SUMMARY
In vertebrates and invertebrates, neurons and glia are generated in a stereotyped manner from neural stem cells, but the purpose of invariant lineages is not understood. We show that two stem cells that produce leg motor neurons in Drosophila also generate neuropil glia, which wrap and send processes into the neuropil where motor neuron dendrites arborize. The development of the neuropil glia and leg motor neurons is highly coordinated. However, although motor neurons have a stereotyped birth order and transcription factor code, the number and individual morphologies of the glia born from these lineages are highly plastic, yet the final structure they contribute to is highly stereotyped. We suggest that the shared lineages of these two cell types facilitate the assembly of complex neural circuits and that the two birth order strategies—hardwired for motor neurons and flexible for glia—are important for robust nervous system development, homeostasis, and evolution.
In Brief
Enriquez et al. show that the glia surrounding each thoracic neuropil come from stem cells that also give rise to leg motor neurons. Unlike the hard-wired birth order and morphology of each motor neuron, the number and morphologies of the glia born from single lineages are variable, yet the structure they contribute to is highly stereotyped.
INTRODUCTION
Cell lineages play a critical role in generating cellular diversity during animal development yet range widely in the number and types of cells they produce. Embryonic stem cells, for example, have the potential to give rise to all cell types, whereas hematopoietic stem cells have much more limited potential, giving rise only to myeloid and lymphoid cells (Seita and Weissman, 2010; Wu et al., 2016). The wide range of developmental potentials is also reflected in the degree to which cell lineages are hardwired or plastic in the types and numbers of cells they can generate. At one extreme, each of the 959 somatic cells of the adult C. elegans hermaphrodite is derived from an invariant lineage that can be traced back to the single-celled zygote (Sulston, 1976). In contrast, cell lineages of the mammalian immune system are plastic and can be modified depending on the environmental challenges that an animal confronts (Boettcher and Manz, 2017). Similarly, intestinal stem cell lineages exhibit plasticity in response to injury and cell death (Apidianakis and Rahme, 2011; Jiang and Edgar, 2011).
Although the capacity of some cell lineages to respond to varying environmental conditions makes sense in light of the cell types these lineages generate, the underlying logic of why some lineages are invariant is usually not understood. For example, although the entire lineage is hardwired in C. elegans, there is almost no correlation between where a cell is born in the lineage and cell-type identity; most sub-lineages contribute broadly to endodermal, mesodermal, and nervous system tissues. Consequently, a single cell type such as a motor neuron can be born from many different sub-lineages in C. elegans (Hobert, 2016). In D. melanogaster, although there are neuroblasts (NBs), stem cells that are dedicated to the generation of neurons, during embryogenesis each NB can generate a mixture of motor neurons, interneurons, and glia that do not share an obvious function (Bossing et al., 1996; Schmidt et al., 1997). These observations raise the question of whether there is a biological purpose for the structure of invariant lineages that may be difficult to discern by only examining the final progeny. Consistent with this notion, in the adult, the neuronal progeny of individual Drosophila NB hemilineages share anatomical features such as stereotyped axon paths and, when activated, the progeny of individual hemilineages can evoke specific behaviors in decapitated flies (Harris et al., 2015; Truman et al., 2004). Thus, by both anatomical and behavioral criteria, there is emerging evidence that individual Drosophila NBs generate functionally related neurons in the adult fly.
Another reason for the existence of hardwired lineages might be to facilitate the assembly of complex structures during development, which is an especially challenging problem for nervous systems. For example, the ~50 motor neurons (MNs) that innervate each leg in adult Drosophila are born from invariant NB lineages and have highly stereotyped birthdates and morphologies (Baek and Mann, 2009; Brierley et al., 2012). These stereotyped morphologies can be seen both in the muscles these motor neurons target and in their elaborate dendritic arbors that establish a very large number of synapses in dense neuropils present in each thoracic neuromere of the ventral nerve cord (VNC) (Court et al., 2017). Although each motor neuron morphology is thought to be determined by unique combinations of morphology transcription factors (mTFs; Enriquez et al., 2015), how these stereotyped morphologies form the correct synaptic connections with interneurons and sensory neurons is not understood. Interestingly, at least one of the major NB lineages that give rise to adult motor neurons in the fly also generates another important cell type in the nervous system, glia (Baek et al., 2013; Lacin and Truman, 2016), which are critical for the establishment of synapses and the maintenance of neuronal activity (Freeman, 2015; Freeman and Rowitch, 2013). The observation that glia and leg motor neurons may be derived from common progenitors (neuroglioblast [NGB]) raised the possibility that their development may be coordinated, and that being born from the same lineages might also play a role in neural circuit assembly. However, in contrast to motor neurons, very little is known about the glial types produced by these NGBs, how their morphologies are genetically specified, and how neurons and glia communicate during development to build a functional CNS.
In other contexts, Drosophila glia have been subdivided into three classes based on their functions and morphologies: surface glia, which isolate the CNS from the hemolymph, cortex glia, which send processes around neuronal cell bodies, and neuropil glia (NG), which surround neuropil and glomeruli (Omoto et al., 2016). NG are further subdivided into astrocytes and ensheathing glia (EG) (Muthukumar et al., 2014; Peco et al., 2016). EG extend flat processes that surround neuropil and glomeruli as well as a small portion of axon bundles as they enter or leave the neuropil (Omoto et al., 2015; Peco et al., 2016). More recently, EG have been shown to extend small protrusions inside the neuropil of the adult brain (Kremer et al., 2017). In the medulla (region of the brain that processes visual information), EG are associated with axons (Kremer et al., 2017) and trachea (Kremer et al., 2017; Pereanu et al., 2007). In contrast, astrocytes send highly ramified processes deep into neuropil regions in close contact with synapses (Awasaki et al., 2008; Muthukumar et al., 2014; Stork et al., 2014). Although NG exhibit complex and diverse morphologies and are generally derived from stem cells that also generate neurons (Awasaki et al., 2008; Omoto et al., 2016), the developmental mechanisms leading to glial diversity remains unknown.
Here, we show that two of the ~11 lineages that generate leg motor neurons in Drosophila also generate all of the NG that populate the adult thoracic neuropils. As a consequence of being born from the same lineages, the development of the NG is physically and temporally coordinated with motor neuron development. However, unlike the hardwired birth order and unique mTF codes of the motor neurons that are born from these NGBs, individual NG do not have a stereotyped morphology, birth order, or unique transcription factor code. Moreover, using a unique mode of division, the gliogenesis phase of these lineages is plastic and highly adaptable: when gliogenesis in one lineage is compromised, other lineages compensate to maintain the correct number of NG. Thus, even though NG and motor neurons come from the same stem cells, and generate adult neuromeres with highly stereotyped structures, there are fundamental differences in how these two cell types are specified. We suggest that the combination of hardwired motor neuron generation and flexible glia production facilitates the robust construction, homeostasis, and evolution of complex neural circuits.
RESULTS
Cellular Organization of the Adult Thoracic Neuropil Glia
In order to characterize the cellular organization of the NG in the adult VNC, we expressed fluorescent reporters under the control of two Gal4 drivers, alrm-Gal4 (astrocyte specific; Doherty et al., 2009) and R56FO3-Gal4 (EG specific; Kremer et al., 2017; Peco et al., 2016; Pfeiffer et al., 2008), which we confirmed are expressed in all adult astrocytes and EG, respectively (Figures 1 and S1). We also used as a marker Distalless (Dll), which we found to be expressed in all NG in the adult VNC (Figure S1). As with the NG in the larval CNS (Beckervordersandforth et al., 2008; Omoto et al., 2015), the number of NG surrounding each thoracic neuropil is very consistent between animals (with a very low SD; Figures 1P and 1Q).
Figure 1. Cellular Organization of the Adult Thoracic NG.
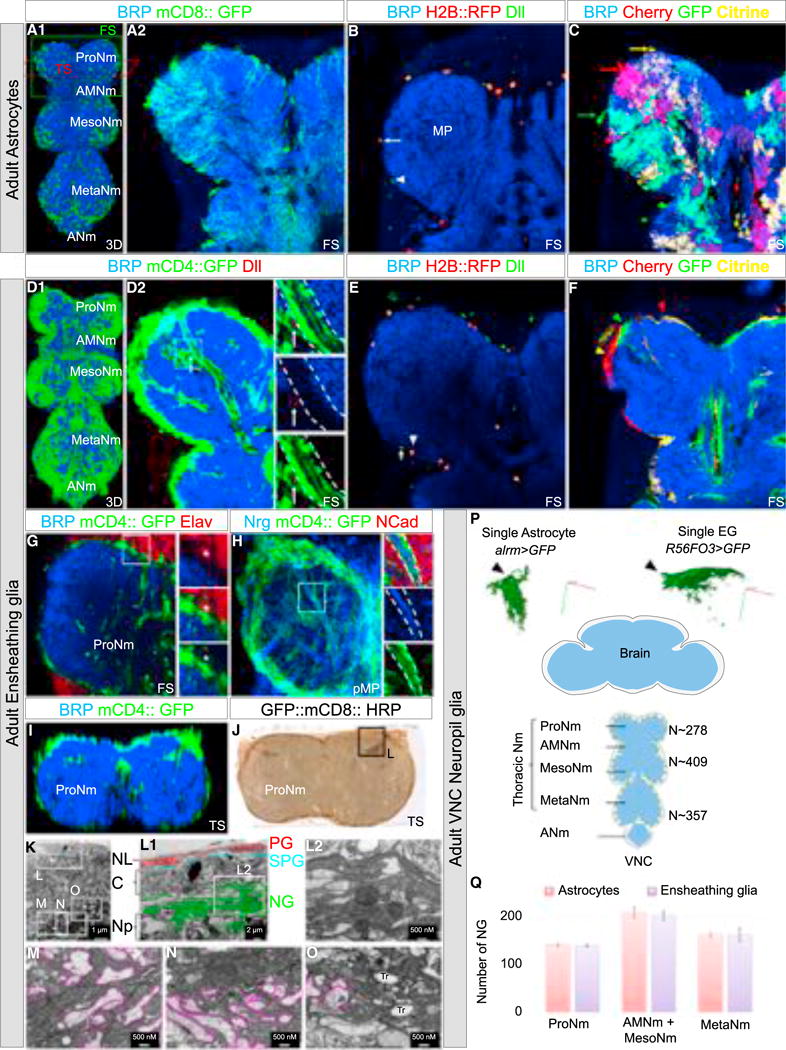
(A–C) Adult VNCs immunostained with anti-BRP (neuropil marker, blue) (A1–C) and anti-Dll (NG marker, green) (B) with astrocytes expressing mCD8::GFP (membrane marker, green) (A1 and A2), H2B::RFP (nuclear marker, red) (B) or the multicolor system FB1.1 (single-cell marker, red, yellow, and green) (C) under the control of alrm-Gal4.
(B) Astrocyte (H2B::RFP+, Dll+): arrow, EG (Dll+): arrowhead.
(C) Different astrocytes with processes occupying different neuropil territories: arrows. See also Figure S1 and Movie S1.
(D–F) Adult VNCs immunostained with anti-BRP (neuropil marker, blue) (D1–F) and anti-Dll (NG marker, green) (E) with EG-expressing mCD4::GFP (membrane marker, green) (D1 and D2), H2B::RFP (nuclear marker, red) (E) or the multicolor system FB1.1 (single-cell marker, red, yellow, and green; Hadjieconomou et al., 2011) (F) under the control of R56F03-Gal4. (E) Astrocyte: arrow (Dll+), EG (H2B::RFP+, Dll+): arrowhead.
(D) arrow: in more than half of the samples analyzed (n = 10) a cell body of an EG (GFP +, Dll+) was observed inside the neuropil, next to axon bundles.
(F) Different EG with processes occupying different neuropil territories: arrows. See also Figure S1 and Movie S1.
(G and H) Prothoracic neuromeres with EG-expressing mCD4::GFP and immunostained with anti-BRP (blue) and anti-Elav (neuron marker, red) (G) or anti-NCad (neuropil marker, red) and Nrg (axon marker, blue) (H).
(G) Asterisk: Elav+ neuron wrapped by an EG. Enlargements of the boxed regions are to the right of each panel. See also Movie S2.
(I and J) Prothoracic neuromeres with EG-expressing mCD4::GFP (I) and GFP::mCD8::HRP (J) immunostained with anti-BRP (blue) (I) or labeled with DAB (brown) (J).
(K) Low-magnification electron microscope image of the boxed region in (J).
(L–O) Enlargement of the boxed regions in (K). (L2) Enlargement of the boxed region in (L1). Note: (L1) and (L2) show the organization of the EG processes around the neuropil while (M)–(O) show the EG processes inside the neuropil. Pink lines outline axons. PG, perineurial glia; SPG, subperineurial glia; NG, neuropil glia; C, cortex; Np, neuropil; NL, neural lamella.
