Abstract
Infective endocarditis (IE) can be diagnosed using the Duke criteria, which cannot be conclusive especially when the results of blood cultures are negative. This study aimed at using real-time polymerase chain reaction (PCR) technique to isolate bacteria present in whole blood samples of patients with definitive IE on the basis of the method designed in this study. This laboratory and test study was conducted on 20 whole blood samples taken from patients with definitive IE. Real-time PCR of the 16s rRNA was utilized to directly analyze whole blood samples to diagnose bacterial IE. Of 20 whole blood samples with definitive IE, only one blood culture (5%) was positive and the isolated bacterium belonged to Streptococci viridans group. Also, 13 whole blood samples were positive using real-time PCR technique. The isolated bacteria were Enterococcus faecalis with seven (35%) cases, Streptococcus gallolyticus with two (10%) cases, Streptococcus mutans with one (5%) case, Streptococcus sanguinis with one (5%) case, Streptococcus salivarius with one (5%) case, and Staphylococcus aureus with one (5%) case. Sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) using real-time PCR technique were 65%, 100%, 100%, and 74%, respectively. The developed real-time PCR method allows us to detect bacteria in whole blood samples and is much more sensitive than culturing method. It also permits the differentiation of the main group of bacteria within a few hours for IE.
Keywords: Infective endocarditis, Whole blood, Real-time PCR
Introduction
Infective endocarditis (IE) is a serious life-threatening condition caused by microbial infection of heart valves and their epithelial tissues. Despite advancements in its treatment management, its nosocomial mortality rate is high (20-40%), so that it demands rapid early diagnosis [1-7]. The diagnosis of this disorder is based on Duke criteria [4] including clinical findings, laboratory results, and echocardiography. The major standard of these criteria is the diagnosis of vegetation on cardiac valves through echocardiography and positive blood culture [8]. Echocardiography is positive in only 50-94% of cases [9], while blood culture is negative in 2.5-31% of cases of IE and, hence, the infection is left undiagnosed. A negative blood culture test may be attributed to cardiac antibiotic therapy, slow-growing or uncultivable microorganisms, fungi, and/or intracellular microorganisms [6, 7, 10], and low number of microorganisms per volume unit [11]. Nonetheless, blood culture tests are rendered as a gold standard for IE [12]. According to the reports by some studies, microorganisms most frequently involved in the etiology of IE are from the Staphylococcus, Streptococcus, and Enterococcus genus in the developed countries, respectively [13, 14]. The identification of the etiologic pathogens incriminated as the causes of endocarditis is important as it can help in antibiotic therapy of the disorder [15]. Hence, many studies have been carried out in recent years concerning the use of molecular methods in diagnosing this condition and the sensitivity and specificity of molecular methods have been investigated [5-9, 11, 15]. Many of these studies have assessed the samples taken from cardiac valve tissue and blood culture, while only the study by Kuhn et al used whole blood samples [16]. Therefore, regarding the low number of studies on whole blood and lack of any study on direct isolation of microorganisms involved in the etiology of IE using whole blood samples, this study used real-time polymerase chain reaction (PCR) technique to isolate bacteria present in whole blood samples of patients with definitive IE.
Materials and Methods
Patients
This laboratory and test study was performed on 20 whole blood samples taken from patients with definitive IE. The inclusion criterion was affliction with definitive IE and the exclusion criteria were probable suspicious endocarditis and rejected endocarditis. The patients with definitive IE were selected using Duke criteria. On this basis, the definitive diagnosis was the presence of the two major criteria, one major criterion along with three minor criteria, or the presence of five minor criteria [4].
Samples
A whole blood sample from the same phlebotomy was drawn for microbiological and molecular analysis in an EDTA tube.
Conventional cultures
Three sets of blood cultures were prepared for each patient with suspected IE. The collected blood was inoculated directly into aerobic (SA) and anaerobic (SN) blood culture containers (BacT/Alert, BioMeriuex, Inc., Durham, NC) and incubated for 7 days.
Bacteria selected for investigating the patients’ whole blood samples and primer designing
The selection and preparation of bacteria involved in IE were done on the basis of the latest published guidelines on IE [17], the common strains involved in IE in Iran [18-21], and financial constraints which prevented the researchers from selecting a greater number of bacteria for this study. As such, the standard selected strains included: Staphylococcus aureus (ATCC 6538), Staphylococcus epidermidis (ATCC 49469), Streptococcus mitis (ATCC 6249), Streptococcus mutans (ATCC 35668), Streptococcus sanguines (ATCC 10556), Streptococcus salivarius (ATCC 13419), Streptococcus pyogenes (ATCC 8668), Streptococcus gallolyticus (ATCC 49147), and Enterococcus faecalis (ATCC 29212). The selected standard strains were cultured in blood agar culture media. In the next stage, 16S rRNA sequences of the bacteria above were obtained from GenBank (http://www.ncbi.nlm.nih.gov /BLAST/) (NCBI) provided in the public domain by the National Center for Biotechnology. So, the forward and reverse primers for 16S rRNA were: 5′-TGGAGCATGTGGTTTAATTCGA-3′ and 5′-TGCGGGACTTAACCCAACA-3′, respectively. Then, the primers for the bacteria above were developed (Table 1) [22].
