Abstract
Objective
Acetaldehyde dehydrogenase 2 (ALDH2) and cytochrome P450 2E1 (CYP2E1) have been associated with hepatocellular carcinoma (HCC) susceptibility and prognosis. The polymorphisms ALDH2 rs671 and CYP2E1 rs2031920 are reportedly correlated with the prevalence of HCC in other countries. The aim of this study was to investigate associations between ALDH2 and CYP2E1, and HCC susceptibility in a population of Guangxi, southern China, an area with a high incidence of HCC.
Patients and methods
The study cohort included 300 HCC cases, 292 healthy controls for HCC susceptibility analysis, and another 20 HCC cases and 10 healthy controls for ascertainment. Genotyping was performed using the polymerase chain reaction-restriction fragment length polymorphism method.
Results
The study results demonstrated that mutant genotypes of ALDH2 (G/A and A/A) led to significant differences in HCC susceptibility, as compared with the wild genotype (G/G) with the same C1/C1 genotype in non-drinking individuals (adjusted P=0.010, OR=0.20, 95% CI=0.06–0.68). The mutant genotypes of CYP2E1 (C1/C2 and C2/C2) brought about significant differences in HCC susceptibility, as compared with the wild genotype (C1/C1) and the same G/G genotype (adjusted P=0.025, OR=0.42, 95% CI=0.20–0.90). Drinking plays a role in HCC susceptibility in the same G/G genotype individuals (adjusted P=0.004, OR=0.32, 95% CI=0.15–0.69), but had no impact when combined with CYP2E1 for analysis (all P>0.05).
Conclusion
These results suggest that the mutant genotypes of ALDH2 and CYP2E1 may be protective factors for HCC susceptibility in Guangxi province, China.
Keywords: hepatocellular carcinoma susceptibility, genetic polymorphism, ALDH2, CYP2E1, Chinese population
Introduction
Hepatocellular carcinoma (HCC), the most common histological subtype of primary liver cancer, is the third most common cause of cancer-related death, with an estimated 745,500 HCC-related deaths worldwide in 2012: about 50% of which occurred in China.1 Etiologically, chronic infections of hepatitis B and C viruses, aflatoxin B1 exposure, and alcohol abuse are the primary risk factors for HCC occurrence.2,3 Alcohol intake varies among cultures, but is prevalent around the world and likely to induce alcohol dependence in heavy drinkers, which account for approximately 14% of the general population,4 approximately 10–15% of whom are at high risk of liver cirrhosis, including a small percentage at an exceptional risk for many types of cancers.5,6 It is well documented that liver cirrhosis can eventually impel the occurrence of HCC. Therefore, identifying alcohol metabolizing genes may help to postpone and even prevent the manifestation of HCC. Although there are many treatment options for HCC, such as hepatectomy, liver transplantation, radiofrequency ablation, transcatheter arterial chemoembolization, percutaneous acid injection, immunotherapy, and percutaneous ethanol injection,7,8 and prognosis has improved remarkably in recent years, prognosis of HCC remains dissatisfactory, especially when diagnosed at advanced stages. Specifically, the 5-year survival rate of HCC is less than 20% in the United States.9 Consequently, the validation of alcohol metabolizing genes on HCC carcinogenesis and progression is of great significance.
Acetaldehyde dehydrogenase 2 (ALDH2) and cytochrome P450 2E1 (CYP2E1) are two principle enzymes involved in alcohol metabolism. Acetaldehyde (AA) is produced from alcohol by alcohol dehydrogenase and CYP2E1, and is then metabolized to acetate by ALDH2.5 CYP2E1, a member of the cytochrome P450 superfamily and microsomal ethanol oxidizing system, is induced by greater amounts of ethanol though CYP2E1 metabolism of ethanol, which is predominantly metabolized by alcohol dehydrogenase, and ranges from 10–30% via the non-alcohol dehydrogenase pathway.10,11 CYP2E1 enzymatic activity is closely associated with the RsaI polymorphism of the CYP2E1 gene, also known as rs2031920, which contains a wild type C1 allele and a variant C2 allele.12–14 Besides, the mutant C2 allele is related to reduced CYP2E1 enzymatic activity.12
ALDH2, a mitochondrial enzyme, is not only primarily responsible for the oxidation of AA, but is also encoded by two alleles (G and A) that are closely correlated with AA metabolism.5,15 Individuals heterozygous or homozygous with a mutated allele metabolize ethanol to AA normally, but metabolize AA poorly, while those homozygous for the variant A allele are completely devoid of ALDH2 activity, and those heterozygous with the G/A phenotype display only 17–50% of normal ALDH2 activity.5,16 Thus, in the population carrying the variant G allele, the capacity to convert AA into acetate is influenced dramatically and leads to an abundance of AA in the circulation, even when consuming a moderate amount of alcohol.17 Notably, about 40% or so of the Eastern Asian population carry the A/G phenotype.15 It is well known that AA, rather than ethanol, is highly toxic, carcinogenic, and mutagenic, and has been identified as the cause of Asian Alcohol Flushing Syndrome, which is characterized by nausea, facial flushing, muscle weakness, tachycardia, palpitation, perspiration, headache, and sleepiness.15,18 Hence, HCC susceptibility could be estimated in individuals with different genotypes by distinguishing the capacity of alcohol ingestion through analysis of drinking habits.
