Abstract
The gene for the large subunit of Rubisco was specifically mutated by transforming the chloroplast genome of tobacco (Nicotiana tabacum). Codon 335 was altered to encode valine instead of leucine. The resulting mutant plants could not grow without atmospheric CO2 enrichment. In 0.3% (v/v) CO2, the mutant and wild-type plants produced similar amounts of Rubisco but the extent of carbamylation was nearly twice as great in the mutants. The mutant enzyme's substrate-saturated CO2-fixing rate and its ability to distinguish between CO2 and O2 as substrates were both reduced to 25% of the wild type's values. Estimates of these parameters obtained from kinetic assays with the purified mutant enzyme were the same as those inferred from measurements of photosynthetic gas exchange with leaves of mutant plants. The Michaelis constants for CO2, O2, and ribulose-1,5-bisphosphate were reduced and the mutation enhanced oxygenase activity at limiting O2 concentrations. Consistent with the reduced CO2 fixation rate at saturating CO2, the mutant plants grew slower than the wild type but they eventually flowered and reproduced apparently normally. The mutation and its associated phenotype were inherited maternally. The chloroplast-transformation strategy surmounts previous obstacles to mutagenesis of higher-plant Rubisco and allows the consequences for leaf photosynthesis to be assessed.
Study of Rubisco in higher plants and its dual role in photosynthesis and photorespiration has been hampered by inability to fold and/or assemble the higher-plant enzyme correctly in bacterial hosts. No eukaryotic Rubisco has been expressed successfully in any foreign host, perhaps because of mismatches between the foreign Rubisco and the host's chaperone system (for reviews, see Gutteridge and Gatenby, 1995; Roy and Andrews, 1999). This has restricted mutagenic analysis of the structure and function of eukaryotic Rubisco to the green alga Chlamydomonas reinhardtii, where well-developed methods exist for mutagenesis and transformation of the chloroplast genome, which encodes Rubisco's large, catalytic subunit (for review, see Spreitzer, 1998). However, some aspects of Rubisco function cannot readily be studied in vivo in an alga. For example, Rubisco's oxygenation reaction and its photorespiratory consequences are suppressed in C. reinhardtii and most other algae by a mechanism that concentrates CO2 at the site of Rubisco (for reviews, see Husic et al., 1987; Spreitzer, 1993).
Development of a method for transforming the chloroplast genome of tobacco (Svab and Maliga, 1993) provides a means for circumventing these frustrations and presents an opportunity for testing of the wealth of information that exists about Rubisco's structure and function (Schreuder et al., 1993; Hartman and Harpel, 1994; Andersson, 1996) within the context of a whole plant. This method has been used to delete the rbcL gene for the Rubisco large subunit from the chloroplast genome, allowing demonstration that its function could be replaced to some extent by introduction to the nucleus of a copy fused to a sequence encoding a chloroplast transit peptide (Kanevski and Maliga, 1994). It has also been used to replace the tobacco rbcL gene with the analogous genes from sunflower and the cyanobacterium Synechococcus PCC6301. However, the resultant Rubiscos (hybrids composed of introduced large subunits and tobacco small subunits) were either not formed (Synechococcus) or too disabled to support photoautotrophic growth (sunflower) (Kanevski et al., 1999). The precise homologous replacement inherent in the transformation mechanism should also facilitate site-specific mutagenesis of the rbcL gene with the bonus that the consequences of the alteration can be assessed not only in vitro, by studying the properties of the mutant protein after isolation, but also in vivo, by measurement of photosynthetic gas exchange by leaves of the mutant plants.
As a first attempt to demonstrate the feasibility of this approach, we aimed to induce an alteration in Rubisco's catalytic properties that would be readily apparent in the leaf's CO2-exchange properties without debilitating the enzyme to such an extent that it would be unable to support the growth of the plant. To achieve this result, we sought a mutation that would impair Rubisco's ability to distinguish between CO2 and O2 (expected to cause an increase in the leaf's CO2-compensation point) without seriously compromising the CO2-saturated rate of carboxylation. CO2 enrichment would largely compensate for such a defect, allowing the mutant plants to photosynthesize and grow.
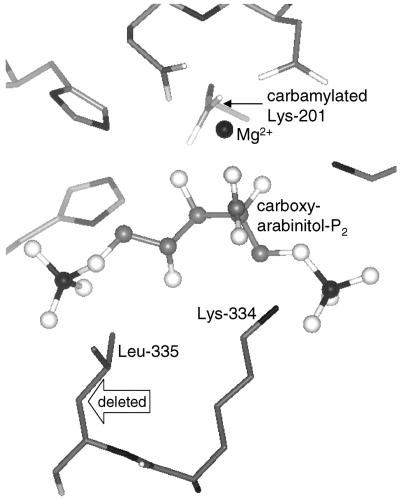
Guided by crystallographic structural information (Schreuder et al., 1993; Andersson, 1996) and previous mutagenesis with prokaryotic Rubiscos (Terzaghi et al., 1986; Lee et al., 1993), we chose Leu-335 and changed it conservatively to Val (Fig. 1). The preceding residue in the primary structure is the catalytically essential Lys-334 that assists in stabilizing the transition state for carboxylation and in promoting specificity for CO2 (Lorimer et al., 1993). However, Leu-335 does not participate directly in catalysis. The side chains of residues 334 and 335 move over 10 Å when a mobile loop closes over the bound substrate (Schreuder et al., 1993). In the final closed position, the side chain of Leu-335 makes van der Waals contact with the phosphate group attached to C5 of the substrate. There are likely to be subtle interactions between the two residues that influence the critical positioning of the ε N atom of Lys-334 (Fig. 1). In the Rubisco from the cyanobacterium, Synechococcus PCC 6301, the Leu to Val substitution at position 335 reduced the CO2/O2 specificity and the substrate-saturated carboxylation rate (Vcmax) to 39% and 40%, respectively, of wild-type values (Lee et al., 1993). We considered that such impairment, if reproduced in the tobacco enzyme, would fulfill our requirements.
Figure 1.
