segmented using the built models and the extracted contours were used to register the T1-w images. Data acquired from 210 patients using free-breathing acquisition protocol were used to train and evaluate the proposed framework. Two independent readers evaluated the quality of the T1 maps before and after correction using a four-point score. Mean Absolute Distance (MAD) and Dice index were used to validate the registration process.
Results
Testing dataset from 180 patients at 5 short axial slices showed a significant decrease of MAD (from 3.3 ± 1.6 to 2.3 ± 0.8 mm, P < 0.001) and increase of Dice (from 0.89 ± 0.08 to 0.94 ± 0.4%, P < 0.001) before and after correction, respectively. T1 maps quality improved in 70 ± 0.3% of the motion-affected maps after correction. Motion-corrupted segments of the myocardium reduced from 21.8% to 8.5% (P < 0.001) of total number of segments after correction.
Conclusion
The proposed method for non-rigid registration of T1-w images allows T1 measurements in more myocardial segments by reducing motion-induced T1 estimation errors in myocardial segments.
Keywords: Myocardial T1 Mapping, Motion Correction, Active Shape Models, Non-rigid registration, MRI
Introduction
Myocardial interstitial diffuse fibrosis and extra-cellular volume expansion are characteristic of many cardiac diseases (1–4) and alter longitudinal relaxation time (T1) values (4–6). Recent improvements in pulse sequence development allow reproducible measurement of myocardial T1 values (7–10). In myocardial T1 mapping, a series of T1-weighted (T1-w) images are acquired with different saturation or inversion times (TI) (11–14) and are used to create T1 maps by voxel-wise fitting through two- or three-parameter fit models (14–16). In the presence of respiratory and cardiac motion, voxels are not aligned in different T1-w images and will cause errors in T1 estimation. Therefore, motion correction is an essential step in myocardial T1 mapping.
To minimize motion artifacts, T1 mapping is often acquired during a breath-hold scan (12,17). Free-breathing T1 mapping sequences have also been developed by using slice-tracking or navigator gating (14), but residual motions can still be detected between different T1-w images due to respiratory drifting or inability of prospective slice-tracking technique to register the images. To overcome this challenge, post-processing motion correction is used to align T1-w images (18,19). Xue et al. (18) proposed a motion correction technique that simulates contrast changes of T1-w images by generating free motion images from an initial T1 estimate. The synthetic images are then matched with the corresponding inversion images to estimate the deformation field and correct the motions, but this framework does not account for T1 variations among different patients at the same TI, where the blood-myocardium contrast can be completely inverted for different cases at the same TI. Roujol et al. used a modified optical flow energy function to estimate the elastic deformation field of the myocardium with an additional term to avoid transient structures from through-plane motions (19). However, estimation of the non-rigid parameters was affected by different signal-to-noise and contrast-to-noise ratios of T1-w images. Additionally, these methods were proposed to register T1 mapping images acquired using breath-holding acquisition protocols, where the respiration-induced cardiac motion is minimized. Furthermore, these intensity-based image registration techniques require expensive computational power and time, so the need to improve motion correction for T1 mapping is still unmet.
Active Shape Models (ASM) allow robust segmentation of myocardial borders and have been used in left and right ventricular (LV/RV) segmentation from cine MR images (20–22). In ASM, a training dataset with pre-delineated contours of the target object (e.g. LV) is used to build shape and appearance models. The shape model is built by estimating the mean shape of the object and inter-shape variations among different patients in the training dataset, as represented by the covariance matrix (21,23). The appearance model is built to capture intensity variations at the LV myocardial borders (21). A matching algorithm is then used to search for the object’s borders in testing images with the built models.
In this study, we propose a new ASM-based framework for non-rigid registration of T1-w images to reduce motion artifacts in free-breathing cardiac T1 mapping. The epi- and endocardial boundaries of the LV are modeled and segmented at different values of TI. Contour-based image registration step is then utilized to estimate rigid and non-rigid parameters from the extracted contours, which are applied to T1-w images to reconstruct motion-corrected myocardial T1 maps. Qualitative and quantitative analyses are performed to evaluate the proposed methods.
Methods
The proposed motion correction technique is comprised of two steps: 1) extraction of the endo- and epicardial contours of all images; and 2) registration of T1-w images using the extracted contours. The first step is based on the active shape and appearance models (21,23,24). General shape and appearance model training is performed only once (offline) and used to extract the epi- and endocardium contours of any given T1-w image. In the second step, the given set of T1-w images is registered using both affine and non-rigid transformations such that the extracted contours of all images are aligned. In the following sections, we describe the steps involved in the proposed motion correction scheme.
Active Shape Model Construction
A modified formulation of the conventional ASM is used to build a shape model that captures shape variations among the LV boundaries in the given training dataset. The LV shape in every image in the training dataset is represented by a vector, x, containing the x- and y- coordinates of each point on the endocardial and epicardial contours:
| [1] |
where (x)epi and (y)epi are the x- and y- coordinates of the epicardial contours, respectively; (x)endo and (y)endo are the x- and y- coordinates of the endocardial contours and L is the number of landmark points in both epicardial and endocardial contour. In order to maintain the point correspondences among the training contours, LV contours are aligned by removing rigid transformations (i.e. translation, rotation and scaling) using Procrustes transformation (23,25). Both epicardial and endocardial contours are aligned simultaneously by applying Procrustes transformation of the x vectors directly.
Having obtained a shape vector, xn, for each image in the training dataset (with n = 1, 2, …, N; where N is the number of images in the training dataset), any given LV shape can be represented by a shape vector, x, (23,24):
| [2] |
where , is the mean-shape of the LV contours in the training dataset, b is the model parameters (associated with the given shape, x) and P is a matrix whose columns represent the principal components of the covariance matrix, . The columns of the matrix P are also referred to as the modes-of-variations (20,21,23) because they contain the most significant variations that can be linearly added to the mean-shape vector to represent a given LV shape. In this work, only the first 12 eigenvectors of C are used as the principal modes-of-variations. This number is determined as the smallest number of eigenvectors whose corresponding eigenvalues represent 99% of the total variations (represented by the summation of all eigenvalues) of C.
