Abstract
Mitogen-activated protein kinase kinase kinase 1 (MAP3K1) regulates numerous intracellular signaling pathways involved in inflammation and apoptosis. We hypothesized that genetic variation in MAP3K1 might be associated with outcomes in patients with acute respiratory distress syndrome (ARDS), and that these variants would alter MAP3K1-mediated changes in inflammation and transcriptional regulation. To test this hypothesis, we genotyped single-nucleotide polymorphisms covering linkage disequilibrium bins in MAP3K1 in 306 subjects with ARDS from the ARDSNet FACTT (Fluid and Catheter Treatment Trial) study, and tested for associations between MAP3K1 single-nucleotide polymorphisms and ventilator-free days (VFDs) and mortality. We then validated these associations in a separate cohort of 241 patients with ARDS from Harborview Medical Center (Seattle, WA). We found the variant allele of rs832582 (MAP3K1906Val) was significantly associated with decreased VFDs using multivariate linear regression (−6.1 d, false discovery rate = 0.06) in the FACTT cohort. In the Harborview Medical Center cohort, subjects homozygous for MAP3K1906Val also had decreased VFDs (−15.1 d, false discovery rate < 0.01), and increased 28-day mortality (all subjects homozygous for the rare allele died). In whole blood stimulated with various innate immune agonists ex vivo, MAP3K1906Val was associated with increased IL-1β, IL-6, IL-8, monocyte chemoattractant protein 1, and TNF-α production. Transcriptome analysis of whole blood stimulated with Toll-like receptor 4 agonist ex vivo demonstrated enrichment of inflammatory gene sets in subjects homozygous for MAP3K1906Val. Our findings show a robust association between the variant allele of rs832582 (MAP3K1906Val) and decreased VFDs in patients with ARDS and suggest that this variant may predispose individuals to a greater inflammatory response.
Keywords: acute respiratory distress syndrome, mitogen-activated protein kinase kinase kinase 1
Clinical Relevance
We have identified a robust association between the variant allele of rs832582 (mitogen-activated protein kinase kinase kinase 1906Val) and decreased ventilator-free days in patients with acute respiratory distress syndrome (ARDS), and demonstrated that this variant may predispose individuals to a greater inflammatory response. These findings may lead to a greater understanding of the pathophysiology in ARDS and ultimately to the identification of therapies directed at the underlying mechanisms of the syndrome.
Acute respiratory distress syndrome (ARDS) is a complex clinical syndrome characterized by the development of increased alveolar–capillary permeability, leakage of proteinaceous fluid into the alveolar space, and an influx of inflammatory cells and mediators into the lung (1). We have previously shown that a commonly occurring FAS (CD95) haplotype is associated with ARDS risk and with higher levels of Fas mRNA in response to LPS stimulation (2). The Fas pathway is involved in the regulation of cellular apoptosis and inflammatory responses in multiple cell types in the lung, including epithelial cells, alveolar macrophages, and neutrophils (3).
Mitogen-activated protein kinase kinase kinase 1 (MAP3K1) (also known as MAP kinase/Erk kinase kinase-1) is a highly conserved member of the MAP kinase family, and plays a central role in orchestrating the cellular response to Fas (4). MAP3K1 regulates the activation of multiple transcription factors involved in inflammation and cell fate, including activator protein 1 (AP-1), NF-κB, and p53 (5). Independent of Fas signaling, MAP3K1 has been shown to coordinate cellular responses to multiple different stimuli including cell stretch, cytoskeletal disruption, and inflammation (6–9), all physiologic processes that have been implicated in ARDS. MAP3K1 has multiple nonsynonymous coding variants that in prior studies have been associated with inflammatory diseases including Crohn’s disease (10), asthma (11), and Langerhans cell histiocytosis (12).
Due to the integral downstream role MAP3K1 plays in Fas signaling, as well as its relationship with multiple inflammatory pathways that are implicated in ARDS, we hypothesized that genetic variation in MAP3K1 might be associated with outcomes in patients with ARDS, and that these variants would alter MAP3K1-mediated changes in inflammation and transcriptional regulation. To test this hypothesis, we performed a two-stage gene association study, and then conducted a functional evaluation of the top hits from those gene association studies. Some of the results of these studies have been previously reported in the form of an abstract (13).
Methods
Study Populations
Fluid and Catheter Treatment Trial subjects
In our discovery cohort, we genotyped white patients with ARDS enrolled in the FACTT (Fluid and Catheter Treatment Trial) (14) who provided informed consent for genetic studies (n = 306). Eligible patients were intubated, receiving positive-pressure ventilation, and qualified for ARDS, as defined by American European Consensus Criteria (15).
Harborview Medical Center Systemic Inflammatory Response Syndrome validation cohort
Subjects were prospectively enrolled from the Harborview Medical Center (HMC) intensive care unit if they met criteria for systemic inflammatory response syndrome (SIRS), and were not admitted with major trauma, intracranial hemorrhage, immunosuppression, or a current diagnosis of cancer. Subjects were genotyped and followed for the development of organ dysfunction, including ARDS (n = 241).
Healthy cohort
Nonsmoking healthy subjects (n = 346) between 18 and 65 years of age were recruited from the metropolitan Seattle area. All studies were approved by the Human Subjects Division at the University of Washington (Seattle, WA).
Identification and Genotyping of Single-Nucleotide Polymorphisms
We selected single-nucleotide polymorphisms (SNPs) in MAP3K1 from multiple databases, and generated a final list of nine linkage disequilibrium (LD) bin (minor allele frequency cutoff = 5%, r2 = 0.8) tagging SNPs (tagSNPs) covering the entire MAP3K1 gene. Complete details on our LD bin tagSNPs approach and genotyping assays can be found in the data supplement.
Statistical Analysis
We used linear regression to test for associations between MAP3K1 tagSNPs and ventilator-free days (VFDs) and logistic regression to test for associations between the tagSNPs and 28-day mortality. Analyses in the discovery set were performed using recessive, additive, and dominant models, and were adjusted for age, sex, ARDS risk factors, and FACTT treatment arm. We used a false discovery rate (FDR) threshold of 0.1 to identify statistical significance (16). Significant associations identified in the discovery cohort were carried forward for analysis in the validation set using the recessive model. Complete details on our statistical analyses can be found in the data supplement.