A view of the active site of spinach Rubisco complexed to carboxyarabinitol-P2 based on the coordinates of the 1.6-Å structure (Andersson, 1996) showing the position of Leu-335. The lower resolution structure of the analogous complex of tobacco Rubisco (Schreuder et al., 1993) appears to be identical in this region. The hollow arrow indicates the carbon atom deleted by the Val substitution. Bound carboxyarabinitol-P2 and Mg2+ are rendered as balls and sticks and amino acid residues as sticks. C, Gray; O, white; N, P, and Mg2+, black.
MATERIALS AND METHODS
Construction of the Transformation Plasmid
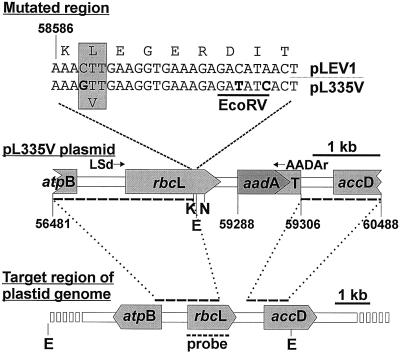
A 985-bp promoterless cassette, comprising the aadA gene and the rps16 terminator sequence, was amplified from pRV112A (Zoubenko et al., 1994) using primers AD5 (5′AAGCGCTTAGATCTAGTTGTAGGGAG3′, introducing the underlined BglII restriction site) and AD3 (5′TGTCAAAGAAGCTTGAATTAATTCAATG3′, HindIII site underlined). During amplification, the BssHII site within aadA was eliminated for purposes unrelated to the present study by including primer ADM (5′pGCTTGGCCTCTCGCGCAGATC3′, silent base change underlined). This cassette was inserted between the 2,805-bp SacI-BamHI fragment of pTB29 and the 1,182-bp BamHI-XhoI fragment of pTB22 (Sugiura et al., 1986) and assembled in pTZ18R (Mead et al., 1986) to produce the transformation plasmid pLEV1 (Fig. 2). Site-specific mutagenesis was conducted on single-stranded DNA derived from the 348-bp KpnI-NruI fragment of pLEV1 inserted in pTZ18R using primer Val-335 (5′pGCCCAAAGTGAtATcTCTTTCACCTTTCAAcTTTACCTACTACGGTACCTGGGATCCTCT3′, introduced EcoRV site and mutated codon 335 underlined, nucleotide changes in lowercase). The italicized T in this primer destroyed a SmaI site in the flanking vector sequence, allowing for selection of mutants (Deng and Nickoloff, 1992). Substitution of the mutant KpnI-NruI fragment into pLEV1 produced pL335V (Fig. 2).
Figure 2.
The pL335V plasmid used for transformation of the tobacco chloroplast genome. This plasmid was based on pLEV1 with the nucleotide substitutions (in bold) required to change codon 335 from Leu to Val and silent substitutions to introduce an adjacent EcoRV site for screening of transformants. The numbers refer to the sequence of the tobacco chloroplast genome (Shinozaki et al., 1986). The dashed bars joined by the dotted lines indicate the homologous flanking regions present in pL335V and available for the crossovers required to introduce the mutated region and the aadA gene into the large single-copy region of the chloroplast genome. The probe used in screening by Southern blotting and the primers for amplifying and sequencing the mutant rbcL are indicated. K, KpnI; N, NruI; E, EcoRV; T, rps16 terminator sequence.
Chloroplast Transformation
The plastid genome of tobacco (Nicotiana tabacum L. cv Petit Havana [N,N]) was transformed with pL335V using the biolistic method (Svab and Maliga, 1993). Leaf tissue from spectinomycin-resistant plantlets was carried through several rounds of regeneration during which the presence of the introduced EcoRV site and homoplasticity were monitored by Southern analysis (Sambrook et al., 1989) of leaf DNA isolated according to the method of Saghai-Maroof et al. (1984) and digested with EcoRV. The probe was a 1,256-bp fragment of rbcL (Fig. 2).
Plant Growth
Regenerating plantlets (homoplastic transformants) were transferred to 10-L pots of soil and grown to maturity in an artificially lit (400 μmol quanta m−2 s−1) growth chamber in an atmosphere of 0.3% (v/v) CO2 in air. Air temperature was 24°C during the 14-h photoperiod, 18°C during darkness, and the pots were watered daily with a complete nutrient solution. At maturity, flowers were allowed to self-pollinate and the progeny were grown through another generation under the same conditions. Some of their flowers were artificially pollinated with wild-type pollen.
Control plants were regenerated from untransformed stem segments and grown and propagated in the same way as the transformants.
Confirmation of Mutation
A 2,144-bp fragment containing rbcL was amplified by PCR from leaf DNA using the primers LSd (5′CACGGAATTCGTGTCGAGTAG3′) and AADAr (5′GAATGTCATTGCGCTGCCATTCTCCA3′) (Fig. 2). The product was ligated into pGEM-T Easy (Promega, Madison, WI) and sequenced using BigDye terminator cycle sequencing (Applied Biosystems, Foster City, CA) using these and several other internal primers.
Segregation of Spectinomycin Resistance and Phenotype
Segregation of spectinomycin resistance was observed by germinating seeds obtained by self-pollinating primary transformants and wild-type regenerants on RMOP agar medium (Svab and Maliga, 1993; Bock, 1998), lacking Suc, and containing 500 μg mL−1 spectinomycin, in an atmosphere containing 0.3% (v/v) CO2. Ability to grow without CO2 supplementation was similarly assessed on spectinomycin-free medium in an atmosphere containing 0.035% (v/v) CO2. To measure growth rate at 0.3% (v/v) CO2, seeds from wild-type regenerants and seeds derived by cross-pollinating flowers of T1 plants with wild-type pollen were germinated in 1-L pots of soil and grown as described above. Twenty-seven days after emergence of the cotyledons, the height and number of leaves of the plants were recorded and the complete shoots were harvested, dried in an oven at 80°C, and weighed.