Therefore, the aim of this study was to investigate the role of the ethanol metabolism genes ALDH2 and CYP2E1 on HCC susceptibility in a Chinese population in Guangxi province in southern China, an area with a notably high incidence of HCC.
Patients and methods
Study population
The study protocol was approved by the Ethical Review Committee of the First Affiliated Hospital of Guangxi Medical University (Nanning, Guangxi, China), and written informed consent was obtained from all enrolled subjects. A total of 300 HCC patients and 292 healthy control subjects were enrolled from the First Affiliated Hospital of Guangxi Medical University from March 2000 to December 2004 for association analysis between HCC susceptibility and expression levels of the ALDH2 and CYP2E1 genes. In addition, 20 HCC patients and 10 healthy control subjects were enrolled from the First Affiliated Hospital of Guangxi Medical University in 2006 to complementary analyses to ascertain the role of ethanol in vitro in patients with different ALDH2 and CYP2E1 genotypes. All cases of HCC were confirmed by pathological diagnosis after hepatectomy. Data regarding the HCC patients were collected from medical records, while information of the control subjects was obtained by questionnaires. Drinking was defined in our research as drinking a high concentration of white wine (ethanol > 40%) at least once in a week, continuing for over half a year.
Collection of HCC tissues and blood samples
HCC tissues were collected within 30 min during surgery and immediately stored at −80°C (Thermo Fisher Scientific, Waltham, MA, USA) until assayed. A 5-mL peripheral venous blood sample was collected from 292 control subjects, mixed well, and then stored in EDTA-coated centrifugation tubes at 4°C (Qingdao Haier Co. Ltd., Qingdao, China). DNA was extracted within 1 week after storage of the blood samples at 4°C and then stored at −20°C (Qingdao Haier Co. Ltd.). A 20-mL peripheral venous blood sample was collected from 30 subjects, of which a 200-μL aliquot was stored in an EDTA-coated centrifugation tube at 4°C for DNA extraction, while the remaining volume was used for lymphocyte extraction (TBDLTS1077).
Lymphocyte identification and growth curve
The blood samples were stained with H&E, then lymphocyte counts were performed under a microscope. The lymphocyte survival rate was determined by the trypan blue method at 3, 6, 12, 24, and 48 h, respectively, and growth curves were constructed.
Identification of ethanol concentration by the MTT method
Various ethanol concentrations (25, 50, 100, 200, 400, 800, and 1600 mmol/L) were used to assay samples from the experimental group. Using the MTT method (MTT xldT0793), optical density was measured at 490 nm (BioTek, Inc, Vernichi, Vermont, USA). Then, the half maximal inhibitory concentration (IC50) was calculated using the Bliss method, and the ethanol concentration was determined. As the maximum ethanol concentration cut-off was 1/3–1/2 of the IC50 value, the ethanol concentration was reasonably set at 200, 100, and 50 mmol/L, respectively.
Identification of ethanol culture time
Using the MTT method, lymphocytes were cultured for various time periods (3, 6, 12, and 24 h, respectively, Gibco 31800-022, FBS, Hangzhou sjq02) in three ethanol gradients (200, 100, and 50 mmol/L, respectively). Accordingly, the cell inhibitory rate was calculated, and the ethanol culture time was determined according to the designed cut-off cell inhibitory rate.
Polymerase chain reaction (PCR) assay
Total RNA was isolated from lymphocytes with the alcohol precipitation method. cDNA was synthesized from mRNA by PCR in a 20.0-μL reaction mixture. Then, PCR amplification of another 20.0-μL reaction mixture was performed. The PCR cycle included a denaturation step at 94°C for 4 min, followed by 30 cycles of denaturing at 94°C for 1 min, annealing at the indicated temperature for 1 min, extension at 72°C for 1 min, and then a final extension step at 72°C for 10 min. The PCR products were identified by electrophoresis at 40 V for 40 min in a 2% agarose gel (xsq01; Biowest, Barcelona, Spain) stained with ethidium bromide (Sigma-Aldrich Co., St Louis, MO, USA) and viewed under ultraviolet light.
Genotyping
ALDH2 rs671 and CYP2E1 rs2031920 polymorphisms were genotyped using PCR-restriction fragment length polymorphism after PCR amplification using the following primers: rs671, forward: 5′-CAA ATT ACA GGG TCA ACT GC-3′/reverse: 5′-CCA CAC TCA CAG TTT TCT CTT-3′ and rs2031920, forward: 5′-CCA GTC GAG TCT ACA TTG TCA-3′/reverse: 5′-TTC ATT CTG TCT TCT AAC TGG-3′.
