ABSTRACT
Aims: Multiple antimicrobial resistance in Escherichia coli of wild vertebrates is a global concern with scarce assessments on the subject from developing countries that have high human-wild species interactions. We studied the ecology of E. coli in a wintering population of Egyptian Vultures in India to understand temporal changes in both E. coli strains and patterns of antimicrobial resistance.
Methods and Results: We ribotyped E. coli strains and assessed antimicrobial resistance from wintering vultures at a highly synanthropic carcass dump in north-west India. Both E. coli occurence (90.32%) and resistance to multiple antimicrobials (71.43%) were very high. Clear temporal patterns were apparent. Diversity of strains changed and homogenized at the end of the Vultures’ wintering period, while the resistance pattern showed significantly difference inter-annually, as well as between arrival and departing individuals within a wintering cycle.
Significance of study: The carcass dump environment altered both E. coli strains and multiple antimicrobial resistance in migratory Egyptian Vultures within a season. Long-distance migratory species could therefore disseminate resistant E. coli strains across broad geographical scales rendering regional mitigation strategies to control multiple antimicrobial resistance in bacteria ineffective.
KEYWORDS: Antimicrobial resistance, carcass dump, Escherichia coli, microbial ecology, Egyptian vulture
Introduction
Escherichia coli occurs widely in the intestines and faeces of all vertebrates, and its prevalence and diversity in wild species varies with host taxonomy, geography, diet, and environmental conditions particularly the degree of synanthropy of the hosts’ surroundings [1–4]. The recognition of wild vertebrate species carrying E. coli is not recent, but the number of wild species and the range of environmental conditions that have been studied to date is biased to a handful of locations globally, with notable paucity of studies from densely-populated, developing countries [5].
Wide-ranging wild species hosting E. coli are subjected to widely varying environments including exposure to human-dominated environments, in a relatively short period of time facilitating development of multiple antimicrobial resistance in E. coli [6]. The prevalence of multiple antimicrobial resistance in commensal E. coli is a strong indicator of selective pressure to use of antimicrobials [7,8]. The level of resistance in bacteria of wild animals correlates well with the degree of association with human activity [5,9]. Also, more synanthropic bird species show increased multidrug resistance E. coli [10,11]. Wild vertebrate species, especially those that use human-dominated landscapes as part of their life history, are therefore likely to be ‘reservoirs and melting pots of bacterial resistance’ with increasing potential of resistant E. coli colonizing intestine and resulting in antimicrobial resistant infections if they end up in urinary tract or the bloodstream adding concern to human health [4,12].
Studies using wild animals have revealed microbial traits of E. coli that indicate its suitability as an indicator organism of multiple ecological aspects. E. coli showed a much greater geographical effect in non-domesticated, resident wild species relative to isolates in humans suggesting that differential movement patterns influence E. coli strains that predominate in a species [3]. Similarly, in wild species, E. coli exhibit very strong geographic effects, which is likely due to a combination of host taxonomy, diet and climate [2,13,14,15]. Phylogeny of E. coli of wild birds varied concomitant to exposure to humans and human-associated vertebrates such as domestic fowl underscoring the influence of anthropogenic components [5,10]. The near-absence of studies of the of E. coli in wild vertebrates in densely-populated countries is worrisome especially given the increased risk of wild species interacting with humans and domestic animals, and are urgently required [4]. In particular, documentation of the temporal and spatial patterns of antimicrobial drug resistance in different habitats and species in developing countries is essential to developing a cogent understanding of wildlife as a source of antimicrobial resistance [16,17]
Two consistent patterns of E. coli ecology are emerging from studies in wild vertebrates: (1) Population structure of E. coli strains strongly reflect the host’s environment and feeding habits, and (2) multiple antimicrobial resistant strains increase in more synanthropic settings. In this study, we tested if these two patterns hold in a unique environmental setting using an explicit temporal study design, and a migrant scavenger species, the Egyptian vulture (Neophron n. percnopterus) in its wintering grounds in north-western India.
The Egyptian vulture is ideal to test both patterns due to its natural history and habits, but microbial studies using this host species are absent to date [18]. The species breeds across a wide geographic range in Europe nesting on cliffs, displaying dietary plasticity that includes both wild and domestic vertebrates [18–20]. The migratory route of the Egyptian vulture can differ annually, but the species travels non-stop and rapidly to wintering grounds, displaying very strong inter-annual site fidelity, with movements in wintering sites restricted to a small area if food is abundant [20]. The population of Egyptian vultures wintering in north-western India stay for several months (October to March), some of them at a highly unusual and synanthropic site, a communal dump for livestock carcasses in Jorbeer, Rajasthan. Livestock carcasses from different clinical settings as well as dairies in Bikaner city are collected, dumped and skinned by resident workers. Such carcasses can be hubs for interactions between different host species that potentially harbour infectious agents [21]. Scavenging raptors’ diet in communal dumps are largely carcasses of farm animals which contain high rates of antimicrobial resistance [22]. The carcass dump, therefore, can facilitate intense interaction of diverse micro-biota including E. coli of human, livestock and multiple congregating, wintering wild bird species, in addition to exposure to unknown quantities and types of veterinary drugs in the carcasses. Conditions were therefore primed to expose Egyptian vultures to different E. coli assemblages relative to their breeding grounds, and also to develop multiple antimicrobial resistance.
The migrating Egyptian Vultures at Jorbeer are therefore exposed to widely varying environmental and synanthropic conditions in their breeding and wintering grounds. In line with the migratory behaviour of the vultures and conditions at Jorbeer, we asked three broad questions in our study: (1) does the population structure of commensal E. coli of the migrating vultures respond to spatial and temporal changes?; (2) could E. coli of the vultures attending communal carcass dumps show multiple antimicrobial resistance and if so; (3) would patterns of such resistance show clear spatial and temporal changes to match with the vultures’ wintering environment? Related to these questions and widely observed patterns of E. coli ecology in wild animals, we framed and tested two specific hypothesis that are conceptualized in the Supplemental Material (Fig. S1): (a) the environmental hypothesis – the diversity of E. coli strains in the vulture population recently arrived at Jorbeer (representing conditions and exposure at breeding grounds) will change as the vulture population spends more time in the wintering grounds, and that there will be increasing homogeneity in commensal E. coli strains just prior to departure reflecting exposure to similar environmental conditions at wintering grounds; and (b) the resistance hypothesis – the highly synanthropic nature of the carcass dump will result in very high incidence of antimicrobial resistance, with patterns of resistance changing progressively over the wintering season reflecting varying exposure to differing drugs and changing diet at the carcass dump. For both aspects, we also hypothesized strong inter-annual variations due to migratory behaviour of the species.