Purification and Assay of Rubisco
Leaf discs (0.79 cm2) from young, fully expanded leaves of mutant and control plants were frozen in liquid N2 and then ground in ice-cold glass homogenizers containing 0.8 mL of extraction buffer (50 mm HEPPS-NaOH, pH 7.8, 0.5 mm EDTA, 5 mm MgCl2, 5 mm DTT, 0.1% [w/v] polyvinylpolypyrrolidone, 1 mm benzamidine, and 0.5 mm PMSF). Following a 30-s centrifugation at 13,000g at 4°C, aliquots were assayed for substrate-saturated ribulose-P2 carboxylase activity (Mate et al., 1993) and Rubisco content and carbamylation status. Content of Rubisco catalytic sites was measured by stoichiometric binding of 14C-carboxyarabinitol-P2 and its carbamylation status by exchanging 14C-carboxyarabinitol-P2 bound loosely at uncarbamylated sites with excess unlabeled carboxyarabinitol-P2 (Butz and Sharkey, 1989; Ruuska et al., 1998). These methods rely on the ligand binding sufficiently tightly so that the Rubisco-ligand complex may be isolated quantitatively. We carried out this isolation by gel filtration as described by Ruuska et al. (1998). We verified that the Leu-335 mutant Rubisco bound carboxyarabinitol-P2 tightly enough for this approach to be valid by observing that its elution profile resembled that of the wild type, showing a peak of bound 14C-label that returned cleanly to baseline without trailing behind the protein peak.
Rubisco was purified from leaf extracts by precipitation with PEG followed by ultracentrifugation through Suc density gradients. Leaves (1.5–2.1 g) were frozen in liquid N2, ground in a mortar and pestle, and added to 200 mL of ice-cold extraction buffer (lacking MgCl2). The extract was filtered through two layers of Miracloth (Calbiochem, San Diego, CA) and 60% (w/v) PEG (Mr 3,350) was added to bring the final concentration to 8%. After 5 min, MgSO4 was added to 20 mm and the extract centrifuged for 20 min at 17,000g at 4°C. Rubisco was precipitated from the supernatant by adding PEG to 18% (w/v) and pelleted by centrifugation. The precipitate was resuspended in 2 mL of gradient buffer (50 mm HEPPS-NaOH, pH 8.3, 1 mm EDTA, and 10 mm MgCl2) and centrifuged at 27,000 rpm for 22 h at 4°C through an exponential density gradient (mixing volume, 24 mL; gradient volume, 35 mL) of 7% to 23.3% (w/v) Suc in gradient buffer using an SW28Ti rotor (Beckman Instruments, Fullerton, CA). Fractions (1.6 mL) were collected from the bottom of the gradient and the Rubisco peak was identified by A280. Peak fractions were frozen in liquid N2 and stored at −80°C. The Michaelis constants (at pH 8.3) for CO2, O2, and ribulose-P2 (Paul et al., 1991) and the CO2/O2 specificity (Kane et al., 1994) were measured using these purified preparations.
Leaf Gas-Exchange Measurements
Plants were brought from the high-CO2 growth cabinet to the laboratory and gas exchange by young, fully expanded leaves was measured using the clamp-on chamber of the portable, flow-through photosynthesis system LI-6400 (LI-COR, Lincoln, NE). Leaf temperature was set at 25°C and illumination (1,000 μmol quanta m−2 s−1) was provided with a tungsten-halogen lamp. To obtain the desired atmospheric gas compositions, N2 and O2 were mixed with mass flow controllers (MKS Type 1179A, MKS Instruments, Andover, MA) and CO2 was varied using the LI-6400 CO2 injection system. Kinetic parameters of Rubisco in vivo were inferred from gas-exchange measurements as described previously (von Caemmerer et al., 1994).
RESULTS
Mutagenesis and Chloroplast Transformation
Mutagenesis of codon 335 of cloned tobacco rbcL was directed using an oligonucleotide that also introduced an EcoRV cleavage site 17 bp downstream (Fig. 2). The mutant rbcL was constructed in a transformation plasmid, pLEV1, which also contained a promoterless aadA gene (conferring resistance to spectinomycin) inserted downstream of rbcL, and homologously recombined into the tobacco plastid genome using the biolistic method (see “Materials and Methods”). Leaf material from the resulting spectinomycin-resistant plantlets was subjected to a further round of regeneration on spectinomycin-containing medium, after which Southern blotting (see “Materials and Methods”) was conducted to determine whether or not the desired mutations had been introduced.
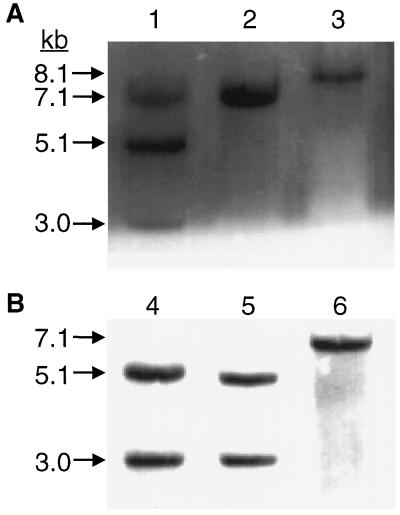
The distribution of EcoRV sites in the rbcL region of the wild-type tobacco plastome (Fig. 2) is such that a single 7.1-kb fragment was detected by the rbcL probe in EcoRV digests of leaf DNA (Fig. 3, lanes 2 and 6). Insertion of the aadA gene in the targeted position 3′ to rbcL increased the size of this fragment to 8.1 kb. If the region exposed to mutagenesis containing the new EcoRV site was also incorporated, the enlarged fragment is divided into 5.1- and 3.0-kb subfragments.
Figure 3.
Southern blots of EcoRV-digested DNA from transformant plants (lanes 1, 3, 4 and 5) probed with the 1,256-bp rbcL sequence (see “Materials and Methods” and Fig. 2). Results for control plants are shown in lanes 2 and 6. A, Plantlets after two rounds of regeneration. Lane 1 shows the results for the single heteroplasmic plantlet that retained the introduced EcoRV site. Lane 3 shows an example of the class of plantlets where loss of the EcoRV site had occurred, followed by sorting out to homoplasmicity. B, Lanes 4 and 5 show results for mature T1 generation plants resulting from cloning and three additional rounds of regeneration of the plantlet analyzed in lane 1, followed by growth to maturity in soil, self-pollination of T0 flowers, and growth of the resulting seed. Progeny of both original homoplastic plantlets derived from regeneration of the lane 1 plantlet are represented.