Table 1. Primer Used in This Study.
| Bacteria | Target gene | Forward primer (5′-3′) | Reverse primer (5′-3′) | Size (bp) | References |
|---|---|---|---|---|---|
| S. aureus | eap | TACTAACGAAGCATCTGCC | TTAAATCGATATCAC TAATACCTC | 230 | [22] |
| S. epidermidis | SEI | CTAAAGAAGAAGCAAATCGTA | TAGATATTGGTTGTGGTGAT | 105 | This study |
| S. pyogenes | sodA | AATATCTTCTGGAATCTTAGTGA | CTTATGTCGCTAATACCAATG | 101 | This study |
| E. faecalis | ddl | CAATAACACGATTGAAATGC | ACTTCCATCTAACATAATATACG | 103 | This study |
| S. mitis | SM1 | TGTCAGATAATGCTAATAAGATG | ATCAGAAAGGTTGTTAATGTC | 197 | This study |
| S. mutans | gtfB | AGAGGAATATACCAATGTTGTG | AAGAACCATCTGTTGAAGAC | 110 | This study |
| S. sanguines | gtfp | GACTGATGAGAAGATTACCA | TGTTGAGATATTCGTTGCTA | 113 | This study |
| S. salivarius | MAP | GGGAAGCATTATGGATTACC | ACACATCAAGGACTGACTTATC | 155 | This study |
| S. gallolyticus | recNSGG | AAAGCTGGTGAGGACCAAGC | CATTTCTTTATAGTCAGCATCA | 189 | This study |
DNA extractions
DNA extraction from the grown colonies of the selected bacteria cultured in culture media and also DNA extraction of whole blood samples using QIAamp DNA mini kit (Qiagen) and the SepsiTest kit (Molzym, Bremen, Germany) were completed following the manufacturer’s protocol, respectively.
DNA purity and concentration
The purity and concentration of total DNA isolates in the samples, that is, the colonies of the selected and prepared bacteria grown in culture media and also whole blood samples were measured using spectrophotometric method at wavelengths of A260 and A280. It was conducted using a NanoDrop machine (Thermo Scientific).
Real-time PCR technique
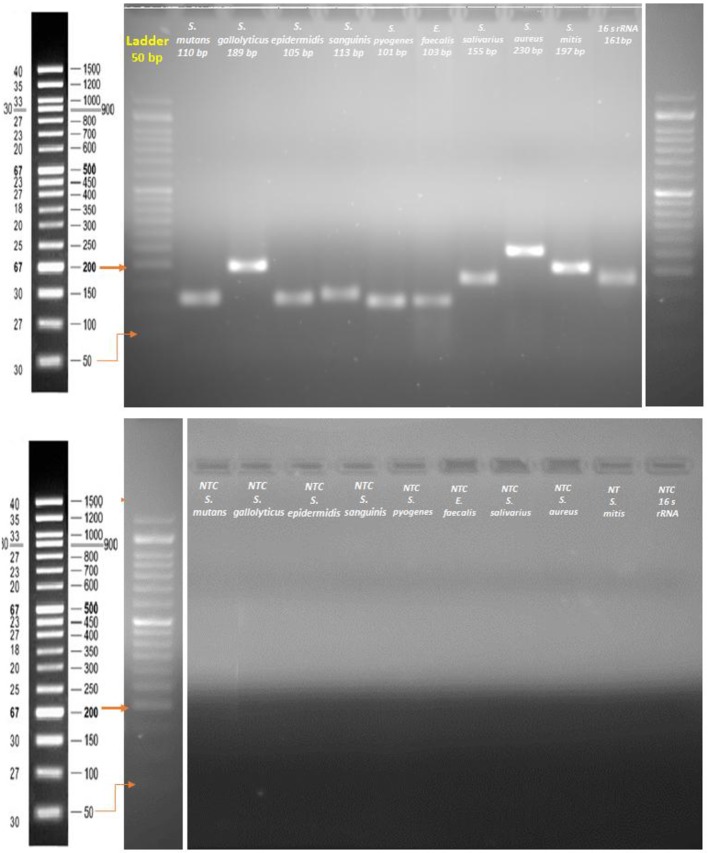
The functioning and optimal annealing temperature of the PCR primers was first examined with gradient PCR (BioRad thermal cycler, USA). The PCR conditions that follow were applied: an initial DNA denaturation at 95 °C for 20 s, primer annealing at 55-65 °C for 20 s, and primer extension at 72 °C for 45 s. This was completed with a final extension step at 72 °C for 5 min. Quantitative PCR was carried out using a StepOnePlus Real-Time PCR System (Applied Biosystems, USA). All PCRs were conducted in duplicate with a volume of 20 µL, using 96-well optical-grade PCR plates and an optical sealing tape (MicroAmp Fast Optical 96-well, Applied Biosystems, USA). EvaGrean dye was used to detect dsDNA synthesis. The thermal cycling conditions included an initial DNA denaturation step at 95 °C for 10 min followed by 40 cycles of denaturation at 95 °C for 15 s, primer annealing at the optimal temperature for 20 s, and extension at 72 °C for 30 s. Eventually, melt-curve analysis was done by slow heating of the PCRs to 95 °C (0.3 per cycle) with concurrent measurement of the EvaGrean signal intensity. To demonstrate the absence of contamination or primer dimmer, a non-template control (NTC) reaction with each primer was conducted on gel electrophoresis (Fig. 1).
