Abstract
Succinic semialdehyde dehydrogenase (SSADH) is one of three enzymes constituting the γ-aminobutyric acid shunt. We have cloned the cDNA for SSADH from Arabidopsis, which we designated SSADH1. SSADH1 cDNA encodes a protein of 528 amino acids (56 kD) with high similarity to SSADH from Escherichia coli and human (>59% identity). A sequence similar to a mitochondrial protease cleavage site is present 33 amino acids from the N terminus, indicating that the mature mitochondrial protein may contain 495 amino acids (53 kD). The native recombinant enzyme and the plant mitochondrial protein have a tetrameric molecular mass of 197 kD. Fractionation of plant mitochondria revealed its localization in the matrix. The purified recombinant enzyme showed maximal activity at pH 9.0 to 9.5, was specific for succinic semialdehyde (K0.5 = 15 μm), and exclusively used NAD+ as a cofactor (Km = 130 ± 77 μm). NADH was a competitive inhibitor with respect to NAD+ (Ki = 122 ± 86 μm). AMP, ADP, and ATP inhibited the activity of SSADH (Ki = 2.5–8 mm). The mechanism of inhibition was competitive for AMP, noncompetitive for ATP, and mixed competitive for ADP with respect to NAD+. Plant SSADH may be responsive to mitochondrial energy charge and reducing potential in controlling metabolism of γ-aminobutyric acid.
The γ-aminobutyric acid (GABA) shunt is a metabolic pathway controlled by three enzymes, the first of which is Glu decarboxylase (GAD) (EC 4.1.1.15), which catalyzes the conversion of Glu to GABA while consuming a proton and releasing CO2. Subsequently, GABA is transaminated by the enzyme GABA transaminase (GABA-T) (EC 2.6.1.19) to succinic semialdehyde (SSA), which is then converted to succinate by the enzyme succinic semialdehyde dehydrogenase (SSADH; EC 1.2.1.24). Succinate then enters the tricarboxylic acid cycle.
GABA metabolism via GABA-shunt enzymes was found in certain prokaryotes (Metzer and Halpern, 1990), as well as in mammals (Tillakaratne et al., 1995) and plants (Bown and Shelp, 1997). However, aside from its occurrence in a wide range of organisms, the GABA shunt has distinct physiological functions and regulatory mechanisms in different organisms. In bacteria, it plays a role in carbon and nitrogen metabolism. Expression of GABA-shunt genes is enhanced by nitrogen deprivation or when succinate replaces Glc as the carbon source (Metzer and Halpern, 1990). In mammals, the GABA shunt is mostly but not exclusively associated with the functions of the inhibitory neurotransmitter GABA in regulating ion channels through GABA receptors (Bormann, 1988). Several functions have been attributed to the GABA shunt in plants, including the control of cytosolic pH, balance between carbon and nitrogen metabolism, and adaptation to stress (for review, see Bown and Shelp, 1997; Snedden and Fromm, 1999). The likelihood of GABA functioning in plants as a signal transmitter has not yet been investigated.
Regarding the regulation of the GABA shunt in plants, the first enzyme of the shunt, GAD, was found to be a calcium/calmodulin-binding protein (Baum et al., 1993, 1996; Snedden et al., 1996). Less is known about the regulation of plant GABA-T and SSADH. We reasoned that cloning of the plant genes coding for these two enzymes and detailed analysis of the purified recombinant proteins would reveal important information about their specific regulation and the functions of the entire GABA shunt. This approach will also provide new tools for employing reverse genetic techniques for studying the GABA shunt in planta. We have focused our studies on SSADH. Although SSADH was purified from plants and characterized to some extent (Yamaura et al., 1988; Satya Narayan and Nair, 1989), the gene for SSADH has to date not been cloned. Only recently was the first full-length sequence of a eukaryotic SSADH cDNA reported (Chambliss et al., 1998). The activity of a recombinant fusion protein of a partial rat SSADH with glutathione S-transferase in crude extracts has been described (Chambliss et al., 1995), but a study of purified recombinant SSADH from eukaryotes has not been reported. Here we describe cloning a cDNA encoding a plant SSADH and detailed kinetic studies of the purified recombinant enzyme.
MATERIALS AND METHODS
SSADH cDNA Cloning and Analysis
A partial cDNA clone from Arabidopsis (accession no. N65241) was obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus), which also supplied a cDNA Lambda ZAP II library of Arabidopsis (CD4-14 and CD4-15, 1- to 3-kb inserts). The cDNA clone N65241 was completely sequenced from both strands. This clone was used to generate a DNA probe (for library screening) by PCR using the upstream sense primer 5′-AGGGTCCACTTATAAATGATG-3′ and the downstream antisense primer 5′-ATGAGTCCTTCGTTCACCCCT-3′. The probe was labeled by random priming using [32P]dCTP and the Klenow enzyme. About 500,000 plaques were screened with this probe. After purification of the recombinant bacteriophages and in vivo excision of the Bluescript SK− plasmid vector with the inserts, the cDNAs were sequenced with T3 and T7 primers from both sides. Additional primers were synthesized to sequence all regions. Computer analysis of the sequence was carried out using Genetics Computer Group programs (Madison, WI). Comparison of the sequence with the GenBank, EMBL, and SwissProt sequence databases was performed using the BLAST (Altschul et al., 1990) network service at the National Center of Biotechnology Information (Bethesda, MD).
Expression of Recombinant Arabidopsis SSADH1 in Bacteria
An Arabidopsis SSADH1-expression clone was constructed by inserting the corresponding cDNA into the bacterial expression vector pET-3d (Novagen, Madison, WI) after amplification by PCR with specific oligonucleotides as primers. The sense primer for PCR amplification contained a NcoI site and began at the second Met of the mature protein. This construct lacked the first two codons (encoding Met-Ser) of the mature protein. An antisense primer located 233 bp downstream of the stop codon was used to create a blunt 3′ end. The pET-3d expression vector was digested with BamHI and the resulting protruding ends were filled by the Klenow reaction. Subsequently, the amplified PCR product and the pET-3d expression vector were digested with NcoI.
