Abstract
MicroRNAs (miRNAs) act as post-transcriptional regulators of gene expression. In sarcoidosis, aberrant miRNA expression may enhance immune responses mounted against an unknown antigenic agent. We tested whether a distinct miRNA signature functions as a diagnostic biomarker and explored its role as an immune modulator in sarcoidosis. The expression of miRNAs in peripheral blood mononuclear cells from subjects who met clinical and histopathologic criteria for sarcoidosis was compared with that observed in matched controls in the ACCESS (A Case Controlled Etiologic Study of Sarcoidosis) study. Signature miRNAs were determined by miRNA microarray analysis and validated by quantitative RT-PCR. Microarray analysis identified 54 mature, human feature miRNAs that were differentially expressed between the groups. Significant feature miRNAs that distinguished subjects with sarcoidosis from controls were selected by means of probabilistic models adjusted for clinical variables. Eight signature miRNAs were chosen to verify the diagnosis of sarcoidosis in a validation cohort, and distinguished subjects with sarcoidosis from controls with a positive predictive value of 88%. We identified both novel and previously described genes and molecular pathways associated with sarcoidosis as targets of these signature miRNAs. Additionally, we demonstrate that signature miRNAs (hsa-miR-150-3p and hsa-miR-342-5p) are significantly associated with reduced lymphocytes and airflow limitations, both of which are known markers of a poor prognosis. Together, these findings suggest that a circulating miRNA signature serves as a noninvasive biomarker that supports the diagnosis of sarcoidosis. Future studies will test the miRNA signature as a prognostication tool to identify unfavorable changes associated with poor clinical outcomes in sarcoidosis.
Keywords: biomarker, gene regulatory network, miRNA, peripheral blood mononuclear cells, sarcoidosis
Clinical Relevance
Sarcoidosis pathogenesis is poorly understood, although the interplay of antigen-presenting cells and T cell subsets in response to an unspecified antigen is observed in the dysregulated immune response. Post-transcriptional modulators in the form of microRNAs may be involved in regulating immune gene expression networks in sarcoidosis and may serve as a clinically applicable circulating diagnostic biomarker in sarcoidosis. As such, a microRNA signature derived from peripheral blood monocytes may also provide prognostic insight into the clinical phenotypes observed in sarcoidosis.
Sarcoidosis immunopathogenesis is poorly understood. The absence of a specific etiologic agent, together with histopathologic dependence and exclusion of multiple diseases often delays diagnosis (1–6). Moreover, biomarkers lack accuracy to diagnose sarcoidosis and cannot reliably predict organ involvement, disease progression, and other clinical outcomes (7–10). To satisfy diagnostic and prognostic needs, efforts have focused on identification of genes and products of cellular activity and metabolism (11–17). Genetic profiling has demonstrated altered expression of pathways related to interferon inducible type 1 T helper (Th1) immune responses, and T cell development and proliferation in lung tissue and peripheral blood linked to granulomatous inflammation (18–20).
Small, noncoding microRNAs (miRNAs) act as post-transcriptional immune modulators involved in gene regulatory networks influencing development and function of either innate or adaptive immune cells (21, 22). In sarcoidosis, the first reported genome-wide microarray analysis of miRNAs obtained from peripheral blood monocytes (PBMCs) identified differential expression of hsa-miR-25, hsa-miR-199a, and hsa-miR-214 compared with controls, and predicted that these target the TGF-β/WNT molecular pathway (23). Subsequently, studies investigating peripheral blood identified dysregulated preselected inflammatory related miRNAs in sarcoidosis compared with controls, with distinct expression profiles linked to sarcoidosis phenotypes (24–27).
We hypothesize that a miRNA signature derived from PBMCs may serve as a circulating diagnostic biomarker in sarcoidosis. Additionally, we examine miRNAs as post-transcriptional immune modulators of pathways that could account for distinct clinical phenotypes and therapeutic targets in sarcoidosis.
Methods
Study Population
Approval from the institutional review board of the University of Illinois at Chicago (Protocol #: 2016–0275) was obtained. The study design and subject enrollment for ACCESS (A Case Controlled Etiologic Study of Sarcoidosis; ClinicalTrials.gov identifier: NCT00005276) were previously described (28). Briefly, peripheral blood samples from Caucasian subjects processed by the RNA core laboratory were analyzed. Cases required tissue confirmation of nonnecrotizing granulomas within 6 months of enrollment. Controls were matched to cases by sex, age, race, and geographic region. Neither cases nor controls had a history of other granulomatous diseases or use of antituberculosis therapy. All matched (n = 25) and unmatched cases (n = 6) were randomly assigned into experimental and validation cohorts.
miRNA Processing and Statistical Analyses
RNA was isolated from PBMCs utilizing the guanidinium thiocyanate phenol-chloroform extraction method (29). Using Affymetrix GeneChip miRNA 4.0 microarrays, mature human miRNAs were annotated against the miRBase database and analyzed using the Genomics Suite 6.6 statistical package (Partek, Inc.) and the R Statistical Environment (version 3.3) (30–34). Differential expression analysis by moderated t-statistics followed by nearest–shrunken centroids feature selection was performed at predetermined false discovery rates (FDRs) of 10% and 1%, respectively (35, 36). We then used regression models to determine signature miRNAs for validation with quantitative RT-PCR (qRT-PCR) assays (Table E1 in the data supplement). Differences in expression were normalized against the ubiquitin C (UBC) housekeeping gene and compared to establish a discriminatory signature in the experimental cohort (37). An optimal threshold identified by receiver operating characteristic (ROC) analysis was used to test the signature’s performance in a validation cohort (38). Clinical data were compared via Student’s t test for continuous data or chi-square analysis for categorical data. Further details regarding the methods used can be found in the data supplement.
Results
Patient Characteristics
The experimental cohort was comprised of Caucasian subjects with sarcoidosis (n = 14), and all but one were matched to controls (n = 13) with no history of granulomatous disease during the recruitment phase of the study. The clinical characteristics of the patients are shown in Table 1. As expected, no significant differences were observed between cases and controls with regard to sex, age, and smoking history.
Table 1.
Baseline Characteristics of Experimental and Validation Cohorts Based on Responses or Results Reported on ACCESS Study Data Forms
| Experimental Cohort (n = 27) |
Validation Cohort (n = 29) |
||||||
|---|---|---|---|---|---|---|---|
| Controls n = 13 (48.15%) | Sarcoidosis Cases n = 14 (51.85%) | P value | Controls n = 12 (41.38%) | Sarcoidosis Cases n = 17 (58.62%) | P value | ||
| Ethnicity (%) | Caucasian | 13 (100) | 14 (100) | 1 | 12 (100) | 17 (100) | 1 |
| Sex (%) | Male | 6 (46.2) | 7 (50.0) | 1 | 4 (33.3) | 6 (35.3) | 1 |
| Female | 7 (53.8) | 7 (50.0) | 8 (66.7) | 11 (64.7) | |||
| Age (%) | <30 years | 1 (7.7) | 1 (7.1) | 0.886 | 1 (8.3) | 3 (17.6) | 0.822 |
| 30–39 years | 4 (30.8) | 5 (35.7) | 6 (50.0) | 6 (35.3) | |||
| 40–49 years | 6 (46.2) | 7 (50.0) | 3 (25.0) | 4 (23.5) | |||
| 50–59 years | 1 (7.7) | 0 (0.0) | 2 (16.7) | 3 (17.6) | |||
| 60–69 years | 1 (7.7) | 1 (7.1) | 0 (0.0) | 1 (5.9) | |||
| Smoking history (%) | Never | 7 (53.8) | 8 (57.1) | 1 | 5 (41.7) | 8 (47.1) | 1 |
| Positive | 6 (46.2) | 6 (42.9) | 7 (58.3) | 9 (52.9) | |||
| On treatment at time of inclusion (%) | No | — | 10 (71.4) | NA | — | 16 (94.1) | NA |
| Yes | — | 4 (28.6) | — | 1 (5.9) | |||
| Definite pulmonary involvement (%) | No | — | 2(14.3) | NA | — | 2 (11.8) | NA |
| Yes | — | 12 (85.7) | — | 15 (88.2) | |||
| Definite extrapulmonary involvement (%) | No | — | 7 (50.0) | NA | — | 9 (52.9) | NA |
| Yes | — | 7 (50.0) | — | 8 (47.1) | |||
| Definite pulmonary and extrapulmonary involvement (%) | No | — | 9 (64.3) | NA | — | 10 (58.8) | NA |
| Yes | — | 5 (35.7) | — | 7 (41.2) | |||
| Scadding stage on chest radiograph (%) | 0 | — | 4 (28.6) | NA | — | 3 (17.6) | NA |
| 1 | — | 5 (35.7) | — | 6 (35.3) | |||
| 2 | — | 5 (35.7) | — | 6 (35.3) | |||
| 3 | — | 0 (0) | — | 2 (11.8) | |||
| White blood cell count (, SD) | Absolute (K/ul) | — | 7.51 (3.39) | NA | — | 5.86 (1.68) | NA |
| PBMC (%) | — | 28.29 (9.34) | NA | — | 31.00 (7.83) | NA | |
| Lymphocyte (%) | — | 21.29 (9.27) | NA | — | 23.47 (7.90) | NA | |
| Baseline spirometry | FEV1 (% predicted) | — | 89.30 (8.54) | NA | — | 92.12 (16.70) | NA |
| FVC (% predicted) | — | 93.75 (10.49) | NA | — | 96.78 (17.03) | NA | |
| FEV1/FVC | — | 76.95 (9.51) | NA | — | 77.15 (6.32) | NA | |
Definition of abbreviations: ACCESS = A Case Controlled Etiologic Study of Sarcoidosis; NA = not applicable; PBMC = peripheral blood monocyte.
