Abstract
Urine is a valuable diagnostic medium and, with the discovery of urinary extracellular vesicles, is viewed as a dynamic bioactive fluid. Extracellular vesicles are lipid-enclosed structures that can be classified into three categories: exosomes, microvesicles (or ectosomes) and apoptotic bodies. This classification is based on the mechanisms by which membrane vesicles are formed: fusion of multivesicular bodies with the plasma membranes (exosomes), budding of vesicles directly from the plasma membrane (microvesicles) or those shed from dying cells (apoptotic bodies). During their formation, urinary extracellular vesicles incorporate various cell-specific components (proteins, lipids and nucleic acids) that can be transferred to target cells. The rigour needed for comparative studies has fueled the search for optimal approaches for their isolation, purification, and characterization. RNA, the newest extracellular vesicle component to be discovered, has received substantial attention as an extracellular vesicle therapeutic, and compelling evidence suggests that ex vivo manipulation of microRNA composition may have uses in the treatment of kidney disorders. The results of these studies are building the case that urinary extracellular vesicles act as mediators of renal pathophysiology. As the field of extracellular vesicle studies is burgeoning, this Review focuses on primary data obtained from studies of human urine rather than on data from studies of laboratory animals or cultured immortalized cells.
Extracellular vesicles are membrane-enclosed particles that can be released by almost any cell. Extracellular vesicles were first described in 1967 in a report describing the release of membrane particles, termed ‘platelet dust’, from activated platelets1. In 1979, alkaline phosphatase activity was ascribed to similar structures derived from placental membrane fragments2. Initially, these structures were regarded as a waste product or, at best, as a curiosity. However, it is now widely accepted that extracellular vesicles play an important part in intercellular communication and represent a potential resource for biomarkers of genitourinary disease3,4. During their formation, extracellular vesicles incorporate various bioactive molecules from their cell of origin, including membrane receptors, soluble proteins, nucleic acids ( mRNAs and microRNAs (miRNAs)) and lipids, which can be transferred to target cells5. Extensive and up-to-date databases of these molecules and associated studies are provided by Exocarta, Vesiclepedia and EVpedia6–10. Extracellular vesicles are a heterogeneous group of particles that are defined by their size, density, composition and site of origin, and include exosomes, microvesicles (also termed ectosomes) and apoptotic bodies11–14 (see below). The establishment of a formal International Society of Extracellular Vesicles ( ISEV) has defined standards for the experimental characterization of extracellular vesicles15. However, currently no consensus exists regarding the nomenclature of extracellular vesicles or the markers that distinguish the cellular origin or type of extracellular vesicles once they have been secreted or shed from the cell16. Therefore, the ISEV has encouraged the use of ‘extracellular vesicle’ as a generic term for all secreted vesicles, and as a keyword in all publications17.
The presence of extracellular vesicles in human urine (urinary extracellular vesicles) was suggested by the proteomic identification of membrane proteins in a pellet of urine that had been subjected to ultracentrifugation18. This result was confirmed in 2004, when these membrane proteins were demonstrated to be present in urinary extracellular vesicles19. Since then, urinary extracellular vesicles have been demonstrated to contain cell-specific marker proteins from every segment of the nephron, including podocytes19,20. Because their content reflects the intracellular composition of the cell of origin, the initial interest in urinary extracellular vesicles was as a potential source of urinary bio markers21. The interest in urinary extracellular vesicles has now expanded beyond their role in disease prognosis or diagnosis and into the field of therapeutics11,22,23. Furthermore, advances are being made in our understanding of the biogenesis, purification, composition and function of extracellular vesicles, including of those in urine — a unique biofluid that can range in pH, osmolality, and composition and concentration of dispersed solutes, even within the same individual and over hours or days. However, the use of urinary extracellular vesicles as biomarkers is relatively recent and these vesicles need to be further characterized. In this Review, we address the evolving nature of the study of urinary extracellular vesicles by highlighting the range of methods and techniques that are used for their purification, focusing on studies of the isolation and characterization of human urinary extracellular vesicles.
Biogenesis of extracellular vesicles
Extracellular vesicles are a heterogeneous group of vesicles, for which the nomenclature is still being defined16. In the past, the classification of extracellular vesicles was based on the source from which they are derived; for example, prostasomes are exosomes isolated from seminal fluid and dexosomes are exosomes released from dendritic cells13. In addition, the terms exosome and microvesicle have been used interchangeably. To address this issue, a nomenclature based on the three known mechanisms by which extracellular membrane vesicles are generated has been proposed13. The release of exosomes results from the fusion of multivesicular bodies with the plasma membrane, whereas budding of vesicles directly from the plasma membrane results in the formation and release of microvesicles, and apoptotic bodies are released from dying cells (FIG. 1, and see below).
Figure 1. Mechanisms of urinary extracellular vesicle formation regulate their composition.
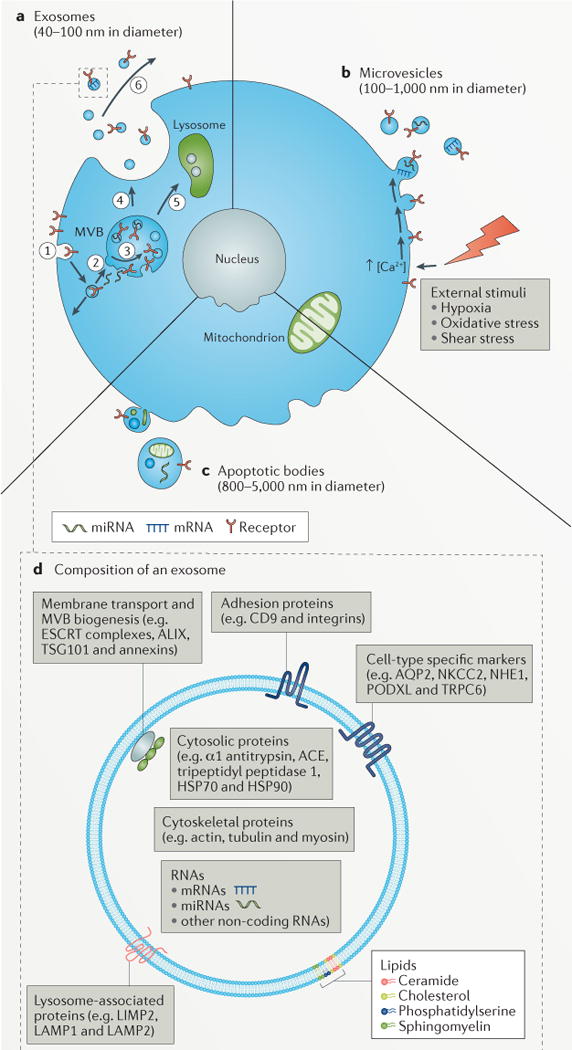
a | Exosomes are small bilayered vesicles (40–100 nm in diameter) that contain proteins, lipids and small molecules that are derived from the plasma membrane, and/or RNA and proteins that are derived from the cytoplasm. In the first step of exosome formation, membrane proteins are internalized (endocytosed) by cells (step 1), resulting in the formation of early endosomes. As these early endosomes mature into late endosomes (step 2), invagination of their delimiting membrane produces intraluminal vesicles, by processes regulated by four distinct endosomal sorting complexes required for transport (ESCRT) complexes (step 3); these late endosomes containing intraluminal vesicles are then referred to as multivesicular bodies (MVBs). MVBs can fuse with the plasma membrane, resulting in the release of intraluminal vesicles (step 4), which are now termed exosomes. Alternatively, MVBs can fuse with lysosomes, which results in the degradation of the contents of the MVB (step 5). Exosomes can participate in molecular signalling events after their release into the urinary space or into the parenchymal interstitial space (step 6). b | Microvesicles are large bilayered vesices (100–1,000 nm in diameter) that contain plasma membrane lipids and proteins, and cytoplasmic lipids, proteins and nucleic acids. Microvesicles form when a stimulus (for example, hypoxia, oxidative stress or shear stress) drives intracellular events such as those mediated by Ca2+ or phospholipid-binding proteins, which cause the shedding and release of microvesicles from the plasma membrane. c | Apoptotic bodies are large bilayered vesicles (800–5,000 nm in diameter) that are highly heterogeneous in both size and composition. The delimiting membrane of apoptotic bodies contains plasma membrane-derived lipids and proteins and encloses cytoplasmic material that includes organelle-specific proteins (for example, those from the nucleus, mitochondria, and so on), nucleic acids and lipids. d | Composition of an exosome. Exosomes contain lipids, nucleic acids and proteins, some of which are unique to the cell type from which the exosomes form. Phospholipids and sterols, such as ceramide, sphingomyelin, phosphatidylserine and cholesterol, are important for the mechanistic and biophysical aspects of bilayer formation, curvature and fluidity, which affect membrane fusion. The mechanistic role for specific lipids (for example, phosphatidylserine) is evident from the redistribution of these lipids between inner and outer leaflets or spatial segregation into the outer leaflet of extracellular vesicles. The membrane of urinary extracellular vesicles also contain integral membrane proteins and membrane-associated proteins, such as adhesion proteins (CD9 and integrins), membrane transport and/or fusion proteins and proteins involved in MVB biogenesis (ESCRT proteins, ALG2-interacting protein X (ALIX), tumour susceptibility gene 101 protein (TSG101) and annexins), lysosomal proteins (lysosome membrane protein 2 (LIMP2), lysosome-associated membrane protein 1 (LAMP1) and LAMP2), whereas the lumen contains soluble proteins (such as α1 antitrypsin, angiotensin-converting enzyme (ACE), tripeptidyl peptidase 1 and the heat shock proteins HSP70 and HSP90), and cytoskeletal proteins (such as actin, tubulin and myosin). Nucleic acids present in the lumen of urinary extracellular vesicles include mRNAs, microRNAs (miRNAs) and long, non-coding RNAs. AQP2, aquaporin 2; NHE1, sodium/hydrogen exchanger 1; NKCC2, Na-K-2Cl cotransporter; PODXL, podocalyxin; TRPC6, short transient receptor potential channel 6.
Exosomes
Exosomes are extracellular vesicles with a diameter of 40–100 nm ( REF. 24) (FIG. 1a), which were first observed as small intracellular vesicles in maturing reticulocytes25. Exosomes can be further characterized by their ability to float on a sucrose gradient at a density of 1.13–1.19 g/ml ( REFS 26–28). Exosomes have a cup-shaped appearance when examined by transmission EM (TEM). However, data from cryoEM studies indicate that their cup-shaped appearance is an artefact of sample preparation29. Exosome formation starts when membrane proteins are endocytosed by inward budding of the cell membrane and transferred to early endosomes19,30 (FIG. 1a, step 1). The early endosomes (FIG. 2a) are either recycled to the plasma membrane or mature into late endosomes (FIG. 2b), which are also known as multivesicular bodies if the limiting membrane of the late endosomes invaginates to form intraluminal vesicles29,31 (FIG. 2c). During invagination, cytosolic proteins, mRNAs and miRNAs are incorporated into the intraluminal vesicles. This process is highly regulated by the endosomal sorting complex required for transport (ESCRT) protein complexes32,33,34 (FIG. 1d). When multivesicular bodies fuse with the plasma membrane, intraluminal vesicles are released into the extracellular space and are then referred to as exosomes35. Exosomes display many of the surface markers from their cell of origin and have differences in the distribution of membrane lipids (such as cholesterol, sphingomyelin and ceramide) with their cell of origin, including enrichment in sphingomyelin and loss of asymmetry in the distribution of phosphatidylserine27,36–39. The composition of exosomes varies according to their cell of origin. However, owing to their endosomal origin, exosomes also contain a number of abundant proteins, including those involved in membrane transport and fusion (GTPases, annexins and flotillins) and multivesicular body biogenesis (ALIX, TSG101 and clathrin), as well as tetraspanins (CD9, CD63, CD81 and CD82)40–42, heat shock proteins (HSC70 and HSP90)12,43, integrins and RAB proteins that regulate docking and membrane fusion of exosomes with target cells.
Figure 2. Overview of exosome formation.
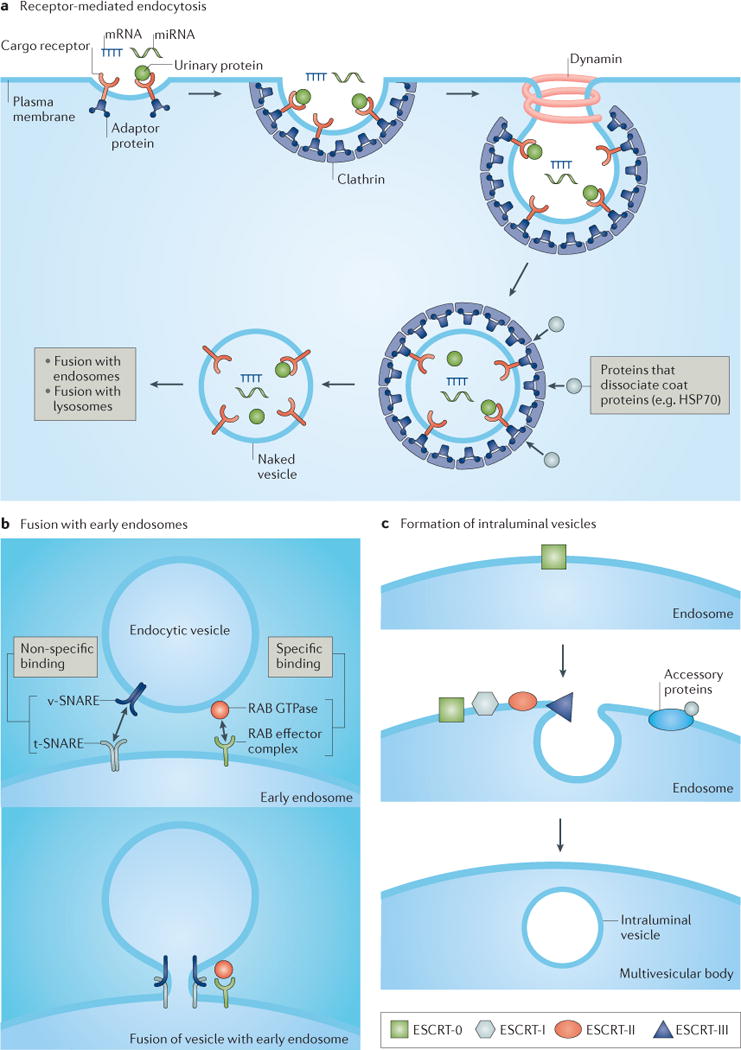
Exosome formation occurs through a multistep process that is initiated by pinocytosis (not shown) or receptor-mediated endocytosis (part a), which involves the binding of urinary proteins to the apical membrane and their internalization, a process that requires the coat protein clathrin and the ATPase dynamin. Invagination of the lipid bilayer results in formation of a small unilamellar vesicle. Proteins such as heat shock protein 70 (HSP70) can dissociate coat proteins (for example, clathrin) to yield a naked vesicle that can fuse with early endosomes (part b), a process that is mediated by soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) and small RAB effector proteins. Intraluminal vesicles form after invagination of the endosomal membrane (part c), a process that is carried out by tetraspanins (not shown) and endosomal sorting complex required for transport (ESCRT) protein complexes, such as ESCRT-0 (which is responsible for cargo clustering), ESCRT-I and ESCRT-II (both are responsible for inducing bud formation), and ESCRT-III (which promotes intraluminal budding of vesicles in endosomes and vesicle scission)171. The dissociation and recycling of the ESCRT machinery is carried out by accessory proteins. miRNA, microRNA; t-SNARE, target SNARE; v-SNARE, vesicle SNARE.