Appearance Model Construction
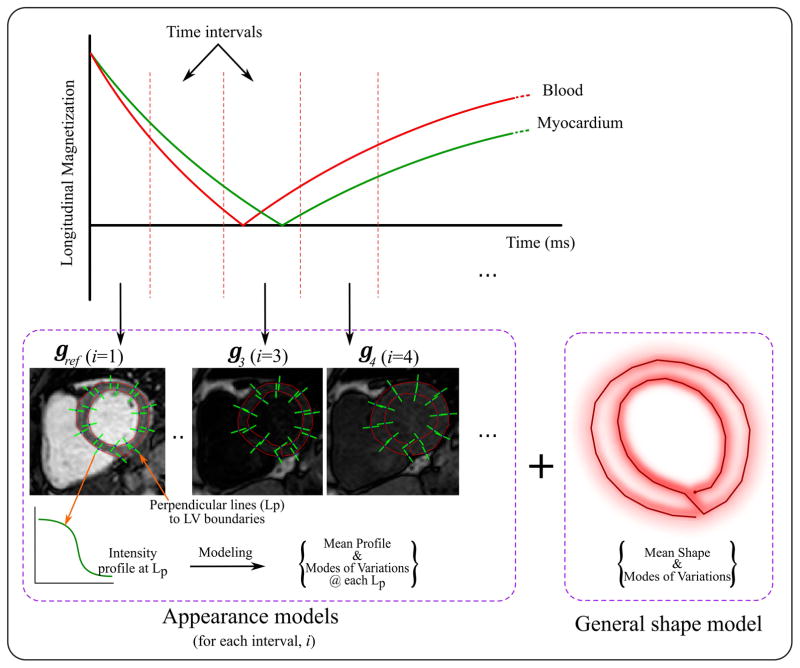
Similarly, an appearance model represents local intensity variations at the LV boundaries is built (21,24). In ASM framework, this is done by modeling the image intensity profile at each landmark point on the given contours. Given the training dataset of the images and the corresponding myocardium contours, each landmark point is traced and a line segment perpendicular to the contour is drawn such that it is centered at this point and extends for a distance of (Z/2) pixel on both sides of the point (Figure 1). The image intensity profile along the line segment is stored as a vector y of length (Z + 1).
Figure 1.
ASM training includes building multiple appearance models for the myocardium at different inversion time intervals. The expected range of inversion times is divided into a number of intervals, i. An appearance model, gi, is built to represent the intensity variations around the myocardial borders in the T1-w images for interval, i. To achieve this, intensity profiles along the perpendicular lines at selected landmark points, Lp, on the myocardial border are captured. The model is then built by calculating the mean intensity profile and covariance matrix within a dataset for each Lp. In addition, a shape model is built for the myocardium, where epicardial and endocardial contours from different subjects are aligned and the mean shape and covariance matrix representing the shape variations are calculated.
At each landmark point, l: l = 1:L, the mean intensity profile and the covariance matrix are computed as and , respectively. The appearance model is then given by,
| [3] |
where gl is the intensity profile captured at the lth landmark point, Sl is a matrix containing the first ρl eigen vectors estimated from the covariance matrix Ql, and hl denotes the appearance model controlling parameters. In this work, the number of eigenvectors, ρl, is selected so that 99% of the intensity variations at landmark l are included in the model.
Given the highly-varying contrast of the training images due to different T1-weighting, the above model may be only used to represent a specific T1-w image; e.g. a landmark in an image at a specific inversion time. Therefore, different appearance models are needed for the different T1-w images. To achieve this, the range of all expected TI is divided into a number of intervals, K, and a separate appearance model is built to represent the images at each interval (Figure 1). First, the T1-w images in the training sets are arranged into K groups depending on their TI. Then, an appearance model is built for each landmark point, l, and each interval, i, using Eqn 4; which is similar to Eqn 3 but with the subscript, i to indicate that there is a separate model for each inversion time interval.
| [4] |
In order to account for the large heart motion caused by patient movement or breathing, a multi-resolution model is considered in this framework (i.e. coarse-to-fine approach). The finest appearance model (level 1) is built from the original full resolution T1-w images while the following coarser levels are built from down-sampled versions of the images (21). Two down-sampling levels are used in this work where a down-sampling ratio of 0.5 per level was employed.
LV Myocardial Segmentation
Given a set of T1-w images with different inversion times, Ii(x, y; TIi), the image with the shortest inversion time; i.e. maximum contrast, is selected as a reference image, Iref(x, y; TI1). The shape model and the appearance model (corresponding to the TIref interval) of Iref are used to extract the myocardial boundaries in Iref. In this step, the initial mean shape of the myocardium is manually deposited on Iref by selecting one point inside the blood pool. The initial contour is then evolved iteratively to delineate the LV myocardium using the standard ASM searching algorithm (21). In each iteration, the matching algorithm uses the appearance models to update the location of the contour points; such that the image intensity profile at each updated contour point is closest to the appearance model (21). In other words, a displacement vector, δ, is estimated to minimize the following error measure:
| [5] |
where Di,l is a diagonal matrix containing the eigenvalues corresponding to the principal components (or modes-of-variations) of the matrix si,l, as estimated in Eqn 4, and pl is the location of the lth landmark point. The searching algorithm is restricted to window, g`i,l of +/− 8 pixels in a direction perpendicular to the contour at the point pl.
Following the conventional ASM framework, the resulting vector, δ, which represents the updated displacement of each contour point, is transformed to the shape model space by removing all rigid parameters following the same alignment procedure as the shape model construction. These updated displacements are projected onto the trained shape model to produce a smooth LV contour (21,23). The previous steps are performed for a fixed number of iterations and during the search algorithm, the number of modes-of-variations is dynamically varied to improve convergence characteristics. Initially, a small number of modes-of-variations (ρl = 5; representing ~97.5% of the total shape variations in the training set) are used for faster searching of a suitable transformation to bring the iterated contour close to the myocardium boundaries. Next, the modes-of-variations are exponentially increased so that the model in the last 5 iterations includes 12 modes-of-variations (representing ~99% of the shape variations in training set) to extract the fine details of the myocardial boundaries. Detailed steps of the whole algorithm are included in Supporting Information S4.
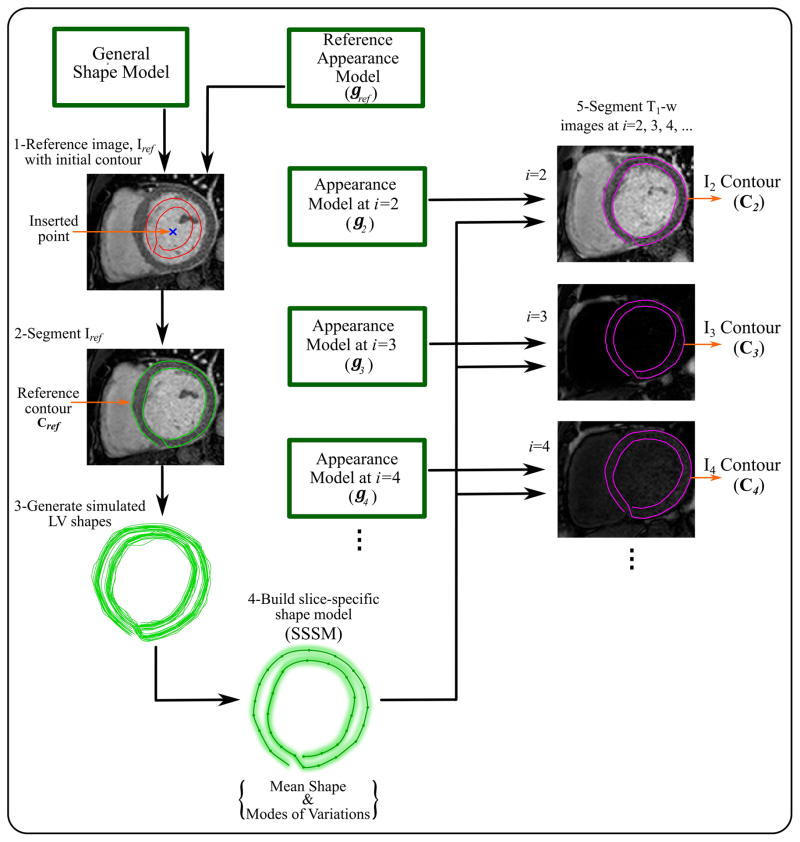
Having segmented the reference image, the extracted reference contour, cref, assists faster and fully automatic segmentation of the remaining T1-w images. First, a new slice-specific shape model (SSSM) is built based on a set of simulated training contours. These training contours are generated by applying scaling and local translations to cref to simulate shape deformations that could be caused by respiratory motion. A simulated contour, xs, is computed by the following equation,
| [6] |
where r is a scaling factor (random Gaussian distribution with mean=1 and empirically estimated standard deviation (SD) of 3.5). Similarly, t is a vector of random displacements (Gaussian distribution with zero mean and SD of 2). In this work, a large number of contours (=500) are generated, smoothed, and used to build the patient-specific model. Using the SSSM and the previously-obtained appearance model, each image, Ii (i = 2:K) is then segmented (Figure 2). The resulting sets of LV contours, ci, are used to determine the myocardium region-of-interest (ROI) and align the set of T1-w images to the reference image.
