Abstract
Aim:
Mutated or overexpressed FLT3 drives about 30% of reported acute myeloid leukemia (AML). Currently, FLT3 inhibitors have shown durable clinical responses but a complete remission of AML with FLT3 inhibitors remains elusive due to mutation-driven resistance mechanisms. The development of FLT3 inhibitors that also target other downstream oncogenic kinases may combat the resistance mechanism.
Results:
4-substituted aminoisoquinoline benzamides potently inhibit Src-family kinases and FLT3, including secondary mutations, such as FLT3D835. Modifications of aminoisoquinoline benzamide to aminoquinoline or aminoquinazoline abrogated FLT3 and Src-family kinase binding.
Conclusion:
The lead aminoisoquinolines potently inhibited FLT3-driven AML cell lines, MV4–11 and MOLM-14. These aminoisoquinoline benzamides represent new kinase scaffolds with high potential to be translated into anticancer agents.
Keywords: : acute myeloid leukemia, FLT3, MOLM-14, multikinase inhibitor, MV4–11, receptor tyrosine kinase, SRC-family kinase
Receptor tyrosine kinases (RTKs) regulate many key biological processes in the cell. Disruption of RTK signaling results in loss of cell homeostasis resulting in cancer and death [1]. For example, RTK-driven cancers are a result of increased expression of many RTKs, activating mutations or fusions that can lead to uncontrolled cellular proliferation [2]. Over the past few years, many tyrosine kinase inhibitors (TKIs) have been identified and used successfully in the clinic to combat different forms of cancers [3]. Imatinib (Gleevec®), an RTK inhibitor (TKI) of BCR-ABL and c-Kit, is a classic example that revolutionized the treatment of chronic myeloid leukemia (CML), Philadelphia chromosome positive (Ph+) acute lymphoblastic leukemia (Ph+ ALL) and gastrointestinal stromal tumors [4]. Response rates to many TKIs are high for the first few months, but over time secondary mutations emerge that render the cancer resistant to the TKI [5]. For example, T315I mutations in ABL1 are resistant to imatinib and many other ABL1 inhibitors [5]. Therefore, there is still a need for newer mutation-resistant generation of TKIs.
We have developed novel inhibitors for the FLT3 receptor, which is an important class III RTK and expressed on hematopoietic progenitor cells to regulate hematopoiesis [6]. The extracellular domain of the FLT3 RTK binds FLT3 ligands and upon binding, the receptor homodimerizes and autophosphorylates (activation) in the plasma membrane [7]. Phosphorylated FLT3 activates numerous cytoplasmic effector molecules, via phosphorylation. These phosphorelays lead to hematopoietic cells differentiation and proliferation [8]. Mutations or the overexpression of FLT3 RTK leads to deregulated hematopoiesis, as seen in acute myeloid leukemia (AML) and some ALL [9,10].
About 30% of AML patients carry the mutated form of FLT3 RTK, thus making FLT3 a prime target for anti-AML therapeutics [11]. In the past decade, there have been concerted efforts by both industry and academia to develop FLT3 inhibitors and several compounds that potently inhibit FLT3 (with nanomolar Kds or IC50 values), such as midostaurin (PKC412), quizartinib, sorafenib, crenolanib and gilteritinib, have been described [12,13]. Some of these compounds have proceeded to clinical trials. Midostaurin performed well in a recently concluded Phase III clinical trial and could become one of the early FLT3 inhibitors to be approved for clinical use. Despite the successes experienced with FLT3 TKIs, none of the tested compounds have achieved a cure or long-term remission of the disease. The majority of the FLT3 inhibitors reported to date manage to clear AML in blood but do not completely eliminate AML in bone marrow [14]. Thus, over time, AML evolves and drug-resistant clones emerge in the bone marrow. Secondary FLT3 mutations, such as D835 and F691, are problematic and resistant to many FLT3 TKI developed to date [15]. In addition to secondary kinase mutations that render targeted TKIs ineffective, the clonal evolution and heterogeneity of AML make it challenging to cure [16]. This contrasts with CML, which has a relatively simple biology and is largely driven by the BCR-ABL and hence can be potentially cured with TKIs.
Results & discussion
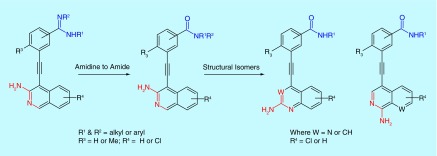
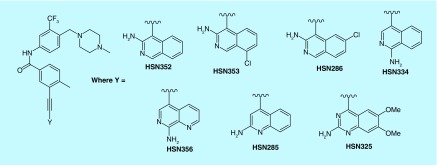
We recently noted that 3-aminoisoquinoline analogs that are substituted at the 4 position with benzamidine, inhibit FLT3 kinase and potently inhibit the proliferation of FLT3-driven AML cell lines, such as MV4–11 and MOLM-14 [17]. Multiple amidine drugs, such as viramidine (a potential antiviral drug [18]), are used clinically or have gone through clinical trials. However, these drugs are generally not orally bioavailable and therefore are typically administered intravenously. For oral bioavailability, amidine drugs are traditionally converted into less basic prodrugs [19]. Benzamides, on the other hand, are orally bioavailable and therefore we embarked on structure–activity relationship (SAR) studies to identify 4-benzamide substituted 3-aminoisoquinoline and structural mimics (Figure 1) as FLT3 inhibitors, which could potentially be translated into anti-AML drugs [20].
Figure 1. . The 4-substituted isoquinolines, quinolones and quinazolines synthesized for this study.