Figure 1.
Agarose gel electrophoresis of primer designing and a non-template control (NTC) reaction.
Results
Of 20 whole blood samples of patients with definitive IE, the result of blood culture of only one patient was positive and the isolated bacterium belonged to the Streptococcus viridans group. Also, 19 patients had negative blood culture test. Sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of blood culture were: 5%, 100%, 100%, and 51%, respectively (Table 2). Moreover, 13 whole blood samples were positive using real-time PCR technique and seven whole blood samples were negative. The bacteria isolated from 13 positive whole blood samples were Enterococcus faecalis with seven (35%) cases, Streptococcus gallolyticus with two (10%) cases, Streptococcus mutans with one (5%) case, Streptococcus sanguines with one (5%) case, Streptococcus salivarius with one (5%) case, and Staphylococcus aureus with one (5%) case (Table 3 and Fig. 2). Sensitivity, specificity, PPV, and NPV of real-time PCR were 65%, 100%, 100%, and 74%, respectively (Table 2). One patient had a positive result of blood culture test with pathogenic bacterium belonging to Streptococcus viridans group. The real-time PCR of this patient was also positive with the pathogenic bacterium belonging to Streptococcus sanguines (Table 3).
Table 2. Sensitivity, Specificity, Positive Predictive Value, and Negative Predictive Value of Blood Culture and Real-Time PCR.
| Diagnostic approach | Blood culture (95% CI) | Real-time PCR (95% CI) |
|---|---|---|
| Sensitivity | 5% (0.13% to 24.87%) | 65% (40.78% to 84.61%) |
| Specificity | 100% (83.16% to 100.00%) | 100% (83.16% to 100.00%) |
| Positive predictive value | 100% | 100% |
| Negative predictive value | 51% (48.77% to 53.79%) | 74% (61.12% to 83.85%) |
Table 3. Microbiologic Findings of Real-Time PCR and Culture of Blood for 20 Patients With Definite IE.
| Patient no. | Previous history of antibiotic therapy | Blood culture | Blood real-time PCR |
|---|---|---|---|
| 1 | Yes | Negative | Negative |
| 2 | Yes | Negative | E. faecalis |
| 3 | Yes | Negative | S. aureus |
| 4 | Yes | Negative | Negative |
| 5 | Yes | Negative | E. faecalis |
| 6 | Yes | Negative | Negative |
| 7 | Yes | Viridans streptocococci | S. sanguines |
| 8 | Yes | Negative | E. faecalis |
| 9 | Yes | Negative | Negative |
| 10 | Yes | Negative | S. gallolyticus |
| 11 | Yes | Negative | Negative |
| 12 | Yes | Negative | S. mutans |
| 13 | Yes | Negative | E. faecalis |
| 14 | Yes | Negative | E. faecalis |
| 15 | Yes | Negative | E. faecalis |
| 16 | Yes | Negative | Negative |
| 17 | Yes | Negative | S. gallolyticus |
| 18 | Yes | Negative | S. salivarius |
| 19 | Yes | Negative | Negative |
| 20 | Yes | Negative | E. faecalis |
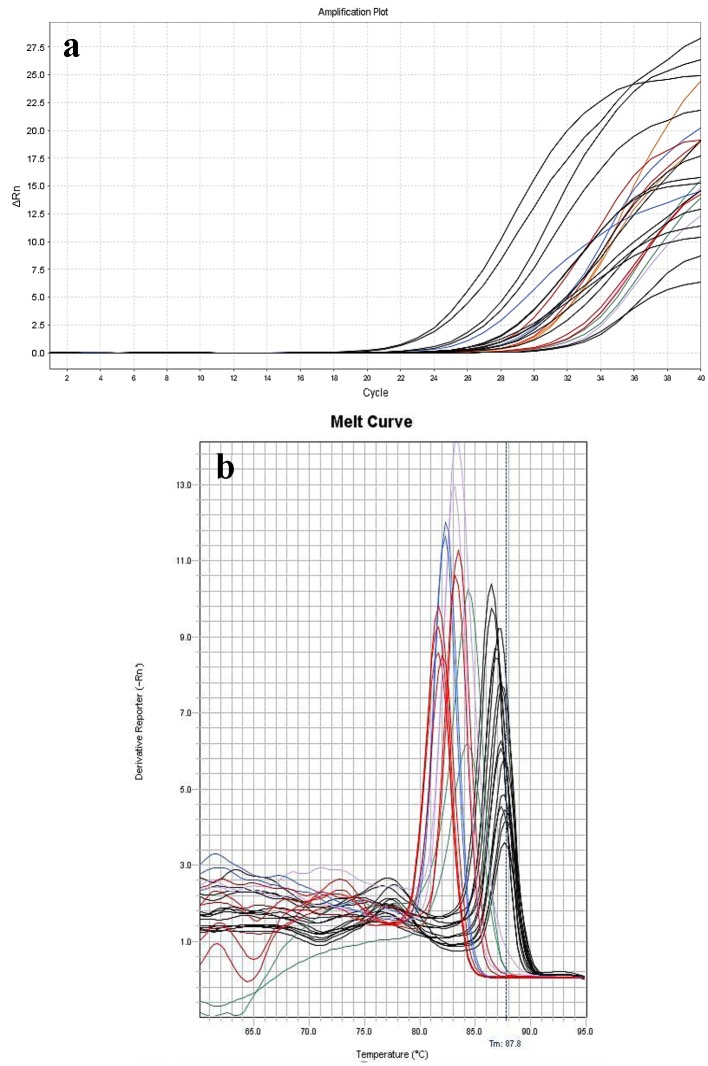
Figure 2.