Expression of the recombinant protein was initiated by adding 0.5 mm isopropyl thio-β-d-galactoside to the bacterial cultures of transformed BL21 cells in 2YT medium. Cultures of 1.5 L were grown for 9 to 11 h at 30°C before harvesting the cells by centrifugation. The bacterial cells were broken in 40 mL of 50 mm sodium phosphate buffer, pH 9.0, 0.1 m NaCl, 1% (v/v) Triton X-100, 1% (v/v) 2-mercaptoethanol, 1 mm EDTA, 3 mm MgCl2, and 1 mm PMSF by high pressure (Yeda press, 2×), and sonicated for 2 min at 0°C. DNase I was added to the homogenate (50 μg mL−1), and the mixture was kept on ice for another 10 min. Cell debris and insoluble protein were removed by centrifugation at 20,000g for 20 min. The supernatant was assayed for SSADH activity and used for purifying the recombinant SSADH.
Purification of Recombinant SSADH
A FPLC system (ÅKTA explorer 100, Pharmacia Biotech, Piscataway, NJ) with a multiple-wavelength detector (UV-900), a conductivity sensor, and a pH meter (pH/C-900) was used. The recombinant SSADH was purified from bacteria in four steps, including ion-exchange chromatography (Q-Sepharose), size-exclusion chromatography (gel filtration), and second ion-exchange and affinity chromatography (Blue Sepharose). Protein was monitored at 280 nm and all operations were performed at 4°C. Attempts to change the order of the purification steps gave less pure protein with lower yields. After each step, active fractions were pooled and adjusted to the conditions of the next step. The supernatant of bacterial extracts that contained active SSADH was dialyzed against buffer Q1 (25 mm sodium phosphate, pH 9.0, and 1 mm DTT) for 2 h to adjust to pH 9.0 and 6.5 mS conductivity. Subsequently, 10 mL of Q-Sepharose beads pre-equilibrated with buffer Q1 were added and stirred for 30 min, and the beads were filtered on a sintered glass funnel.
Bound SSADH was eluted with 15 mL of 60% buffer Q1/40% buffer Q2 (25 mm sodium phosphate, pH 9.0, 1 m NaCl, and 1 mm DTT). The eluted active fractions were concentrated by (NH4)2SO4 precipitation (70% saturation), and centrifuged at 20,000g. The precipitate was dissolved in 5 mL of buffer GF (50 mm sodium phosphate, pH 7.5, 100 mm NaCl, and 1 mm DTT). The clear supernatant was applied to a Sephadex G-200 (16/60) column (120 mL total volume) pre-equilibrated with buffer GF at a flow rate of 1 mL min−1. The pooled active fractions were loaded onto a prepackaged anion-exchange column (5-mL total bed volume, flow rate 5 mL min−1; HiTrap Q, Pharmacia) pre-equilibrated with buffer Q1. For elution, a linear gradient of 0% to 30% buffer Q2 was applied. SSADH was eluted at 23% of buffer Q2 (255 mm Na+). Thirty percent of buffer Q2 was held for 10 mL (2 column volumes), then buffer Q2 was increased to 100% for 30 mL in one step.
The adjusted active samples were applied to a Blue Sepharose 6 Fast-Flow column (1.6 × 4 cm, 8 mL total bed volume) pre-equilibrated with buffer BS (25 mm sodium phosphate, pH 7.2, and 1 mm DTT). The SSADH activity was eluted with 10 mL of 10 mm NAD+ in BS buffer. Active fractions were pooled and concentrated (Mr 30,000) (Fugisep Midi concentrator, Intersep, Workingham, UK). The protein was either stored at 4°C in buffer BS for immediate use or at −20°C in 25% (w/v) glycerol. The purified protein was used for both antibody preparation and for kinetic analysis. In the latter case, the protein was dialyzed to eliminate NAD+. Molecular mass was determined with a Sephadex G-200 (16/60) column (Superdex high load) after calibration according to the manufacturer's instructions (Pharmacia Biotech).
Isolation and Subfractionation of Mitochondria
Mitochondria from dark-grown Arabidopsis cell cultures were prepared as described in Klein et al. (1998) and from potato tubers as outlined in Braun and Schmitz (1995). The mitochondria were isolated by differential centrifugation and purified by Percoll step-gradient centrifugation. The organelles were suspended in 0.4 m mannitol, 0.1% (v/v) BSA, and 10 mm KH2PO4, pH 7.2, and protein was adjusted to 3 mg mL−1. Subfractionation by sonication and final separation of membrane and matrix fractions was achieved by ultracentrifugation at 100,000g for 60 min as described previously (Jaensch et al., 1996). For determining SSADH activity, mitochondria (8–16 mg of protein) were sonicated in 8 mL of sonication buffer (100 mm Na2HPO4/NaH2PO4, pH 7.4, 1 mm DTT, 1 mm EDTA, and 0.1% [w/v] Triton X-100).
PAGE and Immunodetection
Denaturing SDS-PAGE was performed as described previously (Baum et al., 1993). Non-denaturing PAGE was performed on 10% (v/v) acrylamide gels in the same buffers without SDS. The sample buffer contained no SDS or reducing reagent. The molecular mass markers were from Bio-Rad. Proteins were transferred to nitrocellulose membranes (Schleicher & Schuell, Dassel, Germany) by electrophoresis at 135 mA for 30 min. Immunodetection was with polyclonal antibodies raised against the recombinant SSADH after the final chromatography step (described above). Preparation of the antibodies in rabbits was as described previously (Baum et al., 1993). The anti-SSADH1 antibodies detected a single protein band of the expected size in plant mitochondria and in extracts of Escherichia coli expressing the recombinant SSADH, but not in extracts from control bacteria carrying the empty vector.