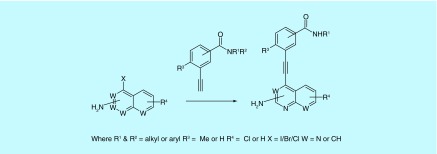
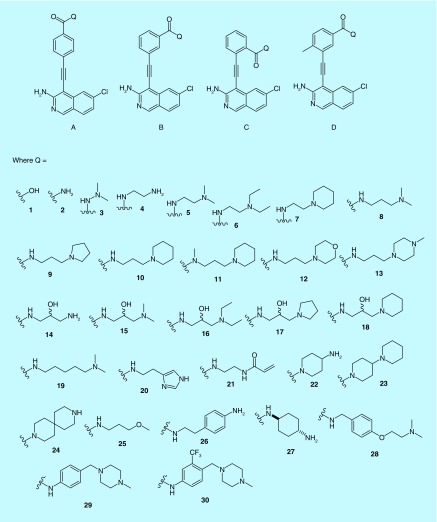
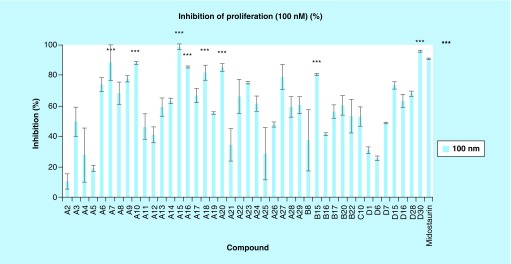
A library of amino isoquinoline, quinoline or quinazoline compounds was synthesized via Sonogashira coupling (Figure 2). Four types of benzamide analogs of 3-aminoisoquinolines were first prepared (Figure 3): amide is para to the alkyne; amide is meta to the alkyne; amide is ortho to the alkyne; and amide is meta to the alkyne and an additional methyl substituent next to the alkyne was also present. With a library of synthesized compounds, we proceeded to the investigation of antiproliferative activities against AML cell line, MV4–11 (a FLT3-driven cell line) and three other solid tumors (MCF7, breast; HCT116, colon and HeLa) (Tables 1 & 2, & Figure 4). From these cell proliferation studies, MV4–11 appeared to be more sensitive to the compounds than the other cell lines (Table 1 & 2). At 1 µM, most of the compounds could inhibit MV4–11 significantly. To identify group of compounds potently inhibiting cancer cell proliferation, we used a lower concentration of compounds (100 nM) to screen against MV4–11 (Figure 4). From these experiments, we selected potent amide compounds A7, A10, A15, A16, A18, A20, B15 and D30 (as indicated by ***, Figure 4). At 100 nM, these selected compounds inhibited MV4–11 at similar levels to midostaurin, a pan kinase inhibitor that recently completely a Phase III clinical trials (Figure 4). Typically amides that contain basic amines are included in compound libraries to improve aqueous solubility but it appears that the presence of a basic amine in the side chain of the compounds also facilitated the actual inhibition of MV4–11 proliferation. For example, compounds A1, A2, A21 and A25, which did not have a basic amine side chain, were inactive against MV4–11 whereas many of the other compounds containing a basic amine chain were active against MV4–11. We currently do not have an explanation for this observation and future structural work, beyond the scope of this report, could shed more light on the role of the basic amine. Stability of the active compounds, in the presence of mouse liver microsomes revealed that compounds with the D substitution pattern (such as D30) preformed much better in the liver microsomal stability assay compared with the other analogs.
Figure 2. . Synthesis of target compounds via Sonogashira coupling.
Condition: Pd(PPh3)2Cl2 (5 mol%), CuI (5 mol%), PPh3 (0.1 equiv.), triethylamine (22 equiv.), 50°C, 12 h.
Figure 3. . Representative examples of compounds synthesized.
See Supplementary Information for a list of all compounds made.
Table 1. . Percent inhibition of cancer cell line proliferation in the presence of compounds (1 µM).
| Compound | HSN210 | HSN204 | HSM1669 | HSM1812 | HSN137 | HSM1673 | HSM1683 | HSN105 | HSM1610 | HSM1674 | HSN177 | HSM1702 | HSN184 | HSM1750 | HSN145 | HSN139 | HSN135 | HSM1773 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Code | A3 | A4 | A5 | A6 | A7 | A9 | A10 | A11 | A12 | A13 | A14 | A15 | A16 | A17 | A20 | A22 | A23 | A24 |
| MV4–11 | 100 | 77 | 83 | 92 | 99 | 98 | 95 | 88 | 90 | 93 | 91 | 99 | 97 | 93 | 99 | 95 | 100 | 93 |
| HCT116 | 6 | 21 | 46 | 17 | 60 | 28 | 10 | 40 | 39 | 14 | 15 | 33 | 38 | 20 | 35 | 32 | 21 | 24 |
| HeLa | 16 | 22 | 20 | 97 | 20 | 38 | 49 | 1 | 20 | 8 | 1 | 17 | 51 | 11 | 11 | 35 | 0 | 24 |
| MCF-7 | 1 | 20 | 98 | 100 | 74 | 56 | 57 | 12 | 29 | 36 | 46 | 100 | 92 | 17 | 6 | 60 | 35 | 43 |
Table 2. . Percent inhibition of cancer cell line proliferation in the presence of compounds (1 µM).
| Compound | HSM1692 | HSN136 | HSN129 | HSN99 | HSM1661 | HSM1688 | HSN161 | HSN189 | HSM1751 | HSN157 | HSN174 | HSM1717 | HSN247 | HSN248 | HSN178 | HSN185 | HSN315 | HSN286 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Code | A25 | A26 | A27 | A28 | A29 | B8 | B15 | B16 | B17 | B20 | B22 | C10 | D6 | D7 | D15 | D16 | D28 | D30 |
| MV-4–11 | 81 | 86 | 99 | 97 | 90 | 74 | 97 | 95 | 91 | 96 | 85 | 82 | 58 | 98 | 98 | 97 | 83 | 99 |
| HCT116 | 29 | 38 | 29 | 20 | 0 | 43 | 18 | 7 | 32 | 12 | 28 | 33 | 54 | 76 | 97 | 67 | 66 | 100 |
| HeLa | 0 | 0 | 22 | 6 | 1 | 64 | 70 | 18 | 23 | 33 | 10 | 2 | 100 | 100 | 100 | 99 | 100 | 100 |
| MCF-7 | 22 | 33 | 37 | 42 | 96 | 33 | 91 | 49 | 47 | 21 | 40 | 76 | 100 | 100 | 100 | 100 | 100 | 100 |
Figure 4. . Percentage inhibition of proliferation in MV4–11 cell line with various analogs (100 nM).