Three of 11 spectinomycin-resistant plantlets displayed only the wild-type 7.1-kb band (not shown). The spectinomycin resistance of these must have derived from spontaneous mutations elsewhere in the plastome or perhaps from illegitimate incorporation of the aadA gene. Of the remaining eight, only one showed the pattern of 5.1- and 3.0-kb bands expected for the desired transformation. However, the 7.1-kb wild-type band was also present, indicating that this plantlet was heteroplasmic, containing both wild-type and transformed plastomes (Fig. 3A, lane 1). Blots of the other seven (an example is shown in Fig. 3A, lane 3) revealed a single hybridizing band of 8.1 kb. The size of this fragment is consistent with insertion of the aadA gene in the expected position but the lack of an internal EcoRV site indicates that it did not contain the mutated region of rbcL. Such a fragment would be produced by crossover between wild-type and mutated plastomes in the region between the mutation site and the aadA gene, resulting in the combination of wild-type rbcL with aadA.
The single heteroplasmic plantlet retaining the internal EcoRV site was cloned and regenerated again. Of the four resulting plantlets, two retained the EcoRV site and showed no trace of the wild-type fragment. The others lost the EcoRV site, again producing the single 8.1-kb hybridizing band (not shown, similar to Fig. 3A, lane 3). The frequency with which this crossover occurred raises suspicion that unimpaired Rubisco function is advantageous, even during growth on a Suc-containing medium, thus applying positive selection for the presence of the wild-type rbcL sequence. Nevertheless, the plantlets retaining the EcoRV site continued to retain it through two additional rounds of cloning and regeneration and subsequent growth to maturity in soil. No trace of the unmutated sequence could be detected in these plants or in their progeny (Fig. 3B, lanes 4 and 5). We concluded that they had become homoplasmic and thus immune to further crossover. A 2,144-bp region encompassing all of the rbcL gene was amplified from mutant plants and sequenced completely (see “Materials and Methods”). This confirmed that the mutation in codon 335 was present and that only the intended changes had been introduced.
Inheritance
When tested as described in “Materials and Methods”, all of the T1 self-progeny (98 tested), but no control progeny (97 tested), were resistant to spectinomycin. On the other hand, no T1 self-progeny (47 tested), but all control progeny (49 tested), were able to grow beyond the cotyledon stage in an atmosphere containing 0.035% (v/v) CO2. This lack of segregation of both spectinomycin resistance and CO2 requirement is consistent with the expected maternal inheritance of the transformed plastid genomes.
As a further control, tobacco was transformed with the pLEV1 construct, which contained the aadA gene but lacked the rbcL mutation (Fig. 2). The three transformed plants that resulted grew normally without CO2 supplementation and appeared to be indistinguishable from the wild type.
Growth Rate at High CO2
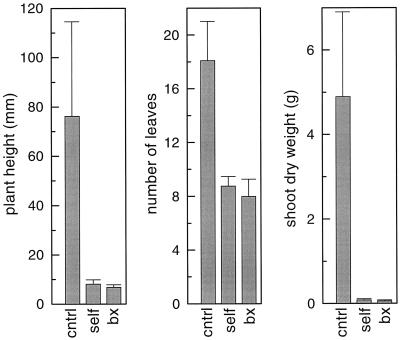
Even in a CO2-enriched atmosphere, the mutant plants grew slower than controls (Fig. 4). This slow-growing phenotype did not segregate in the T1 and T2 generations, even with seed derived from cross-pollinating flowers with wild-type pollen (see “Materials and Methods”). Therefore, the growth retardation appears to be inherited maternally, as well. The average aboveground dry weight 27 d after cotyledon emergence of both self-pollinated and cross-pollinated mutant plants was less than 2% that of the wild type (Fig. 5). Plant height was reduced to one-tenth in the mutants and their number of leaves was reduced 50%.
Figure 4.
A control plant and a T1 generation Val-335 transformant 50 d after emergence of the cotyledons. Both plants were grown in air enriched with CO2 to 0.3% (v/v) as described in “Materials and Methods.”
Figure 5.
Physical characteristics of plants grown in 0.3% (v/v) CO2 in air. The height, total number of leaves, and aboveground dry weight of the plants were measured (±sd) 27 d after emergence of the cotyledons. cntrl, Control (n = 9); self, T1 generation transformants derived by self-pollination of T0 flowers (n = 8); bx, T2 generation transformants derived by pollinating T1 flowers with wild-type pollen (n = 10).
Characteristics of the Mutant Rubisco
The Rubisco content of the mutant plants was similar to that of the controls but approximately twice as many of the mutant enzyme's active sites were carbamylated under the high-CO2 growth conditions (Table I). The Vcmax of the mutant enzyme was reduced to 24% that of the wild type and its Km values for CO2, O2, and ribulose-P2 were reduced to 48%, 17%, and 11%, respectively, of control values. The CO2/O2 specificity of the mutant Rubisco was 25% that of the wild type (Table I).
Table I.
