Abstract
In both vertebrate and invertebrate brains, neurons, glial cells and synapses are plastic, which means that the physiology and structure of these components are modified in response to internal and external stimuli during development and in mature brains. The term plasticity has been introduced in the last century to describe experience-dependent changes in synapse strength and number. These changes result from local functional and morphological synapse modifications; however, these modifications also occur more commonly in pre- and postsynaptic neurons. As a result, neuron morphology and neuronal networks are constantly modified during the life of animals and humans in response to different stimuli. Nevertheless, it has been discovered in flies and mammals that the number of synapses and size and shape of neurons also oscillate during the day. In most cases, these rhythms are circadian since they are generated by endogenous circadian clocks; however, some rhythmic changes in neuron morphology and synapse number and structure are controlled directly by environmental cues or by both external cues and circadian clocks. When the circadian clock is involved in generating cyclic changes in the nervous system, this type of plasticity is called circadian plasticity. It seems to be important in processing sensory information, in learning and in memory. Disruption of the clock may affect major brain functions.
Keywords: circadian rhythm, circadian clock, synaptic plasticity, neural plasticity, Drosophila melanogaster, mice, rat
Learning and Experience-Dependent Plasticity
Over the last few decades, a wide body of experimental data have demonstrated that the structure and physiology of the nervous system change in response to internal and environmental stimuli. This ability of the nervous system to develop lasting changes in neuron properties that enable adaptation to the environment and regeneration and that underlie the mechanism of memory formation and learning is defined as plasticity. Importantly, changes in neuron morphology, neuronal connectivity, biochemical processes or even new neuron generation are observed during development and also occur widely in the adult nervous system (for review, see Bruel-Jungerman et al., 2007; Sweatt, 2016).
The term “plasticity” was introduced to neurobiology at the beginning of the 20th century (reviewed in Stahnisch and Nitsch, 2002); however, modern theories explaining mechanisms of plasticity of the nervous system have their origin in Hebb’s postulate (Hebb, 1949). The first attempts to describe neural plasticity in relation to learning and memory as well as synaptic plasticity were taken later, and since the late 1960s/early 1970s, the terms “plasticity” and “neuroplasticity” have become common in neurobiology (see Berlucchi and Buchtel, 2009; Fuchs and Flügge, 2014). A breakthrough in understanding synaptic plasticity mechanisms was the discovery that brief trains of electrical stimuli result in increased transmission efficiency at the perforant path-granule cell synapse in the rabbit hippocampus (HC), as measured with electrophysiological recording methods, that can last for hours (Bliss and Lømo, 1973; Lømo, 2003). Over the years, this phenomenon, which was termed long-term potentiation (LTP), has generally been accepted as one of the most popular models of the cellular processes underlying memory (reviewed in Bruel-Jungerman et al., 2007; Sweatt, 2016).
The structural modifications accompanying synaptic plasticity fall into two categories: modifications of existing connections and changes in the number of synapses. Pre-existing synapses are regulated in many ways, such as by the phosphorylation of postsynaptic neurotransmitter receptors or the insertion of additional receptor molecules into postsynaptic membranes, to change their efficiency. On the other hand, the LTP-induced increase in the number of functional synapses was also confirmed in several experiments, and it has been found that changes in the number of synapses are correlated with the reorganization of spine and dendrite morphology (reviewed in Bailey and Kandel, 1993; Kondo, 2017).
Recently, it was reported that LTP induction in the rodent HC varies across a 24 h day both in vivo and in vitro (Chaudhury et al., 2005; Bowden et al., 2012; Nakatsuka and Natsume, 2014). It is also important to stress that in humans, time-of-day variations have been described for cognitive domains (Schmidt et al., 2007). For example, chronic jet lag (i.e., caused by traveling across time zones) and shift work disrupt the sleep-wake cycle and produce cognitive disturbances (Cho et al., 2000; Rouch et al., 2005).
In this review, we described a special type of neural plasticity, circadian plasticity, that is driven by the circadian clock and synchronized to daily changes in environmental cues. This type of plasticity has been reported in the nervous system of many vertebrate and invertebrate species; however, in this review we focused on rodents and Drosophila melanogaster (fruit fly) as the most studied animals in the field of circadian plasticity, including our own studies. Nevertheless, the role of biological clocks in brain plasticity has been reviewed recently by e.g., Frank and Cantera (2014), Iyer et al. (2014), Smarr et al. (2014), Bosler et al. (2015) and Frank (2016).
Circadian Clock
Circadian rhythms are periodic oscillations that occur within a span of approximately 24 h. They are present in most animal species and are examples of the biological rhythms that have a profound influence on many functions of an organism, ranging from metabolism to complex behaviors. These rhythms are driven by molecular clocks that evolved to synchronize biological processes with time and cyclic changes in the environment. Circadian rhythms allow organisms to predict the coming day and night and to adapt to different conditions during the day in advance. Although the molecular components of the clock are not conserved across the main divisions of life, the molecular mechanism of the circadian clock in most species is based on negative and positive transcriptional–translational feedback loops, in which transcription factors activate the expression of their own repressors (reviewed i.e., by Rosbash, 2009). Importantly, this molecular clock is composed of core clock genes/proteins and regulates the rhythmic expression of other genes (so-called clock-controlled genes, ccgs) in various tissues (Doherty and Kay, 2010).
Drosophila Clock Neurons
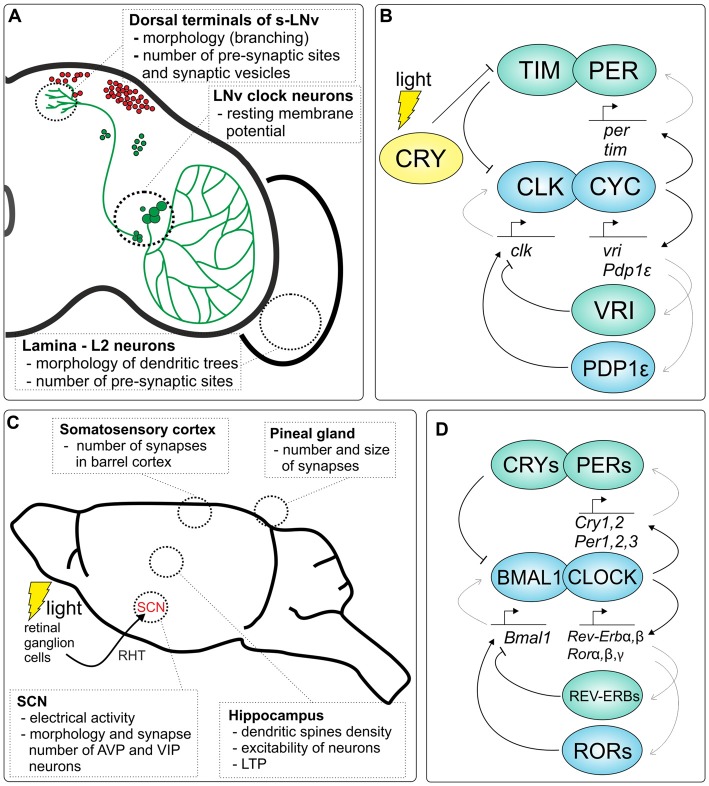
In insects, most of the early work on circadian rhythms and clocks has been done on large non-model species such as cockroaches and crickets (Nishiitsutsuji-Uwo and Pittendrigh, 1968; Page, 1982; Tomioka, 1985). Nevertheless, the clock has been best characterized in D. melanogaster, a common model organism in genetics, development and neurobiology. Approximately 150 neurons, in the fruit fly’s brain, have been identified as clock neurons (Figure 1A), based on the cyclic expression of so-called clock genes and clock proteins (described in Section “Molecular mechanisms of the circadian clock”). These cells are located in distinct clusters and are divided into seven major groups, based on their anatomical position, comprising three groups of dorsal neurons (DN; DN1–3) and four groups of lateral neurons (LNds, LPNs, l-LNvs and s-LNvs). The large and small ventral lateral neurons (l-LNvs and s-LNvs, respectively) are the best-studied clock neurons of D. melanogaster, as they express pigment-dispersing factor (PDF), a neurotransmitter important for clock neuron internal synchronization and for the output pathways of the fly’s circadian clock (Kaneko and Hall, 2000; Helfrich-Förster, 2003, 2005; Rieger et al., 2006; Yoshii et al., 2008; Damulewicz and Pyza, 2011; Damulewicz et al., 2013; Beckwith and Ceriani, 2015).
Figure 1.
Clock-dependent plasticity in the brain of Drosophila and rodents. (A) Examples of circadian plasticity in the Drosophila brain. The localization of DN1–3 clock neurons in the brain is shown in red, while lateral clock neurons and their pigment-dispersing factor (PDF)-positive projections are depicted in green. (B) Molecular mechanism of the fruit fly circadian clock. Two major transcriptional/translational feedback loops are shown, which are regulated by the CLOCK/CYCLE (CLK/CYC) dimer, TIMELESS/PERIOD (TIM/PER) and VRILLE/PAR DOMAIN PROTEIN 1ε (VRI/PDP1ε) loops (activators are marked in blue, while inhibitors are green). Light-activated CRYPTOCHROME (CRY) protein binds TIM to initiate proteasomal degradation which allows to synchronize the clock to external light conditions. (C) A schematic representation of the rodent brain with examples of circadian plasticity in different structures of the brain. The clock in the suprachiasmatic nucleus (SCN) is synchronized by light through the retina; mainly photosensitive retinal ganglion cells (melanopsin cells) are involved in clock light perception via the retinohypothalamic tract (RHT). (D) Molecular clock in mammals. Two major transcriptional/translational feedback loops, PER/CRY and REV-ERB/ROR, are regulated by the CLOCK/BMAL1 dimer (activators are marked in blue, while inhibitors are green).
Circadian Clock in Mammals
Regarding the structure in the mammalian brain responsible for maintaining circadian rhythms, transplantations (Aguilar-Roblero et al., 1986; Lehman et al., 1987; Ralph et al., 1990; Sollars et al., 1995; Sujino et al., 2003) and surgical lesions (Moore and Eichler, 1972; Stephan and Zucker, 1972; Mouret et al., 1978) of the hypothalamus have shown that the primary circadian oscillator is located in the ventral periventricular zone of the hypothalamus, in the suprachiasmatic nucleus (SCN; reviewed in Buhr and Takahashi, 2013). The direct synchronization of this circadian clock with environmental changes in light and darkness (LD) is possible by photosensitive retinal ganglion cells via the retinohypothalamic tract (RHT), which is the retinal projection terminating in the SCN (Hendrickson et al., 1972; Moore and Lenn, 1972; reviewed in Hughes et al., 2015; Figure 1C). Initially, it was believed that the SCN is the only structure capable of integrating environmental cues and driving internally sustained rhythms of cellular activity and gene expression, which is why it was named the “master clock” or “pacemaker”. However, recent studies have revealed that peripheral oscillators located in other tissues in the body are also enmeshed within the regulation of numerous metabolic pathways. These local clocks are able to generate the rhythms even when they are separated from the brain. Thus, recently, it is believed that the SCN does not initiate but rather synchronizes different peripheral oscillators throughout the body to a uniform time (Buhr and Takahashi, 2013).