See Table 3 for IC50 values for selected compounds: D7, D15, D6, A15, D28, D30 and midostaurin.
Compound D30 contains 1-methyl-4-(2-[trifluoromethyl]benzyl)piperazine group, which is found in many kinase inhibitors, including ponatinib [21,22]. Ponatinib, which is used to treat imatinib-resistant CML, has been shown to inhibit FLT3-driven AML [21,22]. Unfortunately, ponatinib causes adverse cardiovascular effects [23] and it is now given with a black box warning and as a last resort drug against CML [24]. In 2014, it was withdrawn from the US market due to blood clotting and cardiovascular adverse effects [24] and therefore it might not be an ideal drug to advance for AML treatment (especially for elderly patients). We were therefore interested in exploring further analogs of D30 to investigate if the kinase selectivity of compounds containing the 1-methyl-4-(2-[trifluoromethyl]benzyl)piperazine group could be modulated via a judicious substitution on the isoquinoline core or use of isoquinoline isomers. Compounds HSN352, HSN353, HSN334, HSN356, HSN285 and HSN325 (containing 3-aminoisoquinoline, 1-aminoisoquinoline, 2-aminoquinoline and 2-aminoquinazoline, Figure 5) were prepared via the Sonogashira coupling. With these compounds in hand, we determined the proliferation inhibition (IC50) against MV4–11 and MOLM-14 (FLT3-driven AML cell lines) (Table 3).
Figure 5. . D30 analogs that were synthesized to investigate the influence of the quinoline/quinazoline/isoquinoline core on anticancer activity and kinase inhibition.
Table 3. . IC50 (proliferation) and FMS-like tyrosine kinase 3 inhibition of activity.
| Compound | % inhibition of FLT3 activity (500 nM) | IC50 (nM) | |
|---|---|---|---|
| MV4–11 | MOLM14 | ||
| HSN248 (D7) | 73% | 181 ± 6.5 | 180 ± 7.8 |
| HSN178 (D15) | 66% | 144 ± 2.6 | 187 ± 5.5 |
| HSN247 (D6) | 74% | 154 ± 7.1 | 129 ± 8.2 |
| HSM1702 (A15) | 66% | 71 ± 1.1 | 102 ± 11 |
| HSN315 (D28) | 32% | 549 ± 7.4 | 523 ± 6.4 |
| HSN285 | 28% | 721 ± 5.8 | 415 ± 6.9 |
| HSN286 (D30) | 97% | 0.49 ± 0.02 | 0.72 ± 0.02 |
| HSN325 | 33% | 135 ± 5.87 | 456 ± 15 |
| HSN334 | 99% | 1.38 ± 0.1 | 1.61 ± 0.02 |
| HSN353 | 94% | 3.45 ± 0.04 | 1.97 ± 0.1 |
| HSN352 | 93% | 3.09 ± 0.1 | 3.01 ± 0.1 |
| HSN356 | 98% | 0.42 ± 0.02 | 0.62 ± 0.02 |
| Midostaurin | 98%† | 18.5 ± 2.5 | 7.37 ± 0.1 |
†% FLT3 inhibition at 412 nM.
FLT3: FMS-like tyrosine kinase 3.
HSN286 had an IC50 against MV4–11 and MOLM-14 of 0.5 and 0.7 nM, respectively (Table 3). The 1-aminoisoquinoline analogs, HSN334 and HSN356, were also potent proliferation inhibitors of MV4–11 and MOLM-14. The degrees of AML proliferation inhibition by the isoquinoline compounds were similar to (or even slightly better than) midostaurin (Table 3). In general, there was a good correlation between the percentage inhibition of FLT3 enzymatic reaction (obtained as percentage inhibition at 500 nM compound, Reaction Biology, PA, USA) and the inhibition of AML cell lines MV4–11 and MOLM-14 proliferation. Not all of the aminoquinoline compounds were potent inhibitors of the AML cell lines proliferation. For example, HSN248, HSN178, HSN247, MXC1702 and HSN315 that also contained the 2-aminoquinoline core were only moderate inhibitors of AML proliferation or FLT3 enzymatic activity. However, the 1-methyl-4-(2-[trifluoromethyl]benzyl)piperazine moiety (Figure 5) is not the sole determinant of FLT3 inhibition. The 2-aminoquinolines and quinazoline analogs, HSN285 and HSN325, both contain this moiety but they were neither potent inhibitors of FLT3 nor active against MV4–11 or MOLM-14 cell lines. Therefore, it appears that the potencies of these compounds are due to the combined or synergistic effects of the 1-methyl-4-(2-[trifluoromethyl]benzyl)piperazine moiety (found in ponatinib) and the aminoisoquinoline moiety.
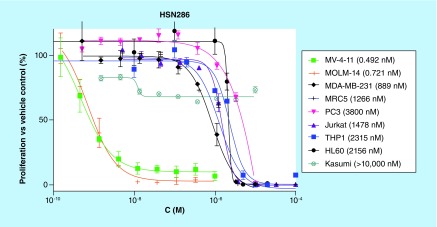
To test for selectivity, HSN286 was tested against various other cancer lines as well as a normal cell line (MRC5, lung fibroblast cell line), Figure 6. When tested against Kasumi-1, a non-FLT3 AML line, the IC50 of HSN286 was determined to be over 10,000 nM. The IC50 values for compound HSN286 against HL60, an acute promyelocytic leukemia cell line and THP1 (another non-FLT3-driven AML line) were 2156 and 2315 nM, respectively (i.e., more than 2000× less active against these cell lines when compared with FLT3-driven cell lines MV4–11 or MOLM-14). In addition, the IC50 of compound HSN286 against MRC5 is 1266 nM (over 1000× less compared with IC50 for MV4–11 or MOLM-14). It therefore appears that HSN286 is selective (at least among the cell lines tested) for FLT3-driven leukemia.
Figure 6. . HSN286 (D30) is active against FMS-like tyrosine kinase 3 driven MV4–11 and MOLM-14 but not other cancer cell lines and normal cell line, MRC5. .