Bioinformatic analysis
A variety of available bioinformatic methods were used to verify the expression patterns of the ADLH2 and CYP2E1 genes. The Genotype-Tissue Expression (GTEx) portal (https://gtexportal.org/home/; accessed July 1, 2017) was used to search for ADLH2 and CYP2E1 expression levels in different tissues and the MERAV website (http://merav.wi.mit.edu/; accessed July 1, 2017) was assessed to retrieve ADLH2 and CYP2E1 expression levels in normal and primary tumor tissues. The GeneMANIA prediction server of biological network integration for gene prioritization and predicting gene function (http://genemania.org/; accessed July 1, 2017) was employed to explore potential interactions between ADLH2 and CYP2E1. The STRING database of known and predicted protein–protein interactions (https://string-db.org/; accessed July 1, 2017) was used to search for biological interactions between the ALDH2 and CYP2E1 proteins. The F-SNP integrated website of the functional effects of single nucleotide polymorphisms (SNPs) (http://compbio.cs.queensu.ca/F-SNP/; accessed July 1, 2017) was used to mimic concrete mechanisms of the two SNP sites. Finally, the SNP function prediction website was employed to explore changes in the two SNPs (https://snpinfo.niehs.nih.gov/snpinfo/snpfunc.html, accessed July 1, 2017).
Statistical analysis
The Hardy–Weinberg equilibrium was calculated by the goodness-of-fit χ2 test. The χ2 test was performed to analyze the association between polymorphisms of ALDH2 and CYP2E1 and HCC susceptibility. The odds ratio (OR) and 95% confidence interval (95% CI) were calculated to estimate relative risk with a binary logistic regression model. The Pearson correlation coefficient was calculated to analyze the correlation between the ethanol concentration and lymphocyte inhibitory rate. All statistical analyses were performed using SPSS version 16.0 software (SPSS, Inc., Chicago, IL, USA). A probability (P) value of <0.05 was considered statistically significant.
Results
Baseline subject characteristics
The genotype frequencies of ALDH2 and CYP2E1 met the Hardy-Weinberg equilibrium in both the HCC and control groups (all P>0.05). The baseline characteristics of the HCC patients and control group subjects are shown in Table 1. Overall, there were no significant differences in age, sex, drinking status, smoking status, and tumor family history between the HCC and control groups (all P>0.05), while there were significant differences in race and HBsAg status (both P<0.001).
Table 1.
Baseline characteristics of the HCC and control groups
| Factors | HCC (%) (n=300) | Control (%) (n=292) | OR (95% CI) | P-value |
|---|---|---|---|---|
| Sex | 1.11 (0.67–1.86) | 0.685 | ||
| Male | 265 (88.3) | 261 (87.0) | ||
| Female | 35 (11.7) | 31 (27.0) | ||
| Age (years) | 1.62 (0.93–2.84) | 0.091 | ||
| ≤60 | 265 (88.3) | 270 (92.5) | ||
| >60 | 35 (11.7) | 22 (7.5) | ||
| Race | 0.44 (0.30–0.64) | <0.001 | ||
| Han | 245 (81.7) | 193 (64.3) | ||
| Minority | 55 (18.3) | 99 (35.7) | ||
| Drinking status | 1.03 (0.72–1.47) | 0.875 | ||
| Yes | 214 (71.3) | 210 (71.9) | ||
| No | 86 (28.7) | 82 (28.1) | ||
| Smoking status | 0.92 (0.67–1.27) | 0.623 | ||
| Yes | 154 (51.3) | 144 (49.3) | ||
| No | 146 (48.7) | 148 (50.7) | ||
| Tumor family history | 0.60 (0.34–1.04) | 0.070 | ||
| Yes | 36 (11.6) | 22 (7.5) | ||
| No | 264 (88.4) | 270 (92.5) | ||
| HBsAg status | 0.02 (0.01–0.03) | <0.001 | ||
| + | 255 (85.0) | 30 (10.3) | ||
| − | 45 (15.0) | 262 (89.7) |
Abbreviations: HCC, hepatocellular carcinoma; OR, odds ratio.
Correlation between ALDH2 and CYP2E1 SNPs and HCC susceptibility
For the ALDH2 rs671 polymorphism, neither the wild-type genotype, GG, nor mutant genotypes (G/A and A/A) gave rise to significant differences in HCC susceptibility (all P>0.05). For the CYP2E1 rs2031920 polymorphism, both the wild-type (C1/C1) and mutant genotypes (C1/C2 and C2/C2) failed to produce significant differences in HCC susceptibility (all P>0.05). Detailed data are presented in Table 2.
Table 2.