Figure 2.
Pipeline for myocardial segmentation from T1-w images. Appearance model, gref, for a selected reference image, Iref, is combined with the general shape model, built in the training step, to segment Iref in the given T1-w images. One point is inserted manually at the middle of the blood pool of Iref to locate the initial LV shape of the general shape model on Iref. The extracted reference contour, cref, is used to generate the number of simulated LV contours. A new slice-specific shape model (SSSM) is built for that set of T1-w images from the simulated contours. Each of the remaining appearance models, gi, built in the training step is combined with the SSSM to segment the corresponding image, Ii, and generate new contour, ci.
Contour-Based Image Registration
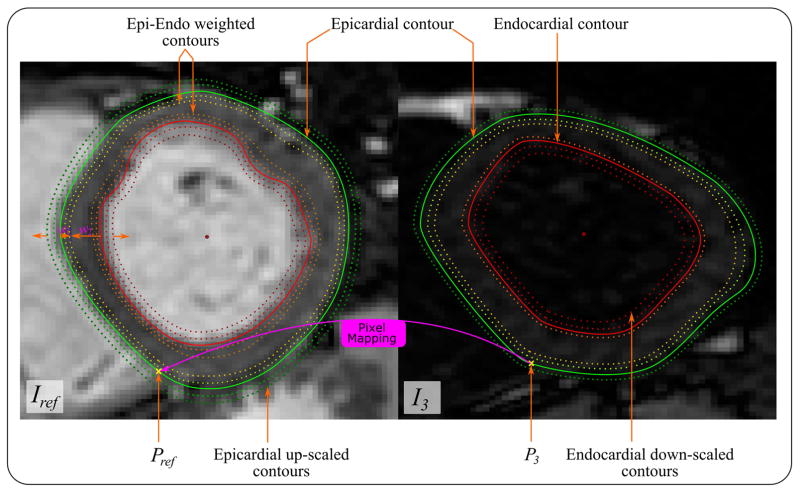
In this step, the given set of T1-w images, Ii, are aligned to the reference image, Iref, based on the estimated displacement vector, d, that aligns the contour ci to cref. The method is based on deforming the images such that the segmented LV contours in each image are aligned to those of the reference image and is achieved through two steps: (1) First, a set of affine transformation parameters is estimated that optimally minimizes the distance between the extracted contour ci and the reference contour, cref via a simple algebraic method (26). The estimated parameters are then used to globally align image Ii to Iref. (2) Secondly, a nonlinear image transformation is applied to Ii such that all the points within the myocardium region are mapped to their counterparts in the reference image. That is, given the displacement vector, d, that maps contour ci to cref, a displacement field (for the entire image, Ii ) is estimated through a linear interpolation algorithm. For this purpose, a mesh of a large number of concentric contours is generated from the segmented epi- and endocardium contours in both Ii and Iref. To generate such mesh, the contours cref and ci are up-sampled at a high rate. Finally, a number of contours are generated according to the following equation,
| [7] |
where, vi is a generated contour on image Ii; and are the endo- and epicardial contours; and w1 and w2 are weighting factors. For a contour generated within the myocardium, 0 < w1 < 1 and w2 = 1 − w1; for a contour within the blood cavity, w1 = 0 and 0 < w2 < 1; and for a contour outside the LV, w2 = 0 and w1 > 1. This results in a mesh of concentric contours (Figure 3). The above meshing operation is done for the image, Ii, and Iref, to obtain two sets of contours (meshes), vi and vref, respectively.
Figure 3.
Non-rigid contour-based registration of a T1-w image (e.g. I3) to its reference image, Iref, starts with defining point correspondence between every pixel on both images. Two insertion points, Pref and P3, are automatically defined at the lower left-right ventricular junction on both Iref and I3 images, respectively, and considered the first points on the extracted contours. A mesh of corresponding points are generated on both images either by: a) the weighted average of both epi- and endocardial contours to generate the myocardial points, b) down-scaling the endocardial contours to generate points in the blood pool, or c) up-scaling the epicardial contour to generate background points. After sampling enough points to cover the whole region of interest, values of I3 are mapped to their corresponding points on Iref, so the registered image with intensity values of I3 and shape of Iref is formed.
The registration process is accomplished by transforming the given image Ii(x, y), into Ĩi(x, y), and aligning it with the reference image. The transformation is represented by,
| [8] |
where W is a B-spline warping that maps the mesh grid vi(x, y) into vref(x, y) (27). The previous steps are applied to all images in the T1-w set.
Algorithm Implementation
The proposed framework has been executed using parallel CPU implementation on MatLab (version 2014b, The MathWorks, Inc., Natick, Massachusetts, United States) using PC of intel-i7 quad-core processor, 16G RAM. In order to maintain the smooth intensity profiles in the segmentation step, each T1-w image in the training and testing phases was convoluted with a Gaussian low-pass filter of size 5×5 pixels and a SD of 2.5. However, after obtaining the myocardial contours, the non-rigid registration step was applied to original T1-w images (i.e. with no smoothing filters applied) to preserve the spatial resolution of the T1 maps. Building the multi-resolution appearance model was done through Bicubic interpolation of the original image to generate half the original image size in the first level of coarse resolution.