No parameter within or between the experimental and validation cohorts demonstrated a statistically significant difference by Student’s t test for continuous data or chi-square test for categorical data.
Ordination, Differential Expression, and Definition of a miRNA Signature in Sarcoidosis
We identified 2,578 mature human miRNA transcripts in the miRBase registry and examined the relationship between clinical grouping (sarcoidosis cases versus matched controls) and the expressed miRNAs within the experimental cohort (Table E2A). Supervised correspondence analysis depicted 2 distinct clusters despite some overlap based on the log2-fold expression of all mature miRNAs identified by microarray analysis (Figure 1). These data suggest an underlying association between clinical grouping and miRNA expression. Among the 2,578 mature human miRNAs we determined those that were differentially expressed between sarcoidosis and matched controls. Employing a moderated t test, 2.7% of miRNAs were differentially expressed at a predetermined significant FDR of 10%. Of these differentially expressed miRNAs, 46 (66.7%) were overexpressed in sarcoidosis cases, while 23 (33.3%) were underexpressed (Figure E1).
Figure 1.
(A) Biplot demonstrating the correspondence analysis performed on microarray expression data, showing distinct clustering of sarcoidosis cases and matched controls. (B) Biplot demonstrating identified miRNAs. Subjects/miRNAs with strong associations are projected in the same direction and with greater distance from the origin. miRNA, microRNA.
To increase the discriminatory accuracy of the differentially expressed miRNAs for class prediction (sarcoidosis versus controls) we applied the nearest shrunken centroids method to select features at a predetermined median FDR of 1% (threshold value = 0.7119) (34, 39). At this threshold, 54 of 69 (78.3%) differentially expressed miRNAs were considered discriminatory features, with a 10-fold cross-validation error rate of 21.9%. Thirty-three of these 54 feature miRNAs (61.1%) were found to be overexpressed in sarcoidosis cases, and 21 (38.9%) were found to be underexpressed in sarcoidosis cases, with an average log2-fold change of 0.947 and −0.681, respectively (Figure 2).
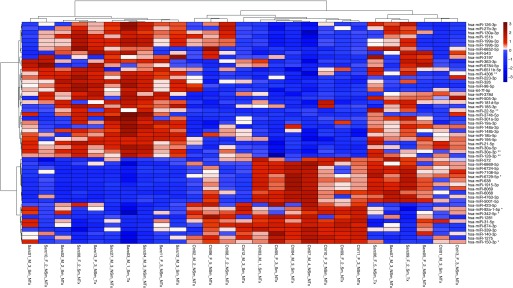
Figure 2.
A supervised heatmap demonstrating clinical characteristics per subject (columns) and the z-score–scaled expression of the 54 feature miRNAs (rows) identified using the nearest shrunken centroids method with 10-fold cross-validation at a median false discovery rate of 1%. Hierarchical clustering was performed using Ward’s agglomerative clustering method. Overexpression in sarcoidosis is depicted in red, and underexpression in sarcoidosis is depicted in blue. Clinical characteristics: male (M), female (F), age group (1: <30 years, 2: 30–39 years, 3: 40–49 years, 4: 50–59 years, 5: >60 years) smoking history (Sm), no smoking history (NSm), immunomodulator therapy (Tx), and no therapy (NTx). **Signature miRNA predictive of sarcoidosis; *signature miRNA predictive of health.
To narrow our target of miRNAs that were obtained through feature selection, we constructed logistic regression models based on individual log2-fold expression level of the miRNAs and clinical variables. All models were adjusted for sex, age, smoking history, and use of immunomodulator therapy at the time of sample acquisition. Upon construction, we observed that 79.6% (43/54) of the miRNAs were significantly associated with the presence or absence of sarcoidosis (P < 0.05). Feature miRNAs with a positive log2-fold change were found to increase the likelihood of sarcoidosis based on regression modeling after adjusting for clinical variables, and thus were deemed to be predictive of sarcoidosis. Conversely, regression modeling demonstrated that feature miRNAs with a negative log2-fold change decreased the likelihood of sarcoidosis. No clinical variable introduced into our regression model significantly affected the likelihood of sarcoidosis (Tables E2B and E2C).
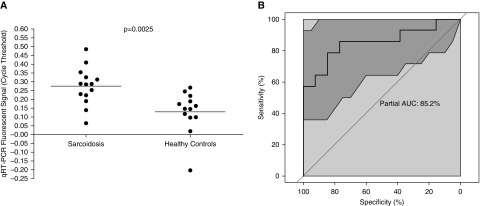
After the model was adjusted, eight miRNAs (hsa-miR-128-3p, hsa-miR-22-5p, hsa-miR-30e-3p, hsa-miR-4306, hsa-miR-92a-1-5p, hsa-miR-150-3p, hsa-miR-6729-5p, and hsa-miR-342-5p) were deemed to have the greatest influence in predicting the presence of sarcoidosis, and these were selected to establish a circulating miRNA signature. Four of these miRNAs were positive predictors of sarcoidosis and four were negative predictors of sarcoidosis (all with P < 0.05 and a log odds ratio (OR) of >20 or <0.05, respectively; Table E3 and Figure E2). Microarray expression data for these eight miRNAs were validated by qRT-PCR. As expected, individual cycle threshold (Ct) values obtained by qRT-PCR for each signature miRNA negatively correlated with microarray expression values. The selected housekeeping gene UBC was invariably expressed in subjects with sarcoidosis and controls in the experimental cohort (Figure E3A), and did not discriminate between the presence and absence of sarcoidosis, with a ROC area under the curve (AUC) of 48.6% (95% CI, 26.37–71.98%; Figure E3B). Among the eight validated miRNAs, the individual qRT-PCR expression ratios of hsa-miR-128-3p, hsa-miR-22-5p, and hsa-miR-150-3p to UBC demonstrated trends toward significance for independently discriminating between sarcoidosis cases and controls (unpaired t test with Welch’s correction; P = 0.0975, 0.0595, and 0.0691, respectively). A signature score was derived from the expression values of positive and negative predictor miRNAs, and a numerical threshold value was determined as described in the data supplement and Table E4. In contrast to UBC expression, a comparison of individual signature scores for sarcoidosis cases and matched controls demonstrated that the subjects with sarcoidosis expressed a significantly higher signature score than controls (unpaired t test with Welch’s correction, P = 0.0025; Figure 3A). Our circulating miRNA signature discriminated sarcoidosis cases from matched controls with an accuracy of 85.2% (95% CI, 68.13–97.8%) by ROC analysis (Figure 3B), which was significantly superior to the housekeeping gene alone for identifying cases of sarcoidosis (bootstrap test, P = 0.0277).
Figure 3.
(A) Comparison of the miRNA signature expression values (Ct) between the sarcoidosis and matched control groups in the experimental cohort. Ct values were significantly greater in the sarcoidosis group compared with the matched controls, indicating reduced expression and supporting the use of these signature miRNAs as a discriminatory test for diagnosing sarcoidosis (unpaired t test with Welch’s correction, P = 0.0025). (B) Receiver operating characteristic curve demonstrating the accuracy of our eight signature miRNAs as a discriminatory test for diagnosing sarcoidosis (AUC = 85.2%) in the experimental cohort. The shaded area represents the 95% CI. AUC = area under the curve; CI = confidence interval; Ct = cycle threshold.