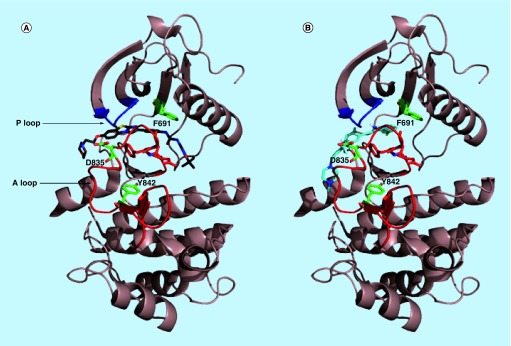
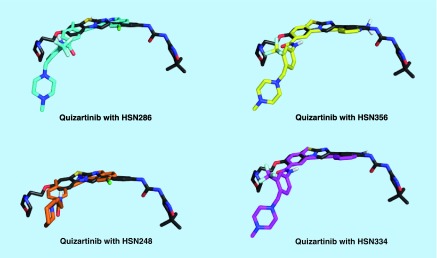
Docking [25] of HSN 286, 334, 356 and 248 to the inactive conformer of FLT3 (a crystal structure of the potent FLT3 inhibitor, quizartinib, was solved in complex with FLT3, PDB # 4xuf) revealed that the compounds bind to both the ATP-binding site and neighboring site (Figure 7). The binding of the compounds partly overlaps with the quizartinib-binding site, but there are some differences in the binding modes (Figures 8 & 9).
Figure 7. . Quizartinib and HSN286 bound to FLT3 kinase.
(A) Docked quizartinib bound to the inactive form of wild-type FLT3; (B) docked HSN286 bound to the inactive form of wild-type FLT3. PDB # 4xuf. Docking was done with AutoDock Vina [24]. The docked quizartinib matched the ligand in the crystal structure of FLT3/quizartinib. Key residues D835, Y842 and F691 which when mutated block binding of FLT3 inhibitors are shown as sticks with green carbons. The DFG motif is represented as residues with red carbons. The A loop and P loop (ATP binding) are colored as red and blue, respectively.
FLT3: FMS-like tyrosine kinase 3.
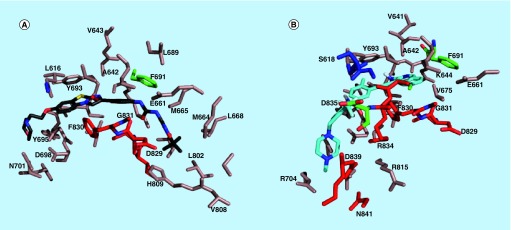
Figure 8. . FLT3 residues that are near the bound ligands.
(A) FLT3 residues that are within 6 Å of quizartinib in the docked structure; (B) FLT3 residues that are within 6 Å of HSN286 in the docked structure. The residues on the A loop including the DFG motif are shown as red while the P loop residues are colored blue.
FLT3: FMS-like tyrosine kinase 3.
Figure 9. . Overlays of FMS-like tyrosine kinase 3 binding mode of quizartinib and HSN analogs.
Although the current drugs on the market or in clinical trials are relatively successful in killing the AML leukemia cells, they currently are unable to deal with secondary mutations that occur after TKI treatment, such as FLT3 (D385V) [26]. We investigated the binding of HSN286, HSN336, HSN334, HSN248 and HSN247 with a few FLT3 secondary mutations (Table 4). Interestingly, HSN356 could bind to FLT3 (D835V) with a Kd of 19 nM, whereas a similar compound, HSN334, was a poor binder of FLT3 (D835V) with a Kd of 120 nM (Table 4).
Table 4. . Kd for isoquinoline analogs binding to FLT3, FLT3-ITD and FLT3 (D835V), determined via DiscoverX Kd Elect service.
| HSN286 | HSN356 | HSN334 | HSN178 | HSN248 | |
|---|---|---|---|---|---|
| FLT3 | 6.5 nM | 1.3 nM | 5.1 nM | 53 nM | 47 nM |
| FLT 3 (ITD) | 27.6 nM | 6.7 nM | 19 nM | 450 nM | 330 nM |
| FLT3 (D835V) | 120 nM | 19 nM | 120 nM | ND | ND |
FLT3: FMS-like tyrosine kinase 3; ITD: Internaltandem duplication; ND: Not determined.
Most kinase inhibitors that have found clinical success have been multikinase inhibitors. For example, sorafenib, used to treat kidney cancer, hepatocellular carcinoma and radioiodine-resistant thyroid cancers, also inhibits several kinases, such as RAF, MEK, ERK, VEGFR, PDGFR, FLT3, c-KIT, FGFR-1 and DDR2 [26]. Dasatinib, another multikinase inhibitor is a first-line treatment for CML and Ph+ ALL, inhibits a multitude of kinases [27]. This includes BCR-ABL, YES, EPHA8, c-KIT, SRC, LCK, DDR2, FRK, FYN, ARG, BTK and HCK [26]. Despite the promiscuity of these multikinase inhibitors, safe and tolerable doses have been found for cancer treatment. The successes of these multikinase inhibitors could be derived, in part, from the simultaneous inhibition of different kinase axes. Therefore, we investigated if HSN286 and its analogs also inhibited other cancer-related kinases. In addition to FLT3, the Src-family kinases (such as BLK, FGR, FYN, HCK, LYN, SRC and YES) have been shown to play critical roles in leukemia [28–31]. Lopez et al. recently demonstrated that CDK6 overexpression in FTL3-ITD positive AML is achieved via the Src-family kinase, HCK [32]. HCK is expressed more in human primary leukemic stem cells than in normal human hematopoietic stem cells. A study showed that when HCK is targeted with small molecules, drug resistance is reduced [29]. Other protein kinases, such as SYK [33], BRAF, p38 (p38MAPK) [34], PDGFRα/β [35], FGFR1 [36], RET [37], FLT4 [38] and Tie2 [39] have also been linked to leukemia. All these data further strengthen the consensus in the field that leukemia is a heterogeneous disease and hence targeting the aforementioned multiple kinase pathways could lead to a better outcome [40]. Therefore, we wanted to test if HSN286 and analogs were also targeting kinases that play critical roles in AML. The kinase screening services Reaction Biology and DiscoverX were used to characterize the inhibition of kinase activity (enzymatic activity in the presence of 500 nM of compounds, Supplementary Information S2). HSN286 and analogs potently inhibit FLT3 and the Src-family kinases but not other kinases, such as Aurora A, CDK6 or PIK3Ca (Table 5 & Supplementary Information). The inhibition of the Src-kinase family could be important clinically because these kinases are downstream of FLT3. In the event of FLT3 mutation, the inhibition of the Src-family kinases could still lead to proliferation inhibition [41].