Materials and methods
Sample collection
Samples were collected throughout the wintering season of the vultures at a large carcass dump (approximate 0.25 Sq. Kms area) known as Jorbeer (N 27°57.958’ E 73°22.598’) in Rajasthan state, north-western India. Focal individuals of Egyptian Vultures were watched, and were approached immediately as they voided faecal lumps. One sample was taken from each individual vulture, samples were collected during mornings for 90 minutes in a single day for a given month of sampling, and were processed the same day within one hour. We minimized the probability of sampling the same individual vulture by moving focus rapidly to a different individual vulture after each sample collection. Since there was always over 500 vultures in the dump, we could not be certain of the landing location of previously sampled individuals. Samples were collected once during the pre-departure period (February 2011), and then sequentially in the subsequent year to reflect the subsequent full wintering period, which included arrival time (October 2011), resident period (November 2011), and departure time (March 2012) (Table 1). The sample therefore had a complete cross-section of one complete wintering period (2011–2012), and representation of the inter-annual variation represented by the bout of sampling in February 2011.
Table 1.
Sampling periods and frequency of isolation of the E. coli.
| Month and year of sampling | No. of samples | Frequency of isolation of E. coli | Frequency of isolation (in percent) |
|---|---|---|---|
| February, 2011 | 13 | 12 | 92.31 |
| October, 2011 | 13 | 12 | 92.31 |
| November, 2011 | 21 | 19 | 90.48 |
| March, 2012 | 15 | 13 | 86.67 |
| Total | 62 | 56 | 90.32 |
Isolating E. coli
In the laboratory, the swabs were inoculated in nutrient broth (Himedia, Mumbai) and incubated for 24 hours at 37°C. and processed further with standard biochemical tests for phenotypic characterization [23].
Isolating E. coli. In the laboratory, the swabs were inoculated in nutrient broth and incubated for 24 hours at 37°C. In short: A loop full of culture was streaked onto MacConkey agar (Himedia, Mumbai) and incubated aerobically at 37°C for 24 hours, pink colonies typical of E. coli were processed further streaked on EMB agar (Himedia, Mumbai) and incubated at 37°C for 24 hours. Colonies showing metallic sheen were randomly picked and streaked on nutrient agar and incubated at 37°C for 24 hours. Gram negative rods from catalase positive and oxidase negative colonies were processed further with standard biochemical tests for phenotypic characterization (23).
Ribotyping
As the design of this experiment focused on study of temporal changes in E. coli strains and not source-tracking, 16S–23S rRNA intergenic spacer region (16S-23S rRNA ISR) polymorphism (Ribotyping) was regarded as a suitable technique for studying intra species phylogeny in bacteria as this is least affected by horizontal gene transfers [24], while the flanking rRNA genes are highly conserved. Also, ribotyping is suitable in epidemiological studies for determining relatedness and discrimination of strains [25,26].
DNA from confirmed E. coli isolates was subsequently processed for ribotyping using 16S-23S r-RNA Intergenic Spacer Region (ISR) polymorphism by PCR. The Microbial Type Culture Collection (MTCC) 443 strain of E. coli was used as a positive control to validate the results. In short the ribotyping involved the following sequential steps:
DNA was extracted, checked for purity, integrity and ribotyped by published protocols [27–29].
Subsequently, the polyacrylamide gel electrophoresis (PAGE) images were analysed by PyElph software [30]. Bands across the lanes were matched based on position and number of bands, and a binary matrix, designating presence of band as ‘1’ and absence as ‘0’, was made using the software.
To confirm validity and reproducibility of the ribotyping process 14 isolates of the E. coli, one each from 14 random faecal samples, picking a different colony other than one used in previous ribotyping, were ribotyped again by 16S-23S r-RNA ISR polymorphism PCR as used in the previous ribotyping, which showed no changes compared to the previous ribotyping.
Antimicrobial susceptibility testing
The antimicrobial resistance pattern of the isolates was determined by disc diffusion method [31] using CLSI standards (2010). All of the pure E. coli cultures were subjected to resistotyping to 13 antimicrobials of different classes. Antimicrobials that were commonly used in veterinary as well as human medicine were included. Eleven classes of commonly used veterinary antimicrobials including combination of ampicillin with sulbactam (a beta-lactamase inhibitor) were chosen to study if the resistance patterns are influenced by the usage. In addition, we selected imipenem specifically as a stand-alone antimicrobial not used in veterinary practices to see if the E. coli could still develop resistance to this antimicrobial as well. This way, a total of 13 antimicrobials were used for antimicrobial susceptibility testing. All agar media and antimicrobial discs were sourced from HiMedia, Mumbai, India.
Control strain MTCC 443 was used as control. Multiple antimicrobial resistance (MAR) was defined as resistance to three or more than three antimicrobials of different chemical classes. A binary matrix (resistant = 1; sensitive = 0) was constructed based on response of the isolates to antimicrobials. The ‘intermediate’ responses were converted either into ‘resistant’ or ‘sensitive’ using lower and higher diameter of zone of inhibition, relative to reference values (of ‘intermediate’ response), respectively for statistical analyses.
Statistical analyses
All analyses were carried out on the R platform [32]. We wanted to estimate relatedness of E. coli strains sampled in each month to ascertain the validity of the environmental hypothesis. For this we used two methods: discriminant analysis of principal components (DAPC), and analysis of molecular variance (AMOVA), using packages adegenet and poppr respectively [33,34].
DAPC is a model-free method that does not assume populations are panmictic, and explores structures of populations by locating individuals into clusters after partitioning the variance of the sample into within- and between- group components. DAPC is sensitive to the number of principal components, so we used the function xvalDapc in package adegenet to select the correct number of components. We used 90% of the data as a training set with 1,000 replicates, and derived the minimum number of principal components as that having the lowest root mean squared value and the highest degree of successful reassignment of the samples. AMOVA was used to determine the degree of differentiation of variations between designated partitions (months and year in this case) using molecular markers. We used a hierarchy of year and month to understand if the E. coli strains varied not just between months of one full wintering period, but also if strains varied between years. Using the package ade4 and 1,000 permutations, we tested significant deviations of the data from a random population structure. In addition to computing significance levels of variations between months and years using AMOVA, we plotted the clustering of the populations using DAPC to visually assess the degree of partitioning of populations.