Amplification (a) and melting curves (b) of 13 positive whole blood samples placed in duplicate manner. The identified bacteria were: S. gallolyticus (red line), S. sanguines (blue line), S. salivarius (violet line), S. aureus (orange line), S. mutans (green line), and E. faecalis (black line).
Discussion
The application of molecular methods to diagnose IE does not have a long history. Indeed, many studies name these methods as innovative procedures for the diagnosing of IE [23, 24]. Goldenberg et al (1997) displayed for the first time the results of their study on IE using PCR. They found that the molecular method enjoys high sensitivity and reliability, and is a quick method of diagnosing microbial IE [25]. Most studies demonstrated that the wide-spectrum bacterial DNA PCR is applicable in many cases such as the following: 1) when a small amount of bacteria is the etiologic cause of IE, 2) when blood culture is positive for solely one case, 3) when replacement of uncontaminated valve results in the histological diagnosis of IE, and 4) when the patient has a positive history of previous antibiotic therapy [25, 26]. Moreover, Millar et al and also Tak et al asserted that molecular diagnostic methods can be added to Duke’s criteria as a gold standard [24, 27]. Most of the molecular studies on IE used cardiac valve tissue as their sample. Only one study [16] used whole blood sample like our research. The results of whole blood sample cultures of 20 patients with definitive IE in our study indicated that only one blood culture was positive with the isolated bacteria belonging to Streptococcus viridans group, while in the study by Kuhn et al [16], three blood cultures were positive and the isolated bacteria were Staphylococcus epidermidis with one case and Enterococcus faecalis with two cases. In our study, all the patients with IE had a positive history of antibiotic therapy before blood sample culture. The blood sample culture is significant because it can reveal the etiologic organism that causes the disease and this eases the appropriate prescription and administration of the required antibiotic predisposing to more effective treatment [15, 28-30]. Several studies have shown that the negative result of blood culture may be attributed to previous antibiotic therapy, the presence of small amounts of microorganisms per volume unit, presence of slow-growing or hard-growing microorganisms such as Actinobacillus actinomycetemcomitans, Coxiella burnetii, Legionella species, Mycoplasma species, Haemophilus species, Tropheryma whipplei, Cardiobacterium hominis, Eikenella species, Chlamydia species and also HACEK group [9, 11, 28-30]. It ought to be pointed out that the culturing method, the number of blood cultures, and vigilant care at the time of blood sampling or phlebotomy can influence the results of blood culture tests [31]. Contrary to our study, most of the previous molecular studies on IE used valvular tissue as the sample. The valvular tissue samples are not rendered as proper since they may be contaminated during surgery and excision. This is because the isolated bacteria may not be the pathogenic organism for IE; rather, they may be transmitted during surgery [5, 32, 33]. Nevertheless, the valvular tissue samples are considered as suitable now to investigate cases of endocarditis with negative blood culture [34] as bacterial DNA may survive in the infected valve for several weeks in spite of antibiotic therapy [9, 35, 36]. The prolonged survival of bacterial DNA in the valve may be related to: first, a persistent infection, second, the original infection not treated fully because of deep infiltration into valve, or since it could not be treated even by appropriate antibiotic therapy, so the bacteria survived even though they could not grow, and third, the PCR method may have detected DNA from the killed bacteria [11, 37]. Marin et al declared in their study that the most important constraint in the use of valve tissue for molecular tests is that it could be applied only for patients who need cardiac valve surgery, whereas it could not be utilized for patients with IE who do not require valve surgery [5]. Our findings demonstrated that the sensitivity of the molecular real-time PCR method was greater than the common conventional methods used in blood sample culturing modalities so that the real-time PCR results of 13 patients were positive. Similar to our study, in the study by Kuhn et al [16], the sensitivity of the PCR molecular method (85%) used was greater than the common conventional method of blood sample culturing so that the results of PCR tests of 17 patients were positive. Additionally, in most studies conducted on IE, the sensitivity of molecular methods was greater than that of the conventional blood culturing methods so that the highest sensitivity pertained to Marin’s study with 