Exosomes seem to play a part in maintaining cellular homeostasis by removing harmful cytoplasmic DNA from cells, as shown both in vitro and in vivo44. Inhibition of exosome release results in an accumulation of genomic DNA in the cell, which leads to apoptotic cell death44.
Microvesicles
Microvesicles are a heterogeneous population of extracellular vesicles, which are generated by direct budding from the plasma membrane45 (FIG. 1b). Microvesicles are 100–1,000 nm in diameter and are usually larger than exosomes, although smaller microvesicles that are of a similar size to exosomes have been described31,46,47. As decribed by Kowal et al.48 in the characterization of extracellular vesicles isolated from dendritic cells in culture, the density values for extracellular vesicles can be influenced by the isolation matrix and the centrifugal force used to fractionate the sample. Microvesicles nonetheless consistently have higher densities than exosomes and bridge between that of exosomes and apoptotic bodies. Consequently, microvesicles and exosomes may overlap in size, especially for extracellular vesicles isolated from bodily fluids49. The release of microvesicles from the plasma membrane increases substantially upon stimulation by hypoxia, oxidative stress or exposure to shear stress50–53. These stimuli result in an increase in cytosolic calcium that not only induces specific membrane changes, such as the appearance of phosphatidylserine in the outer leaflet of the plasma membrane, but also disrupts the cytoskeleton, which results in outward protrusion of the plasma membrane and the formation of microvesicles3,46,54,55. During their formation, microvesicles incorporate cytosolic proteins, mRNAs and miRNAs from the cell of origin56,57. Data from studies also suggest that proteins are enriched in microvesicles by selective incorporation (reviewed in REFS 58,59).
Apoptotic bodies
Apoptotic bodies are released during the late stages of cell death and contain nuclear material, cellular organelles and membrane and cytosolic content60–62 (FIG. 1c). Apoptotic bodies are heterogeneous in size (800–5,000 nm in diameter) and appearance, but are usually larger than other types of extracellular vesicles43,63,64. The density of apoptotic bodies (1.16–1.28 g/ml) overlaps with that of exosomes12. Similarly to microvesicles, apoptotic bodies are characterized by the presence of phosphatidylserine in the outer leaflet of the lipid bilayer. The role of apoptotic bodies in intercellular communication is currently unclear, but apoptotic bodies contain soluble nucleotide factors, chemokines or adhesion molecules, which can act as a chemotactic signal to facilitate phagocytosis65.
Despite apparent differences in the mechanism of biogenesis of the three types of extracellular vesicles, it is difficult to distinguish between the different vesicle types after they are released or secreted from a cell. There are no clear descriptive physical properties or molecular markers that can unambiguously distinguish exosomes from microvesicles66,67,68. It is likely that extracellular vesicles <100 nm in diameter can bud directly from the plasma membrane (that is, microvesicles) and that some extracellular vesicles containing exosome markers are >100 nm16.
Isolating urinary extracellular vesicles
Multiple approaches have been developed for the isolation of urinary extracellular vesicles (FIG. 3), and rely largely on the physicochemical properties of urinary extracellular vesicles for their purification. These approaches include ultracentrifugation19,69–74, density gradient isolation using sucrose or Percoll75–78, antibody-based affinity capture76,79–82, ultrafiltration70,72,78,83 and polymer-based precipitation70,84–86. The choice of a specific isolation technique is probably not only dependent on the type of urine sample (proteinuric or non-proteinuric) (FIG. 3) but also on the type of downstream analyses used for ‘omics’ characterization (for example, transcriptomics or proteomics).
Figure 3. Comparison of approaches to isolate extracellular vesicles from the urine of healthy individuals and from patients with nephrotic syndrome.
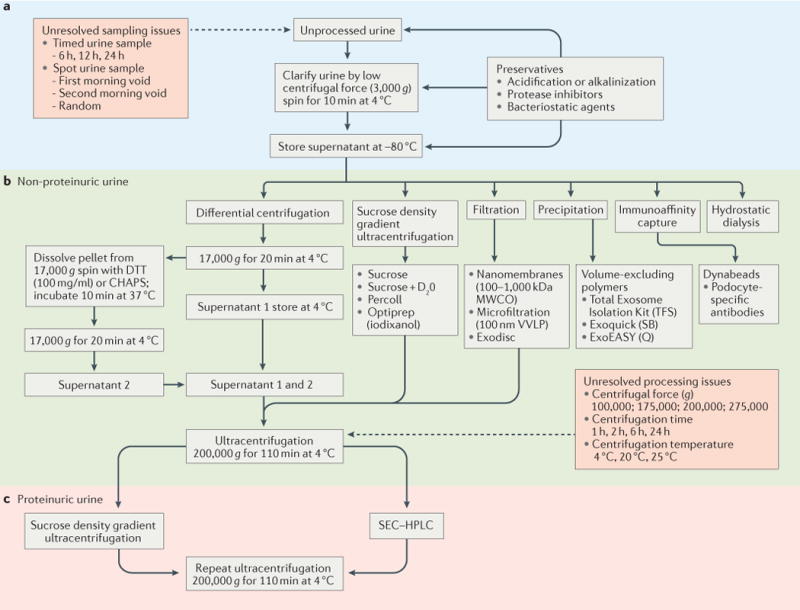
Urine is collected as a spot or timed urine sample (part a). The process of urinary extracellular vesicle isolation begins with a low speed and/or low centrifugal force (3,000 g) centrifugation step for a short time (≤10 min) and at low temperature (4°C) to clarify the urine (that is, remove the flocculent material, which can include bacteria and cells). Ideally, the urine is carried forward (part b) through to the urinary extracellular vesicle isolation step. This step represents a dynamic field of investigation, and includes methods such as differential centrifugation or ultracentrifugation, single step centrifugation using density gradient material (sucrose, Percoll), filtration or ultrafiltration, precipitation (for example, Exoquick), immunoaffinity capture and hydrostatic dialysis. The complex composition of urine from patients with nephrotic syndrome interferes with the isolation of urinary extracellular vesicles, and additional steps (such as density gradient centrifugation or size exclusion chromatography (SEC)) are required (part c) to further purify the urinary extracellular vesicles from contaminating high-molecular-weight protein complexes (such as albumin) that co-isolate with the urinary extracellular vesicles. CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate; D2O, deuterium oxide; DTT, dithiothreitol; HPLC, high performance liquid chromatography; MWCO, molecular weight cut-off; Q, Qiagen; SB, Systems Biosciences; TFS, ThermoFisher Scientific; VVLP, hydrophilic polyvinylidene difluoride.
Given the variable nature of urine (in terms of pH, osmolality, protein concentration, residency time in the bladder, and so on) and its compositional complexity, methods used to isolate extracellular vesicles from urine from a healthy individual will not necessarily be feasible to isolate extracellular vesicles from the urine of a patient with nephrotic syndrome because of the excess protein content16.
Ultracentrifugation
Methodologies that include sequential differential centrifugation and ultracentrifugation have been the most frequently used approach to isolate urinary extracellular vesicles87–89. The sequential approaches are initiated with low centrifugal force (g) spins (3,000 g followed by 17,000 g) to remove cells and debris, and subsequent higher centrifugal force spins (for example, 200,000 g) in an ultracentrifuge to pellet urinary extracellular vesicles from the supernatant19,73. Although most studies use the term exosomes to refer to the vesicles isolated by ultracentrifugation, the pellet obtained by this method contains a mixture of extracellular vesicle types11,16,90. Using EM, we have identified urinary extracellular vesicles of up to 300 nm in diameter in the pellet after ultracentrifugation73. Another study found that up to 40% of urinary extracellular vesicles are retained in the supernatant after ultracentrifugation at 200,000 g ( REF. 69). Although it is difficult to conclude any meaningful differences in these two results given the tendency of the isolation methodology to influence the findings, these results point to a strong need for standard reporting of isolation methods used in any study, as recommended by the ISEV16. A further study reported differing amounts of various protein isoforms in pelleted urinary extracellular vesicles compared with their levels in urinary extracellular vesicles that remained in solution, but did not identify proteins that were exclusive to either fraction91. Before additional isolation steps are advised, further studies should clarify whether the urinary extracellular vesicles that remain in solution contain biomarkers that are not present in pelleted urinary extracellular vesicles91.
Ultracentrifugation is less efficient at isolating extracellular vesicles from the urine of patients with nephrotic syndrome, owing to the nonspecific association of highly abundant soluble proteins with extracellular vesicles in the pellet73, which interfere with the detection of urinary extracellular vesicle proteins. Additional purification methods to remove soluble proteins, such as size-exclusion chromatography (SEC; a widely used technique to separate molecules based on their size) or sucrose density gradients, are advised29,73. SEC can effectively enrich and purify urinary extracellular vesicles from the urine of patients with nephrotic syndrome73. Contaminating proteins can also be removed by taking advantage of the capacity of extracellular vesicles to float on sucrose29. When loaded on top of a linear sucrose gradient, extracellular vesicles enter the sucrose gradient and separate based on their density, whereas contaminants pellet at the bottom of the tube after ultracentrifugation77. This method is also widely used to separate different types of urinary extracellular vesicles. However, sucrose gradients lack sufficient resolution to separate extracellular vesicles that have slightly different densities and are released by different mechanisms90.
As highly abundant proteins in urine do not prohibit RNA extraction, they do not seem to influence RNA profiling of extracellular vesicles that are isolated by ultracentrifugation from the urine of patients with nephrotic syndrome92. Furthermore, some studies suggest that extravesicular RNA does not co-precipitate with urinary extracellular vesicles, which is consistent with the presence of ribonucleases in urine after ultracentrifugation78,92. By contrast, DNA contamination of the extracellular vesicle pellet can occur; DNA should therefore be removed by digestion of the urinary extracellular vesicle pellet with a DNase78.
Uromodulin (also known as Tamm–Horsfall urinary glycoprotein (THP)) is a membrane protein that is synthesized exclusively in the thick ascending limb of the loop of Henle and is the most abundant protein in urine under physiological conditions. In addition to interfering with detection of vesicular proteins, uromodulin can form polymeric networks that entrap urinary extracellular vesicles in the 17,000 g pellet. The addition of the reducing agent dithiothreitol (DTT) or the zwitterionic detergent 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) to the pellet can disrupt the polymeric uromodulin network and increase the yield of urinary extracellular vesicles93,94. A technical report has challenged the value of uromodulin removal95 — DTT treatment of the pellet only slightly increased the yield of exosomes, as evaluated by immunoblotting for an exosomal marker. Furthermore, the yield of RNA from urinary extracellular vesicles does not increase after treatment with DTT. Therefore, these researchers consider DTT treatment unnecessary for transcriptomic studies of urinary extracellular vesicles95.
Filtration and ultrafiltration
Although ultracentrifugation is effective for the isolation of urinary extracellular vesicles, this technique is labour-intensive and requires expensive equipment. Ultrafiltration represents a faster and simpler method to isolate urinary extracellular vesicles, and usually involves the use of a polyethersulfone nanomembrane filter with an approximately 100 kDa molecular mass cut-off83. The nanomembrane concentrator enables isolation of extracellular vesicles of the size of exosomes from small volumes of urine (0.5 ml) as effectively as the standard ultracentrifugation method. However, a subset of the extracellular vesicles containing aquaporin 2 and TSG101 adhere to the nanomembrane and can only be recovered from the nanomembrane using heated Laemmli buffer containing 400 mM DTT. Unfortunately, this buffer is not always compatible with downstream proteomic analyses, such as liquid chromatography–mass spectrometry. The ultrafiltration method yields similar RNA concentrations from urinary extracellular vesicles compared with the ultracentrifugation method78. If future studies show that RNA profiles of samples from both methods are similar, then the filtration concentrator may be a good alternative to ultracentrifugation, at least for non-proteinuric urine.
Unfortunately, in addition to urinary extracellular vesicles, the ultrafiltration method retains and concentrates soluble proteins that are present in urine96. Therefore, nanomembrane ultrafiltration is not an efficient method to isolate urinary extracellular vesicles from the urine of patients with nephrotic syndrome. The high concentration of soluble proteins present in this urine obstructs the nanomembrane during ultrafiltration73. As a result, ultrafiltration efficiency is reduced and soluble proteins are still present in sufficient quantity after ultrafiltration, and interfere with the detection of less abundant urinary extracellular vesicle proteins by matrix-assisted laser desorption/ionization time of flight (MALDI-TOF)-TOF tandem mass spectrometry. The efficiency of ultrafiltration might be improved by using membranes that have low protein-binding capacity, such as hydrophilic polyvinylidene difluoride VVLP membranes. In comparison to polyethersulfone membranes, VVLP membranes show equivalent recovery of extracellular vesicles from normal urine, but co-purification of abundant soluble proteins is reduced72. Other advances in the filtration approach include the development of integrated, multistage filtration steps in microfluidic devices, which are initiated with a microfiltration step (using micrometer pore size molecular filters) followed by a nanofiltration step (using nanometer pore size molecular filters)97,98. These microfluidic devices have substantial promise for the purification of exosomes and microvesicles using sequential molecular filters, allowing for separate elution. The use of these devices still requires sequential clarifying centrifugation steps to remove cells, cellular debris, cellular casts and bacteria from the urine. After clarification, these devices can purify extracellular vesicles within 0.5–4 h, using positive hydrostatic pressure or a tabletop centrifugal microfluidic system. Whether these higher throughput nanofiltration approaches can perform efficiently with substantial levels of uromodulin or with the urine from patients with nephrotic syndrome remains to be demonstrated.