Data Acquisition and Evaluation
An informed consent was obtained from each subject and the imaging protocol was approved by the Institutional Review Board. Imaging was performed using a 1.5T Philips Achieva system (Philips Healthcare, Best, The Netherlands) with a 32-channel cardiac coil. T1 mapping was performed in 210 consecutive patients (134 male; age 57 ± 14 years) with known or suspected cardiovascular diseases referred for a clinical cardiac MR exam (available online at https://cardiacmr.hms.harvard.edu/downloads-0). The imaging protocol included free-breathing, respiratory-navigated, slice-interleaved T1 mapping (STONE) sequence (14) with the following parameters: TR/TE = 2.7/1.37 ms, FOV = 360×351 mm2, acquisition matrix = 172×166, voxel size = 2.1×2.1 mm2, linear ordering, SENSE factor = 1.5, slice thickness = 8 mm, bandwidth = 1845 Hz/pixel, diastolic imaging, and flip angle = 70°. Each patient dataset comprises five short axial slices covering the LV from base to apex. At each slice location, eleven T1-w images were acquired at different inversion times, TIi, with i = 1:K, where K = 11, equal to (TIi = ∞, 135, 135+RR, 135+2 RR, …, 135+4 RR, 350, 350+RR, … 350+4 RR. ms) with 3 sec rest-periods between the two inversions, and RR as the duration of the cardiac cycle (14).
The epicardial and endocardial boundaries of all the images in the database (=11550 images) were manually delineated, with each contour starting from a myocardium point closest to the anterior insertion of the right ventricle into the LV. This unified beginning of each contour allowed inherent alignment of the contours and facilitated the contour handling in the training and testing phases as will be described. Each contour was then resampled to a fixed number of points, L = 40, and was subsequently stored for training and testing. For model training purposes, a training dataset of 30 patients (~14% of the whole database) was randomly selected and used to train the model while the remaining 180 patients (testing dataset) were used to evaluate the proposed method.
The manually segmented contours of the T1-w images are used as the reference for evaluating the proposed registration framework by applying the estimated image transformation, W(x, y), to the manually segmented contour, ci, and obtaining the registered contour, c̃i, for comparison to cref.
The Mean Absolute Distance (MAD) and Dice similarity index are used as quantitative measures for the accuracy of the registration process (28,29). The MAD is calculated between the registered myocardial contours, c̃i, and reference contours, cref, as:
| [10] |
where d(p̃l, cref) is the minimum Euclidean distance between the landmark point, p̃l, and cref; p̃l is the lth landmark point in c̃i. Dice similarity index for LV myocardial area is calculated as:
| [11] |
where Hafter and Href refer to the set of pixels within the myocardial area in the T1-w images after registration and the reference image, respectively.
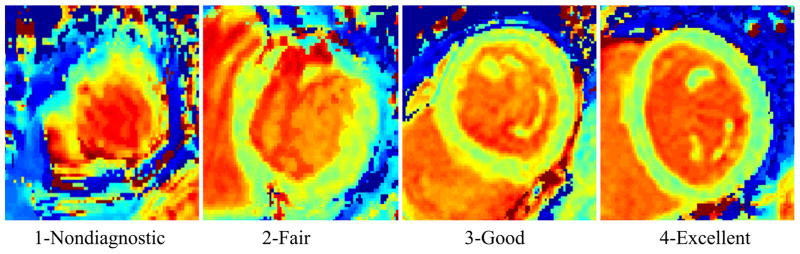
Subjective T1 maps quality was assessed to evaluate the performance of registration. T1 maps were reconstructed before and after motion correction using two-parameter curve fitting of the T1-w images (19). Two experienced readers independently assessed the image quality using a 1–4 score for each segment (19): Score 1- Non-diagnostic/severe motion artifacts: where the T1 map at the myocardium in this score should be completely distorted and T1 cannot be measured at any of its segments; Score 2- Fair/large motion artifacts: the myocardial T1 map could be partially distorted or disappeared but can still be used for diagnosis in some segments; Score 3- Good/small motion artifacts: where the myocardial T1 map completely appeared but a small blur still exists. It is important to differentiate between the blurring caused by the motion artifact and that caused by partial volume effect; the latter can be discovered by checking the T1-w images; Score 4- Excellent/no motion artifacts: the myocardial T1 map should be clear with sharp edges in this score. Figure 4 shows an example of images scored by readers. In this evaluation, all T1 maps have been anonymized and each reader has separately been asked to give a score to each map and determine the corrupted segments in each map, based on the 16-segment model.
Figure 4.
Sample images with different assessment scores of image quality.
Accuracy and precision of T1 mapping within each segment are assessed before and after motion correction w.r.t. T1 values measured using ROI. To calculate the ROI-based T1 values, the manually segmented LV contours are used to automatically select ROI from each myocardial segment in T1-w images. The ROI-based T1 values are estimated by fitting the ROIs pixels within each segment across different T1-w images. Similarly, ROIs are selected from T1 maps before and after correction at each segment to be compared to the ROI-based T1 values. Accuracy and precision of T1 values are calculated as the mean and SD of Diff(T1), respectively; where Diff(T1) is the difference between T1 values in the ROI-based T1 and corrected (or uncorrected) maps. Only T1 values within the range of the myocardial relaxation time (i.e. 900 < T1 < 1400 ms) are included in this analysis (14).
Statistical Analysis
The average number of T1 Maps in each quality score from both readers before and after motion correction, as well as the mean SD, was statistically compared using a paired Student’s t-test. Inter-reader variability of T1 maps scores before and after motion correction was tested using intraclass correlation coefficients. Statistical significance was defined at P-value < 0.05. The average of MAD and Dice index measures before and after motion correction were also compared using a paired Student’s t-test.
Results
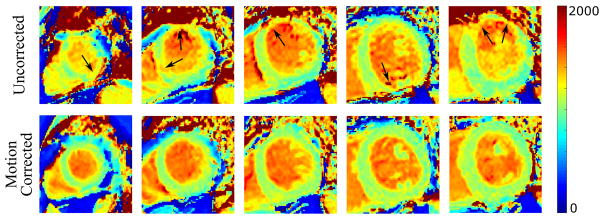
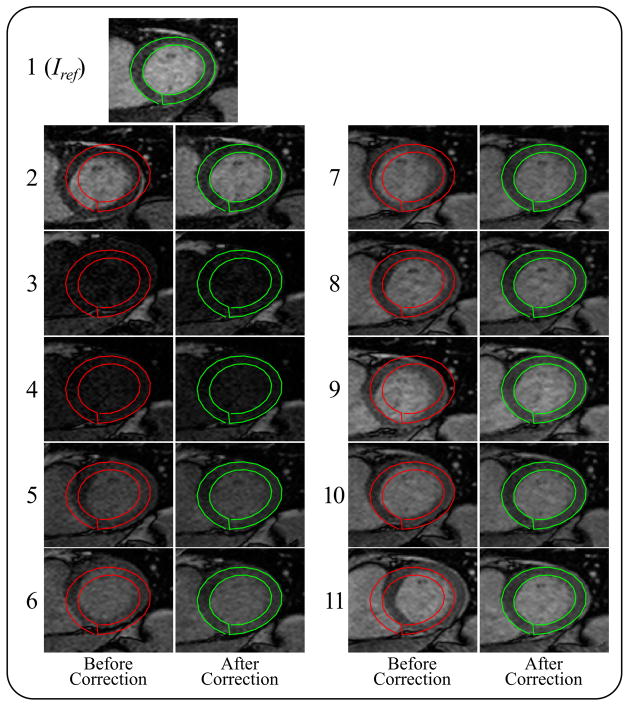
Figure 5 shows an example of T1 maps at five short axial slices before and after motion correction. The corrupted segments of myocardial T1 maps were restored after applying motion correction despite the vague myocardial borders and large motion at apical slice, as indicated by the corrupted T1 map before correction. Registration of eleven T1-w images of the heart at basal slice is illustrated in Figure 6 (and Supporting Figure S1). Inner and outer myocardial contours of the reference image are copied to all other T1-w images in both uncorrected and corrected sets. Improved correspondence of myocardial pixels of all T1-w images showed improved registration, and the proposed framework also showed a consistent performance at images of low myocardial contrast, where displacement and orientation of the myocardium was preserved as indicated in the 3rd and 4th images in Figure 6.