Molecular Mechanisms of the Circadian Clock
The core molecular components that underlie the generation of circadian oscillations in animals are best described in mammals and the fruit fly (Figures 1B,D). The current model of the clock at the molecular level is based on two main interconnected transcriptional/translational feedback loops that function together to generate cyclic gene expression. At the heart of the molecular clock, CLOCK (or its analog NPAS2) and BMAL1 in mammals, and CLOCK (CLK) and CYCLE (CYC) in Drosophila have been identified as transcriptional activators.
In Drosophila CLK and CYC form heterodimers and activate the expression of period (per) and timeless (tim) which encode their own repressors PER and TIM proteins in the primary feedback loop. CLK/CYC heterodimers also regulate their own expression via the second loop by the activation of vrille (vri) and Par domain protein 1ε (Pdp1ε) transcription. Both VRI and PDP1ε proteins bind to the promoter region of Clk, however VRI inhibits, while PDP1ε activates, Clk transcription. In addition, these transcriptional feedback loops are additionally regulated posttranslationally by numerous cytoplasmic proteins, e.g., kinases, that together generate endogenous circadian rhythms at the level of mRNAs and proteins of the molecular oscillator. In addition, CLK/CYC regulates the expression of other genes, so-called ccgs, which drive circadian rhythms in physiological and cellular processes. The circadian clock of flies also possesses its own photoreceptor, the CRYPTOCHROME (CRY) that is involved in light entrainment.
In mammals, in the first loop, after dimerization and translocation to the nucleus, CLOCK/BMAL1 initiates the transcription of Per1, Per2, Per3 and Cry1, Cry2 encoding their own repressors: PERs and CRYs. In the second loop, CLOCK/BMAL1 regulates their own transcription by controlling the expression of two other genes, Rorα/β/γ and Rev-erbα/β, that encode retinoid acid receptor-related orphan receptors (RORs) and orphan nuclear receptors (REV-ERBs), respectively. Both, RORs and REV-ERBs regulate the expression of Bmal1; however, RORs act as activators while REV-ERBs are repressors. As in Drosophila CLOCK/BMAL1 also regulates the expression of ccgs.
For the most recent and more detailed reviews on the molecular mechanism of the circadian clock in mammals, see Buhr and Takahashi (2013), and for the fly’s clock, see Özkaya and Rosato (2012) or Tataroglu and Emery (2015).
The existence of output pathways of the circadian clock enables the regulation of many genes. Genome-wide studies have identified numerous rhythmically expressed genes in non-neuronal tissues (Storch et al., 2002) and the nervous system. In the human brain (Li et al., 2013) and different regions of the mouse brain, including the HC (Renaud et al., 2015), prefrontal cortex (Yang et al., 2007), cerebellum, hypothalamus and brain stem (Zhang et al., 2014), dozens of genes exhibit daily rhythms of transcriptional activity. Significant differences in the level of many transcripts during the day and night have also been identified in the cerebral cortex (Cirelli et al., 2004) and in the rat pineal gland (Fukuhara and Tosini, 2008; Bailey et al., 2009). Diurnal and circadian expression of several genes has also been confirmed in insect heads (Claridge-Chang et al., 2001; McDonald and Rosbash, 2001; Ceriani et al., 2002; Ueda et al., 2002; Wijnen et al., 2006; Keegan et al., 2007; Naeger et al., 2011; Rodriguez-Zas et al., 2012). Interestingly, many of these genes are involved in remodeling neuronal circuits and/or synapses and might be directly involved in the regulation of brain plasticity.
Drosophila Melanogaster as a Model Species to Study Circadian Plasticity in the Brain
The fruit fly’s brain, despite its simplicity, shows pronounced plasticity, being continuously remodeled in response to environmental stimuli and by intrinsic circadian inputs (summarized in Figure 1A). Due to a large number of available genetic tools and the high homology of genes and proteins between Drosophila and mammals, this species is exceptionally useful in research on circadian rhythm and sleep, as well as complex behaviors such as learning and memory, and in research on circadian plasticity (Heisenberg et al., 1995; Greenspan and Dierick, 2004; Olsen and Wilson, 2008; Pyza, 2013).
Clock Neuron Circadian Plasticity
Neurons that comprise the central clock not only regulate the circadian plasticity in the brain but also undergo circadian remodeling of their own processes. These morphological changes, besides the circadian release of PDF (Park et al., 2000), contribute to the downstream transmission of information. Indeed, the dorsal terminals of s-LNvs are more complex in the morning than during the early night. These changes are maintained in constant darkness (DD) but are abolished in per01 and tim01 clock mutants, confirming their circadian origin (Fernández et al., 2008). Recently, it has been found that the retraction of s-LNv axonal terminals at dusk depends on actin and myosin light-chain phosphorylation, which in turn is regulated by clock-controlled Rho1 protein activity (Petsakou et al., 2015). Circadian oscillations in the morphology of neurites have also been detected in the housefly‘s clock neurons (Pyza and Meinertzhagen, 1997), while daily structural changes have been observed in the PDF-immunoreactive fibers of the cockroach Leucophaea maderae (Wei and Stengl, 2011).
Circadian changes in the morphology of PDF-positive fruit fly neurons are accompanied by changes in the number of synapses. The number of active zones labeled by antibody against Bruchpilot, a presynaptic scaffolding protein, and the number of vesicles detected by anti-Synaptotagmin serum are increased at the beginning of the subjective day in DD. Moreover, s-LNvs seem to form synaptic contacts with different partners, which allows these neurons to regulate different processes in the brain during the day and night. These results revealed that circadian network activity in the brain can be regulated not only by modulating synapse strength between different cells but also by changes in the number of synaptic contacts between particular groups of neurons (Gorostiza et al., 2014).
Clock neuron plasticity is accompanied by rhythmic changes in cell membrane excitability that are critical for maintaining their function. Electrical silencing of the pacemaker neurons by targeting K+ channel expression inhibits the clock and leads to arrhythmic behavior (Nitabach et al., 2002). On the other hand, whole-cell recordings from a large subset of LNvs have shown that their resting membrane potential is modulated by both the clock and light input (Park and Griffith, 2006). Other studies also confirmed the circadian origin of membrane excitability, as well as the pattern and action potential firing rate of lateral ventral clock neurons (Cao and Nitabach, 2008; Sheeba et al., 2008).
The circadian remodeling of s-LNv terminals and the ability of s-LNvs to change axonal arbor conformation in response to neuronal firing requires myocyte enhancer factor 2 (Mef2), and the Mef2 gene is a direct target of the master circadian regulator dimer CLK/CYC. Therefore, it seems that circadian information is transmitted to the machinery involved in neuronal remodeling via the Mef2 protein (Sivachenko et al., 2013). Interestingly, in the mammalian nervous system, MEF2 suppresses the number of excitatory synapses and thus regulates the strength of neuronal connections (Flavell et al., 2006).
Circadian Plasticity in the Visual System
Since the function of cells in the visual system of D. melanogaster is similar to the function of cells in the retina of vertebrates, the visual system of the fruit fly is a good model for studying various processes, e.g., circadian rhythms, at the cellular level (Pyza, 2002). The visual system of D. melanogaster, as in other insect species, is composed of the retina and three optic neuropils: the lamina, medulla and lobula complex. Circadian changes in neuronal morphology in the nervous system were first described by Pyza and Meinertzhagen (1993, 1995) in the housefly (Musca domestica), the blowfly (Calliphora vicina, Pyza and Cymborowski, 2001) and also in the fruit fly (Pyza and Meinertzhagen, 1999).
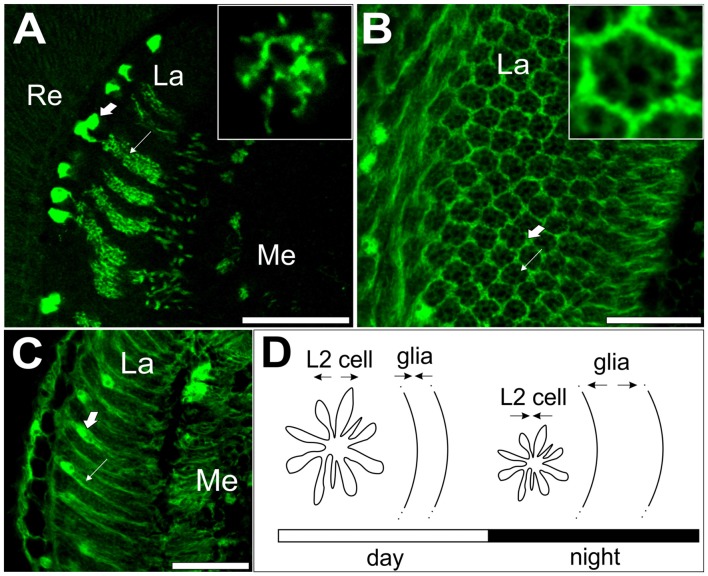
For example, in the retina of D. melanogaster, the enzyme hemeoxygenase that metabolizes heme to carbon monoxide, ferrous iron and biliverdin affects the molecular mechanism of the circadian clock in photoreceptors (Damulewicz et al., 2017). However, the most robust circadian rhythms have been described in the lamina. The cross-sectional area of L1 and L2 monopolar cells, the first order interneurons in the lamina, increases twice a day: in the morning and in the evening (Pyza and Meinertzhagen, 1999). Similarly, analysis of GFP-labeled L2 cells in the lamina revealed the daily changes in the L2 dendrite length, with longest dendrites at the beginning of both day and night (Figures 2A,D; Górska-Andrzejak et al., 2005). Rhythmic changes in the size of the L2 dendritic tree are controlled by the circadian clock since the rhythm is still present in constant darkness, while in the null per01 clock mutants, the rhythm is abolished, and the dendrites are shorter than those in wild-type flies. Moreover, in cryb mutants, the morning peak of the L2 dendritic tree perimeter is shifted, which indicates that CRY is important for setting the phase of the circadian oscillation in L2 dendrite morphology (Weber et al., 2009). The involvement of core clock genes in neuronal morphology regulation has also been described for s-LNvs. In tim01 mutants, the axons of these neurons are shorter and hyperbranched, while in per01 they are longer with less complicated branching (Fernández et al., 2008).
Figure 2.
L2 interneurons and glial cells showing circadian plasticity in the visual system of Drosophila melanogaster. (A) GFP-labeled L2 monopolar cells, first order interneurons, in the optic lobe lamina. Large arrow indicates L2 somata; small arrow indicates L2 axons with dendrites. Insert: cross section of an L2 axon with dendrites. (B) Cross section of the lamina with GFP-labeled epithelial glial cells (large arrow) surrounding lamina cartridges (small arrow). Insert: a single cartridge surrounded by three epithelial glial cells. (C) GFP-labeled epithelial glial cells in the lamina. Large arrow indicates the soma; small arrow indicates processes of epithelial glial cells. (D) A schematic representation of circadian plasticity in the L2 dendritic tree and epithelial glial cells. Changes in neuron size are offset by changes in glia size. Re, retina, La, lamina, Me, medulla of the optic lobe. Scale bar: 20 μm (Pyza and Górska-Andrzejak, 2004; Weber et al., 2009).