Precipitation methods
Commercial polymer-based precipitation mixtures to precipitate urinary extracellular vesicles using low-speed ultracentrifugation (<20,000 g) are now available99. These polymer mixtures can precipitate exosomes by a method known as volume exclusion, which was first developed for the purification of viruses100. However, the vesicles recovered by this method contain large amounts of CD9 but low amounts of CD63, which are both markers of exosomes, suggesting that this procedure is not specific for exosomes and may also recover other membranous organelles90.
The standard Exoquick polymer-based exosome precipitation method and a modified version of this method have been compared to differential ultracentrifugation and nanomembrane ultrafiltration70. The modified Exoquick method, which incorporates an ultracentrifugation step and includes DTT in the isolation buffer, yielded the highest quantity and quality of exosomal miRNA and mRNA, compared with the other methods. By contrast, ultracentrifugation resulted in the highest yield of the exosomal proteins ALIX and TSG101. Total protein yield from urine was substantially higher with nanomembrane ultrafiltration than with the Exoquick method, whereas the levels of exosomal proteins were low or undetectable. This outcome is probably explained by interference from abundant soluble proteins that were retained by the nanomembrane concentrator70,96. The standard Exoquick protocol did not perform well owing to the recovery of abundant soluble proteins, including uromodulin.
For RNA analysis, a commercial isolation kit has become available that binds urinary exosomes to a proprietary resin and enriches exosomal RNA by lysing the bound exosomes. Isolation is quicker than other methods because it does not require ultracentrifugation95. Almost four times the amount of miRNA was isolated with this kit compared with ultracentrifugation methods. However, the exosome miRNA profile obtained from the isolation kit differed from the profile obtained from urinary extracellular vesicles isolated by ultracentrifugation. Four of the 10 most abundant miRNAs were also found in the cell-free urine, indicating that some non-exosomal miRNA co-precipitates with exosomes obtained using the isolation kit.
Characterizing extracellular vesicles
Measurement of the size distribution and quantification of urinary extracellular vesicles facilitates their characterization; different size distributions and/or abundances of urinary extracellular vesicles could reflect different disease states and could be of clinical relevance as biomarkers of disease.
Extracellular vesicles were first visualized in urine by using TEM19, which can detect the smallest vesicles that are present101. However, analysis of the abundance of urinary extracellular vesicles by TEM can be affected by sample loss during preparation29. Furthermore, preparation of the sample includes fixation and dehydration, which can cause extracellular vesicles to shrink and thereby affect their morphology and influence measurement of their size distribution102. Fixation and dehydration can be avoided by using cryoEM. Rapid freezing better preserves the morphology of extracellular vesicles, revealing their spherical shape with a visible lipid bilayer instead of the cup-shaped morphology that is observed with TEM102,103.
In addition to the ‘gold standard’, TEM, other techniques are available to characterize extracellular vesicles. Flow cytometry is a commonly used technique that is based on particles passing through a laser beam and thereby scattering light to detectors. A major advantage of flow cytometry compared with TEM is that it is a high-throughput method. Flow cytometry can measure both the concentration and size of extracellular vesicles in a sample, but its main limitation is the size of the extracellular vesicles that can be detected, as conventional flow cytometers can only measure extracellular vesicles that are >270 nm in diameter. The resolving power is better with newer flow cytometers, which can detect extracellular vesicles of approximately 150 nm in diameter101. A major advantage of flow cytometry is its ability to simultaneously quantify multiple markers in a sample using different fluorescent labels, which can be conjugated antibodies or ligands. Other commonly used techniques for characterizing extracellular vesicles are resistive pulse sensing (RPS), nanoparticle tracking analysis (NTA), dynamic light scattering (DLS) and enzyme-linked immunosorbent assay (ELISA) (summarized in TABLE 1, and reviewed elsewhere103,104–106).
Table 1.
Methods for the characterization of urinary extracellular vesicles*
| Detection method‡ | Measurement technique | Detection limit | Concentration obtained? | Specific labelling possible? | Remarks |
|---|---|---|---|---|---|
| Transmission EM (TEM) | Scattered electron beam | ~1 nm | No | Yes (immunogold) | EV size distribution can be influenced by shrinkage during preparation of samples99 |
| Flow cytometry | Forward and sideward scattering of visible and/or fluorescent light | 270–600 nm | Yes | Yes (fluorescent) |
|
| Resistive pulse sensing (RPS) | Measurement of resistance pulses caused by particles moving through a pore | 70–100 nm | Yes | No | |
| Nanoparticle tracking analysis (NTA) | Trajectories of individual scattering objects are analysed by tracking of the Brownian motion of particles in solution | 70–90 nm | Yes | No |
|
| Dynamic light scattering (DLS; also known as photon correlation spectroscopy) | Analysis of intensity fluctuations of light scattered by particles undergoing Brownian motion in solution | 5–10 nm | No | No | More reliable data about vesicle size can be obtained when only one type of vesicle is present in the suspension; the method is less accurate for suspensions containing EVs of different sizes (when larger EVs are present, the detection of smaller EVs will be reduced)100 |
| Enzyme-linked immunosorbent assay (ELISA) | Binding of antibodies (for example, CD9) | NA | Yes | Yes (antibody) | Quantification is based on extracellular vesicle marker proteins, such as CD9. A second marker can be detected (for example, AQP2) |
AQP2, aquaporin 2; EV, extracellular vesicle; NA, not applicable.
The most commonly used methods for the characterization of urinary extracellular vesicles are listed and have been reviewed elsewhere103–106.
A size distribution of extracellular vesicles is obtainable for all detection methods except ELISA.
A comparison of TEM, flow cytometry, NTA and RPS demonstrated that each technique yields a different size distribution and a different concentration of urinary extracellular vesicles from the same sample101. The disparities are primarily caused by differences in the minimum size of vesicles that are detectable by each technique. Therefore, combining techniques is recommended when studying urinary extracellular vesicles.
Newer techniques for studying urinary extracellular vesicles include atomic force microscopy, single-particle interferometric reflectance imaging sensor and the nanofluidic optical fibre platform106–108. Further investigation and evaluation of these techniques is needed before they can be used in research and/or clinical practice.
Urinary extracellular vesicle proteome
Early proteomics studies of urinary extracellular vesicles had the same overarching goal as most expression proteomics experiments of that time109 — to establish a comprehensive index of proteins and to determine relative protein abundance using label-free quantification approaches. An early study used two-dimensional polyacrylamide electrophoresis and MALDI-TOF mass spectrometry to compare the proteome of urine subjected to acetone precipitation or ultracentrifugation18. A small number of proteins were identified in the pellet obtained after ultracentrifugation of urine pooled from five individuals, and a substantial fraction were integral membrane proteins and membrane-associated proteins18. Subsequently, a seminal publication in 2004 ( REF. 19) definitively demonstrated the presence of exosomes in urine by TEM, and the vesicular nature and dimensions of the isolated urine particles were characterized. Proteomic analysis of the particles detected 265 proteins19. Gene ontology analysis demonstrated that 24.7% of these proteins were of endosomal origin (such as ESCRT proteins and ALIX), 16.3% were integral membrane proteins and 2.7% were glycosylphosphatidylinositol (GPI)-anchored proteins, thus suggesting that these vesicles were exosomes. These and more recent studies are the subject of several excellent reviews110,111 and, as such, these studies will not be considered in detail. Here, we focus on an analysis of the available proteomics datasets of extracellular vesicles from human urine.
Databases containing proteomics data on extracellular vesicles
The majority of studies of extracellular vesicles utilize physicochemical (TEM) or immunological analysis of urinary extracellular vesicles to confirm their identity. Whereas the size or diameter of an extracellular vesicle is related to its origin and mechanism of formation, the protein content of an extracellular vesicle is associated with the cell-type of origin or the nature of the pathophysiology for the sample source. To determine whether extracellular vesicle marker proteins exist that are specific for extracellular vesicles in urine compared with, for example, those in serum, we compared the available data on abundant extracellular vesicle proteins and urinary extracellular vesicle datasets. Several databases (Exocarta, Vesiclepedia and EVpedia) and the US National Heart, Lung and Blood Institute (NHLBI) Kidney Systems Biology Project (NHLBI-KSBP) have developed online resources that index extracellular vesicle data, including proteomics data. Although Vesiclepedia and EVpedia provide a comprehensive registry of proteomics and transcriptomics studies of extracellular vesicles, only Exocarta and the NHLBI-KSBP provide indexed proteomes specifically from human urine. Together, these two sites provide a protein dataset of 1,593 urinary extracellular vesicle proteins, of which 165 are unique to Exocarta and 88 are unique to the NHLBI-KSBP. These data can be used in hypothesis-driven experiments by laboratories that do not have ready access to proteomics facilities. Publications on urinary extracellular vesicles over the last 5 years have identified >5,000 urinary extracellular vesicle proteins, but these proteins are not available online as an indexed resource. Data that are available online at EVpedia and Vesiclepedia for the 100 most common extracellular vesicle proteins (Top100 extracellular vesicle proteins) were benchmarked against 15 published urinary extracellular vesicle proteomic datasets and the NHLBI-KSBP database, which includes 200 or more proteins 19,20,72,76,84,112–120. The goal of these comparisons was twofold: first, to evaluate reproducibility in the proteins identified in urinary extracellular vesicle samples to discover possible loading control proteins or targets for affinity purification and, second, to identify proteins that are enriched in extracellular vesicles from blood or cerebrospinal fluid but are not in those from the renal parenchyma. The 100 most prevalent extracellular vesicle proteins were separated into quintiles based on prevalence across urinary extracellular vesicle studies (TABLE 2). Owing to the marked increase in the sensitivity of proteomics techniques since the first publications in 2002, it is difficult to posit the relevance of the proteins that are present in the middle three quintiles. Eight proteins are highly abundant in most urinary extracellular vesicle preparations: annexin A1 (ANXA1), ANXA4, ANXA5, chloride intracellular channel 1 (CLIC1), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), lactate dehydrogenase B (LDHB), RAS-related C3 botulinum toxin substrate 1 (RAC1) and stomatin (STOM). Annexins are membrane-associated proteins that require Ca2+ for membrane binding and have important roles in exocytosis and in regulating coagulation and immune responses. CLIC1, RAC1 and STOM are integral membrane proteins or membrane-associated proteins that can affect ion transport and thereby membrane potential, either directly by regulating chloride transport (CLIC1) or indirectly by regulating sodium (STOM) or hydrogen ion (RAC1) transporting proteins. GAPDH and LDHB have oxidoreductase activities that might contribute to maintaining the activation– inactivation state of the urinary extracellular vesicle during intercellular diffusion by affecting both local pH and redox potential. Eight proteins in the 100 most prevalent extracellular vesicle proteins were undetectable or present at very low levels in urinary extracellular vesicles: bagisin (BSG), histone H4 (HIST1H4B), HIST2H4A, integrin α6 (ITGA6), monocarboxylate transporter 1 (MCT1), prostaglandin F2 receptor negative regulator (PTGFRN), transferrin receptor protein 1 (TFR1) and tubulin α1B chain (TUBA1B). BSG, MCT1 and ITGA6 are known or predicted to physically interact. BSG targets MCT1 to the plasma membrane of platelets, where it can interact with ITGA6. PTGFRN and TFR1 are both membrane proteins that are known to be shed in extracellular vesicles in the blood. These proteins provide a list of possible candidates to use as positive or negative loading controls of urinary extracellular vesicles, as well as possible targets for the affinity purification of urinary extracellular vesicles.
Table 2.
Prevalence of Top100 EV proteins* in proteomics datasets of uEVs
| Quintile (prevalence) | Proteins |
|---|---|
| 5 (81–100%) | ANXA1, ANXA4, ANXA5, CLIC1, GAPDH, LDHB, RAC1, STOM |
| 4 (61–80%) | Serum albumin, ALDOA, ANXA2, ANXA6, ANXA11, CD81, CFL1, eEF1A1, EHD4, ENO1, ezrin, FLOT1, GDI2, GNAI2, GNAS, GNB2, HSPA8, LAMP2, LDHA, LGALS3BP, MSN, MYH9, PDCD6IP, PFN1, PGK1, PPIA, PRDX1, RAB5C, RAP1B, SDCBP, SLC3A2, TPI1, TSG101, YWHAE, YWHAQ, YWHAZ |
| 3 (41–60%) | A2M, ACLY, ACTB, ACTN4, ATP1A1, CCT2, CD63, CDC42, CLTC, EEF2, FASN, GNB1, HSP90AA1, HSP90AB1, HSPA5, ITGB1, PKM, PRDX2, RAB14, RAB1A, RAB5B, RAB7A, RHOA, UBA1, VCP, YWHAB, YWHAG |
| 2 (21–40%) | ACTG1, AHCY, ARF1, CCT3, CCT5, CD9, FLNA, HIST1H4A, HSPA1A, KPNB1, MFGE8, MVP, RAB5A, RAB8A, RAN, TCP1, THBS1, TKT, TUBA1A, TUBA1C, YWHAH |
| 1 (1–20%) | BSG‡, HIST1H4B§, HIST2H4A§, ITGA6§, PTGFRN§, SLC16A1§, TFRC§, TUBA1B‡ |
Top100 EV proteins, 100 most abundant extracellular vesicle proteins; uEVs, urinary EVs.
As defined by the EVpedia and Exocarta databases, taking into account all available proteomics datasets for urine, blood and cell culture medium.
Detected in three of 18 mass spectrometry datasets of urinary extracellular vesicles.
Not detected in any of the 18 mass spectrometry datasets of urinary extracellular vesicles.
In addition to documenting proteins that are present in extracellular vesicles, quantification of the relative abundance of proteins is also important. Although the three extracellular vesicle databases provide an immense amount of descriptive information, lack of information on the relative abundance of the proteins is a shortcoming. Of note, an important aspect of the NHLBI-KSBP site is the availability of relative urinary extracellular vesicle quantification data, albeit spectral counting data. Improvements in the collection of mass spectrometry datasets using high-resolution mass spectrometers will no doubt lead to improvements in the deposited urinary extracellular vesicle data files to include intensity-based relative quantification values. These values will be necessary to use the archived data for interpretation of changes in protein abundance between different datasets.