We tested the resistance hypothesis, or if patterns of antimicrobial resistance varied between months and years, using three different methods, each for a different resolution: non-metric multidimensional scaling (NMDS), dissimilarity between months using Jaccard’s index, and Sørensen’s β-diversity. These were implemented in R packages vegan, simba and betapart respectively [35–37]. Using package vegan, we implemented NMDS to assess if patterns of antimicrobial resistance exhibited by E. coli varied across months. NMDS is a gradient analyses that uses a rank–based approach to represent as closely as possible the dissimilarity between objects in a reduced number of dimensions. Original distance data is substituted by ranks, which does not require to meet assumptions regarding distributions of data sets. NMDS uses an iterative process, and produces stress values that can be used to assess the fit of the algorithm, with stress values < 0.2 indicating a reasonable to good fit, and values > 0.3 indicating that the ordination is arbitrary. We produced an NMDS plot to visually assess dissimilarities of microbial resistance of E. coli in different months. We carried out a statistical test using presence/absence matrices of the resistance pattern obtained each month using the multi-response permutation procedure (MRPP) also using package vegan. The MRPP statistic δ evaluates if there is a difference between two or more groups of sampling units, and represents the weighted-mean of within-group means of pairwise dissimilarities in the sampling units. A significant value would indicate considerable differences in antimicrobial resistance patterns between months. We then assessed pair-wise monthly differences with Jaccard’s indices using the function com.sim in package simba. To overcome sample size limitations, we ran 1,000 permutations of the data and derived statistical significance values to assess if variation was significant between months. This additional analysis was required since month-wise variation using only NMDS plots can be difficult to interpret directly owing to the arbitrary scales of the axis, and the reduction of dimensions.
The overall diversity of antimicrobial resistance of the E. coli occurring in wintering vultures at Jorbeer could be due to two different processes. The first is ‘turnover’, indicating acquirement of new resistance by E. coli incrementally as the wintering period progressed. Turnover would show sequential and consistent changes in resistance patterns over a period of time. The second is ‘nestedness’, where a large part or all of the resistance pattern is acquired in bursts over relatively short periods. This would suggest very rapid change in resistance patterns of the E. coli corresponding to a particular sampling period, with few to no additional new resistance being added as time lapsed. We assessed which pattern contributes to the overall β-diversity (or the Sørensen’s dissimilarity equivalent, βSOR), that can be parsed into two additive components [37]. The first is the level of turn-over (βSIM; or Simpson’s dissimilarity, which is independent of the number of antimicrobials), and the second was the level of nestedness (βNSE). These three measures, βSOR = βSIM + βNSE, would therefore provide not only the diversity of antimicrobial resistance experienced by vultures at Jorbeer, but also indicate the process that contributed to the observed resistance pattern. We used the function beta.sample to resample the data set 1,000 times to avoid systematic biases that can arise due to sample sizes, and obtained means and standard deviations of all three measures.
Finally, we assessed if resistance to an individual antimicrobial occurred more than by random chance in one or more months using the ‘indicator species analysis’ with package indicspecies [38]. We computed a single-antimicrobial single-month analyses to check how many of the antimicrobials showed significant variation in resistance patterns much more than by random chance in a single month. We also assessed if resistance of individual antimicrobials varied in more than one month significantly more than by random chance [39]. In short, while β-diversity provided metrics useful to understand the overall pattern and processes, the indicator species analysis identified individual antimicrobials whose non-random variation contributed to the overall patterns.
Results
Environmental hypothesis
The occurence of E. coli was high with very low variation throughout the sampling period (90.32 ± 2.66 (SD) % of samples tested positive; range = 86.67 – 92.31%). All the E. coli isolates (N = 56) showed variable patterns consisting of bands ranging from 800 to 3000 bp in the ribotyping visually displaying 16S-23S rRNA ISR polymorphism. The most parsimonious number of principal components for DAPC was seven (Supporting Material Figure S2), and the scatterplot confirmed our hypothesis of E. coli populations changing over the winter, and homogenizing in the later part of the wintering season (Figure. 1). The E. coli community in October was distinctly different from the other months, and were most similar in the months of November and March (Figure. 1). AMOVA confirmed significant difference within and between samples (P = 0.001 in each case; Supporting Material Figure S3). However, E. coli communities were not significantly different between years (P = 0.50) despite a considerable portion of the spread for February non-overlapping with other months as visible on the scatterplot (Figure. 1).
Figure 1.
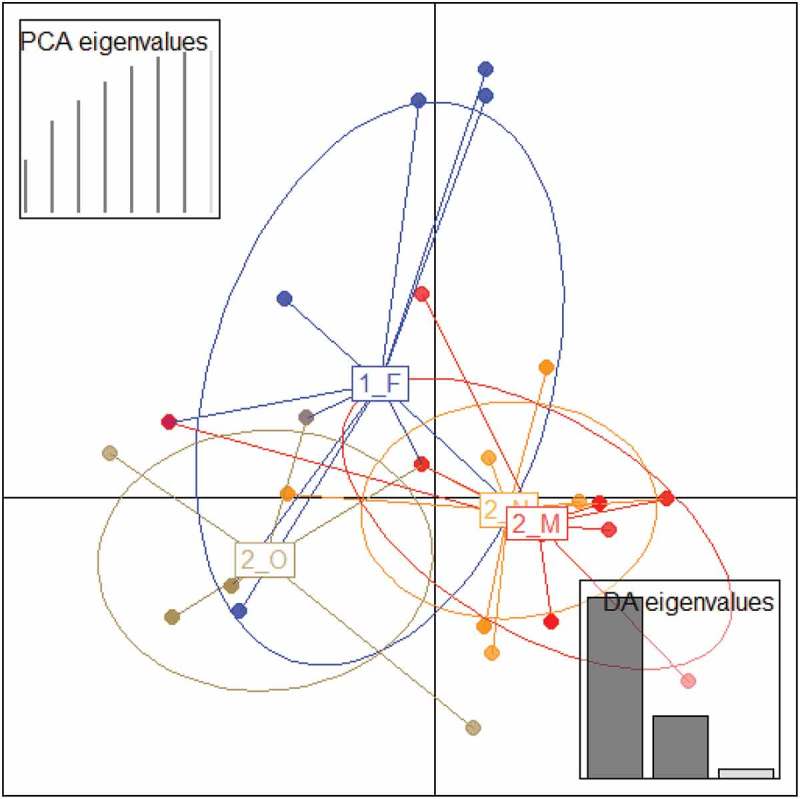
Scatterplot from discriminant analysis of principal components (DAPC) of the first two principal components discriminating E. coli communities colour-coded by month (F – February; O – October; N – November; M – March) across two wintering periods (1 – 2010-2011; 2 – 2011-2012). Each point represents actual data, and lines represent each population’s membership. Ellipses represent 67% confidence intervals based on a bivariate normal distribution. Number of principal components at which the maximal reassignment of samples occurred are depicted as black lines the PCA graph on the top-left corner, with subsequent components in grey line.