Similar to the circadian plasticity of the PDF-positive neurons of the central clock, changes in the morphology of non-clock interneurons in the visual system of D. melanogaster seem to be correlated with the circadian oscillation in the number of synapses formed between photoreceptors and postsynaptic cells, including L1 and L2 interneurons. In the lamina, two types of synapses, tetrad and feedback, have been examined during the 24 h cycle. Tetrad synapses, formed between the photoreceptor terminal R1-R6 and four postsynaptic cells, constitute the majority of synapses in the lamina, while feedback synapses, between L2 back onto R1-R6, are less numerous. The presynaptic profiles of tetrad synapses exhibit a bimodal rhythm, with two peaks in the morning and in the evening, whereas the feedback synapse frequency peaks at night (Woźnicka et al., 2015). Remodeling of these synaptic contacts is correlated with daily or circadian fluctuations in synaptic protein levels, such as Bruchpilot (Górska-Andrzejak et al., 2013), Synapsin and Disc large (Krzeptowski et al., 2014).
Structural circadian plasticity in the visual system of the fruit fly is modulated by PDF and ion transport peptide (ITP) produced by the 5th small LNv that projects to the lamina (Damulewicz and Pyza, 2011; Damulewicz et al., 2013, 2015). Moreover, it appears that clock-controlled activity in the D. melanogaster visual system is regulated by a mechanism based on Na+/K+-ATPase (Górska-Andrzejak et al., 2009; Damulewicz et al., 2013, 2015). This type of clock-controlled regulation has also been described in the mammalian SCN (Wang and Huang, 2004).
Circadian Changes in Motor Terminals
Another example of circadian-dependent neural plasticity described in D. melanogaster has been found in the MN5 motor neuron that innervates two longitudinal indirect flight muscles. In flies reared in LD conditions, the size of MN5 terminal boutons is significantly larger in the middle of the day than in the middle of the night, and this rhythm is maintained in constant darkness, which proves its regulation by the circadian clock. These findings have been confirmed in per and tim clock gene mutants, which lacked daily changes in the size of MN5 boutons (Mehnert et al., 2007). Similar to the results obtained studying PDF-positive s-LNv terminals (Fernández et al., 2008), per01 mutants presented with significantly fewer branches than wild-type flies, while the tim mutation causes hyperbranching of axonal projections. This type of plasticity in D. melanogaster seems to be regulated locally by a peripheral clock located in the thoracic ganglia, as the rhythm remains intact in decapitated flies (Mehnert and Cantera, 2008). The most recent study revealed rhythmic changes in the number of motor neuron synapses in both LD and DD conditions and light-dependent mechanisms in the regulation of synapse number throughout the day (Ruiz et al., 2013), similar to those discovered in the lamina (Górska-Andrzejak et al., 2013; Woźnicka et al., 2015).
Memory Formation in D. melanogaster
It has been reported that short-term associative memory formation is modulated by the circadian clock in Drosophila. Lyons and Roman (2008) found that a particular time of day improves memory formation and the performance peak occurs during the early night/subjective night in both LD and DD. Moreover, this rhythm is absent in per01 and tim01 mutants as well as in constant light conditions (LL); however, this rhythm is present in cryb flies. These results indicate that the central clock, not peripheral oscillators or the perception of stimuli, regulates the formation of short-term associative memory (Lyons and Roman, 2008). Such modulation of memory formation might be controlled by the s-LNvs that have projections near the calyx of mushroom bodies, which are memory-processing structures in the insect’s brain (Heisenberg, 1998; Helfrich-Förster et al., 2002).
Cyclic Plasticity in the Mammalian Brain
Several studies have demonstrated that rapid changes in the efficiency of existing synapses and modifications to already formed neural networks are important for nervous system plasticity in mammals. This process can be regulated, among other factors, by environmental light/dark cycles and by the endogenous circadian clock. Importantly, these daily or circadian structural changes in the adult nervous system have been described not only in the SCN but also in other regions of the brain, e.g., the HC, and in the retina (summarized in Figure 1C).
Circadian Plasticity of the SCN
As mentioned above, the SCN is a master clock in mammals that synchronizes circadian peripheral clocks throughout the body and, like in the fruit fly, undergoes daily activity and structure reorganization. In the SCN, circadian rhythms in neural activity seem to be crucial for proper central clock function (Brown and Piggins, 2007; Colwell, 2011). Multiunit in vivo recordings of diurnal electrical activity patterns in the rat SCN and experiments conducted in constant darkness (Inouye and Kawamura, 1979, 1982), as well as the spontaneous firing rate of rat SCN neurons recorded in vitro in hypothalamic slice preparations (Green and Gillette, 1982; Groos and Hendriks, 1982; Shibata et al., 1982), have demonstrated the circadian nature of the mentioned rhythms.
Indeed, rhythmic oscillations of the membrane potential in the SCN are essential for neuropeptide release and thus contribute to clock-based plasticity (reviewed in Iyer et al., 2014). Recent studies focused on the rearrangement of two main sources of the SCN efferents: neurons expressing arginine vasopressin (AVP) and vasoactive intestinal peptide (VIP). Becquet et al. (2008) reported a decrease in the mean coverage of somata and VIP neuron dendrites during night-time and, conversely, an increase in the extent of somata and dendritic membrane appositions involving AVP neurons. In turn, the results obtained by Girardet et al. (2010) showed that synaptic contacts on neurons expressing VIP increase during the day but AVP neuron connections remain unchanged. These data are similar to those observed in the clock neurons of Drosophila (described in the previous paragraph) and support the hypothesis that the SCN structure undergoes circadian plasticity, which is necessary for the central clock to synchronize with the LD cycle.
Circadian Changes in the Hippocampus
The SCN is connected directly and indirectly to many brain regions, including the HC, a structure that is crucial for learning and memory formation. Early in vivo experiments performed by Barnes et al. (1977), revealed that basal synaptic transmission, measured as the amplitude of the excitatory postsynaptic potential (EPSP) evoked in the rat dentate gyrus by afferent stimulation, is larger during the night. In contrast to rats, which are nocturnal animals, monkeys exhibit a larger response amplitude during the day. On the other hand, West and Deadwyler (1980) did not find circadian variations in dentate gyrus synaptic currents in rats, but they reported that the amplitude of the population spikes elicited by perforant path axon stimulation is greater during the light phase of the LD cycle. Importantly, these changes were not induced by the intensity of the behavioral arousal or circulating corticosterone level (West and Deadwyler, 1980), indicating that the excitability of granule cells may vary in a circadian-dependent manner.
Harris and Teyler (1983) found that in the rat hippocampal CA1 area, LTP is evoked more readily during the day than during the night, whereas the dentate gyrus develops more robust LTP during the dark phase of the LD cycle. In the CA1 area of hippocampal slices from Syrian hamsters, more robust LTP was also detected during the day (Raghavan et al., 1999). However, in mice, LTP in the CA1 hippocampal area was significantly greater during the night (Chaudhury et al., 2005). The variability in these data is likely a result of differences in experimental approaches, including the recording of either field EPSPs or population spikes and different afferent stimulation paradigms (Nakatsuka and Natsume, 2014). In vivo recording results from rat CA1 pyramidal cells suggest that firing rates go through a sinusoidal cycle on a time scale with a 25-h period (Munn and Bilkey, 2012). This may be related to the observed diurnal differences in LTP magnitude in CA1 when population spike amplitudes are measured. Moreover, recently, it was found that the suppression of local inhibitory connections during the dark phase contributes to larger LTP during that phase (Nakatsuka and Natsume, 2014).
The circadian regulation of neural plasticity is associated with fluctuations in neuron morphology and synaptic contact remodeling. Dendritic spines, carrying postsynaptic elements of synapses, regulate the synaptic strength of neuronal networks. The process of dendritic spine generation and elimination is dynamic and occurs even without any specific environmental stimulation, and spine density fluctuates during the day, as was observed in the Siberian hamster HC (Ikeno et al., 2014). Diurnal changes in the spine density of CA1 pyramidal neurons were also detected in vivo with fixed rat HC, and compared with the sleep state, the highest density was observed upon waking (Ikeda et al., 2015). Interestingly in the mouse HC, adult neurogenesis occurs under the control of clock genes (Bouchard-Cannon et al., 2013; Schnell et al., 2014).
Neural and Synaptic Clock-Dependent Plasticity in Other Parts of the Brain
Other brain regions also appear to be regulated by the circadian clock. For example, dendritic morphology of rat infralimbic cortex neurons changes with diurnal phases of resting and activity. During the animal active period, dendrites are longer and have more spines than during the resting period (Perez-Cruz et al., 2009).
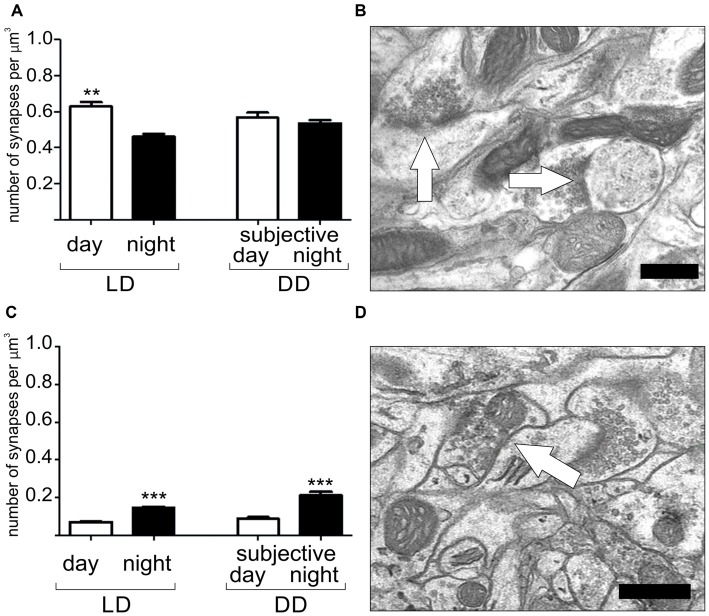
Robust circadian plasticity has been observed in the somatosensory cortex of mice (Figure 3). During the night, when mice are active, sensory input signals, received by whiskers, are transmitted to barrels in the fourth layer of the somatosensory cortex, the whisker representation region. Under LD conditions, the number of excitatory synapses in the barrels is highest when mice are resting (light phase of the LD cycle), while inhibitory synapse density increases during their locomotor activity (dark phase of the LD cycle). Inhibitory synapse density is controlled endogenously by the circadian clock, as the robust rhythm of the number of this type of synapse is also maintained in constant darkness. In contrast, the number of excitatory synapses is influenced by light because their density is constant during both active and rest times in animals kept in DD. These results indicate that the sensory input associated with the circadian rhythm of animal rest and activity regulates daily and circadian rhythms in synapse density in the mouse somatosensory cortex (Jasinska et al., 2014, 2015). In addition to the circadian rhythm in synapse density, Jasinska et al. (2015) have also reported changes in the number of single and double synapse dendritic spines. The increased number of double synapse spines during the dark phase of the LD cycle suggests that, during the night, inhibitory synapses can be added to existing dendritic spines carrying one excitatory synapse.
Figure 3.
Daily and circadian rhythms in the number of excitatory (A,B) and inhibitory synapses (C,D) in the barrel cortex of the mouse somatosensory cortex. Light and darkness (LD) 12:12–12 h of light and 12 h of darkness, DD–constant darkness. Scale bar: 0.5 μm. Graphs show means ± SD, **P < 0.01, ***P < 0.001 (Jasinska et al., 2015).