Proteomics studies of human urine
Whereas many studies of extracellular vesicles using cell culture or animal models of human kidney diseases have been published, to date only 37 proteomics studies of human urine have been published73,83,121–133 (summarized in TABLE 3). The data from these studies suggest that the composition of urinary extracellular vesicles vary with renal pathophysiology. We do not address the individual studies here, as most studies are small and cross-sectional, with the exception of a small series of studies addressing optimal methods for the enrichment of urinary extracellular vesicles (including the use of N-linked glycohydrolases to improve exosomal proteome datasets), identifying a possible role for urinary extracellular vesicle signalling through primary cilia on proximal tubule cells, localization of urinary extracellular vesicle proteins throughout the nephron and a small study of patients with mutations in PKD1 or PKD2 correlating exosomal proteins with height-adjusted total kidney volume76,118,125. Our goal in using this aggregated data is to identify important elements of the data that might be useful for guiding future studies of urinary extracellular vesicles. The data are arranged and presented to review the disease types analysed, the consistency in the approach to urine collection and processing, and to highlight experiments addressing single-stage versus multistage urinary extracellular vesicle isolation steps (TABLE 3). The majority of these studies have addressed methods development and comparisons using urine samples from healthy individuals (n = 28, 76%). Studies have also included urine samples from patients with renal diseases or complications: acute kidney injury (AKI, n = 3) with and without sepsis, autosomal dominant polycystic kidney disease (ADPKD; n = 5), idiopathic focal segmental glomerulosclerosis (FSGS, n = 4), nephrotic syndrome (n = 1), Bartter syndrome (n = 2), idiopathic membranous nephropathy (n = 2), bladder cancer (n = 1), prostate cancer (n = 1), diabetic nephropathy (n = 2), cystinuria (n = 1), Gitelman syndrome (n = 1), renal transplantation (n = 2), primary aldosteronism (n = 1), IgA nephropathy (n = 1) and thin basement membrane disease (n = 1). One study on sex differences in urinary extracellular vesicles from living donor kidneys has had a broad impact on the design and interpretation of future studies of urinary extracellular vesicles130. The goal of that study was to identify age-related and early disease-related changes within the kidney by evaluating the association of urinary extracellular vesicle markers with histology from renal biopsy samples. Urine samples were collected at random from individuals before living kidney donation with concomitant biopsy collection. The biopsy samples were histologically evaluated for glomerular and tubular hypertrophy and nephrosclerosis. Urinary extracellular vesicles were characterized using flow cytometry and markers of extracellular vesicle origin (microvesicles using annexin A5; exosomes using CD63) and renal parenchymal cell origin (parietal epithelial cells (using claudin 1 and cytokeratin 8); podocytes (using nephrin and podocin); mesangial cells (using transgelin); juxtaglomerular cells (using β1 adrenergic receptor); proximal tubular epithelium cells (using megalin and urate anion exchanger 1); distal tubular epithelium cells (using prominin 2 and thiazide-sensitive sodium–chloride cotransporter); the descending limb (using urea transporter 2 and aquaporin 1) and ascending limb (using epidermal growth factor receptor and uromodulin) of the loop of Henle; the collecting duct (using aquaporin 2 and vacuolar ATPase) and the renal pelvis (using cytokeratin 19 and cytokeratin 20)). Substantially higher levels of both microvesicles and exosomes were observed in the urine of females than in that of males, and increased levels of markers of urinary extracellular vesicles from mesangial and parietal epithelial cells were detected. Spearman correlations for relative abundances of these markers with donor age suggested that the production of urinary exosomes, in particular from juxtaglomerular cells and podocytes, decreases with age. Biopsy samples from donors with renal hypertrophy had decreased levels of urinary extracellular vesicle markers associated with inflammation (assayed using MCP1; mesangial cells, parietal cells, descending limb of the loop of Henle and collecting duct), whereas biopsay samples with evidence of nephrosclerosis had decreased levels of urinary extracellular vesicle markers associated with cell adhesion (assayed using ICAM1; juxtaglomerular cells, podocytes, parietal cells, proximal and distal tubular cells and collecting duct). These data firmly support the idea that future studies involving urinary extracellular vesicle biomarker discovery should consider both sex and age as important variables.
Table 3.
Proteomic analysis of extracellular vesicles in human urine
| Study | Sample origin | Sampling method | Isolation* (force, time and temperature) | uEV proteome analysis | |
|---|---|---|---|---|---|
| First step | Second step; third step | ||||
| Thongboonkerd et al. (2002)18 | Healthy | Spot urine | UC* (2 h, 4 °C) | NA; NA | Discovery |
| Pisitkun et al. (2004)19 | Healthy | Spot urine | UC* (1 h, 4 °C) | NA; NA | Discovery |
| Zhou et al. (2006)121 | Healthy | Spot urine | UC* (1 h, 4 °C) | NA; NA | Discovery |
| Zhou et al. (2006)122 | AKI with sepsis, AKI without sepsis | Spot urine | UC* (1 h, 4 °C) | 1.3 M DTT then UC (1 h, 4 °C); NA | Discovery |
| Cheruvanky et al. (2007)83 | Healthy, FSGS | Spot urine |
|
NA; NA | Targeted immunoblot |
| Zhou et al. (2008)123 | Healthy, AKI, FSGS | Spot urine Timed | UC* (1 h, 4 °C) | 200 mg/ml DTT then UC (1 h, 4 °C); NA | Targeted immunoblot |
| Smalley et al. (2008)112 | Healthy, bladder cancer | Random urine sample | UC (1 h) | NA; NA | Discovery |
| Gonzales et al. (2009)71 | Healthy | Spot urine | UC (1 h, 25 °C) | 1.3 M DTT then UC (1 h, 25 °C); NA | Discovery |
| Hogan et al. (2009)76 | Healthy, ADPKD | Spot urine (1st morning void) | UC (150,000 g, 1 h, 4 °C) | Sucrose/D2O density gradient UC (24 h, 4 °C); UC (150,000 g, 1 h, 4 °C) for each of 14 sucrose/D2O fractions | Targeted immunoblot |
| Gonzales et al. (2009)20 | Healthy, Bartter syndrome type 1 | Spot urine | UC (1 h, 25 °C) | 1.3 M DTT then UC (1 h, 25 °C); NA | Discovery, phosphoproteomics |
| Merchant et al. (2010)72 | Healthy | Spot urine |
|
NA; NA | Discovery |
| Rood et al. (2010)73 | Healthy, iMN and iFSGS | Spot urine |
|
|
Discovery |
| Moon et al. (2011)114 | Healthy, IgA nephropathy, thin basement membrane nephropathy | Spot urine (mid-morning void) | UC* (1 h, 4 °C) | 60 mg/ml DTT then UC (10 min, 60 °C), then sucrose density gradient UC (270,000 g, 16 h, 4 °C); UC (200,000 g, 1 h) of sucrose fractions | Discovery |
| Stamer et al. (2011)113 | Healthy | Random urine sample | UC (100,000 g, 1 h, 4 °C) | NA; NA | Discovery |
| Wang et al. (2012)124 | Healthy | Spot urine | UC (1 h, 25 °C) | 1.3 M DTT then UC (1 h, 25 °C); NA | Discovery |
| Raj et al. (2012)115 | Healthy | Spot urine (2nd morning void) | UC (1 h, 4 °C) to isolate crude uEV preparation |
|
Discovery |
| Fraser et al. (2013)117 | Healthy, unknown | Random urine sample | UC (unknown g, time, temperature) | NA; NA | Discovery |
| Zhou et al. (2008)123 | Healthy, FSGS, SSNS | Spot urine | UC* (1 h, 4 °C) | NA; NA | Targeted immunoblot |
| Hogan et al. (2013)77 | Membranous GN | Spot urine | UC (150,000 g, 1 h, 4 °C) | Sucrose/D2O density gradient UC (24 h, 4 °C); NA | Discovery |
| Prunotto et al. (2013)79 | Healthy | Spot urine (1st morning void) |
|
Immunoaffinity enrichment using immobilized anti-CR1 antibody; NA | Targeted immunoblot or immunfluorescence |
| Gerlach et al. (2013)125 | Healthy, ADPKD | Spot urine (1st morning void) |
|
|
Flow cytometry and lectin microarrays |
| Principe et al. (2013)116 | Healthy, low grade, organ-confined, Gleason 6 prostate cancer | Spot urine | UC* (100,000 g, 5 h) | NA; NA | Discovery |
| Zubiri et al. (2014)84 | Healthy, DN with CKD stage 3–5 | Spot urine |
|
NA; NA | Discovery |
| Hogan et al. (2014)77 | ADPKD | Spot urine (1st morning void) | Sucrose/D2O density gradient UC (1 h, 4 °C) | NA; NA | Discovery |
| Corbetta et al. (2014)126 | Gitelman syndrome, Bartter syndrome | Spot urine (2nd morning void) | UC (1 h, 4 °C) | NA; NA | Targeted immunoblot |
| Hiemstra et al. (2014)74 | Healthy | Spot urine | Centrifugation (17,000 g, 20 min, 0.22 μm sterile filtration) | UC (234,000g, 135 min, 4 °C); NA | Discovery |
| Esteva-Font et al. (2014)127 | Kidney transplant with or without ciclosporin A treatment | Timed urine collection (24 h) | UC (2 h, 4 °C) | NA; NA | Targeted immunoblot |
| Saraswat et al. (2014)128 | Healthy | Spot urine | UC (2 h, room temperature) | NA; NA | Discovery |
| Kosanovic & Jankovic (2014)129 | Healthy | Spot urine (1st morning void) |
|
|
Lectin-binding assays, SEM, TEM |
| Bourderioux et al. (2015)119 | Healthy, cystinuria | Spot urine (1st morning void) | UC (1 h, 20 °C) | 200 mg/ml DTT then UC (1 h, room temperature); NA | Discovery |
| Hogan et al. (2015)118 | ADPKD–PKD1 | Spot urine (1st morning void) | Sucrose/D2O density gradient UC (1 h, 4 °C) | NA; NA | Discovery, targeted immunoblot |
| Rood et al. (2015)120 | Healthy, iMN, iFSGS | Spot urine (2nd morning void) | UC* (2 h, 4 °C) | SEC-HPLC (RT); UC (2 h, 4 °C) | Discovery, targeted immunoblot |
| Salih et al. (2016)81 | Healthy, ADPKD, non-ADPKD CKD | Spot urine (2nd morning void) | UC* (200,000 g, 2 h, 4 °C) | NA; NA | Discovery, targeted immunoblot |
| Turco et al. (2016)130 | Before and after kidney transplant | Spot urine | Low-speed centrifugation to clarify cells and debris | NA; NA | Fluorescence-activated cell sorting |
| Panich et al. (2017)131 | AKI | Spot urine | UC (1 h, temperature unknown) | NA; NA | Targeted immunoblot |
| Wolley et al. (2017)132 | Primary aldosteronism | Spot urine (2nd morning void) | UC (1 h, 20 °C) | 200 mg/ml DTT then UC (1 h, 4 °C); NA | Targeted immunoblot |
| Lytvyn et al. (2017)133 | Healthy, normotensive type 1 diabetics | Biorepository spot urine sample | Differential centrifugation (20,000 g) | NA; NA | Fluorescence-activated cell sorting |
ADPKD, autosomal dominant polycystic kidney disease; AKI, acute kidney injury; CKD, chronic kidney disease; CR1, complement receptor type 1; DN, diabetic nephropathy; DTT, dithiothreitol; FSGS, focal segmental glomerulosclerosis; GN, glomerulonephritis; iFSGS, idiopathic FSGS; iMN, idiopathic membranous nephropathy; LN, lupus nephritis; MF, microfiltration; MWCO, molecular weight cut-off; NA, not applicable; NF, nanofiltration; P1, procedure 1; PBS, phosphate-buffered saline; PKD1, polycystin 1; RT, room temperature; SEC–HPLC, size-exclusion chromatography–high performance liquid chromatography; SEM, scanning electron microscopy; SSNS, steroid-sensitive nephrotic syndrome; TEM, transmission electron microscopy; UC, ultracentrifugation; uEV, urinary extracellular vesicle; UF, ultrafiltration.
Urine precleared of large particulate or flocculate material, usually by two sequential centrifugation steps, a short (5–30 min), low speed (1,500–3,000 g) step and a longer (20–60min), higher speed (14,000–17,500 g) step.
Protocols for urine collection for isolating urinary extracellular vesicles
Protocols for urine sample handling are available online from the European Kidney and Urine Proteomics group (EuroKUP) or have been published134,135, including for exosome preparation29,71. Use of the spot urine sampling method seems to be prevalent, as it was used in 35 of 37 studies in TABLE 3. These spot urine samples have been fruitful for comparisons of various isolation methods and for pilot studies for candidate biomarker discovery. Whereas some researchers have argued convincingly that spot urine samples accurately associate with trends in renal function136, a weakness of this method is the inability to normalize or standardize acquired urinary extracellular vesicle data to urine protein excretion rates, fractional excretion rates and calculated measures of renal function. It is likely that these standardizations will be important for experimental rigour as the number of future clinical proteomics studies incorporating 24 h timed urine collections to address ethnicity, sex and geographic variation in urinary extracellular vesicle composition increases. Whereas the composition of urinary extracellular vesicles may not change markedly within a 24 h period in healthy individuals, it is unclear whether the quantity, yield or even the composition of urinary extracellular vesicles in patients with non-proteinuric renal diseases or in patients with proteinuric renal diseases will have a time-dependent compositional bias. Therefore, trends in urinary extracellular vesicle biology as a function of spot urine versus timed urine collection must be compared.
Ultracentrifugation is the most widely implemented approach for isolating urinary extracellular vesicles (it is used at some stage in 32 of the 37 studies in TABLE 3). However, other methods, such as nanofiltration, microfiltration or precipitation approaches, might be more easily implemented in a clinical laboratory setting. Another issue that has been highlighted is the lack of consistency in isolation methods137 — in particular, studies use different relative centrifugal forces, temperatures and durations during centrifugation steps. It will be important to standardize these variables as the field progresses from the simple isolation and shotgun comparison of urinary extracellular vesicles to the isolation of specific types of urinary extracellular vesicles (exosomes versus microvesicles versus apoptotic bodies) that is needed to differentiate discovery. These structure-specific isolation approaches are being explored using fluorescence-activated cell sorting (FACS) methods133 and are likely to become important for the discovery of urinary extracellular vesicles that are able to differentiate renal diseases with associated podocyturia, in which possible differences may exist in urinary extracellular vesicles secreted from intact podocytes versus those undergoing foot process effacement or from the glomerular basement membrane through microvesicle formation or membrane blebbing138.