Resistance hypothesis
Results confirmed that E. coli in vultures that feed in carcass dumps had high incidence of MAR (72%) (Table 2). No resistance to imipenem and gentamicin was observed. Distinct variation in resistance patterns of cephotaxime and cefepime between February 2011 and October 2011 was apparent (Table 2). None of the E. coli was sensitive to cefepime in February 2011, but all isolates from March 2012 were sensitive to cefepime. The resistance patterns of chloramphenicol in February 2011 and March 2012 were identical (100% sensitivity). Overall, the E. coli showed temporally variable resistance patterns to 11 out of 13 antimicrobials used (Table 2).
Table 2.
Antimicrobial resistance pattern of the E. coli in four sampling efforts.
| Sr No. | Name of Antimicrobial | Response of the E. coli | February, 2011 (N = 12) |
October, 2011 (N = 12) |
November, 2011 (N = 19) |
March, 2012 (N = 13) |
Overall (N = 56) |
|---|---|---|---|---|---|---|---|
| 1 | Ampicillin | Sensitive | 0.00 | 0.00 | 0.00 | 7.69 | 1.79 |
| Intermediate | 33.33 | 0.00 | 0.00 | 0.00 | 7.14 | ||
| Resistance | 66.67 | 100.00 | 100.00 | 92.31 | 91.07 | ||
| 2 | Gentamicin | Sensitive | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 |
| Intermediate | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||
| Resistance | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||
| 3 | Imipenem | Sensitive | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 |
| Intermediate | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||
| Resistance | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||
| 4 | Ampicillin+ Sulbactam |
Sensitive | 100.00 | 83.33 | 68.42 | 53.85 | 75.00 |
| Intermediate | 0.00 | 0.00 | 21.05 | 46.15 | 17.86 | ||
| Resistance | 0.00 | 16.67 | 10.53 | 0.00 | 7.14 | ||
| 5 | Cephotaxime | Sensitive | 0.00 | 91.67 | 42.11 | 53.85 | 46.43 |
| Intermediate | 0.00 | 0.00 | 52.63 | 0.00 | 17.86 | ||
| Resistance | 100.00 | 8.33 | 5.26 | 46.15 | 35.71 | ||
| 6 | Cefepime | Sensitive | 0.00 | 91.67 | 78.95 | 84.62 | 66.07 |
| Intermediate | 25.00 | 0.00 | 5.26 | 15.38 | 10.71 | ||
| Resistance | 75.00 | 8.33 | 15.79 | 0.00 | 23.21 | ||
| 7 | Ofloxacin | Sensitive | 75.00 | 91.67 | 47.37 | 69.23 | 67.86 |
| Intermediate | 0.00 | 0.00 | 21.05 | 7.69 | 8.93 | ||
| Resistance | 25.00 | 8.33 | 31.58 | 23.08 | 23.21 | ||
| 8 | Erythromycin | Sensitive | 0.00 | 0.00 | 47.37 | 0.00 | 16.07 |
| Intermediate | 0.00 | 50.00 | 0.00 | 53.85 | 23.21 | ||
| Resistance | 100.00 | 50.00 | 52.63 | 46.15 | 60.71 | ||
| 9 | Co-trimoxazole | Sensitive | 58.33 | 75.00 | 73.68 | 53.85 | 66.07 |
| Intermediate | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||
| Resistance | 41.67 | 25.00 | 26.32 | 46.15 | 33.93 | ||
| 10 | Rifampicin | Sensitive | 91.67 | 58.33 | 73.68 | 53.85 | 69.64 |
| Intermediate | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||
| Resistance | 8.33 | 41.67 | 26.32 | 46.15 | 30.36 | ||
| 11 | Chloramphenicol | Sensitive | 100.00 | 83.33 | 78.95 | 100.00 | 89.29 |
| Intermediate | 0.00 | 0.00 | 5.26 | 0.00 | 1.79 | ||
| Resistance | 0.00 | 16.67 | 15.79 | 0.00 | 8.93 | ||
| 12 | Ciprofloxacin | Sensitive | 66.67 | 83.33 | 52.63 | 46.15 | 60.71 |
| Intermediate | 8.33 | 8.33 | 10.53 | 38.46 | 16.07 | ||
| Resistance | 25.00 | 8.33 | 36.84 | 15.38 | 23.21 | ||
| 13 | Tetracyclin | Sensitive | 41.67 | 16.67 | 10.53 | 7.69 | 17.86 |
| Intermediate | 16.67 | 41.67 | 31.58 | 7.69 | 25.00 | ||
| Resistance | 41.67 | 41.67 | 57.89 | 84.62 | 57.14 |
Results of NMDS had an overall stress of 0.137 and a nearly linear stress plot (R2 = 0.98; Supporting Material, Figure S4). The NMDS graph showed clear separation between years (as depicted by the non-overlapping polygon for February), and also revealed varying levels of overlaps within the wintering period of 2011–2012 (October and November 2011, and March 2012; see Figure. 2). MRPP confirmed an unambiguous differentiation of antimicrobial resistance patterns between months (δ = 0.001, chance corrected within-group agreement A = 0.172, 1,000 permutations). Pair-wise comparison of mean dissimilarity using Jaccard’s index revealed that the E. coli from most months had significantly different resistance patterns (Table 3). The Jaccard’s index and NMDS clearly suggest that the most diverse resistance patterns of E. coli was in November. Resistance pattern in October was most similar to other months. The Sørensen’s β-diversity (βSOR = 0.554 ± 0.05) was almost equally partitioned between turnover (βSIM = 0.328 ± 0.09) and nestedness (βNSE = 0.226 ± 0.07). Indicator-species analysis revealed that rifampicin and tetracyclin displayed temporal variations in resistance patterns occurring more than expected by random chance in February 2011 (Table 4). Cefotaxime and cefepime also showed this variation in October 2011 (Table 4(a)), and within the wintering cycle (October, November 2011 and March, 2012; Table 4(b)).
Figure 2.