In vertebrates, cyclic plasticity also concerns so-called ribbon synapses in the pineal gland, the retina and other sensory organs. Analyses of ribbon size by both transmission electron microscopy and fluorescent labeling of synaptic proteins revealed that ribbon length varies in LD conditions. In the pineal gland of rats or guinea pigs, ribbon number and size increase during the night (Vollrath, 1973; Jastrow et al., 1997), while mouse photoreceptor synaptic ribbons also change their shape during the day/night cycle but exhibit the reverse pattern, being longest during the day under LD conditions (Adly et al., 1999; Balkema et al., 2001). Striking changes in the number and shape of ribbon synapses indicate that the number and morphology of these synapses are remodeled during the day.
Circadian synaptic plasticity has also been described in other vertebrate species (Elbaz et al., 2013). For example, in zebrafish larvae, the number of presynaptic elements in hypocretin/orexin (HCRT) neurons exhibits circadian rhythmicity (Appelbaum et al., 2010), and circadian rhythmicity in the synaptic ribbons of goldfish pineal glands was described by McNulty (1981).
Mechanisms of Circadian Plasticity of the Brain
The mechanisms of clock-based brain plasticity are not yet fully recognized. Circadian changes observed in the nervous system probably rely on intracellular fluctuations in circadian clock proteins, the activity of enzymes in clock cells and circadian changes in the expression of small molecules known to participate in neural plasticity that are controlled by the central and peripheral clock system. Each of these molecules might be regulated in different ways.
In mammals, the expression of core clock genes has been confirmed not only in the SCN but also in other brain structures, including the cortex and HC (Namihira et al., 1999; Reick et al., 2001; Shieh, 2003; Jilg et al., 2010; Ono et al., 2015; Riddle et al., 2016; Yamaguchi et al., 2016). Moreover, several studies have shown that clock gene mutations disrupt synaptic plasticity, memory formation and learning. For example, Per1 knockout mice display defects in LTP recorded in vivo (Rawashdeh et al., 2014) and in the HC-dependent long-term spatial learning paradigm (Jilg et al., 2010), while Per2 mutant mice exhibit abnormal hippocampal LTP and deficits in trace recall (Wang et al., 2009). Mutations in other core clock genes also affect brain plasticity, as Npas2 mutant mice show impairments in cued and contextual fear memory (Garcia et al., 2000), while Cry1−/− Cry2−/− mice fail to learn time-place associations, and their electroretinogram is changed, indicating abnormalities in retinal visual processing (Cameron et al., 2008; Van der Zee et al., 2008). In turn, Kondratova et al. (2010) found alterations in short- and long-term memory formation in Bmal1−/− mutants, while Wardlaw et al. (2014) found defects in learning and spatial memory and a reduced LTP magnitude in hippocampal slices from the same mouse strain.
The results obtained by Rawashdeh et al. (2014) highlighted another important issue, namely, daytime deficits in the working memory performance of Per1 knockout mice, which was correlated with a loss of CREB phosphorylation circadian rhythm in the HC. Interestingly, hippocampal plasticity is modulated by effectors also involved in SCN plasticity, such as CREB, BDNF and γ-aminobutyric acid (GABA; Girardet et al., 2013; Schildt et al., 2013; Martin-Fairey and Nunez, 2014; Nakatsuka and Natsume, 2014; Hwang et al., 2016; Rawashdeh et al., 2016; Albers et al., 2017). For example, hippocampal-based learning seems to be strictly correlated with the circadian-controlled regulation of mitogen-activated protein kinase (MAPK) cascade activity, which is crucial for inducing hippocampal LTP and the formation of long-term memory. Moreover, circadian oscillations in cAMP and MAPK activity in the HC are absent in memory-deficient mice that lack calmodulin-dependent adenylyl cyclase activity. In general, physiological or pharmacological disruptions in hippocampal MAPK activity oscillations impair memory persistence (Eckel-Mahan et al., 2008; Eckel-Mahan, 2012).
The existence of common molecular pathways in the SCN and other brain structures, such as the HC, make it difficult to determine whether changes in brain plasticity and learning are regulated by the SCN or by local peripheral clocks. Some results suggest that SCN lesions abolish rhythms of passive avoidance in rats (Stephan and Kovacevic, 1978; Ruby et al., 2008). Furthermore, in arrhythmic Siberian hamsters, the daily variance in novel object learning is abolished (Ruby et al., 2008). However, the most recent reports suggest that the SCN is not necessary for learning. For example, in golden hamsters, after the removal of the SCN, the circadian modulation of conditioned place preference was unaffected, which suggests an extra-SCN origin of the rhythm (Cain and Ralph, 2009). This conclusion is supported by the presence of cyclic Per2 gene expression in the isolated hippocampal slices of those animals (Wang et al., 2009). In turn Storch et al. (2007) showed that a lack of Bmal1 expression in the retina leads to the disruption of retinal electrical responses to light, similar to results in Bmal1−/− mutant mice. Finally, circadian plasticity regulation by peripheral clocks has also been confirmed in the structure of MN5 motor neurons in D. melanogaster (Mehnert and Cantera, 2008). On the other hand, in the HC, the circadian-based regulation of long-term memory via the cAMP/MAPK/CREB transcriptional pathway seems to be maintained by the SCN, as electrolytic lesions of this structure ablate circadian oscillations in hippocampal MAPK pathway activation and lead to long-term memory deficits (Phan et al., 2011). In addition, circadian oscillations in glucocorticoid activity under normal physiological conditions are correlated with the formation and stabilization of new dendritic spines in the HC (Liston et al., 2013). These results indicate that the circadian clock in the SCN may regulate only specific aspects of brain plasticity and that local oscillators also contribute to this process.
Indeed, over the past few years, it has become clear that the central clock (pacemaker) in the brain is a part of the larger network of clocks, including peripheral clocks, which interact with each other and with many other systems within the organism. The coordination of rhythmicity among different cells is a complicated process in which many factors seem to be involved. These neurons might communicate with each other by synaptic and paracrine mechanisms and/or by gap junctions. In the fruit fly, PDF plays a major role in the synchronization and modulation of oscillations between clock neurons and in the transmission of circadian information downstream from clock neurons. It is also possible that additional mechanisms are involved in maintaining circadian rhythms since disrupting fast synaptic transmission in PDF-positive clock cells does not affect the circadian rhythms in behavior (Renn et al., 1999; Kaneko et al., 2000; Park et al., 2000; Blanchardon et al., 2001; Johard et al., 2009; Im and Taghert, 2010; Yao and Shafer, 2014; reviewed also in Muraro et al., 2013; Beckwith and Ceriani, 2015). In the SCN, vasointestinal peptide and other peptides are involved in circadian signaling (Harmar, 2003; O’Neill and Reddy, 2012). Interestingly, despite the same clock core genes and proteins existing in the SCN and peripheral clocks, which are involved in maintaining circadian oscillations, the regulation of their expression might be slightly different. For example, the Per1 and Per2 mRNA levels are controlled by transcription factors other than CLOCK/BMAL1, and their activity is under the control of systemic signals, such as hormones, temperature and secondary messengers [(Travnickova-Bendova et al., 2002; So et al., 2009; Hanyu et al., 2011; Tamaru et al., 2011; Chappuis et al., 2013), reviewed also in an excellent article on circadian synaptic plasticity (Frank, 2016)].
The circadian multioscillatory system is even more complicated since glial cells have also been proposed to act as circadian oscillators in both mammals (Prolo et al., 2005; Yagita et al., 2010) and insects (Siwicki et al., 1988; Zerr et al., 1990; Ewer et al., 1992; Suh and Jackson, 2007). Recently, it has been confirmed by qPCR methods that per and tim genes have rhythmic expression in the glial cells of fruit flies (Damulewicz et al., 2015). Glial cells seem to also be involved in the regulation of neuron circadian plasticity. In the housefly lamina, epithelial glial cells undergo daily changes in morphology (Figures 2B–D; Pyza and Górska-Andrzejak, 2004). More interestingly, glial metabolism disruption or blocking gap junctions affects circadian plasticity in L2 interneurons (Pyza and Górska-Andrzejak, 2004), while disrupting the clock in glia changes the daily pattern of BRP expression in the lamina (Górska-Andrzejak et al., 2013). In mammals, it has been suggested that the circadian clock mechanism within microglia surrounding synapses drives the circadian modulation of dendritic spine density and synaptic strength via cathepsin S (Hayashi et al., 2013). While a detailed description of the role of glia in the regulation of circadian plasticity is outside the scope of this review, readers are encouraged to read the following reviews: Jackson (2011), Jackson et al. (2015) and Chi-Castañeda and Ortega (2016).
Conclusion
During day and night, the brain undergoes significant functional and morphological changes, and some of these rhythms are generated by endogenous mechanisms driven by circadian clocks. These cyclical changes in the efficiency and number of synapses or in the morphology of neurons and glial cells seem to be correlated with locomotor activity in both vertebrates and invertebrates. However, the most common problem in interpreting the results of experiments on circadian-controlled brain functions is the fact that it is not always obvious if the observed rhythm is clock- or state-driven. Recently, a “state-clock” model, which integrates clock- and state-driven changes in synaptic plasticity, has been introduced. In this model, clock outputs regulate circadian plasticity processes, and global changes observed across sleep and wake cycles are driven by clocks and not by the state of the brain (Frank and Cantera, 2014; Frank, 2016). Nevertheless, future studies are necessary to better understand these interactions and the regulation of circadian plasticity in the brain.
Author Contributions
WK, GH and EP contributed to the writing, literature review and revision of this article. EP proposed the concept of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer SS and handling Editor declared their shared affiliation.
Footnotes
Funding. This work was supported by the K/ZDS/007356 grant from the Jagiellonian University in Kraków and by the European Union FP7-People-2012-ITN INsecTIME project nr 316790 to EP.