Joint-effects analysis between ALDH2 or CYP2E1 and drinking status on HCC susceptibility
| Groups | Genotype
|
Whole
|
Drinking
|
Non-drinking
|
Crude OR (95% CI) | Crude P-value | Adjusted OR (95% CI) | Adjusted P-value* | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALDH2 | CYP2E1 | HCC | Control | HCC | Control | HCC | Control | |||||
| G/G | 149 | 152 | 1.00 | 0.469 | 1.00 | 0.999 | ||||||
| G/A | 121 | 119 | 1.04 (0.74–1.46) | 0.833 | 0.99 (0.59–1.65) | 0.967 | ||||||
| A/A | 30 | 21 | 1.46 (0.80–2.66) | 0.220 | 1.00 (0.40–2.48) | 0.996 | ||||||
| G/A+A/A | 151 | 140 | 1.10 (0.80–1.52) | 0.561 | 0.99 (0.61–1.62) | 0.972 | ||||||
| C1/C1 | 203 | 196 | 1.00 | 0.546 | 1.00 | 0.381 | ||||||
| C1/C2 | 87 | 81 | 1.04 (0.72–1.49) | 0.843 | 0.67 (0.38–1.19) | 0.171 | ||||||
| C2/C2 | 10 | 15 | 0.64 (0.28–1.47) | 0.295 | 0.77 (0.22–2.70) | 0.684 | ||||||
| C1/C2+C2/C2 | 97 | 96 | 0.98 (0.69–1.38) | 0.888 | 0.69 (0.40–1.18) | 0.170 | ||||||
| i | G/G | C1/C1 | 104 | 96 | 1.00 | 0.397 | 1.00 | 0.160 | ||||
| ii | G/G | C1/C2+C2/C2 | 45 | 56 | 0.74 (0.46–1.20) | 0.223 | 0.42 (0.20–0.90) | 0.025 | ||||
| iii | G/A+A/A | C1/C1 | 99 | 100 | 0.91 (0.62–1.35) | 0.653 | 0.72 (0.40–1.31) | 0.287 | ||||
| iiii | G/A+A/A | C1/C2+C2/C2 | 52 | 40 | 1.20 (0.73–1.97) | 0.472 | 0.83 (0.39–1.77) | 0.626 | ||||
| 1 | G/G | C1/C1 | 70 | 79 | 1.00 | 0.067 | 1.00 | 0.137 | ||||
| 2 | G/G | C1/C2+C2/C2 | 32 | 45 | 0.80 (0.46–1.40) | 0.438 | 0.45 (0.19–1.06) | 0.068 | ||||
| 3 | G/A+A/A | C1/C1 | 74 | 63 | 1.33 (0.83–2.11) | 0.235 | 1.13 (0.56–2.26) | 0.740 | ||||
| 4 | G/A+A/A | C1/C2+C2/C2 | 38 | 23 | 1.87 (1.01–3.43) | 0.045 | 1.33 (0.54–3.28) | 0.538 | ||||
| ! | G/G | C1/C1 | 34 | 17 | 1.00 | 0.043 | 1.00 | 0.053 | ||||
| ! ! | G/G | C1/C2+C2/C2 | 13 | 11 | 0.59 (0.22–1.59) | 0.299 | 0.35 (0.07–1.72) | 0.197 | ||||
| ! ! ! | G/A+A/A | C1/C1 | 25 | 37 | 0.34 (0.16–0.73) | 0.006 | 0.20 (0.06–0.68) | 0.010 | ||||
| ! ! ! ! | G/A+A/A | C1/C2+C2/C2 | 14 | 17 | 0.41 (0.17–1.03) | 0.058 | 0.20 (0.04–0.96) | 0.045 | ||||
Note:
P adjustment for race and HBsAg.
Abbreviations: HCC, hepatocellular carcinoma; OR, odds ratio; CI, confidence interval.
Joint-effects analysis of combining the ALDH2 and CYP2E1 genes on HCC susceptibility
Joint-effects analysis of the ALDH2 and CYP2E1 genes combined on HCC susceptibility showed significant difference between group 2 (GG + C1C2/C2C2) and group 1 (GG + C1C1) (adjusted P=0.025), between the non-drinking populations of group 3 (G/A + A/A + C1/C1) and group 1 (G/G + C1/C1) (adjusted P=0.010), and between group 4 (GA/AA + C1C2/C2C2) and group 1 (adjusted P=0.045). Detailed data are shown in Table 2.
Joint-effects analysis of combining drinking status with ALDH2 or CYP2E1 on HCC susceptibility
Joint-effects analysis of combining drinking status with ALDH2 or CYP2E1 on HCC susceptibility showed significant differences between drinking (group 3) and non-drinking individuals with the same GG genotype (adjusted P=0.004). Among the non-drinking individuals, there were significant differences between mutant genotypes (group 2) and the wild genotype (adjusted P=0.014), while there were no significant differences between the other groups (all P>0.05).
For joint-effects analysis of combining drinking status with CYP2E1 genotypes, there were no significant differences between any groups (all P>0.05). Specific results are presented in Table 3.
Table 3.