The predictive potential of the signature circulating miRNA biomarker was tested on a randomly selected validation cohort (n = 29). The clinical characteristics of the validation cohort were similar to those of the experimental cohort (Table 1). We evaluated the performance of our circulating miRNA signature in the validation cohort. Expression of the eight selected signature miRNAs and UBC in the validation cohort by qRT-PCR was assessed blindly by two researchers (C.S. and Y.H.) after randomization was performed. To test the evenness of miRNA expression obtained by qRT-PCR, we compared the Ct values for UBC expression between the experimental and validation cohorts, and found that there was no significant difference in UBC expression (unpaired t test with Welch’s correction, P = 0.8243; Figure E4). The circulating signature score was calculated for all samples in the validation cohort. Applying a strict decision rule, we deemed all validation cohort samples to be sarcoidosis cases if the circulating signature score was greater than the determined threshold (Figure E5). When we compared clinical–pathologic grouping and biomarker grouping at this cutoff, our circulating miRNA biomarker proved to have a sensitivity and specificity of 68.18% and 71.43%, respectively, with a positive predictive value (PPV) of 88.24% and an OR of 5.36 (Table E5). Evaluation of the discriminatory capacity of the circulating signature score by ROC analysis of all samples revealed an accuracy of 74.8% (P = 0.0015; 95% CI, 61.84–87.84%; Figure E6), supporting its potential use for diagnosing sarcoidosis.
Underexpression of hsa-miR-150-3p and hsa-miR-342-5p Correlates with Reduced Lymphocytes and Is Associated with Airflow Limitations in Sarcoidosis
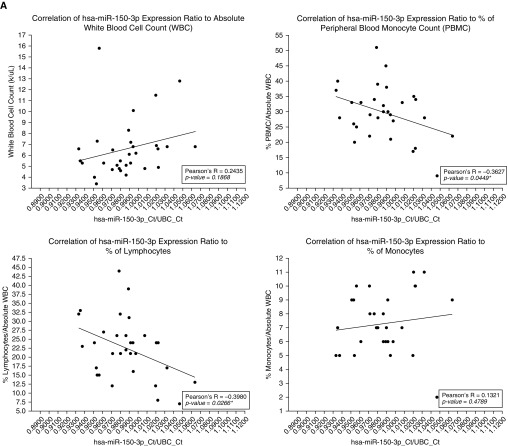
We evaluated the potential role of miRNAs in phenotypic manifestations of sarcoidosis by examining the relationship between miRNA expression profiles and baseline clinical parameters (white blood cell count, spirometric values, and Scadding stage on chest radiograph) that were previously related to worse prognosis and disease severity (40, 41). To identify specific miRNAs that could be involved in either protecting against or promoting unfavorable changes in these clinical parameters, we compared the individual qRT-PCR expression ratios of all eight signature miRNAs. No significant differences in expression of any of the four positive predictors of sarcoidosis were found between cases (n = 31) and controls (n = 25) (Figure E7). In contrast, hsa-miR-92a-1-5p, hsa-miR-150-3p, hsa-miR-6729-5p, and hsa-miR-342-5p (all negative predictors) were observed to be significantly underexpressed in sarcoidosis (unpaired t test with Welch’s correction; P = 0.0101, 0.0009, 0.0480, and 0.0192, respectively) (Figure E8). Among these, both hsa-miR-150-3p and hsa-miR-342-5p demonstrated a significant negative correlation with the percentage of PBMCs (Pearson’s R = −0.3627 and −0.3582; P = 0.045 and 0.048, respectively) in sarcoidosis cases. These significant negative correlations were due exclusively to the relationship between the percentage of lymphocytes in peripheral blood (but not in monocytes) and hsa-miR-150-3p and hsa-miR-342-5p (Pearson’s R = −0.3980 and −0.4053; P = 0.027 and 0.024, respectively) (Figures 4A and 4B).
Figure 4.
Correlations between miRNA expression profiles (adjusted for UBC) and white blood cell parameters. Pearson’s correlation coefficient depicts the strength of the relationships (*P < 0.05 was considered significant). (A) Correlation between hsa-miR-150-3p expression and white blood cell parameters (absolute white blood cell count, PBMC [%], lymphocyte [%], monocyte [%]). A significant negative correlation was found between hsa-miR-150-3p expression and PBMC (%) and lymphocyte (%). (B) Correlation between hsa-miR-342-5p expression and white blood cell parameters (absolute white blood cell count, PBMC [%], lymphocyte [%], and monocyte [%]). A significant negative correlation was found between hsa-miR-342-5p expression and PBMC (%) and lymphocyte (%). PBMC = peripheral blood monocyte; UBC = ubiquitin C.
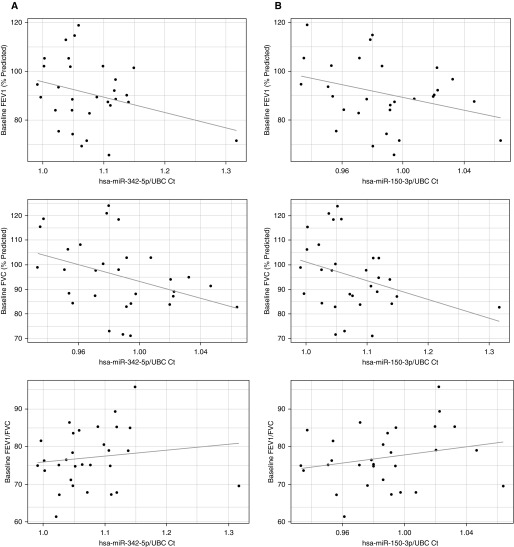
To further assess the impact on disease severity, we examined the relationship between baseline respiratory status and hsa-miR-150-3p and hsa-miR-342-5p in sarcoidosis cases by linear regression after adjusting for variables capable of influencing spirometric values (age, sex, smoking history, immunomodulator use, and Scadding stage). Although it was significantly associated with percent predicted forced expiratory volume in 1 second (FEV1%) and percent predicted forced vital capacity (FVC%) (P = 0.026 and 0.38; β = −99.57 and −84.76, respectively), the effect of hsa-miR-342-5p (Figure 5A) on FVC was significantly and inversely impacted by immunomodulator use (P = 0.027; β = 13.56). Reduced expression of hsa-miR-150-3p was significantly and independently associated with both decreased FEV1% and FVC% (P = 0.0423 and 0.0498; β = −183.83 and −161.03, respectively) (Figure 5B). Taken together, these results indicate that reduced levels of either of these miRNAs may play a role in the development of peripheral lymphopenia. Additionally, the underexpression of hsa-miR-150-3p and to a lesser extent hsa-miR-342-5p in this setting may be indicative of a clinical phenotype with airflow limitations.
Figure 5.
General linear models depicting associations between miRNA expression profiles (adjusted for UBC) and spirometric values (spirometric parameter ∼ age + sex + smoking history + Scadding + treatment + miRNA expression). (A) Association between hsa-miR-342-5p expression and baseline spirometric parameters (FEV1 [% predicted], FVC [% predicted], FEV1/FVC). A significant and independent relationship between hsa-miR-342-5p and FEV1 (P = 0.026; β = −99.57) was demonstrated. Although it was significant, the association between hsa-miR-342-5p and FVC was not independent and was impacted by treatment status (P = 0.026, 0.027; β = −84.76 and 13.56, respectively). No significant relationship was found between FEV1/FVC and hsa-miR-342-5p expression. (B) Association between hsa-miR-150-3p expression and baseline spirometric parameters (FEV1 [% predicted], FVC [% predicted], and FEV1/FVC). Significant and independent relationships between hsa-miR-150-3p expression and both FEV1 and FVC (P = 0.04232 and 0.04982; β = −183.83 and −161.03, respectively) were identified. No statistically significant association was found between hsa-miR-150-3p expression and FEV1/FVC.
miRNA Enrichment Analysis Demonstrates an Association with Cell Proliferation and Death
The miRNet miRNA-target algorithm was employed for functional enrichment analysis of miRNA–gene interactions obtained from the Kyoto Encyclopedia of Genes and Genomes (KEGG) to elucidate the target pathways of the eight signature miRNAs comprising the circulating biomarker. On the miRNet algorithm, no filtering of experimental methods used to validate miRNA-gene interactions was performed. KEGG pathways were considered significant at a P-value threshold of <0.05 using the algorithm’s hypergeometric approach adjusted for FDR (42).