(P) Bottom, schematic of adult CNS (blue: neuropils, gray: cortex); top, single astrocyte and EG labeled with mCD8::GFP under the control of alrm-Gal4 and R56F03-Gal4 using MARCM. Axes: green (posterior), blue (dorsal), red (medial).
(Q) Average number of NG in the adult VNC, in which EG or astrocytes were expressing H2B::RFP under the control of R56F03-Gal4 or alrm-Gal4, respectively, and immunostained with anti-Dll. Number of samples = 4/genotype. Error bars indicate SD.
ProNm, Prothoracic neuromere; AMesoNm, Accessory mesothoracic neuromere; MesoNm, Mesothoracic neuromere; MetaNm, Metathoracic neuromere; ANm, Abdominal neuromeres; FS, frontal cross-section; TS, transverse cross-section; 3D, 3-dimensional reconstruction of confocal image stack; pMP, partial maximum projection.
As described for other regions of the CNS (Awasaki et al., 2008; Muthukumar et al., 2014), adult thoracic astrocytes send processes deep into the neuropil that respect each other’s territory, thus exhibiting a tiling-like phenomenon (Figures 1A and 1C; Movie S1). EG extend processes that surround each neuropil and also respect each other’s territory with very infrequent overlaps (Figures 1D and 1F; Movie S1). In addition, the adult thoracic EG feature a more complex morphology than previously described for larval EG. First, adult EG wrap the cell bodies of neurons (Elav+) that are localized next to the neuropil (Figure 1G; Movie S2) as well as astrocyte cell bodies (Figure S1). Second, they send processes inside the neuropil where they wrap axon bundles (Neuroglian+) where no synapses are formed (BRP-negative, a marker of the active presynaptic zone) (Figures 1G and 1H; Movie S2). Interestingly, EG only wrap these axons inside the neuropil; outside of the neuropil these axons are surrounded by a distinct class of wrapping glia (Figure 1H; data not shown). Thus, EG isolate nerves inside neuropil before they make synapses with other neurons.
We next performed electron microscopy of transverse sections of prothoracic neuromeres (ProNs) (Figures 1K–1O). EG wrap the thoracic neuropils very densely with several layers (Figure 1N). These results, in combination with the tiling results described above, suggest that one EG can send processes organized in layers around the neuropil. Inside the neuropil, axon bundles and single axons can be wrapped by layers of EG processes (Figures 1M–1O). Interestingly, each nerve also contains trachea bundles that are surrounded by EG, forming a structure with all three cell types: EG, axons, and trachea (Figure 1O). Notably, axons close to the midline are not wrapped by EG (data not shown).
Developmental Origins of the Adult Thoracic NG
As described above, the adult thoracic neuropils are complex structures containing glia processes and axons/dendrites in close juxtaposition to each other, raising the question of how these structures develop. A previous study suggested that pockets of glia, marked by the general glia marker Repo, surround the immature leg neuropils in late third-instar larvae (just prior to the onset of metamorphosis) and produce adult astrocytes (Li et al., 2014), while another study suggests that these glia express Dll and may migrate to the leg to form wrapping glia, which are distinct from EG and astrocytes (Plavicki et al., 2016). In order to definitively determine what type of adult cells these Repo+ glia give rise to, we used a Dll-Gal4 enhancer trap and a Dll antibody to label these cells during development (Figure 2).
Figure 2. Developmental Origin of the Adult NG.
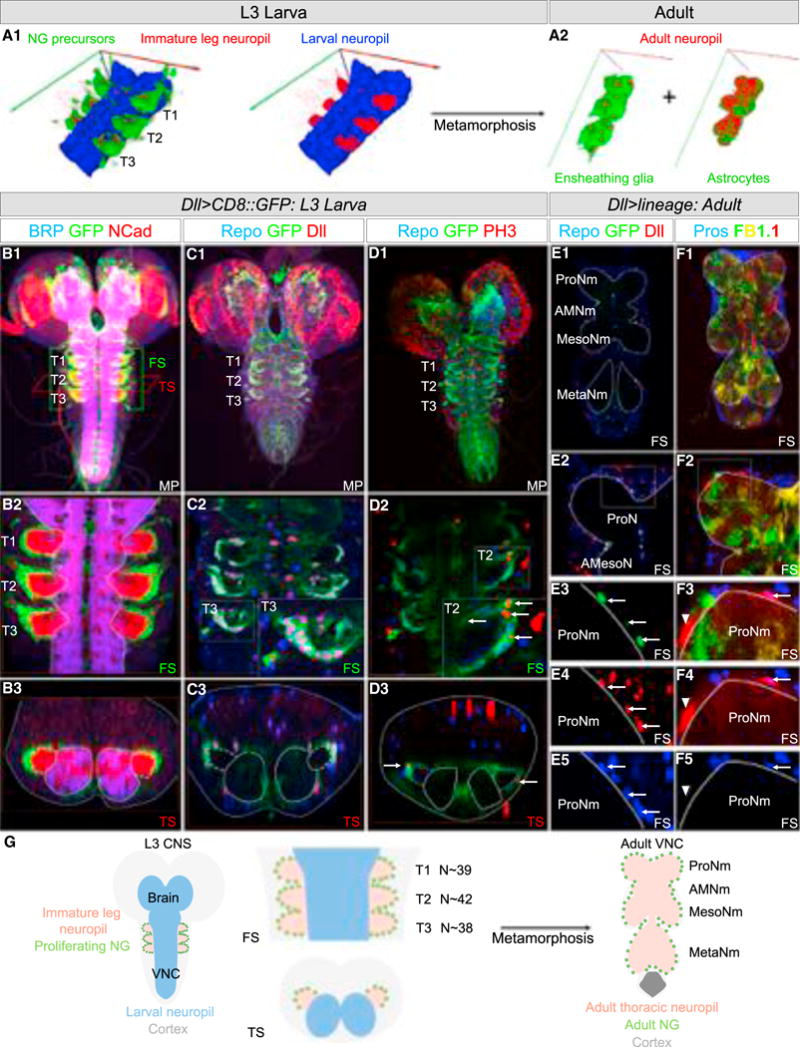
(A) 3D rendering of the thoracic segments of a L3 VNC (A1) labeled with anti-BRP (mature neuropil marker, blue) and anti-NCad (mature and immature neuropil marker, red). The glia surrounding the six immature leg neuropils express mCD8::GFP (green, left image only) under the control of Dll-Gal4. (T1, T2, and T3 indicate the three thoracic segments.) (A2) Following metamorphosis, 3D rendering of adult VNCs with astrocytes and EG-expressing GFP under the control of alrm-Gal4 (right) and R56F03-Gal4 (left) and costained with anti-BRP (red).
(B–D) L3 CNSs in which glia surrounding the six immature leg neuropils express mCD8::GFP under the control of Dll-Gal4 and co-stained with anti-BRP (blue), anti-NCad (red) (B1–B3), anti-Repo (blue), anti-Dll (red) (C1–C3), or anti-Repo (blue), anti-PH3 (red) (D1–D3). (D2 and D3) Arrows point to glia expressing PH3. (E and F) Adult VNCs with NG expressing nuclear GFP (green) (E1–E5) or the multicolor Flybow system FB1.1 (yellow, red, and green) (see STAR Methods for details) and immunostained with anti-Repo (blue), anti-Dll (red) (E1–E5), or anti-Pros (blue) (F1–F5). (E1–E5) Arrow points to adult NG-expressing GFP. (F1–F5) Arrow: astrocyte (mCherry+, Pros+), arrowhead: EG (mCherry+, Pros−). See also Figure S2.
(G) Left: schematic of an L3 CNS. The immature leg neuropils and proliferating NG are indicated. Right: schematic of an adult VNC with the adult leg NG. N, average number of proliferating NG in each L3 thoracic hemisegment.
In third-instar larvae (L3), there were ~40 Repo+ Dll+ cells surrounding each immature leg neuropil (Figures 2B, 2C, and 2G), where nascent motor neuron axons enter and then exit (Figure 4A). If these cells are the progenitors of the adult NG, they should be mitotically active. To test this idea, we first confirmed that they express phosphorylated histone H3 (pH3), a marker for mitotically active cells (Figure 2D).
Figure 4. NGBs Produce Leg Motor Neurons and All Thoracic NG.
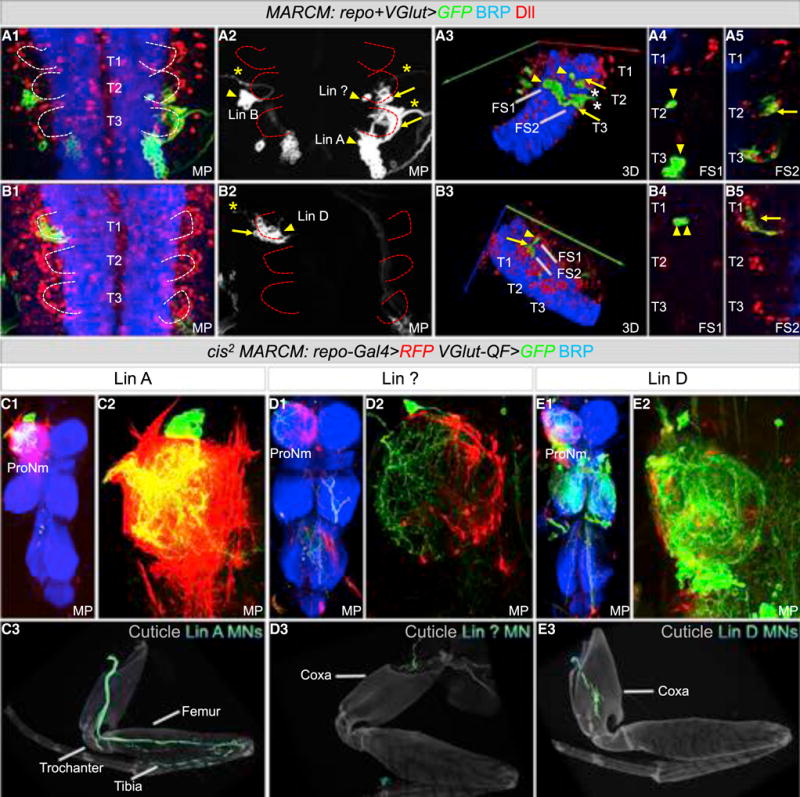
(A and B) Thoracic segments of an L3 VNC labeled with anti-BRP (blue) and anti-Dll (red) containing MARCM clones expressing mCD8::GFP under the control of both VGlut-Gal4 and repo-Gal4. Lin A, Lin ?, and Lin D clones produce motor neurons (arrowhead) and glia (arrow). (A1–A5) Sample with Lin A, Lin B and Lin ? clones; note that Lin B does not produce glia. (B1–B5) Sample with a Lin D clone. Asterisks mark motor neuron axons exiting the VNC.
(C–E) Lin A (C1–C3), Lin ? (D1–D3) and Lin D (E1–E3) cis2 MARCM clones in a ProN labeled with anti-BRP (blue) (C1–C2, D1–D2, E1–E2) and in a T1 Leg (C3, D3, E3) expressing mCD8::RFP in NG (red) and mCD8::GFP (green) under the control of repo-Gal4 and VGlut-QF, respectively.
Although Dll protein is expressed in adult NG, Dll-Gal4 is only active in immature NG (data not shown), allowing us to mark the progeny of Dll-Gal4-expressing larval cells in lineage tracing experiments (see STAR Methods). In the first experiment, we labeled the adult progeny of Dll-Gal4-expressing L3 glia with nuclear GFP (UAS-nGFP) and co-stained with an antibody against Repo and Dll (Figure 2E). Only the adult Repo+ Dll+ NG were labeled with nGFP, demonstrating that the Dll-Gal4-expressing cells in the larval CNS only generate adult NG. In a second experiment, we used Dll-Gal4 to activate the Flybow system (UAS-FB1.1), which permanently labels cells with different fluorescent markers (Hadjieconomou et al., 2011). We co-stained the resulting adult VNCs with Prospero, which is expressed in astrocytes but not in EG (Griffiths and Hidalgo, 2004; Peco et al., 2016) (Figures 2F and S1). Based on Prospero expression and morphology, we found that both astrocytes and EG were labeled. Significantly, neither experiment labeled other classes of glia, such as surface glia or cortex glia, arguing that these glia subtypes are not derived from Dll-Gal4-expressing cells. We confirmed this result with another driver (R31F10-Gal4) that is also expressed in immature NG (Figure S2).