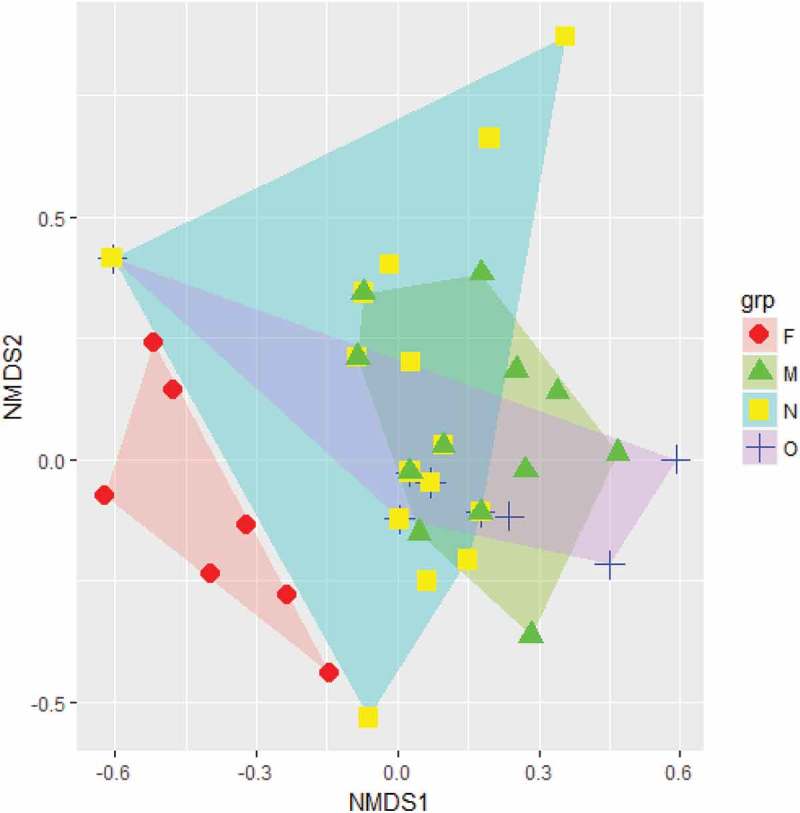
Non-metric multidimensional scaling (NMDS) plot representing resistance to varied antimicriobials by E. coli in Egyptian vultures across different months (F – February; O – October; N – November; M – March) in two-dimensional space.
Table 3.
Pair-wise comparisons of dissimilarity in E. coli ribotypes in a wintering Egyptian Vulture population in Jorbeer using Jaccard’s index. (* – P < 0.05; n.s. – not significant).
| October 2011 | November 2011 | March 2012 | |
|---|---|---|---|
| February 2011 | n.s. | * | * |
| October 2011 | * | n.s. | |
| November 2011 | * |
Table 4.
Results of the indicator species analyses showing antimicrobials with temporal variation more than by random chance in (a) single months, and (b) multiple months.
| A. Single-antimicrobial, single-month (Indicator values, with P-value in parenthesis) | |||||
|---|---|---|---|---|---|
| Antimicrobial | February, 2011 | October, 2011 | November, 2011 | March, 2012 | |
| Cephotaxime | 0.602 (0.015) | ||||
| Cefepime | 0.596 (0.015) | ||||
| Rifampicin | 0.568 (0.015) | ||||
| Tetracyclin |
0.527 (0.01) |
|
|
|
|
| B. Single-antimicrobial, multiple-months. | |||||
| Antimicrobial |
Indicator Value (P) |
February, 2011 |
October, 2011 |
November, 2011 |
March, 2012 |
| Cephotaxime | 0.953 (0.005) | + | + | + | |
| Cefepime | 0.965 (0.005) | + | + | + | |
Discussion
Prevalence
Information on prevalence of E. coli is not known in Egyptian vultures from other locations, but similarly high levels of prevalence was observed in the scavenging Eurasian griffon (90%) and other captive birds of prey (89%) [40,41]. High prevalence, however, was not common across all vultures (36–38% in cinereous vultures) or all raptors (15% in common buzzards) suggesting that species-level investigations are necessary to evaluate E. coli prevalence in this group of birds [42]. Contrary to our observations, effect of spatial differences were insignificant on the prevalence of E. coli in red kites, also a highly synanthropic species [43]. High occurence of E. coli strains matches well with the strongly synanthropic habit of the Egyptian vultures in their wintering grounds. This in turn matches past observations of higher prevalence in more synanthropic host species [11,44].
Temporal variation in E. coli
DAPC was very useful to reveal temporal variations in the E. coli community found in Egyptian vultures. The strong overlapping of November and March (see Figure. 1) suggests that homogenization of E. coli communities occur relatively early in the wintering season. Our results show that despite a statistical non-significance, inter-annual variations in E. coli communities were visible. However, it will be necessary to obtain data that completely cover full wintering periods for multiple years to understand if inter-annual variations are significant, and if they occur across multiple years. It would be useful to conduct similar studies with a temporal design in both migratory and non-migratory species to understand if patterns obtained in our study are shared across different environmental conditions and species, or if these patterns occur only in highly synanthropic species.
Patterns of MAR
Results confirmed our hypotheses that E. coli in Egyptian vultures feeding in carcass dumps will have a very high incidence of MAR (72%). Raptors have been known to harbour higher incidence of MAR relative to other bird species [45]. High MAR incidence during November to February reflects the unique conditions at the carcass dump, but high incidence in October reflects conditions in vultures’ breeding areas. This is suggestive of Egyptian Vultures being exposed to treated carcasses throughout their life cycle. This level of occurrence reflects the unique conditions present at the carcass dump – exceedingly high levels of human activity, and the presence of mostly pre-treated animal carcasses for vultures to feed on. Studies on wild birds have unfortunately not always noted environmental conditions, including an objective measurement of synanthropy, making it difficult to draw ecological conclusions using merely observed incidence values of MAR in past studies [5].
Gentamicin, an aminoglycoside, was the only commonly used antimicrobial that all samples of E. coli showed sensitivity to (see Table 2). This is significant in view of more than 50 aminoglycoside-modifying enzymes conferring widespread resistance with E. coli harbouring ‘key-resistance’ to aminoglycosides [46,47].
Determinants of resistance to tetracyclines, specifically plasmid borne gram-negative efflux genes are widespread [48]. Inter-species transfer of these genes can take place between different species of bacteria as well.