Inflammatory Cytokine Production and Transcriptome Analysis in Subjects with MAP3K1 Variants
Healthy subjects (n = 346) were genotyped using an Illumina Human 1M Beadchip array (Illumina). Whole-blood samples procured from these subjects were treated with the following Toll-like receptor (TLR) ligands ex vivo: Pam3CSK4 (TLR1/2 agonist), Yersinia pestis LPS (TLR4 agonist), Salmonella minnesota Re595 LPS (TLR4 agonist), R848 (TLR7/8 agonist), and control media. Concentrations of IL-1β, IL-1 receptor antagonist (IL-1ra), IL-6, IL-8, monocyte chemoattractant protein (MCP)-1, and TNF-α were measured in supernatants by fluorescent bead–based immunoassays. RNA was purified in a subset of the samples (n = 252), and gene expression was measured using Illumina Quad660 gene expression microarrays. Complete details on our cytokine and gene expression assays can be found in the data supplement.
Site-Directed Mutagenesis of MAP3K1 and Functional Analysis
We performed transfection assays of human embryonic kidney cells 293 (HEK-293) using plasmids coding for MAP3K1 variants, and measured MAP3K1 transcription, protein expression, cell viability, and metabolism. For reporter gene assays, cotransfection assays of HEK-293 cells were performed with plasmids coding for MAP3K1 variants and luciferase reporter plasmids for p53, NF-κB, and AP-1. Complete details on cell culturing, plasmid constructs, site-directed mutagenesis, transfections, luciferase reporter experiments, and functional assays can be found in the data supplement.
Results
Characteristics of Patients and Genotype Frequencies
Characteristics of subjects in the discovery set from FACTT and the validation set from HMC SIRS are shown in Table 1. In the discovery set, 306 subjects were genotyped; however, eight were removed because of a low genotyping rate (<80% valid genotype calls). In the validation set, all 241 subjects with ARDS were genotyped for the MAP3K1 nonsynonymous coding SNP, rs832582, and were subsequently included in the final analysis. The genotype frequencies of subjects included in our final analyses are shown in Table 2.
Table 1.
Patient Demographics and Clinical Characteristics
| Characteristic | FACTT (n = 306) | HMC SIRS (n = 241) |
|---|---|---|
| Patient age, mean (SD) | 50 (15) | 56 (15) |
| Male patients, n (%) | 157 (51.3) | 172 (71.4) |
| VFDs, mean (SD) | 14 (10) | 13 (11) |
| VFDs, median (IQR) | 18 (0–22) | 17 (0–23) |
| 28-d mortality, n (%) | 56 (18.3) | 59 (24.5) |
| APACHE III mean score (SD) | 92 (29) | 77 (25) |
| ARDS risk factor, n (%)* | ||
| Aspiration | 52 (17.0) | 53 (22.0) |
| Pneumonia | 138 (45.1) | 97 (40.3) |
| Sepsis | 67 (21.9) | 208 (86.3) |
| Trauma | 24 (7.8) | 0 |
| Multiple Transfusions | 4 (1.3) | 8 (3.3) |
| Other | 21 (6.8) | 26 (10.8) |
| Comorbidities, n (%) | ||
| Diabetes | 55 (18) | 62 (26) |
| HIV or AIDS | 15 (5) | 0 |
| Cirrhosis | 7 (2) | 36 (15) |
| Solid tumors | 4 (1) | 0 |
| Leukemia/Lymphoma | 6 (2) | 0 |
| Immunosuppression | 23 (7) | 0 |
Definition of abbreviations: APACHE = Acute Physiology and Chronic Health Evaluation; ARDS = acute respiratory distress syndrome; FACTT = Fluid and Catheter Treatment Trial; HMC = Harborview Medical Center; IQR = interquartile range; SIRS = systemic inflammatory response syndrome; VFDs = ventilator-free days.
Recorded as primary risk factor in FACTT and as present or not in HMC SIRS.
Table 2.
Genomic Marker Information and Statistics in Subjects from the Fluid and Catheter Treatment Trial and Harborview Medical Center Systemic Inflammatory Response Syndrome Cohorts
| Genotype Counts* |
||||||||
|---|---|---|---|---|---|---|---|---|
| SNP | Function | Amino Acid† | n (%) | MAF FACTT | HWE χ2P Value | MAF HapMap | ||
| FACTT cohort | ||||||||
| rs832582 | Exon | Val (V) | GG, 16 (5.4) | AG, 89 (30.2) | AA, 190 (64.4) | 0.21 | 0.20 | 0.16 |
| rs832583 | Exon | Arg (R) | AA, 14 (4.8) | CA, 85 (29.0) | CC, 194 (66.2) | 0.20 | 0.24 | 0.16 |
| rs702689 | Exon | Asp (D) | GG, 33 (11.1) | AG, 122 (41.1) | AA, 142 (47.8) | 0.32 | 0.38 | 0.28 |
| rs726501 | Intron | AA, 3 (1.0) | GA, 51 (17.8) | GG, 232 (81.1) | 0.10 | 0.92 | 0.13 | |
| rs866223 | Intron | AA, 47 (17.2) | GA, 136 (49.6) | GG, 91 (33.2) | 0.42 | 0.75 | 0.34 | |
| rs1867731 | Intron | GG, 47 (15.8) | CG, 138 (46.5) | CC, 112 (37.7) | 0.39 | 0.68 | 0.34 | |
| rs832575 | Exon | Thr (T) | GG, 9 (3.1) | AG, 67 (23.0) | AA, 215 (73.9) | 0.15 | 0.19 | 0.11 |
| rs33330 | Intron | AA, 18 (6.3) | GA, 138 (47.9) | GG, 132 (45.8) | 0.30 | 0.02 | 0.41 | |
| rs33323 | Intron | CC, 46 (15.9) | GC, 132 (45.7) | GG, 111 (38.4) | 0.39 | 0.52 | 0.34 | |
| HMC SIRS cohort | ||||||||
| rs832582 | Exon | Val (V) | GG, 5 (2.1) | AG, 73 (31.3) | AA, 155 (66.5) | 0.18 | 0.28 | 0.16 |
Definition of abbreviations: Arg = arginine; Asp = aspartate; FACTT = Fluid and Catheter Treatment Trial; HMC = Harborview Medical Center; HWE = Hardy–Weinberg equilibrium; MAF = minor allele frequency; HapMap = CEU population from HapHap Phase 3 CEU population; SIRS = systemic inflammatory response syndrome; SNP = single-nucleotide polymorphism; Thr = threonine; Val = valine.
n = 298 for the FACTT cohort and n = 241 for the HMC SIRS cohort.
Amino acid represents the amino acid corresponding with the minor allele.