Taken together, we conclude that the !40 Repo+ Dll+ Dll-Gal4-expressing cells surrounding each immature leg neuropil in L3 are the precursors of the >280 NG that surround each adult thoracic neuropil.
Coordinated Development of the Adult NG and Neuropil
To characterize the development of the adult neuropil, we conducted several experiments to trace the origins of this complex structure using BRP to label the larval neuropils, N-cadherin to label the immature adult neuropils, and Dll-Gal4; UAS-GFP (Dll>GFP) to label the proliferating NG precursors (Figure 3; Movie S3).
Figure 3. Coordinated Development of NG and Adult Neuropil.
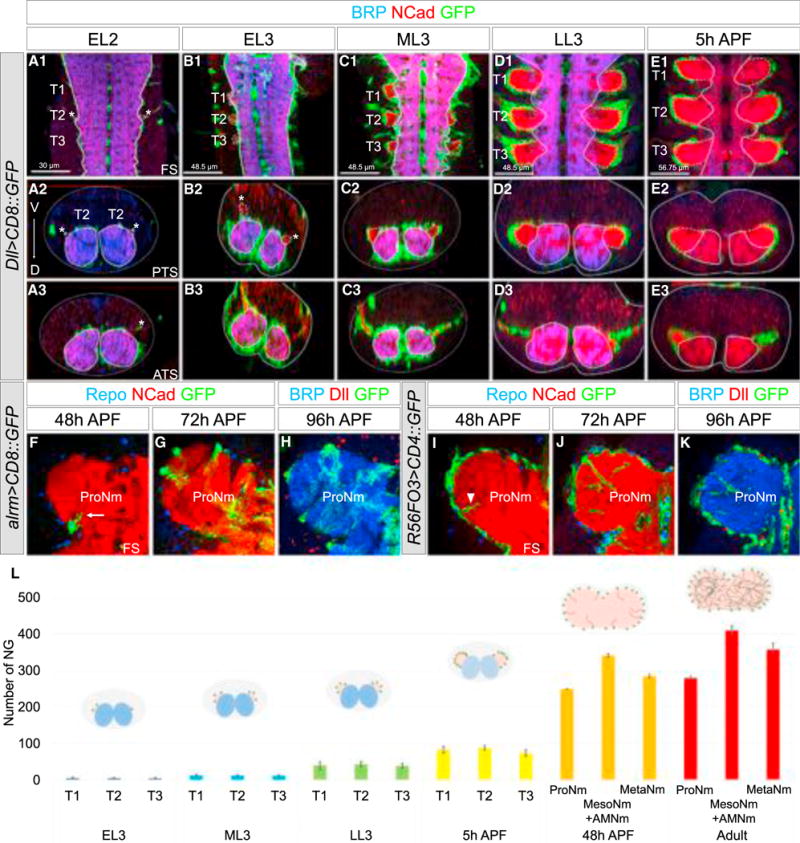
(A–E) Thoracic segments of larval VNCs at different stages (A, early L2; B, early L3; C, mid L3; D, late L3, E, 5 hr after pupal formation) with proliferating NG-expressing mCD8::GFP under the control of Dll-Gal4 and labeled with anti-BRP (blue), anti-NCad (red). Asterisks indicate axons of incoming leg sensory neurons. See also Figure S3 and Movies 3 and 4. (E1–E3) At this stage, the expression of BRP is almost undetectable, probably a consequence of the remodeling and/or degradation of the larval neuropils.
(F–K) ProNm of an pupal VNC with astrocytes (F–H) or EG (I–K) expressing a membrane-tagged GFP under the control of alrm-Gal4 and R56F03-Gal4, respectively, and co-stained with anti-Repo (blue) and anti-NCad (red) (F, G, I, and J), anti-BRP (blue), and anti-Dll (red) (H and K). (F and I) Processes of astrocytes (arrow) and EG (arrowhead) are first observed invading the neuropil.
(L) Average number of NG at different stages (number of samples analyzed at each stage = 4) with schematics of transverse sections of T1 or ProNm. NG (green), larval neuropils (blue), and leg neuropils (red) are indicated. Error bars indicate SD.
ATS, anterior transverse section; PTS, posterior transverse section; APF, after pupa formation.
In early second-instar larvae (EL2), most of the adult leg motor neurons are not yet born (Baek and Mann, 2009) and no Dll > GFP+-proliferating NG are visible. However, at this stage incoming leg sensory axons are visible and mark the location where the adult neuropil will form (Figure 3A). In early L3 (EL3; 72 hr after egg laying, AEL), 0–10 proliferating NG are observed per thoracic hemisegment (Figures 3B, 3L, and S3), and their numbers gradually expand through subsequent larval and pupal stages (Figures 3C–3E and S3). As proliferating NG are produced, they send processes that surround the growing neuropil and wrap the leg nerves (Figures 3A–3E and S3). By the mid pupal stage (48 hr APF), most of EG and astrocytes have been generated and start to differentiate by sending processes inside the neuropil (Figures 3F–3I). By the late pupal stage (96 hr APF), the adult neuropils are fully invaded by astrocyte processes and wrapped by EG (Figures 3H and 3K).
These results suggest that the adult thoracic NG are produced from early L3 to mid pupa. These glia create a structure that encompasses the immature adult neuropil (Movies S3 and S4); by L3, the motor neuron neurites are completely surrounded by the processes of the immature NG. During metamorphosis, the dendrites and axons of the neurons establish their final morphologies within the glia-defined neuropil.
The Adult Thoracic NG Are Produced by NGBs that Also Generate Leg Motor Neurons
Previous work established that the larval NG are born from a single glioblast in each hemisegment of the larval CNS (Becker-vordersandforth et al., 2008; Ito et al., 1995; Jacobs et al., 1989) and from a single glioblast or NGB in each hemisphere of the brain (Hartenstein et al., 1998; Omoto et al., 2015), while in the adult brain NG are derived from a group of uncharacterized NGBs (Omoto et al., 2015). To characterize the lineages that give rise to the adult thoracic NG, we carried out several clonal analysis experiments. Previous observations demonstrated that NB lineages such as Lin A (also called Lin 15), produce both leg motor neurons and glial cells (Baek et al., 2013; Truman et al., 2004). This observation raised the intriguing possibility that NG and leg motor neurons may share common developmental origins.
First, we used MARCM to express the membrane reporter UASmCD8::GFP with two different drivers, repo-Gal4 (expressed in all glia, Sepp and Auld, 2003) and VGlut-Gal4 (expressed in all motor neurons and some interneurons, Mahr and Aberle, 2006). We induced these clones in L1 larvae and dissected late L3 CNS and co-stained for Dll, to mark immature adult NG, and BRP to mark the mature larval neuropil. If a stem cell produces both motor neurons and NG, individual MARCM clones should contain both cell types. We identified glial cells by their position and expression of Dll, and motor neurons by their morphology, particularly motor neuron axons, which exit the CNS and target the leg imaginal discs (Baek and Mann, 2009; Truman et al., 2004). 62/62 clones that labeled NG also labeled motor neurons (Figure 4). We found 3 types of clones based on the number of motor neurons they contained. Type 1 clones (n = 36) contained ~28 leg motor neurons and ~20 immature adult NG. Based on motor neuron morphology, this lineage is Lin A (Figure 4A). Type 2 clones (n = 23) contained 2 motor neurons and ~10 immature NG (Figure 4B). In type 3 (n = 3), the clones contained 1 motor neuron and ~7 glia (Figure 4A). The total number of glia (~37) produced by these three NGBs is very close to the total number of NG progenitors that surround each neuropil at L3 (~40), suggesting that these three lineages produce most, and perhaps all, of the adult thoracic NG.
In a second approach, we generated clones using a modified QMARCM/MARCM method (cis2-MARCM) in which the two transcriptional repressors, GAL80 and QS, are recombined on the same chromosome arm (see STAR Methods for details). We used two drivers, VGlut-QF and repo-Gal4, allowing us to visualize VGlut+ motor neurons and Repo+ glia derived from the same stem cell with two different fluorescent markers. Using this method, and analyzing the adult legs and VNCs, we confirmed that there are three NGBs that produce leg motor neurons and adult NG. Lin A clones produce NG and ~28 motor neurons targeting the trochanter, femur, and tibia (n = 14, in ProN) (Figures 4C and 4F). Type 2 clones labeled NG and 2 motor neurons that targeted the coxa; based on the morphology of these motor neurons, we recognized this lineage as Lin D (Baek and Mann, 2009) (n = 2) (Figure 4E). Type 3 clones labeled NG and a single motor neuron that targeted a body wall muscle at the base of the coxa (n = 16) (Figure 4F). Notably, this motor neuron was previously described to be derived from Lin A (Brierley et al., 2012), a conclusion that we confirm and revise below.
The Lin A NGB Initially Produces Glioblasts and Postmitotic Motor Neurons
Because Lin A generates both glia and a stereotyped set of motor neurons, we used this lineage to investigate the birth order of these two types of progeny. In the first set of experiments we used a lineage tracing system that is restricted to Dpn+ NBs (Awasaki et al., 2014). When combined with 10C12-Gal4, which is expressed early in the Lin A lineage, this system specifically labels all Lin A progeny with GFP (Lacin and Truman, 2016). We immunostained the resulting larval CNSs with Repo, Elav, and Dpn to visualize glia, neurons, and NBs, respectively, and dissected larva at different time points during L3 (Figures 5A–5J). As a consequence of NB division patterns, progeny further from the NB tend to be older compared to progeny closer to the NB, thus potentially providing birth order information.
Figure 5. Lin A Initially Generates Postmitotic Motor Neurons and Proliferating Glia and Then Only Motor Neurons.
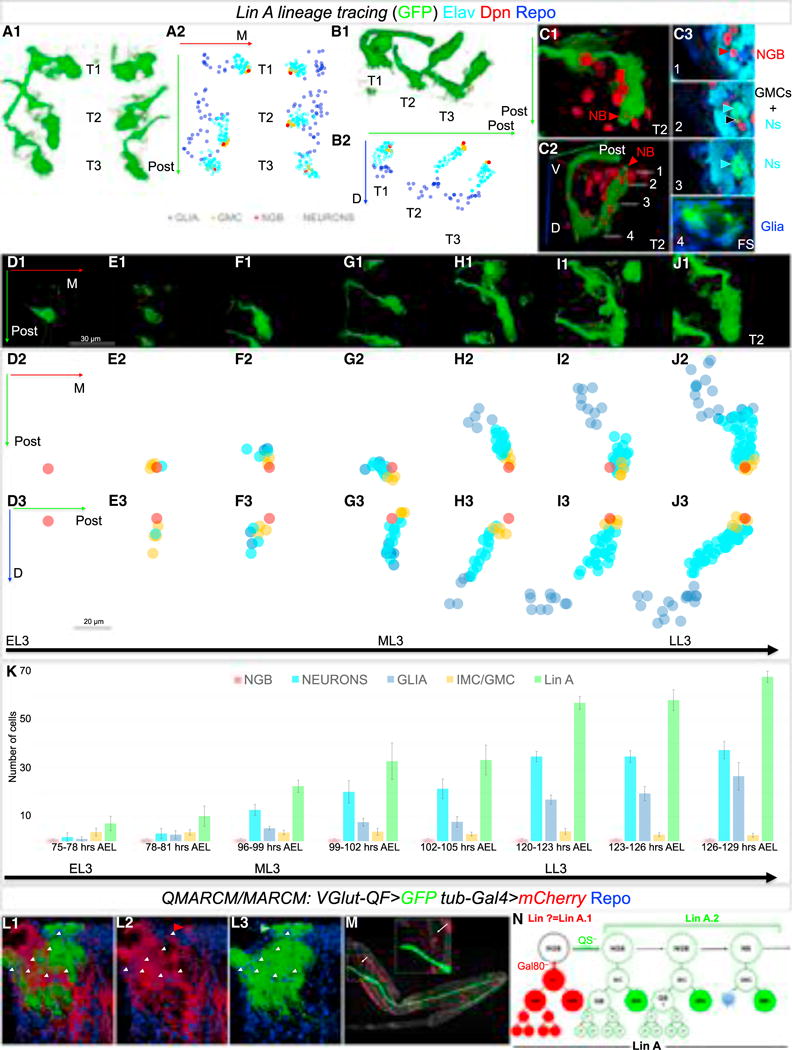
(A–C) 3D reconstruction of all six (A1), three (B1), or a single (C) Lin A clones labeled with myr::GFP (green) generated by a Lin A tracing system in the thoracic segments of a late L3 VNC, and immunostained with anti-Dpn (red), anti-Elav (cyan), and anti-Repo (blue); A1 and C1 are ventral views, B1 and C2 are lateral views. C3 shows a series of frontal cross-sections (FS) of Lin A from ventral (1) to dorsal (4). NB: Lin A NB/NGB, red arrowhead, GMC: Lin A GMC, black arrowhead, Ns: Lin A neurons, cyan arrowhead. Axes in A2, B2: green (posterior, Post), blue (dorsal, D), red (medial, M). A2 and B2 show plots of Lin A progeny as seen in A1 and B1, respectively, color coded according to cell type. Blue, Repo+; red, Dpn+; cyan, Elav+, orange: (GMC/IMC, ganglion mother cell/intermediate mother cell, Dpn−, Elav−, Repo−); note: some of these GMC/IMC could be immature neurons or glia.