Figure 5.
Myocardial T1 maps at five short axial slices of the left ventricle, from apex to base, before and after motion correction. Black arrows point to corrupted myocardial segments with motion artifacts that have been restored after motion correction.
Figure 6.
A T1-weighted set of eleven images before motion correction (red contours) and after motion correction (green contours). Outer and inner myocardial contours of the reference image Iref are copied to each of the T1-w images to show correspondence with the LV myocardium. The automatic segmentation of the LV myocardium in the reference T1-w image is shown in the top image.
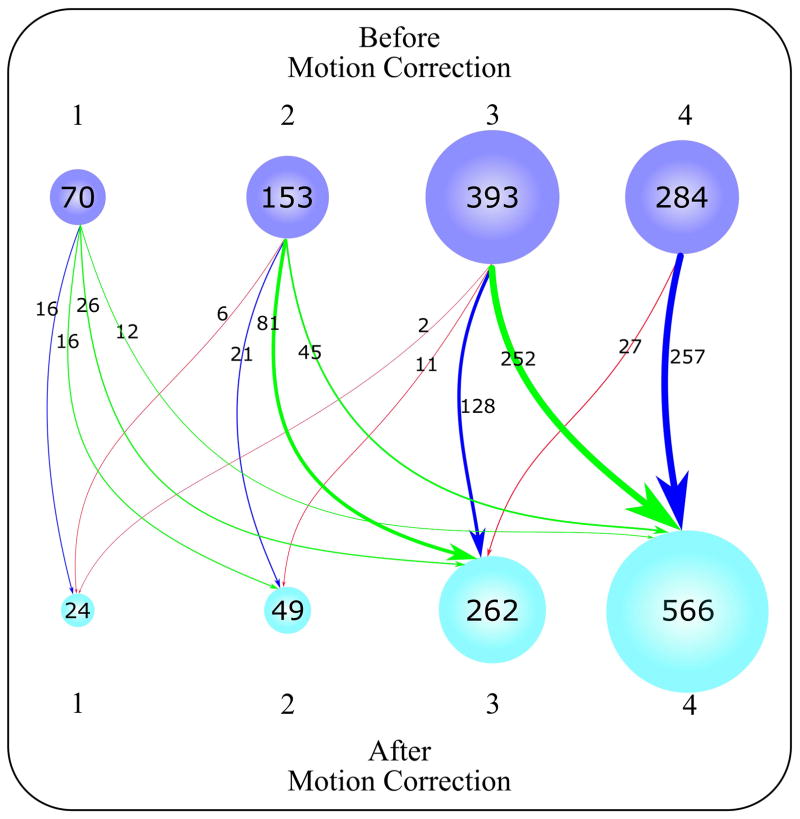
Figure 7 illustrates the qualitative assessment of 900 myocardial T1 maps (Supporting Figure S2). Before motion correction, 68 ± 0.2% of the T1 maps were considered motion-affected maps (i.e. scores of 1, 2 and 3); with 7.5 ± 3.5% of the maps with severe motions artifacts, 17 ± 0.0% with large motions and 43.1 ± 17% with small motions. After motion correction, 37% (P < 0.001) of T1 maps were considered motion-affected maps with only 2.6 ± 1.4% (P < 0.001) with severe motions, 5.5 ± 0.2% (P < 0.001) with large motions, and 29 ± 8.5% (P < 0.001) with small motions. Motion-corrected T1 maps showed improvement in 70 ± 0.2% of the motion-affected maps than before motion correction, while no change occurred in 27 ± 0.3% of the T1 maps before and after correction, and T1 map quality was also degraded for 3.2 ± 0.2% of the maps after correction. T1 map quality at apical slices contributed to 59 ± 10.7% of the non-diagnostic score after motion correction mainly due to large motions and increased partial volume artifacts at LV apex. Intraclass correlation coefficients between both readers of T1 maps scores were 0.86 and 0.82 before and after motion correction, respectively.
Figure 7.
Graphical illustration of T1 map quality distribution (total of 900 T1 maps) based on a four-point scoring system for quality evaluation averaged from two independent readers. Scores 1, 2, 3, and 4 indicate non-diagnostic, fair, good, and excellent T1 map quality, respectively. The number of T1 maps at each score is displayed in purple and cyan for before and after motion correction, respectively. The status of T1 maps before and after correction is represented by arrows of varying thickness, according to the number of T1 maps moving in a given direction. The enhanced T1 maps are represented by green arrows (i.e. T1 maps moved from a lower score to a higher score). The T1 maps that moved from a higher score to a lower score are represented by red arrows, and T1 maps whose scores did not change are represented by blue arrows.
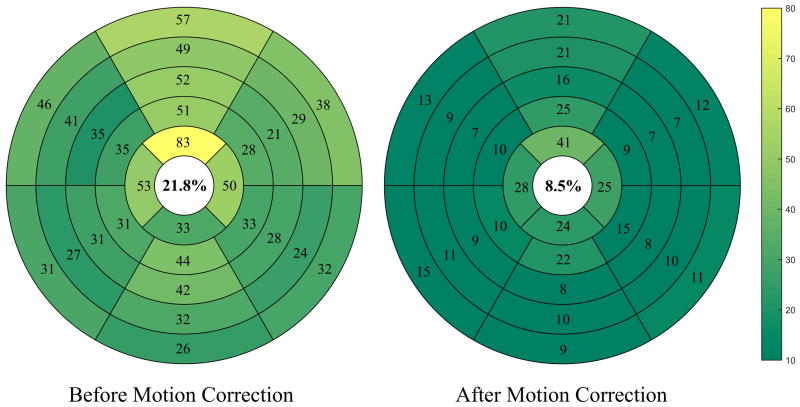
Figure 8 demonstrates the regional analysis of T1 maps before and after motion correction. The bullseye depiction of myocardial segments for five short axial slices indicates a higher number of motion-corrupted myocardial segments before correction, 21.8 ± 10.4% of total segments versus 8.5 ± 4.8% (P < 0.001) after correction. An increased number of corrupted segments are observed at apical slices versus basal or mid-cavity slices before and after correction. The number of corrupted T1 segments at apical slice was significantly decreased after motion correction to 15.6 ± 7% from 30 ± 8.6% (P < 0.001). In addition, the number of corrupted segments in basal and mid-cavity slices significantly decreased to 6.7 ± 4.2% from 19.7 ± 10.8% (P < 0.001) after correction.