Content, carbamylation, and kinetic parameters for Leu-335 (wild type) and Val-335 Rubiscos measured in vivo and in vitro
| Parameter | In Vivo
|
In Vitro
|
||
|---|---|---|---|---|
| Leu-335 | Val-335a | Leu-335b | Val-335a | |
| Measured parameters | ||||
| Rubisco content (μmol m−2)c | 30.3 ± 1.6b | 33.0 ± 5.6 | n.a. | n.a. |
| % Carbamylationc | 48.1 ± 5.6b | 89.9 ± 5.4 | n.a. | n.a. |
| Vcmax (s−1) | 3.53d | 1.02 | 3.43 ± 0.12 | 0.81 ± 0.04 |
| Km(CO2) (μm) | 8.6de–13.5df | 8.6e–10.6f | 10.7 ± 0.6 | 5.1 ± 0.8 |
| Ki(O2) (μm) | 226de–313df | 55e–70f | 295 ± 71 | 48.9 ± 11.5 |
| Km (ribulose-P2) (μm)g | n.a. | n.a. | 18.8 ± 3.2 | 2.1 ± 0.2 |
| Sc/o | 102.0d | 27.0h | 81.1 ± 1.6 | 20.1 ± 1.5 |
| Calculated parameters | ||||
| Vomax (s−1)i | n.a. | n.a. | 1.17 | 0.39 |
| Vcmax/Kc (mm−1 s−1) | n.a. | n.a. | 321 | 159 |
| Vomax/Ko (mm−1 s−1) | n.a. | n.a. | 3.95 | 7.90 |
| Net CO2 uptake in air (s−1)j | n.a. | n.a. | 0.82 | 0.04 |
| Net CO2 uptake in 0.3% (v/v) CO2 (s−1)j | n.a. | n.a. | 2.80 | 0.57 |
n.a., Not applicable.
Measurements with T0 transformants.
Measurements with wild-type regenerants.
Measured with leaves sampled under the 0.3% (v/v) CO2 growth conditions.
Measurements with anti-rbcS tobacco cv W38 taken from von Caemmerer et al. (1994).
Assuming a value of 0.3 mol m−2 s−1 bar−1 for the conductance for CO2 transfer between the intercellular air spaces and the sites of carboxylation (von Caemmerer et al., 1994).
Assuming infinite conductance for CO2 transfer between the intercellular air spaces and the sites of carboxylation.
For the carboxylase reaction.
Estimated from the slope of the line in the Figure 6B legend.
Vomax = (Vcmax · Ko)/(Kc · Sc/o).
Net CO2 uptake (per carbamylated Rubisco site) from combined photosynthesis and photorespiration (neglecting nonphotorespiratory CO2 release, i.e. Rd = 0) calculated at 25°C from the in vitro data using Equation 1 and assuming that the CO2 concentration in the chloroplast is 8 μm in air and 94 μm in 0.3% (v/v) CO2.
Photosynthetic Gas-Exchange Characteristics
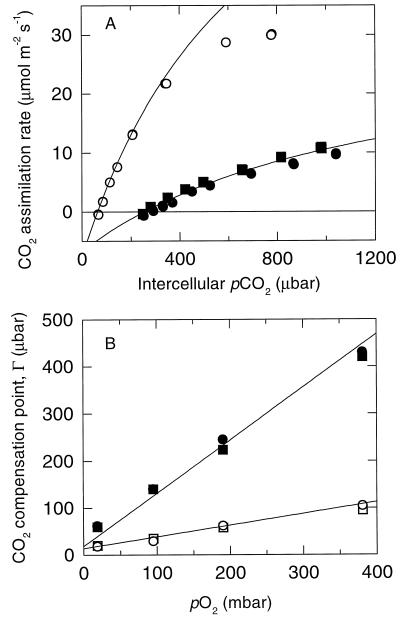
CO2 assimilation by leaves of the mutant plants was severely reduced. The CO2-compensation point in 21% (v/v) O2 was increased 4-fold relative to the controls and assimilation remained limited by Rubisco activity at all CO2 concentrations, unlike assimilation in the control, which became limited by light-dependent ribulose-P2 regeneration above 300 μbar CO2, as expected (Fig. 6A). CO2-response curves were measured at different O2 concentrations and the kinetic parameters of the mutant Rubisco in vivo were calculated from these data with a model for photosynthetic CO2 exchange (Farquhar et al., 1980; von Caemmerer et al., 1994). The increased slope of the dependency of the CO2-compensation point on O2 concentration accords with the poorer CO2/O2 specificity of the mutant Rubisco (Fig. 6B; Table I).
Figure 6.
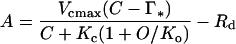 |
1 |
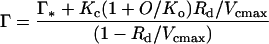 |
2 |
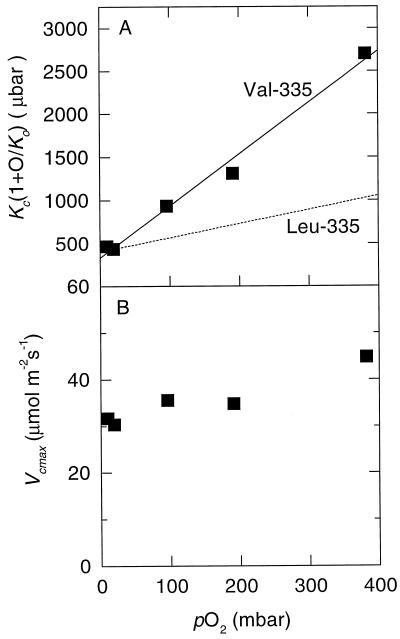
Since photosynthesis in the control plants becomes limited by ribulose-P2 regeneration above 300 μbar CO2 (Fig. 6A), these plants are not suitable for measurement of the wild-type Rubisco's kinetic properties in vivo. However, a useful comparison can be made with previous data for tobacco with a reduced content of wild-type Rubisco induced by a nuclear anti-rbcS gene. Photosynthesis in these plants was also limited by Rubisco activity rather than by ribulose-P2 regeneration, even at high CO2 concentrations (von Caemmerer et al., 1994). The increased slope of the dependency of the apparent Km for CO2 on O2 concentration observed with plants with the mutant Rubisco, compared with the previous observations, revealed the reduction in Km for O2 (Fig. 7A; Table I). Vcmax, on the other hand, was little affected by O2 concentration, as expected if CO2 and O2 are competitive alternate substrates (Fig. 7B).
Figure 7.
The responses of the apparent Km for CO2 (A) and the maximal carboxylation rate (Vcmax) (B) of the Val-335 Rubisco to pO2 inferred from gas-exchange data. The data were calculated from CO2-response curves similar to those in Figure 6A measured at different pO2 as described by von Caemmerer et al. (1994). An infinite conductance for CO2 transfer between the intercellular air spaces and the sites of carboxylation was assumed. The solid line in A is a linear fit to the data for one transformant plant (▪). For comparison, the dashed line shows the in vivo response of the wild-type Rubisco (Leu-335) calculated from the data of von Caemmerer et al. (1994) for Rubisco-deficient anti-rbcS tobacco.