(D–J) Ventral view of a 3D reconstruction of single Lin A clones in a T2 segment at different time points during L3 (D1–J1) and graphs of each Lin A cell from two perspectives (D2–J2 and D3–J3). Axes: green (posterior, Post), blue (dorsal, D), red (medial, M). Shown are: (D) 75–78 hr AEL, (E) 96–99 hr AEL, (F) 99–102 hr AEL, (G) 102–105 hr AEL, (H) 120–123 hr AEL, (I) 123–126 hr AEL, and (J) 126–129 hr AEL.
(K) Average number of Lin A cell types at different L3 stages. Number of samples analyzed at each stage: (75-to 78-hr AEL): n = 16; (78-to 81-hr AEL): n = 16; (96-to 99-hr AEL): n = 8; (99-to 102-hr AEL): n = 10; (102-to 105-hr AEL): n = 7; (120-to 123-hr AEL): n = 9; (123-to 126-hr AEL): n = 8; (126-to 129-hr AEL: n = 7. Error bars indicate SD. AEL, after egg laying. “Lin A” refers to all cells labeled by GFP.
(L and M) Lin A QMARCM/MARCM clone in a ProNm generated before the first post-embryonic division and labeled with Repo (blue). Motor neurons produced by one daughter cell are labeled with mCD8::GFP under the control of VGlut-QF, and all the cells produced by the sister daughter cell are labeled with RFP under the control of tubulin-Gal4 (L1, L2). RFP labels a single motor neuron targeting a body wall muscle (M) and NG when the clones are induced at early L3 or earlier. Number of samples analyzed: n > 30.
(N) Inferred pattern of Lin A divisions. Lin A.1 produces at least one glioblast (GB) and one motor neuron targeting the body wall; Lin A.2 also produces at least one glioblast and the remaining Lin A motor neurons. We refer to the mother of a glioblast and motor neuron as an intermediate mother cell (IMC) and the progeny of the Glioblast as proliferating glia (pG) Later cell divisions of Lin A.2 generate a GMC, which produce one motor neuron and a sister cell that undergoes apoptosis (blue star; Truman et al., 2010).
Using this approach, we found that Repo+ glia and Elav+ neurons are first observed at about the same time, as early as 75–78 hr AEL (Figures 5F and 5K). In some samples at this early time point, only a single Dpn+ NGB or clones with a NGB surrounded by cells not marked with Repo, Elav, or Dpn are observed (Figures 5D, 5E, and 5K). These unlabeled cells are always close to the NB and therefore may be ganglion mother cells (GMCs) or another type of intermediate cell type. In addition, due to the limitations of labeling recently born cells, they may be immature glia or neurons. As development proceeds, the cells close to the NGB are either unlabeled or Elav+ neurons, suggesting that starting at mid L3 (~96 hr AEL) the Lin A stem cell produces only neurons (Figures 5G–5K). Although the number of Repo+ glia increases over time, they are far from the NGB, consistent with the idea (and supported by the pH3 staining shown in Figure 2D) that early born glia continue to divide. We also note that as development proceeds, the Repo+ glia appear to migrate, such that by late L3 they surround the immature adult neuropil. These results suggest that the Lin A stem cell initially produces mitoticially active glioblasts and a small number of motor neurons, while at later stages it transitions to a NB that only produces neurons.
To complement these lineage tracing experiments, we used the QMARCM/MARCM twin spot system (Potter et al., 2010; see STAR Methods for details) in which both daughter lineages from a single cell division are permanently labeled with RFP and GFP, respectively. In these experiments, we induced clones during L1 to capture the first post-embryonic division and labeled motor neurons with GFP, allowing us to recognize Lin A due to the stereotyped morphology of Lin A dendrites. Sister progeny were marked with tub>RFP to label all cell types. Remarkably, in twin clones where GFP labels Lin A motor neurons, the RFP+ progeny are composed of Repo+ NG and a single motor neuron that targets a body wall muscle at the base of the coxa (Figures 5L–5M), the same motor neuron that was observed in our cis2 MARCM experiments (Figure 4D). This motor neuron has also been described previously to be part of Lin A (Brierley et al., 2012). Similar results were obtained when Lin A QMARCM/MARCM clones were generated in L2 or early L3, consistent with the timing inferred from our lineage tracing experiments (data not shown).
Taken together, these results suggest that the first post-embryonic division of the Lin A NGB results in two sub-lineages (Figure 5N): one, which we refer to as Lin A.1, generates a glioblast and a single post-mitotic motor neuron, and a second (Lin A.2) generates at least one additional glioblast and the remaining 28 Lin A motor neurons. To our knowledge, this mode of division, in which a NGB generates an intermediate mother cell (IMC) that divides to produce a post-mitotic motor neuron and a glioblast, has not been observed previously (Figure 5N).
Lin A.1 and A.2 Generate Both Astrocytes and EG
To determine which types of NG are produced by Lin A and Lin D, we repeated the cis2-MARCM experiments with alrm-Gal4 or R56FO3-Gal4 (instead of repo-Gal4), to label astrocytes and EG, respectively (Table S2). We found that Lin A.1 (n = 33) produces both types of NG, while Lin D (n = 4) only generates astrocytes (Figures 6A–6I; Movie S5; Table S2). Lin A.2 also produces both types of NG (Figures 6A–6I).
Figure 6. Lin A.1 and Lin A.2 Produce Astrocytes and EG, While Lin D Produces Only Astrocytes.
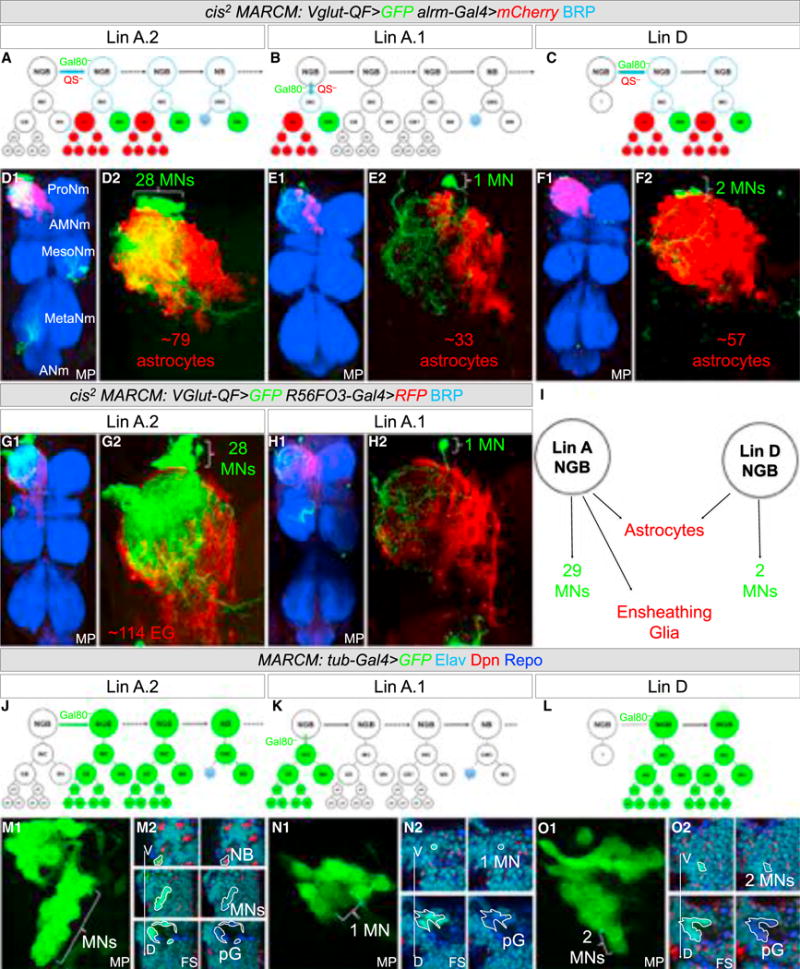
(A–C) Schematic representation of the cis2 MARCM experiments labeling Lin A.2 (A), Lin A.1 (B), and Lin D (C).
(D–H) Lin A.2 (D and G), Lin A.1 (E and H), and Lin D (F) cis2 MARCM clones in a ProNm stained with anti-BRP (blue), with motor neurons expressing mCD8::GFP, astrocytes expressing mCherry, and EG-expressing mCD8::RFP, under the control of VGlut-QF, and alrm-Gal4 or R56F03-Gal4, respectively. Number of VNCs analyzed: Lin A astrocyte: n = 10, Lin A EG: n = 4, Lin D astrocyte: n = 4, Lin Z astrocytes: n = 13. See also Figure S4 and Movie S5.
(I) Summary of progeny produced by Lin A and Lin D.
(J–L) Schematic representation of tub-Gal4 MARCM experiments in (M)–(O) labeling Lin A.2 (J), Lin A.1 (K), and Lin D (L).
(M–O) Lin A.2 (M), Lin A.1 (N), Lin D (O) MARCM clones labeled with mCD8::GFP and nGFP under the control of tub-Gal4 in a thoracic segment of an L3 VNC and immunostained with anti-Dpn (red), anti-Elav (cyan), and anti-Repo (blue). (M2, N2, O2) are successive frontal cross-sections (FS) from ventral (V) to dorsal (D). pG, proliferating glia (Repo+).
To determine whether Lin A and Lin D generate only motor neurons and NG, we repeated the MARCM experiments using a tubulin driver (tub-Gal4), to label all cell types. We stained the resulting L3 larvae with Elav, Dpn, and Repo (Figures 6J–6O). The results confirmed that Lin A.2 generates motor neurons and NG. Lin A.2 clones also usually included two cells that did not stain for either Elav or Repo. Because of their proximity to the NB, they are likely to be GMCs (Figures 6J and 6M; data not shown). All non-Lin A.2 clones (n = 4) that generated Repo+ NG labeled only 1 (Lin A.1) or 2 (Lin D) Elav+ neurons (Figure 6K). Together with our earlier experiments, these results suggest that, post-embryogenesis, Lin A and Lin D give rise to only leg motor neurons and NG. Notably, at late L3, a Dpn+ NB remains associated with Lin A, but not with Lin D.
We conclude that in each thoracic hemisegment, all of the adult NG are generated from only 2 NGBs, which also give rise only to leg motor neurons.
Unlike Motor Neurons, Individual NG Born from the Same Lineage Are Not Stereotyped
Previous studies have revealed that each of the 47 leg motor neurons have a specific birth date and morphology, characterized by their axonal targeting, dendritic arbor, and cell body position (Baek and Mann, 2009; Brierley et al., 2012). We recently characterized Lin B, a lineage that produces only 7 motor neurons, and described a post-mitotic code of mTFs that controls individual motor neuron morphologies (Enriquez et al., 2015). We identified these TFs by an expression screen using ~250 antibodies, yet, surprisingly, none of these TFs are differentially expressed in subsets of thoracic NG (data not shown). Instead, we found TFs, such as Dll, expressed in all NG. The one exception is Prospero (Pros), which is expressed in all astrocytes but not EG (Griffiths and Hidalgo, 2004; Peco et al., 2016) (Figure S1). If no single-cell TF code exists for NG, it follows that the number and morphology of the NG progeny from the same lineage may differ from animal to animal. Below we used the cis2-MARCM technique to test this prediction (Figures 7A–7R; Table S3).
Figure 7. Gliogenesis Is Not Stereotyped and Depends on Inter-lineage Competition.