References
- Adly M. A., Spiwoks-Becker I., Vollrath L. (1999). Ultrastructural changes of photoreceptor synaptic ribbons in relation to time of day and illumination. Invest. Ophthalmol. Vis. Sci. 40, 2165–2172. [PubMed] [Google Scholar]
- Aguilar-Roblero R., García-Hernández F., Aguilar R., Arankowsky-Sandoval G., Drucker-Colín R. (1986). Suprachiasmatic nucleus transplants function as an endogenous oscillator only in constant darkness. Neurosci. Lett. 69, 47–52. 10.1016/0304-3940(86)90412-x [DOI] [PubMed] [Google Scholar]
- Albers H. E., Walton J. C., Gamble K. L., McNeill J. K., IV., Hummer D. L. (2017). The dynamics of GABA signaling: revelations from the circadian pacemaker in the suprachiasmatic nucleus. Front. Neuroendocrinol. 44, 35–82. 10.1016/j.yfrne.2016.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelbaum L., Wang G., Yokogawa T., Skariah G. M., Smith S. J., Mourrain P., et al. (2010). Circadian and homeostatic regulation of structural synaptic plasticity in hypocretin neurons. Neuron 68, 87–98. 10.1016/j.neuron.2010.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey M. J., Coon S. L., Carter D. A., Humphries A., Kim J. S., Shi Q., et al. (2009). Night/day changes in pineal expression of >600 Genes: central role of adrenergic/cAMP signaling. J. Biol. Chem. 284, 7606–7622. 10.1074/jbc.M808394200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey C. H., Kandel E. R. (1993). Structural changes accompanying memory storage. Annu. Rev. Physiol. 55, 397–426. 10.1146/annurev.ph.55.030193.002145 [DOI] [PubMed] [Google Scholar]
- Balkema G. W., Cusick K., Nguyen T. H. (2001). Diurnal variation in synaptic ribbon length and visual threshold. Vis. Neurosci. 18, 789–797. 10.1017/s0952523801185123 [DOI] [PubMed] [Google Scholar]
- Barnes C. A., McNaughton B. L., Goddard G. V., Douglas R. M., Adamec R. (1977). Circadian rhythm of synaptic excitability in rat and monkey central nervous system. Science 197, 91–92. 10.1126/science.194313 [DOI] [PubMed] [Google Scholar]
- Beckwith E. J., Ceriani M. F. (2015). Communication between circadian clusters: the key to a plastic network. FEBS Lett. 589, 3336–3342. 10.1016/j.febslet.2015.08.017 [DOI] [PubMed] [Google Scholar]
- Becquet D., Girardet C., Guillaumond F., François-Bellan A.-M., Bosler O. (2008). Ultrastructural plasticity in the rat suprachiasmatic nucleus. Possible involvement in clock entrainment. Glia 56, 294–305. 10.1002/glia.20613 [DOI] [PubMed] [Google Scholar]
- Berlucchi G., Buchtel H. A. (2009). Neuronal plasticity: historical roots and evolution of meaning. Exp. Brain Res. 192, 307–319. 10.1007/s00221-008-1611-6 [DOI] [PubMed] [Google Scholar]
- Blanchardon E., Grima B., Klarsfeld A., Chélot E., Hardin P. E., Préat T., et al. (2001). Defining the role of Drosophila lateral neurons in the control of circadian rhythms in motor activity and eclosion by targeted genetic ablation and PERIOD protein overexpression. Eur. J. Neurosci. 13, 871–888. 10.1046/j.0953-816x.2000.01450.x [DOI] [PubMed] [Google Scholar]
- Bliss T. V., Lømo T. (1973). Long-lasting potentiation of synaptic transmission in the dentate area of the anaesthetized rabbit following stimulation of the perforant path. J. Physiol. 232, 331–356. 10.1113/jphysiol.1973.sp010273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosler O., Girardet C., Franc J.-L., Becquet D., François-Bellan A.-M. (2015). Structural plasticity of the circadian timing system. An overview from flies to mammals. Front. Neuroendocrinol. 38, 50–64. 10.1016/j.yfrne.2015.02.001 [DOI] [PubMed] [Google Scholar]
- Bouchard-Cannon P., Mendoza-Viveros L., Yuen A., Kærn M., Cheng H.-Y. M. (2013). The circadian molecular clock regulates adult hippocampal neurogenesis by controlling the timing of cell-cycle entry and exit. Cell Rep. 5, 961–973. 10.1016/j.celrep.2013.10.037 [DOI] [PubMed] [Google Scholar]
- Bowden J. B., Abraham W. C., Harris K. M. (2012). Differential effects of strain, circadian cycle and stimulation pattern on LTP and concurrent LTD in the dentate gyrus of freely moving rats. Hippocampus 22, 1363–1370. 10.1002/hipo.20972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown T. M., Piggins H. D. (2007). Electrophysiology of the suprachiasmatic circadian clock. Prog. Neurobiol. 82, 229–255. 10.1016/j.pneurobio.2007.05.002 [DOI] [PubMed] [Google Scholar]
- Bruel-Jungerman E., Davis S., Laroche S. (2007). Brain plasticity mechanisms and memory: a party of four. Neuroscience 13, 492–505. 10.1177/1073858407302725 [DOI] [PubMed] [Google Scholar]
- Buhr E. D., Takahashi J. S. (2013). Molecular components of the Mammalian circadian clock. Handb. Exp. Pharmacol. 217, 3–27. 10.1007/978-3-642-25950-0_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain S. W., Ralph M. R. (2009). Circadian modulation of conditioned place avoidance in hamsters does not require the suprachiasmatic nucleus. Neurobiol. Learn. Mem. 91, 81–84. 10.1016/j.nlm.2008.10.005 [DOI] [PubMed] [Google Scholar]
- Cameron M. A., Barnard A. R., Hut R. A., Bonnefont X., van der Horst G. T. J., Hankins M. W., et al. (2008). Electroretinography of wild-type and Cry mutant mice reveals circadian tuning of photopic and mesopic retinal responses. J. Biol. Rhythms 23, 489–501. 10.1177/0748730408325874 [DOI] [PubMed] [Google Scholar]
- Cao G., Nitabach M. N. (2008). Circadian control of membrane excitability in Drosophila melanogaster lateral ventral clock neurons. J. Neurosci. 28, 6493–6501. 10.1523/JNEUROSCI.1503-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceriani M. F., Hogenesch J. B., Yanovsky M., Panda S., Straume M., Kay S. A. (2002). Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J. Neurosci. 22, 9305–9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappuis S., Ripperger J. A., Schnell A., Rando G., Jud C., Wahli W., et al. (2013). Role of the circadian clock gene Per2 in adaptation to cold temperature. Mol. Metab. 2, 184–193. 10.1016/j.molmet.2013.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury D., Wang L. M., Colwell C. S. (2005). Circadian regulation of hippocampal long-term potentiation. J. Biol. Rhythms 20, 225–236. 10.1177/0748730405276352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi-Castañeda D., Ortega A. (2016). Clock genes in glia cells: a rhythmic history. ASN Neuro 8:1759091416670766. 10.1177/1759091416670766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho K., Ennaceur A., Cole J. C., Suh C. K. (2000). Chronic jet lag produces cognitive deficits. J. Neurosci. 20:RC66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C., Gutierrez C. M., Tononi G. (2004). Extensive and divergent effects of sleep and wakefulness on brain gene expression. Neuron 41, 35–43. 10.1016/s0896-6273(03)00814-6 [DOI] [PubMed] [Google Scholar]
- Claridge-Chang A., Wijnen H., Naef F., Boothroyd C., Rajewsky N., Young M. W. (2001). Circadian regulation of gene expression systems in the Drosophila head. Neuron 32, 657–671. 10.1016/s0896-6273(01)00515-3 [DOI] [PubMed] [Google Scholar]
- Colwell C. S. (2011). Linking neural activity and molecular oscillations in the SCN. Nat. Rev. Neurosci. 12, 553–569. 10.1038/nrn3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damulewicz M., Loboda A., Bukowska-Strakova K., Jozkowicz A., Dulak J., Pyza E. (2015). Clock and clock-controlled genes are differently expressed in the retina, lamina and in selected cells of the visual system of Drosophila melanogaster. Front. Cell. Neurosci. 9:353. 10.3389/fncel.2015.00353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damulewicz M., Loboda A., Jozkowicz A., Dulak J., Pyza E. (2017). Interactions between the circadian clock and heme oxygenase in the retina of Drosophila melanogaster. Mol. Neurobiol. 54, 4953–4962. 10.1007/s12035-016-0026-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damulewicz M., Pyza E. (2011). The clock input to the first optic neuropil of Drosophila melanogaster expressing neuronal circadian plasticity. PLoS One 6:e21258. 10.1371/journal.pone.0021258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damulewicz M., Rosato E., Pyza E. (2013). Circadian regulation of the Na+/K+-ATPase α subunit in the visual system is mediated by the pacemaker and by retina photoreceptors in Drosophila melanogaster. PLoS One 8:e73690. 10.1371/journal.pone.0073690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty C. J., Kay S. A. (2010). Circadian control of global gene expression patterns. Annu. Rev. Genet. 44, 419–444. 10.1146/annurev-genet-102209-163432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Mahan K. L. (2012). Circadian oscillations within the hippocampus support memory formation and persistence. Front. Mol. Neurosci. 5:46. 10.3389/fnmol.2012.00046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Mahan K. L., Phan T., Han S., Wang H., Chan G. C.-K., Scheiner Z. S., et al. (2008). Circadian oscillation of hippocampal MAPK activity and cAMP: implications for memory persistence. Nat. Neurosci. 11, 1074–1082. 10.1038/nn.2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaz I., Foulkes N. S., Gothilf Y., Appelbaum L. (2013). Circadian clocks, rhythmic synaptic plasticity and the sleep-wake cycle in zebrafish. Front. Neural Circuits 7:9. 10.3389/fncir.2013.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewer J., Frisch B., Hamblen-Coyle M. J., Rosbash M., Hall J. C. (1992). Expression of the period clock gene within different cell types in the brain of Drosophila adults and mosaic analysis of these cells’ influence on circadian behavioral rhythms. J. Neurosci. 12, 3321–3349. 10.1523/JNEUROSCI.12-09-03321.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández M. P., Berni J., Ceriani M. F. (2008). Circadian remodeling of neuronal circuits involved in rhythmic behavior. PLoS Biol. 6:e69. 10.1371/journal.pbio.0060069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell S. W., Cowan C. W., Kim T.-K., Greer P. L., Lin Y., Paradis S., et al. (2006). Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science 311, 1008–1012. 10.1126/science.1122511 [DOI] [PubMed] [Google Scholar]
- Frank M. G. (2016). Circadian regulation of synaptic plasticity. Biology 5:31. 10.3390/biology5030031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M. G., Cantera R. (2014). Sleep, clocks, and synaptic plasticity. Trends Neurosci. 37, 491–501. 10.1016/j.tins.2014.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E., Flügge G. (2014). Adult neuroplasticity: more than 40 years of research. Neural Plast. 2014:541870. 10.1155/2014/541870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara C., Tosini G. (2008). Analysis of daily and circadian gene expression in the rat pineal gland. Neurosci. Res. 60, 192–198. 10.1016/j.neures.2007.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia J. A., Zhang D., Estill S. J., Michnoff C., Rutter J., Reick M., et al. (2000). Impaired cued and contextual memory in NPAS2-deficient mice. Science 288, 2226–2230. 10.1126/science.288.5474.2226 [DOI] [PubMed] [Google Scholar]