Figure 8.
Bullseye representation of the number of corrupted segments of five short axial slices before and after motion correction. The myocardium at basal and mid-cavity slices is divided into six standard segments, while the apical slice is divided into four segments. We see a significant decrease in the number of corrupted T1 myocardial segments (represented by dark color) after motion correction than before motion correction (represented by bright color).
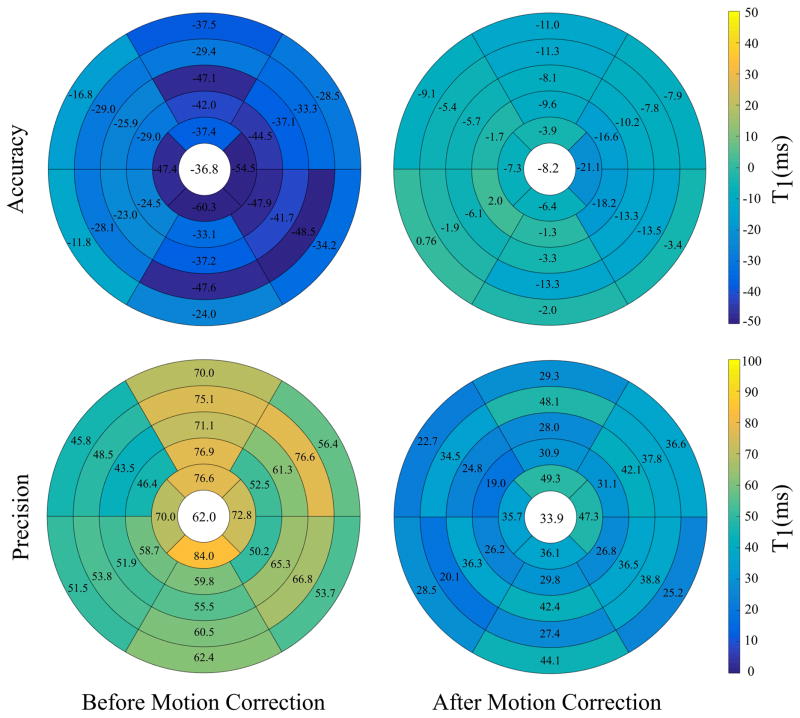
Figure 9 shows the accuracy and precision of the estimated T1 values before and after motion correction w.r.t. ROI-based T1 values within each segment. Motion-corrected T1 maps showed significant increased T1 accuracy and precision compared to uncorrected maps (−8.2±33.9 and −36.8±62 ms, respectively; P < 0.001). MAD distance between extracted and reference myocardial contours significantly decreased from 3.3 ± 1.6 mm to 2.3 ± 0.8 mm (P < 0.001) after motion correction. In addition, the Dice similarity index significantly increased from 0.89 ± 0.08 before correction to 0.94 ± 0.4 (P < 0.001) after the correction. The computation time of the proposed method to register T1-w set of 11 images was ~5s.
Figure 9.
Accuracy and precision of the estimated T1 values, w.r.t. ROI-based T1 values, for each myocardial segment at five slices before and after motion correction. Accuracy and precision of T1 mapping is calculated as the mean and STD of T1 differences between corrected/uncorrected maps and ROI-based T1 at each segment, respectively. Both accuracy and precision are reported for each segment.
Discussion
In this work, we introduced an ASM-based framework for motion correction of myocardial T1 mapping. The proposed framework utilizes a two-step algorithm: segmentation of the myocardial boundaries followed by a contour-based registration of the acquired set of T1-w images. Myocardial segmentation is automatically achieved by applying ASM, which incorporates prior knowledge from trained shape and appearance models. In ASM, the appearance model guides an iterative search process for the myocardium borders in the T1-w images. In each iteration, myocardial contours are estimated and then smoothed through projection on the trained shape model. The computation time of this registration process is 5 sec/11 T1-w images, which is less than that of current conventional intensity-based registration methods (=110 or 10 sec/9 or 8 T1-w images as reported in (19) and (18), respectively).
The high blood-myocardium contrast and brightness variations at different TIs and among different patients make the registration process of T1-w images challenging. In intensity-based methods, a matching algorithm is applied to look for similar intensity patterns across the T1-w images; however, contrast/brightness variations among T1-w images hinder its performance. The variations in brightness at different TIs have been previously addressed by applying variable-brightness tracking of feature points on the myocardium (19); nevertheless, the contrast/brightness also varies for different patients at the same inversion time. In the proposed framework, both types of contrast/brightness variations are handled through two steps: a) building multiple appearance models at different TIs to take into account the variations among different T1-w images, and b) building each appearance model from actual patient data to capture the intra-patient T1 variations.
Different patterns of blood-myocardium contrast can still be noticed at the same TI interval (i.e. intervals near zero-crossing of MR recovery, e.g. at i =3 or 4) in different patients: brighter blood than myocardium, fainter blood than myocardium, or blood and myocardium with equal brightness. Modeling such contrast patterns using one appearance model is challenging. However, ASM is able to represent them in different modes-of-variation vectors in the training phase. Additionally, the search algorithm, which depends on projecting the intensity profiles of testing images onto the appearance model, successfully recognizes these different patterns during segmentation. This projection-based matching criteria shows better performance than correlation and other edge detection methods in previous studies (21,23).
In our study, we chose a training dataset of 30 patients (1650 T1 images). The optimal size of training dataset for ASM is not fixed and depends on the complexity and inter-patient variations of the LV. In a pilot study, we investigated the impact of choosing different sizes of training datasets on registration performance. The proposed models were constructed with a dataset representing 10 to 60 patients and evaluated the performance of the registration using MAD and Dice index. The resulting indices show that the performance reaches a plateau with 30 patients and there is no significance improvement in performance (Supporting Figure S3). Further studies are warranted to investigate the optimal size of the training datasets.
The contour-based image registration allows efficient utilization of both rigid and non-rigid transformations estimated from the extracted contours. However, it is crucial to preserve the point-correspondence among all extracted myocardial contours of T1-w images. ASM maintains a consistent arrangement of landmark points similar to that used in training (21,23). Since all the manually delineated contours extracted in training have the same contour point arrangement, the resulting contours in the testing step have the same arrangement. In this work, we used the lower insertion point of LV and RV as a starting point in the manual delineation, due to its fixed anatomical characteristics, and a fixed number of landmark points selected with equidistant steps from each contour.