The Variant Allele of rs832582 Is Associated with Decreased VFDs
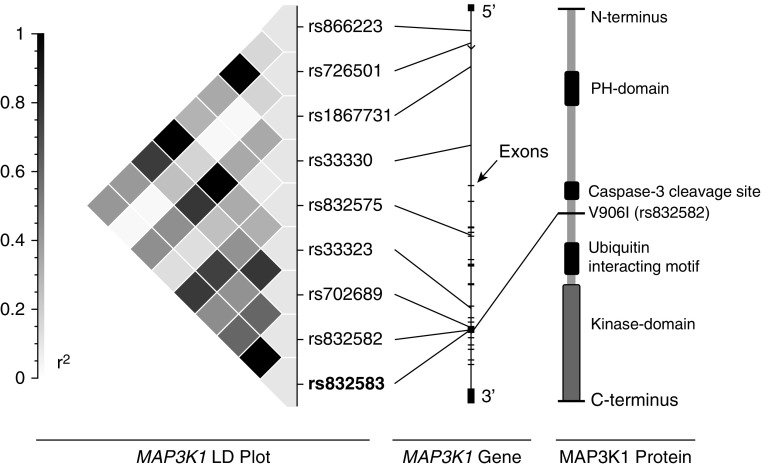
We first tested whether genetic variation in MAP3K1 was associated with VFDs in patients with ARDS. We genotyped nine coding and noncoding tagSNPs covering the MAP3K1 gene in subjects with ARDS from FACTT, and tested for associations between these SNPs and VFDs using linear regression (using recessive, additive, and dominant models). Subjects in FACTT who were homozygous for the minor allele (genotype: GG, MAP3K1906Val) of rs832582 experienced six fewer VFDs compared with subjects who carried at least one copy of the major allele (genotype: AG or AA, MAP3K1906Ile) (Table 3). The strongest associations were identified assuming a recessive model (analyses using additive and dominant statistical models are shown in Tables E1 and E2 in the data supplement). rs832582 is a nonsynonymous SNP that results in an amino acid substitution of valine for isoleucine at amino acid position 906 (Figure 1), and has a minor allele frequency of 0.16 in the HapMap CEU population.
Table 3.
Mitogen-activated Protein Kinase Kinase Kinase 1 Single-Nucleotide Polymorphism Genotype Associations with Ventilator-Free Days in the Fluid and Catheter Treatment Trial and Harborview Medical Center Systemic Inflammatory Response Syndrome Cohorts
| SNP | VFDs Rare Allele Mean (±SD) | VFDs Common Allele Mean (±SD) | β (±SD) | P Value (Recessive) | FDR |
|---|---|---|---|---|---|
| FACTT cohort | |||||
| rs832582 | 8.3 (9.1) | 14.2 (9.7) | −6.1 (2.4) | 0.01 | 0.06 |
| rs832583 | 9.5 (9.2) | 14.1 (9.7) | −4.7 (2.6) | 0.07 | 0.24 |
| rs702689 | 11.8 (9.6) | 14.2 (9.7) | −2.5 (1.8) | 0.16 | 0.41 |
| rs726501 | 20.3 (2.6) | 13.9 (9.8) | 7.4 (5.5) | 0.18 | 0.36 |
| rs866223 | 13.3 (9.6) | 14.2 (9.7) | −0.6 (1.5) | 0.68 | 1.00 |
| rs1867731 | 13.3 (9.6) | 14.0 (9.8) | −0.6 (1.5) | 0.71 | 1.00 |
| rs832575 | 12.6 (9.6) | 14.1 (9.7) | −0.9 (3.2) | 0.78 | 0.98 |
| rs33330 | 15.9 (9.4) | 14.1 (9.6) | 0.3 (2.3) | 0.90 | 1.00 |
| rs33323 | 13.6 (9.5) | 14.0 (9.8) | −0.2 (1.5) | 0.92 | 0.92 |
| HMC SIRS cohort | |||||
| rs832582 | 0 (0) | 13.5 (10.4) | −15.1 (4.6) | < 0.01 | < 0.01 |
Definition of abbreviations: β = change in ventilator-free days of homozygotes for the rare allele from carriers of the common allele (recessive model); FACTT = Fluid and Catheter Treatment Trial; FDR = false discovery rate; HMC = Harborview Medical Center; SIRS = Systemic Inflammatory Response Syndrome; SNP = single-nucleotide polymorphism; VFD = ventilator-free days.
Figure 1.
Location of the nonsynonymous coding single-nucleotide polymorphism (SNP) rs832582 in mitogen-activated protein kinase kinase kinase 1 (MAP3K1). Left: linkage disequilibrium (LD) plot showing r2 values for pairwise LD analysis for the FACTT (Fluid and Catheter Treatment Trial) white cohort (plot created using SVS, Golden Helix, Bozeman, MT). Center: diagram of the MAP3K1 gene with exons and the locations of the nine LD bin tagging SNP (tagSNPs) that we identified. Right: cartoon of the MAP3K1 protein with the coding SNP identified along the noncatalytic regulatory segment of the protein. MAP3K1 contains a Pleckstrin homology (PH) domain as well as a kinase domain.
We next sought to validate the association between rs832582 and decreased VFDs that we observed in the FACTT subjects in an independent cohort of patients with ARDS (HMC SIRS). We observed that subjects who were homozygous for the MAP3K1906Val allele at rs832582 again had fewer VFDs (slope, −15.1; SE, ±4.61; FDR < 0.01) compared with subjects that carried at least one copy of the major allele, MAP3K1906Ile (Table 3).
MAP3K1 Genotype and Mortality in Patients with ARDS
We next tested for associations between rs832582 and 28-day mortality in subjects with ARDS. The overall mortality of patients genotyped in the FACTT and HMC SIRS cohorts was 18.3 and 24.5%, respectively (Table 1). In FACTT, subjects who were homozygous for the MAP3K1906Val allele at rs823582 had a numerically but not statistically significant increase in mortality (Table 4). In HMC SIRS, all five subjects who were homozygous MAP3K1906Val allele at rs823582 died, precluding the calculation of an association (Table 4).
Table 4.
Associations between rs832582 and 28-Day Mortality in the Fluid and Catheter Treatment Trial and Harborview Medical Center Systemic Inflammatory Response Syndrome Cohorts
| SNP | OR (95% CI) | P Value (Recessive) | FDR |
|---|---|---|---|
| FACTT cohort | |||
| rs832582 | 1.70 (0.45–6.41) | 0.45 | 0.57 |
| HMC SIRS cohort | |
||
| rs832582 | All five subjects homozygous for the rare allele died | ||
Definition of abbreviations: CI = confidence interval; FACTT = Fluid and Catheter Treatment Trial; FDR = false discovery rate; HMC = Harborview Medical Center; OR = odds ratio using logistic regression with recessive model; SIRS = systemic inflammatory response syndrome; SNP = single-nucleotide polymorphism.