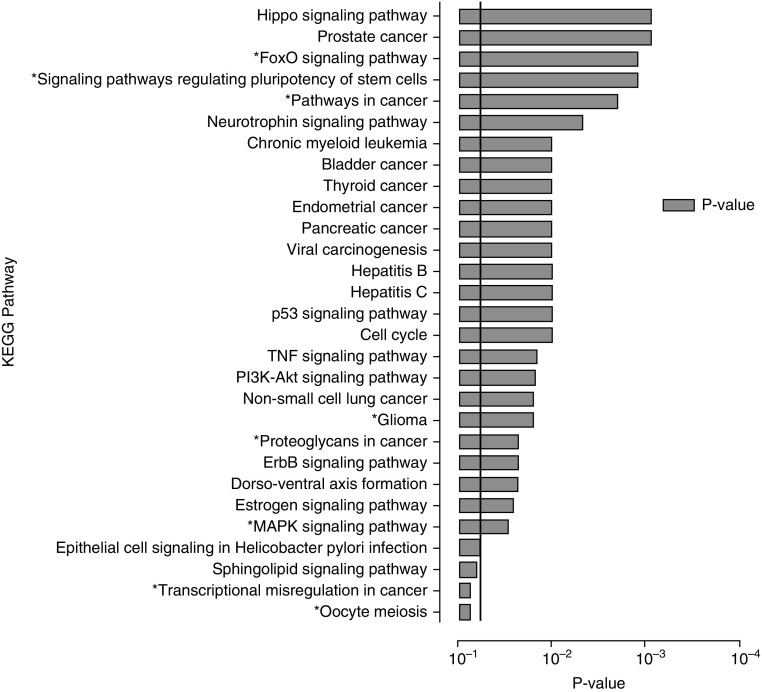
The combination of these eight miRNAs demonstrated a large network of interactions resulting in 1,100 targeted genes in 265 KEGG pathways (Table E6 and Figure S9), with a total of 29 (10.9%) pathways that met the predetermined level of significance (Figure 6). Within the network, we identified 70 genes that were predicted to be modulated by at least two miRNAs, and constructed a reduced interaction network to highlight the target genes on which miRNA action may converge (Figure 7). Among these targeted genes, ALDH9A1, NUFIP2, and ZNF385A were found to be likely hubs of interaction. Notably, these genes were all targeted by a combination of three of four miRNAs (hsa-miR-128-3p, hsa-miR-4306, hsa-miR-30e-3p, and hsa-miR-342-5p) that appear to be key miRNAs given their degree of interactions within the network. Of these four miRNAs, hsa-miR-128-3p directly targeted all three genes, highlighting its central role in the regulatory network.
Figure 6.
KEGG pathways enriched by the eight miRNAs that comprise our circulating miRNA signature identified by the miRNet miRNA-target algorithm. Significance was established with a P-value threshold of <0.05. *Pathways targeted by both hsa-miR-150-3p and hsa-miR-342-5p. KEGG = Kyoto Encyclopedia of Genes and Genomes.
Figure 7.
Reduced miRNA-KEGG pathway interaction network, showing all genes within pathways that were targeted by at least two of the eight circulating signature miRNAs. Square nodes represent miRNAs, and circles represent target genes. The major hubs of interaction within the network of targeted KEGG pathways appear to be hsa-miR-128-3p, hsa-miR-4306, hsa-miR-30e-3p, and hsa-miR-342-5p through ALDH9A1, NUFIP2, and ZNF385A (each with three degrees of interaction).
Among the significantly enriched pathways, the majority of pathways were involved in molecular interaction and reaction networks pertaining to human diseases (cancers, 41.4%), environmental information processing (signal transduction, 24.14%), and cellular processes (13.8%, mainly cell growth and death). Multiple significantly enriched pathways, such as the TNF, PIK3-AKT, MAPK, and p53 signaling pathways that have been previously implicated in the immunopathogenesis of sarcoidosis (43–46), were identified as targets of our eight-miRNA signature. Interestingly, within the large interaction network, the most significantly enriched pathway identified was the HIPPO signaling pathway (P = 0.0006), which to our knowledge has not been previously associated with sarcoidosis. In addition, upon assessment of the reduced network exclusively involving genes predicted to be targeted by two or more miRNAs, only the p53 signaling pathway was identified as significantly enriched (P = 0.0218).
The KEGG pathways and genes targeted by hsa-miR-150-3p and hsa-miR-342-5p were further analyzed to identify key attributes that could account for their association with either circulating lymphocyte counts or pulmonary function. Individually, hsa-miR-150-3p is predicted to modulate 29 genes in 35 KEGG pathways, whereas hsa-miR-342-5p interacts with 93 genes in 101 KEGG pathways through the miRNet algorithm. Interestingly, no overlap of target genes was identified between these two miRNAs within the regulatory network. However, 15% of the targeted KEGG pathways were predicted to be modulated by both miRNAs, and eight of these pathways were among the significantly enriched targets of the circulating signature (denoted with an asterisk in Figure 6). Notably, the FOXO and MAPK signaling pathways, as well as several pathways pertaining to human cancers, were among the significantly enriched pathways. The interactions of our eight signature miRNAs, gene targets, and corresponding KEGG pathways are suggestive of a close relationship between inflammation, dysregulated cell proliferation or death and sarcoidosis severity and prognosis.
Discussion
There are no biomarkers specific enough to establish a diagnosis of sarcoidosis without histopathologic confirmation. Expert opinion emphasizes that a diagnosis of sarcoidosis is never certain and is arbitrarily made only after excluding alternative diagnoses, thus underscoring the need to identify reliable early biomarkers suggestive of sarcoidosis (47). In this study, we employed an unbiased, high-throughput approach to identify aberrantly expressed miRNAs, which act as key components of gene regulatory networks in sarcoidosis. Our goal was to use these miRNAs as a novel genetic signature with potential diagnostic and prognostic value. We observed significantly altered miRNA expression between our cohort of Caucasians with recently diagnosed sarcoidosis and matched controls. Feature miRNAs that were significantly differentially expressed established a clinically applicable circulating signature based on the over- or underexpression of eight miRNAs that determined the presence of sarcoidosis with an accuracy of 74.8% (95% CI, 61.84–87.84%) (Figure E6). When compared with the diagnostic utility of angiotensin-converting enzyme (ACE) levels in a population-based study comprised mainly of Caucasians, our signature demonstrated a smaller degree of accuracy (74.8% versus 86.21%). However, our circulating miRNA signature demonstrated an enhanced PPV when compared with ACE (circulating miRNA signature PPV = 88.24% versus ACE PPV = 25.43%), supporting its value as an initial noninvasive diagnostic test for sarcoidosis (48). Pertaining to the use of miRNAs, a prior study examined the accuracy of select miRNAs derived from extracellular RNA as clinical biomarkers to distinguish between subjects with sarcoidosis with/without Löfgren’s syndrome and matched controls (26). The study showed an overall significant discriminatory capacity with a fair predictive power (72% when using a six-miRNA model and 65% with a seven-miRNA model) that is comparable to our results. The diagnostic utility of different combinations of gene expression signatures to discriminate between subjects with sarcoidosis and healthy individuals has shown sensitivities and specificities ranging from 83 to 92% (20, 49). Our signature’s reduced accuracy compared with gene expression signatures may be due to the lower expression density of miRNAs (50). To increase the signature’s clinical applicability as a biomarker, we may have lost discriminatory power in this study by selecting only eight of 54 miRNAs that were identified as significantly differentially expressed features by microarray analysis and were found to have an overall error rate of 21.9%. Despite this tradeoff, the signature’s ability to differentiate between sarcoidosis cases and matched controls was significant. It may be advantageous to use the signature in combination with other gene expression profiles to support a diagnosis of sarcoidosis.