The qualitative assessment showed motion artifacts in 68% of the T1 maps in our dataset before correction. After applying the proposed methods, the number of motion-affected maps significantly decreased. T1 maps with severe- and large-motions showed a significant decrease after motion correction, which indicates the ability of the proposed method to capture large motions of the myocardium. Additionally, 63 ± 0.3% of the small-motion scored maps completely recovered and were assigned the no-motion score after correction, indicating the ability of the proposed methods to correct for fine myocardial deformation caused by cardiac motion in the through-plan direction. However, small motion artifacts were noticed after correction in 10.6 ± 4.5% of the uncorrected motion-free maps (i.e. represents ~28 T1 maps) due to inaccurate segmentation of the LV myocardium. Qualitative analysis of myocardial segments also showed significant decrease of the number of motion-corrupted segments down to 8.5% of all segments, with about 38.4 ± 5.3% of the corrupted segments at the apical slice. The increased number of motion- corrupted segments at apical slices is mainly due to the increased myocardial motions at the LV apex and degraded myocardial contrast caused by partial volume artifacts. We also noticed an increased average number of motion-corrupted segments for both corrected and uncorrected maps at LV inferior-wall in all slices: basal, mid-cavity, and apical as previously reported (10).
The high variability of myocardial morphology caused by different diseases (e.g. hypertrophic and dilated cardiomyopathies) poses a challenge in building the shape model. To circumvent this problem, patients with different cardiac diseases should be sufficiently represented in the training dataset. In the proposed registration approach, the contours for epicardium and endocardium borders are extended and aligned instead of pixel-by-pixel alignment. One potential disadvantage of this approach, compared to intensity based image registration, is that only contour information is used in registration. The registration of contours does not automatically guarantee alignment of myocardial pixels and may cause registration error within the myocardium. Also, in the proposed model-based framework, new training of shape and appearance models is needed for images acquired with different orientations (e.g. long axis view) or different ranges of T1 (e.g. post-contrast T1 mapping). Although, the multi-resolution implementation of the proposed framework alleviates large motions of the myocardium, there are still trade-offs in the model flexibility for capturing the fine variations of the myocardium and capturing the large motions. The parallel implementation of this framework is found to be effective because every T1-w image within a given case (except for the reference image) can be processed independently from others. Thus, all T1-w images are segmented and registered simultaneously on multiple processing cores, leading to a shorter processing time by a factor of the number of CPU cores (i.e. one forth in our experiments) to the regular implementation time.
Conclusions
The proposed method for non-rigid registration of T1-w images allows T1 measurements in more myocardial segments by eliminating motion-induced T1 estimation errors in the myocardium segments.
Supplementary Material
Supporting Figure S1. A T1-weighted set of eleven images before (with red contours) and after (with green contours) motion correction. Outer and inner myocardial contours of the reference image Iref are copied to each of the T1-w images to show correspondence with the LV myocardium. The automatic segmentation of the LV myocardium in the reference T1-w image is shown in the top image.
Supporting Figure S2. Graphical illustration of T1 map quality distribution (total of 900 T1 maps) based on a four-point scoring system for quality evaluation averaged from two independent readers. Scores 1, 2, 3, and 4 indicate non-diagnostic, fair, good, and excellent T1 map quality, respectively. The number of T1 maps at each score is displayed in purple and cyan for before and after motion correction, respectively. The status of T1 maps before and after correction is represented by arrows of varying thickness, according to the number of T1 maps moving in a given direction. The enhanced T1 maps are represented by green arrows (i.e. T1 maps moved from a lower score to higher score). The T1 maps that moved from higher score to lower score are represented by red arrows, and T1 maps whose scores did not change are represented by blue arrows.
Supporting Figure S3. Effect of increasing training dataset size on the performance of the proposed model. Mean and standard deviation for Dice index and Mean Absolute Distance (MAD) are shown on both vertical axes with increasing training dataset size from 10 to 60 patients. Thirty patients were selected as the optimal size of the training dataset as no significant increase of the model’s performance is recorded with larger size.
Acknowledgments
Research reported in this publication was supported by National Institutes of Health under award numbers: 1R21HL127650, 1R01HL129185, 1R01HL129157, and AHA 15EIA22710040. The authors would like to thank Jennifer Rodriguez for editorial correction.
References
- 1.Weber KT, Brilla CG. Pathological hypertrophy and cardiac interstitium. Fibrosis and renin-angiotensin-aldosterone system. Circulation. 1991;83:1849–65. doi: 10.1161/01.cir.83.6.1849. [DOI] [PubMed] [Google Scholar]
- 2.Lin L-Y, Wu C-K, Juang J-MJ, Wang Y-C, Su M-YM, Lai L-P, Hwang J-J, Chiang F-T, Tseng W-YI, Lin J-L. Myocardial Regional Interstitial Fibrosis is Associated With Left Intra-Ventricular Dyssynchrony in Patients With Heart Failure: A Cardiovascular Magnetic Resonance Study. Sci Rep. 2016;6:20711. doi: 10.1038/srep20711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beltrami CA, Finato N, Rocco M, Feruglio GA, Puricelli C, Cigola E, Sonnenblick EH, Olivetti G, Anversa P. The cellular basis of dilated cardiomyopathy in humans. J Mol Cell Cardiol. 1995;27:291–305. doi: 10.1016/s0022-2828(08)80028-4. [DOI] [PubMed] [Google Scholar]
- 4.Iles L, Pfluger H, Phrommintikul A, Cherayath J, Aksit P, Gupta SN, Kaye DM, Taylor AJ. Evaluation of Diffuse Myocardial Fibrosis in Heart Failure With Cardiac Magnetic Resonance Contrast-Enhanced T1 Mapping. J Am Coll Cardiol. 2008;52:1574–1580. doi: 10.1016/j.jacc.2008.06.049. [DOI] [PubMed] [Google Scholar]