SNPs in MAP3K1 Are Associated with Whole-Blood Cytokine Production Ex Vivo
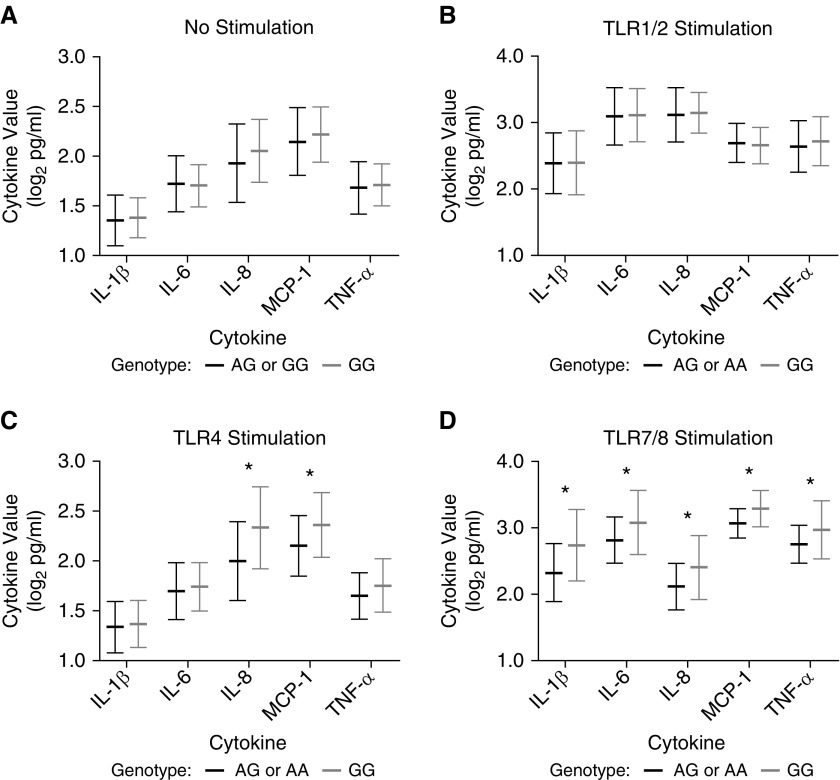
Given our finding of a robust association between MAP3K1906Val and decreased VFDs, we next focused on the identification of potential functional effects of rs832582. We tested for associations between rs832582 and innate immune agonist–induced production of inflammatory mediators relevant to ARDS pathophysiology in a cohort of healthy subjects. We performed array-based genotyping, and measured the whole-blood cytokine responses to various TLR agonists ex vivo in these subjects. Notably, homozygosity for the minor allele of rs832582 (genotype: GG, MAP3K1906Val; using a recessive statistical model) was associated with increased IL-8 and MCP-1 production in whole blood stimulated with a TLR4 agonist, and increased IL-1β, IL-6, IL-8, MCP-1, and TNF-α production in whole blood stimulated with a TLR7/8 agonist (Figure 2). We did not observe any associations between MAP3K1 variants and levels of the cytokines/chemokines that we measured after whole-blood stimulation of TLR1/2 (Figure 2).
Figure 2.
Whole-blood cytokine levels after ex vivo stimulation with Toll-like receptor (TLR) agonists by rs832582 genotype. Whole blood was stimulated for 6 hours on 96-well plates coated with stimulating agents or media alone. Cytokine levels were measured by cytometric bead–based immunoassays, adjusted for monocyte counts, and log2 transformed. Summary plots showing mean (±SD) cytokine levels in: (A) whole blood after stimulation with normal media based on rs832582 genotype (AG or AA: n = 328 versus GG: n = 18); (B) whole blood after stimulation with Pam3CysSerLys4 (Pam3CSK4) (TLR1/2 agonist) based on rs832582 genotype (AG or AA: n = 328 versus GG: n = 18); (C) whole blood after stimulation with Yersinia pestis LPS (TLR4 agonist) based on rs832582 genotype (AG or AA: n = 169 versus GG: n = 10); and (D) whole blood after stimulation with R848 (TLR7/8) based on rs832582 genotype (AG or AA: n = 169 versus GG: n = 10). P values for all comparisons were generated using the unpaired Student’s t test, and significant comparisons are designated with an asterisk. MCP-1 = monocyte chemoattractant protein-1.
SNPs in MAP3K1 Are Associated with Enrichment of Inflammatory Gene Sets in Whole-Blood Transcriptome
We next tested whether rs832582 was associated with transcriptional responses after innate immune activation. A subset of the blood samples (n = 252) from the healthy subjects cohort underwent RNA purification after stimulation with LPS (S. minnesota Re595), a TLR4 agonist. To identify transcriptional programs activated in subjects homozygous for the minor allele of rs832582 (genotype: GG, MAP3K1906Val) compared with subjects who carried at least one copy of the major allele (genotype AG or AA, MAP3K1906Ile), we applied Gene Set Enrichment Analysis (GSEA) using 50 Hallmark gene sets curated from the Molecular Signature Database (17). Notably, the only gene set that was significantly enriched in subjects homozygous for the minor allele of rs832582 (genotype: GG, MAP3K1906Val) with a FDR less than 0.05 was the HALLMARK_INFLAMMATORY_RESPONSE gene set (Table 5). There were no gene sets enriched with an FDR less than 0.05 in subjects with at least one copy of the common allele of rs832582 (genotype: AG or AA, MAP3K1906Ile). The full HALLMARK INFLAMMATORY RESPONSE gene list can be found on the Molecular Signature Database Web site (http://software.broadinstitute.org/gsea/msigdb), and is composed of genes defining the human inflammatory response. To further capture the biological themes that this gene set represented, we compiled the leading-edge subset of genes from the GSEA enrichment plot (Figure E1), and ranked them based on the degree of overrepresentation that each gene had within the gene set in subjects homozygous for the minor allele of rs832582 (genotype: GG, MAP3K1906Val) compared with subjects who carried at least one copy of the major allele (genotype: AG or AA, MAP3K1906Ile) (Table 6). The MAP3K1 transcript levels between the two groups (genotype: AG or AA, MAP3K1906Ile versus genotype: GG, MAP3K1906Val) were unchanged under either unstimulated or TLR4-stimulated conditions (Figure E2).
Table 5.
Gene Set Enrichment Analysis of Whole Blood Transcriptome after Toll-Like Receptor 4 Stimulation per rs832582 Mitogen-activated Protein Kinase Kinase Kinase 1 Genotypes
| Gene Set | NES | P Value | FDR |
|---|---|---|---|
| HALLMARK_INFLAMMATORY_RESPONSE | 1.79 | <0.001 | 0.013 |
| HALLMARK_INTERFERON_GAMMA_RESPONSE | 1.47 | <0.001 | 0.115 |
| HALLMARK_TNFA_SIGNALING_VIA_NFKB | 1.35 | <0.001 | 0.165 |
| HALLMARK_REACTIVE_OXYGEN_SPECIES_PATHWAY | 1.27 | 0.104 | 0.234 |
| HALLMARK_NOTCH_SIGNALING | 1.26 | 0.151 | 0.197 |
Definition of abbreviations: FDR = false discovery rate; NES = normalized enrichment score.