Urinary extracellular vesicle transcriptome
The transcriptome can be defined as the complete collection of transcribed elements of the genome present at any given moment in a cell or tissue, and includes mRNA, miRNA and other non-coding RNAs. Intact miRNAs are enriched in urinary extracellular vesicles compared with those in the cell pellet and the cell-free component of urine95. miRNAs are small, non-coding, single-stranded RNAs that regulate mRNA processing at the transcriptional and post-transcriptional level. Specifically, miRNAs reduce the stability and translation of mRNAs, thereby downregulating gene expression139. The importance of miRNAs in kidney physiology and diseases has been extensively reviewed140.
Most efforts to establish the composition and role of the urinary extracellular vesicles transcriptome have focused on mRNAs and miRNAs. Exosomes that are released from a mast cell line contain both mRNAs and miRNAs5; the gene profile of the mRNAs in these exosomes is different to that of the cellular mRNA from the donor cells. A similar observation was made for exosomes isolated from HeLa cell culture medium141. The cellular RNA preparation had a higher amount of ribosomal RNA (rRNA) compared with that in exosomes, whereas in comparison to rRNA, increased fractional levels of miRNA, mRNA and tRNA were observed in the exosomal RNA preparation. In addition, the composition of RNA molecules was different between cells and exosomes in human serum samples141. Data from both studies suggest that RNA molecules are selectively incorporated into extracellular vesicles.
Interestingly, transcripts in urinary extracellular vesicles can be delivered to other cells5. Transfer of mouse exosomes to human mast cells resulted in translation of mouse proteins in the recipient human cells. Transrenal communication has been observed with microvesicular miRNA isolated from endothelial progenitor cells delivered by intravenous injection to peritubular capillaries and tubular cells in a rat model of ischaemia–reperfusion injury, with corresponding decreases in serum creatinine, blood urea nitrogen values, and improved histologic scoring of rat kidneys, suggesting enhanced regenerative responses142. Importantly, these effects were lost upon RNase treatment of microvesicles or knockdown of Dicer in progenitor cells. Exosomes labelled with fluorescent exosome-specific fusion proteins from multiple proximal tubule cell lines were taken up by distal tubule cells and collecting duct cells143. Using dopamine receptor-specific agonists (fenoldopam), these exosomes reduced the generation of reactive oxygen species in distal tubular and collecting duct cells, although transfer of RNA was not examined143. These observations indicate that communication exists between neighbouring cells through exosome-mediated delivery of transcripts and the ability to modify protein production and gene expression in the recipient cell. Based on these findings, the RNA in exosomes has also been termed exosomal shuttle RNA (esRNA)5.
Urinary extracellular vesicles with the same density range as exosomes have been shown to contain mRNA from all regions of the nephron78,144. The RNA in these urinary extracellular vesicles was better preserved compared with that in whole cells isolated from urine, suggesting that RNA in urinary extracellular vesicles is protected from degradation. Similarly, miRNAs are highly enriched in extracellular vesicles obtained from the urine of healthy individuals95. RNA molecules incorporated in urinary extracellular vesicles represent an attractive source of biomarkers owing to their stability. As mentioned earlier, on a methodological note, extraneous DNA contaminates isolated urinary extracellular vesicles and needs to be removed before nucleic acid analysis78.
To date, many urinary biomarkers have been investigated in AKI, including an esRNA145. In a mouse model of AKI, the levels of activating transcription factor 3 (Atf3) mRNA in urinary exosomes increases within 1 h of reperfusion (after a period of ischaemia). The ATF3 mRNA level in urinary extracellular vesicles is also higher in patients with AKI in the intensive care unit compared with levels in healthy individuals. However, total urinary ATF3 mRNA levels are not different, suggesting that the increased level of ATF3 mRNA in extracellular vesicles reflects a protective signalling effect through extracellular vesicle uptake and activation of ATF3-responsive gene expression programmes in affected cells.
Several studies have examined the feasibility of using RNA molecules from urinary extracellular vesicles — including podocyte-related mRNAs in urinary extracellular vesicles (which have an average diameter of 65 nm) — as potential biomarkers146. Levels of the mRNA encoding CD2-associated protein (CD2AP) are significantly lower in patients with glomerular disease than in healthy individuals. miRNAs have also been investigated as biomarkers of renal dysfunction. In a small group of patients with FSGS, diabetic nephropathy or IgA nephropathy, miR-29c and miR-200c levels were 2.0-fold and 2.3-fold higher, respectively, in patients with IgA nephropathy than in patients with diabetic nephropathy, although this difference was not statistically significant92. In the same patient group, miR-29c levels correlated positively with estimated glomerular filtration rate (eGFR) and negatively with the extent of tubulointerstitial fibrosis147.
Exosome miRNA expression has also been examined in animal models of autoimmune nephritis and in patients with lupus nephritis148. In patients with lupus nephritis or IgA nephropathy, miR-26a levels in exosomes in the glomeruli were significantly lower compared with those in healthy individuals. By contrast, miR-26a levels in urinary extracellular vesicles were significantly increased in patients with lupus nephritis compared with those in healthy individuals. Silencing of miR-26a expression decreased the expression of genes encoding podocyte proteins in immortalized mouse podocytes. The elevated level of miR-26a in urinary extracellular vesicles in patients with lupus nephritis could therefore be a marker of podocyte injury148.
In microalbuminuric patients with type 1 diabetes mellitus, levels of miR-155 and miR-424 in urinary extracellular vesicles were significantly lower, whereas levels of miR-130a and miR-145 in urinary extracellular vesicles were significantly higher, compared with levels in normoalbuminuric patients with type 1 diabetes mellitus149. The increase in miRNA-145 levels was also detected in glomeruli and in urinary extracellular vesicles of diabetic mice. Exposure of mesangial cells to high levels of glucose induced a similar increase in whole-cell and extracellular vesicle miR-145 levels, suggesting that hyperglycaemia induces miR-145 overexpression. In type 2 diabetes mellitus, exosomal miRNAs that could serve as biomarkers for diabetic nephropathy include miR-15b, miR-34a, miR-636 and miR-192 ( REFS 150,151). Although further validation will be required, data from these studies suggest that urinary extracellular vesicles are a promising source of transcripts for biomarker discovery. In particular, urinary extracellular vesicles with selectively incorporated transcripts, including miRNAs, may have high specificity for pathological processes based on their cellular origin, extracellular vesicular packaging or the direct action of miRNAs on gene activation.
In light of this growing interest in the miRNA content of urinary extracellular vesicles, different methods for the isolation of urinary extracellular vesicles have been developed (TABLE 4). Furthermore, different methods exist for the extraction and analysis of miRNAs. It is important that these methods are standardized so that data from different research centres can be compared16. Importantly, the isolation of extracellular vesicles from serum using Exoquick or ultracentrifugation recovers different subsets of miRNAs152. For urine, different miRNAs (analysed by deep sequencing) are obtained by ultracentrifugation compared with a commercial ‘exosome to RNA isolation kit’ (from Norgen Biotek). In all other studies, only the yield and quality of miRNAs has been investigated, and miRNAs were investigated by targeted sequencing instead of deep sequencing70,78,85,95.
Table 4.
Transcriptomic analysis of extracellular vesicles in human urine
| Study | Urine source | Urine sample collection | uEV isolation* (force, duration and temperature) | Isolation of total RNA or uEV RNA? | uEV nucleic acid analysis |
|---|---|---|---|---|---|
| Miranda et al. (2010)78 | Healthy | Timed (24 h) urine collection |
|
|
Targeted real-time PCR of mRNA for podocyte proteins |
| Alvarez et al. (2012)70 | Healthy | Spot urine (1st morning void) |
|
AllPrep DNA/RNA/Protein Mini kit and RNeasy MinElute Cleanup kit (Qiagen) | Targeted real-time PCR for miRNA and mRNA of renal proteins |
| Cheng et al. (2013)95 | Healthy | Spot urine (1st morning void) |
|
|
Deep sequencing of RNA |
| Barutta et al. (2013)149 | Healthy, DN | Timed (overnight) urine collection | UC (200,000 g, 75 min, 4 °C) | Total RNA isolation using Trizol reagent (Invitrogen) | Targeted miRNA analysis using real-time PCR |
| Lv et al. (2014)147 | Healthy, DN, FSGS, IgA | Spot urine (1st morning void) | UC (2 h, temperature not specified) | uEV-RNeasy kit (Qiagen) | Targeted real-time PCR of mRNA for podocyte proteins |
| Chen et al. (2014)145 | Healthy, AKI | Spot urine | UC (200,000 g, 1 h, 4 °C) | uEV-QiAMP kit (Qiagen) | Targeted real-time PCR of mRNA for podocyte proteins |
| Channavajjhala et al. (2014)85 | Healthy | Spot urine (1st morning void) adjusted to pH 7 and protease inhibitors added |
|
|
Targeted real-time PCR of mRNA for specific miRNA |
| Murakami et al. (2014)144 | Healthy | Timed (12 h) urine collection |
|
Proprietary N-lauroylsarcosine-based lysis buffer | Targeted real-time PCR of mRNA for renal proteins |
| Ichii et al. (2014)148 | Healthy, LN | Spot urine | UC (75 min, 4 °C) | Total RNA isolation using miRNeasy Mini kit (Qiagen) | Targeted miRNA using qPCR |
| Eissa et al. (2016)150 | Type 2 diabetes | Spot urine | Direct analysis of exosome miRNA by extraction | Urine Exosome RNA Isolation Kit (Norgen Biotek) | Targeted miRNA using qPCR |
| Khurana et al. (2017)172 | Healthy, CKD | Spot urine | UC (187,000 g, 90 min, 25 °C) | Total RNA isolation using TRI Reagent (Sigma) | Deep sequencing of RNA |
| Gracia et al. (2017)168 | Healthy | Spot urine (2nd morning void) | UC (235,000 g, 135 min, 4 °C) | miRNeasy Mini kit with RNeasy MinElute Cleanup Kit (Qiagen) | RNA-seq, targeted miRNA qPCR and targeted protein immunoblotting |
AKI, acute kidney injury; CKD, chronic kidney disease; DN, diabetic nephropathy; DTT, dithiothreitol; FSGS, focal segmental glomerulosclerosis; IgA, IgA nephropathy; LN, lupus nephritis; miRNA, microRNA; MWCO, molecular weight cut-off; P1, procedure 1; qPCR, quantitative real-time PCR; UC, ultracentrifugation; uEV, urinary extracellular vesicle; UF, ultrafiltration.
Urine precleared of large particulate or flocculate material, usually by two sequential centrifugation steps, a short (5–30 min), low speed (1,500–3,000g) step and a longer (20–60 min), higher speed (14,000–17,500g) step.
Urinary extracellular vesicle lipidome
The fluid mosaic model of biological membrane structure strongly links membrane bilayer physicochemical properties (for example, curvature, fluidity or polarity) to lipid composition153. Lipids contribute integrally to the mechanisms of extracellular vesicle formation, and small but important differences exist between exosomes, microvesicles and apoptotic bodies in their lipid composition (the lipidome)38,154,155. Exosome release occurs by the regulated fusion of multivesicular bodies with the plasma membrane (possibly regulated by the neutral sphingolipid ceramide156), and exosomes are enriched for neutral cholesterols and sphingomyelin, and the saturated phospholipids phosphatidylcholine and phosphatidylethanolamine37,157. Whereas the lipid composition of microvesicles is similar to the plasma membrane from which they originate, the normal asymmetry of acidic phospholipids, such as phosphatidylethanolamine and phosphatidylserine, is lost, and these lipids are no longer limited to the inner membrane leaflet46,158. Regulation of microvesicle shedding might involve tight regulation of the transition from lipid microdomains to functional states that are yet to be determined, and may involve proteins that are responsible for transferring phospholipids between the inner and outer membrane leaflets (for example, ADP-ribosylation factor 6, proteases or phospholipid-binding or membrane-binding enzymes, such as flippases, floppases and scramblases)46,54,159. Phosphatidylserine in the apical plasma membrane, together with annexin A5 and Ca2+, have important roles in apoptosis and regulated cell death160. Methods for the efficient isolation of all three categories of urinary extracellular vesicles will be vital in expanding our understanding of the urinary extracellular vesicle lipidome.
A review of the published literature and the websites mentioned above (Vesicalpedia, Exocarta, EVpedia) identified two research studies addressing human urinary extracellular vesicle lipidome analysis (TABLE 5). In one study, ultracentrifugation was used to isolate urinary extracellular vesicles from the urine samples of eight healthy individuals and eight patients with clear cell renal cell carcinoma (RCC)161. The lipids were extracted from the resulting pellets using an organic solvent system (4:1 tetrahydrofuran:water mixture). Following liquid chromatography–mass spectrometry data acquisition and analysis, the researchers identified a total of 413 molecular species that were common to the urinary extracellular vesicle pellets from the urine samples of healthy individuals and patients with RCC. Importantly for biomarker discovery purposes, 95 lipid mass spectrometry features were enriched (64 uniquely) in the urine samples from patients with RCC and 102 were enriched (51 uniquely) in the urine samples from healthy individuals. The largest fraction of differentially abundant urinary extracellular vesicle phospholipids identified were lysophospholipids, phosphatidic acid, phosphatidylcholine and phosphatidylethanolamine. In a second study119, a combined proteomics and lipidomics analysis of urinary extracellular vesicles from healthy individuals and patients with cystinuria was carried out. Whereas the proteomics analysis detected a pattern of robust differences in the abundance of proteins between the healthy individuals and the patients with cystinuria, the lipidomics analysis was less conclusive. Urinary extracellular vesicles isolated from healthy individuals were extracted using a Bligh–Dyer-based method using chloroform and methanol, samples were separated using a TLC approach and visualized using fluorescent dyes (cyanine dyes). The tentative assignment of lipid composition was based on a comparison of the migration of samples versus lipid standards, including cholesterol, phosphatidylethanolamine, phosphatidylserine, phosphatidylcholine and sphingomyelin. The main conclusion from the lipidomics analysis was that the urinary extracellular vesicles from the urine samples of healthy individuals had high concentrations of cholesterol, sphingomyelin and phosphatidylserine, consistent with the literature12. Of note, KIM1 belongs to the hepatitis A virus cellular receptor family; this receptor family has been demonstrated to act as a phosphatidylserine receptor162. Therefore, we expect a robust field of inquiry to develop addressing the roles of extracellular vesicles and KIM1 in the development of renal disease. The limited but promising data on urinary extracellular vesicle lipidomics and the suggestion of differences in lipid concentrations between healthy and disease samples suggests that this is a robust field of research for the future.