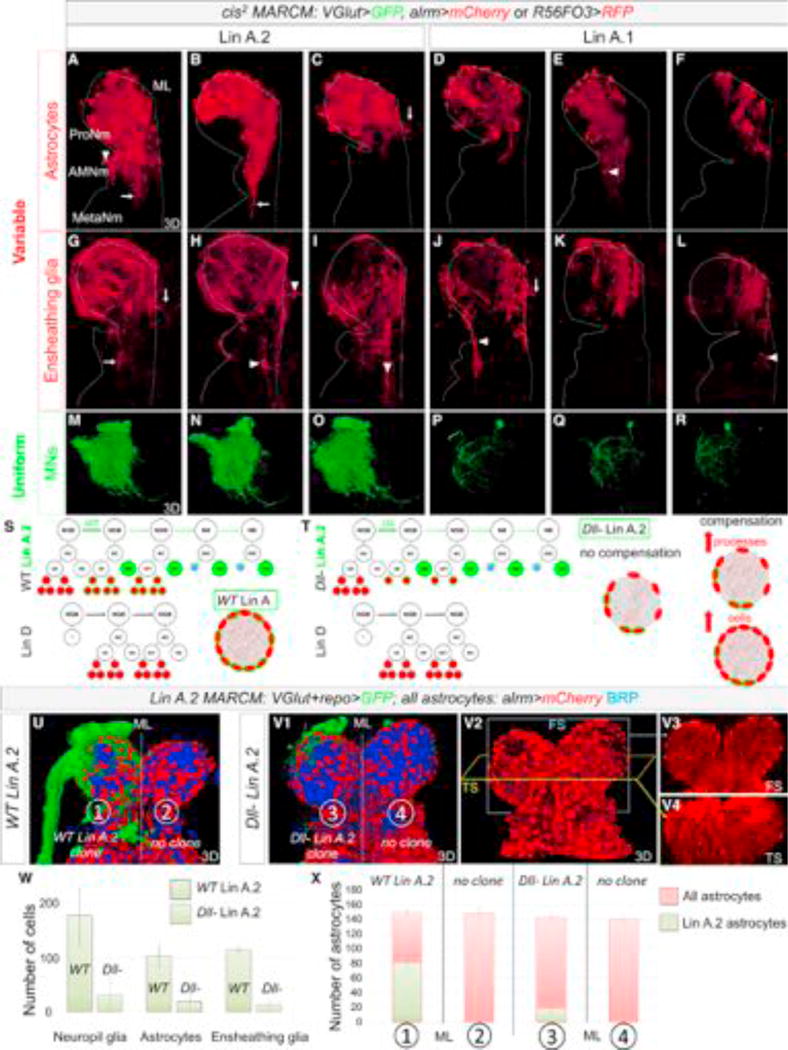
(A–R) Three examples each of Lin A.2 (A–C, G–I, M–O) and Lin A.1 (D–F, J–L, P–R) cis2 MARCM clones expressing alrm > mCherry (A–F) or R56F03 > mCD8:RFP (G–L) and VGlut > mCD8::GFP (M–R). Astrocyte and EG clone morphology is variable from animal to animal, while motor neuron clones have a uniform, lineage-specific morphology. (A–G) Arrows point to NG processes and arrowheads point to NG cell bodies that extend into or are located in a different neuromere than the ProNm in which the clone originated.
(S) Schematic summarizing the Lin A and Lin D NGBs and their progeny. Green indicates a Lin A.2 MARCM clone induced in the NGB; red indicates all proliferating glia. Bottom right: schematic of a thoracic neuromere, showing all (red) or just Lin A.2 (green outline) astrocytes with their processes entering the neuropil.
(T) Same as (S) showing three possible outcomes for a Dll− Lin A.2 MARCM clone: no compensation, an increase in astrocyte processes, or an increase in astrocyte number.
(U and V) WT (U) and Dll− (V) Lin A.2 MARCM clones in the right ProNm, labeled with mCD8::GFP (green) under the control of VGlut-Gal4 and repo-Gal4, and co-stained with anti-BRP (blue). All astrocytes are labeled with mCherry under the control alrm-QF. Frontal (FS) (V3) and transverse (TS) (V4) sections showing that both thoracic neuropils appear to be equally invaded by astrocyte processes even if the neuropil contains a Dll− Lin A.2 clone.
(W) Average number of repo > H2B:RFP+ NG, alrm > H2B:RFP+ astrocytes and R56F03 > H2B:RFP+ EG in WT and Dll− cis2 MARCM Lin A.2 clones. Error bars indicate SD. See also Figure S6. Number of samples analyzed = 4
(X) Average number of astrocytes in WT and Dll− MARCM Lin A.2 clones (GFP+, mCherry+), compared to the total number of astrocytes (GFP−, mCherry+). Error bars represent SD. Number of samples analyzed for each type of neuromere = 4.
Lin A.2, D and A.1 give rise on average to 79 (n = 10), 57 (n = 4), and 33 (n = 13) astrocytes, respectively, but with a very high SD, suggesting that each lineage has the potential to generate a variable number of NG (Figure S5). We also find that the final position of NG progeny can vary from animal to animal: for each of the three lineages born in a T1 hemisegment, astrocyte processes mostly invade the ProNp (Prothoracic Neuropil) but, depending on the sample, can also send processes to the Accessory Mesothoracic Neuropil (AMesoNp, where wing motor neuron dendrites innervate), the mesothoracic neuropil (MesoNp), and occasionally cross the mid-line to populate the contralateral neuropil (Figures 7A–7L). Similarly, although most T1-born astrocyte cell bodies remain in the ProNm (Prothoracic Neuromere), they can also end up in the Accessory Mesothoracic Neuromere (AMesoNm) and occasionally in the Mesothoracic Neuromere (MesoNm) (Figures 7A–7L, data not shown for Lin D, Table S3). Lin A astrocytes born in a T2 and T3 hemisegment have the same ability to populate neighboring neuromeres (Figure S5). Thus, unlike the highly stereotyped morphology of motor neurons born from the same lineage (Figures 7M–7R), the number, final cell body position, and neuropil regions invaded by astrocyte processes born from individual NGBs are variable from animal to animal (Figures 7 and S5; Table S3).
Although we were unable to count the number of EG produced by Lin A.1 (see STAR Methods), Lin A.2 generates ~114 EG (n = 4) with a low SD (±6), suggesting that the number of EG produced by Lin A is similar in different animals. However, as with astrocytes, the neuropil region surrounded and invaded by Lin A EG is variable from animal to animal, and these glia can populate neighboring neuromeres and cross the midline (Figures 7 and S5; Table S3).
In summary, an individual NGB produces a variable number of NG with different morphologies and final locations, but the same number of motor neurons, each with a stereotyped morphology.
Final Astrocyte Number Depends on Inter-lineage Competition
Although the number of NG produced by a single NGB (e.g., Lin A) can be highly variable, the total number of NG produced by all lineages is very constant from animal to animal (Figures 1 and S5). In addition, the non-stereotyped morphology and final positions of NG derived from single NGBs led us to hypothesize that NG may compete during development to ensure that each neuropil is fully wrapped and innervated. We tested this hypothesis for astrocytes, by generating Dll mutant Lin A.2 MARCM clones, which results in a severely compromised number of astrocytes born from this lineage without affecting motor neuron number or morphology (Figures 7W and S6). Dll− Lin A.2 clones were generated in a background where all astrocytes were labeled, thus allowing us to assess whether other lineages could compensate for the fewer number of Lin A.2-derived astrocytes. We considered three possible outcomes (Figures 7S and 7T): (1) no compensation, in which WT (non-Lin A.2) astrocytes do not respond to the reduced number of astrocytes derived from Lin A; (2) compensation where WT astrocytes respond to the lower number of Lin A.2 astrocytes by increasing the number or extent of processes invading the neuropil; and (3) compensation where WT lineages generate more astrocytes to maintain the total final number.
To distinguish between these possibilities, we generated WT or Dll mutant Lin A MARCM clones expressing mCD8::GFP under the control of repo-Gal4 and VGlut-Gal4. In this genetic background, we labeled all astrocytes with a cytoplasmic QUAS-mCherry under the control of alrm-QF. As a result, astrocytes produced by Lin A are labeled by mCherry and mCD8::GFP while astrocytes produced by other lineages express only mCherry. Astrocytes were counted in a ProNm containing a single Lin A MARCM clone and in the contralateral neuromere that had no clones, which served as an internal WT control. Although Dll mutant Lin A clones (n = 4) produced on average ~18 astrocytes (compared to ~80 for WT Lin A clones), the total number of astrocytes in these neuromeres was normal (~140) (Figures 7U–7X). Altogether, these results suggest that Lin D and Lin A.1 compensate for the reduced number of astrocytes produced by Lin A.2.
To determine when during development NG compensation is likely to occur, we generated Dll− Lin A.2 MARCM clones (n = 4) and counted the number of Dll− Repo+ cells in late L3 larva. We found that the number was similar to the number in WT clones (~20) (Figure S6), arguing that the absence of Dll does not affect the generation of astrocytes until the pupal stage, when NG number continue to increase (Figure 3L). It follows that compensation from WT lineages also occurs during this pupal amplification phase, long after the glioblasts are born.
These results suggest that the production of NG is extremely plastic during development, and that their progenitors continue to divide until the entire neuropil is occupied.
DISCUSSION
The Logic of Lineages Producing Multiple, Functionally Related Cell Types
Here, we show that two NGBs per thoracic larval hemisegment give rise to leg motor neurons, astrocytes, and EG. All three of these cell types are key components of each thoracic neuromere: motor neurons extend axons to innervate leg muscles and elaborate complex dendritic arbors inside the neuropil, EG wrap the developing neuropil and incoming sensory axons, and astrocytes send processes inside the neuropil where they associate with synapses between motor neuron dendrites, interneurons, and sensory neurons. Together, these cells develop in a coordinated manner to form a complex functional unit that comprises the neural circuitry used for many adult behaviors such as walking.
Glial cells are a major component of nervous systems, critical not only for the activity of neurons, but also for their development (Freeman, 2015; Freeman and Rowitch, 2013). Neurons and glia are always associated with each other and exist in all bilateria, even in primitive phyla such as flatworms (Hartline, 2011). As nervous system complexity increases, there is an increase in both neuronal diversity and in the diversity of glia types and morphologies (Paredes et al., 2016). Given this close relationship, it is striking that in the thorax of the adult fly all NG are derived from lineages that also—and only—give rise to leg motor neurons. In the absence of extensive cell migration, being born from the same stem cell results in an anatomical proximity that may be important for subsequent steps in development. In particular, we propose that the shared lineages of the leg motor neurons, astrocytes, and EG facilitates the assembly of anatomically complex neuropils and neural circuits. This idea helps explain why lineage relationships appear to play less of a role in simpler nervous systems such as in C. elegans. Consistent with the idea that anatomical proximity of functionally related cell types is important for building complex nervous systems, in mammals, astrocytes and oligodendrocytes are born from the same progenitor domains as motor neurons and interneurons (Ravanelli and Appel, 2015; Rowitch and Kriegstein, 2010). Although it is currently unclear whether these cell types are derived from the same lineages in vertebrates, our results provide a striking precedent and compelling reasons that this may be the case. Alternatively, vertebrate nervous systems may have solved the neuropil assembly problem differently, by having distinct progenitors born in the same domain but at different times that give rise to either neurons or glia.
Divergent Mechanisms for Generating Motor Neuron and Glia Stereotypy
One of the most striking conclusions stemming from our findings is how cell lineages can use very different mechanisms to produce stereotyped outcomes. On the one hand, each motor neuron is morphologically distinct and born from a specific lineage and with a specific birth order, properties that are a consequence of the unique TF code that they express. At the other extreme, based on the hundreds of TFs we have surveyed, individual astrocytes or EG appear to share the same TF code as all other astrocytes or EG, respectively, and can end up with different morphologies in multiple positions within or even in neighboring neuropils. Yet, despite this plasticity, the end result—~280 NG evenly distributed throughout each thoracic leg neuropil—is highly stereotyped. Nevertheless, these two very different modes of achieving stereotypy—hardwired (for motor neurons) and plastic (for NG)—arise from the same stem cells. We also found that, although the total number of NG in each thoracic neuropil is stereotyped (~140 astrocytes and ~140 EG), the individual contributions from the two NGB lineages can vary from animal to animal. Moreover, when one lineage is compromised the others can compensate to maintain the correct number of total NG. Although the mechanism regulating final NG number is currently unknown, we suggest that it is analogous to what has been referred to as neutral competition in mammalian and Drosophila gut homeostasis (de Navascués et al., 2012; Snippert et al., 2010). It is possible that this competition occurs via direct communication between NG. This idea is supported by the observation that individual astrocytes and EG respect each other’s territory and thus exhibit tiling-like behavior. Alternatively, communication between motor neuron dendrites (or other components of the thoracic neuropil) and NG may be responsible for NG stereotypy. For example, astrocytes and EG may be able to sense and adapt to the scaffold generated by motor neuron dendrites in order to fully invade and wrap each thoracic neuropil. Accordingly, NG stereotypy, both cell number and morphology, would be an indirect consequence of the mTF codes that generate the unique morphologies of motor neurons and likely other neurons.