Joint-effects analysis between ALDH2 or CYP2E1 and drinking status on HCC susceptibility
| Groups | Drinking status | Genotype | HCC (%) (n=300) | Control (%) (n=292) | Crude OR (95% CI) | Crude P-value | Adjusted OR (95% CI) | Adjusted P-value* |
|---|---|---|---|---|---|---|---|---|
| 1 | − | G/G | 47 | 28 | 1.00 | 0.006 | 1.00 | 0.022 |
| 2 | − | G/A+A/A | 39 | 54 | 0.43 (0.23–0.80) | 0.008 | 0.32 (0.13–0.79) | 0.014 |
| 3 | +/++ | G/G | 102 | 124 | 0.49 (0.29–0.84) | 0.009 | 0.32 (0.15–0.69) | 0.004 |
| 4 | +/++ | G/A+A/A | 112 | 86 | 0.78 (0.45–1.34) | 0.362 | 0.49 (0.23–1.07) | 0.075 |
| I | + | G/G | 55 | 77 | 1.00 | 0.004 | 1.00 | 0.360 |
| II | + | G/A+A/A | 69 | 68 | 1.42 (0.88–2.30) | 0.153 | 1.75 (0.85–3.58) | 0.127 |
| III | ++ | G/G | 47 | 47 | 1.40 (0.82–2.38) | 0.215 | 1.53 (0.68–3.42) | 0.306 |
| IV | ++ | G/A+A/A | 43 | 18 | 3.34 (1.75–6.41) | <0.001 | 2.00 (0.78–5.12) | 0.148 |
| a | − | C1/C1 | 59 | 54 | 1.00 | 0.982 | 1.00 | 0.338 |
| b | − | C1/C2+C2/C2 | 27 | 28 | 0.88 (0.46–1.68) | 0.704 | 0.69 (0.25–1.85) | 0.456 |
| c | +/++ | C1/C1 | 144 | 142 | 0.93 (0.60–1.44) | 0.737 | 0.71 (0.37–1.37) | 0.307 |
| d | +/++ | C1/C2+C2/C2 | 70 | 68 | 0.94 (0.57–1.55) | 0.815 | 0.49 (0.23–1.05) | 0.067 |
| A | + | C1/C1 | 88 | 98 | 1.00 | 0.094 | 1.00 | 0.534 |
| B | + | C1/C2+C2/C2 | 36 | 47 | 0.85 (0.51–1.44) | 0.550 | 0.61 (0.28–1.34) | 0.215 |
| C | ++ | C1/C1 | 56 | 44 | 1.42 (0.87–2.31) | 0.162 | 1.14 (0.55–2.37) | 0.728 |
| D | ++ | C1/C2+C2/C2 | 34 | 21 | 1.80 (0.97–3.34) | 0.060 | 1.00 (0.39–2.57) | 0.995 |
Notes:
P adjustment for race and HBsAg. −, Not drinking; +, drinking little; ++, drinking a lot.
Abbreviations: HCC, hepatocellular carcinoma; OR, odds ratio; CI, confidence interval.
Lymphocyte growth curve and growth inhibitory curve, and identification of ethanol intervention time
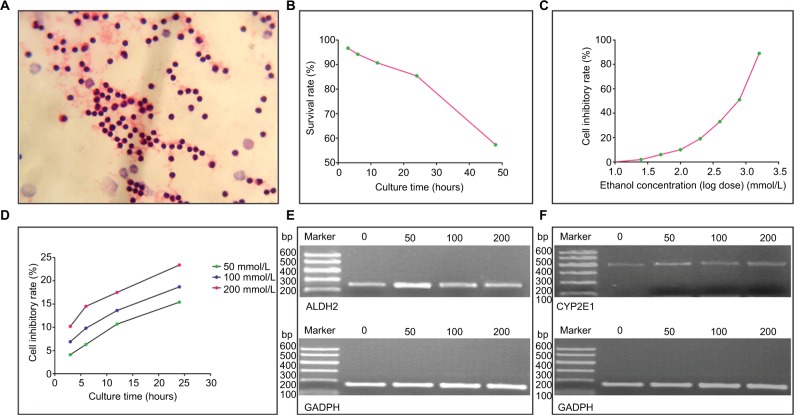
Lymphocytes stained by the H&E method are presented in Figure 1A. A growth curve of lymphocytes is presented in Figure 1B. In order to eliminate hindrance from lymphocytes, the lymphocyte intervention time was set at 4 h before ethanol intervention. Pearson correlation analysis was conducted to identify correlations between lymphocyte inhibitory rates and ethanol concentrations (r=0.993, P<0.0001). A lymphocyte inhibitory rate curve (logarithm dose conversion) is shown in Figure 1C.
Figure 1.
Lymphocytes and ALDH2/CYP2E1 mRNA expression profiles. (A) Lymphocytes stained by the H&E method. (B) Curve of lymphocyte survival rate–culture time. (C) Curve of lymphocyte inhibitory rate–ethanol concentration. (D) Curves of lymphocyte inhibitory rate–culture time at different ethanol concentrations. (E and F) ALDH2, CYP2E1, and GAPDH mRNA expression levels at different ethanol concentrations.