Table 5. . Kd (nM) determined via DiscoverX Kd Elect service.
| Kinase | HSN286 | HSN356 |
|---|---|---|
| AURKA | >30,000 | 5100 |
| BLK | 3.9 | 1.2 |
| CDK6 | 28,000 | ND |
| CDK9 | 2300 | ND |
| FAK | 5100 | 4700 |
| FGR | 7 | 7.9 |
| FLT3 | 7.2 | 1.3 |
| FYN | 28 | 38 |
| HCK | 3.6 | 2.5 |
| KIT | 42 | 7.2 |
| LYN | 11 | 5.4 |
| PIK3CA | 26,000 | >30,000 |
| PIM1 | 11,000 | 19,000 |
| PLK1 | >30,000 | 9600 |
| SRC | 16 | 15 |
ND: Not determined.
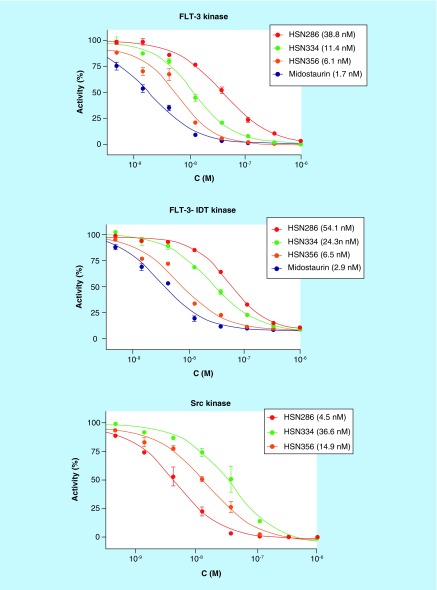
In addition to the binding assays (Table 5), IC50 values for the inhibition of FLT3 kinase enzymatic activity by midostaurin, HSN286, 334 and 356 were determined. All of the four compounds could inhibit FLT3 with low nanomolar values, although midostaurin and HSN356 were better than HSN286 and HSN334 (Figure 10). Midostaurin and HSN356 inhibited FLT3 and FLT3 ITD with single digit IC50 values. However in the case of Src kinase, HSN286 was the most potent (IC50 of 4.5 nM). HSN356 is also an effective inhibitor of Src kinase with an IC50 of 14.9 nM.
Figure 10. . Enzymatic inhibition of FLT3, FLT3 ITD and Src kinases by select compounds.
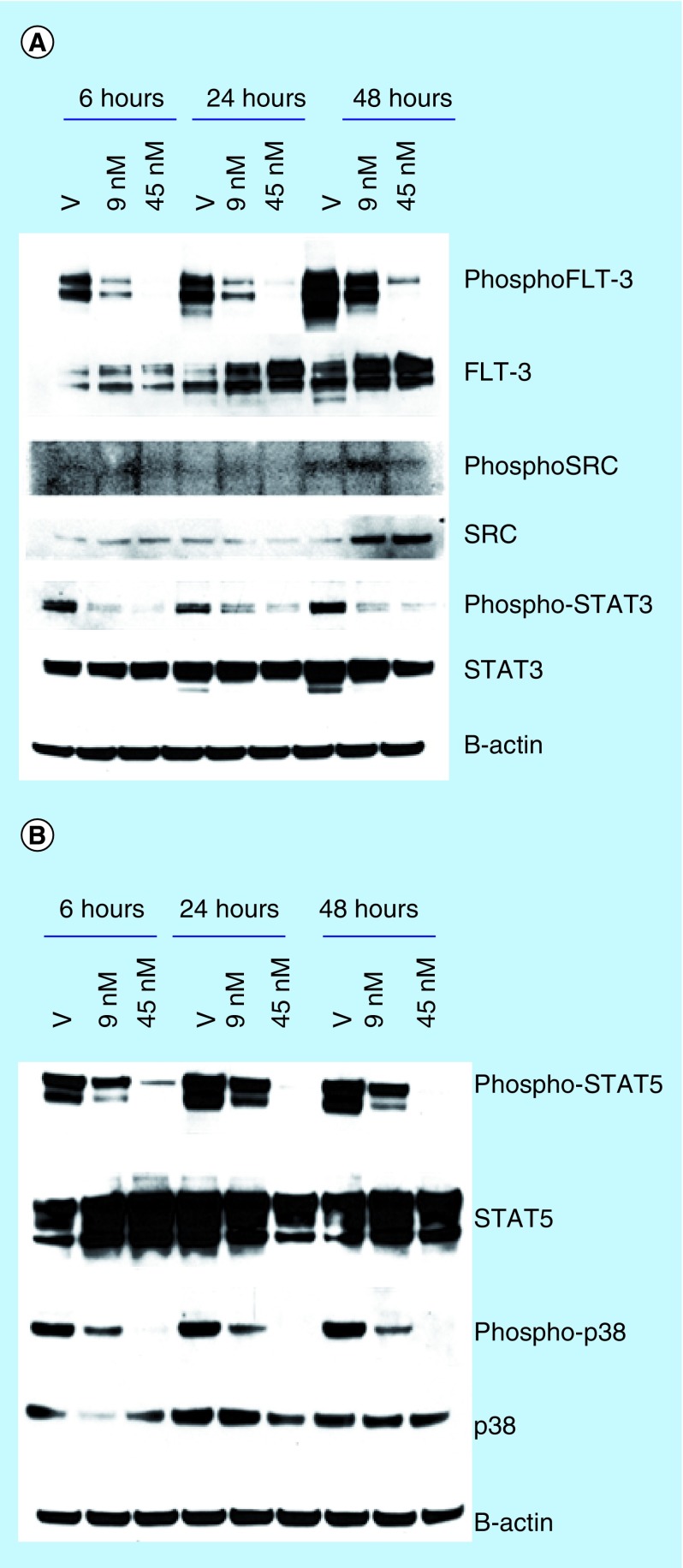
To determine if the phosphorylation of FLT3, SRC kinase and downstream effectors or kinases (such as STAT3 [42], STAT5 [43] and p-38 [44]) could be affected by the compounds, we performed western blot analysis of total protein obtained from MV4–11 treated with HSN286 for FLT3/phosphorylated FLT3, STAT5/phosphorylated STAT5, STAT3/phosphorylated STAT3 as well as SRC kinase/phosphorylated SRC and p-38/phosphorylated p-38 to ascertain that one could indeed target FLT3 and associated signaling axis in cells (target engagement or confirmation, Figure 11). In line with the in vitro kinase inhibition data, the phosphorylation of FLT3, STAT5, STAT3 and p-38 could be inhibited by HSN286 (Figure 11). The level of SRC kinase increased over time (amount at the 48-h time point is greater than at the 6- and 24-h time points). Unfortunately, the bands for the phosphorylated SRC kinase are blurred despite repeated attempts to get clearer bands. Despite the technical challenge with the phospho-SRC western, it is conclusive that at the 48-h time point, the level of unphosphorylated SRC kinase increases as the compound HSN286 is added (compare the band corresponding to vehicle [V] to bands when cells were treated with 9 and 45 nM HSN286 in Figure 11). From the above western analyses, we confirm that the proliferation inhibition of MV4–11 is due to the inhibition of FLT3 signaling axis.