What might be the reason for the existence of such different strategies to achieve stereotypy? One answer may be developmental robustness, which is especially challenging for establishing neural circuits. While motor neurons need to make precise connections (both in the CNS and in the legs), and therefore may require precise TF codes to achieve this precision, our results suggest that astrocytes and EG may have a more generic role in neuropil development. Nevertheless, NG are likely to be just as critical for neuropil development and function. However, instead of specifying each glia on a single-cell basis, the system has evolved a different strategy to achieve robustness: a generic TF code, but the ability to communicate with each other to ensure that the entire neuropil is appropriately and evenly populated by the correct number of NG. There are several potential advantages to such a strategy. One is that as the number of motor neurons or size and complexity of nervous systems vary during evolution, the number of NG can readily adapt in response. Second, NG plasticity may be crucial during normal development to adjust to natural variations in neuronal morphology. For example, in the antennal lobe of Drosophila the morphology and connectivity of local interneurons can vary between animals and may require NG to respond accordingly (Chou et al., 2010). Finally, NG may also play an important role in maintaining axons and in monitoring and pruning synapses to ensure the proper number and type of synaptic connections, as they do in invertebrates (O’Connor et al., 2017) and vertebrates (Risher et al., 2014), respectively, during normal development and after injury (Burda et al., 2016; He and Jin, 2016; Stephan et al., 2012). Glial developmental plasticity may be essential to achieve robustness and maintain homeostasis by readily adapting to these and perhaps other variations in nervous system size and morphology.
STAR*METHODS
Detailed methods are provided in the online version of this paper and include the following:
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| mouse anti BRP | DSHB | RRID:AB_528108 |
| mouse anti-repo | DSHB | RRID:AB_528448 |
| rat anti-Elav | DSHB | RRID:AB_528218 |
| mouse anti-Nrg | DSHB | RRID:AB_10804674 |
| mouse anti-Pros | DSHB | RRID:AB_528440 |
| rabbit anti-PH3 | Abcam | RRID:AB_2164915 |
| rat anti-NCad | DSHB | RRID:AB_528121 |
| guinea-pig anti-Dll | Estella and Mann, 2008 | N/A |
| guinea-pig anti-Dpn (gift from Jim skeath) | Gift from Jim Skeath | N/A |
| Goat anti-mouse Alexa 647 | Invitrogen | cat# A-32728 |
| Goat anti-guinea pig Alexa 488 | Invitrogen | cat# A-11073 |
| Goat anti-rat Alexa 555 | Abcam | cat# ab150199 |
| Goat anti-rabbit Alexa 555 | Invitrogen | cat# A-32732 |
| Goat anti-rat Alexa 405 | Abcam | cat# ab175673 |
| Goat anti-guinea pig Alexa 555 | Invitrogen | cat# A-21435 |
| Goat anti-mouse Alexa 488 | Invitrogen | cat# A-32723 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| formaldehyde | Thermo Scientific | cat# 28908 |
| PBS | Dutscher | cat# X0515-500 |
| triton | Sigma | cat# T8787-100mL |
| BSA | Sigma | cat# A7906-500 g |
| Vectashield mounting medium | Vector Laboratories | cat# H1000 |
| DAB substrate kit | ThermoFisher | cat# 34002 |
| pCR8/GW/TOPO/VGlut vector | Enriquez et al., 2015 | N/A |
| pattb-QF-hsp70 vector | Addgene | cat# 24368 |
| pattb-QF-VGlut-hsp70 vector | This work | N/A |
| Experimental Models: Organisms/Strains | ||
| Alrm-GAl4 (3rd chromosome) | BDSC | RRID:BDSC_67031 |
| UAS-mCD8::GFP (2nd chromosome) | BDSC | RRID:BDSC_60707 |
| UAS-mCD8::GFP (3rd chromosome) | BDSC | RRID:BDSC_60726 |
| UAS-nGFP | BDSC | RRID:BDSC_4776 |
| tub-QS | BDSC | RRID:BDSC_30132 |
| UAS-FLP | BDSC | BDSC_4539 |
| y, w, hs-Flp1.22, tub-Gal4, UAS-nGFP | Gary Struhl | N/A |
| act > CD2 > Gal4 | BDSC | BDSC_4779 |
| FRT42D | BDSC | RRID:BDSC_1802 |
| UAS-H2B::GFP (3rd chromosome) | Mayer et al., 2005 | N/A |
| UAS-FB1.1 (2nd chromosome) | BDSC | RRID:BDSC_35537 |
| R56F03-Gal4 (attP2 insertion) | BDSC | RRID:BDSC_39157 |
| UAS-GFP::CD8::HRP (3rd chromosome) | Alexandre et al., 2014 | N/A |
| R31F10-GAL4 (attP2 insertion) | BDSC | RRID:BDSC_49685 |
| UAS-CD4-tdGFP (3rd chromosome) | BDSC | RRID:BDSC_35839 |
| Dll-Gal4 | Calleja et al., 1996 | RRID:BDSC_3038 |
| UAS-mFlp5 (X chromosome) | Enriquez et al., 2015 | N/A |
| UAS-mFlp5 (2nd chromosome) | Enriquez et al., 2015 | N/A |
| w, hs-Flp1.22, FRT82B tub-Gal80 | Gary Struhl | N/A |
| y, w, hs-Flp1.22, tub-Gal80, FRT19A | Gary Struhl | N/A |
| y, w, hs-Flp1.22, FRT42D tub-Gal80 | Gary Struhl | N/A |
| tub-QS#0, FRT19A | BDSC | RRID:BDSC_30129 |
| Mhc-RFP (2nd chromosome) | BDSC | RRID:BDSC_38464 |
| VGlut-Gal4 (also called OK371) (3rd chromosome) | BDSC | RRID:BDSC_26160 |
| repo-gal4 (3rd chromosome) | BDSC | RRID:BDSC_7415 |
| R22C10-GAL4 (attP2 insertion) | BDSC | RRID:BDSC_47841 |
| UAS-mCD8::RFP (2nd chromosome) | BDSC | RRID:BDSC_27398 |
| UAS-mCD8::RFP (3rd chromosome) | BDSC | RRID:BDSC_27399 |
| VGlut-QF (attp 86f) | This work | N/A |
| Dll-(Dll[SA1]) | Cohen and Jürgens, 1989 | N/A |
| tub-Gal4, QUAS-mCD8::GFP/TM6b | BDSC | not available anymore, SN:BDSC_30031 |
| QUAS-mCD8::GFP (2nd chromosome) | BDSC | RRID:BDSC_30002 |
| QUAS-mcherry(attP2 insertion) | BDSC | RRID:BDSC_52270 |
| UAS-mcherry(attP2 insertion) | BDSC | RRID:BDSC_52268 |
| alrm-QF (3rd chromosome) | BDSC | RRID:BDSC_66464 |
| elav-Gal80 | Rideout et al., 2010 | N/A |
| cha-Gal80 | Kitamoto, 2002 | N/A |
| dpn > KDRT > Cre//; act > LoxP > LexA, LexA-myr::GFP//; UAS-Kd// | Awasaki et al., 2014 | N/A |
| Recombinant DNA | ||
| pCR8/GW/TOPO/VGlut vector | Enriquez et al., 2015 | N/A |
| pattb-QF-hsp70 vector | Addgene | cat# 24368 |
| pattb-QF-VGlut-hsp70 vector | This work | N/A |
| Software and Algorithms | ||
| Amira 3D software | version 6.2 | https://www.fei.com/ |
| ImageJ | version 1.48 | https://imagej.nih.gov/ij/ |
| Microsoft Excel | version 2016 | https://products.office.com/en-US/ |
| Microsoft Power Point | version 2016 | https://products.office.com/en-US/ |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Richard Mann (rsm10@columbia.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All in vivo experiments were carried out using standard laboratory strains of D. melanogaster.
With the execpetion of R56F03-Gal4, UAS-CD4-tdGFP recombinant (Peco et al., 2016), all genotypes used to make recombinants and fly stocks are listed on the REAGENT or RESOURCE sheet.
Unless otherwise noted, fly stocks were obtained from the Bloomington Stock Center: alrm-Gal4-III, UAS-mCD8::GFP-III, UAS-mCD8::GFP-II UAS-H2B::RFP-III (Mayer et al., 2005), UAS-FB1.1-II, R56F03-Gal4; UAS-GFP::CD8::HRP (Alexandre et al., 2014); R56F03-Gal4, UAS-CD4-tdGFP recombinant (Peco et al., 2016), UAS-mFlp5 (Enriquez et al., 2015); dll-Gal4, R31F10-Gal4, w hs-Flp1.22 FRT82B tub-Gal80 and FRT42D tub-Gal80 and y, w, hs-Flp1.22, tub-Gal80, FRT19A (Gary Struhl), tub-QS#0, FRT19A, Mhc-RFP, VGlut-Gal4 (also called OK371-Gal4), tub-QS, repo-Gal4, 10C12-Gal4, UAS-mCD8::RFP, VGlut-QF (this work), Dll-(Dll[SA1]) (Cohen and Jürgens, 1989), tub-Gal4, QUAS-mCD8::GFP/TM6b, QUAS-mCD8::GFP-II, QUAS-mcherry-III, UAS-mcherry-III, alrm-QF-III, elav-Gal80-I, cha-Gal80 (Kitamoto, 2002), dpn > KDRT > Cre//; act > LoxP > LexA, LexA-myr::GFP//; UAS-Kd//(Awasaki et al., 2014).
METHOD DETAILS
Immunostaining of L3 Larva
Inverted L3 larvae were fixed in 4% formaldehyde with PBS for 20 minutes. L3 larval CNS were dissected in PBS triton and incubated with primary antibodies for two days and secondary antibodies for one day at 4°C. Fresh PBT (PBS with 0.1% Triton X-100 0,3%, 1% BSA) was used for the blocking, incubation, and washing steps: five times for 20 minutes at room temperature after fixation and after primary/secondary antibodies. L3 CNS mounted onto glass slides using Vectashield mounting medium (Vector Labs).
Immunostaining of adult VNC
After removing the abdominal and head segments, the thoraces of the flies were opened and fixed in 4% formaldehyde with PBS for 20 minutes at room temperature and blocked in the blocking buffer for one hour. After dissection, adult VNC were incubated with primary antibodies for two days and secondary antibodies for one day at 4°C. Fresh PBT (PBS with 0.1% Triton X-100, 1% BSA) was used for the blocking, incubation, and washing steps: five times for 20 minutes at room temperature after fixation and after primary/secondary antibodies. VNCs were dissected and mounted onto glass slides using Vectashield mounting medium (Vector Labs).
Primary and secondary antibodies
Unless otherwise noted primary antibodies were obtained from Hybridoma Bank: mouse anti-BRP, mouse anti-repo, rat anti-Elav, mouse anti-Nrg (neuronal specific form), mouse anti-Pros, rat anti-NCad, guinea-pig anti-Dll (Estella and Mann, 2008), guinea-pig anti-Dpn (gift from Jim Skeath). Secondary antibodies: Alexa 555, 647 and Alexa 405 (Invitrogen and Abcam).
Time course of NG development
We expressed a membrane reporter (UASmCD8::GFP) with Dll-Gal4 and dissected larva and pupae at different development time points (Figure 3; Movie S3). In order to specifically visualize the immature leg neuropil, larval CNS were co-stained with anti-BRP (which labels mature neuropils) and anti-N-Cadherin (NCad, which labels mature and immature neuropils at high and low levels, respectively [Kurusu et al., 2012) (Figures 3A–3E). Dll-Gal4 is also expressed in adult leg sensory neurons, which allowed us to visualize where sensory axons first enter the CNS and was used as a reference point in these developing CNS. We also stained larval CNS for Dll and BRP or Repo and Dll in order to quantify the number of glia at each development stage (Figures 3F–3J and data not shown). Finally, we used alrm-Gal4 and R56FO3-Gal4 to drive the expression of mCD8::GFP to determine when EG and astrocytes morphologically differentiate.
EM protocol
After removal of abdominal and head segments, the thorax of the flies was opened and fixed in 4% glutaraldehyde, washed 5 times quickly in PBS, incubated in PBS with 0.03% Triton X-100 for 30 mins and washed 5 times over 20 mins in PBS. To specifically recognize the EG in EM we expressed UAS-GFP::mCD8::HRP (HRP: horseradish peroxidase) and DAB (3,3′-Diaminobenzidine) was added to catalyze the HRP to form an electron dense product. HRP expression was revealed overnight using the DAB substrate kit (Thermofisher).