- Girardet C., Blanchard M.-P., Ferracci G., Lévêque C., Moreno M., François-Bellan A.-M., et al. (2010). Daily changes in synaptic innervation of VIP neurons in the rat suprachiasmatic nucleus: contribution of glutamatergic afferents. Eur. J. Neurosci. 31, 359–370. 10.1111/j.1460-9568.2009.07071.x [DOI] [PubMed] [Google Scholar]
- Girardet C., Lebrun B., Cabirol-Pol M.-J., Tardivel C., François-Bellan A.-M., Becquet D., et al. (2013). Brain-derived neurotrophic factor/TrkB signaling regulates daily astroglial plasticity in the suprachiasmatic nucleus: electron-microscopic evidence in mouse. Glia 61, 1172–1177. 10.1002/glia.22509 [DOI] [PubMed] [Google Scholar]
- Gorostiza E. A., Depetris-Chauvin A., Frenkel L., Pírez N., Ceriani M. F. (2014). Circadian pacemaker neurons change synaptic contacts across the day. Curr. Biol. 24, 2161–2167. 10.1016/j.cub.2014.07.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Górska-Andrzejak J., Keller A., Raabe T., Kilianek L., Pyza E. (2005). Structural daily rhythms in GFP-labelled neurons in the visual system of Drosophila melanogaster. Photochem. Photobiol. Sci. 4, 721–726. 10.1039/b417023g [DOI] [PubMed] [Google Scholar]
- Górska-Andrzejak J., Makuch R., Stefan J., Görlich A., Semik D., Pyza E. (2013). Circadian expression of the presynaptic active zone protein Bruchpilot in the lamina of Drosophila melanogaster. Dev. Neurobiol. 73, 14–26. 10.1002/dneu.22032 [DOI] [PubMed] [Google Scholar]
- Górska-Andrzejak J., Salvaterra P. M., Meinertzhagen I. A., Krzeptowski W., Görlich A., Pyza E. (2009). Cyclical expression of Na+/K+-ATPase in the visual system of Drosophila melanogaster. J. Insect Physiol. 55, 459–468. 10.1016/j.jinsphys.2009.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green D. J., Gillette R. (1982). Circadian rhythm of firing rate recorded from single cells in the rat suprachiasmatic brain slice. Brain Res. 245, 198–200. 10.1016/0006-8993(82)90361-4 [DOI] [PubMed] [Google Scholar]
- Greenspan R. J., Dierick H. A. (2004). ‘Am not I a fly like thee?’ From genes in fruit flies to behavior in humans. Hum. Mol. Genet. 13, R267–R273. 10.1093/hmg/ddh248 [DOI] [PubMed] [Google Scholar]
- Groos G., Hendriks J. (1982). Circadian rhythms in electrical discharge of rat suprachiasmatic neurones recorded in vitro. Neurosci. Lett. 34, 283–288. 10.1016/0304-3940(82)90189-6 [DOI] [PubMed] [Google Scholar]
- Hanyu R., Hayata T., Nagao M., Saita Y., Hemmi H., Notomi T., et al. (2011). Per-1 is a specific clock gene regulated by parathyroid hormone (PTH) signaling in osteoblasts and is functional for the transcriptional events induced by PTH. J. Cell. Biochem. 112, 433–438. 10.1002/jcb.22957 [DOI] [PubMed] [Google Scholar]
- Harmar A. J. (2003). An essential role for peptidergic signalling in the control of circadian rhythms in the suprachiasmatic nuclei. J. Neuroendocrinol. 15, 335–338. 10.1046/j.1365-2826.2003.01005.x [DOI] [PubMed] [Google Scholar]
- Harris K. M., Teyler T. J. (1983). Age differences in a circadian influence on hippocampal LTP. Brain Res. 261, 69–73. 10.1016/0006-8993(83)91284-2 [DOI] [PubMed] [Google Scholar]
- Hayashi Y., Koyanagi S., Kusunose N., Okada R., Wu Z., Tozaki-Saitoh H., et al. (2013). The intrinsic microglial molecular clock controls synaptic strength via the circadian expression of cathepsin S. Sci. Rep. 3:2744. 10.1038/srep02744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebb D. O. (1949). The Organization of Behavior. New York, NY: Wiley. [Google Scholar]
- Heisenberg M. (1998). What do the mushroom bodies do for the insect brain? An introduction. Learn. Mem. 5, 1–10. 10.1080/13534640701781297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisenberg M., Heusipp M., Wanke C. (1995). Structural plasticity in the Drosophila brain. J. Neurosci. 15, 1951–1960. 10.1523/JNEUROSCI.15-03-01951.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Förster C. (2003). The neuroarchitecture of the circadian clock in the brain of Drosophila melanogaster. Microsc. Res. Tech. 62, 94–102. 10.1002/jemt.10357 [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C. (2005). Organization of endogenous clocks in insects. Biochem. Soc. Trans. 33, 957–961. 10.1042/bst20050957 [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C., Wulf J., de Belle J. S. (2002). Mushroom body influence on locomotor activity and circadian rhythms in Drosophila melanogaster. J. Neurogenet. 16, 73–109. 10.1080/01677060213158 [DOI] [PubMed] [Google Scholar]
- Hendrickson A. E., Wagoner N., Cowan W. M. (1972). An autoradiographic and electron microscopic study of retino-hypothalamic connections. Z. Zellforsch. Mikrosk. Anat. 135, 1–26. 10.1007/bf00307084 [DOI] [PubMed] [Google Scholar]
- Hughes S., Jagannath A., Hankins M. W., Foster R. G., Peirson S. N. (2015). “Chapter six—photic regulation of clock systems,” in Methods in Enzymology, ed. Sehgal A. (Cambridge, MA: Academic Press; ) 125–143. [DOI] [PubMed] [Google Scholar]
- Hwang D. S., Kwak H. B., Ko I. G., Kim S. E., Jin J. J., Ji E. S., et al. (2016). Treadmill exercise improves memory function depending on circadian rhythm changes in mice. Int. Neurourol. J. 20, S141–S149. 10.5213/inj.1632738.369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M., Hojo Y., Komatsuzaki Y., Okamoto M., Kato A., Takeda T., et al. (2015). Hippocampal spine changes across the sleep-wake cycle: corticosterone and kinases. J. Endocrinol. 226, M13–M27. 10.1530/JOE-15-0078 [DOI] [PubMed] [Google Scholar]
- Ikeno T., Weil Z. M., Nelson R. J. (2014). Timing of light pulses and photoperiod on the diurnal rhythm of hippocampal neuronal morphology of Siberian hamsters. Neuroscience 270, 69–75. 10.1016/j.neuroscience.2014.04.002 [DOI] [PubMed] [Google Scholar]
- Im S. H., Taghert P. H. (2010). PDF receptor expression reveals direct interactions between circadian oscillators in Drosophila. J. Comp. Neurol. 518, 1925–1945. 10.1002/cne.22311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S. T., Kawamura H. (1979). Persistence of circadian rhythmicity in a mammalian hypothalamic “island” containing the suprachiasmatic nucleus. Proc. Natl. Acad. Sci. U S A 76, 5962–5966. 10.1073/pnas.76.11.5962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S. T., Kawamura H. (1982). Characteristics of a circadian pacemaker in the suprachiasmatic nucleus. J. Comp. Physiol. 146, 153–160. 10.1007/bf00610233 [DOI] [Google Scholar]
- Iyer R., Wang T. A., Gillette M. U. (2014). Circadian gating of neuronal functionality: a basis for iterative metaplasticity. Front. Syst. Neurosci. 8:164. 10.3389/fnsys.2014.00164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson F. R. (2011). Glial cell modulation of circadian rhythms. Glia 59, 1341–1350. 10.1002/glia.21097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson F. R., Ng F. S., Sengupta S., You S., Huang Y. (2015). Glial cell regulation of rhythmic behavior. Meth. Enzymol. 552, 45–73. 10.1016/bs.mie.2014.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinska M., Grzegorczyk A., Jasek E., Litwin J. A., Kossut M., Barbacka-Surowiak G., et al. (2014). Daily rhythm of synapse turnover in mouse somatosensory cortex. Acta Neurobiol. Exp. (Wars) 74, 104–110. [DOI] [PubMed] [Google Scholar]
- Jasinska M., Grzegorczyk A., Woznicka O., Jasek E., Kossut M., Barbacka-Surowiak G., et al. (2015). Circadian rhythmicity of synapses in mouse somatosensory cortex. Eur. J. Neurosci. 42, 2585–2594. 10.1111/ejn.13045 [DOI] [PubMed] [Google Scholar]
- Jastrow H., von Mach M. A., Vollrath L. (1997). The shape of synaptic ribbons in the rat pineal gland. Cell Tissue Res. 287, 255–261. 10.1007/s004410050750 [DOI] [PubMed] [Google Scholar]
- Jilg A., Lesny S., Peruzki N., Schwegler H., Selbach O., Dehghani F., et al. (2010). Temporal dynamics of mouse hippocampal clock gene expression support memory processing. Hippocampus 20, 377–388. 10.1002/hipo.20637 [DOI] [PubMed] [Google Scholar]
- Johard H. A. D., Yoishii T., Dircksen H., Cusumano P., Rouyer F., Helfrich-Förster C., et al. (2009). Peptidergic clock neurons in Drosophila: ion transport peptide and short neuropeptide F in subsets of dorsal and ventral lateral neurons. J. Comp. Neurol. 516, 59–73. 10.1002/cne.22099 [DOI] [PubMed] [Google Scholar]
- Kaneko M., Hall J. C. (2000). Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J. Comp. Neurol. 422, 66–94. [DOI] [PubMed] [Google Scholar]
- Kaneko M., Park J. H., Cheng Y., Hardin P. E., Hall J. C. (2000). Disruption of synaptic transmission or clock-gene-product oscillations in circadian pacemaker cells of Drosophila cause abnormal behavioral rhythms. J. Neurobiol. 43, 207–233. [DOI] [PubMed] [Google Scholar]
- Keegan K. P., Pradhan S., Wang J.-P., Allada R. (2007). Meta-analysis of Drosophila circadian microarray studies identifies a novel set of rhythmically expressed genes. PLoS Comput. Biol. 3:e208. 10.1371/journal.pcbi.0030208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M. (2017). Molecular mechanisms of experience-dependent structural and functional plasticity in the brain. Anat. Sci. Int. 92, 1–17. 10.1007/s12565-016-0358-6 [DOI] [PubMed] [Google Scholar]
- Kondratova A. A., Dubrovsky Y. V., Antoch M. P., Kondratov R. V. (2010). Circadian clock proteins control adaptation to novel environment and memory formation. Aging 2, 285–297. 10.18632/aging.100142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzeptowski W., Górska-Andrzejak J., Kijak E., Görlich A., Guzik E., Moore G., et al. (2014). External and circadian inputs modulate synaptic protein expression in the visual system of Drosophila melanogaster. Front. Physiol. 5:102. 10.3389/fphys.2014.00102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman M. N., Silver R., Gladstone W. R., Kahn R. M., Gibson M., Bittman E. L. (1987). Circadian rhythmicity restored by neural transplant. Immunocytochemical characterization of the graft and its integration with the host brain. J. Neurosci. 7, 1626–1638. 10.1523/JNEUROSCI.07-06-01626.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. Z., Bunney B. G., Meng F., Hagenauer M. H., Walsh D. M., Vawter M. P., et al. (2013). Circadian patterns of gene expression in the human brain and disruption in major depressive disorder. Proc. Natl. Acad. Sci. U S A 110, 9950–9955. 10.1073/pnas.1305814110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston C., Cichon J. M., Jeanneteau F., Jia Z., Chao M. V., Gan W.-B. (2013). Circadian glucocorticoid oscillations promote learning-dependent synapse formation and maintenance. Nat. Neurosci. 16, 698–705. 10.1038/nn.3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lømo T. (2003). The discovery of long-term potentiation. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 358, 617–620. 10.1098/rstb.2002.1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons L. C., Roman G. (2008). Circadian modulation of short-term memory in Drosophila. Learn. Mem. 16, 19–27. 10.1101/lm.1146009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Fairey C. A., Nunez A. A. (2014). Circadian modulation of memory and plasticity gene products in a diurnal species. Brain Res. 1581, 30–39. 10.1016/j.brainres.2014.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald M. J., Rosbash M. (2001). Microarray analysis and organization of circadian gene expression in Drosophila. Cell 107, 567–578. 10.1016/s0092-8674(01)00545-1 [DOI] [PubMed] [Google Scholar]