96% sensitivity [5], whereas the PCR results in some cases have been even negative. Marin et al and Miller et al relate the negative results of PCR test to the presence of low amounts of microorganisms in the sample or to the high possibility of sampling error [5, 15]. Kotilainen et al reported in their study that negative PCR results do not mean that the patients have responded properly to antibiotics [35]. Gauduchon introduces three reasons for the negative results of PCR test: 1) the long time interval between antibiotic treatment and surgery, 2) the use of an inappropriate valve section for PCR, and 3) the presence of PCR inhibitors (though extraction and amplification controls yielded the expected results, and amplification of the beta-globin gene was positive) [8]. Moreover, Vollmer et al reported in their study that their previous study with unpublished data explored whole blood samples of 33 patients with IE using 23rDNA PCR assay. Yet, the PCR results were not successful. They explained that their failure was due to low number of bacteria in whole blood samples [11]. Fournier et al postulated that highly sensitive PCR assays that specifically target bacteria may be valuable detection instruments for blood specimens in the future [28]. The pathogenic bacteria incriminated as the etiologic causes of IE in whole blood samples of patients were isolated by primers developed in this study for nine bacteria including: Enterococcus faecalis with seven cases, Streptococcus gallolyticus with two cases, Streptococcus mutans with one case, Streptococcus sanguines with one case, Streptococcus salivarius with one case, and Staphylococcus aureus with one case. In the study by Kuhn et al [16], the pathogenic bacteria involved in the etiology of IE isolated from whole blood samples of the patients using PCR included: Enterococcus faecalis with four cases, Streptococcus gallolyticus with three cases, Streptococcus gordonii with three cases, Streptococcus epidermidis with three cases, E. faecium with one case, Streptococcus dysgalactiae with one case, and Escherichia coli with one case. Moreover, on the basis of previous studies, 45% to 60% of cases of IE were induced by Streptococci and enterococci [23, 38-41], 5% to 15% by D group such as Streptococcus bovis [23, 40, 42], 17% to 41% by Streptococci viridans (such as Streptococcus sanguinis, Streptococcus salivarius and Streptococcus mutans) [23, 38, 40, 43, 44], and 5% to 10% by Enterococcus faecalis [23, 38, 40]. Staphylococci are regarded as the second group of microorganisms that cause IE, so that 15% to 23% of cases are brought about by Staphylococcus aureus, while 3% to 8% of cases are caused by coagulase-negative Staphylococcus [23, 38, 40, 45]. Other causative organisms predisposing to IE include different bacteria such as Enterobacteriaceae or fungi such as Candida spp. Moreover, some bacteria such as HACEK (Haemophilus parainfluenzae, Haemophilusa phrophilus, and Haemophilus paraphrophilus, Actinobacillus actinomycescomitans, Cardiobacterium hominis, Eikenella corrodens and Kingella kingae) are averagely responsible for 4% of cases [23, 38, 40, 44]. On the basis of the results of the etiologic microorganisms of IE referred to in the studies above, it could be inferred that gram positive bacteria accounted for most cases of IE. However, different other microorganisms are incriminated in IE as well, and the incidence of this disorder cannot be attributed to merely one specific group of microorganisms [23]. The sole reason for the dominant role of gram positive bacteria in the etiology of this disorder is their fast growth compared to other microorganisms, specifically the fastidious bacteria [46].
Conclusion
It could be asserted on the basis of our findings that considering the low copy number of bacteria in whole blood samples of patients with IE and high negative results of blood culturetests of these patients on one hand, and regarding the high sensitivity and capacity of real-time PCR technique in diagnosing and demonstrating various pathogens even with low number of cases on the other hand, real-time PCR can be used today to directly analyze whole blood samples in patients with IE in a shorter time compared to the conventional culturing method.
Acknowledgments
The authors of this article give their special thanks to Yazd Cardiovascular Research Center and Shahid Sadoughi University of Medical Sciences, Yazd, Iran.
Financial Support
This thesis was presented for PhD degree and supported by Shahid Sadoughi University of Medical Sciences in Yazd, Iran. The study was not supported by industry.
Conflict of Interest
There was no financial conflict of interest.
Ethical Approval
This study was reviewed and approved by the Committee of Ethics in Research at Shahid Sadoughi University of Medical Sciences in Yazd, Iran (Ethics ID: IR.SSU.MEDICINE.REC.1394.39).
