Abstract
Objective
This pilot study compared eSource-enabled versus traditional manual data transcription (non-eSource methods) for the collection of clinical registry information. The primary study objective was to compare the time spent completing registry forms using eSource versus non-eSource methods The secondary objectives were to compare data quality associated with these two data capture methods and the flexibility of the workflows. This study directly addressed fundamental questions relating to eSource adoption: what time-savings can be realized, and to what extent does eSource improve data quality.
Materials and methods
The study used time and motion methods to compare eSource versus non-eSource data capture workflows for a single center OB/GYN registry. Direct observation by industrial engineers using specialized computer software captured keystrokes, mouse clicks and video recordings of the study team in their normal work environment completing real-time data collection.
Results
The overall average data capture time was reduced with eSource versus non-eSource methods (difference, 151 s per case; eSource, 1603 s; non-eSource, 1754 s; p = 0.051). The average data capture time for the demographic data was reduced (difference, 79 s per case; eSource, 133 s; non-eSource, 213 s; p < 0.001). This represents a 37% time reduction (95% confidence interval 27% to 47%). eSourced data field transcription errors were also reduced (eSource, 0%; non-eSource, 9%).
Conclusion
The use of eSource versus traditional data transcription was associated with a significant reduction in data entry time and data quality errors. Further studies in other settings are needed to validate these results.
Keywords: Data accuracy, Data collection, Data retrieval, Information extraction, Registries, Time-motion analysis
1. Introduction
For more than 60 years, the cost of conducting biomedical research has increased exponentially while net productivity has declined [1,2]. The greatest cost increases have occurred in late phase clinical trials where > 65% of total costs are site-related (for site management and site trial work) and the complexity of protocol-mandated activities has escalated [3,4]. Several initiatives are investigating ways to reduce clinical trial costs without compromising their scientific validity. These efforts have focused on reducing costs by monitoring only high risk tasks and studies (risk-based monitoring) and the secondary use of existing registry and billing data [5–8]. However, none of these initiatives address an area of high cost and great inaccuracy in clinical research studies, namely the collection and transcription of study data from the patient’s health record into the clinical study’s electronic case report form (eCRF).
2. Background and significance
There is growing interest in utilizing EHR data for clinical research. According to the American Medical Informatics Association (AMIA), “Secondary use of health data can enhance healthcare experiences for individuals, expand knowledge about disease and appropriate treatments, strengthen understanding about the effectiveness and efficiency of our healthcare systems, support public health and security goals, and aid businesses in meeting the needs of their customers” [9]. Typically, EHR data is manually abstracted and then entered into electronic clinical research forms (eCRF). The next step in making EHR data available for clinical research is to directly link the EHR and eCRF systems. The Federal Drug Administration (FDA) coined the term “eSource” to represent the secondary use of EHR data for completing eCRFs using interoperability standards [10,11].
The Retrieve Form Data Capture (RFD) standard provides eSource capability. RFD is an Integrating the Healthcare Enterprise (IHE) standard that allows for secure interoperability between systems by providing a window into the EHR so that the eCRF form can be auto-populated using previously mapped EHR data elements [12]. RFD also enables other study-specific data elements to be entered directly into the eCRF at the point of care and from within the EHR, posting the eCRF data into the study database and not the EHR itself.
This study tests the hypothesis that eSource data management reduces time and transcription errors, with the implication of savings in study costs, particularly if widely adopted in the most expensive late phase clinical trials.
3. Objective
The primary study objective is to compare the time spent completing the eCRF using traditional (non-eSource) and eSource-enabled workflows. The secondary objective is to compare data quality associated with these data capture methods. This study directly addresses fundamental questions relating to eSource adoption: what time-savings can be realized, and to what extent does eSource improve data quality.