- McNulty J. A. (1981). Synaptic ribbons in the pineal organ of the goldfish: circadian rhythmicity and the effects of constant light and constant darkness. Cell Tissue Res. 215, 491–497. 10.1007/bf00233525 [DOI] [PubMed] [Google Scholar]
- Mehnert K. I., Beramendi A., Elghazali F., Negro P., Kyriacou C. P., Cantera R. (2007). Circadian changes in Drosophila motor terminals. Dev. Neurobiol. 67, 415–421. 10.1002/dneu.20332 [DOI] [PubMed] [Google Scholar]
- Mehnert K. I., Cantera R. (2008). A peripheral pacemaker drives the circadian rhythm of synaptic boutons in Drosophila independently of synaptic activity. Cell Tissue Res. 334, 103–109. 10.1007/s00441-008-0670-0 [DOI] [PubMed] [Google Scholar]
- Moore R. Y., Eichler V. B. (1972). Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res. 42, 201–206. 10.1016/0006-8993(72)90054-6 [DOI] [PubMed] [Google Scholar]
- Moore R. Y., Lenn N. J. (1972). A retinohypothalamic projection in the rat. J. Comp. Neurol. 146, 1–14. 10.1002/cne.901460102 [DOI] [PubMed] [Google Scholar]
- Mouret J., Coindet J., Debilly G., Chouvet G. (1978). Suprachiasmatic nuclei lesions in the rat: alterations in sleep circadian rhythms. Electroencephalogr. Clin. Neurophysiol. 45, 402–408. 10.1016/0013-4694(78)90191-8 [DOI] [PubMed] [Google Scholar]
- Munn R. G. K., Bilkey D. K. (2012). The firing rate of hippocampal CA1 place cells is modulated with a circadian period. Hippocampus 22, 1325–1337. 10.1002/hipo.20969 [DOI] [PubMed] [Google Scholar]
- Muraro N. I., Pírez N., Ceriani M. F. (2013). The circadian system: plasticity at many levels. Neuroscience 247, 280–293. 10.1016/j.neuroscience.2013.05.036 [DOI] [PubMed] [Google Scholar]
- Naeger N. L., Van Nest B. N., Johnson J. N., Boyd S. D., Southey B. R., Rodriguez-Zas S. L., et al. (2011). Neurogenomic signatures of spatiotemporal memories in time-trained forager honey bees. J. Exp. Biol. 214, 979–987. 10.1242/jeb.053421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuka H., Natsume K. (2014). Circadian rhythm modulates long-term potentiation induced at CA1 in rat hippocampal slices. Neurosci. Res. 80, 1–9. 10.1016/j.neures.2013.12.007 [DOI] [PubMed] [Google Scholar]
- Namihira M., Honma S., Abe H., Tanahashi Y., Ikeda M., Honma K. (1999). Daily variation and light responsiveness of mammalian clock gene, Clock and BMAL1, transcripts in the pineal body and different areas of brain in rats. Neurosci. Lett. 267, 69–72. 10.1016/s0304-3940(99)00324-9 [DOI] [PubMed] [Google Scholar]
- Nishiitsutsuji-Uwo J., Pittendrigh C. S. (1968). Central nervous system control of circadian rhythmicity in the cockroach. Z. Vgl. Physiol. 58, 1–13. 10.1007/bf00302433 [DOI] [Google Scholar]
- Nitabach M. N., Blau J., Holmes T. C. (2002). Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell 109, 485–495. 10.1016/s0092-8674(02)00737-7 [DOI] [PubMed] [Google Scholar]
- O’Neill J. S., Reddy A. B. (2012). The essential role of cAMP/Ca2+ signalling in mammalian circadian timekeeping. Biochem. Soc. Trans. 40, 44–50. 10.1042/BST20110691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen S. R., Wilson R. I. (2008). Cracking neural circuits in a tiny brain: new approaches for understanding the neural circuitry of Drosophila. Trends Neurosci. 31, 512–520. 10.1016/j.tins.2008.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono D., Honma K., Honma S. (2015). Circadian and ultradian rhythms of clock gene expression in the suprachiasmatic nucleus of freely moving mice. Sci. Rep. 5:12310. 10.1038/srep12310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özkaya Ö., Rosato E. (2012). The circadian clock of the fly: a neurogenetics journey through time. Adv. Genet. 77, 79–123. 10.1016/B978-0-12-387687-4.00004-0 [DOI] [PubMed] [Google Scholar]
- Page T. L. (1982). Transplantation of the cockroach circadian pacemaker. Science 216, 73–75. 10.1126/science.216.4541.73 [DOI] [PubMed] [Google Scholar]
- Park D., Griffith L. C. (2006). Electrophysiological and anatomical characterization of PDF-positive clock neurons in the intact adult Drosophila brain. J. Neurophysiol. 95, 3955–3960. 10.1152/jn.00117.2006 [DOI] [PubMed] [Google Scholar]
- Park J. H., Helfrich-Förster C., Lee G., Liu L., Rosbash M., Hall J. C. (2000). Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc. Natl. Acad. Sci. U S A 97, 3608–3613. 10.1073/pnas.070036197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Cruz C., Simon M., Flügge G., Fuchs E., Czéh B. (2009). Diurnal rhythm and stress regulate dendritic architecture and spine density of pyramidal neurons in the rat infralimbic cortex. Behav. Brain Res. 205, 406–413. 10.1016/j.bbr.2009.07.021 [DOI] [PubMed] [Google Scholar]
- Petsakou A., Sapsis T. P., Blau J. (2015). Circadian rhythms in rho1 activity regulate neuronal plasticity and network hierarchy. Cell 162, 823–835. 10.1016/j.cell.2015.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T. X., Phan T. H., Chan G. C.-K., Sindreu C. B., Eckel-Mahan K. L., Storm D. R. (2011). The diurnal oscillation of MAP (mitogen-activated protein) kinase and adenylyl cyclase activities in the hippocampus depends on the suprachiasmatic nucleus. J. Neurosci. 31, 10640–10647. 10.1523/JNEUROSCI.6535-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prolo L. M., Takahashi J. S., Herzog E. D. (2005). Circadian rhythm generation and entrainment in astrocytes. J. Neurosci. 25, 404–408. 10.1523/JNEUROSCI.4133-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyza E. M. (2002). Dynamic structural changes of synaptic contacts in the visual system of insects. Microsc. Res. Tech. 58, 335–344. 10.1002/jemt.10141 [DOI] [PubMed] [Google Scholar]
- Pyza E. M. (2013). Plasticity in invertebrate sensory systems. Front. Physiol. 4:226. 10.3389/fphys.2013.00226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyza E., Cymborowski B. (2001). Circadian rhythms in behaviour and in the visual system of the blow fly, Calliphora vicina. J. Insect Physiol. 47, 897–904. 10.1016/s0022-1910(01)00062-2 [DOI] [Google Scholar]
- Pyza E., Górska-Andrzejak J. (2004). Involvement of glial cells in rhythmic size changes in neurons of the housefly’s visual system. J. Neurobiol. 59, 205–215. 10.1002/neu.10307 [DOI] [PubMed] [Google Scholar]
- Pyza E., Meinertzhagen I. A. (1993). Daily and circadian rhythms of synaptic frequency in the first visual neuropile of the housefly’s (Musca domestica L.) optic lobe. Proc. Biol. Sci. 254, 97–105. 10.1098/rspb.1993.0133 [DOI] [PubMed] [Google Scholar]
- Pyza E., Meinertzhagen I. A. (1995). Monopolar cell axons in the first optic neuropil of the housefly, Musca domestica L., undergo daily fluctuations in diameter that have a circadian basis. J. Neurosci. 15, 407–418. 10.1523/JNEUROSCI.15-01-00407.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyza E., Meinertzhagen I. A. (1997). Neurites of period-expressing PDH cells in the fly’s optic lobe exhibit circadian oscillations in morphology. Eur. J. Neurosci. 9, 1784–1788. 10.1111/j.1460-9568.1997.tb01537.x [DOI] [PubMed] [Google Scholar]
- Pyza E., Meinertzhagen I. A. (1999). Daily rhythmic changes of cell size and shape in the first optic neuropil in Drosophila melanogaster. J. Neurobiol. 40, 77–88. [DOI] [PubMed] [Google Scholar]
- Raghavan A. V., Horowitz J. M., Fuller C. A. (1999). Diurnal modulation of long-term potentiation in the hamster hippocampal slice. Brain Res. 833, 311–314. 10.1016/s0006-8993(99)01523-1 [DOI] [PubMed] [Google Scholar]
- Ralph M. R., Foster R. G., Davis F. C., Menaker M. (1990). Transplanted suprachiasmatic nucleus determines circadian period. Science 247, 975–978. 10.1126/science.2305266 [DOI] [PubMed] [Google Scholar]
- Rawashdeh O., Jilg A., Jedlicka P., Slawska J., Thomas L., Saade A., et al. (2014). PERIOD1 coordinates hippocampal rhythms and memory processing with daytime. Hippocampus 24, 712–723. 10.1002/hipo.22262 [DOI] [PubMed] [Google Scholar]
- Rawashdeh O., Jilg A., Maronde E., Fahrenkrug J., Stehle J. H. (2016). Period1 gates the circadian modulation of memory-relevant signaling in mouse hippocampus by regulating the nuclear shuttling of the CREB kinase pP90RSK. J. Neurochem. 138, 731–745. 10.1111/jnc.13689 [DOI] [PubMed] [Google Scholar]
- Reick M., Garcia J. A., Dudley C., McKnight S. L. (2001). NPAS2: an analog of clock operative in the mammalian forebrain. Science 293, 506–509. 10.1126/science.1060699 [DOI] [PubMed] [Google Scholar]
- Renaud J., Dumont F., Khelfaoui M., Foisset S. R., Letourneur F., Bienvenu T., et al. (2015). Identification of intellectual disability genes showing circadian clock-dependent expression in the mouse hippocampus. Neuroscience 308, 11–50. 10.1016/j.neuroscience.2015.08.066 [DOI] [PubMed] [Google Scholar]
- Renn S. C., Park J. H., Rosbash M., Hall J. C., Taghert P. H. (1999). A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99, 791–802. 10.1016/s0092-8674(00)81676-1 [DOI] [PubMed] [Google Scholar]
- Riddle M., Mezias E., Foley D., LeSauter J., Silver R. (2016). Differential localization of PER1 and PER2 in the brain master circadian clock. Eur. J. Neurosci. 45, 1357–1367. 10.1111/ejn.13441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger D., Shafer O. T., Tomioka K., Helfrich-Förster C. (2006). Functional analysis of circadian pacemaker neurons in Drosophila melanogaster. J. Neurosci. 26, 2531–2543. 10.1523/JNEUROSCI.1234-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Zas S. L., Southey B. R., Shemesh Y., Rubin E. B., Cohen M., Robinson G. E., et al. (2012). Microarray analysis of natural socially regulated plasticity in circadian rhythms of honey bees. J. Biol. Rhythms 27, 12–24. 10.1177/0748730411431404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosbash M. (2009). The implications of multiple circadian clock origins. PLoS Biol. 7:e62. 10.1371/journal.pbio.1000062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouch I., Wild P., Ansiau D., Marquié J.-C. (2005). Shiftwork experience, age and cognitive performance. Ergonomics 48, 1282–1293. 10.1080/00140130500241670 [DOI] [PubMed] [Google Scholar]