The IC50 value was calculated to be 539.646. Accordingly, ethanol concentrations were set at 200, 100, and 50 mmol/L. A lymphocyte inhibitory curve after intervention for 3, 6, 12, and 24 h is depicted in Figure 1D. The ethanol intervention time was determined as 2 h.
ALDH2 and CYP2E1 mRNA expression levels of the HCC and control groups
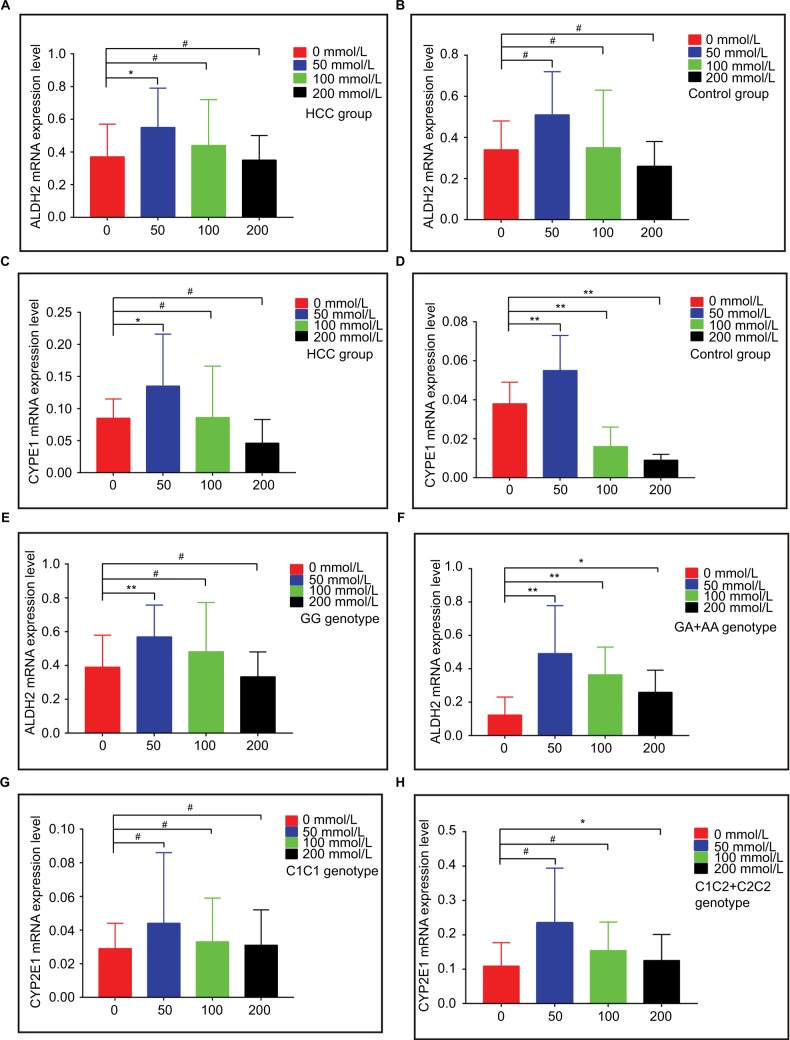
The mRNA expression levels of ALDH2 and CYP2E1 after intervention with different ethanol concentrations for 2 h are shown in Figure 1E and F, respectively. ALDH2 mRNA expression levels increased in both the HCC and control groups, as compared with non-ethanol intervention, but then decreased with increasing ethanol concentrations. Ethanol at 50 mmol/L showed a significant difference in the HCC group (P<0.05; Figure 2A), while no concentration led to a significant difference in the control group (all P>0.05; Figure 2B).
Figure 2.
mRNA expression levels of ALDH2 and CYP2E1 in different groups. (A and B) ALDH2 mRNA expression levels in HCC and control groups. (C and D) CYP2E1 mRNA expression levels in HCC and control groups. (E and F) ALDH2 mRNA expression levels of the GG and GA+AA genotypes. (G and H) CYP2E1 mRNA expression levels of the C1/C1 and C1/C2+C2/C2 genotypes.
Note: * P<0.05; ** P<0.01; # P>0.05.
Abbreviation: HCC, hepatocellular carcinoma.
For CYP2E1 in both the HCC and control groups, mRNA expression levels increased, as compared with non-ethanol intervention, but then decreased with increasing ethanol concentrations. Peculiarly, only 50 mmol/L ethanol intervention showed a significant difference in the HCC group (P<0.05; Figure 2C), and all control groups showed statistically significant P-values (all P<0.05; Figure 2D).
mRNA expression levels of the ALDH2 rs671 and CYP2E1 rs2031920 polymorphisms
For the ALDH2 rs671 polymorphism, mRNA expression levels increased, as compared with non-ethanol intervention, but then decreased with increasing ethanol concentrations for all genotypes. For the G/G genotype, only 50 mmol/L ethanol intervention brought about a significant difference (P<0.05; Figure 2E). For the G/A + A/A genotypes, all three ethanol concentrations gave rise to significant differences (50 and 100 mmol/L, P<0.01; 200 mmol/L, P<0.05; Figure 2F).