SSADH Enzyme Assay
The standard enzyme assay contained 0.1 m sodium phosphate buffer (pH 9.0), 1 mm DTT, 0.1 mm SSA, and 0.5 mm NAD+. Measurements of the initial activity of SSADH (5 s to 2 min) were carried out spectrophotometrically at 340 nm (UVIKON 930, Kontron Instruments, Milan) in a 0.5-mL reaction vessel at 24°C with 460 μL of reaction buffer and up to 40 μL of SSADH-containing sample. For testing multiple fractions during the purification procedure of the protein, a kinetics software program (MR5000, Dynatech, Burlington, MA) was used. Up to 40 fractions could be checked for SSADH activity at once. The pH dependence of SSADH1 was determined in 0.1 m Na2HPO4/NaH2PO4 buffer containing 1 mm DTT at different pH values. The initial velocity kinetics were linearized by the method of Hanes (Bisswanger, 1994). Initially, Michaelis-Menten kinetics were assumed. The noncompetitive inhibitor (I) binds either to the free enzyme (E) or to the enzyme substrate complex (ES), and the dissociation constants are Kic = [E][I]/[EI] and Kiu = [ES][I]/[ESI].
The values of Kic and Kiu were determined from secondary plots (the slopes or the intercepts of linearized plots versus inhibitor concentration). In case of partial inhibition, these secondary plots are nonlinear. Dead-end inhibitions display linear secondary plots. Kinetic studies in the presence of inhibitors with respect to NAD+ were performed at several constant inhibitor concentrations while the NAD+ concentration was varied. Kinetics studies were repeated at least three times with different preparations of the purified recombinant SSADH.
RESULTS
Cloning of an Arabidopsis cDNA for SSADH
The Arabidopsis EST database was searched with a partial human SSADH sequence (accession no. P51649; 323 bp) as a query using BLASTp (Altschul et al., 1990). Several Arabidopsis ESTs were identified as SSADH homologs, and all of them appeared to be partial clones. One partial cDNA clone (accession no. N65241; 325 bp), corresponding to the presumed C terminus of SSADH, was used as a probe to screen an Arabidopsis cDNA library. The screening resulted in the isolation of a longer cDNA clone with an ORF of 1485 bp encoding a polypeptide of 495 amino acid residues. Further analysis of the Arabidopsis EST database with this sequence as a query revealed another clone (accession no. T21534) whose 5′ determined sequence perfectly matched the nucleotide sequence of the previous clone, but extended beyond it by 33 codons with an ATG codon in-frame.
This clone was completely sequenced and the encoded protein was named SSADH1. The ORF contains an RQMS sequence that has been shown to be a site for mitochondrial matrix protease (Gavel and von Heinje, 1990) (Fig. 1). In addition, the 33-amino acid N-terminal segment contains basic amino acids and lacks acidic residues, which is characteristic of mitochondrial targeting sequences (Gavel and von Heinje, 1990). Thus, the full sequence of the SSADH1 cDNA encodes a 528-amino acid protein (56.42 kD), whereas the presumed mature protein, lacking the N-terminal 33 amino acid residues, contains 495 amino acids (53.15 kD) (Fig. 1). Furthermore, antibodies against the recombinant protein detected a mitochondrial protein of an apparent molecular mass of 53 kD, which was indistinguishable in mobility on SDS-PAGE from that of the recombinant, mature protein.
Figure 1.
Comparison of Arabidopsis SSADH1 with SSADH from bacteria and mammals. Alignment of the deduced amino acid sequences of the full-length SSADH from Arabidopsis (A), E. coli (E), and human (H) (Chambliss et al., 1998) (GenBank accession nos. AF117335, M8834, and Y11192, respectively), and a partial rat (R) SSADH (Chambliss et al., 1995) (GenBank accession no. P51650). Identical amino acids are boxed with a gray background. The ADH motifs discussed in the text are underlined. Arrows indicate the predicted cleavage sites of plant and human (Chambliss et al., 1998) SSADH by mitochondrial proteases.
Comparison of At-SSADH1 with SSADH from Mammals and Prokaryotes
BLASTp searches of databases with the Arabidopsis SSADH1 amino acid sequence revealed the highest similarity to SSADH from several organisms (Fig. 1) and less similarity to other types of aldehyde dehydrogenases (ADHs). Arabidopsis SSADH1 shares 59.2%, 58.2%, and 59.5% amino acid sequence identity with SSADH from E. coli, rat, and human SSADH, respectively. Among the different SSADHs, the sequences near the C-termini show higher similarity than those near their N termini. Several ADH-specific motifs are present in the primary structure of Arabidopsis SSADH1. For example, the ADH Glu active site (Chambliss et al., 1995) LELGGNAP corresponds to residues 296 through 303 (residues 263–270 in the mature protein). The consensus motif FRNSGQTCVCAN, containing the important ADH Cys active site (Chambliss et al., 1995), is present in SSADH1 at residues 324 through 335 (residues 291–302 in the mature protein). In addition, invariant Gly residues present at positions 245 and 250 of the mature protein are engaged in NAD+ binding in cytosolic ADHs (Hempel et al., 1993; Chambliss et al., 1995; Vedadi et al., 1997). In contrast, Arabidopsis SSADH1 is different from other SSADH proteins in several positions. It contains an EEIFGP sequence motif near the C terminus (residues 384–389 in the mature protein), like most other ADHs (Chambliss et al., 1998), instead of the EETFGP motif found in SSADH from E. coli, rat, and humans. SSADH1 also possesses an additional Cys residue at position 53 of the mature protein. Acidic residues 326 and 332 replace basic amino acids present in the corresponding positions of E. coli, rat, and human SSADH, and replacement of basic with nonpolar amino acids is found at positions 344 and 405 of the mature protein. Thus, although plant SSADH is very similar to bacterial and mammalian SSADH, it has certain unique residues in its sequence.