FDR threshold of 0.05 was used to identify statistical significance and is designated by italics. Gene set enrichment in GG (n = 15) versus AG or AA (n = 237).
Table 6.
Leading-Edge Subset of Genes Driving the Association Between Mitogen-activated Protein Kinase Kinase Kinase 1906Val and the HALLMARK_INFLAMMATORY_RESPONSE Gene Set
| Gene Symbol | Rank in Gene List | Running ES | Gene Symbol | Rank in Gene List | Running ES | Gene Symbol | Rank in Gene List | Running ES |
|---|---|---|---|---|---|---|---|---|
| CCL22 | 403 | 0.009 | KIF1B | 1,549 | 0.155 | NFKB1 | 2,961 | 0.249 |
| AXL | 477 | 0.017 | TACR3 | 1,570 | 0.162 | CSF3R | 3,340 | 0.240 |
| SELS | 565 | 0.024 | TLR2 | 1,609 | 0.168 | DCBLD2 | 3,355 | 0.244 |
| PTGIR | 580 | 0.033 | RGS16 | 1,676 | 0.173 | NDP | 3,390 | 0.248 |
| ATP2C1 | 586 | 0.043 | GPR132 | 1,693 | 0.180 | AHR | 3,439 | 0.252 |
| IL15RA | 587 | 0.053 | ITGA5 | 1,744 | 0.186 | KLF6 | 3,443 | 0.257 |
| CSF3 | 695 | 0.058 | PLAUR | 1,974 | 0.184 | CD70 | 3,472 | 0.261 |
| TNFAIP6 | 701 | 0.068 | ADRM1 | 1,983 | 0.191 | HRH1 | 3,514 | 0.265 |
| IL8 | 771 | 0.075 | ATP2A2 | 2,004 | 0.197 | IL6 | 3,630 | 0.266 |
| ABCA1 | 780 | 0.084 | EMR1 | 2,053 | 0.202 | ICOSLG | 3,765 | 0.265 |
| NLRP3 | 796 | 0.093 | KCNMB2 | 2,090 | 0.207 | IL2RB | 3,841 | 0.267 |
| PVR | 825 | 0.101 | GCH1 | 2,102 | 0.214 | NAMPT | 3,921 | 0.269 |
| P2RX4 | 952 | 0.105 | OSM | 2,129 | 0.220 | EMP3 | 3,926 | 0.274 |
| TNFSF15 | 961 | 0.114 | HBEGF | 2,258 | 0.221 | SCARF1 | 3,960 | 0.277 |
| LPAR1 | 1,144 | 0.115 | CCL17 | 2,329 | 0.225 | CXCL11 | 4,068 | 0.278 |
| CD40 | 1,196 | 0.121 | LIF | 2,492 | 0.225 | CDKN1A | 4,125 | 0.280 |
| IL15 | 1,316 | 0.125 | CXCL9 | 2,553 | 0.229 | TAPBP | 4,147 | 0.284 |
| CLEC5A | 1,403 | 0.129 | FZD5 | 2,615 | 0.233 | RELA | 4,225 | 0.285 |
| CD14 | 1,447 | 0.136 | OPRK1 | 2,626 | 0.239 | FPR1 | 4,255 | 0.289 |
| IL4R | 1,449 | 0.144 | APLNR | 2,728 | 0.241 | MEFV | 4,293 | 0.292 |
| MSR1 | 1,449 | 0.144 | MMP14 | 2,833 | 0.243 | MEP1A | 4,312 | 0.296 |
| EBI3 | 1,468 | 0.151 | BEST1 | 2,955 | 0.244 | IL10RA | 4,346 | 0.299 |
Definition of abbreviation: ES = enrichment score.
Site-directed Mutagenesis of MAP3K1 and Functional Analysis
Our clinical association studies suggested that subjects homozygous for the minor allele of rs832582 (genotype: GG, MAP3K1906Val) had significantly fewer VFDs as compared with subjects with one or two copies of the major allele (genotype: AA or AG, MAP3K1906Ile). Furthermore, our data suggested that rs832582 might influence levels of inflammatory cytokines in response to TLR4 and TLR7/8 stimuli, as well as impact the transcriptome after innate immune activation. Therefore, we next sought to identify a mechanism by which these MAP3K1 coding variants might influence cell function using a cell culture model for cell signaling.
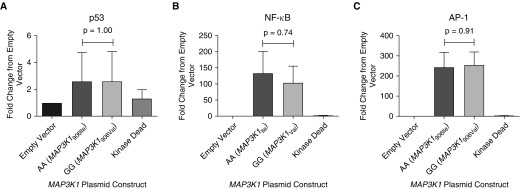
We created two different MAP3K1 cDNA constructs containing either the common allele of rs832582 (genotype: AA, MAP3K1906Ile) or the variant allele (genotype: GG, MAP3K1906Val), and then overexpressed those constructs in HEK-293 cells. MAP3K1 mRNA and MAP3K1 protein production were unchanged between cells transfected with either the rare allele (MAP3K1906Val) or the common allele (MAP3K1906Ile) (Figures E3 and E4). We also evaluated the effects of mutated MAP3K1 on HEK-293 cell metabolism and viability, and did not observe differences when comparing MAP3K1 variant constructs (data not shown). Finally, we did not observe any statistically significant change in reporter activity of p-53, NF-κB, or AP-1 activity, as measured by promoter-luciferase reporter plasmids cotransfected with the various MAP3K1 plasmids in HEK-293 cells (Figure 3).
Figure 3.
Luciferase reporter activity for p53 (n = 3) (A), NF-κB (n = 3) (B), and activator protein 1 (AP-1) (n = 4) (C) in human embryonic kidney cells 293 (HEK-293) are not influenced by coding variants in MAP3K1 SNP rs832582. All samples were processed in triplicate. The experimental output was relative luminescence units, which were derived by dividing firefly luciferase luminescence for each reporter construct by Renilla (control) luminescence at 48 hours after transfection with a MAP3K1 construct. Data are shown with each construct normalized to the empty vector value. P values for all pair-wise comparisons were generated using the Mann–Whitney test on normalized data.