The differential expression of miRNAs in peripheral blood from subjects with sarcoidosis has been characterized with the use of selective and unbiased strategies, limiting the comparability of relevant findings (25). When mapped against other reported miRNA signature sequences (23, 25, 26, 51), the expression of several closely related mature miRNA sequences coincided with signature miRNAs in our study. For example, concordant overexpression of hsa-miR-22-5p and hsa-miR-30e-3p was previously reported based on microarray data (51). Notably, two differentially expressed miRNAs within our signature, hsa-miR-6729-5p and hsa-miR-4306, represent novel associations whose role in sarcoidosis has not been previously elucidated. In contrast, the predominant hub of interactions in our cohort, hsa-miR-128-3p, was previously depicted as underexpressed (23, 51). In addition, hsa-miR-128a was previously linked to immune cells through a negative correlation with the CD4:CD8 ratio in peripheral blood (25). Furthermore, hsa-miR-150-3p and hsa-miR-342-5p, which correlated with lymphocytes and demonstrated a significant association with spirometric parameters in our study, have also been reported to be underexpressed (51). Evaluations of hsa-miR-150-3p by qRT-PCR have been inconsistent, although it was previously reported to be underexpressed and capable of differentiating between sarcoidosis cases without Löfgren’s syndrome and matched controls (24, 26). Overall, the differences between this work and previous studies with regard to miRNA expression may stem from different patient or microarray characteristics. Nevertheless, our analyses suggest the presence of a core gene regulatory network in sarcoidosis, as evidenced by the analogous aberrant expression of several miRNAs.
To determine biological functions that may relate to the underlying immunopathogenesis of sarcoidosis, we assessed predicted targets of differentially under- and overexpressed miRNAs in subjects with sarcoidosis compared with matched controls. When mapped against significantly enriched gene transcript signatures, several previously annotated transcripts were predicted to be targets of miRNAs in our miRNA signature. Not surprisingly, among these were several related to cell-cycle progression, including ATF3, MAZ, CACNA1E, CCND2, ZNF614, and TAOK1 (18, 20, 49, 51, 52). Similarly, previous studies that evaluated miRNA expression in peripheral blood also predicted targeting of KEGG pathways associated with cancer and cell proliferation, such as the TGF-β/WNT and Pathways in Cancer signaling pathways (23, 26). Both of these pathways were identified in our cohort, although only the latter was found to be significantly enriched. Furthermore, dysregulation of other significantly enriched KEGG pathways, including the HIPPO, FOXO, TNF, PIK3-AKT, MAPK, and p53 signaling pathways, may promote the complex maladaptive inflammatory response involving T cell subsets with heightened local inflammation and peripheral anergy that is found in sarcoidosis (53–55).
In the present study, we demonstrate that underexpression of hsa-miR-150-3p and hsa-miR-342-5p in subjects with sarcoidosis correlates significantly with a reduced percentage of lymphocytes and reduced FEV1% and FVC%. Although the process is poorly understood, apoptosis and disequilibrium between effector and regulatory T cells have been proposed as mechanisms that lead to peripheral lymphopenia involving T cell subsets and correlate with disease severity (40, 54, 56, 57). In accordance with previous evidence (57), our study suggests that apoptotic pathways and genes are related to lymphopenia. Additionally, we show that a key component of our reduced gene regulatory network (targeting various genes, such as: PTEN, p53, MDM2, MDM4) is the significantly enriched p53 pathway, known to induce cell cycle arrest or apoptosis (58). We also demonstrate that miRNA gene targets, namely, ALDH9A1, NUFIP2, and ZNF385A, are modulated by multiple miRNAs and serve as hubs of interaction in the reduced regulatory network. Interestingly, it was previously shown that when bound, ZNF385A and p53 modulate an autoregulatory feedback loop that results in preferential transactivation of proarrest rather than proapoptotic p53 target genes (59). We speculate that in sarcoidosis, the p53 signaling pathway is critical for stimulating apoptosis and the subsequent lymphopenia that contributes to peripheral anergy, possibly through direct modulation (unbinding) of ZNF385A provoked by low expression of hsa-miR-342-5p, as well as high expression of hsa-miR-128-3p and hsa-miR-4306. The role of hsa-miR-150-3p is less clear, although its gene targets (SOD2 and MKNK2) within our reduced interaction network suggest that modulation of the MAPK and FOXO signaling pathways may lead to alterations in CD4+, CD8+, and T regulatory cell differentiation, proliferation, and function (60–62). These observations lend further support to the notion that peripheral anergy and exaggerated local inflammation play a role in the immunopathogenesis of sarcoidosis (54, 57, 63, 64). Moreover, the predominance of miRNA interactions with multiple KEGG pathways related to cancer, cellular proliferation, and inflammation in PBMCs points to a molecular mechanism supporting the hypothesis that immune dysregulation in sarcoidosis significantly increases the risk of hematologic malignancies such as leukemia, Hodgkin’s lymphoma, and non-Hodgkin’s lymphoma (65). Thus, it is possible that in the presence of aberrant expression of our signature miRNAs, chronic inflammation and a state of compromised immune surveillance generate an imbalance of proapoptotic and prosurvival signaling pathways that results in neoplastic expansion (64).
We note several limitations to this study that may be addressed in future work. In our study, we analyzed clearly defined sarcoidosis cases and well-matched controls in an experimental and validation cohort derived from a single center in the ACCESS study. Despite this rigorous characterization, our findings are rooted in a small cohort of Caucasian subjects with sarcoidosis (n = 31) without evidence of advanced (fibrotic) disease, whereas in the United States, African Americans exhibit a greater incidence of sarcoidosis with more severe clinical manifestations and worse prognosis (66–68). Consequently, these findings may not be universally applicable and will need to be expanded upon in a larger, multicenter, more diverse cohort with subjects exhibiting early and advanced disease. Our analysis was also limited to controls without a history of granulomatous disease; however, overlapping signatures from other granulomatous disorders, such as tuberculosis, have been described, and thus further comparisons to establish the exclusiveness of our miRNA signature in sarcoidosis will be required (18, 51). Additionally, our study characterizes a miRNA regulatory network at a single time point early in the course of the disease. Although this early time point may provide relevant insight into the pathogenesis of phenotypic features, a longitudinal assessment is required to validate clinical outcomes in subjects who express our miRNA signature. Also, details pertaining to disease-modifying therapies and some clinical parameters were not made publicly available. Therefore, our study may be underpowered with regard to the true effect of these factors on immune regulatory networks, although the early time point at which samples were obtained makes therapy a less likely confounder. Furthermore, we explored the interactions of mature miRNAs derived from PBMCs by using a commercial microarray chip that may not have fully examined a more complex regulatory process involving distinct cell types and other gene regulatory elements underlying the immunopathogenesis of sarcoidosis. Lastly, we were also limited by the use of data stemming from databases that use computational approaches and statistical probabilities to ascertain miRNA interactions. In the future, we plan to construct complete gene regulatory networks by incorporating the full spectrum of transcriptomic profiles, and to develop in vivo analyses to validate these interactions.
In summary, our study illustrates the use of select aberrantly expressed miRNAs derived from analysis of high-throughput data as a potential biomarker intricately involved in the dysregulated immune response in sarcoidosis. Moreover, we analyzed the regulatory nature of the miRNAs that comprise our biomarker and demonstrated that they may serve as early prognostication tools to portend unfavorable changes associated with poor clinical outcomes in sarcoidosis.
Acknowledgments
Acknowledgments
The authors thank Hongzhen He, M.D., for her contributions to the RNA isolation and processing, and Zarema Arbieva, Ph.D., and the Core Genomics Facility at the University of Illinois at Chicago for their help in processing the miRNA microarray chips.
ACCESS (A Case Control Etiologic Study of Sarcoidosis) Research Group Clinical Centers
Beth Israel Deaconess Medical Center (Boston, MA): Steven E. Weinberger, M.D.; Patricia W. Finn, M.D.; Erik Garpestad, M.D.; Allison Moran, R.N.
Georgetown University Medical Center (Washington, DC): Henry Yeager, Jr., M.D.; David L. Rabin, M.D.; Susan Stein, M.A.
Case Western Reserve University–Henry Ford Health Sciences Center (Detroit, MI): Michael C. Iannuzzi, M.D.; Benjamin A. Rybicki, Ph.D.; Marcie Major, R.N.; Mary Maliarik, Ph.D.; John Popovich, Jr., M.D.
Johns Hopkins University School of Medicine (Baltimore, MD): David R. Moller, M.D.; Carol J. Johns, M.D.†; Cynthia Rand, Ph.D.; Joanne Steimel, R.N.
Medical University of South Carolina (Charleston, SC): Marc A. Judson, M.D.; Susan D’Alessandro, R.N.; Nancy Heister, R.N.; Theresa Johnson, R.N.; Daniel T. Lackland, Dr.P.H.; Janardan Pandey, Ph.D.; Steven Sahn, M.D.; Charlie Strange, M.D.