Purification of the Recombinant SSADH1 to Homogeneity and Immunodetection of a Plant SSADH in Mitochondria
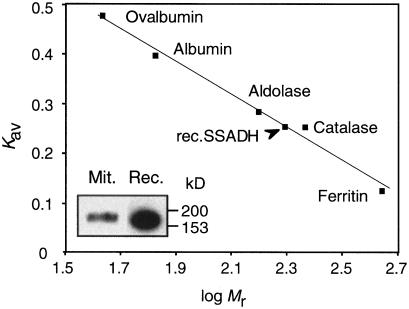
A recombinant protein corresponding to the mature Arabidopsis SSADH1 (but lacking the first Met-Ser residues) was expressed in E. coli and purified from the soluble fraction by a four-step procedure (see “Materials and Methods”). On SDS-PAGE, a single band of approximately 53 kD was stained with Coomassie Blue (Fig. 2, lane 4). Table I shows the data from a typical purification experiment. Non-denaturing PAGE of the purified recombinant enzyme gave a single band with mobility near 200 kD, which was indistinguishable from the mobility of the plant mitochondrial protein detected with anti-SSADH antibodies (Fig. 3, inset). The apparent molecular mass of SSADH1 was confirmed by gel filtration (Fig. 3), suggesting that the active Arabidopsis SSADH1 is a homotetramer of a 53-kD subunit. The activity of the purified recombinant enzyme was 23 units mg−1 protein. This is higher than SSADH of purified barley (2.7 units mg−1 protein), potato (6.5 units mg−1 protein), or human (2.7 units mg−1 protein) (Yamaura et al., 1988; Satya Narayan and Nair, 1989; Chambliss and Gibson, 1992).
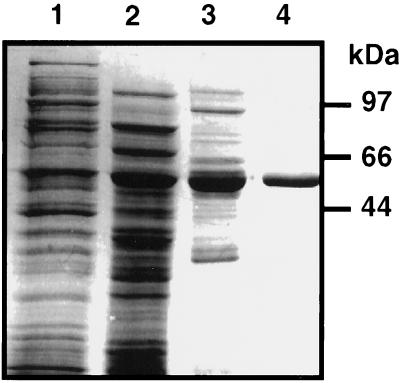
Figure 2.
Purification of recombinant Arabidopsis SSADH1. Coomassie Blue-stained SDS-PAGE of different chromatography fractions during purification of Arabidopsis SSADH1. Lane 1, Total soluble fraction; lane 2, pooled active fractions of the first Q-Sepharose; lane 3, pooled active fractions of the second Q-Sepharose; lane 4, pooled active fractions of the Blue Sepharose. Molecular mass markers are shown on the right.
Table I.
Purification of the recombinant Arabidopsis SSADH1
| Step | Total Protein | Total Activity | Specific Activity | Yield | Purification |
|---|---|---|---|---|---|
| mg | units | units mg−1 protein | % | fold | |
| Soluble protein | 1230.0 | 195 | 0.16 | 100 | 1.0 |
| Q-Sepharose I | 72.8 | 33 | 0.45 | 17 | 2.9 |
| Size exclusion | 41.1 | 29 | 0.71 | 15 | 4.5 |
| Q-Sepharose II | 11.3 | 16 | 1.39 | 8 | 8.8 |
| Blue Sepharose | 0.6 | 14 | 22.70 | 7 | 143.0 |
Figure 3.
Arabidopsis SSADH1 is a homotetramer under nondenaturing conditions. Molecular mass of the purified recombinant SSADH1 (rec.SSADH) was determined by gel filtration. Inset, Nondenaturing-PAGE separation of the purified recombinant SSADH1 (Rec.) and mitochondrial SSADH (Mit.) from potato tubers. Proteins were immunodetected with polyclonal antibodies raised against the purified recombinant Arabidopsis SSADH1 protein.
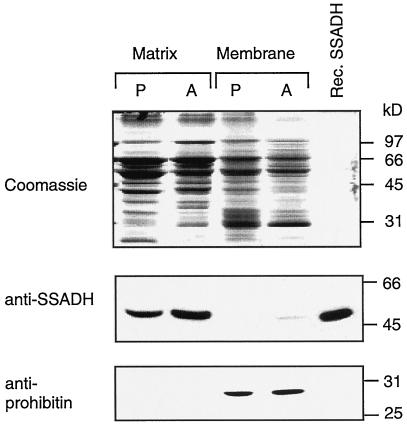
The localization of the SSADH protein in plant mitochondria is consistent with recent evidence for the association of soybean SSADH activity exclusively in mitochondria (Breitkreuz and Shelp, 1995). To investigate SSADH submitochondrial localization, purified mitochondria exhibiting SSADH activity (approximately 0.2 unit mg−1 total mitochondrial protein) were further subfractionated into membrane and matrix proteins. SSADH activity was detected exclusively in the matrix fraction (0.3–0.39 unit mg−1 protein for potato and Arabidopsis). On SDS-PAGE, a single immunoreactive protein band was detected predominantly in the matrix fraction of both Arabidopsis and potato mitochondria (Fig. 4). Its electrophoretic mobility was indistinguishable from that of the 53-kD recombinant SSADH1 (Fig. 4). In contrast, prohibitin, another mitochondrial protein (Snedden and Fromm, 1997), was detected predominantly in the mitochondrial membrane fraction. In addition, recombinant SSADH1 displayed maximal activity between pH 9.0 and 10. Below pH 6.0, its activity dropped to less than 11% of maximal activity (data not shown), in agreement with the pH optimum of SSADH from other sources (Callewaert et al., 1973; Ryzlak and Pietruszko, 1988). These results strongly suggest that plant SSADH is a mitochondrial matrix enzyme.
Figure 4.
Detection of a 53-kD SSADH in the plant mitochondrial matrix. Matrix and membrane fractions of mitochondria from potato tubers (P) and Arabidopsis cell cultures (A), and a sample of the recombinant Arabidopsis SSADH1 (Rec. SSADH) were separated on SDS-PAGE and either stained with Coomassie Blue or tested with antibodies against Arabidopsis SSADH1 or prohibitin (Snedden and Fromm, 1997).
Substrate and Cofactor Specificity of Arabidopsis SSADH1
Several substrates in the concentration range of 10 μm to 10 mm were tested. This analysis revealed that Arabidopsis SSADH1 is highly specific for SSA (Table II) and could not utilize formaldehyde, propionaldehyde, glycerine aldehyde, lactate, or ethanol. Very low activity (0.2% of SSA) was found with 100 μm acetaldehyde as a substrate. Glutaraldehyde (GA) was slowly oxidized by SSADH1. The kinetics showed a substrate inhibition at [GA] higher than 2.5 mm. The K0.5 value for GA was 32-fold higher than that for SSA (500 μm, and 15 ± 5 μm, respectively), which corresponds to a lower affinity of SSADH to GA. Substrate inhibition by SSA was found at concentrations higher than 0.15 mm (data not shown). Thus, with respect to the substrates GA and SSA, SSADH1 does not follow Michaelis-Menten kinetics. Therefore, K0.5 values instead of Km values are used (Bisswanger, 1994) to describe the substrate concentration at which half of the binding sites are occupied. SSADH1 exclusively utilized NAD+ as a cofactor. No activity was found with NADP+ as a cofactor. The Km value for NAD+ was 130 ± 77 μm (Table II), depending on the preparation, which is in between the value for barley and potato SSADH. The K0.5 value for SSA was on the same order of magnitude as for barley, pig, and potato SSADHs (Table II).