DISCUSSION
Heteroplasmy Makes Directed Partial Disablement of the Rubisco Large Subunit Difficult But Not Impossible
Following transformation of the chloroplast genome, sorting-out between transformed and untransformed genomes occurs by a mechanism currently not well understood (Maliga, 1993). Eventually, the tissue may become homoplasmic for the transformed plastome, which encodes resistance to the selective agent spectinomycin, but during the sorting-out process, which may take several rounds of regeneration in tissue culture, the two genomes co-exist. Site-directed mutagenesis of an existing chloroplast-encoded gene requires linkage between the necessary base substitution and an introduced aadA gene, which encodes spectinomycin resistance. Some homology between the transforming DNA construct and the wild-type plastome in the region between the mutagenic substitution and the aadA gene is unavoidable. Therefore, the potential exists for recombination in this intervening region between the wild-type and mutant plastomes while they coexist. The product of such a recombination event will encode a wild-type copy of the target gene together with the aadA gene. The problem is exacerbated if the transformation results in partial disablement of an essential chloroplast gene. Then there is active selection for such recombinants. These proved to be the most common outcome of our transformations. However, despite this difficulty, we ultimately succeeded in obtaining a transformant that retained the desired directed mutation in rbcL. Its stability through several generations of plants and lack of detectable copies of the wild-type plastome lead us to conclude that it is homoplasmic.
The low frequency of recovery of the desired transformants might have been circumvented if we had transformed a tobacco line with a deletion in the region of rbcL targeted by our mutation, such as that engineered by Kanevski and Maliga (1994). However, such an approach would have had to surmount difficulties associated with the propagation of photosynthetically disabled plants and with the use of a selective agent other than spectinomycin (since the aadA gene replaced the deleted rbcL gene).
The Phenotype Is Maternally Inherited
Plastid transformation, like any procedure requiring repeated cycles of tissue culture and regeneration, carries with it a risk of introducing unwanted nuclear mutations. These can be removed by repeated back-crossing with wild-type pollen and we are currently proceeding with this. However, it is unlikely that such nuclear mutations, if they occurred, can be contributing to the phenotype and the changed Rubisco properties that our transformants display. Both resistance to spectinomycin and inability to grow without CO2 enrichment appear to be maternally inherited. Furthermore, the slower growth rate of transformant progeny compared with the wild type under CO2 enrichment shows no signs of segregating, even with progeny resulting from back-crossing with wild-type pollen (Fig. 5). Thus, the impaired growth also appears to be a maternally inherited characteristic, consistent with the engineered change in the plastid rbcL gene.
Kinetic Properties of the Val-335 Mutant Rubisco Are Consistent with a Disturbance of the Position or Orientation of the Adjacent Residue, Lys-334
Substitution of Val for Leu at residue 335 of Rubisco's large subunit caused changes in the catalytic properties of tobacco Rubisco (Table I) similar to those seen previously with Synechococcus Rubisco (Lee et al., 1993) but a little more severe. For example, the mutation reduced both the maximal activity and CO2/O2 specificity of the cyanobacterial enzyme by approximately 60% but, for the tobacco enzyme, the reductions were 75%. The Km for ribulose-P2 of both enzymes was also reduced. However, while the mutation nearly doubled the Km for CO2 of the cyanobacterial enzyme, it halved it for the tobacco enzyme. The increased inhibition of the mutant tobacco enzyme by O2 was particularly obvious in the reduction of its Ki for O2 to one-sixth that of the wild type (Table I).
The changes in kinetic parameters induced by the Val-335 mutation are consistent with this side chain influencing the position and orientation and, therefore, the reactivity of the adjacent Lys-334 residue. Contact between the terminal methyl groups of the side chain of residue 335 and the P2 phosphate group of the substrate is likely to regulate the depth to which the ε amino group of Lys-334 penetrates into the active-site region when the flexible loop that contains both of these residues (loop 6) closes over the bound substrate and reaction intermediates (Fig. 1). Shortening the side chain of residue 335 by one carbon atom would allow the terminal N atom of the Lys residue to penetrate too far into the active site, perhaps leading to distortion or deflection of the lysyl side chain and a change in the critically important orientation of the N atom. This possibility is supported by a structural study with a mutant of Synechococcus Rubisco in which Lys-334 was replaced by Met, which showed deflection of this side chain (Newman, 1992).
The ε N atom of residue 334 makes hydrogen-bonding contact with one of the O atoms of the nascent carboxyl group formed when CO2 attacks the enediol form of ribulose-P2. This interaction is likely to lower the energy of the transition state associated with this process, thus facilitating the carboxylation reaction. This interaction is not duplicated in the analogous attack of O2 on the enediol (for review, see Roy and Andrews, 1999). Therefore, perturbation of the position and alignment of the ε N atom would have more serious consequences for carboxylation than for oxygenation, which is consistent with our observations.
If our interpretation that the effects of the Val-335 mutation are indirect manifestations of perturbation of the critical Lys-334 side chain is correct, our data allow a further conclusion to be drawn about the role of Lys-334. Whereas the mutation decreases the Vmax/Km value for carboxylation by 50%, it doubles the analogous quotient for oxygenation (Table I). This means that the mutant enzyme is twice as effective as the wild type in handling limiting concentrations of its substrate, O2, presumably because the transition state associated with O2 addition to the enediol has a lower energy in the mutant's active site than in the wild type's. Therefore, correct position and alignment of Lys-334 must not only facilitate attack by CO2 on the enediol intermediate but also actively impede attack by O2.