Figure 11. . Western blot analysis after treating MV4–11 with HSN286.
(A) Phospho-FLT3/FLT3, phosphoSRC/SRC and phospho-STAT3/STAT3; (B) phospho-STAT5/STAT5 and phospho-p-38/p-38. Cells were treated with DMSO vehicle (V), HSN286 (9 and 45 nM) for 6, 24 and 48 h. Western blot with antirabbit or antimouse antibody. Scanned images were analyzed using image J software.
FLT3: FMS-like tyrosine kinase 3.
Conclusion
We have developed novel FLT3 inhibitors that can inhibit the problematic D835 secondary mutants. Specifically, 4-alkyne substituted aminoisoquinolines (readily obtained via Sonogashira coupling) but not the related quinolines, were potent FLT3 and SRC-kinase inhibitors. Extensive SAR studies revealed that the FLT3 inhibition profiles and antiproliferative activities against FLT3-driven cancer cell lines, MV4–11 and MOLM-14 were dictated by the substitution pattern and nature of benzamide. These SAR studies have led to the identification of 3-amino and 1-aminoisoquinoline benzamides, compounds D30 (HSN286), HSN334 and HSN356 as potent FLT3 kinase inhibitors. Some of these novel kinase inhibitors also inhibit the proliferation of FLT3-driven AML cell lines at concentrations as low as 500 pM. In addition to FLT3, we identified compounds that also inhibit the Src-family kinases and FGFR kinases (Supplementary Information). We also show that one could combine different isoquinolines with 1-methyl-4-(2-[trifluoromethyl]benzyl)piperazine group (a privileged moiety in kinase inhibitors) to develop analogs that have different kinase selectivities (a kind of ‘plug-and-play’ strategy). This work has unveiled a new class of aminoisoquinoline benzamide kinase inhibitors, which have a high potential for clinical translation. Future studies that investigate the efficacy, pharmacokinetics/pharmacodynamics (PK/PD) of the selected potent inhibitors and its analogs on animal cancer models will be done to assess their potential as clinical lead compounds.
Future perspective
About 30% of AML patients carry mutated FLT3 kinase (such as FLT3-ITD) and hence efforts have been directed at the development of FLT3 inhibitors. Despite the availability of potent and selective FLT3 inhibitors, such as quizartinib, a complete cure for FLT3-driven AML has not been found [12]. Secondary mutations in FLT3, such as D835, cause resistance to FLT3-directed therapies. In addition, compensatory activated pathways account for resistance to FLT3 inhibitors. Perhaps successful AML therapies that will achieve complete cure would have to inhibit FLT3-ITD and secondary mutations, such as D835V/H/Y and F691I as well as downstream or compensatory signaling axes. Indeed, FLT3 inhibitors that also inhibit other pathways are beginning to emerge and such compounds could achieve the elusive complete cure for FLT3-driven AML, especially for older patients [45,46].
Summary points.
Mutated or overexpressed FLT3, drives about 30% of reported acute myeloid leukemia (AML).
Many FLT3 inhibitors have shown durable clinical responses but a complete cure of AML with FLT3 inhibitors remains elusive.
The lead aminoisoquinolines, HSN286, -334 and -356 potently inhibited FLT3-driven AML cell lines, MV4–11 and MOLM-14 (single-digit or subnanomolar IC50 values).
Some of the tested aminoisoquinolines inhibited kinases with secondary mutations, such as FLT3 (D835V), which have been traditionally difficult to inhibit with tyrosine kinase inhibitors.
Supplementary Material
Footnotes
Financial & competing interests disclosure
We thank Purdue University for financial support. NMR and MS data were acquired by the NMR and MS facilities supported by NIH P30 CA023168. X Ma thanks China Scholarship Council (no. 201406140119) for financial support. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141(7):1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fleuren EDG, Zhang LX, Wu JM, Daly RJ. The kinome ‘at large’ in cancer. Nat. Rev. Cancer. 2016;16(2):83–98. doi: 10.1038/nrc.2015.18. [DOI] [PubMed] [Google Scholar]
- 3.Fabbro D, Cowan-Jacob SW, Moebitz H. Ten things you should know about protein kinases: IUPHAR Review 14. Br. J. Pharmacol. 2015;172(11):2675–2700. doi: 10.1111/bph.13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heinrich MC, Corless CL, Demetri GD, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J. Clin. Oncol. 2003;21(23):4342–4349. doi: 10.1200/JCO.2003.04.190. [DOI] [PubMed] [Google Scholar]; •• One of the early reports to document relationship between mutations in a cancer-driven kinase and clinical response to a kinase inhibitor.