Table II.
Kinetic constants of recombinant Arabidopsis SSADH1 compared with SSADH from other organisms
| Kinetic Constant | Arabidopsis | Pig | Barley | Potato |
|---|---|---|---|---|
| μm | ||||
| K0.5, SSA | 15 ± 5 | 11 | 15.9 | 4.6 |
| Km, NAD+ | 130 ± 77 | 310 | 166.0 | 31.0 |
| Ki, NADH | 122 ± 86 | 310 | ||
| K0.5, GA | 500 | — | — | — |
The kinetic constants are compared to those published for pig SSADH (Blaner and Churchich, 1980), barley SSADH (Yamaura et al., 1988), and potato SSADH (Satya Narayan and Nair, 1989).
The effect of reversible binding of substrate analogs on SSADH activity was tested with 3-hydroxybenzaldehyde, 4-hydroxybenzaldehyde, and p-pyridoxal. Inhibition of SSADH by these components has been previously described (Chambliss and Gibson, 1992). The recombinant plant SSADH was pre-incubated with the respective analogs (100 μm and 10 mm) for 1 min, and then 100 μm SSA was added and activity was monitored. SSADH1 activity was inhibited by 82% in the presence of 10 mm 4-hydroxybenzaldehyde and by 25% with 10 mm p-pyridoxal.
Regulation of Arabidopsis SSADH1 by Nucleotides
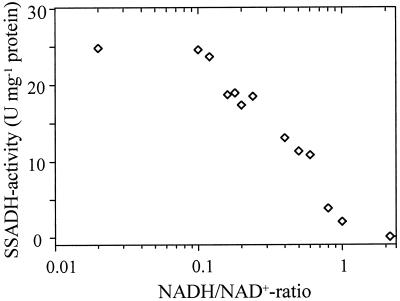
Compared with succinyl-CoA-synthetase, which provides the Krebs cycle with succinate and ATP, mitochondrial SSADH supplies succinate and NADH. An inhibitory effect of NADH on SSADH activity is known for animal SSADH (Duncan and Tipton, 1971; Blaner and Churchich, 1980; Rivett and Tipton, 1981). We found a similar inhibition of Arabidopsis SSADH1. With increasing NADH concentrations, SSADH1 activity decreased significantly (Fig. 5). At a NADH/NAD+ ratio of 1, SSADH1 activity was reduced to less than 10%, and SSADH1 was completely inhibited at a NADH/NAD+ ratio of 2. The inhibition was competitive for NADH with respect to NAD+. The affinity of the enzyme for NAD+ and NADH was the same (Table II, kinetics not shown).
Figure 5.
Control of SSADH1 activity by the NADH/NAD+ ratio. SSADH1 activity was determined in the presence of increasing NADH concentrations, while the NAD+ concentration was held constant at 500 μm. The SSA concentration was 100 μm.
We then tested the regulation of SSADH1 activity by adenine nucleotides. Inhibition of SSADH by AMP has been reported previously (Rivett and Tipton, 1981; Satya Narayan and Nair, 1989), but the effects of ADP and ATP on SSADH activity have not. We found an inhibitory effect for all three adenine nucleotides on SSADH1 activity. The inhibition was competitive, mixed-competitive, and noncompetitive for AMP, ADP, and ATP, respectively. AMP was a dead-end competitive inhibitor with respect to NAD+ (data not shown), in agreement with previous studies (Rivett and Tipton, 1981), and had a Ki value of 2.9 mm. Secondary intercepts and/or slope re-plots were linear.
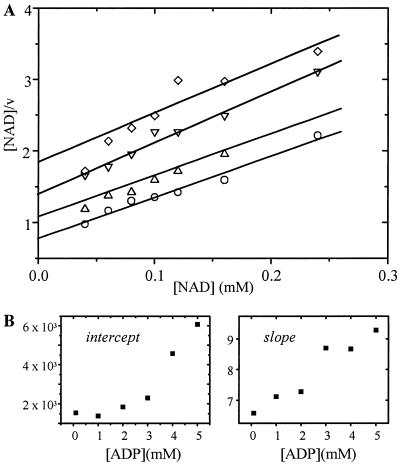
Inhibition of SSADH1 by ADP was mixed-competitive with respect to NAD+ (Fig. 6A). The Kic value was lower than the Kiu value, and both were in the millimolar range. Most adenine-dependent enzymes use the nucleotides complexed with Mg2+. However, no difference in ADP inhibition of SSADH1 activity was found in the presence or absence of 5 mm Mg2+. Thus, binding of NAD+ was hindered by ADP and vice versa. The second-order plots were not linear (Fig. 6B), indicating a partial rather than a dead-end inhibition.
Figure 6.
Inhibition of SSADH1 by ADP determined by Hanes-plot (A) and secondary plots (B). The concentration of NAD+ varied between 0.01 and 0.3 mm, while the ADP concentration was held constant at the following values: 1 mm ADP (○); 2 mm ADP (▵); 3 mm ADP (▿); 4 mm ADP (⋄). The SSA concentration was 100 μm.