4. Materials and methods
4.1. Product design
Duke University Office of Research Informatics developed middleware, called RADaptor, that uses the RFD standard to electronically call a study eCRF from a REDCap [13] database into Duke’s Epic (Epic Inc, Verona, WI) EHR. This use of the RFD standard is part of Epic’s model research functionality, and configuration for a study to use RADaptor can be accomplished within Epic’s normal research configuration in the research (RSH) record. Study team members using RADaptor open the EHR, and “call” the eCRF. Once security is authorized, the eCRF will appear within the EHR window. Data points that have been previously mapped will auto-populate in the eCRF based upon EHR data availability. For the present study, data points mapped with RADaptor included those contained within the EHR’s continuity of care document (CCD) [14]. Data elements that are not mapped appear as unanswered (study-specific) eCRF sections. Data can be edited or study-specific data can be entered directly into the eCRF form. Edited or study-specific data in the eCRF will not be stored in the EHR, being posted directly to the study’s REDCap database.
4.2. Study design
This is a single site, observational comparative effectiveness study that examines the impact of Duke’s RADaptor on eCRF completion workflows. The study protocol employs time-motion methods to compare eSource and non-eSource workflows for a single center OB/GYN study.
Study data collected included: process time, motion, mouse clicks, and keystrokes. Typically mouse click and keystroke research data are collected manually in a laboratory environment using pseudo patients. This is a tedious, error prone process. To address this problem, we augmented the direct observations of the industrial engineers with software that documented keystrokes, mouse clicks and took video recordings in the actual work setting. The Duke University School of Medicine Institutional Review Board (IRB) approved the study protocol on December 1, 2015 (PRO00068189).
4.3. Information security
At the recommendation of the Duke Information Security Office (ISO), two desktop computers and two laptop computers were acquired and configured with keylogging and study software. These desktop computers were set up and enabled in the study team members’ normal workspace at the start of each observation period. At the end of each observation period, study desktops were removed, the study teams’ passwords reset and the original work computers replaced.
4.4. Study participants
The study included a convenience sample of Duke University clinical research staff working on a Prematurity Prevention registry. Subjects had access to the institution’s EHR, Epic, and were eligible to access the primary study’s eCRF, REDCap. Study data were collected and managed using Research Electronic Data Capture (REDCap) electronic data capture tools hosted at Duke University. REDCap is a secure, web-based application designed to support data capture for research studies, providing 1) an intuitive interface for validated data entry; 2) audit trails for tracking data manipulation and export procedures; 3) automated export procedures for seamless data downloads to common statistical packages; and 4) procedures for importing data from external sources [15]. This software tool was developed and supported by the US National Institute of Health’s National Center for Advancing Translational Sciences, is intended to support smaller investigator-initiated studies, and is considered the premier software tool in this niche.
The clinical study team is proficient in the use of REDCap and has completed multiple studies using this system and requested the Prematurity Prevention Registry to be built in REDCap. The eCRF was built by the Duke Office of Clinical Research (DOCR) and is considered to be a typical registry eCRFs for principal investigator initiated studies in terms of complexity.
The eCRF was created by the Duke Office of Clinical Research (DOCR) and its complexity is typical of investigator initiated registry eCRFs at our institution. This eCRF contains 401 data elements relating to a mother’s pregnancy and birth of the infant; however, most of these data elements are not entered for each case. The present study is limited to the eCRF’s demographic section. The eSource methodology allowed for 7 of 14 demographic data elements to be auto-populated into the demographics form. Abstracting in the present study was limited to the copying of information from patient electronic medical records to the eCRF demographics section.
4.5. Observation
Three study participants were observed completing two different workflows in their Duke work environment.
4.5.1. Non-eSource observation
Fig. 1 shows the non-eSource workflow performed by a Clinical Research Coordinator (CRC) and a Data Entry Technician completing registry data collection. A CRC initializes the workflow by opening a patient’s record in the Epic EHR, then transcribes the data from the EHR onto the paper case report form (CRF). The CRC gives the completed paper CRF to a data entry technician to transfer the information from the paper CRF to the eCRF.
Fig. 1.