- 5.Jellis CL, Kwon DH. Myocardial T1 mapping: modalities and clinical applications. Cardiovasc Diagn Ther. 2014;4:126–37. doi: 10.3978/j.issn.2223-3652.2013.09.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Germain P, El Ghannudi S, Jeung M-Y, Ohlmann P, Epailly E, Roy C, Gangi A. Native T1 mapping of the heart - a pictorial review. Clin Med Insights Cardiol. 2014;8:1–11. doi: 10.4137/CMC.S19005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roujol S, Weingärtner S, Foppa M, Chow K, Kawaji K, Ngo LH, Kellman P, Manning WJ, Thompson RB, Nezafat R. Accuracy, Precision, and Reproducibility of Four T1 Mapping Sequences: A Head-to-Head Comparison of MOLLI, ShMOLLI, SASHA, and SAPPHIRE. Radiology. 2014;272:683–689. doi: 10.1148/radiol.14140296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shao J, Liu D, Sung K, Nguyen K-L, Hu P. Accuracy, precision, and reproducibility of myocardial T1 mapping: A comparison of four T1 estimation algorithms for modified look-locker inversion recovery (MOLLI) Magn Reson Med. 2016 doi: 10.1002/mrm.26565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pica S, Sado DM, Maestrini V, et al. Reproducibility of native myocardial T1 mapping in the assessment of Fabry disease and its role in early detection of cardiac involvement by cardiovascular magnetic resonance. J Cardiovasc Magn Reson. 2014;16:99. doi: 10.1186/s12968-014-0099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bellm S, Basha TA, Shah RV, Murthy VL, Liew C, Tang M, Ngo LH, Manning WJ, Nezafat R. Reproducibility of myocardial T1 and T 2 relaxation time measurement using slice-interleaved T 1 and T 2 mapping sequences. J Magn Reson Imaging. 2016;44:1159–1167. doi: 10.1002/jmri.25255. [DOI] [PubMed] [Google Scholar]
- 11.Messroghli DR, Radjenovic A, Kozerke S, Higgins DM, Sivananthan MU, Ridgway JP. Modified Look-Locker inversion recovery (MOLLI) for high-resolutionT1 mapping of the heart. Magn Reson Med. 2004;52:141–146. doi: 10.1002/mrm.20110. [DOI] [PubMed] [Google Scholar]
- 12.Piechnik SK, Ferreira VM, Dall’Armellina E, Cochlin LE, Greiser A, Neubauer S, Robson MD. Shortened Modified Look-Locker Inversion recovery (ShMOLLI) for clinical myocardial T1-mapping at 1.5 and 3 T within a 9 heartbeat breathhold. J Cardiovasc Magn Reson. 2010;12:69. doi: 10.1186/1532-429X-12-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chow K, Flewitt JA, Green JD, Pagano JJ, Friedrich MG, Thompson RB. Saturation recovery single-shot acquisition (SASHA) for myocardial T1 mapping. Magn Reson Med. 2014;71:2082–2095. doi: 10.1002/mrm.24878. [DOI] [PubMed] [Google Scholar]
- 14.Weingärtner S, Roujol S, Akçakaya M, Basha TA, Nezafat R. Free-breathing multislice native myocardial T1 mapping using the slice-interleaved T1 (STONE) sequence. Magn Reson Med. 2015;74:115–124. doi: 10.1002/mrm.25387. [DOI] [PubMed] [Google Scholar]
- 15.Nekolla S, Gneiting T, Syha J, Deichmann R, Haase A. T1 maps by K-space reduced snapshot-FLASH MRI. J Comput Assist Tomogr. 16:327–32. doi: 10.1097/00004728-199203000-00031. [DOI] [PubMed] [Google Scholar]
- 16.Kellman P, Hansen MS. T1-mapping in the heart: accuracy and precision. J Cardiovasc Magn Reson. 2014;16:2. doi: 10.1186/1532-429X-16-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marks B, Mitchell DG, Simelaro JP. Breath-holding in healthy and pulmonary-compromised populations: effects of hyperventilation and oxygen inspiration. J Magn Reson Imag. 7:595–7. doi: 10.1002/jmri.1880070323. [DOI] [PubMed] [Google Scholar]
- 18.Xue H, Shah S, Greiser A, Guetter C, Littmann A, Jolly M-P, Arai AE, Zuehlsdorff S, Guehring J, Kellman P. Motion correction for myocardial T1 mapping using image registration with synthetic image estimation. Magn Reson Med. 2012;67:1644–55. doi: 10.1002/mrm.23153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roujol S, Foppa M, Weingärtner S, Manning WJ, Nezafat R. Adaptive registration of varying contrast-weighted images for improved tissue characterization (ARCTIC): Application to T 1 mapping. Magn Reson Med. 2015;73:1469–1482. doi: 10.1002/mrm.25270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ordas S, Boisrobert L, Huguet M, Frangi AF. Computers in Cardiology. IEEE; 2003. Active shape models with invariant optimal features (IOF-ASM) application to cardiac MRI segmentation; pp. 633–636. [Google Scholar]
- 21.Van Ginneken B, Frangi AF, Staal JJ, Ter Haar Romeny BM, Viergever MA. Active shape model segmentation with optimal features. IEEE Trans Med Imaging. 2002;21:924–933. doi: 10.1109/TMI.2002.803121. [DOI] [PubMed] [Google Scholar]
- 22.El-Rewaidy H, Ibrahim E-S, Fahmy AS. Segmentation of the right ventricle in MRI images using a dual active shape model. IET Image Process. 2016;10:717–723. [Google Scholar]
- 23.Cootes TFT, Taylor CCJ, Cooper DDH, Graham J. Active Shape Models-Their Training and Application. Comput Vis Image Underst. 1995;61:38–59. [Google Scholar]
- 24.Cootes TF, Taylor CJ. Active Shape Model Search using Local Grey-Level Models: A Quantitative Evaluation. Procedings of the British Machine Vision Conference; 1993; British Machine Vision Association; 1993. pp. 64.1–64.10. [Google Scholar]
- 25.Gower JC. Generalized procrustes analysis. Psychometrika. 1975;40:33–51. [Google Scholar]
- 26.Meserve B. Fundamental concepts of geometry. Cambridge Mass: Addison-Wesley; 1953. [Google Scholar]
- 27.Rueckert D, Sonoda LI, Hayes C, Hill DLG, Leach MO, Hawkes DJ. Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans Med Imaging. 1999;18:712–21. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- 28.Dice LR. Measures of the Amount of Ecologic Association Between Species. Ecology. 1945;26:297–302. [Google Scholar]
- 29.Babalola KO, Patenaude B, Aljabar P, Schnabel J, Kennedy D, Crum W, Smith S, Cootes T, Jenkinson M, Rueckert D. An evaluation of four automatic methods of segmenting the subcortical structures in the brain. Neuroimage. 2009;47:1435–1447. doi: 10.1016/j.neuroimage.2009.05.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Figure S1. A T1-weighted set of eleven images before (with red contours) and after (with green contours) motion correction. Outer and inner myocardial contours of the reference image Iref are copied to each of the T1-w images to show correspondence with the LV myocardium. The automatic segmentation of the LV myocardium in the reference T1-w image is shown in the top image.
Supporting Figure S2. Graphical illustration of T1 map quality distribution (total of 900 T1 maps) based on a four-point scoring system for quality evaluation averaged from two independent readers. Scores 1, 2, 3, and 4 indicate non-diagnostic, fair, good, and excellent T1 map quality, respectively. The number of T1 maps at each score is displayed in purple and cyan for before and after motion correction, respectively. The status of T1 maps before and after correction is represented by arrows of varying thickness, according to the number of T1 maps moving in a given direction. The enhanced T1 maps are represented by green arrows (i.e. T1 maps moved from a lower score to higher score). The T1 maps that moved from higher score to lower score are represented by red arrows, and T1 maps whose scores did not change are represented by blue arrows.
Supporting Figure S3. Effect of increasing training dataset size on the performance of the proposed model. Mean and standard deviation for Dice index and Mean Absolute Distance (MAD) are shown on both vertical axes with increasing training dataset size from 10 to 60 patients. Thirty patients were selected as the optimal size of the training dataset as no significant increase of the model’s performance is recorded with larger size.