In an EM microwave (Pelco model 3451 system with coldspot), specimens were post-fixed with 1% osmium tetroxide in 0.1M phosphate buffer (PB) for 2 × 40 s (with each 40 s exposure in fresh osmium), and then washed 3 × 10 minutes in 0.1M PB. Specimens were dehydrated in ethanol grades of 50%, 70%, 95% (1 × 40 s for each grade), and 100% (2 × 40 s). Dehydrated tissue was then infiltrated with epoxy resin (Electron Microscopy Sciences Embed 812) and 100% ethanol (1:1 mixture) for 15 minutes, and then in 100% epoxy resin (2 × 15 minutes with fresh resin each time).
Specimens were mounted between 2 plastic slides with 100% epoxy resin and polymerized overnight at 60°C. The next day the polymerized resin wafers were separated from the plastic slides and placed flat on a glass slide to choose specimens for thin sectioning. An entire specimen sample was then cut out of the wafer using a razor blade and carefully placed on its side in a Dykstra flat embedding mold with the protoracic neuromere (PN) end (region of interest) closest to the beveled tip in one of the cavities of the mold. Small slits were made at the tip and floor of the silicon mold to secure the perpendicularly oriented tissue. This orientation is used so that thin sections will be cut transversely through the specimen starting at the PN. The mold was then filled with 100% epoxy resin and polymerized for 18–24 hours at 60°C.
After polymerization, the block was removed from the cavity and trimmed down at the beveled tip in the shape of a 2mm square with the specimen near the center of the blockface. With an ultramicotome, 10 micron thick sections were cut and collected serially using a diamond Histo-knife (Diatome) and then mounted and coverslipped on a glass slide with immersion oil. The 10 micron sections were then observed with a light microscope and sections with the best areas of interest are photographed. The coverslip was carefully removed and the sections of interest are washed in 95% ethanol to remove any oil by passing them through the ethanol in 3 wells for 15 s in each well. Each 10 micron section was then carefully placed on a piece of lens tissue to dry and then remounted on a tiny drop of epoxy resin on a blank polymerized Beem capsule resin block (blockface should be flat and smooth) and polymerized overnight at 60°C; a spring tension apparatus was used to keep the 10 micron section flat against a plastic slide. The polymerized blockface with the 10 micron section was trimmed down to size and 60 nanometer thin sections were cut with an ultramicrotome using a Diatome diamond knife. The ultrathin sections were collected onto formvar coated slot grids and then stained with uranyl acetate and lead citrate, and examined on a JEOL 1200EX electron microscope.
Fly genetics
Crosses or stocks used to visualize NG (Figure 1, 2, 3, S1, S3):
UAS-mCD8::GFP-II, alrm-Gal4-III
alrm-Gal4, UAS-FB1.1-III crossed with UAS-mFlp5-II
alrm-Gal4, UAS-H2B::RFP-III
UAS-mCD8::GFP-II, R56F03-Gal4-III
R56F03, UAS-FB1.1-III crossed with UAS-mFlp5-II
R56F03, UAS-H2B::RFP-III
R56F03-Gal4-III crossed with UAS-GFP::CD8::HRP
R56F03-Gal4, UAS-nGFP-III
R56F03-Gal4, UAS-nGFP-III crossed with alrm-Gal4
Dll-Gal4, UAS-mCD8::GFP-II
Lineage tracing systems (Figures 2 and S2):
Elav-Gal80; Dll-Gal4, UAS-Flp1/cyo; cha-Gal80/MKRS crossed with act > cd2 > Gal4, UAS-nGFP-III. Note: since Dll-Gal4 is expressed in sensory neurons we used cha-Gal80 and elav-Gal80 to inhibit Gal4 in these neurons.
Elav-Gal80; Dll-Gal4, UAS-Flp1/Cyo; cha-gal80/MKRS crossed with UAS-mFlp5; UAS-FB1.1//; act > CD2 > Gal4 R31F10-Gal4 crossed with UAS-mFlp5-X, UAS-Flybow-I.I
MARCM (Figures 4, 6, 7, and S6):
Y, w, hs-Flp1.22; FRT42 tub-gal80//; repo-Gal4, UAS-mCD8::GFP/MKRS crossed with
y, w, hs-Flp1.22; VGlut-Gal4, UAS-mCD8::GFP, Mhc-RFP FRT42D//; repo-Gal4, UAS-mCD8::GFP/Tm6b or
y, w, hs-Flp1.22; VGlut-Gal4, UAS-mCD8::GFP, Mhc-RFP FRT42D Dll-//; repo-Gal4, UAS-mCD8::GFP/Tm6b
y, w, hs-Flp1.22, tub-Gal4; sp/cyo; FRT82B tub-Gal80 crossed with
y, w, hs-Flp1.22, UAS-mCD8::GFP//; FRT82B//
y, w, hs-Flp1.22; FRT42D tub-Gal80//; alrm-QF, QUAS-mcherry/MKRS crossed with
y, w, hs-Flp1.22; VGlut-Gal4, UAS-mCD8::GFP, Mhc-RFP FRT42D Dll-//; repo-Gal4, UAS-mCD8::GFP/Tm6b or
y, w, hs-Flp1.22; VGlut-Gal4, UAS-mCD8::GFP, Mhc-RFP FRT42D//; repo-Gal4, UAS-mCD8::GFP/Tm6b
y, w, hs-Flp1.22; FRT42 tub-Gal80, tub-QS/cyo; repo-Gal4, UAS-mCD8::RFP/TM6b or
y, w, hs-Flp1.22; FRT42 tub-Gal80, tub-QS/cyo; R56F03-Gal4, UAS-mCD8::RFP/TM6b or
y, w, hs-Flp1.22; FRT42 tub-Gal80, tub-QS/cyo; alrm-Gal4, UAS-mCherry/TM6b or
y, w, hs-Flp1.22; FRT42 tub-Gal80, tub-QS/cyo; repo-Gal4, UAS-H2B::RFP, UAS-mCD8::GFP/TM6b or
y, w, hs-Flp1.22; FRT42 tub-Gal80, tub-QS/cyo; R56F03-Gal4, UAS-H2B::RFP/tm6b or
y, w, hs-Flp1.22; FRT42 tub-Gal80, tub-QS/cyo; alrm-Gal4, UAS-H2B::RFP/tm6b crossed with
y, w, hs-Flp1.22; QUAS-mCD8:GFP FRT42D//; VGlut-QF/MKRS or
y, w, hs-Flp1.22; QUAS-mCD8:GFP FRT42D Dll-//; VGlut-QF/MKRS
Note: cis2 MARCM and MARCM clones where generated by heat shocks of first instar larva.
QMARCM/MARM (Figure 5):
y,w, hs-Flp1.22, tub-Gal80, FRT19A; QUAS-mCD8::GFP, UAS-mCD8::RFP/Cyo; tub-Gal4, QUAS-mCD8::GFP/TM6b crossed with
tub-QS#0, FRT19A; QUAS-mCD8::GFP, UAS-mCD8::RFP/Cyo; DVGlut-QF::hsp70term (attp86F)/MKRS
Note: MARCM/MARCM clones where generated by heat shocks from late embryogenesis to early third instar larva.
Lin A tracing system (Figure 5; Lacin and Truman, 2016)
10C12-Gal4 crossed with dpn KDRT > Stop > KDRT Cre//; act LoxP > Stop > LoxP-LexA, LexA-myr::GFP//; UAS-Kd//
Cloning
pattb-VGlut-QF-hsp70. A SpeI-VGlut-Spe1 fragment containing the 5.9 kb sequence upstream of the VGlut translation start site was taken from a pCR8/GW/TOPO (Enriquez et al., 2015) vector and clone in a pattb-QF-hsp70 vector (addgene). The resulting pattb-VGlut-QF-hsp70 construct was inserted in position 86f on chromosome III by injection into embryos carrying attP-86f landing site.
Leg imaging
See also Figure 4. Legs were dissected and fixed overnight at 4°C, washed five times for 20 minutes at room temperature in PBS withTriton X-100 (0.3%) and mounted onto glass slides using Vectashield mounting medium (Vector Labs). Due to their large size, final leg images may be a composite of more than one image.
Microscopy and 2D Imaging
Multiple 1-μm-thick sections in the z axis (dorsoventral for L3 CNS or adult VNC and mediolateral for adult legs) were imaged with a Leica TCS SP5 II or a Zeiss LSM 780 confocal microscope. Binary images for z stack images were generated using NIH ImageJ and Photoshop (Adobe Systems).
3D Leg analysis
See also Figure 4. Z stacks were generated using NIH ImageJ. Specific cuticle background was generated using the Argon laser 488 and by placing the confocal detector out of the range of the GFP emission wavelength. This cuticle background of the leg was used to remove the nonspecific signal from the GFP channel using ImageJ. Z stacks were used to generate 3D reconstructions of legs with Amira 3D software. The color codes used were Volrengreen to visualize mCD8::GFP, color used for cuticles was gray.
3D images of adult VNC and larval CNS
Ventral to dorsal Z stacks were generated using NIH ImageJ. and were used to generate 3D reconstructions using the Volren or volume rendering in Amira 3D software. The color codes used were constant color in correlation with the fluorescent proteins or the secondary Alexa used.
QUANTIFICATION AND STATISTICAL ANALYSIS
All bar plots were generated with Microsoft Excel. All bar errors represent standard deviation of a minimum of 4 samples. The total number of samples used for each genotype is indicated in the text and/or the figures legends.
All Lin A cell plots were generated with Microsoft Excel. All bar errors represent standard deviation of a minimum of 7 samples. The total number of samples used for each genotype is indicated in the Figure 5 legend. The spatial coordinates were assigned to each cell using the cell counter plug-in of NIH ImageJ software. In Figure 5 (D2-J3) the coordinates of each cell were normalized with Microsoft Excel in order to have the positon of Lin A NB at the origin of the plot graph.
Supplementary Material
Highlights.
The development of the neuropil glia and leg motor neurons is highly coordinated
All adult thoracic neuropil glia are born from two lineages per hemisegment
These lineages also give rise only to leg motor neurons
Unlike neurons, glia generation is plastic and exhibits interlineage compensation
Acknowledgments
We thank members of the Mann lab for comments and suggestions and Jeremy Dasen, Oliver Hobert, Laura Johnston, Emilie Peco, and Alain Vincent for comments on the manuscript. This work was supported by the Ellison Medical Foundation grant AG-SS-2945-12 and NIH grant NS070644 to R.S.M. and funding from the ALS Association (#256) and FRM (#AJE20170537445) to J.E.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures, three tables, and five movies and can be found with this article online at https://doi.org/10.1016/j.neuron. 2018.01.007.
AUTHOR CONTRIBUTIONS
Conceptualization, J.E. and R.S.M.; Methodology, J.E. and R.S.M.; Investigation, J.E., L.Q.R., R.B., S.B., and P.G.; Writing – Original Draft, J.E. and R.S.M.; Writing – Review & Editing, J.E. and R.S.M.; Funding Acquisition, J.E. and R.S.M.; Resources, J.E., C.M., and R.S.M.; Supervision, J.E. and R.S.M.
DECLARATION OF INTERESTS
The authors declare no competing interests.