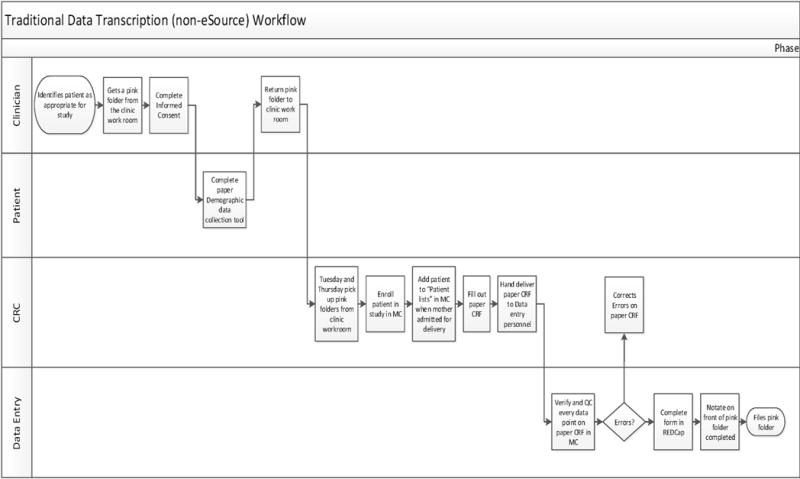
Traditional Data Transcription (non-eSource) workflow.
4.6. ESource observation
The eSource workflow requires one CRC to complete data collection (Fig. 2). A CRC initializes the file by opening the patient’s medical records and the eCRF, all within Epic’s hyperspace (initialization phase). The CRC then verifies the eSourced variables, (demographic information), that were pre-populated with data extracted from the EHR utilizing the RFD standard, and simply clicks the “Save” button (eSourced phase). The CRC then finds the necessary information in the EHR to manually complete the remainder of the fields in the supplemental form (supplemental phase) which appears within the EHR; appearing to the end user as if it is a part of the EHR.
Fig. 2.
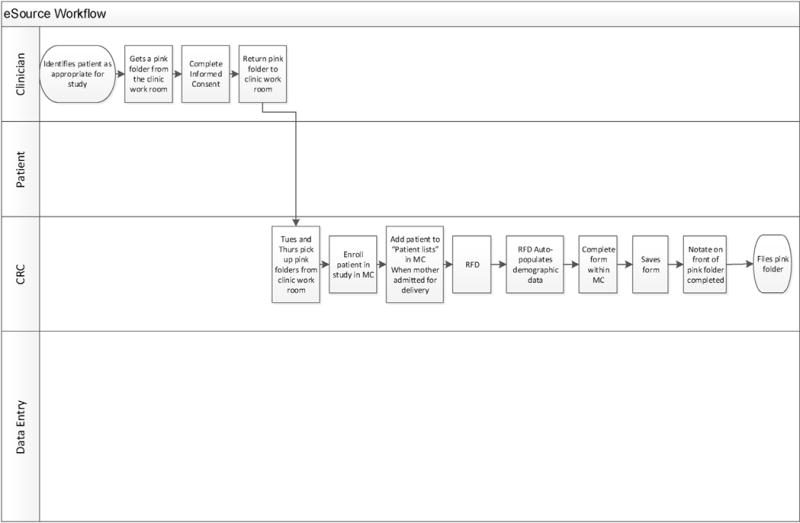
eSource workflow.
4.7. Data collection
Study workflow measurements were obtained through the use of specialized software that contained recording, observing and management components [16]. The recording component captured study participants’ mouse clicks, keystrokes, mouse scrolls, video recorded study participants’ screens, and measured workflow milestones. The observing component was connected to the recording component, recorded milestone attainment and created an Excel file containing milestone related time data. The managing component was used to consolidate study recordings, allowing the study team to view and analyze keystrokes, mouse clicks, and mouse scrolls that occurred in each case, and throughout the study.
4.8. Data analysis
Three workflow sections, (initiation, RFD sourced variables (demographic information), and supplemental manually completed information), were analyzed separately and cumulatively. For each section, the primary outcome variable was the time spent on the task. Secondary outcomes involved data quality (RFD sourced variables only), key strokes and various mouse movements. Using observations obtained prior to study initiation, the study team estimated that the use of eSource versus non-eSource methods would reduce demographic data entry time by 50% with a standard deviation equal to the effect size. Assuming a two-sided alpha of 0.05 and a beta of 0.20, the estimated sample size was 17 cases per group [17].