Grouping the resistance to ampicillin, cephotaxime and cefepime, the highest resistance was observed in the E. coli sampled in February, 2011 (67, 100, and 75% respectively). However, no resistance to combination of ampicillin and sulbactam was observed in this period suggesting relatively high proportion of extended spectrum β-lactamases (ESBL) producing strains. Cephotaxime and cefepime displayed most conspicuous and statistically significant changes in pattern of resistance both at the end of the 2010–2011 wintering season in February 2011 as well as across the 2011–2012 wintering period from October 2011 to March 2012. (Table 4(a,b)). This translates into resistance to cephotaxime and cefepime more likely to be temporally structured and that the change of habitat could have affected the associated resistance determinants. This variation could possibly be attributed to more than 400 wide-spread horizontally transferred genes coding β-lactamases in E. coli and permissibility of genome of E. coli to such genes, though chromosomal genes providing intrinsic resistance are not uncommon [49]. While many studies report ESBL in E. coli of wild species, the response of migration on cephalopsporin resistance determinants is poorly known [17].
NMDS analysis indicated that most antimicrobials showed inter-annual variation in resistance patterns, except the ones the E. coli were completely resistant or susceptible to throughout the study. This variability was high for cephotaxime, cefepime, erythromycin rifampicin, very low for cotrimoxazole, ofloxacin and absent in chloramphenicol (Table 2). The highest diversity of the resistotypes observed in November 2011 (Figure. 2) is suggestive of vultures arriving from different breeding grounds (See Supporting Material Figure S1), potentially leading to intense and increased interactions of E. coli and associated resistance determinants. This is also, therefore, the time period that also likely most closely reflects resistance profiles relating to their breeding grounds. Conducting studies simultaneously in the breeding and wintering grounds over multiple years can provide undisputable insights related to variation in resistance profile patterns, and to ascertain if some antimicrobials show consistent inter-annual variations more than others. Such an investigation is critical in management strategies to identify antimicrobial classes more prone to dissemination through wild migratory species.
We are not aware of other attempts to understand the processes driving overall diversity (Sørensen’s β-diversity in our case) of resistance profiles of E. coli. A nearly-equal partitioning of overall diversity to turnover and nestedness shows that our resistance hypothesis was substantiated, but also that there were episodic increases in resistance to novel antimicrobials. The strong dissimilarity of resistance patterns using Jaccard’s index, and the identification of two individual antimicrobials (cephotaxime and cefepime) with marked variations in resistance profile in a few months than could be expected by chance, provide additional evidence of multiple active processes by which E. coli in Egyptian vultures are gaining MAR.
The observed distinctive temporal patterns (Figure. 2) of these strains suggest that, apart from imparting resistance determinants, the dump did act as a ‘melting pot’ of antimicrobial resistance with the potential to disseminate resistant E. coli to multiple geographical locations (breeding grounds of the vultures) corroborating the contention of [12].
It seems that resistotypes are more sensitive to the effect of temporal and geographical variations compared to the E. coli isolates in this study. This points to the involvement of extra-nuclear genetic interactions governing resistance factors (resistance plasmids and integrons), which could additionally provide variation in resistance patterns along with chromosomal DNA compared to exclusive chromosomal DNA based changes accounted for clonal differences in E. coli. Antimicrobial agents contribute to selection pressure in commensals E. coli [50]. While some phylogenetic groups (A and some D group strains) of E. coli are more permissive to develop resistance to third generation cephalosporins whereas group B2 strains are less likely to develop resistance compared to the other groups [51–53]. This variability influences the strain structures of E. coli in hosts frequently exposed to antimicrobials [4]. Clearly, MAR E. coli strains are surviving, persisting and propagating in the gut of the vultures. Combining the observed patterns of changes in population structure (ribotypes) and MAR patterns provide further insights into E. coli ecology. For example, selection pressure may have contributed to the homogenization of the E. coli strains observed at the end of the migratory period 2011–2012. The recurrence of some strains in multiple years (see Figure. 1) suggests that selection pressures working over multiple years could favour persistence of MAR E. coli strains over relatively susceptible ones.
These findings are significant from an epidemiological point of view in that, while places like the Jorbeer carcass dump are important food sources to a number of raptor species of global conservation concern, they are also likely locations where diet (livestock carcasses with antimicrobials) and selection pressure can produce dangerously high levels of MAR E. coli in migratory bird species. Apart from MAR E. coli, Egyptain vultures could carry other potentially pathogenic and virulent MAR bacteria like Klebsiella pneumoniae and Pseudomonas aeruginosa [54,55] as well suggesting the spectrum of MAR bacteria of public health significance associated with vultures (and other migratory birds) is bigger than presently known.
MAR bacterial strains could be dispersed and colonized at new locations due to migratory bird species [56,57]. Environmental conditions and presence of high levels of antimicrobials as found in Jorbeer are likely common in many developing countries that have high human and livestock densities. Studies on antimicrobial resistance in bacteria of wild species are very rare in such countries [5]. Coverage of varied conditions and wild vertebrate species using robust spatial and temporal designs are entirely absent. This lacuna of even basic information requires to be urgently rectified given the unregulated and injudicious usage of antimicrobials, intensive livestock presence, high human densities, combined with very high degree of synanthropy shown by wild bird species in south Asia. We advocate multi-disciplinary approaches involving ecological, veterinary and microbiological components. Vultures render immense ecological benefit by disposing livestock carcasses without human resources investment, and the conservation of these endangered species is exceedingly important. However, the role of such migratory species in future remedial strategies to control spread of antimicrobial resistance should also be carefully looked into.
This study shows that Egyptian vultures feeding in a livestock carcass dump in north-west India: (1) had high occurence (90.32%) of E. coli, with strains showing host-species restriction and limited diversity with clear temporal structuring, and (2) E. coli showed 71.43% MAR, with the resistance patterns showing strong spatial and temporal structuring. E. coli in migrating Egyptian vultures, therefore, displayed excellent ability to incorporate and reflect resistance determinants prevailing both at the dump and the period of sampling. This strongly suggests that solely local or regional measures to spread of antimicrobial resistance is unlikely to be adequate, and we echo previous calls to execute more comprehensive and interdisciplinary mitigation methods concomitant with the ecology of migratory species such as Egyptian vultures [5].