References
- 1.Duval X, Millot S, Chirouze C, Selton-Suty C, Moby V, Tattevin P, Strady C. et al. Oral streptococcal endocarditis, oral hygiene habits, and recent dental procedures: a case-control study. Clin Infect Dis. 2017;64(12):1678–1685. doi: 10.1093/cid/cix237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lagier JC, Aubry C, Delord M, Michelet P, Tissot-Dupont H, Million M, Brouqui P. et al. From expert protocols to standardized management of infectious diseases. Clin Infect Dis. 2017;65(suppl_1):S12–S19. doi: 10.1093/cid/cix403. [DOI] [PubMed] [Google Scholar]
- 3.Fernandez-Hidalgo N, Almirante B, Tornos P, Pigrau C, Sambola A, Igual A, Pahissa A. Contemporary epidemiology and prognosis of health care-associated infective endocarditis. Clin Infect Dis. 2008;47(10):1287–1297. doi: 10.1086/592576. [DOI] [PubMed] [Google Scholar]
- 4.Kim MS, Chang J, Kim MN, Choi SH, Jung SH, Lee JW, Sung H. Utility of a direct 16S rDNA PCR and sequencing for etiological diagnosis of infective endocarditis. Ann Lab Med. 2017;37(6):505–510. doi: 10.3343/alm.2017.37.6.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marin M, Munoz P, Sanchez M, del Rosal M, Alcala L, Rodriguez-Creixems M, Bouza E. et al. Molecular diagnosis of infective endocarditis by real-time broad-range polymerase chain reaction (PCR) and sequencing directly from heart valve tissue. Medicine (Baltimore) 2007;86(4):195–202. doi: 10.1097/MD.0b013e31811f44ec. [DOI] [PubMed] [Google Scholar]
- 6.Le Guern R, Loiez C, Armand S, Marceau L, Courcol R, Wallet F. Infective endocarditis: does a new 16S rDNA set of primers improve the microbiological diagnosis? Infect Dis (Lond) 2015;47(12):896–901. doi: 10.3109/23744235.2015.1075661. [DOI] [PubMed] [Google Scholar]
- 7.El-Kholy AA, El-Rachidi NG, El-Enany MG, AbdulRahman EM, Mohamed RM, Rizk HH. Impact of serology and molecular methods on improving the microbiologic diagnosis of infective endocarditis in Egypt. Infection. 2015;43(5):523–529. doi: 10.1007/s15010-015-0761-2. [DOI] [PubMed] [Google Scholar]
- 8.Bosshard PP, Kronenberg A, Zbinden R, Ruef C, Bottger EC, Altwegg M. Etiologic diagnosis of infective endocarditis by broad-range polymerase chain reaction: a 3-year experience. Clin Infect Dis. 2003;37(2):167–172. doi: 10.1086/375592. [DOI] [PubMed] [Google Scholar]
- 9.Lang S, Watkin RW, Lambert PA, Bonser RS, Littler WA, Elliott TS. Evaluation of PCR in the molecular diagnosis of endocarditis. J Infect. 2004;48(3):269–275. doi: 10.1016/S0163-4453(03)00102-6. [DOI] [PubMed] [Google Scholar]
- 10.Blumental S, Reynders M, Willems A, Biarent D, Duttman R, Lepage P, Vergison A. Enteroviral infection of a cardiac prosthetic device. Clin Infect Dis. 2011;52(6):710–716. doi: 10.1093/cid/ciq189. [DOI] [PubMed] [Google Scholar]
- 11.Vollmer T, Piper C, Horstkotte D, Korfer R, Kleesiek K, Dreier J. 23S rDNA real-time polymerase chain reaction of heart valves: a decisive tool in the diagnosis of infective endocarditis. Eur Heart J. 2010;31(9):1105–1113. doi: 10.1093/eurheartj/ehp600. [DOI] [PubMed] [Google Scholar]
- 12.Lamas CC, Eykyn SJ. Blood culture negative endocarditis: analysis of 63 cases presenting over 25 years. Heart. 2003;89(3):258–262. doi: 10.1136/heart.89.3.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandez-Hidalgo N, Almirante B, Gavalda J, Gurgui M, Pena C, de Alarcon A, Ruiz J. et al. Ampicillin plus ceftriaxone is as effective as ampicillin plus gentamicin for treating enterococcus faecalis infective endocarditis. Clin Infect Dis. 2013;56(9):1261–1268. doi: 10.1093/cid/cit052. [DOI] [PubMed] [Google Scholar]
- 14.Le T, Bayer AS. Combination antibiotic therapy for infective endocarditis. Clin Infect Dis. 2003;36(5):615–621. doi: 10.1086/367661. [DOI] [PubMed] [Google Scholar]
- 15.Miller RJ, Chow B, Pillai D, Church D. Development and evaluation of a novel fast broad-range 16S ribosomal DNA PCR and sequencing assay for diagnosis of bacterial infective endocarditis: multi-year experience in a large Canadian healthcare zone and a literature review. BMC Infect Dis. 2016;16:146. doi: 10.1186/s12879-016-1476-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuhn C, Disque C, Muhl H, Orszag P, Stiesch M, Haverich A. Evaluation of commercial universal rRNA gene PCR plus sequencing tests for identification of bacteria and fungi associated with infectious endocarditis. J Clin Microbiol. 2011;49(8):2919–2923. doi: 10.1128/JCM.00830-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Habib G, Lancellotti P, Antunes MJ, Bongiorni MG, Casalta JP, Del Zotti F, Dulgheru R. et al. 2015 ESC Guidelines for the management of infective endocarditis: The Task Force for the Management of Infective Endocarditis of the European Society of Cardiology (ESC). Endorsed by: European Association for Cardio-Thoracic Surgery (EACTS), the European Association of Nuclear Medicine (EANM) Eur Heart J. 2015;36(44):3075–3128. doi: 10.1093/eurheartj/ehv319. [DOI] [PubMed] [Google Scholar]