Mount Sinai Medical Center (New York, NY): Alvin S. Teirstein, M.D.; Louis DePalo, M.D.; Sheldon Brown, M.D.; Marvin Lesser, M.D.; Maria L. Padilla, M.D.; Marilyn Marshall
National Jewish Medical and Research Center (Denver, CO): Lee S. Newman, M.D., M.A.; Cecile Rose, M.D., M.P.H.; Juliana Barnard, M.A.
University of Cincinnati Medical Center (Cincinnati, OH): Robert P. Baughman, M.D.; Elyse E. Lower, M.D.; Donna B. Winget
University of Iowa College of Medicine (Iowa City, IA): Geoffrey McLennan, M.D., Ph.D.; Gary Hunninghake, M.D.; Chuck Dayton, B.S.Pharm.; Linda Powers, M.S.
University of Pennsylvania and Medical College of Pennsylvania–Hahnemann Medical Centers (Philadelphia, PA): Milton D. Rossman, M.D.; Eddy A. Bresnitz, M.D.; Ronald Daniele, M.D.; Jackie Regovich, M.P.H.; William Sexauer, M.D.
National Heart, Lung, and Blood Institute (Bethesda, MD): Study Chairman: Reuben Cherniack, M.D.; Study Co-Chairman: Lee Newman, M.D.; Robert Musson, Ph.D.; Joanne Deshler; Paul Sorlie, Ph.D.; Margaret Wu, Ph.D.
Clinical Coordinating Center
Clinical Trials & Surveys Corp.: Genell L. Knatterud, Ph.D.; Michael L. Terrin, M.D.; Bruce W. Thompson, Ph.D.; Kathleen Brown, Ph.D.; Margaret Frederick, Ph.D.; Frances LoPresti, M.S.; Patricia Wilkins, B.S.; Martha Canner, M.S.; Judy Dotson
Central Repository
McKesson Bioservices (September 1996 to November 1998): Steve Lindenfelser
BBI-Biotech Research Laboratories (December 1988 to present): Mark Cosentino, Ph.D.
Central Laboratories
DNA Core Laboratory: Mary Maliarik, Ph.D.
BAL Central Laboratory: Robert Baughman, M.D.
HLA Class II Typing Laboratory: Milton Rossman, M.D.; Dimitri Monos, Ph.D.; Chung Wha Lee, Ph.D.; Boyana Cizman, Ph.D.
Etiologic Antigen in Kveim Reagent Laboratory: David Moller, M.D.
Immunogenetics Laboratory: Janardan Pandey, Ph.D.
L-Forms Core Laboratory: Peter Almenoff, M.D.; Ian Brett; Sheldon Brown, M.D.; Marvin Lesser, M.D.
Pathogenic T Cells Laboratory: Lee Newman, M.D.; Brian Kotzin, M.D.
Ribosomal DNA Core Laboratory: Geoffrey McLennan, M.D., Ph.D.; Gary Hunninghake, M.D.
RNA Core Laboratory: Patricia Finn, M.D.
Random Digit Dialing Interview Group
Telesurveys Research Associates: Richard D. Jaffe, M.A.
Executive Committee
Reuben Cherniack, M.D. (Chair); Robert P. Baughman, M.D. (9/1/98–8/31/99); Joanne Deshler; Michael C. Iannuzzi, M.D. (9/1/96–8/31/97; 9/1/00–6/30/01); Marc A. Judson, M.D. (9/1/96–8/31/97); 9/1/00–6/30/01); Genell L. Knatterud, Ph.D.; Geoffrey McLennan, M.D. (9/1/97–8/31/98); David R. Moller, M.D. (9/1/95–3/31/96, 9/1/99–8/31/00); Robert A Musson, Ph.D.; Lee S. Newman, M.D.; Milton D. Rossman, M.D. (8/1/95–3/31/86, 9/1/99–8/31/00); Alvin S. Teirstein, M.D. (9/1/97–8/31/98); Michael L. Terrin, M.D., M.P.H.; Steven E. Weinberger, M.D. (9/1/97–3/31/98); Henry Yeager, Jr., M.D. (9/1/98–8/31/99)
Data Safety and Monitoring Board
William Martin, M.D. (Chair); Takamaru Ashikaga, Ph.D.; David B. Coultas, M.D.; Gerald S. Davis, M.D.; Fred Gifford, Ph.D.; James J. Schlesselman, Ph.D.; Diane Stover, M.D.
Ex Officio
Reuben Cherniack, M.D.; Genell L. Knatterud, Ph.D.; Robert Musson, Ph.D.; Lee Newman, M.D
Footnotes
This study was supported by National Institutes of Health grants T32HL082547 and F30HL137267-01 to the Division of Pulmonary, Critical Care, Sleep and Allergy Department of Medicine, University of Illinois at Chicago.
Author Contributions: C.A., Y.H., and C.S. conceived, designed, and performed experiments. Specimens and clinical data were collected by the ACCESS Research Group. C.A., C.S., B.A.T., and A.M. analyzed data. D.L.P. and P.W.F. contributed reagents and materials. C.A., D.L.P., and P.W.F. wrote the manuscript. All authors reviewed and approved the manuscript.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2017-0207OC on August 16, 2017
Author disclosures are available with the text of this article at www.atsjournals.org.
Contributor Information
Collaborators: the ACCESS Research Group, Steven E. Weinberger, Patricia Finn, Erik Garpestad, Allison Moran, Henry Yeager, David L. Rabin, Susan Stein, Michael C. Iannuzzi, Benjamin A. Rybicki, Marcie Major, Mary Maliarik, John Popovich, David R. Moller, Carol J. Johns, Cynthia Rand, Joanne Steimel, Marc A. Judson, Susan D’Alessandro, Nancy Heister, Theresa Johnson, Daniel T. Lackland, Janardan Pandey, Steven Sahn, Charlie Strange, Alvin S. Teirstein, Louis DePalo, Sheldon Brown, Marvin Lesser, Maria L. Padilla, Marilyn Marshall, Lee S. Newman, Cecile Rose, Juliana Barnard, Robert P. Baughman, Elyse E. Lower, Donna B. Winget, Geoffrey McLennan, Gary Hunninghake, Chuck Dayton, Linda Powers, Milton D. Rossman, Eddy A. Bresnitz, Ronald Daniele, Jackie Regovich, William Sexauer, Robert Musson, Joanne Deshler, Paul Sorlie, Margaret Wu, Reuben Cherniack, Lee Newman, Genell L. Knatterud, Michael L. Terrin, Bruce W. Thompson, Kathleen Brown, Margaret Frederick, Frances LoPresti, Patricia Wilkins, Martha Canner, Judy Dotson, McKesson Bioservices, Steve Lindenfelser, Mark Cosentino, Mary Maliarik, Robert Baughman, Milton Rossman, Dimitri Monos, Chung Wha Lee, Boyana Cizman, David Moller, Janardan Pandey, Peter Almenoff, Ian Brett, Sheldon Brown, Marvin Lesser, Lee Newman, Brian Kotzin, Geoffrey McLennan, Gary Hunninghake, Patricia Finn, Richard D. Jaffe, Reuben Cherniack, Robert P. Baughman, Joanne Deshler, Michael C. Iannuzzi, Marc A. Judson, Genell L. Knatterud, Geoffrey McLennan, David R. Moller, Robert A Musson, Lee S. Newman, Milton D. Rossman, Alvin S. Teirstein, Michael L. Terrin, teven E. Weinberger, Henry Yeager, William Martin, Takamaru Ashikaga, David B. Coultas, Gerald S. Davis, Fred Gifford, James J. Schlesselman, Diane Stover, Reuben Cherniack, Genell L. Knatterud, Robert Musson, and Lee Newman
References
- 1.Grunewald J, Spagnolo P, Wahlström J, Eklund A. Immunogenetics of disease-causing inflammation in sarcoidosis. Clin Rev Allergy Immunol. 2015;49:19–35. doi: 10.1007/s12016-015-8477-8. [DOI] [PubMed] [Google Scholar]
- 2.Iannuzzi MC, Rybicki BA, Teirstein AS. Sarcoidosis. N Engl J Med. 2007;357:2153–2165. doi: 10.1056/NEJMra071714. [DOI] [PubMed] [Google Scholar]
- 3.Huang H, Lu Z, Jiang C, Liu J, Wang Y, Xu Z. Imbalance between Th17 and regulatory T-cells in sarcoidosis. Int J Mol Sci. 2013;14:21463–21473. doi: 10.3390/ijms141121463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramstein J, Broos CE, Simpson LJ, Ansel KM, Sun SA, Ho ME, et al. IFN-γ-producing T-helper 17.1 cells are increased in sarcoidosis and are more prevalent than T-helper type 1 cells. Am J Respir Crit Care Med. 2016;193:1281–1291. doi: 10.1164/rccm.201507-1499OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hunninghake GW, Costabel U, Ando M, Baughman R, Cordier JF, du Bois R, et al. ATS/ERS/WASOG statement on sarcoidosis. American Thoracic Society/European Respiratory Society/World Association of Sarcoidosis and other Granulomatous Disorders. Sarcoidosis Vasc Diffuse Lung Dis. 1999;16:149–173. [PubMed] [Google Scholar]