Acknowledgments
We thank the Department of Forests, Government of Rajasthan, especially Partap Kataria, for rapidly processing the sampling permits. PS thanks the Department of Microbiology and Biotechnology, Rajasthan University of Veterinary and Animal sciences for infrastructural support and encouragement. KSGS thanks the Nature Conservation Foundation and the International Crane Foundation for institutional support.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1]. Souza V, Rocha M, Valera A, et al. Genetic structure of natural populations of Escherichia coli in wild hosts on different continents. Appl Environ Microbiol. 1999;65:3373–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2]. Gordon DM, Cowling A.. The distribution and genetic structure of Escherichia coli in Australian vertebrates: host and geographic effects. Microbiology. 2003;149:3575–3586. [DOI] [PubMed] [Google Scholar]
- [3]. Escobar-Paramo P, Le Menac’h A, Le Gall T, et al. Identification of forces shaping the commensal Escherichia coli genetic structure by comparing animal and human isolates. Environ Microbiol. 2006;8:1975–1984. [DOI] [PubMed] [Google Scholar]
- [4]. Tenaillon O, Skurnik D, Picard B, et al. The population genetics of commensal Escherichia coli . Nat Rev Microbiol. 2010;8:207–217. [DOI] [PubMed] [Google Scholar]
- [5]. Vittecoq M, Durand P, Brazier L, et al. Antimicrobial resistance in wildlife. J Appl Ecol. 2016;53:519–529. [Google Scholar]
- [6]. Williams N, Sherlock C, Jones T, et al. The prevalence of antimicrobial-resistant Escherichia coli in sympatric wild rodents varies by season and host. J Appl Microbiol. 2011;110:962‒970. [DOI] [PubMed] [Google Scholar]
- [7]. Fluckey WM, Loneragan WGH, Warner R, et al. Antimicrobial drug resistance of Salmonella and Escherichia coli isolates from cattle feces, hides, and carcasses. J Food Prot. 2007;70:551–556. [DOI] [PubMed] [Google Scholar]
- [8]. Salehi TZ, Bonab SF. Antibiotics susceptibility pattern of Escherichia coli strains isolated from chickens with Colisepticemia in Tabriz province, Iran. Int J Poult Sci. 2006;5:677–684. [Google Scholar]
- [9]. Skurnik D, Ruimy R, Andremont A, et al. Effect of human vicinity on antimicrobial resistance and integrons in animal faecal Escherichia coli . J Antimicrob Chemother. 2006;57:1215–1219. [DOI] [PubMed] [Google Scholar]
- [10]. Blyton MDJ, Pi H, Vangchhia B, et al. Genetic structure and antimicrobial resistance of Escherichia coli and cryptic clades in birds with diverse human associations. Appl Environ Microbiol. 2015;81:5123–5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11]. Allen HK, Donato J, Wang HH, et al. Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol. 2010;8:251–259. [DOI] [PubMed] [Google Scholar]
- [12]. Radhouani H, Silva N, Poeta P, et al. Potential impact of antimicrobial resistance in wildlife, environment, and human health. Front Microbiol. 2014;5:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13]. Gordon DM. Geographical structure and host specificity in bacteria and the implications for tracing the source of coliform contamination. Microbiol. 2001;147:1079–1085. [DOI] [PubMed] [Google Scholar]
- [14]. Sherley M, Gordon D, Cillingnon P. Variations in antibiotic resistance profiles of Enterobacteriaceae isolated from wild Australian mammals. Env Microbiol. 2000;2:620–631. [DOI] [PubMed] [Google Scholar]
- [15]. Carlos C, Pires MM, Stoppe NC, et al. Escherichia coli phylogenetic group determination and its application in the identification of the major animal source of fecal contamination. BMC Microbiol. 2010;10:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16]. Hernandez J, Bonnedahl J, Eliasson I, et al. Globally disseminated human path- ogenic Escherichia coli of O25b- ST131 clone, harbouring blaCTX- M-15, found in glaucous-winged gull at remote Commander Islands, Russia. Environ Microbiol Rep. 2010;2:329–332. [DOI] [PubMed] [Google Scholar]
- [17]. Guenther S, Ewers C, Wieler LH, et al. Extended-spectrum beta-lactamases producing E. coli in wildlife, yet another form of environmental pollution? Front Microbiol. 2011;2:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18]. Iñigo A, Barov B, Orhun C, et al. Action plan for the Egyptian vulture Neophron percnopterus in the European union. BirdLife International for the European Commission 2008;62 pp. .
- [19]. Margalida A, García D, Cortéz-Avizanda A. Factors influencing the breeding density of Bearded vultures, Egyptian vultures and Eurasian griffon vultures in Catalonia (NE Spain): management implications. Anim Biodiv Conserv. 2000;30:189–200. [Google Scholar]
- [20]. López-López P, García-Ripollés C, Urios V. Individual repeatability in timing and spatial flexibility of migration routes of trans-Saharan migratory raptors. Curr Zool. 2014;60:642‒652. [Google Scholar]
- [21]. Ragg J, Mackintosh C, Moller H. The scavenging behaviour of ferrets (Mustela furo), feral cats (Felis domesticus), possums (Trichosurus vulpecula), hedgehogs (Erinaceus europaeus) and harrier hawks (Circus approximans) on pastoral farmland in New Zealand: implications for bovine tuberculosis transmission. N Z Vet J. 2000;48:166–175. [DOI] [PubMed] [Google Scholar]
- [22]. Kang H, Jeong Y, Oh J, et al. Characterization of antimicrobial resistance and class 1 integrons found in Escherichia coli isolates from humans and animals in Korea. J Antimicrob Chemother. 2005;55:639–644. [DOI] [PubMed] [Google Scholar]
- [23]. Barrow GI, Feltham RKA. Cowan and Steel’s manual for the identification of medical bacteria. London: Cambridge University Press; 2004. [Google Scholar]
- [24]. Cohen O, Gophan U, Pupko T. The complexity hypothesis revisited: connectivity rather than function constitutes a barrier to horizontal gene transfer. Mol Biol Evol. 2011;28:1481–1489. [DOI] [PubMed] [Google Scholar]
- [25]. Seurinck S, Verstraete W, Siciliano SD. Use of 16S-23S rRNA intergenic spacer region PCR and repetitive extragenic palindromic PCR analyses of Escherichia coli isolates to identify nonpoint fecal sources. Appl Environ Microbiol. 2003;69:4942–4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26]. Buchan A, Alber M, Hodson RE. Strain-specific differentiation of environmental Escherichia coli isolates via denaturing gradient gel electrophoresis (DGGE) analysis of the 16S −23S intergenic spacer region. FEMS Microbiol Ecol. 2001;35:313–321. [DOI] [PubMed] [Google Scholar]
- [27]. Chen W, Kuo T. A simple and rapid method for the preparation of gram- negative bacterial genomic DNA. Nucleic Acids Res. 1993;21:2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28]. Sambrook J, Russell DW. Molecular Cloning: A laboratory manual. 3rd ed. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- [29]. Kostman JR, Edlind TD, LiPuma JJ, et al. Molecular epidemiology of Pseudomonas cepacia determined by polymerase chain reaction ribotyping. J Clin Microbiol. 1992;30:2084–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30]. Pavel AB, Vasile CI. PyElph - a software tool for gel images analysis and phylogenetics. BMC Bioinformatics. 2012;13:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31]. Bauer A, Kirby W, Sherris J, et al. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45:493–496. [PubMed] [Google Scholar]
- [32]. R Core Team R: A language and environment for statistical computing [Computer Program]. R Foundation for Statistical Computing: Vienna; 2016. ISBN 3-900051-07-0, Available from: http://www.R-project.org/.