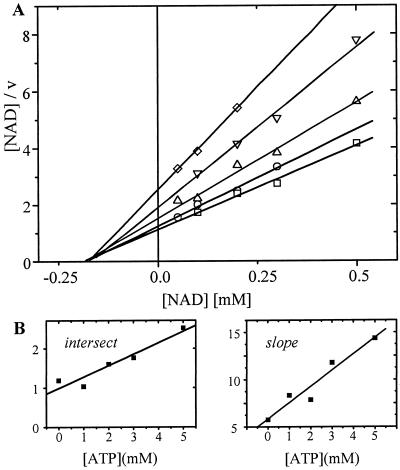
ATP was a noncompetitive dead-end inhibitor with respect to NAD+ (Fig. 7A). The Kic value was 2.5 mm and the Kiu value was 8.0 mm (Fig. 7B). No difference was found in the presence or absence of 5 mm Mg2+. Therefore, binding of NAD+ is hindered by ATP and vice versa. ATP is assumed to bind to the free enzyme and the enzyme-substrate complex. In the presence of the inhibitor, the maximal velocity was not reached even with high NAD+ concentrations.
Figure 7.
Inhibition of SSADH1 by ATP determined by Hanes-plot (A) and secondary plots (B). The concentration of NAD+ varied between 0.05 and 0.5 mm, while the ATP concentration was held constant at the following values: 0 mm ATP (□); 1 mm ATP (○); 2 mm ATP (▵); 3 mm ATP (▿); 5 mm ATP (⋄). The SSA concentration was 100 μm.
DISCUSSION
In this study, we describe the biochemical analysis of the first cloned SSADH from plants. Antibodies raised against purified recombinant Arabidopsis SSADH1 revealed the localization of plant SSADH in the mitochondria of potato tubers and Arabidopsis cell cultures. The purified recombinant protein is similar to other multimeric eukaryotic SSADH enzymes. The native 53-kD protein was assembled into a 197-kD homotetramer, indistinguishable in its electrophoretic mobility from that of SSADH from potato (Fig. 3), and similar to SSADH from barley and animals (Blaner and Churchich, 1980; Ryzlak and Pietruszko, 1988; Chambliss and Gibson, 1992). A mammalian brain SSADH reported by Ryzlak and Pietruszko (1988), however, was a heterotetrameric protein composed of different-sized subunits (61–63 kD). The apparent size of a previously reported homotetrameric SSADH from potato mitochondria (Satya Narayan and Nair, 1989) was substantially smaller (145 kD).
A four-amino acid sequence motif resembling a mitochondrial protease recognition site was found 33 amino acid residues from the N terminus of the full-length Arabidopsis SSADH1-coding region. This suggests that transport of SSADH1 into mitochondria involves cleavage of the N terminus, resulting in a mature SSADH subunit of 53 kD. Indeed, SSADH from potato and Arabidopsis mitochondria displayed the same electrophoretic mobility as that of the 53-kD recombinant SSADH1 (Fig. 4), which is clearly smaller than the full-length SSADH1 (56 kD). The fact that all reported eukaryotic SSADHs displayed an optimum of activity at a pH above 9.0 is also characteristic of their mitochondrial localization. In addition, we showed that plant mitochondrial SSADH is present predominantly in the matrix fraction, which is consistent with its calculated average hydrophobicity of 0, with a uniform distribution throughout the protein.
Like other eukaryotic SSADHs, Arabidopsis SSADH1 requires NAD+ rather than NADP+, which is typically used by bacterial SSADH. We further investigated the possible regulation of SSADH by intermediates of the Krebs cycle and the GABA shunt. GABA, Glu, pyruvate, and succinate had no effect at concentrations from 1 μm to 10 mm. Because Ca2+ was found to be a regulator of certain mitochondrial dehydrogenases (Nichols and Denton, 1995), we tested the activity of SSADH in the presence of several cations, including Ca2+, Mg2+, Zn2+, Hg2+, Pb2+, Cu2+, and Ni2+. None of these had any effect at concentrations as great as 0.1 mm.
In addition to succinyl-CoA-synthetase, SSADH also supplies the Krebs cycle with succinate (Fig. 8). However, in the former reaction succinate and ATP are formed, whereas in the latter succinate and NADH are produced. Product inhibition by NADH was shown for SSADH from animals (Blaner and Churchich, 1980; Rivett and Tipton, 1981), and our study now provides similar data on plant SSADH, which exhibits a relatively high affinity for NADH (Ki = 122 ± 86 μm). Adenine nucleotides are well known as regulatory components in mitochondrial energy metabolism (e.g. succinate dehydrogenase is activated by ATP; Singer et al., 1973). However, the effects of ADP and ATP on SSADH activity have not been investigated. With initial-rate studies of SSADH kinetics, we found that AMP, ADP and ATP inhibited the enzyme. The type of inhibition was competitive, mixed-competitive, and noncompetitive, respectively, with respect to NAD+.
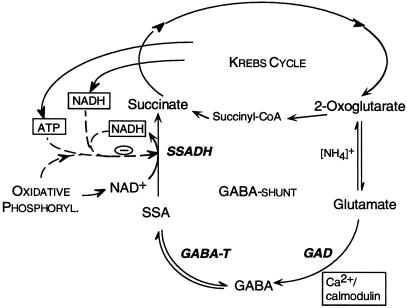
Figure 8.
Model for the regulation of the GABA shunt in plants. The three enzymes of the GABA shunt, GAD (cytosolic), GABA-T (mitochondrial), and SSADH (mitochondrial) are in bold letters. Dashed lines, Effectors; solid lines, substrates and products. Phosphoryl., Phosphorylation.
The effect of AMP (and less so for ADP) depended on the substrate (NAD+) concentration, whereas the inhibitory effect of ATP was independent of it. Vmax was not reached even under a high substrate (NAD+) concentration in the presence of ATP. The dissociation constants (Kiu and Kic) of SSADH for ADP and ATP were in the millimolar range. Certain enzymes of the Krebs cycle in plants are affected by adenine nucleotides (ATP) in the millimolar range (for review, see Raymond et al., 1987). However, concentrations of nucleotides in plant mitochondria have not been determined definitively. For animal mitochondria, concentrations of total adenine nucleotides vary from less than 0.5 to 6 mm (Pradet and Raymond, 1983; Hutson et al., 1989). NAD(H) is present in the millimolar range in some plants (Wigge et al., 1993). The redox state of endogenous mitochondrial pyridine nucleotides strongly depends on the respiratory state of mitochondria. NADH/NAD+ ratios were reported to vary from 0.065 to 0.33 in state 3 and state 4, respectively (Kroemer and Heldt, 1991). These values are well within the responsive range of SSADH1 (Fig. 5).