Thirty-three cases were completed for each workflow (eSource and non-eSource). Twelve cases were eliminated due to incomplete data capture caused by the study team’s unfamiliarity with the data capture software. This left 21 pairs of complete eSource and non-Sourced entered records in each workflow available for analysis. Raw data was extracted from the recorder and observer software applications and merged into a single file containing record, user, action, stage, and timestamp. All keystroke data was replaced with a placeholder ‘X’ value and timestamps were converted to elapsed seconds to de-identify the data. The resulting data set was then imported to IBM SPSS and SAS for analysis. Tables were assembled to summarize total time and motion by each user in each study section. Time saving first were calculated as seconds and then translated into percentages (i.e., by dividing by the mean time in the eSource period). Statistical comparisons were made using 2-sample paired t-tests and Wilcoxon signed rank tests.
5. Results
5.1. Primary objective − efficiency
5.1.1. Time results
The overall average data capture time was reduced with eSource versus non-eSource methods (difference, 151 s per case; eSource, 1603 s; non-eSource, 1754 s; p = 0.051). The average data capture time for the demographic data was reduced (difference, 79 s per case; eSource, 133 s; non-eSource, 213 s; p < 0.001). This represents a 37% time reduction (95% confidence interval 27% to 47%). There was no change in non-eSourced data average data capture time (difference, 28.2 s per case; eSource 1447.9 s; non-eSource, 1476.1 s; p = 0.708) (Table 1).
Table 1.
Data entry Time Comparison.
| Phase | N= | Non-eSourcea | eSourcea | Difference (95%CI) |
p-valueb |
|---|---|---|---|---|---|
| Initiation | 21 | 66.3 (50.5) | 21.3 (19.6) | 45.0 (19.7, 70.4) | 0.001 |
| Demographic | 21 | 212.5 (49.4) | 133.5 (38.1) | 79.1 (56.7,101.4) | 0.000 |
| non-e-Sourced | 21 | 1476.1 (406.7) | 1447.9 (463.2) | 28.2 (−126.6,183.1) | 0.708 |
| Total Time | 21 | 1755.0 (396.5) | 1602.6 (470.0) | 152.3 (−1.1,305.7) | 0.051 |
Mean (Std. Dev).
Paired Samples T−Test.
5.1.2. Keyboard and mouse motion results
Three workflow sections, (initiation, RFD sourced variables (demographic information), and supplemental manually completed information), were analyzed separately and cumulatively in terms of process motions (keystrokes, mouse clicks, etc).
There was a 65% reduction, from 1120 to 392, in eSource workflow keystrokes and a 30% reduction, from 11469 to 8004, in total eSource process motions (less scrolling data) (Table 2). We opted to present these data minus the scrolling counts because of inter-participant variability in computer mouse use. Some participants used the scroll button on the mouse and others dragged and dropped for the same function.
Table 2.
Keyboard and Mouse Movement Results.
| Method | Scroll Button Motion Included | Initiation | Demographicb | Supplemental | Total |
|---|---|---|---|---|---|
| e-Source | Yes | 337 | 1152 | 6974 | 8463 |
| Manuala | Yes | 931 | 2308 | 23533 | 26772 |
| e-Source | No | 337 | 1038 | 6629 | 8004 |
| Manuala | No | 929 | 1695 | 8845 | 11469 |
Manual method includes combined effort from CRC and data entry personnel.
The demographic section contains e−Sourced fields in the database. (This is the only section that contains auto−populated fields).
5.2. Data quality
All eSourced variables (entered by eSource and non-eSource methods) were manually reviewed and adjudicated by the lead clinical research coordinator. Data discrepancies that did not affect the integrity of the data (such as St. abbreviated for street) were not counted as errors. Nine percent of eSourced auto-populated fields were transcribed in error when using non-eSource methods. Errors appeared in critical information such as patient name and medical record number (MRN). There were no eSource data transcription errors.
6. Discussion
This was a proof of concept study. We found improved data entry time and quality for eSource vs. non-eSource data entry methods, and similar data entry time in the supplemental period. (That is, abstraction was faster and more accurate, consistent with the results for the keyboard and mouse times). When the demographic and supplemental periods were combined, the eSource versus non-eSource results were statistically significant. In practice the eSourced elements do not need to be limited to demographics and, indeed, can eventually expand to include the entire EHR.