Discussion
In this study, we sought to determine whether common genetic variants in MAP3K1, a key regulator of cell apoptotic and inflammatory responses, would be associated with outcomes in ARDS. Using a two-stage gene association study approach in two independent populations of patients with ARDS, we discovered that subjects homozygous for the minor allele (genotype: GG, MAP3K1906Val) of rs832582 had fewer VFDs compared with subjects who carried at least one copy of the major allele (genotype: AA or AG, MAP3K1906Ile), with an FDR of 0.06 (Table 3). The magnitude of the decrease in VFDs for homozygous carriers of MAP3K1906Val was substantial, and may have been due in part to increased death among homozygous carriers of MAP3K1906Val. Although there was not a statistically significant difference in mortality in the FACTT cohort, all the HMC cohort subjects with ARDS who were homozygous for MAP3K1906Val died (Table 4). Furthermore, we determined that this SNP was also associated with increased concentrations of multiple cytokines/chemokines in whole blood simulated ex vivo by TLR4 and TLR7/8 ligands, adding a potential mechanistic link to our gene association findings (Figure 2). Taken together, our results support the hypothesis that common genetic variation in the MAP3K1 gene is associated with outcomes in patients with ARDS, and that one potential mechanism driving this association may be related to MAP3K1’s regulation of inflammation.
Our finding that genetic variation in MAP3K1 is associated with an increased inflammatory response and, in turn, outcomes in ARDS is consistent with prior evidence that supports an important role for inflammation in ARDS. Significantly increased levels of IL-1β, IL-6, IL-8, MCP-1, and TNF-α have all been found in the alveolar fluid of subjects with ARDS compared with control subjects without ARDS, and elevated levels of IL-1β and MCP-1 have been associated with ARDS-related death (18, 19). Serum IL-6 and IL-8 levels strongly distinguish between “hyperinflammatory” and “nonhyperinflammatory” subphenotypes of ARDS, with higher concentrations of these two cytokines associated with the “hyperinflammatory” subphenotype that correlates with worse clinical outcomes (20). In whole blood procured from healthy subjects, we found that basal levels of IL-1ra were increased in subjects who were homozygous for the minor allele of rs832582 (Figure E5), and that levels of IL-1β, IL-6, IL-8, MCP-1, and TNF-α were elevated in response to TLR 4 and TLR7/8 stimulation ex vivo (Figure 2).
We also found that rs832582 was associated with transcriptional responses after innate immune activation. Transcriptome analysis of whole blood stimulated with TLR4 agonists ex vivo demonstrated enrichment of inflammatory gene sets in subjects homozygous for MAP3K1906Val compared with subjects carrying at least one copy of the major allele (genotype: AG or AA, MAP3K1906Ile) (Table 5). GSEA is a powerful analytical method that allows for an interpretation of the entire transcriptome, and accounts for the inherent complexity of biological pathways that is often missed through individual gene-based comparisons (17). Our finding that subjects homozygous for the minor allele of rs832582 (genotype: GG, MAP3K1906Val) had an augmented proinflammatory transcriptional program in response to TLR4 stimulation corroborates our cytokine findings above, and supports the overall finding that MAP3K1906Val predisposes individuals toward a hyperinflammatory response.
There are many potential mechanisms by which the common genetic variants at rs832582 might influence MAP3K1 protein function and, in turn, downstream activation of key transcription factors that influence cytokine expression and ARDS-associated clinical outcomes. The amino acid coded by rs832582 resides outside the kinase moiety (Figure 1). However, there are multiple sequences in the regulatory domain of MAP3K1 that are known to interact with scaffold proteins (5, 21), and the nonkinase domain of MAP3K1 has been shown to influence transcriptional activity in certain pathways independent of the MAP3K1 phosphorylation cascade (22). Activity of MAP3K1 can be regulated in various subcellular compartments by caspase cleavage of the nonkinase domain (23, 24), and proposed caspase cleavage sites lie in close proximity, with rs832582 at amino acid Asp874 (Figure 1) (7, 25). Finally, in silico analysis has predicted that mutations in rs832582 (at position 906) likely impact a putative phosphorylation site (26). Taken together, there are many ways in which variations in the amino acid sequence in the noncatalytic domain influenced by rs832582 might affect the structure and function of MAP3K1 in a manner that impacts transcription factor activation of cytokine-related genes.
Thus, we were surprised that we did not observe any differential regulation of three key transcription factors involved in inflammation and apoptosis based on the MAP3K1 coding variants that we studied using in vitro transfection assays (Figure 3). Nor did we find any association between the coding variants that we studied and cell viability or metabolism. Some of the earliest studies describing MAP3K1’s function demonstrated its relationship with p53 (27), NF-κB (6, 28, 29) (though the specifics of this relationship are controversial [30–32]), and AP-1 (33). Moreover, IL-1β, IL-6, IL-8, MCP-1, and TNF-α are some of the most important target genes for up-regulation by NF-κB (34), whereas AP-1 and p53 are important regulators of cell cycle and apoptotic genes (35). Subtle changes in transcription factor activity can have profound effect sizes on cytokine levels, and it is possible that our transfection assays lacked the granularity to detect very small molecular differences between the groups. MAP3K1 is known to regulate many other transcription factors other than the ones we tested, such as nuclear factor of activated T-cells 4 (NF-AT4), signal transducer and activator of transcription 3 (Stat3), and Smad2 (reviewed in Ref. 5). We cannot exclude the possibility that coding variants in MAP3K1 may impact the regulation of transcription factors that we were not able to detect, and that differential regulation of those transcription factors might explain the variable outcomes in ARDS that are associated with MAP3K1 genetic variants. Finally, it is possible that other SNPs in tight LD with rs832582 may regulate p53, NF-κB, or AP-1 activity independently of the direct influence the variant allele of rs832582 has on MAP3K1 protein structure. Indeed, rs832582 is in tight LD with several SNPs we did not test that are predicted to have strong regulatory and epigenetic influences on gene expression and phenotype (Table E3).
Our study has several strengths. First, our two-stage study design greatly reduces the chance that our findings represent a type I error. The samples that we genotyped in our discovery cohort were taken from a well characterized group of patients with ARDS taken from the multicentered NHLBI ARDSnet FACTT study, and we validated these data with samples taken from a completely independent cohort of critically ill patients (HMC SIRS). Second, our LD bin tagSNP approach is a well validated method of providing a comprehensive list of common genetic variation targets within a gene. Finally, we provide a potential mechanistic link between the minor allele (genotype: GG, MAP3K1906Val) of rs832582 and worse outcomes in ARDS: increased inflammatory responses to innate immune agonists.