- 5.Carter TA, Wodicka LM, Shah NP, et al. Inhibition of drug-resistant mutants of ABL, KIT, and EGF receptor kinases. Proc. Natl Acad. Sci. USA. 2005;102(31):11011–11016. doi: 10.1073/pnas.0504952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grafone T, Palmisano M, Nicci C, Storti S. An overview on the role of FLT3-tyrosine kinase receptor in acute myeloid leukemia: biology and treatment. Oncol. Rev. 2012;6(1):e8. doi: 10.4081/oncol.2012.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verstraete K, Savvides SN. Extracellular assembly and activation principles of oncogenic class III receptor tyrosine kinases. Nat. Rev. Cancer. 2012;12(11):753–766. doi: 10.1038/nrc3371. [DOI] [PubMed] [Google Scholar]
- 8.Grafone T, Palmisano M, Nicci C, et al. Monitoring of FLT3 phosphorylation status and its response to drugs by flow cytometry in AML blast cells. Hematol. Oncol. 2008;26(3):159–166. doi: 10.1002/hon.854. [DOI] [PubMed] [Google Scholar]
- 9.Levis M, Small D. FLT3: ITDoes matter in leukemia. Leukemia. 2003;17(9):1738–1752. doi: 10.1038/sj.leu.2403099. [DOI] [PubMed] [Google Scholar]
- 10.Birg F, Courcoul M, Rosnet O, et al. Experssion of the FMS/KIT-like gene FLT3 in human acute leukemia of the myeloid and lymphoid lineages. Blood. 1992;80:2584–2593. [PubMed] [Google Scholar]; •• One of the early reports that revealed that FMS-like tyrosine kinase 3 (FLT3) played a role in hematopoietic progenitor cells, setting the stage for investigating FLT3 as a therapeutic target of acute myeloid leukemia (AML).
- 11.Radomska HS, Basseres DS, Zheng R, et al. Block of C/EBP alpha function by phosphorylation in acute myeloid leukemia with FLT3 activating mutations. J. Exp. Med. 2006;203(2):371–381. doi: 10.1084/jem.20052242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kindler T, Lipka DB, Fischer T. FLT3 as a therapeutic target in AML: still challenging after all these years. Blood. 2010;116(24):5089–5102. doi: 10.1182/blood-2010-04-261867. [DOI] [PubMed] [Google Scholar]
- 13.Wander SA, Levis MJ, Fathi AT. The evolving role of FLT33 inhibitors in acute myeloid leukemia: quizartinib and beyond. Ther. Adv. Hematol. 2014;5(3):65–77. doi: 10.1177/2040620714532123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pemmaraju N, Kantarjian H, Ravandi F, et al. FLT3 inhibitors in the treatment of acute myeloid leukemia the start of an era? Cancer. 2011;117(15):3293–3304. doi: 10.1002/cncr.25908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiyoi H. FLT3 inhibitors: recent advances an problems for clinical application. Nagoya J. Med. Sci. 2015;77(1–2):7–17. [PMC free article] [PubMed] [Google Scholar]
- 16.Grimwade D, Ivey A, Huntly JPB. Molecuar landscape of acute myeloid leukemia in younger adults and its clinical relevance. Blood. 2016;127(1):29–41. doi: 10.1182/blood-2015-07-604496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma X, Zhou J, Wang C, et al. Identification of new FLT3 inhibitors that potently inhibit AML cell lines, via an azo click-it/staple-it approach. ACS Med. Chem. Lett. 2017 doi: 10.1021/acsmedchemlett.6b00468. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Youxi Z, Yikun G, Wen X, et al. Current prodrug strategies for improving oral absorption of nucleoside analogues. Asian J. Pharmcol. 2014;9(2):65–74. [Google Scholar]
- 19.Riggs JR, Kolesnikov A, Hendrix J, et al. Factor VIIa inhibitors: a prodrug strategy to improve oral bioavailability. Bioorg. Med. Chem. Lett. 2006;16(8):2224–2228. doi: 10.1016/j.bmcl.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 20.Gao M, Duan L, Luo J, et al. Discovery and optimization of 3-(2-(Pyrazolo[1,5-a]pyrimidin-6-yl)ethynyl)benzamides as novel selective and orally bioavailable discoidin domain receptor 1 (DDR1) inhibitors. J. Med. Chem. 2013;56(8):3281–3295. doi: 10.1021/jm301824k. [DOI] [PubMed] [Google Scholar]
- 21.Gozgit JM, Wong MJ, Wardwell S, et al. Potent activity of ponatinib (AP24534) in models of FLT3-driven acute myeloid leukemia and other hematologic malignancies. Mol. Cancer Ther. 2011;10(6):1028–1035. doi: 10.1158/1535-7163.MCT-10-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith CC, Lasater EA, Zhu X, et al. Activity of ponantinib against clinically- relevent AC220- resistant kinase domain mutants of FLT3-IDT. Blood. 2013;121(16):3165–3171. doi: 10.1182/blood-2012-07-442871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gainor JF, Chabner BS. Ponatinib: accelerated disapproval. Oncologist. 2015;20(8):847–848. doi: 10.1634/theoncologist.2015-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Talbert DR, Doherty KR, Trusk PB, Moran DM, Shell SA, Bacus S. A multi-parameter in vitro screen in human stem cell-derived cardiomyocytes identifies ponantinib- inducted structrual and functional cardiac toxicity. Toxicol. Sci. 2015;143(1):147–155. doi: 10.1093/toxsci/kfu215. [DOI] [PubMed] [Google Scholar]
- 25.Trott O, Olson AJ. Software news and update AutoDock Vina: improving the speed and accuract of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kitagawa D, Yokota K, Gouda M, et al. Activity-based kinase profiling of approced tyrosine kinase inhibitors. Genes Cells. 2013;18:110–122. doi: 10.1111/gtc.12022. [DOI] [PubMed] [Google Scholar]
- 27.Alpaz M, Shah NP, Kantarjian H, et al. Dasatinib in imatinib-resistance Philadelphia chromosome- postive lekemias. N. Engl. J. Med. 2006;354(24):2531–2541. doi: 10.1056/NEJMoa055229. [DOI] [PubMed] [Google Scholar]
- 28.Santos CD, Demur C, Bardet V, Prade-Houdellier N, Payrastre B, Recher C. A critical role for Lyn in acute myeloid Leukemia. Blood. 2008;111:2269–2279. doi: 10.1182/blood-2007-04-082099. [DOI] [PubMed] [Google Scholar]
- 29.Saito Y, Yuki H, Kuratani M, et al. A pyrrolo-pyrimidine kinase inhibitor, RK-20449, eliminates chemotherapy-resistant human primary AML stem cells in vivo . Sci. Transl. Med. 2013;5(181):181. doi: 10.1126/scitranslmed.3004387. [DOI] [PubMed] [Google Scholar]
- 30.Dos Santos C1, McDonald T, Ho YW, et al. The Src and c-Kit kinase inhibitor dasatinib enhances p53-mediated targeting of human acute myeloid leukemia stem cells by chemotherapeutic agents. Blood. 2013;122(11):1900–1913. doi: 10.1182/blood-2012-11-466425. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Demonstrated that SRC-family kinases are overexpressed and activated in AML and that inhibiting these kinases with small molecules could increase p53 activity and eliminate AML stem cells.