Table 5.
Lipidomic analysis of extracellular vesicles in human urine
| Study | Urine source | Urine sample collection | Urinary extracellular vesicle isolation (force, duration and temperature) | Lipid extraction | Lipid analysis | ||
|---|---|---|---|---|---|---|---|
| Step 1 | Step 2 | Step 3 | |||||
| Del Boccio (2012)161 | Healthy, RCC | Unknown | 1,000 g, 10 min, 4 °C* | 17,000 g, 15 min, 4 °C | 200,000 g, 60 min, 4 °C |
|
Lipid samples were separated using reversed-phase chromatography and lipidomic data were captured by mass spectrometry using a Q-TOF Ultima instrument (Micromass). Data were processed using MarkerLynx software (Waters) and lipids were identified using the LipidMaps and Human Metabolome databases |
| Bourderioux (2015)119 | Healthy, Cys | Spot urine (1st morning void)* | 17,000 g, 30 min, 20 °C | 200,000 g, 180 min, 20 °C |
|
|
Lipid samples were separated by HP-TLC to separately resolve cholesterol and the main classes of phospholipids. Lipids were visualized by heating copper sulfate-treated HP-TLC plates to 185 °C for 5 min |
Cys, cystinuria; DTT, dithiothreitol; HP-TLC, high performance thin-layer chromatography; Q-TOF, quadrupole time-of-flight; RCC, renal cell carcinoma.
Urine precleared of large particulate or flocculate material, usually by two sequential centrifugation steps, a short (5–30 min), low speed (1,500–3,000 g) step and a longer (20–60 min), higher speed (14,000–17,500 g) step.
Urinary exosomes as renal therapeutics
As noted above, an increase in exosomal miR-145 has been detected in patients with early diabetic nephropathy and microalbuminuria, as well as in animal models of diabetes149. Cell culture experiments demonstrated that high glucose levels induced mesangial cell expression of miR-145, probably though increased levels of transforming growth factor β1 (TGFβ1). Interestingly, miR-145 suppressed vascular smooth muscle cell expression of TGFβ receptor 2 and suppressed extracellular matrix synthesis163. Regulation of the extracellular matrix by the TGFβ pathway is well-recognized to have a role in the pathogenesis of diabetic nephropathy, suggesting that urinary miR-145 may be a useful therapeutic target. Exosomal mRNA has also been examined as a source of non-mutated RNA to repair damaged or diseased kidneys. Exosomes from healthy Sprague–Dawley rats could transfer wild-type Pkhd1 RNA to polycystic kidney cells in vitro and in vivo, and thereby restrict cyst formation and improve renal structure and function164. Exosomes can also affect acute kidney injury (AKI), even when they are not derived from renal cells. Indeed, exosomes derived from bone marrow mesenchymal stem cells contribute to kidney repair and preservation of renal function in several different models of AKI165–167. In humans, a phase I trial investigating the effect of microvesicles derived from cell-free cord blood on β cell mass in type 1 diabetes mellitus is ongoing (NCT02138331168). Considered together, these findings indicate that exosomes from urine and other biofluids may provide a source of therapeutic agents and targets for the treatment of renal diseases.
Conclusions
Over a decade and a half has passed since the first report describing and characterizing urinary exosomes and their protein content. Since then, the other types of extracellular vesicles — namely, microvesicles and apoptotic bodies — have been detected in urine, and their contents have been found to include RNA and metabolites. Although the aim of many early studies was to discover disease biomarkers, more recent investigations have focused on the role of extracellular vesicles in intercellular communication. Our focus in this Review has been to address studies of human urinary extracellular vesicles and the approaches that are used for their isolation and characterization (TABLE 1,3). The roles of urinary extracellular vesicles in the healthy kidney and in renal disease pathophysiology have been reviewed elsewhere110.
An outcome of these studies is the ability to discern differences in the omics composition of urinary extracellular vesicles, correlate observed differences to health and acute or chronic renal disease, and use these observations to develop testable hypotheses addressing disease-specific mechanisms74,118,130,132,133,169,170. Furthermore, the utilization in these studies of second morning voids and spot urine samples to yield reproducible data suggests that the future for urinary extracellular vesicle diagnostics is encouraging. The use of high-throughput, high-sensitivity omics methods is enhancing our molecular understanding of disease-specific urinary extracellular vesicles and has enabled mining of urinary extracellular vesicles for biomarkers. The resultant knowledge base will aid researchers in generating testable hypotheses about the mechanistic roles of urinary extracellular vesicles in the development of AKI (for example, the roles of podocyte-derived urinary extracellular vesicle ATF3 in AKI123) or ageing-related chronic renal disease (for example, the effect of diminished podocyte-derived urinary extracellular vesicles with age130).
Despite significant progress, many questions remain unanswered about urinary extracellular vesicles in general, and about exosomes in particular. From an analytical perspective, further work is needed on how to optimize the methods for isolation and storage of urinary extracellular vesicles, how to characterize urinary extracellular vesicles in the urine of patients with nephrotic proteinuria and how to normalize protein and gene expression levels across studies. From a biological perspective, much work is needed to elucidate the roles of extracellular vesicles in intercellular communication and in the development or progression of chronic kidney disease, with or without the modifying effects of age or sex. Finally, more work is needed to validate candidate urinary extracellular vesicle biomarkers of renal dysfunction that were initially discovered using small cross-sectional patient groups utilizing spot urine samples. The stringent approaches used in interventional clinical trials need to be applied for the validation of these biomarkers.
Key points.
Urinary extracellular vesicles comprise a wide range of biologically distinct structures with contents that are a snapshot of the life of a cell
Urine is a dynamic biofluid, which changes over hours and days within an individual; therefore, at present, no single approach for the isolation of urinary extracellular vesicles is likely to comprehensively distinguish between healthy and disease states
Alterations in the composition of urinary extracellular vesicles are useful experimentally and may provide information about disease pathophysiology as well as provide diagnostic end points for the study of renal disease
Perhaps the greatest promise of this ‘extracellular organelle’ is to open a window for science into a greater understanding of cellular therapeutics
Acknowledgments
J.B.K. and M.L.M. are supported in part by grants R01DK110077 (J.B.K.), R01DK091584 (M.L.M.), P50AA024337 (M.L.M.) and P20GM113226 (MLM) from the US NIH. I.M.R. is supported by an AGIKO grant from ZonMw-NWO (grant 92003587).
Glossary
- mRNAs
Messenger RNAs, which are transcripts of DNA
- MicroRNAs
(miRNAs). Small, non-coding RNAs that regulate gene expression post-transcriptionally by targeting specific mRNAs for inhibition or degradation through complementary base pairing
- Exosomes
Extracellular vesicles that are formed by inward budding of the cell membrane, followed by fusion with a multivesicular body (MVB) and formation of intraluminal vesicles inside the MVB. The intraluminal vesicles that are released by fusion of the MVB with the cell plasma membrane are called exosomes
- Microvesicles
Extracellular vesicles that are formed by direct budding from the cell plasma membrane
- Apoptotic bodies
Extracellular vesicle that are released during the late stages of cell death
- Transcriptomics
The study of the complete set of RNA transcripts (the transcriptome) that is encoded by the genome, underspecific circumstances or in a specific cell
- Proteomics
Large scale studies of proteins involving the systematic identification and quantification of the complete set of proteins (the proteome) of a biological system (cell, tissue, organ, biological fluid or organism) at a specific point in time. Mass spectrometry is the technique most often used for proteomic analysis
- Nephrotic syndrome
A constellation of symptoms characterized by heavy proteinuria (>3–3.5 g/24 h), hypoalbuminuria (<30 g/l), peripheral oedema and hyperlipidaemia
Footnotes
Author contributions
All authors researched data for the article. M.L.M, I.M.R, J.K.J.D. and J.B.K made substantial contributions to discussions of the content. M.L.M, I.M.R and J.K.J.D. wrote the article and M.L.M., I.M.R., J.K.J.D. and J.B.K. reviewed and/or edited the manuscript before submission.
Competing interests statement
The authors declare no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
DATABASES
Clinicaltrials.gov: http://www.clinicaltrials.gov
EVpedia: http://evpedia.info
ExoCarta: http://www.exocarta.org
Vesiclepedia: http://www.microvesicles.org
FURTHER INFORMATION
The Cancer Genome Atlas: http://cancergenome.nih.gov
EuroKUP: http://www.eurokup.org/
ISEV: https://www.isev.org/
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Wolf P. The nature and significance of platelet products in human plasma. Br J Haematol. 1967;13:269–288. doi: 10.1111/j.1365-2141.1967.tb08741.x. [DOI] [PubMed] [Google Scholar]
- 2.Taylor DD, Doellgast GJ. Quantitation of peroxidase-antibody binding to membrane fragments using column chromatography. Anal Biochem. 1979;98:53–59. doi: 10.1016/0003-2697(79)90704-8. [DOI] [PubMed] [Google Scholar]
- 3.Ratajczak J, Wysoczynski M, Hayek F, Janowska-Wieczorek A, Ratajczak MZ. Membrane-derived microvesicles: important and underappreciated mediators of cell-to-cell communication. Leukemia. 2006;20:1487–1495. doi: 10.1038/sj.leu.2404296. [DOI] [PubMed] [Google Scholar]
- 4.Street JM, et al. Exosomal transmission of functional aquaporin 2 in kidney cortical collecting duct cells. J Physiol. 2011;589:6119–6127. doi: 10.1113/jphysiol.2011.220277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 6.Simpson RJ, Kalra H, Mathivanan S. ExoCarta as a resource for exosomal research. J Extracell Vesicles. 2012;1:18374. doi: 10.3402/jev.v1i0.18374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathivanan S, Fahner CJ, Reid GE, Simpson RJ. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012;40:D1241–D1244. doi: 10.1093/nar/gkr828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalra H, et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012;10:e1001450. doi: 10.1371/journal.pbio.1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim DK, et al. EVpedia: a community web portal for extracellular vesicles research. Bioinformatics. 2015;31:933–939. doi: 10.1093/bioinformatics/btu741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim DK, et al. EVpedia: an integrated database of high-throughput data for systemic analyses of extracellular vesicles. J Extracell Vesicles. 2013;2:20384. doi: 10.3402/jev.v2i0.20384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dear JW, Street JM, Bailey MA. Urinary exosomes: a reservoir for biomarker discovery and potential mediators of intrarenal signalling. Proteomics. 2013;13:1572–1580. doi: 10.1002/pmic.201200285. [DOI] [PubMed] [Google Scholar]
- 12.Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 13.Simpson RJ, Mathivanan S. Extracellular microvesicles: the need for internationally recognised nomenclature and stringent purification criteria. J Proteomics Bioinf. 2012;5:1–2. [Google Scholar]
- 14.Simpson RJ, Jensen SS, Lim JW. Proteomic profiling of exosomes: current perspectives. Proteomics. 2008;8:4083–4099. doi: 10.1002/pmic.200800109. [DOI] [PubMed] [Google Scholar]
- 15.Lotvall J, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles. 2014;3:26913. doi: 10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Witwer KW, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;2:20360. doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gould SJ, Raposo G. As we wait: coping with an imperfect nomenclature for extracellular vesicles. J Extracell Vesicles. 2013;2:20389. doi: 10.3402/jev.v2i0.20389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thongboonkerd V, McLeish KR, Arthur JM, Klein JB. Proteomic analysis of normal human urinary proteins isolated by acetone precipitation or ultracentrifugation. Kidney Int. 2002;62:1461–1469. doi: 10.1111/j.1523-1755.2002.kid565.x. [DOI] [PubMed] [Google Scholar]
- 19.Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci USA. 2004;101:13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzales PA, et al. Large-scale proteomics and phosphoproteomics of urinary exosomes. J Am Soc Nephrol. 2009;20:363–379. doi: 10.1681/ASN.2008040406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moon PG, You S, Lee JE, Hwang D, Baek MC. Urinary exosomes and proteomics. Mass Spectrom Rev. 2011;30:1185–1202. doi: 10.1002/mas.20319. [DOI] [PubMed] [Google Scholar]
- 22.Dear JW. Urinary exosomes join the fight against infection. J Am Soc Nephrol. 2014;25:1889–1891. doi: 10.1681/ASN.2014020204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borges FT, et al. TGF-β1-containing exosomes from injured epithelial cells activate fibroblasts to initiate tissue regenerative responses and fibrosis. J Am Soc Nephrol. 2013;24:385–392. doi: 10.1681/ASN.2012101031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raimondo F, Morosi L, Chinello C, Magni F, Pitto M. Advances in membranous vesicle and exosome proteomics improving biological understanding and biomarker discovery. Proteomics. 2011;11:709–720. doi: 10.1002/pmic.201000422. [DOI] [PubMed] [Google Scholar]
- 25.Pan BT, Teng K, Wu C, Adam M, Johnstone RM. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J Cell Biol. 1985;101:942–948. doi: 10.1083/jcb.101.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raposo G, et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183:1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 28.van Niel G, et al. Intestinal epithelial cells secrete exosome-like vesicles. Gastroenterology. 2001;121:337–349. doi: 10.1053/gast.2001.26263. [DOI] [PubMed] [Google Scholar]