References
- Alexandre C, Baena-Lopez A, Vincent JP. Patterning and growth control by membrane-tethered Wingless. Nature. 2014;505:180–185. doi: 10.1038/nature12879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apidianakis Y, Rahme LG. Drosophila melanogaster as a model for human intestinal infection and pathology. Dis Model Mech. 2011;4:21–30. doi: 10.1242/dmm.003970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasaki T, Lai SL, Ito K, Lee T. Organization and postembryonic development of glial cells in the adult central brain of Drosophila. J Neurosci. 2008;28:13742–13753. doi: 10.1523/JNEUROSCI.4844-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasaki T, Kao CF, Lee YJ, Yang CP, Huang Y, Pfeiffer BD, Luan H, Jing X, Huang YF, He Y, et al. Making Drosophila lineage-restricted drivers via patterned recombination in neuroblasts. Nat Neurosci. 2014;17:631–637. doi: 10.1038/nn.3654. [DOI] [PubMed] [Google Scholar]
- Baek M, Mann RS. Lineage and birth date specify motor neuron targeting and dendritic architecture in adult Drosophila. J Neurosci. 2009;29:6904–6916. doi: 10.1523/JNEUROSCI.1585-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek M, Enriquez J, Mann RS. Dual role for Hox genes and Hox co-factors in conferring leg motoneuron survival and identity in Drosophila. Development. 2013;140:2027–2038. doi: 10.1242/dev.090902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckervordersandforth RM, Rickert C, Altenhein B, Technau GM. Subtypes of glial cells in the Drosophila embryonic ventral nerve cord as related to lineage and gene expression. Mech Dev. 2008;125:542–557. doi: 10.1016/j.mod.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Boettcher S, Manz MG. Regulation of inflammation- and infection-driven hematopoiesis. Trends Immunol. 2017;38:345–357. doi: 10.1016/j.it.2017.01.004. [DOI] [PubMed] [Google Scholar]
- Bossing T, Udolph G, Doe CQ, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. I Neuroblast lineages derived from the ventral half of the neuroectoderm. Dev Biol. 1996;179:41–64. doi: 10.1006/dbio.1996.0240. [DOI] [PubMed] [Google Scholar]
- Brierley DJ, Rathore K, VijayRaghavan K, Williams DW. Developmental origins and architecture of Drosophila leg motoneurons. J Comp Neurol. 2012;520:1629–1649. doi: 10.1002/cne.23003. [DOI] [PubMed] [Google Scholar]
- Burda JE, Bernstein AM, Sofroniew MV. Astrocyte roles in traumatic brain injury. Exp Neurol. 2016;275:305–315. doi: 10.1016/j.expneurol.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calleja M, Moreno E, Pelaz S, Morata G. Visualization of gene expression in living adult Drosophila. Science. 1996;274:252–255. doi: 10.1126/science.274.5285.252. [DOI] [PubMed] [Google Scholar]
- Chou YH, Spletter ML, Yaksi E, Leong JC, Wilson RI, Luo L. Diversity and wiring variability of olfactory local interneurons in the Drosophila antennal lobe. Nat Neurosci. 2010;13:439–449. doi: 10.1038/nn.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SM, Jürgens G. Proximal-distal pattern formation in Drosophila: Cell autonomous requirement for Distal-less gene activity in limb development. EMBO J. 1989;8:2045–2055. doi: 10.1002/j.1460-2075.1989.tb03613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court RC, Armstrong JD, Borner J, Card G, Costa M, Dickinson M, Duch C, Korff W, Mann R, Merritt D, et al. A systematic nomenclature for the Drosophila ventral nervous system. bioRxiv. 2017 doi: 10.1016/j.neuron.2020.08.005. https://doi.org/10.1101/122952. [DOI] [PMC free article] [PubMed]
- de Navascués J, Perdigoto CN, Bian Y, Schneider MH, Bardin AJ, Martínez-Arias A, Simons BD. Drosophila midgut homeostasis involves neutral competition between symmetrically dividing intestinal stem cells. EMBO J. 2012;31:2473–2485. doi: 10.1038/emboj.2012.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty J, Logan MA, Taşdemir OE, Freeman MR. Ensheathing glia function as phagocytes in the adult Drosophila brain. J Neurosci. 2009;29:4768–4781. doi: 10.1523/JNEUROSCI.5951-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enriquez J, Venkatasubramanian L, Baek M, Peterson M, Aghayeva U, Mann RS. Specification of individual adult motor neuron morphologies by combinatorial transcription factor codes. Neuron. 2015;86:955–970. doi: 10.1016/j.neuron.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estella C, Mann RS. Logic of Wg and Dpp induction of distal and medial fates in the Drosophila leg. Development. 2008;135:627–636. doi: 10.1242/dev.014670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman MR. Drosophila central nervous system glia. Cold Spring Harb Perspect Biol Published online February. 2015;26:2015. doi: 10.1101/cshperspect.a020552. https://doi.org/10.1101/cshperspect.a020552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman MR, Rowitch DH. Evolving concepts of gliogenesis: A look way back and ahead to the next 25 years. Neuron. 2013;80:613–623. doi: 10.1016/j.neuron.2013.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths RL, Hidalgo A. Prospero maintains the mitotic potential of glial precursors enabling them to respond to neurons. EMBO J. 2004;23:2440–2450. doi: 10.1038/sj.emboj.7600258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjieconomou D, Rotkopf S, Alexandre C, Bell DM, Dickson BJ, Salecker I. Flybow: Genetic multicolor cell labeling for neural circuit analysis in Drosophila melanogaster. Nat Methods. 2011;8:260–266. doi: 10.1038/nmeth.1567. [DOI] [PubMed] [Google Scholar]
- Harris RM, Pfeiffer BD, Rubin GM, Truman JW. Neuron hemilineages provide the functional ground plan for the Drosophila ventral nervous system. eLife. 2015;4 doi: 10.7554/eLife.04493. Published online July 20, 2015. https://doi.org/10.7554/eLife.04493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstein V, Nassif C, Lekven A. Embryonic development of the Drosophila brain. II. Pattern of glial cells. J Comp Neurol. 1998;402:32–47. [PubMed] [Google Scholar]
- Hartline DK. The evolutionary origins of glia. Glia. 2011;59:1215–1236. doi: 10.1002/glia.21149. [DOI] [PubMed] [Google Scholar]
- He Z, Jin Y. Intrinsic control of axon regeneration. Neuron. 2016;90:437–451. doi: 10.1016/j.neuron.2016.04.022. [DOI] [PubMed] [Google Scholar]
- Hobert O. A map of terminal regulators of neuronal identity in Caenorhabditis elegans. Wiley Interdiscip. Rev Dev Biol. 2016;5:474–498. doi: 10.1002/wdev.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Urban J, Technau GM. Distribution, classification, and development of Drosophila glial cells in the late embryonic and early larval ventral nerve cord. Rouxs Arch Dev Biol. 1995;204:284–307. doi: 10.1007/BF02179499. [DOI] [PubMed] [Google Scholar]
- Jacobs JR, Hiromi Y, Patel NH, Goodman CS. Lineage, migration, and morphogenesis of longitudinal glia in the Drosophila CNS as revealed by a molecular lineage marker. Neuron. 1989;2:1625–1631. doi: 10.1016/0896-6273(89)90051-2. [DOI] [PubMed] [Google Scholar]
- Jiang H, Edgar BA. Intestinal stem cells in the adult Drosophila midgut. Exp Cell Res. 2011;317:2780–2788. doi: 10.1016/j.yexcr.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamoto T. Conditional disruption of synaptic transmission induces male-male courtship behavior in Drosophila. Proc Natl Acad Sci USA. 2002;99:13232–13237. doi: 10.1073/pnas.202489099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer MC, Jung C, Batelli S, Rubin GM, Gaul U. The glia of the adult Drosophila nervous system. Glia. 2017;65:606–638. doi: 10.1002/glia.23115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurusu M, Katsuki T, Zinn K, Suzuki E. Developmental changes in expression, subcellular distribution, and function of Drosophila N-cadherin, guided by a cell-intrinsic program during neuronal differentiation. Dev Biol. 2012;366:204–217. doi: 10.1016/j.ydbio.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacin H, Truman JW. Lineage mapping identifies molecular and architectural similarities between the larval and adult Drosophila central nervous system. eLife. 2016;5:e13399. doi: 10.7554/eLife.13399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HH, Kroll JR, Lennox SM, Ogundeyi O, Jeter J, Depasquale G, Truman JW. A GAL4 driver resource for developmental and behavioral studies on the larval CNS of Drosophila. Cell Rep. 2014;8:897–908. doi: 10.1016/j.celrep.2014.06.065. [DOI] [PubMed] [Google Scholar]
- Mahr A, Aberle H. The expression pattern of the Drosophila vesicular glutamate transporter: A marker protein for motoneurons and glutamatergic centers in the brain. Gene Expr Patterns. 2006;6:299–309. doi: 10.1016/j.modgep.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Mayer B, Emery G, Berdnik D, Wirtz-Peitz F, Knoblich JA. Quantitative analysis of protein dynamics during asymmetric cell division. Curr Biol. 2005;15:1847–1854. doi: 10.1016/j.cub.2005.08.067. [DOI] [PubMed] [Google Scholar]
- Muthukumar AK, Stork T, Freeman MR. Activity-dependent regulation of astrocyte GAT levels during synaptogenesis. Nat Neurosci. 2014;17:1340–1350. doi: 10.1038/nn.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor RM, Stone EF, Wayne CR, Marcinkevicius EV, Ulgherait M, Delventhal R, Pantalia MM, Hill VM, Zhou CG, McAllister S, et al. A Drosophila model of Fragile X syndrome exhibits defects in phagocytosis by innate immune cells. J Cell Biol. 2017;216:595–605. doi: 10.1083/jcb.201607093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoto JJ, Yogi P, Hartenstein V. Origin and development of neuropil glia of the Drosophila larval and adult brain: Two distinct glial populations derived from separate progenitors. Dev Biol. 2015;404:2–20. doi: 10.1016/j.ydbio.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoto JJ, Lovick JK, Hartenstein V. Origins of glial cell populations in the insect nervous system. Curr Opin Insect Sci. 2016;18:96–104. doi: 10.1016/j.cois.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes MF, Sorrells SF, Garcia-Verdugo JM, Alvarez-Buylla A. Brain size and limits to adult neurogenesis. J Comp Neurol. 2016;524:646–664. doi: 10.1002/cne.23896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peco E, Davla S, Camp D, Stacey SM, Landgraf M, van Meyel DJ. Drosophila astrocytes cover specific territories of the CNS neuropil and are instructed to differentiate by Prospero, a key effector of Notch. Development. 2016;143:1170–1181. doi: 10.1242/dev.133165. [DOI] [PubMed] [Google Scholar]
- Pereanu W, Spindler S, Cruz L, Hartenstein V. Tracheal development in the Drosophila brain is constrained by glial cells. Dev Biol. 2007;302:169–180. doi: 10.1016/j.ydbio.2006.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci USA. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plavicki JS, Squirrell JM, Eliceiri KW, Boekhoff-Falk G. Expression of the Drosophila homeobox gene, Distal-less, supports an ancestral role in neural development. Dev Dyn. 2016;245:87–95. doi: 10.1002/dvdy.24359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CJ, Tasic B, Russler EV, Liang L, Luo L. The Q system: A repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell. 2010;141:536–548. doi: 10.1016/j.cell.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanelli AM, Appel B. Motor neurons and oligodendrocytes arise from distinct cell lineages by progenitor recruitment. Genes Dev. 2015;29:2504–2515. doi: 10.1101/gad.271312.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rideout EJ, Dornan AJ, Neville MC, Eadie S, Goodwin SF. Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nat Neurosci. 2010;13:458–466. doi: 10.1038/nn.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risher WC, Patel S, Kim IH, Uezu A, Bhagat S, Wilton DK, Pilaz LJ, Singh Alvarado J, Calhan OY, Silver DL, et al. Astrocytes refine cortical connectivity at dendritic spines. eLife Published online December. 2014;17:2014. doi: 10.7554/eLife.04047. https://doi.org/10.7554/eLife.04047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowitch DH, Kriegstein AR. Developmental genetics of vertebrate glial-cell specification. Nature. 2010;468:214–222. doi: 10.1038/nature09611. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Rickert C, Bossing T, Vef O, Urban J, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. II. Neuroblast lineages derived from the dorsal part of the neuroectoderm. Dev Biol. 1997;189:186–204. doi: 10.1006/dbio.1997.8660. [DOI] [PubMed] [Google Scholar]
- Seita J, Weissman IL. Hematopoietic stem cell: Self-renewal versus differentiation. Wiley Interdiscip. Rev Syst Biol Med. 2010;2:640–653. doi: 10.1002/wsbm.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepp KJ, Auld VJ. Reciprocal interactions between neurons and glia are required for Drosophila peripheral nervous system development. J Neurosci. 2003;23:8221–8230. doi: 10.1523/JNEUROSCI.23-23-08221.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, Clevers H. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- Stephan AH, Barres BA, Stevens B. The complement system: An unexpected role in synaptic pruning during development and disease. Annu Rev Neurosci. 2012;35:369–389. doi: 10.1146/annurev-neuro-061010-113810. [DOI] [PubMed] [Google Scholar]
- Stork T, Sheehan A, Tasdemir-Yilmaz OE, Freeman MR. Neuron-glia interactions through the Heartless FGF receptor signaling pathway mediate morphogenesis of Drosophila astrocytes. Neuron. 2014;83:388–403. doi: 10.1016/j.neuron.2014.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE. Post-embryonic development in the ventral cord of Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1976;275:287–297. doi: 10.1098/rstb.1976.0084. [DOI] [PubMed] [Google Scholar]
- Truman JW, Schuppe H, Shepherd D, Williams DW. Developmental architecture of adult-specific lineages in the ventral CNS of Drosophila. Development. 2004;131:5167–5184. doi: 10.1242/dev.01371. [DOI] [PubMed] [Google Scholar]
- Truman JW, Moats W, Altman J, Marin EC, Williams DW. Role of Notch signaling in establishing the hemilineages of secondary neurons in Drosophila melanogaster. Development. 2010;137:53–61. doi: 10.1242/dev.041749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Yamauchi T, Izpisua Belmonte JC. An overview of mammalian pluripotency. Development. 2016;143:1644–1648. doi: 10.1242/dev.132928. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