- 6.Valeyre D, Bernaudin JF, Uzunhan Y, Kambouchner M, Brillet PY, Soussan M, et al. Clinical presentation of sarcoidosis and diagnostic work-up. Semin Respir Crit Care Med. 2014;35:336–351. doi: 10.1055/s-0034-1381229. [DOI] [PubMed] [Google Scholar]
- 7.Chopra A, Kalkanis A, Judson MA. Biomarkers in sarcoidosis. Expert Rev Clin Immunol. 2016;12:1191–1208. doi: 10.1080/1744666X.2016.1196135. [DOI] [PubMed] [Google Scholar]
- 8.Judson MA, Baughman RP, Thompson BW, Teirstein AS, Terrin ML, Rossman MD, et al. ACCESS Research Group. Two year prognosis of sarcoidosis: the ACCESS experience. Sarcoidosis Vasc Diffuse Lung Dis. 2003;20:204–211. [PubMed] [Google Scholar]
- 9.Judson MA, Baughman RP. Worsening of pulmonary sarcoidosis. Curr Opin Pulm Med. 2014;20:508–516. doi: 10.1097/MCP.0000000000000082. [DOI] [PubMed] [Google Scholar]
- 10.Paone G, Leone A, Batzella S, Conti V, Belli F, De Marchis L, et al. Use of discriminant analysis in assessing pulmonary function worsening in patients with sarcoidosis by a panel of inflammatory biomarkers. Inflamm Res. 2013;62:325–332. doi: 10.1007/s00011-012-0585-9. [DOI] [PubMed] [Google Scholar]
- 11.Casanova N, Zhou T, Knox KS, Garcia JGN. Identifying novel biomarkers in sarcoidosis using genome-based approaches. Clin Chest Med. 2015;36:621–630. doi: 10.1016/j.ccm.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gurrieri C, Bortoli M, Brunetta E, Piazza F, Agostini C. Cytokines, chemokines and other biomolecular markers in sarcoidosis. Sarcoidosis Vasc Diffuse Lung Dis. 2005;22:S9–S14. [PubMed] [Google Scholar]
- 13.Katchar K, Eklund A, Grunewald J. Expression of Th1 markers by lung accumulated T cells in pulmonary sarcoidosis. J Intern Med. 2003;254:564–571. doi: 10.1111/j.1365-2796.2003.01230.x. [DOI] [PubMed] [Google Scholar]
- 14.Mirsaeidi M, Banoei MM, Nienow CK, Abassi T, Hakim A, Schraufnagel D, et al. Plasma metabolomic profile in fibrosing pulmonary sarcoidosis. Sarcoidosis Vasc Diffuse Lung Dis. 2016;33:29–38. [PubMed] [Google Scholar]
- 15.Patterson KC, Franek BS, Müller-Quernheim J, Sperling AI, Sweiss NJ, Niewold TB. Circulating cytokines in sarcoidosis: phenotype-specific alterations for fibrotic and non-fibrotic pulmonary disease. Cytokine. 2013;61:906–911. doi: 10.1016/j.cyto.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sekiya M, Ohwada A, Miura K, Takahashi S, Fukuchi Y. Serum vascular endothelial growth factor as a possible prognostic indicator in sarcoidosis. Lung. 2003;181:259–265. doi: 10.1007/s00408-003-1028-8. [DOI] [PubMed] [Google Scholar]
- 17.Ziegenhagen MW, Rothe ME, Schlaak M, Müller-Quernheim J. Bronchoalveolar and serological parameters reflecting the severity of sarcoidosis. Eur Respir J. 2003;21:407–413. doi: 10.1183/09031936.03.00010403. [DOI] [PubMed] [Google Scholar]
- 18.Bloom CI, Graham CM, Berry MP, Rozakeas F, Redford PS, Wang Y, et al. Transcriptional blood signatures distinguish pulmonary tuberculosis, pulmonary sarcoidosis, pneumonias and lung cancers. PLoS One. 2013;8:e70630. doi: 10.1371/journal.pone.0070630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crouser ED, Culver DA, Knox KS, Julian MW, Shao G, Abraham S, et al. Gene expression profiling identifies MMP-12 and ADAMDEC1 as potential pathogenic mediators of pulmonary sarcoidosis. Am J Respir Crit Care Med. 2009;179:929–938. doi: 10.1164/rccm.200803-490OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koth LL, Solberg OD, Peng JC, Bhakta NR, Nguyen CP, Woodruff PG. Sarcoidosis blood transcriptome reflects lung inflammation and overlaps with tuberculosis. Am J Respir Crit Care Med. 2011;184:1153–1163. doi: 10.1164/rccm.201106-1143OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cushing L, Jiang Z, Kuang P, Lü J. The roles of microRNAs and protein components of the microRNA pathway in lung development and diseases. Am J Respir Cell Mol Biol. 2015;52:397–408. doi: 10.1165/rcmb.2014-0232RT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ebrahimi A, Sadroddiny E. MicroRNAs in lung diseases: recent findings and their pathophysiological implications. Pulm Pharmacol Ther. 2015;34:55–63. doi: 10.1016/j.pupt.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Crouser ED, Julian MW, Crawford M, Shao G, Yu L, Planck SR, et al. Differential expression of microRNA and predicted targets in pulmonary sarcoidosis. Biochem Biophys Res Commun. 2012;417:886–891. doi: 10.1016/j.bbrc.2011.12.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jazwa A, Kasper L, Bak M, Sobczak M, Szade K, Jozkowicz A, et al. Differential inflammatory microRNA and cytokine expression in pulmonary sarcoidosis. Arch Immunol Ther Exp (Warsz) 2015;63:139–146. doi: 10.1007/s00005-014-0315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiszałkiewicz J, Piotrowski WJ, Pastuszak-Lewandoska D, Górski P, Antczak A, Górski W, et al. Altered miRNA expression in pulmonary sarcoidosis. BMC Med Genet. 2016;17:2. doi: 10.1186/s12881-016-0266-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novosadova E, Chabronova A, Kolek V, Petrek M, Navratilova Z. The serum expression of selected miRNAs in pulmonary sarcoidosis with/without Löfgren’s syndrome. Mediators Inflamm. 2016;2016:1246129. doi: 10.1155/2016/1246129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crouser ED, Hamzeh NY, Maier LA, Julian MW, Gillespie M, Rahman M, et al. Exosomal microRNA for detection of cardiac sarcoidosis Am J Respir Crit Care Med [online ahead of print] 28 Feb 2017; DOI: 10.1164/rccm.201611-2183LE [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Group AR ACCESS Research Group. Design of a case control etiologic study of sarcoidosis (ACCESS) J Clin Epidemiol. 1999;52:1173–1186. doi: 10.1016/s0895-4356(99)00142-0. [DOI] [PubMed] [Google Scholar]
- 29.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 30.Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.R Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2016. Available from: http://www.r-project.org/. [Google Scholar]
- 34.Scicluna BP, Klein Klouwenberg PM, van Vught LA, Wiewel MA, Ong DS, Zwinderman AH, et al. A molecular biomarker to diagnose community-acquired pneumonia on intensive care unit admission. Am J Respir Crit Care Med. 2015;192:826–835. doi: 10.1164/rccm.201502-0355OC. [DOI] [PubMed] [Google Scholar]
- 35.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hastie T, Tibshirani R, Balasubramanian N, Chu G. PAM: prediction analysis for microarrays. [Updated 2015 Feb 20; accessed 2015 Oct 14]. Available from: https://cran.r-project.org/web/packages/pamr/pamr.