- 29.Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;30:3.22.1–3.22.29. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 30.Fevrier B, Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr Opin Cell Biol. 2004;16:415–421. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 31.Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 32.Williams RL, Urbe S. The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- 33.Hurley JH. ESCRT complexes and the biogenesis of multivesicular bodies. Curr Opin Cell Biol. 2008;20:4–11. doi: 10.1016/j.ceb.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 35.Keller S, Sanderson MP, Stoeck A, Altevogt P. Exosomes: from biogenesis and secretion to biological function. Immunol Lett. 2006;107:102–108. doi: 10.1016/j.imlet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 36.Laulagnier K, et al. Mast cell- and dendritic cell-derived exosomes display a specific lipid composition and an unusual membrane organization. Biochem J. 2004;380:161–171. doi: 10.1042/BJ20031594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skotland T, Sandvig K, Llorente A. Lipids in exosomes: current knowledge and the way forward. Prog Lipid Res. 2017;66:30–41. doi: 10.1016/j.plipres.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 38.Record M, Carayon K, Poirot M, Silvente-Poirot S. Exosomes as new vesicular lipid transporters involved in cell-cell communication and various pathophysiologies. Biochim Biophys Acta. 2014;1841:108–120. doi: 10.1016/j.bbalip.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 39.Record M, Poirot M, Silvente-Poirot S. Emerging concepts on the role of exosomes in lipid metabolic diseases. Biochimie. 2014;96:67–74. doi: 10.1016/j.biochi.2013.06.016. [DOI] [PubMed] [Google Scholar]
- 40.Bard MP, et al. Proteomic analysis of exosomes isolated from human malignant pleural effusions. Am J Respir Cell Mol Biol. 2004;31:114–121. doi: 10.1165/rcmb.2003-0238OC. [DOI] [PubMed] [Google Scholar]
- 41.Escola JM, et al. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J Biol Chem. 1998;273:20121–20127. doi: 10.1074/jbc.273.32.20121. [DOI] [PubMed] [Google Scholar]
- 42.Chaput N, et al. The potential of exosomes in immunotherapy of cancer. Blood Cells Mol Dis. 2005;35:111–115. doi: 10.1016/j.bcmd.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 43.Thery C, et al. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001;166:7309–7318. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- 44.Takahashi A, et al. Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat Commun. 2017;8:15287. doi: 10.1038/ncomms15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heijnen HF, Schiel AE, Fijnheer R, Geuze HJ, Sixma JJ. Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and α-granules. Blood. 1999;94:3791–3799. [PubMed] [Google Scholar]
- 46.Hugel B, Martinez MC, Kunzelmann C, Freyssinet JM. Membrane microparticles: two sides of the coin. Physiology (Bethesda) 2005;20:22–27. doi: 10.1152/physiol.00029.2004. [DOI] [PubMed] [Google Scholar]
- 47.McConnell RE, et al. The enterocyte microvillus is a vesicle-generating organelle. J Cell Biol. 2009;185:1285–1298. doi: 10.1083/jcb.200902147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kowal J, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci USA. 2016;113:E968–E977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Pol E, Boing AN, Harrison P, Sturk A, Nieuwland R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev. 2012;64:676–705. doi: 10.1124/pr.112.005983. [DOI] [PubMed] [Google Scholar]
- 50.Barry OP, FitzGerald GA. Mechanisms of cellular activation by platelet microparticles. Thromb Haemost. 1999;82:794–800. [PubMed] [Google Scholar]
- 51.Redman CW, Sargent IL. Microparticles and immunomodulation in pregnancy and pre-eclampsia. J Reprod Immunol. 2007;76:61–67. doi: 10.1016/j.jri.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 52.Miyazaki Y, et al. High shear stress can initiate both platelet aggregation and shedding of procoagulant containing microparticles. Blood. 1996;88:3456–3464. [PubMed] [Google Scholar]
- 53.Horstman LL, Jy W, Jimenez JJ, Bidot C, Ahn YS. New horizons in the analysis of circulating cell-derived microparticles. Keio J Med. 2004;53:210–230. doi: 10.2302/kjm.53.210. [DOI] [PubMed] [Google Scholar]
- 54.Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 55.Pilzer D, Gasser O, Moskovich O, Schifferli JA, Fishelson Z. Emission of membrane vesicles: roles in complement resistance, immunity and cancer. Springer Semin Immunopathol. 2005;27:375–387. doi: 10.1007/s00281-005-0004-1. [DOI] [PubMed] [Google Scholar]
- 56.Dean WL, Lee MJ, Cummins TD, Schultz DJ, Powell DW. Proteomic and functional characterisation of platelet microparticle size classes. Thromb Haemost. 2009;102:711–718. doi: 10.1160/TH09-04-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hunter MP, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.D’Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 59.Donaldson JG. Multiple roles for Arf6: sorting, structuring, and signaling at the plasma membrane. J Biol Chem. 2003;278:41573–41576. doi: 10.1074/jbc.R300026200. [DOI] [PubMed] [Google Scholar]
- 60.Beyer C, Pisetsky DS. The role of microparticles in the pathogenesis of rheumatic diseases. Nat Rev Rheumatol. 2010;6:21–29. doi: 10.1038/nrrheum.2009.229. [DOI] [PubMed] [Google Scholar]
- 61.Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Turturici G, Tinnirello R, Sconzo G, Geraci F. Extracellular membrane vesicles as a mechanism of cell-to-cell communication: advantages and disadvantages. Am J Physiol Cell Physiol. 2014;306:C621–C633. doi: 10.1152/ajpcell.00228.2013. [DOI] [PubMed] [Google Scholar]
- 63.Hristov M, Erl W, Linder S, Weber PC. Apoptotic bodies from endothelial cells enhance the number and initiate the differentiation of human endothelial progenitor cells in vitro. Blood. 2004;104:2761–2766. doi: 10.1182/blood-2003-10-3614. [DOI] [PubMed] [Google Scholar]
- 64.Turiak L, et al. Proteomic characterization of thymocyte-derived microvesicles and apoptotic bodies in BALB/c mice. J Proteomics. 2011;74:2025–2033. doi: 10.1016/j.jprot.2011.05.023. [DOI] [PubMed] [Google Scholar]
- 65.Poon IK, Lucas CD, Rossi AG, Ravichandran KS. Apoptotic cell clearance: basic biology and therapeutic potential. Nat Rev Immunol. 2014;14:166–180. doi: 10.1038/nri3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bobrie A, Colombo M, Raposo G, Thery C. Exosome secretion: molecular mechanisms and roles in immune responses. Traffic. 2011;12:1659–1668. doi: 10.1111/j.1600-0854.2011.01225.x. [DOI] [PubMed] [Google Scholar]
- 67.Choi DS, Kim DK, Kim YK, Gho YS. Proteomics of extracellular vesicles: exosomes and ectosomes. Mass Spectrom Rev. 2014;34:474–490. doi: 10.1002/mas.21420. [DOI] [PubMed] [Google Scholar]
- 68.Simons M, Raposo G. Exosomes — vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 69.Musante L, Saraswat M, Ravida A, Byrne B, Holthofer H. Recovery of urinary nanovesicles from ultracentrifugation supernatants. Nephrol Dial Transplant. 2013;28:1425–1433. doi: 10.1093/ndt/gfs564. [DOI] [PubMed] [Google Scholar]
- 70.Alvarez ML, Khosroheidari M, Kanchi Ravi R, DiStefano JK. Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int. 2012;82:1024–1032. doi: 10.1038/ki.2012.256. [DOI] [PubMed] [Google Scholar]
- 71.Gonzales PA, et al. Isolation and purification of exosomes in urine. Methods Mol Biol. 2010;641:89–99. doi: 10.1007/978-1-60761-711-2_6. [DOI] [PubMed] [Google Scholar]
- 72.Merchant ML, et al. Microfiltration isolation of human urinary exosomes for characterization by MS. Proteomics Clin Appl. 2010;4:84–96. doi: 10.1002/prca.200800093. [DOI] [PubMed] [Google Scholar]
- 73.Rood IM, et al. Comparison of three methods for isolation of urinary microvesicles to identify biomarkers of nephrotic syndrome. Kidney Int. 2010;78:810–816. doi: 10.1038/ki.2010.262. [DOI] [PubMed] [Google Scholar]
- 74.Hiemstra TF, et al. Human urinary exosomes as innate immune effectors. J Am Soc Nephrol. 2014;25:2017–2027. doi: 10.1681/ASN.2013101066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen CY, Hogan MC, Ward CJ. Purification of exosome-like vesicles from urine. Methods Enzymol. 2013;524:225–241. doi: 10.1016/B978-0-12-397945-2.00013-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hogan MC, et al. Characterization of PKD protein-positive exosome-like vesicles. J Am Soc Nephrol. 2009;20:278–288. doi: 10.1681/ASN.2008060564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hogan MC, et al. Subfractionation, characterization, and in-depth proteomic analysis of glomerular membrane vesicles in human urine. Kidney Int. 2013;85:1225–1237. doi: 10.1038/ki.2013.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Miranda KC, et al. Nucleic acids within urinary exosomes/microvesicles are potential biomarkers for renal disease. Kidney Int. 2010;78:191–199. doi: 10.1038/ki.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Prunotto M, et al. Proteomic analysis of podocyte exosome-enriched fraction from normal human urine. J Proteomics. 2013;82:193–229. doi: 10.1016/j.jprot.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 80.Hildonen S, Skarpen E, Halvorsen TG, Reubsaet L. Isolation and mass spectrometry analysis of urinary extraexosomal proteins. Sci Rep. 2016;6:36331. doi: 10.1038/srep36331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Salih M, et al. Proteomics of urinary vesicles links plakins and complement to polycystic kidney disease. J Am Soc Nephrol. 2016;27:3079–3092. doi: 10.1681/ASN.2015090994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Salih M, Fenton RA, Zietse R, Hoorn EJ. Urinary extracellular vesicles as markers to assess kidney sodium transport. Curr Opin Nephrol Hypertens. 2016;25:67–72. doi: 10.1097/MNH.0000000000000192. [DOI] [PubMed] [Google Scholar]
- 83.Cheruvanky A, et al. Rapid isolation of urinary exosomal biomarkers using a nanomembrane ultrafiltration concentrator. Am J Physiol Renal Physiol. 2007;292:F1657–F1661. doi: 10.1152/ajprenal.00434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zubiri I, et al. Diabetic nephropathy induces changes in the proteome of human urinary exosomes as revealed by label-free comparative analysis. J Proteomics. 2014;96:92–102. doi: 10.1016/j.jprot.2013.10.037. [DOI] [PubMed] [Google Scholar]
- 85.Channavajjhala SK, et al. Optimizing the purification and analysis of miRNAs from urinary exosomes. Clin Chem Lab Med. 2014;52:345–354. doi: 10.1515/cclm-2013-0562. [DOI] [PubMed] [Google Scholar]
- 86.Rider MA, Hurwitz SN, Meckes DG., Jr ExtraPEG: a polyethylene glycol-based method for enrichment of extracellular vesicles. Sci Rep. 2016;6:23978. doi: 10.1038/srep23978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang D, Sun W. Urinary extracellular microvesicles: isolation methods and prospects for urinary proteome. Proteomics. 2014;14:1922–1932. doi: 10.1002/pmic.201300371. [DOI] [PubMed] [Google Scholar]
- 88.Salih M, Zietse R, Hoorn EJ. Urinary extracellular vesicles and the kidney: biomarkers and beyond. Am J Physiol Renal Physiol. 2014;306:F1251–F1259. doi: 10.1152/ajprenal.00128.2014. [DOI] [PubMed] [Google Scholar]
- 89.Gardiner C, et al. Techniques used for the isolation and characterization of extracellular vesicles: results of a worldwide survey. J Extracell Vesicles. 2016;5:32945. doi: 10.3402/jev.v5.32945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bobrie A, Colombo M, Krumeich S, Raposo G, Thery C. Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation. J Extracell Vesicles. 2012;1:18397. doi: 10.3402/jev.v1i0.18397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jacquillet G, Hoorn EJ, Vilasi A, Unwin RJ. Urinary vesicles: in splendid isolation. Nephrol Dial Transplant. 2013;28:1332–1335. doi: 10.1093/ndt/gfs599. [DOI] [PubMed] [Google Scholar]
- 92.Lv LL, et al. Isolation and quantification of microRNAs from urinary exosomes/microvesicles for biomarker discovery. Int J Biol Sci. 2013;9:1021–1031. doi: 10.7150/ijbs.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fernandez-Llama P, et al. Tamm-Horsfall protein and urinary exosome isolation. Kidney Int. 2010;77:736–742. doi: 10.1038/ki.2009.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Musante L, et al. Biochemical and physical characterisation of urinary nanovesicles following CHAPS treatment. PLoS ONE. 2012;7:e37279. doi: 10.1371/journal.pone.0037279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cheng L, Sun X, Scicluna BJ, Coleman BM, Hill AF. Characterization and deep sequencing analysis of exosomal and non-exosomal miRNA in human urine. Kidney Int. 2014;86:433–444. doi: 10.1038/ki.2013.502. [DOI] [PubMed] [Google Scholar]
- 96.Gonzales P, Pisitkun T, Knepper MA. Urinary exosomes: is there a future? Nephrol Dial Transplant. 2008;23:1799–1801. doi: 10.1093/ndt/gfn058. [DOI] [PubMed] [Google Scholar]
- 97.Woo HK, et al. Exodisc for rapid, size-selective, and efficient isolation and analysis of nanoscale extracellular vesicles from biological samples. ACS Nano. 2017;11:1360–1370. doi: 10.1021/acsnano.6b06131. [DOI] [PubMed] [Google Scholar]
- 98.Liang LG, et al. An integrated double-filtration microfluidic device for isolation, enrichment and quantification of urinary extracellular vesicles for detection of bladder cancer. Sci Rep. 2017;7:46224. doi: 10.1038/srep46224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Taylor DD, Zacharias W, Gercel-Taylor C. Exosome isolation for proteomic analyses and RNA profiling. Methods Mol Biol. 2011;728:235–246. doi: 10.1007/978-1-61779-068-3_15. [DOI] [PubMed] [Google Scholar]
- 100.Albertsson PA, Frick G. Partition of virus particles in a liquid two-phase system. Biochim Biophys Acta. 1960;37:230–237. doi: 10.1016/0006-3002(60)90228-6. [DOI] [PubMed] [Google Scholar]
- 101.van der Pol E, et al. Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis, and resistive pulse sensing. J Thromb Haemost. 2014;12:1182–1192. doi: 10.1111/jth.12602. [DOI] [PubMed] [Google Scholar]
- 102.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rupert DL, Claudio V, Lasser C, Bally M. Methods for the physical characterization and quantification of extracellular vesicles in biological samples. Biochim Biophys Acta. 2017;1861:3164–3179. doi: 10.1016/j.bbagen.2016.07.028. [DOI] [PubMed] [Google Scholar]