Effect of the Val-335 Mutation on Carbamylation
At high CO2, the potential Rubisco activity in wild-type plants substantially exceeds the rate of electron transport and CO2 assimilation becomes limited by electron transport. This can be seen as the progressively larger shortfall of the measured assimilation rate, as compared with that modeled, for the control as the CO2 concentration increased (Fig. 6A). The shortfall would be larger and start to occur at lower CO2 concentrations under the growth illumination (400 μmol quanta m−2 s−1) than under the higher measurement illumination (1,000 μmol quanta m−2 s−1). When potential Rubisco activity is excessive, actual activity is reduced by reducing Rubisco carbamylation by a control mechanism involving the response of Rubisco activase activity to the abundance of electron-transport products or to the size of the transthylakoid pH gradient.
The low carbamylation status of Rubisco in the wild type under the high-CO2 growth conditions (Table I) is therefore the expected response to the scarcity of electron-transport products (or low ΔpH) induced by Calvin-cycle overcapacity at high CO2. By contrast, assimilation by plants with the Val-335 mutant Rubisco never encounters the electron-transport limitation, even at high CO2 concentration (Fig. 6A). As a result, electron-transport products remain abundant (and ΔpH remains high) at high CO2, contributing to the observed near-complete Rubisco carbamylation (Table I).
The Physiological Phenotype Is Consistent with the Changes in Rubisco Properties
Except for the modest reduction in Km(CO2) seen with the mutant enzyme in vitro, all Rubisco kinetic parameters calculated from gas-exchange measurements agreed well with those measured in vitro with the isolated enzyme (Table I). This consistency, in the face of the gross perturbation of the balance between photosynthesis and photorespiration caused by the Val-335 mutation, validates the concepts on which the photosynthesis/photorespiration model (Farquhar et al., 1980) is based. The net rate of photosynthetic/photorespiratory CO2 assimilation (per carbamylated Rubisco site) in air calculated from the in vitro data for the plant with the mutant Rubisco was only 5% that of the wild type (Table I). This would be barely sufficient to compensate for nonphotorespiratory CO2 release, leading to a CO2-compensation point for the mutant plant near current atmospheric CO2 concentration, as observed by gas exchange (Fig. 6). Therefore, the mutant plants would be barely able to maintain positive carbon balance in air, explaining their inability to grow.
Even though CO2 enrichment to 0.3% (v/v) strongly suppresses the oxygenation activity of the mutant Rubisco, it cannot compensate for the large reduction in Vcmax. The mutant plant's calculated net rate of photosynthetic/photorespiratory CO2 assimilation (per carbamylated Rubisco site) in 0.3% (v/v) CO2 was only 21% that of the control plant under similar conditions (Table I). Therefore, the impairment of the mutant plants' growth, relative to that of the controls under CO2-enriched conditions (Figs. 4 and 5) is to be expected. Other tobacco lines with reduced Rubisco capacity induced by antisense suppression of Rubisco or Rubisco activase content also grew more slowly than the wild type both with and without CO2 supplementation (Fichtner et al., 1993; Masle et al., 1993; Mate et al., 1993; He et al., 1997).
CONCLUSION
Inability to conduct site-specific mutagenesis with the higher plant Rubisco has been a major hindrance to the study of its structure and function. The strategy we have used surmounts this barrier and opens the way for further detailed investigations. Provision of the mutant Rubisco in its natural stromal environment for physiological study is an outstanding bonus.
ACKNOWLEDGMENTS
We thank J. Jones for supplying seeds of tobacco cv Petit Havana (N,N), M. Sugiura for supplying plasmids pTB22 and pTB29, P. Maliga for supplying plasmid pRV112A, and P. Maliga and J.-W. Liu for advice about the biolistic technique. Some of these data were communicated at the XIth International Congress on Photosynthesis (Budapest, August 17–22, 1998).
Footnotes
This work was supported by the Australian National University's Centre for Molecular Structure and Function.
LITERATURE CITED
- Andersson I. Large structures at high resolution: the 1.6Å crystal structure of spinach ribulose-1,5-bisphosphate carboxylase/oxygenase complexed with 2-carboxyarabinitol bisphosphate. J Mol Biol. 1996;259:160–174. doi: 10.1006/jmbi.1996.0310. [DOI] [PubMed] [Google Scholar]
- Bock R. Analysis of RNA editing in plastids. Methods. 1998;15:75–83. doi: 10.1006/meth.1998.0607. [DOI] [PubMed] [Google Scholar]
- Butz ND, Sharkey TD. Activity ratios of ribulose-1,5-bisphosphate carboxylase accurately reflect carbamylation ratios. Plant Physiol. 1989;89:735–739. doi: 10.1104/pp.89.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng WP, Nickoloff JA. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal Biochem. 1992;200:81–88. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- Farquhar GD, von Caemmerer S, Berry JA. A biochemical model of photosynthetic CO2 assimilation in leaves of C3 species. Planta. 1980;149:78–90. doi: 10.1007/BF00386231. [DOI] [PubMed] [Google Scholar]
- Fichtner K, Quick WP, Schulze E-D, Mooney HA, Rodermel SR, Bogorad L, Stitt M. Decreased ribulose-1,5-bisphosphate carboxylase-oxygenase in transgenic tobacco transformed with “antisense” rbcS. V. Relationship between photosynthetic rate, storage strategy, biomass allocation and vegetative plant growth at three different nitrogen supplies. Planta. 1993;190:1–9. [Google Scholar]
- Gutteridge S, Gatenby AA. Rubisco synthesis, assembly, mechanism, and regulation. Plant Cell. 1995;7:809–819. doi: 10.1105/tpc.7.7.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman FC, Harpel MR. Structure, function, regulation, and assembly of d-ribulose-1,5-bisphosphate carboxylase/oxygenase. Annu Rev Biochem. 1994;63:197–234. doi: 10.1146/annurev.bi.63.070194.001213. [DOI] [PubMed] [Google Scholar]