- 18.Badiee P, Amirghofran AA, Ghazi Nour M, Shafa M, Nemati MH. Incidence and outcome of documented fungal endocarditis. Int Cardiovasc Res J. 2014;8(4):152–155. [PMC free article] [PubMed] [Google Scholar]
- 19.Hosseini SM, Bakhshian R, Moshkani Farahani M, Abdar Esfahani M, Bahrami A, Sate A. An observational study on infective endocarditis: a single center experience. Res Cardiovasc Med. 2014;3(4):e18423. doi: 10.5812/cardiovascmed.18423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moshkani Farahani M, Rostami Z, Einollahi B, Khosravi A, Nemati E, Lessan Pezeshki M, Pourfarziani V. et al. Infective endocarditis after renal transplantation. Nephrourol Mon. 2014;6(1):e12326. doi: 10.5812/numonthly.12326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shoar MG, Zomorodian K, Saadat F, Hashemi MJ, Tarazoei B. Fatal endocarditis due to Aspergillus flavus in Iran. J Pak Med Assoc. 2004;54(9):485–486. [PubMed] [Google Scholar]
- 22.Hussain M, von Eiff C, Sinha B, Joost I, Herrmann M, Peters G, Becker K. eap Gene as novel target for specific identification of Staphylococcus aureus. J Clin Microbiol. 2008;46(2):470–476. doi: 10.1128/JCM.01425-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gauduchon V, Chalabreysse L, Etienne J, Celard M, Benito Y, Lepidi H, Thivolet-Bejui F. et al. Molecular diagnosis of infective endocarditis by PCR amplification and direct sequencing of DNA from valve tissue. J Clin Microbiol. 2003;41(2):763–766. doi: 10.1128/JCM.41.2.763-766.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Millar B, Moore J, Mallon P, Xu J, Crowe M, McClurg R, Raoult D. et al. Molecular diagnosis of infective endocarditis—a new Duke’s criterion. Scand J Infect Dis. 2001;33(9):673–680. doi: 10.1080/00365540110026764. [DOI] [PubMed] [Google Scholar]
- 25.Goldenberger D, Kunzli A, Vogt P, Zbinden R, Altwegg M. Molecular diagnosis of bacterial endocarditis by broad-range PCR amplification and direct sequencing. J Clin Microbiol. 1997;35(11):2733–2739. doi: 10.1128/jcm.35.11.2733-2739.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breitkopf C, Hammel D, Scheld HH, Peters G, Becker K. Impact of a molecular approach to improve the microbiological diagnosis of infective heart valve endocarditis. Circulation. 2005;111(11):1415–1421. doi: 10.1161/01.CIR.0000158481.07569.8D. [DOI] [PubMed] [Google Scholar]
- 27.Tak T, Shukla SK. Molecular diagnosis of infective endocarditis: a helpful addition to the Duke criteria. Clin Med Res. 2004;2(4):206–208. doi: 10.3121/cmr.2.4.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fournier PE, Casalta JP, Habib G, Messana T, Raoult D. Modification of the diagnostic criteria proposed by the Duke Endocarditis Service to permit improved diagnosis of Q fever endocarditis. Am J Med. 1996;100(6):629–633. doi: 10.1016/S0002-9343(96)00040-X. [DOI] [PubMed] [Google Scholar]
- 29.Li JS, Sexton DJ, Mick N, Nettles R, Fowler VG Jr, Ryan T, Bashore T. et al. Proposed modifications to the Duke criteria for the diagnosis of infective endocarditis. Clin Infect Dis. 2000;30(4):633–638. doi: 10.1086/313753. [DOI] [PubMed] [Google Scholar]
- 30.Harris KA, Yam T, Jalili S, Williams OM, Alshafi K, Gouliouris T, Munthali P. et al. Service evaluation to establish the sensitivity, specificity and additional value of broad-range 16S rDNA PCR for the diagnosis of infective endocarditis from resected endocardial material in patients from eight UK and Ireland hospitals. Eur J Clin Microbiol Infect Dis. 2014;33(11):2061–2066. doi: 10.1007/s10096-014-2145-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nunes MC, Gelape CL, Ferrari TC. Profile of infective endocarditis at a tertiary care center in Brazil during a seven-year period: prognostic factors and in-hospital outcome. Int J Infect Dis. 2010;14(5):e394–398. doi: 10.1016/j.ijid.2009.06.024. [DOI] [PubMed] [Google Scholar]