Despite these strengths, there are several important limitations of this study. First, our study subjects were all white; thus, the extent to which our findings can be generalized to other populations is not known. Second, our analysis linking MAP3K1 genotype and cytokine expression are exploratory, and were not corrected for multiple hypothesis testing. Thus, we cannot exclude the possibility of type I error. Finally, we performed our cell culture studies using only HEK-293 cells. We chose this cell type based on prior successful transfection studies for MAP3K1 using this cell line (22, 27, 36, 37), but our approach did not observe differential apoptosis, cell viability, or cell metabolism rates after transfection with MAP3K1 variants. This may be due to compensatory gene and/or protein expression of MAP3K1 from the endogenous gene. Alternatively, as mentioned previously here, rs832582 may only represent a proxy for a causative variant that functions in a way not measured by our assay, such as splicing differences or epigenetic modifications (Table E3).
In summary, our study identified a coding SNP in MAP3K1 (rs832582) that associates with severity of outcomes in ARDS. We observed that this coding SNP was linked with IL-8 and MCP-1 levels in whole blood after TLR4 stimulation, and IL-1β, IL-6, IL-8, MCP-1, and TNF-α levels in whole blood after TLR7/8 stimulation. Genome-wide expression analysis revealed an association between this coding SNP and transcriptional response to TLR4 stimulation. Further research is needed to better elucidate how the variant in MAP3K1 that we have identified influences pathophysiology in ARDS.
Acknowledgments
Acknowledgments
The authors acknowledge the work by the Fluid and Catheter Treatment Trial, without which our work would not have been possible. Derek J. Schulte provided excellent technical assistance to this project.
Footnotes
This work was supported by National Institutes of Health grants T32 HL007287 and P50 HL073996.
This article was prepared using Fluid and Catheter Treatment Trial research materials obtained from the National Heart, Lung, and Blood Institute (NHLBI) Biologic Specimen and Data Repository Information Coordinating Center, and does not necessarily reflect the opinions or views of the investigators who performed this trial or the NHLBI.
Author Contributions: M.M.W. contributed to the conception and design of the work; E.D.M., D.S.O’M., B.J.G., S.H.-B., C.N., S.G., A.A., F.R., G.R., R.A.B., and M.M.W. contributed to the acquisition, analysis, and interpretation of the data for the work; E.D.M., D.S.O’M., and M.M.W. drafted and revised the manuscript for important intellectual content; E.D.M., D.S.O’M., B.J.G., S.H.-B., C.N., S.G., A.A., F.R., G.R., R.A.B., and M.M.W. significantly contributed to and approved the final version of the manuscript for publication; E.D.M., D.S.O’M., B.J.G., S.H.-B., C.N., S.G., A.A., F.R., G.R., R.A.B., and M.M.W. agree to be accountable for all aspects of the work.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2017-0030OC on August 31, 2017
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Matthay MA, Zimmerman GA. Acute lung injury and the acute respiratory distress syndrome: four decades of inquiry into pathogenesis and rational management. Am J Respir Cell Mol Biol. 2005;33:319–327. doi: 10.1165/rcmb.F305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glavan BJ, Holden TD, Goss CH, Black RA, Neff MJ, Nathens AB, et al. ARDSnet Investigators. Genetic variation in the FAS gene and associations with acute lung injury. Am J Respir Crit Care Med. 2011;183:356–363. doi: 10.1164/rccm.201003-0351OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin TR, Nakamura M, Matute-Bello G. The role of apoptosis in acute lung injury. Crit Care Med. 2003;31(4 suppl):S184–S188. doi: 10.1097/01.CCM.0000057841.33876.B1. [DOI] [PubMed] [Google Scholar]
- 4.Deak JC, Cross JV, Lewis M, Qian Y, Parrott LA, Distelhorst CW, et al. Fas-induced proteolytic activation and intracellular redistribution of the stress-signaling kinase MEKK1. Proc Natl Acad Sci USA. 1998;95:5595–5600. doi: 10.1073/pnas.95.10.5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hagemann C, Blank JL. The ups and downs of MEK kinase interactions. Cell Signal. 2001;13:863–875. doi: 10.1016/s0898-6568(01)00220-0. [DOI] [PubMed] [Google Scholar]
- 6.Lee FS, Hagler J, Chen ZJ, Maniatis T. Activation of the IκB α kinase complex by MEKK1, a kinase of the JNK pathway. Cell. 1997;88:213–222. doi: 10.1016/s0092-8674(00)81842-5. [DOI] [PubMed] [Google Scholar]
- 7.Yujiri T, Sather S, Fanger GR, Johnson GL. Role of MEKK1 in cell survival and activation of JNK and ERK pathways defined by targeted gene disruption. Science. 1998;282:1911–1914. doi: 10.1126/science.282.5395.1911. [DOI] [PubMed] [Google Scholar]
- 8.Sadoshima J, Montagne O, Wang Q, Yang G, Warden J, Liu J, et al. The MEKK1-JNK pathway plays a protective role in pressure overload but does not mediate cardiac hypertrophy. J Clin Invest. 2002;110:271–279. doi: 10.1172/JCI14938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tricker E, Arvand A, Kwan R, Chen GY, Gallagher E, Cheng G. Apoptosis induced by cytoskeletal disruption requires distinct domains of MEKK1. PLoS One. 2011;6:e17310. doi: 10.1371/journal.pone.0017310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hancock L, Beckly J, Geremia A, Cooney R, Cummings F, Pathan S, et al. Clinical and molecular characteristics of isolated colonic Crohn’s disease. Inflamm Bowel Dis. 2008;14:1667–1677. doi: 10.1002/ibd.20517. [DOI] [PubMed] [Google Scholar]
- 11.Szczepankiewicz A, Sobkowiak P, Rachel M, Bręborowicz A, Schoneich N, Bruce K, et al. Multilocus analysis of candidate genes involved in neurogenic inflammation in pediatric asthma and related phenotypes: a case–control study. J Asthma. 2012;49:329–335. doi: 10.3109/02770903.2012.669442. [DOI] [PubMed] [Google Scholar]
- 12.Nelson DS, van Halteren A, Quispel WT, van den Bos C, Bovée JVMG, Patel B, et al. MAP2K1 and MAP3K1 mutations in Langerhans cell histiocytosis. Genes Chromosomes Cancer. 2015;54:361–368. doi: 10.1002/gcc.22247. [DOI] [PubMed] [Google Scholar]