We anticipated that: (a) in the initiation period the eSource group would require less time, because two people were logging into the system rather than one; (b) in the demographic period the eSource group would require less time because some data fields were being pre-populated; and (c) in the supplemental period the times would be similar because even though different individuals were performing the abstraction they all possessed a similar skill level. (Indeed, observing similar times for the supplemental period would provide substantial reassurance in this regard). It should be noted that the abstraction tasks in the supplemental period weren’t exactly identical – in the non eSource group two screens were open at once whereas in the eSource group they were not – and thus, assuming that the abstractors were comparable, observing similar times for this task would suggest to us that the differences in protocol during the supplemental period were not significant.
Observing similar times in the supplemental period indicated the ability to change workflow processes (from eSource to supplemental data entry) without negatively impacting productivity. These findings indicate the ability to reduce the resources needed to compete data collection by removing the non-value added paper CRF step. (Of note, the data entry person in this study was promoted to a CRC I at study completion).
The study demonstrated a 30% reduction, from 11469 to 8004, in total motion movements (without scrolling data) for the eSource process. We believe that these findings indicate a reduction in effort necessary to complete data collection using the eSource process. We anticipate the reduction in effort will become greater as more variables are auto-populated using eSource.
Another secondary objective compared eSource versus non-eSource workflow flexibility. The non-eSource workflow requires that study monitors access the EHR directly to verify data entry accuracy. The eSource workflow provides a web-based audit trail that could be extended to remote study monitors, removing the need to travel to the site to access the EHR. The current, onsite monitoring process is a resource drain on the sponsor for monitor travel time and on the site for study team member time. We believe there is a potential for increased efficiency through the use of a remote web −based portal but this requires further study.
The eSource process does not prevent or limit the original use of the EDC which is of particular importance in consideration of downtime use as well as scalability. Sites can use either the eSource or non-eSource process for collecting eCRF data.
7. Limitations
Our study has several limitations. The sample size was modest, and reduced further by problems with the recording instruments. This reduced the statistical power for detecting eSource versus non-eSource measurement differences. Different abstractors were used for the eSource and non-eSource methods, and thus an alternative explanation is that the eSource abstractor was more proficient than the non-eSource abstractor − although the similarity in supplemental period results provides substantial reassurance this is not the case. An alternative design might have used the same personnel for eSource and non-eSource workflow, randomized their order and used a larger sample size (which would have the additional advantage of reducing the impact of memory on the abstraction process). The trade-off for such an alternative design would be between internal and external validity − in particular, what would have been lost was the more realistic context under which abstraction took place.
8. Conclusion
This proof of concept study of eSource clinical effectiveness indicates that this method is more efficient, requires less effort to complete data collection, provide an opportunity to redefine data collection workflows and results in better data quality. The eSource process both maintains the flexibility to return to the current manual data transcription process when required and introduces the opportunity for an additional web based monitoring tool. However, further research in different contexts is required to validate our findings.
Supplementary Material
What we knew
It was generally accepted that the eSource method would require less time but there was not a clear consistently quantify because most comparisons were based on manual keystroke logging models which are difficult to replicate.
It was generally accepted that the data quality using eSource would improve but there is limited quantifiable data to support this.
Data quality for manually transcribed data ranged between 2 and 30% error rate.
Not all the data would be able to be auto-populated.
What we learned
A 37% reduction in time was found following the eSource workflow.
The eSource workflow required one less full time employee.
Time savings was identified in the eSource workflow in auto-populated as well as the non auto-populated fields.
Auto-populated data fields have 100% data quality. Manually transcription of those same fields’ results in a 9% error rate that included key data points such as patient name and medical record number.
Even with less than 10 out of 400 fields auto-populated, there was a 37% reduction in time to completion.
Using computer software to capture keystroke and mouse clicks to quantify keystroke logging models is scalable and reproducible.
Acknowledgments
We would like to acknowledge Simona Farcas, Research Practice Manager for the Duke Clinical Research Institute, without her help with the regulatory process this study would be still just an idea, the Duke University Health System Information Security Office: Chuck Kessler, Shelley Epps, and Kendall Lewis for balancing the need to keep our data safe with the desire for innovation and Stephanie Hamre for not only ensuring we had the appropriate hardware to conduct the study but her willingness to set it up and break it down for every observation period and Audrey Brown for kick starting the team on data collection and analysis. The team is grateful for Greg Samsa, PhD who was instrumental in statistical analysis of the data and provided mentorship for the team.