- 32.Campbell WN, Tsai W, Mispireta LA. Evaluation of the practice of routine culturing of native valves during valve replacement surgery. Ann Thorac Surg. 2000;69(2):548–550. doi: 10.1016/S0003-4975(99)01354-5. [DOI] [PubMed] [Google Scholar]
- 33.Giladi M, Szold O, Elami A, Bruckner D, Johnson BL Jr. Microbiological cultures of heart valves and valve tags are not valuable for patients without infective endocarditis who are undergoing valve replacement. Clin Infect Dis. 1997;24(5):884–888. doi: 10.1093/clinids/24.5.884. [DOI] [PubMed] [Google Scholar]
- 34.Podglajen I, Bellery F, Poyart C, Coudol P, Buu-Hoi A, Bruneval P, Mainardi JL. Comparative molecular and microbiologic diagnosis of bacterial endocarditis. Emerg Infect Dis. 2003;9(12):1543–1547. doi: 10.3201/eid0912.030229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kotilainen P, Heiro M, Jalava J, Rantakokko V, Nikoskelainen J, Nikkari S, Rantakokko-Jalava K. Aetiological diagnosis of infective endocarditis by direct amplification of rRNA genes from surgically removed valve tissue. An 11-year experience in a Finnish teaching hospital. Ann Med. 2006;38(4):263–273. doi: 10.1080/07853890600622119. [DOI] [PubMed] [Google Scholar]
- 36.Rovery C, Greub G, Lepidi H, Casalta JP, Habib G, Collart F, Raoult D. PCR detection of bacteria on cardiac valves of patients with treated bacterial endocarditis. J Clin Microbiol. 2005;43(1):163–167. doi: 10.1128/JCM.43.1.163-167.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horstkotte D, Follath F, Gutschik E, Lengyel M, Oto A, Pavie A, Soler-Soler J. et al. Guidelines on prevention, diagnosis and treatment of infective endocarditis executive summary; the task force on infective endocarditis of the European society of cardiology. Eur Heart J. 2004;25(3):267–276. doi: 10.1016/j.ehj.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 38.Hoen B, Alla F, Selton-Suty C, Beguinot I, Bouvet A, Briancon S, Casalta JP. et al. Changing profile of infective endocarditis: results of a 1-year survey in France. JAMA. 2002;288(1):75–81. doi: 10.1001/jama.288.1.75. [DOI] [PubMed] [Google Scholar]
- 39.Prendergast BD. The changing face of infective endocarditis. Heart. 2006;92(7):879–885. doi: 10.1136/hrt.2005.067256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delahaye F, Goulet V, Lacassin F, Ecochard R, Selton-Suty C, Hoen B, Etienne J. et al. Characteristics of infective endocarditis in France in 1991. A 1-year survey. Eur Heart J. 1995;16(3):394–401. doi: 10.1093/oxfordjournals.eurheartj.a060923. [DOI] [PubMed] [Google Scholar]
- 41.Wilson WR, Karchmer AW, Dajani AS, Taubert KA, Bayer A, Kaye D, Bisno AL. et al. Antibiotic treatment of adults with infective endocarditis due to streptococci, enterococci, staphylococci, and HACEK microorganisms. American Heart Association. JAMA. 1995;274(21):1706–1713. doi: 10.1001/jama.1995.03530210060032. [DOI] [PubMed] [Google Scholar]
- 42.Duval X, Delahaye F, Alla F, Tattevin P, Obadia JF, Le Moing V, Doco-Lecompte T. et al. Temporal trends in infective endocarditis in the context of prophylaxis guideline modifications: three successive population-based surveys. J Am Coll Cardiol. 2012;59(22):1968–1976. doi: 10.1016/j.jacc.2012.02.029. [DOI] [PubMed] [Google Scholar]
- 43.van der Meer JT, van Vianen W, Hu E, van Leeuwen WB, Valkenburg HA, Thompson J, Michel MF. Distribution, antibiotic susceptibility and tolerance of bacterial isolates in culture-positive cases of endocarditis in The Netherlands. Eur J Clin Microbiol Infect Dis. 1991;10(9):728–734. doi: 10.1007/BF01972497. [DOI] [PubMed] [Google Scholar]
- 44.Goulet V, Etienne J, Fleurette J, Netter R. [Infectious endocarditis in France. Epidemiological characteristics] Presse Med. 1986;15(37):1855–1858. [PubMed] [Google Scholar]
- 45.Bayliss R, Clarke C, Oakley CM, Somerville W, Whitfield AG, Young SE. The microbiology and pathogenesis of infective endocarditis. Br Heart J. 1983;50(6):513–519. doi: 10.1136/hrt.50.6.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grijalva M, Horvath R, Dendis M, Erny J, Benedik J. Molecular diagnosis of culture negative infective endocarditis: clinical validation in a group of surgically treated patients. Heart. 2003;89(3):263–268. doi: 10.1136/heart.89.3.263. [DOI] [PMC free article] [PubMed] [Google Scholar]