The concentrations of adenine nucleotides are controversial. In one study the amount of ATP and ADP was found to be double that of NAD(H), and the ATP/ADP ratio was approximately 2 (Roberts et al., 1997). Therefore, it is possible that under specific physiological conditions the concentrations of NADH, ADP, and ATP in plant mitochondria are in the range that affects SSADH activity. Because increasing [NAD+] cannot abolish the effect of ATP on SSADH, high concentrations of ATP will attenuate SSADH activity.
Our study supports the model that plant SSADH is highly sensitive to changes in the NADH/NAD+ ratio and adenylate composition. Stress situations (e.g. oxygen deprivation) affect mitochondrial metabolism and are accompanied by higher ratios of NADH to NAD+ (Wigge et al., 1993) and ADP to ATP (Shelp et al., 1995). Responsiveness of SSADH to these changes will influence not only the supply of succinate to the Krebs cycle, but also the backward reaction from SSA to GABA (Fig. 8). In response to several stress situations, the first enzyme of the GABA shunt, GAD, is stimulated (for review, see Snedden and Fromm, 1999), leading to an increase in GABA synthesis. However, the fate of the synthesized GABA is not necessarily the same under all stress conditions. Under hypoxia, for example, oxidative phosphorylation is attenuated and GABA accumulates (Snedden and Fromm, 1999). Under these conditions, a fairly high reduction potential (Wigge et al., 1993) in mitochondria will attenuate the metabolism of GABA via SSADH, causing its accumulation (Fig. 8). In summary, we propose that in plants the GABA shunt is regulated by calcium signaling via calmodulin in the cytosol and by the energy charge and reducing potential in the mitochondria.
ACKNOWLEDGMENTS
We thank Drs. Wayne Snedden and Jacob Piehler for critical reading of the manuscript and Profs. Amnon Horovitz and Hans Bisswanger for discussions concerning kinetics. We also thank Dr. H.P. Braun (University of Hannover, Germany) for providing results of SSADH1 localization in Arabidopsis mitochondria, and the Arabidopsis Biological Resource Center at Ohio State University for providing cDNA clones.
Footnotes
This work was supported by a grant (to H.F.) from the Leo and Julia Forchheimer Center for Molecular Genetics, Weizmann Institute of Science. K.B. was the recipient of a MINERVA postdoctoral fellowship.
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Baum G, Chen Y, Arazi T, Takutsuji H, Fromm H. A plant glutamate decarboxylase containing a calmodulin binding domain: cloning, sequence, and functional analysis. J Biol Chem. 1993;268:19610–19617. [PubMed] [Google Scholar]
- Baum G, Lev-Yadun S, Fridmann Y, Arazi T, Katsnelson H, Zik M, Fromm H. Calmodulin binding to glutamate decarboxylase is required for regulation of glutamate and GABA metabolism and normal development in plants. EMBO J. 1996;15:2988–2996. [PMC free article] [PubMed] [Google Scholar]
- Bisswanger H. Enzymkinetik. Weinheim, Germany: VHC; 1994. [Google Scholar]
- Blaner WS, Churchich J. The binding of NADH to succinic semialdehyde dehydrogenase. Eur J Biochem. 1980;109:431–437. doi: 10.1111/j.1432-1033.1980.tb04812.x. [DOI] [PubMed] [Google Scholar]
- Bormann J. Electrophysiology of GABAA and GABAB receptor subtypes. Trends Neurosci. 1988;11:112–116. doi: 10.1016/0166-2236(88)90156-7. [DOI] [PubMed] [Google Scholar]
- Bown AW, Shelp BJ. The metabolism and function of γ-aminobutyric acid. Plant Physiol. 1997;115:1–5. doi: 10.1104/pp.115.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun HP, Schmitz UK. Molecular structure of the 8 kD subunit of cytochrome c reductase from potato and its ΔΨ dependent import into isolated mitochondria. Bioenerg Biomembr. 1995;27:423–436. [Google Scholar]
- Breitkreuz KE, Shelp BJ. Subcellular distribution of the 4-aminobutyrate shunt in developing soybean cotyledon protoplasts. Plant Physiol. 1995;108:99–103. doi: 10.1104/pp.108.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callewaert DM, Rosemblatt MS, Suzuki K, Tchen TT. Succinic semialdehyde dehydrogenase from a Pseudomonas species. I. Purification and chemical properties. J Biol Chem. 1973;248:6009–6013. [PubMed] [Google Scholar]
- Chambliss KL, Caudle DL, Hinson DD, Moomaw CR, Slaughter CA, Jakobs C, Gibson KM. Molecular cloning of the mature NAD+-dependent succinic semialdehyde dehydrogenase from rat and human. J Biol Chem. 1995;270:461–467. doi: 10.1074/jbc.270.1.461. [DOI] [PubMed] [Google Scholar]
- Chambliss KL, Gibson KM. Succinic semialdehyde dehydrogenase from mammalian brain: subunit analysis using polyclonal antiserum. Int J Biochem. 1992;24:1493–1499. doi: 10.1016/0020-711x(92)90077-e. [DOI] [PubMed] [Google Scholar]
- Chambliss KL, Hinson DD, Trettel F, Malaspina P, Novelletto A, Jakobs C, Gibson KM. Two exon-skipping mutations as the molecular basis of succinic semialdehyde dehydrogenase deficiency (4-hydroxybutyric aciduria) Am J Hum Genet. 1998;63:399–408. doi: 10.1086/301964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan RJS, Tipton KF. The kinetics of pig brain aldehyde dehydrogenase. Eur J Biochem. 1971;22:538–543. doi: 10.1111/j.1432-1033.1971.tb01574.x. [DOI] [PubMed] [Google Scholar]