- [33]. Jombart T, Lyon D, Biome LD. adegenet : a R package for the multivariate analysis of genetic markers. Bioinformatics Appl Note. 2008;24:1403–1405. [DOI] [PubMed] [Google Scholar]
- [34]. Kamvar ZN, Tabima JF, Grünwald NJ. Poppr : an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ. 2014;2:e281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35]. Oksanen J, Blanchet F, Friendly M, et al. vegan: community ecology package [Computer Program]. R package version 2.4-1. 2016. Available from: https://CRAN.R-project.org/package=vegan.
- [36]. Jurasinski G, Retzer V. simba: A collection of functions for similarity analysis of vegetation data [Computer program]. R package version 0.3-5. 2012. Available from: https://CRAN.R-project.org/package=simba.
- [37]. Baselga A, Orme C. betapart: an R package for the study of beta diversity. Methods Ecol Evol. 2012;3:802–812. [Google Scholar]
- [38]. Dufrêne M, Legendre P. Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol Monogr. 1997;67:354–366. [Google Scholar]
- [39]. De Cáceres M, Legendre P, Wiser S, et al. Using species combinations in indicator value analyses. Methods Ecol Evol. 2012;3:973–982. [Google Scholar]
- [40]. Kocijan I, Prukner-Radovčić E, Beck R, et al. Microflora and internal parasites of the digestive tract of Eurasian griffon vultures (Gyps fulvus) in Croatia. Eur J Wildl Res. 2009;55:71–74. [Google Scholar]
- [41]. Bangert RL, Ward AC, Stauber EH, et al. A survey of the aerobic bacteria in the feces of captive raptors. Avian Dis. 1988;32:53–62. [PubMed] [Google Scholar]
- [42]. Radhouani H, Pinto L, Coelho C, et al. Detection of Escherichia coli harbouring extended-spectrum {beta}-lactamases of the CTX-M classes in faecal samples of common buzzards (Buteo buteo). J Antimicrob Chemother. 2010;65:171–173. [DOI] [PubMed] [Google Scholar]
- [43]. Blanco G, Lemus JA, Grande J. Faecal bacteria associated with different diets of wintering red kites: influence of livestock carcass dumps in microflora alteration and pathogen acquisition. J Appl Ecol. 2006;43:990–998. [Google Scholar]
- [44]. Bonnedahl J, Järhult JD. Antibiotic resistance in wild birds. Ups J Med Sci. 2014;119:113–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45]. Guenther S, Grobbel M, Lubke- Becker A, et al. Antimicrobial resistance profiles of Escherichia coli from common European wild bird species. Vet Microbiol. 2010;144:219–225. [DOI] [PubMed] [Google Scholar]
- [46]. Schmitz F, Fluit A. Mechanisms of resistance In: Armstrong D, Cohen S, editors. Infectious Diseases. London: Mosby Limited; 1999. p. 7.2.1–7.2.14. [Google Scholar]
- [47]. Alekshun MN, Levy SB. Review molecular mechanisms of antibacterial multidrug resistance. Cell. 2007;128:1037–1050. [DOI] [PubMed] [Google Scholar]
- [48]. Levy SB. Tetracycline resistance determinants are widespread. ASM News. 1988;54:418–421. [Google Scholar]
- [49]. Bush K, Palzkill T. [Internet] Beta-lactamase classification and amino acid sequences for TEM, SHV and OXA extended-spectrum and inhibitor resistant enzymes. [cited 2017 May29] Available from: www.lahey.org/studies/
- [50]. van den Bogaard AE, Stobberingh EE. Epidemiology of resistance to antibiotics. Links between animals and humans. Int J Antimicrob Agents. 2000;14:327–335. [DOI] [PubMed] [Google Scholar]
- [51]. Mammeri H, Galleni M, Nordmann P. Role of the Ser- 287-Asn replacement in the hydrolysis spectrum extension of AmpC betalactamases in Escherichia coli . Antimicrob Agents Chemother. 2009;53:323–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52]. Deschamps C, Clermont O, Hipeaux MC, et al. Multiple acquisitions of CTX-M plasmids in the rare D2 genotype of Escherichia coli provide evidence for convergent evolution. Microbiology. 2009;155:1656–1668. [DOI] [PubMed] [Google Scholar]
- [53]. Johnson JR, Orskov I, Orskov F, et al. O, K, and H antigen predict virulence factors, carboxylesterase B pattern, antimicrobial resistance, and host compromise among Escherichia coli strains causing urosepsis. J Infect Dis J Infect Dis. 1994;169:119–126. [DOI] [PubMed] [Google Scholar]
- [54]. Sharma S, Mudgal N, Sharma P, et al. Phenotypic and genotypic characterization of Klebsiella pneumoniae obtained from Egyptian vultures and Steppes Eagles from India. Isr J Vet Med. 2015;69:122–128. [Google Scholar]
- [55]. Sharma P, Faridi F, Mir IA, et al. Characterization of exo-s, exo-u, and alg virulence factors and antimicrobial resistance in Pseudomonas aeruginosa isolated from migratory Egyptian vultures from India. Infect Ecol Epidemiol. 2014;4:24553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56]. Pedro JG, García De Los Ríos JE, Pozuelo MJ, et al. β-lactam resistance in Escherichia coli isolates from raptors in Spain In: Mendez-Vilas A, editor. Current research topics in applied microbiology and microbial biotechnology. Singapore: World Scientific; 2009. p. 483–486. [Google Scholar]
- [57]. Literak I, Micudova M, Tausova D, et al. Plasmid-mediated quinolone resistance genes in fecal bacteria from rooks commonly wintering throughout Europe. Microb Drug Resist. 2012;18:567–573. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