- Ruby N. F., Hwang C. E., Wessells C., Fernandez F., Zhang P., Sapolsky R., et al. (2008). Hippocampal-dependent learning requires a functional circadian system. Proc. Natl. Acad. Sci. U S A 105, 15593–15598. 10.1073/pnas.0808259105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz S., Ferreiro M. J., Menhert K. I., Casanova G., Olivera A., Cantera R. (2013). Rhythmic changes in synapse numbers in Drosophila melanogaster motor terminals. PLoS One 8:e67161. 10.1371/journal.pone.0067161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schildt S., Endres T., Lessmann V., Edelmann E. (2013). Acute and chronic interference with BDNF/TrkB-signaling impair LTP selectively at mossy fiber synapses in the CA3 region of mouse hippocampus. Neuropharmacology 71, 247–254. 10.1016/j.neuropharm.2013.03.041 [DOI] [PubMed] [Google Scholar]
- Schmidt C., Collette F., Cajochen C., Peigneux P. (2007). A time to think: circadian rhythms in human cognition. Cogn. Neuropsychol. 24, 755–789. 10.1080/02643290701754158 [DOI] [PubMed] [Google Scholar]
- Schnell A., Chappuis S., Schmutz I., Brai E., Ripperger J. A., Schaad O., et al. (2014). The nuclear receptor REV-ERBα regulates Fabp7 and modulates adult hippocampal neurogenesis. PLoS One 9:e99883. 10.1371/journal.pone.0099883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeba V., Gu H., Sharma V. K., O’Dowd D. K., Holmes T. C. (2008). Circadian- and light-dependent regulation of resting membrane potential and spontaneous action potential firing of Drosophila circadian pacemaker neurons. J. Neurophysiol. 99, 976–988. 10.1152/jn.00930.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S., Oomura Y., Kita H., Hattori K. (1982). Circadian rhythmic changes of neuronal activity in the suprachiasmatic nucleus of the rat hypothalamic slice. Brain Res. 247, 154–158. 10.1016/0006-8993(82)91041-1 [DOI] [PubMed] [Google Scholar]
- Shieh K.-R. (2003). Distribution of the rhythm-related genes rPERIOD1, rPERIOD2, and rCLOCK, in the rat brain. Neuroscience 118, 831–843. 10.1016/s0306-4522(03)00004-6 [DOI] [PubMed] [Google Scholar]
- Sivachenko A., Li Y., Abruzzi K. C., Rosbash M. (2013). The transcription factor Mef2 links the Drosophila core clock to Fas2, neuronal morphology, and circadian behavior. Neuron 79, 281–292. 10.1016/j.neuron.2013.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siwicki K. K., Eastman C., Petersen G., Rosbash M., Hall J. C. (1988). Antibodies to the period gene product of Drosophila reveal diverse tissue distribution and rhythmic changes in the visual system. Neuron 1, 141–150. 10.1016/0896-6273(88)90198-5 [DOI] [PubMed] [Google Scholar]
- Smarr B. L., Jennings K. J., Driscoll J. R., Kriegsfeld L. J. (2014). A time to remember: the role of circadian clocks in learning and memory. Behav. Neurosci. 128, 283–303. 10.1037/a0035963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So A. Y.-L., Bernal T. U., Pillsbury M. L., Yamamoto K. R., Feldman B. J. (2009). Glucocorticoid regulation of the circadian clock modulates glucose homeostasis. Proc. Natl. Acad. Sci. U S A 106, 17582–17587. 10.1073/pnas.0909733106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollars P. J., Kimble D. P., Pickard G. E. (1995). Restoration of circadian behavior by anterior hypothalamic heterografts. J. Neurosci. 15, 2109–2122. 10.1523/JNEUROSCI.15-03-02109.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahnisch F. W., Nitsch R. (2002). Santiago Ramón y Cajal’s concept of neuronal plasticity: the ambiguity lives on. Trends Neurosci. 25, 589–591. 10.1016/s0166-2236(02)02251-8 [DOI] [PubMed] [Google Scholar]
- Stephan F. K., Kovacevic N. S. (1978). Multiple retention deficit in passive avoidance in rats is eliminated by suprachiasmatic lesions. Behav. Biol. 22, 456–462. 10.1016/s0091-6773(78)92565-8 [DOI] [PubMed] [Google Scholar]
- Stephan F. K., Zucker I. (1972). Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc. Natl. Acad. Sci. U S A 69, 1583–1586. 10.1073/pnas.69.6.1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch K.-F., Lipan O., Leykin I., Viswanathan N., Davis F. C., Wong W. H., et al. (2002). Extensive and divergent circadian gene expression in liver and heart. Nature 417, 78–83. 10.1038/nature744 [DOI] [PubMed] [Google Scholar]
- Storch K.-F., Paz C., Signorovitch J., Raviola E., Pawlyk B., Li T., et al. (2007). Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell 130, 730–741. 10.1016/j.cell.2007.06.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh J., Jackson F. R. (2007). Drosophila ebony activity is required in glia for the circadian regulation of locomotor activity. Neuron 55, 435–447. 10.1016/j.neuron.2007.06.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sujino M., Masumoto K., Yamaguchi S., van der Horst G. T. J., Okamura H., Inouye S.-I. T. (2003). Suprachiasmatic nucleus grafts restore circadian behavioral rhythms of genetically arrhythmic mice. Curr. Biol. 13, 664–668. 10.1016/s0960-9822(03)00222-7 [DOI] [PubMed] [Google Scholar]
- Sweatt J. D. (2016). Neural plasticity and behavior—sixty years of conceptual advances. J. Neurochem. 139, 179–199. 10.1111/jnc.13580 [DOI] [PubMed] [Google Scholar]
- Tamaru T., Hattori M., Honda K., Benjamin I., Ozawa T., Takamatsu K. (2011). Synchronization of circadian Per2 rhythms and HSF1-BMAL1:CLOCK interaction in mouse fibroblasts after short-term heat shock pulse. PLoS One 6:e24521. 10.1371/journal.pone.0024521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tataroglu O., Emery P. (2015). The molecular ticks of the Drosophila circadian clock. Curr. Opin. Insect Sci. 7, 51–57. 10.1016/j.cois.2015.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomioka K. (1985). Residual circadian rhythmicity after bilateral lamina-medulla removal or optic stalk transection in the cricket, Gryllus bimaculatus. J. Insect Physiol. 31, 653–657. 10.1016/0022-1910(85)90065-4 [DOI] [Google Scholar]
- Travnickova-Bendova Z., Cermakian N., Reppert S. M., Sassone-Corsi P. (2002). Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc. Natl. Acad. Sci. U S A 99, 7728–7733. 10.1073/pnas.102075599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H. R., Matsumoto A., Kawamura M., Iino M., Tanimura T., Hashimoto S. (2002). Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. J. Biol. Chem. 277, 14048–14052. 10.1074/jbc.c100765200 [DOI] [PubMed] [Google Scholar]
- Van der Zee E. A., Havekes R., Barf R. P., Hut R. A., Nijholt I. M., Jacobs E. H., et al. (2008). Circadian time-place learning in mice depends on Cry genes. Curr. Biol. 18, 844–848. 10.1016/j.cub.2008.04.077 [DOI] [PubMed] [Google Scholar]
- Vollrath L. (1973). Synaptic ribbons of a mammalian pineal gland circadian changes. Z. Zellforsch. Mikrosk. Anat. 145, 171–183. 10.1007/bf00307386 [DOI] [PubMed] [Google Scholar]
- Wang L. M., Dragich J. M., Kudo T., Odom I. H., Welsh D. K., O’Dell T. J., et al. (2009). Expression of the circadian clock gene Period2 in the hippocampus: possible implications for synaptic plasticity and learned behaviour. ASN Neuro 1:e00012. 10.1042/AN20090020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.-Y., Huang R.-C. (2004). Diurnal modulation of the Na+/K+-ATPase and spontaneous firing in the rat retinorecipient clock neurons. J. Neurophysiol. 92, 2295–2301. 10.1152/jn.00061.2004 [DOI] [PubMed] [Google Scholar]
- Wardlaw S. M., Phan T. X., Saraf A., Chen X., Storm D. R. (2014). Genetic disruption of the core circadian clock impairs hippocampus-dependent memory. Learn. Mem. 21, 417–423. 10.1101/lm.035451.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber P., Kula-Eversole E., Pyza E. (2009). Circadian control of dendrite morphology in the visual system of Drosophila melanogaster. PLoS One 4:e4290. 10.1371/journal.pone.0004290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H., Stengl M. (2011). Light affects the branching pattern of peptidergic circadian pacemaker neurons in the brain of the cockroach Leucophaea maderae. J. Biol. Rhythms 26, 507–517. 10.1177/0748730411419968 [DOI] [PubMed] [Google Scholar]
- West M. O., Deadwyler S. A. (1980). Circadian modulation of granule cell response to perforant path synaptic input in the rat. Neuroscience 5, 1597–1602. 10.1016/0306-4522(80)90023-8 [DOI] [PubMed] [Google Scholar]
- Wijnen H., Naef F., Boothroyd C., Claridge-Chang A., Young M. W. (2006). Control of daily transcript oscillations in Drosophila by light and the circadian clock. PLoS Genet. 2:e39. 10.1371/journal.pgen.0020039.eor [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woźnicka O., Görlich A., Sigrist S., Pyza E. (2015). BRP-170 and BRP190 isoforms of Bruchpilot protein differentially contribute to the frequency of synapses and synaptic circadian plasticity in the visual system of Drosophila. Front. Cell. Neurosci. 9:238. 10.3389/fncel.2015.00238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagita K., Yamanaka I., Emoto N., Kawakami K., Shimada S. (2010). Real-time monitoring of circadian clock oscillations in primary cultures of mammalian cells using Tol2 transposon-mediated gene transfer strategy. BMC Biotechnol. 10:3. 10.1186/1472-6750-10-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Okada K., Mizuno T., Ota T., Yamada H., Doi M., et al. (2016). Real-time recording of circadian Per1 and Per2 expression in the suprachiasmatic nucleus of freely moving rats. J. Biol. Rhythms 31, 108–111. 10.1177/0748730415621412 [DOI] [PubMed] [Google Scholar]
- Yang S., Wang K., Valladares O., Hannenhalli S., Bucan M. (2007). Genome-wide expression profiling and bioinformatics analysis of diurnally regulated genes in the mouse prefrontal cortex. Genome Biol. 8:R247. 10.1186/gb-2007-8-11-r247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z., Shafer O. T. (2014). The Drosophila circadian clock is a variably coupled network of multiple peptidergic units. Science 343, 1516–1520. 10.1126/science.1251285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii T., Todo T., Wülbeck C., Stanewsky R., Helfrich-Förster C. (2008). Cryptochrome is present in the compound eyes and a subset of Drosophila’s clock neurons. J. Comp. Neurol. 508, 952–966. 10.1002/cne.21702 [DOI] [PubMed] [Google Scholar]
- Zerr D. M., Hall J. C., Rosbash M., Siwicki K. K. (1990). Circadian fluctuations of period protein immunoreactivity in the CNS and the visual system of Drosophila. J. Neurosci. 10, 2749–2762. 10.1523/JNEUROSCI.10-08-02749.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Lahens N. F., Ballance H. I., Hughes M. E., Hogenesch J. B. (2014). A circadian gene expression atlas in mammals: implications for biology and medicine. Proc. Natl. Acad. Sci. U S A 111, 16219–16224. 10.1073/pnas.1408886111 [DOI] [PMC free article] [PubMed] [Google Scholar]