- He Z, von Caemmerer S, Hudson GS, Price GD, Badger MR, Andrews TJ. Ribulose-1,5-bisphosphate carboxylase/oxygenase activase deficiency delays senescence of ribulose-1,5-bisphosphate carboxylase/oxygenase but progressively impairs its catalysis during tobacco leaf development. Plant Physiol. 1997;115:1569–1580. doi: 10.1104/pp.115.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husic DW, Husic HD, Tolbert NE. The oxidative photosynthetic carbon cycle or C2 cycle. CRC Crit Rev Plant Sci. 1987;5:45–100. [Google Scholar]
- Kane HJ, Viil J, Entsch B, Paul K, Morell MK, Andrews TJ. An improved method for measuring the CO2/O2 specificity of ribulosebisphosphate carboxylase-oxygenase. Aust J Plant Physiol. 1994;21:449–461. [Google Scholar]
- Kanevski I, Maliga P. Relocation of the plastid rbcL gene to the nucleus yields functional ribulose-1,5-bisphosphate carboxylase in tobacco chloroplasts. Proc Natl Acad Sci USA. 1994;91:1969–1973. doi: 10.1073/pnas.91.5.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanevski I, Maliga P, Rhoades DF, Gutteridge S. Plastome engineering of ribulose-1,5-bisphosphate carboxylase/oxygenase in tobacco to form a sunflower large subunit and tobacco small subunit hybrid. Plant Physiol. 1999;119:133–141. doi: 10.1104/pp.119.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GJ, McDonald KA, McFadden BA. Leucine 332 influences the CO2/O2 specificity factor of ribulose-1,5-bisphosphate carboxylase/oxygenase from Anacystis nidulans. Protein Sci. 1993;2:1147–1154. doi: 10.1002/pro.5560020709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorimer GH, Chen Y-R, Hartman FC. A role for the ε-amino group of lysine-334 of ribulose-1,5-bisphosphate carboxylase in the addition of carbon dioxide to the 2,3-enediol(ate) of ribulose 1,5-bisphosphate. Biochemistry. 1993;32:9018–9024. doi: 10.1021/bi00086a006. [DOI] [PubMed] [Google Scholar]
- Maliga P. Towards plastid transformation in flowering plants. Trends Biotechnol. 1993;11:101–107. [Google Scholar]
- Masle J, Hudson GS, Badger MR. Effects of ambient CO2 concentration on growth and nitrogen use in tobacco (Nicotiana tabacum) plants transformed with an antisense gene to the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase. Plant Physiol. 1993;103:1075–1088. doi: 10.1104/pp.103.4.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mate CJ, Hudson GS, von Caemmerer S, Evans JR, Andrews TJ. Reduction of ribulose bisphosphate carboxylase activase levels in tobacco (Nicotiana tabacum) by antisense RNA reduces ribulose bisphosphate carboxylase carbamylation and impairs photosynthesis. Plant Physiol. 1993;102:1119–1128. doi: 10.1104/pp.102.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead DA, Szczesna-Skorupa E, Kemper B. Single-stranded DNA “blue” T7 promoter plasmids: a versatile tandem promoter system for cloning and protein engineering. Protein Eng. 1986;1:67–74. doi: 10.1093/protein/1.1.67. [DOI] [PubMed] [Google Scholar]
- Newman J. Structural studies of ribulose-1,5-bisphosphate carboxylase/oxygenase from the cyanobacterium Synechococcus PCC 6301. PhD Thesis. Uppsala: Swedish University of Agricultural Sciences; 1992. [Google Scholar]
- Paul K, Morell MK, Andrews TJ. Mutations in the small subunit of ribulosebisphosphate carboxylase affect subunit binding and catalysis. Biochemistry. 1991;30:10019–10026. doi: 10.1021/bi00105a029. [DOI] [PubMed] [Google Scholar]
- Roy H, Andrews TJ. Rubisco: assembly and mechanism. In: Leegood RC, Sharkey TD, von Caemmerer S, editors. Photosynthesis: Physiology and Metabolism. Dordrecht, the Netherlands: Kluwer Academic Publishers; 1999. , in press. [Google Scholar]
- Ruuska SA, Andrews TJ, Badger MR, Hudson GS, Laisk A, Price GD, von Caemmerer S. The interplay between limiting processes in C3 photosynthesis studied by rapid-response gas exchange using transgenic tobacco impaired in photosynthesis. Aust J Plant Physiol. 1998;25:859–870. [Google Scholar]
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci USA. 1984;81:8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schreuder HA, Knight S, Curmi PMG, Andersson I, Cascio D, Brändén C-I, Eisenberg D. Formation of the active site of ribulose-1,5-bisphosphate carboxylase/oxygenase by a disorder-order transition from the unactivated to the activated form. Proc Natl Acad Sci USA. 1993;90:9968–9972. doi: 10.1073/pnas.90.21.9968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zoutu N, Sugiura M. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreitzer RJ. Genetic dissection of Rubisco structure and function. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:411–434. [Google Scholar]
- Spreitzer RJ. Genetic engineering of Rubisco. In: Rochaix J-D, Goldschmidt-Clermont M, Merchant S, editors. The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. [Google Scholar]
- Sugiura M, Shinozaki K, Zoutu N, Kusuda M, Kumano M. Clone bank of the tobacco (Nicotiana tabacum) chloroplast genome as a set of overlapping restriction endonuclease fragments: mapping of eleven ribosomal protein genes. Plant Sci. 1986;44:211–216. [Google Scholar]
- Svab Z, Maliga P. High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc Natl Acad Sci USA. 1993;90:913–917. doi: 10.1073/pnas.90.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi BE, Laing WA, Christeller JT, Petersen GB, Hill DF. Ribulose 1,5-bisphosphate carboxylase: effect on the catalytic properties of changing methionine-330 to leucine in the Rhodospirillum rubrum enzyme. Biochem J. 1986;235:839–846. doi: 10.1042/bj2350839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Caemmerer S, Evans JR, Hudson GS, Andrews TJ. The kinetics of ribulose-1,5-bisphosphate carboxylase/oxygenase in vivo inferred from measurements of photosynthesis in leaves of transgenic tobacco. Planta. 1994;195:88–97. [Google Scholar]
- Zoubenko OV, Allison LA, Svab Z, Maliga P. Efficient targeting of foreign genes into the tobacco plastid genome. Nucleic Acids Res. 1994;22:3819–3824. doi: 10.1093/nar/22.19.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]