For the CYP2E1 rs2031920 polymorphism, mRNA expression levels increased, as compared with non-ethanol intervention, but then decreased with increasing ethanol concentrations for all genotypes. For the C1/C1 genotype, no concentration was associated with a significant difference (all P>0.05; Figure 2G). For the C1/C2 + C2/C2 genotypes, only 200 mmol/L ethanol intervention produced a significant difference (P<0.05; Figure 2H). Specific results are shown in Table 4.
Table 4.
Changes in ALDH2 and CYP2E1 mRNA expression levels after ethanol exposures
| Gene | Group | Genotype | Subject (n=30) | Ethanol concentration (mmol/L)
|
|||
|---|---|---|---|---|---|---|---|
| 0 | 50 | 100 | 200 | ||||
| ALDH2 | HCC | 20 | 0.37±0.20 | 0.55±0.24* | 0.44±0.28 | 0.35±0.15 | |
| Control | 10 | 0.34±0.14 | 0.51±0.21 | 0.35±0.28 | 0.26±0.12 | ||
| CYP2E1 | HCC | 20 | 0.09±0.03 | 0.14±0.08* | 0.09±0.08 | 0.05±0.04 | |
| Control | 10 | 0.04±0.01 | 0.055±0.02** | 0.02±0.01** | 0.01±0.00** | ||
| ALDH2 | G/G | 18 | 0.39±0.19 | 0.57±0.19** | 0.48±0.29 | 0.33±0.15 | |
| G/A+A/A | 12 | 0.12±0.11 | 0.49±0.29** | 0.36±0.17** | 0.26±0.13* | ||
| CYP2E1 | C1/C1 | 20 | 0.03±0.02 | 0.04±0.04 | 0.03±0.03 | 0.03±0.02 | |
| C1/C2+C2/C2 | 10 | 0.11±0.07 | 0.24±0.16* | 0.15±0.08 | 0.13±0.08 | ||
Note:
P<0.05,
P<0.01; comparisons were done between before ethanol intervention (0 mmol/L) and different ethanol intervention concentrations (50, 100, 200 mmol/L); data are presented as the mean±standard deviation.
Abbreviation: HCC, hepatocellular carcinoma.
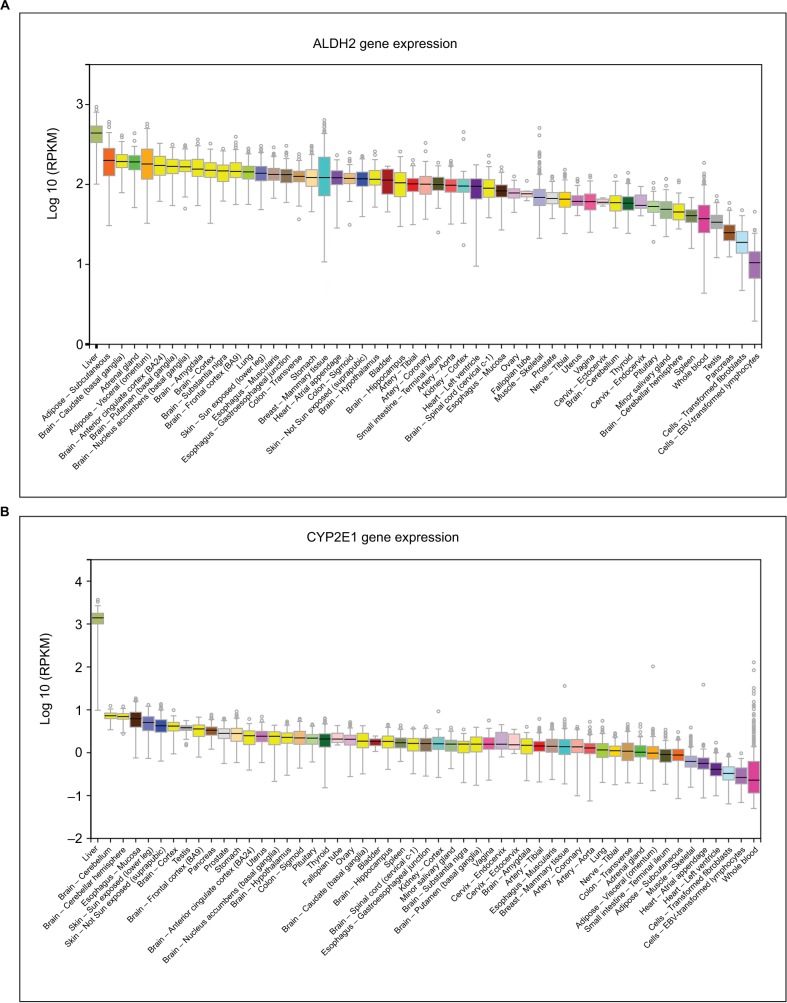
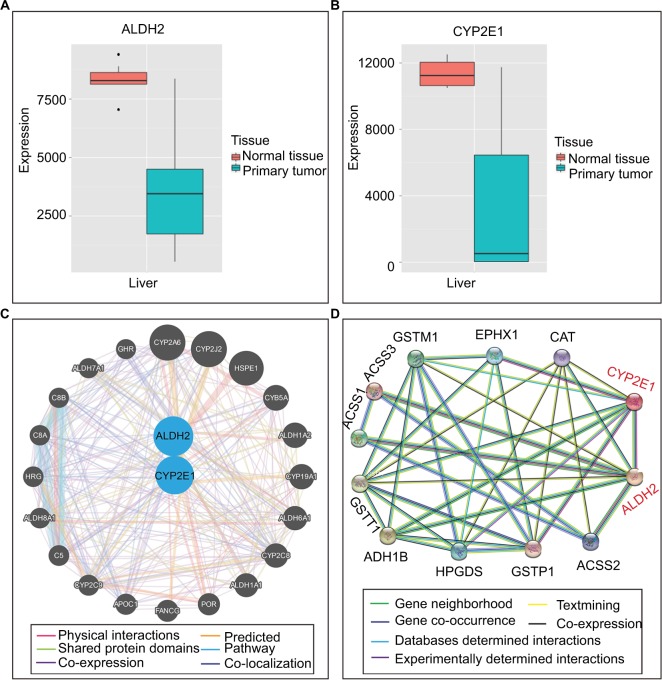
Bioinformatic analysis
Expression levels of ALDH2 and CYP2E1 in different tissues were presented in Figure 3. Besides, Figure 4A and 4B showed the expression levels of ALDH2 and CYP2E1 genes in the primary liver tumor tissue and normal liver tissue. Furthermore, interaction networks of them are shown in Figure 4C and D in gene and protein level, respectively.
Figure 3.
Expression levels of ALDH2 and CYP2E1 in different tissues. (A) ALDH2 expression levels in different tissues. (B) CYP2E1 expression levels in different tissues.
Figure 4.
Gene expression levels and protein interactions. (A and B) ALDH2 and CYP2E1 gene expression levels in normal and primary tumor tissues. (C) Interactions of ALDH2 and CYP2E1 with other genes. (D) Interactions of ALDH2 and CYP2E1 with other proteins.