Funding: Supported in part by Duke’s CTSA grant (UL1TR001117) Summary Points
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.ijmedinf.2017.04.015.
References
- 1.Scannell JW, Blanckley A, Boldon H, et al. Diagnosing the decline in pharmaceutical R & D efficiency. Nat Rev Drug Discov. 2012;11(3):191–200. doi: 10.1038/nrd3681. [DOI] [PubMed] [Google Scholar]
- 2.Munos B. Lessons from 60 years of pharmaceutical innovation. Nat Rev Drug Discov. 2009;8(12):959–968. doi: 10.1038/nrd2961. [DOI] [PubMed] [Google Scholar]
- 3.Getz K. Improving protocol design feasibility to drive drug development economics and performance. Int J Environ Res Public Health. 2014;12(11(5)):5069–5080. doi: 10.3390/ijerph110505069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eisenstein EL, Collins R, Cracknell BS, et al. Sensible approaches for reducing clinical trial costs. Clin Trials. 2008;5(1):75–84. doi: 10.1177/1740774507087551. [DOI] [PubMed] [Google Scholar]
- 5.Morrison BW, Cochran CJ, White JG. Monitoring the quality of conduct of clinical trials: a survey of current practices. Clin Trials. 2011;8(3):342–349. doi: 10.1177/1740774511402703. [DOI] [PubMed] [Google Scholar]
- 6.Lobach DF, Kawamoto K, Anstrom KJ, et al. A randomized trial of population-based clinical decision support to manage health and resource use for medicaid beneficiaries. J Med Syst. 2013;37(1):9922. doi: 10.1007/s10916-012-9922-3. Feb 2013. [DOI] [PubMed] [Google Scholar]
- 7.Frobert O, Lagerqvist B, Olivecrona GK, et al. Thrombus aspiration during ST-segment elevation myocardial infarction. N Engl J Med. 2013;369(17):1587–1597. doi: 10.1056/NEJMoa1308789. Oct 24. [DOI] [PubMed] [Google Scholar]
- 8.Lauer MS, D’Agostino RB., Sr The randomized registry trial—the next disruptive technology in clinical research? N Engl J Med. 2013;369(17):1579–1581. doi: 10.1056/NEJMp1310102. [DOI] [PubMed] [Google Scholar]
- 9.Botsis T, Hartvigsen G, Chen F, et al. Secondary use of EHR: data quality issues and informatics opportunities. Summit Trans Bioinf. 2010;2010:1–5. [PMC free article] [PubMed] [Google Scholar]
- 10.Office of the National Coordinator for Health Information Technology (ONC) Office of the Secretary, United States Department of Health and Human Services. Federal health IT strategic plan 2015–2020. Department of Health and Human Services; Washington D. C: 2015. [Google Scholar]
- 11.U.S. Department of Health and Human Services, Food and Drug Administration, Center for Drug Evaluation and Research (CDER) et al. Use of Electronic Health Record Data in Clinical Investigations. 2016 http://www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm501068.pdf.
- 12.ITI Technical Committee. IHE IT infrastructure technical framework supplement retrieve form for data capture. IHE International, Inc; 2010. http://www.ihe.net/Technical_Framework/upload/IHE_ITI_Suppl_RFD_Rev2-1_TI_2010-08-10.pdf. [Google Scholar]
- 13.Fadly A, Rance B, Lucas N, et al. Integrating clinical research with the healthcare enterprise: from the RE-USE project to the EHR4CR platform. J Biomed Inform. 44(2011):S94–S102. doi: 10.1016/j.jbi.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 14.Ferranti J, Musser R, Kawamoto K, et al. The clinical document architecture and the continuity of care record: a critical analysis. J Am Med Inform Assoc. 13(2006):245–252. doi: 10.1197/jamia.M1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris P, Taylor R, Thielke R, et al. Research electronic data capture (REDCap)—A metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009 Apr;42(2):377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Techsmith Morae. 2017 https://www.techsmith.com/morae.html.
- 17.Hulley SB, Cummings SR, Browner WS, Grady D, Hearst N, Newman TB. Designing Clinical Research: An Epidemiological Approach, Lippincott Williams & Wilkins, 2001. 2017 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