- 31.Gu T, Nardone J, Wang Y, et al. Survey of activated FLT3 signaling in leukemia. PloS ONE. 2011;6(4):e19169. doi: 10.1371/journal.pone.0019169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopez S, Voisset E, Tisserand JC, et al. An essential pathway links FLT3-ITD, HCK and CDK6 in acute myeloid leukemia. Oncotarget. 2016;7(23):51163–51173. doi: 10.18632/oncotarget.9965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puissant A, Fenouille N, Alexe G, et al. SYK is a critical regulator of FLT3 in acute myeloid leukemia. Cancer Cell. 2014;25(2):226–242. doi: 10.1016/j.ccr.2014.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carey A, Garg S, Cleary MM, et al. p38MAPK inhibition blocks inflammatory signaling in acute myeloid leukemia. Blood. 2015;126:2603. [Google Scholar]
- 35.Byrgazov K, Kastner R, Gorna M, et al. NDEL1-PDGFRB fusion gene in a myeloid malig-nancy with eosinophilia associated with resistance to tyrosine kinase inhibitors. Leukemia. 2016;31:237–240. doi: 10.1038/leu.2016.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee H, Kim M, Lim J, et al. Acute myeloid leukemia associated with FGGR1 abnormalities. Int. J. Hemtol. 2013;97(6):808–812. doi: 10.1007/s12185-013-1337-5. [DOI] [PubMed] [Google Scholar]
- 37.Rudat S, Pfaus A, Buhler C, et al. The RET receptor tyrosine kinase promotes acute mye-loid leukemia through protection of FLT3-ITD mutants from autophagic degradation. Blood. 2016;128:2849. [Google Scholar]
- 38.Fielder W, Graeven U, Ergun S, et al. Expression of FLT4 and its ligand VEGF-C in acture myeloid leukemia. Leukemia. 1997;11(8):1234–1237. doi: 10.1038/sj.leu.2400722. [DOI] [PubMed] [Google Scholar]
- 39.Hauang H, Bhat A, Woodnutt G, Lappe R. Targeting the ANGPT-TIE2 pathway in malignancy. Nat. Rev. Cancer. 2010;10:575–585. doi: 10.1038/nrc2894. [DOI] [PubMed] [Google Scholar]
- 40.ter Elst A, Kampen KR, Diks SH, et al. Targeting multiple active kinase pathways in 11q23 translocated pediatric acute myeloid leukemia using a VEGFR2 antibody together with a MEK inhibitor. Blood. 2010;116:3626. [Google Scholar]
- 41.Ozawa Y, Williams AH, Estes ML, et al. Src family kinases promote AML cell survival through activation of signal transducers and activators of transcription (STAT) Leuk. Res. 2008;32(6):893–903. doi: 10.1016/j.leukres.2007.11.032. [DOI] [PubMed] [Google Scholar]; • Demonstrated that inhibiting Src-family kinases surppressed STAT5 and decreased viability.
- 42.Zhou J, Bi C, Janakakumara JV, et al. Enhanced activation of STAT pathways and overexpression of survivin confer resistance to FLT3 inhibitors and could be therapeutic targets in AML. Blood. 2009;113(17):4052–4062. doi: 10.1182/blood-2008-05-156422. [DOI] [PubMed] [Google Scholar]
- 43.Choudhary C, Brandts C, Schwable J, et al. Activation mechanism of STAT5 by oncogenic Flt3-ITD. Blood. 2007;110(1):370–374. doi: 10.1182/blood-2006-05-024018. [DOI] [PubMed] [Google Scholar]
- 44.Odgerel T, Kikuchi J, Wada T, Shimizu R, Kano Y, Furukawa Y. MSK1 activiation in acute myeloid leukemia cells with FLT3 mutations. Leukemia. 2010;24:1087–1090. doi: 10.1038/leu.2010.48. [DOI] [PubMed] [Google Scholar]
- 45.Li Z, Wang X, Eksterowicz J, et al. Discovery of AMG 925, a FLT3 and CDK dual kinase inhibitor with preferential affinity for the activated state of FLT3. J. Med. Chem. 2014;57(8):3430–3449. doi: 10.1021/jm500118j. [DOI] [PubMed] [Google Scholar]; •• One of the important reports that teaches that dual inhibition of FLT3 and other kinases, such as cyclin-dependent kinase, could lead to a better response in AML therapy.
- 46.Zhang W, Borthakur G, Gao C, et al. The dual MEK/FLT3 inhibitor E6201 excerts cytotox-ic activity against acute myeloid leukemia cells harboring resistance-conferring FLT3 mutations. Cancer Res. 2016;76(6):1528–1537. doi: 10.1158/0008-5472.CAN-15-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.