- Gavel Y, von Heinje G. Cleavage-site motifs in mitochondrial targeting peptides. Protein Eng. 1990;4:33–37. doi: 10.1093/protein/4.1.33. [DOI] [PubMed] [Google Scholar]
- Hempel J, Nicholas H, Lindahl R. Aldehyde dehydrogenases: widespread structural and functional diversity within a shared framework. Protein Sci. 1993;2:1890–1900. doi: 10.1002/pro.5560021111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutson SM, Berkich D, Williams GD, LaNoue KF, Briggs RW. 31P-NMR visibility and characterization of rat liver mitochondrial matrix adenine nucleotides. Biochemistry. 1989;28:4325–4332. doi: 10.1021/bi00436a030. [DOI] [PubMed] [Google Scholar]
- Jaensch L, Kruft V, Schmitz UK, Braun HP. New insights into the composition, molecular mass and stoichiometry of the protein complexes of plant mitochondria. Plant J. 1996;9:357–368. doi: 10.1046/j.1365-313x.1996.09030357.x. [DOI] [PubMed] [Google Scholar]
- Klein M, Binder S, Brennicke A. Purification of mitochondria from Arabidopsis. Methods Mol Biol. 1998;82:49–53. doi: 10.1385/0-89603-391-0:49. [DOI] [PubMed] [Google Scholar]
- Kroemer S, Heldt HW. Respiration of pea leaf mitochondria and redox transfer between the mitochondrial and extramitochondrial compartment. Biochim Biophys Acta. 1991;1057:42–50. [Google Scholar]
- Metzer E, Halpern YS. In vivo cloning and characterization of the gabCTDP gene cluster of Escherichia coli K-12. J Bacteriol. 1990;172:3250–3256. doi: 10.1128/jb.172.6.3250-3256.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols BJ, Denton RM. Towards the molecular basis for the regulation of mitochondrial dehydrogenases by calcium ions. Mol Cell Biochem. 1995;149–150:203–212. doi: 10.1007/BF01076578. [DOI] [PubMed] [Google Scholar]
- Pradet A, Raymond P. Adenine nucleotide ratios and adenylate energy charge in energy metabolism. Annu Rev Plant Physiol. 1983;34:199–224. [Google Scholar]
- Raymond P, Gidrol X, Salon C, Pradet A. Control involving adenine nucleotides and pyridine nucleotides. In: Stump PK, Conn EE, editors. The Biochemistry of Plants. Vol. 11. New York: Academic Press; 1987. pp. 129–176. [Google Scholar]
- Rivett AJ, Tipton KF. Kinetic studies with rat-brain succinic-semialdehyde dehydrogenase. Eur J Biochem. 1981;117:187–193. doi: 10.1111/j.1432-1033.1981.tb06319.x. [DOI] [PubMed] [Google Scholar]
- Roberts JKM, Aubert S, Gout E, Bligny R, Douce R. Cooperation and competition between adenylate kinase, nucleoside diphosphokinase, electron transport, and ATP synthase in plant mitochondria studied by 31P-nuclear magnetic resonance. Plant Physiol. 1997;113:191–199. doi: 10.1104/pp.113.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryzlak MT, Pietruszko R. Human Brain “high Km” aldehyde dehydrogenase: purification, characterization, and identification as NAD+-dependent succinic semialdehyde dehydrogenase. Arch Biochem Biophys. 1988;266:386–396. doi: 10.1016/0003-9861(88)90270-6. [DOI] [PubMed] [Google Scholar]
- Satya Narayan V, Nair PM. Potato tuber succinate dehydrogenase: purification and characterization. Arch Biochem Biophys. 1989;275:469–477. doi: 10.1016/0003-9861(89)90393-7. [DOI] [PubMed] [Google Scholar]
- Shelp BJ, Walton CS, Snedden WA, Tuin LG, Oresnik IJ, Layzell DB. Gaba shunt in developing soybean seeds is associated with hypoxia. Physiol Plant. 1995;94:219–228. [Google Scholar]
- Singer TP, Oestreicher G, Hogue P, Contreiras J, Brandao I. Regulation of succinate dehydrogenase in higher plants. Plant Physiol. 1973;52:616–621. doi: 10.1104/pp.52.6.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snedden WA, Fromm H. Characterization of the plant homologue of prohibitin, a gene associated with antiproliferative activity in mammalian cells. Plant Mol Biol. 1997;33:753–756. doi: 10.1023/a:1005737026289. [DOI] [PubMed] [Google Scholar]
- Snedden WA, Fromm H. Regulation of the γ-aminobutyrate-synthesizing enzyme, glutamate decarboxylase, by calcium-calmodulin: a mechanism for rapid activation in response to stress. In: Lerner HR, editor. Plant Responses to Environmental Stresses: From Phytohormones to Genome Reorganization. New York: Marcel Dekker; 1999. pp. 549–574. [Google Scholar]
- Snedden WA, Koutsia N, Baum G, Fromm H. Activation of a recombinant petunia glutamate decarboxylase by calcium/calmodulin or by a monoclonal antibody which recognizes the calmodulin binding domain. J Biol Chem. 1996;271:4148–4153. doi: 10.1074/jbc.271.8.4148. [DOI] [PubMed] [Google Scholar]
- Tillakaratne NJK, Medina-Kauwe L, Gibson KM. γ-Aminobutyric acid (GABA) metabolism in mammalian neural and nonneural tissues. Comp Biochem Physiol. 1995;11:247–263. doi: 10.1016/0300-9629(95)00099-2. [DOI] [PubMed] [Google Scholar]
- Vedadi M, Vrielenk A, Meighen E. Involvement of conserved glycine residues, 229 and 234, of Vibrio harveyi aldehyde dehydrogenase in activity and nucleotide binding. Biochem Biophys Res Commun. 1997;238:448–451. doi: 10.1006/bbrc.1997.7300. [DOI] [PubMed] [Google Scholar]
- Wigge B, Kroemer S, Gardestroem P. The redox levels and subcellular distribution of pyridine nucleotides in illuminated barley leaf protoplasts studied by rapid fractionation. Physiol Plant. 1993;88:10–18. [Google Scholar]
- Yamaura I, Matsumoto T, Funatsu M, Shinohara T. Purification and some properties of succinic semialdehyde dehydrogenase from barley seeds. Agric Biol Chem. 1988;52:2929–2930. [Google Scholar]