- 104.Szatanek R, et al. The methods of choice for extracellular vesicles (EVs) characterization. Int J Mol Sci. 2017;18:E1153. doi: 10.3390/ijms18061153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Erdbrugger U, Lannigan J. Analytical challenges of extracellular vesicle detection: a comparison of different techniques. Cytometry A. 2016;89:123–134. doi: 10.1002/cyto.a.22795. [DOI] [PubMed] [Google Scholar]
- 106.Coumans FAW, et al. Methodological guidelines to study extracellular vesicles. Circ Res. 2017;120:1632–1648. doi: 10.1161/CIRCRESAHA.117.309417. [DOI] [PubMed] [Google Scholar]
- 107.Daaboul GG, et al. Digital detection of exosomes by interferometric imaging. Sci Rep. 2016;6:37246. doi: 10.1038/srep37246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Faez S, et al. Fast, label-free tracking of single viruses and weakly scattering nanoparticles in a nanofluidic optical fiber. ACS Nano. 2015;9:12349–12357. doi: 10.1021/acsnano.5b05646. [DOI] [PubMed] [Google Scholar]
- 109.McLeish KR, Merchant ML, Klein JB, Ward RA. Technical note: proteomic approaches to fundamental questions about neutrophil biology. J Leukoc Biol. 2013;94:683–692. doi: 10.1189/jlb.1112591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Erdbrugger U, Le TH. Extracellular vesicles in renal diseases: more than novel biomarkers? J Am Soc Nephrol. 2016;27:12–26. doi: 10.1681/ASN.2015010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang W, et al. Extracellular vesicles in diagnosis and therapy of kidney diseases. Am J Physiol Renal Physiol. 2016;311:F844–F851. doi: 10.1152/ajprenal.00429.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Smalley DM, Sheman NE, Nelson K, Theodorescu D. Isolation and identification of potential urinary microparticle biomarkers of bladder cancer. J Proteome Res. 2008;7:2088–2096. doi: 10.1021/pr700775x. [DOI] [PubMed] [Google Scholar]
- 113.Stamer WD, Hoffman EA, Luther JM, Hachey DL, Schey KL. Protein profile of exosomes from trabecular meshwork cells. J Proteomics. 2011;74:796–804. doi: 10.1016/j.jprot.2011.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Moon PG, et al. Proteomic analysis of urinary exosomes from patients of early IgA nephropathy and thin basement membrane nephropathy. Proteomics. 2011;11:2459–2475. doi: 10.1002/pmic.201000443. [DOI] [PubMed] [Google Scholar]
- 115.Raj DA, Fiume I, Capasso G, Pocsfalvi G. A multiplex quantitative proteomics strategy for protein biomarker studies in urinary exosomes. Kidney Int. 2012;81:1263–1272. doi: 10.1038/ki.2012.25. [DOI] [PubMed] [Google Scholar]
- 116.Principe S, et al. In-depth proteomic analyses of exosomes isolated from expressed prostatic secretions in urine. Proteomics. 2013;13:1667–1671. doi: 10.1002/pmic.201200561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fraser KB, et al. LRRK2 secretion in exosomes is regulated by 14-3-3. Hum Mol Genet. 2013;22:4988–5000. doi: 10.1093/hmg/ddt346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hogan MC, et al. Identification of biomarkers for PKD1 using urinary exosomes. J Am Soc Nephrol. 2015;26:1661–1670. doi: 10.1681/ASN.2014040354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bourderioux M, et al. A new workflow for proteomic analysis of urinary exosomes and assessment in cystinuria patients. J Proteome Res. 2015;14:567–577. doi: 10.1021/pr501003q. [DOI] [PubMed] [Google Scholar]
- 120.Rood IM, et al. Increased expression of lysosome membrane protein 2 in glomeruli of patients with idiopathic membranous nephropathy. Proteomics. 2015;15:3722–3730. doi: 10.1002/pmic.201500127. [DOI] [PubMed] [Google Scholar]
- 121.Zhou H, et al. Collection, storage, preservation, and normalization of human urinary exosomes for biomarker discovery. Kidney Int. 2006;69:1471–1476. doi: 10.1038/sj.ki.5000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhou H, et al. Exosomal fetuin-A identified by proteomics: a novel urinary biomarker for detecting acute kidney injury. Kidney Int. 2006;70:1847–1857. doi: 10.1038/sj.ki.5001874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhou H, et al. Urinary exosomal transcription factors, a new class of biomarkers for renal disease. Kidney Int. 2008;74:613–621. doi: 10.1038/ki.2008.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang Z, Hill S, Luther JM, Hachey DL, Schey KL. Proteomic analysis of urine exosomes by multidimensional protein identification technology (MudPIT) Proteomics. 2012;12:329–338. doi: 10.1002/pmic.201100477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gerlach JQ, et al. Surface glycosylation profiles of urine extracellular vesicles. PLoS ONE. 2013;8:e74801. doi: 10.1371/journal.pone.0074801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Corbetta S, et al. Urinary exosomes in the diagnosis of Gitelman and Bartter syndromes. Nephrol Dial Transplant. 2015;30:621–630. doi: 10.1093/ndt/gfu362. [DOI] [PubMed] [Google Scholar]
- 127.Esteva-Font C, et al. Renal sodium transporters are increased in urinary exosomes of cyclosporine-treated kidney transplant patients. Am J Nephrol. 2014;39:528–535. doi: 10.1159/000362905. [DOI] [PubMed] [Google Scholar]
- 128.Saraswat M, et al. N-linked (N−) glycoproteomics of urinary exosomes. [Corrected] Mol Cell Proteomics. 2015;14:263–276. doi: 10.1074/mcp.M114.040345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kosanovic M, Jankovic M. Isolation of urinary extracellular vesicles from Tamm-Horsfall protein-depleted urine and their application in the development of a lectin-exosome-binding assay. Biotechniques. 2014;57:143–149. doi: 10.2144/000114208. [DOI] [PubMed] [Google Scholar]
- 130.Turco AE, et al. Specific renal parenchymal-derived urinary extracellular vesicles identify age-associated structural changes in living donor kidneys. J Extracell Vesicles. 2016;5:29642. doi: 10.3402/jev.v5.29642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Panich T, et al. Urinary exosomal activating transcriptional factor 3 as the early diagnostic biomarker for sepsis-induced acute kidney injury. BMC Nephrol. 2017;18:10. doi: 10.1186/s12882-016-0415-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wolley MJ, et al. In primary aldosteronism, mineralocorticoids influence exosomal sodium-chloride cotransporter abundance. J Am Soc Nephrol. 2017;28:56–63. doi: 10.1681/ASN.2015111221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lytvyn Y, et al. Assessment of urinary microparticles in normotensive patients with type 1 diabetes. Diabetologia. 2017;60:581–584. doi: 10.1007/s00125-016-4190-2. [DOI] [PubMed] [Google Scholar]
- 134.Hogan MC, et al. Strategy and rationale for urine collection protocols employed in the NEPTUNE study. BMC Nephrol. 2015;16:190. doi: 10.1186/s12882-015-0185-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Thomas CE, Sexton W, Benson K, Sutphen R, Koomen J. Urine collection and processing for protein biomarker discovery and quantification. Cancer Epidemiol Biomarkers Prev. 2010;19:953–959. doi: 10.1158/1055-9965.EPI-10-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rodby RA. Timed urine collections for albumin and protein: “the king is dead, long live the king!”. Am J Kidney Dis. 2016;68:836–838. doi: 10.1053/j.ajkd.2016.06.025. [DOI] [PubMed] [Google Scholar]
- 137.Musante L, Tataruch DE, Holthofer H. Use and isolation of urinary exosomes as biomarkers for diabetic nephropathy. Front Endocrinol (Lausanne) 2014;5:149. doi: 10.3389/fendo.2014.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sir Elkhatim R, Li JY, Yong TY, Gleadle JM. Dipping your feet in the water: podocytes in urine. Expert Rev Mol Diagn. 2014;14:423–437. doi: 10.1586/14737159.2014.908122. [DOI] [PubMed] [Google Scholar]
- 139.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 140.Trionfini P, Benigni A, Remuzzi G. MicroRNAs in kidney physiology and disease. Nat Rev Nephrol. 2015;11:23–33. doi: 10.1038/nrneph.2014.202. [DOI] [PubMed] [Google Scholar]
- 141.Schageman J, et al. The complete exosome workflow solution: from isolation to characterization of RNA cargo. Biomed Res Int. 2013;2013:253957. doi: 10.1155/2013/253957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cantaluppi V, et al. Microvesicles derived from endothelial progenitor cells protect the kidney from ischemia-reperfusion injury by microRNA-dependent reprogramming of resident renal cells. Kidney Int. 2012;82:412–427. doi: 10.1038/ki.2012.105. [DOI] [PubMed] [Google Scholar]
- 143.Gildea JJ, et al. Exosomal transfer from human renal proximal tubule cells to distal tubule and collecting duct cells. Clin Biochem. 2014;47:89–94. doi: 10.1016/j.clinbiochem.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Murakami T, et al. Development of glomerulus-, tubule-, and collecting duct-specific mRNA assay in human urinary exosomes and microvesicles. PLoS ONE. 2014;9:e109074. doi: 10.1371/journal.pone.0109074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chen HH, et al. Exosomal ATF3 RNA attenuates pro-inflammatory gene MCP-1 transcription in renal ischemia-reperfusion. J Cell Physiol. 2014;229:1202–1211. doi: 10.1002/jcp.24554. [DOI] [PubMed] [Google Scholar]
- 146.Lv LL, et al. CD2AP mRNA in urinary exosome as biomarker of kidney disease. Clin Chim Acta. 2014;428:26–31. doi: 10.1016/j.cca.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 147.Lv LL, et al. MicroRNA-29c in urinary exosome/microvesicle as a biomarker of renal fibrosis. Am J Physiol Renal Physiol. 2013;305:F1220–F1227. doi: 10.1152/ajprenal.00148.2013. [DOI] [PubMed] [Google Scholar]
- 148.Ichii O, et al. Decreased miR-26a expression correlates with the progression of podocyte injury in autoimmune glomerulonephritis. PLoS ONE. 2014;9:e110383. doi: 10.1371/journal.pone.0110383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Barutta F, et al. Urinary exosomal microRNAs in incipient diabetic nephropathy. PLoS ONE. 2013;8:e73798. doi: 10.1371/journal.pone.0073798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Eissa S, Matboli M, Aboushahba R, Bekhet MM, Soliman Y. Urinary exosomal microRNA panel unravels novel biomarkers for diagnosis of type 2 diabetic kidney disease. J Diabetes Complications. 2016;30:1585–1592. doi: 10.1016/j.jdiacomp.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 151.Jia Y, et al. miRNAs in urine extracellular vesicles as predictors of early-stage diabetic nephropathy. J Diabetes Res. 2016;2016:7932765. doi: 10.1155/2016/7932765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Rekker K, et al. Comparison of serum exosome isolation methods for microRNA profiling. Clin Biochem. 2014;47:135–138. doi: 10.1016/j.clinbiochem.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 153.Nicolson GL. The fluid-mosaic model of membrane structure: still relevant to understanding the structure, function and dynamics of biological membranes after more than 40 years. Biochim Biophys Acta. 2014;1838:1451–1466. doi: 10.1016/j.bbamem.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 154.Kreimer S, et al. Mass-spectrometry-based molecular characterization of extracellular vesicles: lipidomics and proteomics. J Proteome Res. 2015;14:2367–2384. doi: 10.1021/pr501279t. [DOI] [PubMed] [Google Scholar]
- 155.Gyorgy B, et al. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell Mol Life Sci. 2011;68:2667–2688. doi: 10.1007/s00018-011-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Trajkovic K, et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 157.Llorente A, et al. Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochim Biophys Acta. 2013;1831:1302–1309. doi: 10.1016/j.bbalip.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 158.Subra C, Laulagnier K, Perret B, Record M. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie. 2007;89:205–212. doi: 10.1016/j.biochi.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 159.Muralidharan-Chari V, et al. ARF6-regulated shedding of tumor cell-derived plasma membrane microvesicles. Curr Biol. 2009;19:1875–1885. doi: 10.1016/j.cub.2009.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Agmon E, Stockwell BR. Lipid homeostasis and regulated cell death. Curr Opin Chem Biol. 2017;39:83–89. doi: 10.1016/j.cbpa.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Del Boccio P, et al. A hyphenated microLC-Q-TOF-MS platform for exosomal lipidomics investigations: application to RCC urinary exosomes. Electrophoresis. 2012;33:689–696. doi: 10.1002/elps.201100375. [DOI] [PubMed] [Google Scholar]
- 162.Ichimura T, et al. Kidney injury molecule–1 is a phosphatidylserine receptor that confers a phagocytic phenotype on epithelial cells. J Clin Invest. 2008;118:1657–1668. doi: 10.1172/JCI34487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhao N, et al. MicroRNA miR145 regulates TGFBR2 expression and matrix synthesis in vascular smooth muscle cells. Circ Res. 2015;116:23–34. doi: 10.1161/CIRCRESAHA.115.303970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kelly KJ, et al. Improved structure and function in autosomal recessive polycystic rat kidneys with renal tubular cell therapy. PLoS ONE. 2015;10:e0131677. doi: 10.1371/journal.pone.0131677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Collino F, et al. AKI recovery induced by mesenchymal stromal cell-derived extracellular vesicles carrying microRNAs. J Am Soc Nephrol. 2015;26:2349–2360. doi: 10.1681/ASN.2014070710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bruno S, et al. Microvesicles derived from mesenchymal stem cells enhance survival in a lethal model of acute kidney injury. PLoS ONE. 2012;7:e33115. doi: 10.1371/journal.pone.0033115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Bruno S, et al. Mesenchymal stem cell-derived microvesicles protect against acute tubular injury. J Am Soc Nephrol. 2009;20:1053–1067. doi: 10.1681/ASN.2008070798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.US National Library of Medicine. ClinicalTrials.gov. 2014 https://clinicaltrials.gov/ct2/show/NCT02138331.
- 169.Gracia T, et al. Urinary exosomes contain microRNAs capable of paracrine modulation of tubular transporters in kidney. Sci Rep. 2017;7:40601. doi: 10.1038/srep40601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Burger D, et al. Urinary podocyte microparticles identify prealbuminuric diabetic glomerular injury. J Am Soc Nephrol. 2014;25:1401–1407. doi: 10.1681/ASN.2013070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 172.Khurana R, et al. Identification of urinary exosomal noncoding RNAs as novel biomarkers in chronic kidney disease. RNA. 2017;23:142–152. doi: 10.1261/rna.058834.116. [DOI] [PMC free article] [PubMed] [Google Scholar]