- 13.O’Mahony DS, Glavan BJ, Strout J, Holden TD, Rona G, Harju-Baker S, et al. A common non-synonymous coding SNP In MEKK1 increases AP1 transcription activity and is associated with mortality in ALI. Presented at the 2012 American Thoracic Society International Conference. May 20, 2012, San Francisco, CA. Abstract 185, p. A1967. [Google Scholar]
- 14.Wiedemann HP, Wheeler AP, Bernard GR, Thompson BT, Hayden D, deBoisblanc B, et al. National Heart, Lung, and Blood Institute Acute Respiratory Distress Syndrome (ARDS) Clinical Trials Network. Comparison of two fluid-management strategies in acute lung injury. N Engl J Med. 2006;354:2564–2575. doi: 10.1056/NEJMoa062200. [DOI] [PubMed] [Google Scholar]
- 15.Bernard GR, Artigas A, Brigham KL, Carlet J, Falke K, Hudson L, et al. The American–European Consensus Conference on ARDS: definitions, mechanisms, relevant outcomes, and clinical trial coordination. Am J Respir Crit Care Med. 1994;149:818–824. doi: 10.1164/ajrccm.149.3.7509706. [DOI] [PubMed] [Google Scholar]
- 16.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodman RB, Strieter RM, Martin DP, Steinberg KP, Milberg JA, Maunder RJ, et al. Inflammatory cytokines in patients with persistence of the acute respiratory distress syndrome. Am J Respir Crit Care Med. 1996;154:602–611. doi: 10.1164/ajrccm.154.3.8810593. [DOI] [PubMed] [Google Scholar]
- 19.Lin W-C, Lin C-F, Chen C-L, Chen C-W, Lin Y-S. Prediction of outcome in patients with acute respiratory distress syndrome by bronchoalveolar lavage inflammatory mediators. Exp Biol Med (Maywood) 2010;235:57–65. doi: 10.1258/ebm.2009.009256. [DOI] [PubMed] [Google Scholar]
- 20.Calfee CS, Delucchi K, Parsons PE, Thompson BT, Ware LB, Matthay MA NHLBI ARDS Network. Subphenotypes in acute respiratory distress syndrome: latent class analysis of data from two randomised controlled trials. Lancet Respir Med. 2014;2:611–620. doi: 10.1016/S2213-2600(14)70097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Avruch J. MAP kinase pathways: the first twenty years. Biochim Biophys Acta. 2007;1773:1150–1160. doi: 10.1016/j.bbamcr.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Islam MR, Jimenez T, Pelham C, Rodova M, Puri S, Magenheimer BS, et al. MAP/ERK kinase kinase 1 (MEKK1) mediates transcriptional repression by interacting with polycystic kidney disease-1 (PKD1) promoter-bound p53 tumor suppressor protein. J Biol Chem. 2010;285:38818–38831. doi: 10.1074/jbc.M110.145284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cardone MH, Salvesen GS, Widmann C, Johnson G, Frisch SM. The regulation of anoikis: MEKK-1 activation requires cleavage by caspases. Cell. 1997;90:315–323. doi: 10.1016/s0092-8674(00)80339-6. [DOI] [PubMed] [Google Scholar]
- 24.Zebrowski DC, Alcendor RR, Kirshenbaum LA, Sadoshima J. Caspase-3 mediated cleavage of MEKK1 promotes p53 transcriptional activity. J Mol Cell Cardiol. 2006;40:605–618. doi: 10.1016/j.yjmcc.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 25.Widmann C, Gerwins P, Johnson NL, Jarpe MB, Johnson GL. MEK kinase 1, a substrate for DEVD-directed caspases, is involved in genotoxin-induced apoptosis. Mol Cell Biol. 1998;18:2416–2429. doi: 10.1128/mcb.18.4.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Savas S, Taylor IW, Wrana JL, Ozcelik H. Functional nonsynonymous single nucleotide polymorphisms from the TGF-β protein interaction network. Physiol Genomics. 2007;29:109–117. doi: 10.1152/physiolgenomics.00226.2006. [DOI] [PubMed] [Google Scholar]
- 27.Fuchs SY, Adler V, Pincus MR, Ronai Z. MEKK1/JNK signaling stabilizes and activates p53. Proc Natl Acad Sci USA. 1998;95:10541–10546. doi: 10.1073/pnas.95.18.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakano H, Shindo M, Sakon S, Nishinaka S, Mihara M, Yagita H, et al. Differential regulation of IκB kinase α and β by two upstream kinases, NF-κB–inducing kinase and mitogen-activated protein kinase/ERK kinase kinase-1. Proc Natl Acad Sci USA. 1998;95:3537–3542. doi: 10.1073/pnas.95.7.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer CF, Wang X, Chang C, Templeton D, Tan T-H. Interaction between c-Rel and the mitogen-activated protein kinase kinase kinase 1 signaling cascade in mediating κB enhancer activation. J Biol Chem. 1996;271:8971–8976. doi: 10.1074/jbc.271.15.8971. [DOI] [PubMed] [Google Scholar]
- 30.Xia Y, Makris C, Su B, Li E, Yang J, Nemerow GR, et al. MEK kinase 1 is critically required for c-Jun N-terminal kinase activation by proinflammatory stimuli and growth factor–induced cell migration. Proc Natl Acad Sci USA. 2000;97:5243–5248. doi: 10.1073/pnas.97.10.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yujiri T, Ware M, Widmann C, Oyer R, Russell D, Chan E, et al. MEK kinase 1 gene disruption alters cell migration and c-Jun NH2-terminal kinase regulation but does not cause a measurable defect in NF-κB activation. Proc Natl Acad Sci USA. 2000;97:7272–7277. doi: 10.1073/pnas.130176697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karin M, Delhase M. JNK or IKK, AP-1 or NF-κB, which are the targets for MEK kinase 1 action? Proc Natl Acad Sci USA. 1998;95:9067–9069. doi: 10.1073/pnas.95.16.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karin M. The regulation of AP-1 activity by mitogen-activated protein kinases. J Biol Chem. 1995;270:16483–16486. doi: 10.1074/jbc.270.28.16483. [DOI] [PubMed] [Google Scholar]
- 34.Pahl HL. Activators and target genes of Rel/NF-κB transcription factors. Oncogene. 1999;18:6853–6866. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- 35.Shaulian E, Karin M. AP-1 in cell proliferation and survival. Oncogene. 2001;20:2390–2400. doi: 10.1038/sj.onc.1204383. [DOI] [PubMed] [Google Scholar]
- 36.Chipps E, Protzman A, Muhi MZ, Ando S, Calvet JP, Islam MR. Nuclear localization signal and p53 binding site in MAP/ERK kinase kinase 1 (MEKK1) J Cell Biochem. 2015;116:2903–2914. doi: 10.1002/jcb.25238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buschmann T, Potapova O, Bar-Shira A, Ivanov VN, Fuchs SY, Henderson S, et al. Jun NH2-terminal kinase phosphorylation of p53 on Thr-81 is important for p53 stabilization and transcriptional activities in response to stress. Mol Cell Biol. 2001;21:2743–2754. doi: 10.1128/MCB.21.8.2743-2754.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]