- 37.Oturai DB, Søndergaard HB, Börnsen L, Sellebjerg F, Christensen JR. Identification of suitable reference genes for peripheral blood mononuclear cell subset studies in multiple sclerosis. Scand J Immunol. 2016;83:72–80. doi: 10.1111/sji.12391. [DOI] [PubMed] [Google Scholar]
- 38.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci USA. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sweiss NJ, Salloum R, Gandhi S, Alegre ML, Sawaqed R, Badaracco M, et al. Significant CD4, CD8, and CD19 lymphopenia in peripheral blood of sarcoidosis patients correlates with severe disease manifestations. PLoS One. 2010;5:e9088. doi: 10.1371/journal.pone.0009088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Viskum K, Vestbo J. Vital prognosis in intrathoracic sarcoidosis with special reference to pulmonary function and radiological stage. Eur Respir J. 1993;6:349–353. [PubMed] [Google Scholar]
- 42.Fan Y, Siklenka K, Arora SK, Ribeiro P, Kimmins S, Xia J. miRNet—dissecting miRNA-target interactions and functional associations through network-based visual analysis. Nucleic Acids Res. 2016;44:W135–141. doi: 10.1093/nar/gkw288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ziegenhagen MW, Benner UK, Zissel G, Zabel P, Schlaak M, Müller-Quernheim J. Sarcoidosis: TNF-α release from alveolar macrophages and serum level of sIL-2R are prognostic markers. Am J Respir Crit Care Med. 1997;156:1586–1592. doi: 10.1164/ajrccm.156.5.97-02050. [DOI] [PubMed] [Google Scholar]
- 44.Mirsaeidi M, Gidfar S, Vu A, Schraufnagel D. Annexins family: insights into their functions and potential role in pathogenesis of sarcoidosis. J Transl Med. 2016;14:89. doi: 10.1186/s12967-016-0843-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rastogi R, Du W, Ju D, Pirockinaite G, Liu Y, Nunez G, et al. Dysregulation of p38 and MKP-1 in response to NOD1/TLR4 stimulation in sarcoid bronchoalveolar cells. Am J Respir Crit Care Med. 2011;183:500–510. doi: 10.1164/rccm.201005-0792OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Celada LJ, Rotsinger JE, Young A, Shaginurova G, Shelton D, Hawkins C, et al. Programmed Death-1 inhibition of phosphatidylinositol 3-kinase/AKT/mechanistic target of rapamycin signaling impairs sarcoidosis CD4(+) T cell proliferation. Am J Respir Cell Mol Biol. 2017;56:74–82. doi: 10.1165/rcmb.2016-0037OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baughman RP, Culver DA, Judson MA. A concise review of pulmonary sarcoidosis. Am J Respir Crit Care Med. 2011;183:573–581. doi: 10.1164/rccm.201006-0865CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ungprasert P, Carmona EM, Crowson CS, Matteson EL. Diagnostic utility of angiotensin-converting enzyme in sarcoidosis: a population-based study. Lung. 2016;194:91–95. doi: 10.1007/s00408-015-9826-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou T, Zhang W, Sweiss NJ, Chen ES, Moller DR, Knox KS, et al. Peripheral blood gene expression as a novel genomic biomarker in complicated sarcoidosis. PLoS One. 2012;7:e44818. doi: 10.1371/journal.pone.0044818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang B, Xi Y. Challenges for microRNA microarray data analysis. Microarrays (Basel) 2013;2:34–50. doi: 10.3390/microarrays2020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maertzdorf J, Weiner J, III, Mollenkopf HJ, Bauer T, Prasse A, Müller-Quernheim J, et al. TBornotTB Network. Common patterns and disease-related signatures in tuberculosis and sarcoidosis. Proc Natl Acad Sci USA. 2012;109:7853–7858. doi: 10.1073/pnas.1121072109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singla S, Zhou T, Javaid K, Abbasi T, Casanova N, Zhang W, et al. Expression profiling elucidates a molecular gene signature for pulmonary hypertension in sarcoidosis. Pulm Circ. 2016;6:465–471. doi: 10.1086/688316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wahlström J, Katchar K, Wigzell H, Olerup O, Eklund A, Grunewald J. Analysis of intracellular cytokines in CD4+ and CD8+ lung and blood T cells in sarcoidosis. Am J Respir Crit Care Med. 2001;163:115–121. doi: 10.1164/ajrccm.163.1.9906071. [DOI] [PubMed] [Google Scholar]
- 54.Miyara M, Amoura Z, Parizot C, Badoual C, Dorgham K, Trad S, et al. The immune paradox of sarcoidosis and regulatory T cells. J Exp Med. 2006;203:359–370. doi: 10.1084/jem.20050648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Facco M, Cabrelle A, Teramo A, Olivieri V, Gnoato M, Teolato S, et al. Sarcoidosis is a Th1/Th17 multisystem disorder. Thorax. 2011;66:144–150. doi: 10.1136/thx.2010.140319. [DOI] [PubMed] [Google Scholar]
- 56.Hunninghake GW, Crystal RG. Pulmonary sarcoidosis: a disorder mediated by excess helper T-lymphocyte activity at sites of disease activity. N Engl J Med. 1981;305:429–434. doi: 10.1056/NEJM198108203050804. [DOI] [PubMed] [Google Scholar]
- 57.Rutherford RM, Kehren J, Staedtler F, Chibout SD, Egan JJ, Tamm M, et al. Functional genomics in sarcoidosis—reduced or increased apoptosis? Swiss Med Wkly. 2001;131:459–470. doi: 10.4414/smw.2001.09808. [DOI] [PubMed] [Google Scholar]
- 58.Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 59.Das S, Raj L, Zhao B, Kimura Y, Bernstein A, Aaronson SA, et al. Hzf determines cell survival upon genotoxic stress by modulating p53 transactivation. Cell. 2007;130:624–637. doi: 10.1016/j.cell.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kerdiles YM, Stone EL, Beisner DR, McGargill MA, Ch’en IL, Stockmann C, et al. Foxo transcription factors control regulatory T cell development and function. Immunity. 2010;33:890–904. doi: 10.1016/j.immuni.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Du X, Shi H, Li J, Dong Y, Liang J, Ye J, et al. Mst1/Mst2 regulate development and function of regulatory T cells through modulation of Foxo1/Foxo3 stability in autoimmune disease. J Immunol. 2014;192:1525–1535. doi: 10.4049/jimmunol.1301060. [DOI] [PubMed] [Google Scholar]
- 62.Mulroy T, Sen J. p38 MAP kinase activity modulates alpha beta T cell development. Eur J Immunol. 2001;31:3056–3063. doi: 10.1002/1521-4141(2001010)31:10<3056::aid-immu3056>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 63.Xu P, Guo M, Hay BA. MicroRNAs and the regulation of cell death. Trends Genet. 2004;20:617–624. doi: 10.1016/j.tig.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 64.Tana C, Giamberardino MA, Di Gioacchino M, Mezzetti A, Schiavone C. Immunopathogenesis of sarcoidosis and risk of malignancy: a lost truth? Int J Immunopathol Pharmacol. 2013;26:305–313. doi: 10.1177/039463201302600204. [DOI] [PubMed] [Google Scholar]
- 65.Bonifazi M, Bravi F, Gasparini S, La Vecchia C, Gabrielli A, Wells AU, et al. Sarcoidosis and cancer risk: systematic review and meta-analysis of observational studies. Chest. 2015;147:778–791. doi: 10.1378/chest.14-1475. [DOI] [PubMed] [Google Scholar]
- 66.Rybicki BA, Major M, Popovich J, Jr, Maliarik MJ, Iannuzzi MC. Racial differences in sarcoidosis incidence: a 5-year study in a health maintenance organization. Am J Epidemiol. 1997;145:234–241. doi: 10.1093/oxfordjournals.aje.a009096. [DOI] [PubMed] [Google Scholar]
- 67.Westney GE, Judson MA. Racial and ethnic disparities in sarcoidosis: from genetics to socioeconomics. Clin Chest Med. 2006;27:453–462, vi. doi: 10.1016/j.ccm.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 68.Mirsaeidi M, Machado RF, Schraufnagel D, Sweiss NJ, Baughman RP. Racial difference in sarcoidosis mortality in the United States. Chest. 2015;147:438–449. doi: 10.1378/chest.14-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]