At a functional significance (FS) score of 0.856, ALDH2 rs671 was depicted in the following three functional categories: protein coding, splicing regulation, and post-translation modification. In regard to protein coding, most prediction tools brought about damaging effects, and a nonsynonymous mutation was confirmed by Ensembl-NS. In regard to splicing regulation, the results of the four prediction tools were consistently changing. In regard to post-translation modification, both OGPET and Sulfinator gave rise to non-existent results. In a FS score of 0.398, CYP2E1 rs2031920 was depicted in the functional category of transcriptional regulation. Ensembl-NS indicated a frameshift result in the coding sequence, while GoldenPath showed no role in regulation, and the Ensembl-TR results confirmed the result of a regulatory region. Detailed data are presented in Table 5. SNP functional prediction results showed that a mutation to rs671 was correlated with splicing, exonic splicing enhancer (ESE), or exonic splicing silencer (ESS), as well as a nonsynonymous mutation, and a mutation in rs2031920 that was associated with the transcription factor binding site (TFBS).
Table 5.
Functional information of ALDH2 rs671 and CYP2E1 rs2031920 genetic polymorphisms
| Gene site | FS score | Functional category | Prediction tool | Prediction result |
|---|---|---|---|---|
| ALDH2 | 0.856 | Protein coding | PolyPhen | Probably damaging |
| rs671 | SIFT | Damaging | ||
| SNP effect | Benign | |||
| LS-SNP | Benign | |||
| SNPs3D | Deleterious | |||
| Ensembl-NS | Nonsynonymous | |||
| Splicing regulation | ESEfinder | Changed | ||
| ESRSearchi | Changed | |||
| PESX | Changed | |||
| RESCUE_ESE | Changed | |||
| Post translation | OGPET | Not exist | ||
| Sulfinator | Not exist | |||
| CYP2E1 | 0.398 | Transcriptional regulation | Ensembl-NS | Frameshift coding |
| GoldenPath | Not exist | |||
| Ensembl-TR | Regulatory region |
Abbreviation: FS, functional significance.
Discussion
In the present study, we first investigated the association of different genotypes of ALDH2 and CYP2E1 with HCC susceptibility in the Guangxi Zhuang Autonomous Region, China. The results of this study demonstrated that, in non-drinking individuals, the mutant genotypes of ALDH2 (G/A and A/A) had significantly different effects on HCC susceptibility, as compared with the wild genotype (G/G) in the same C1/C1 genotype individuals. The mutant genotypes of CYP2E1 (C1/C2 and C2/C2) resulted in significant differences in HCC susceptibility, as compared with the wild genotype (C1/C1) in the same G/G genotype individuals. Meanwhile, the joint-effects analysis of drinking status and ALDH2 showed that alcohol consumption led to significant interactions with HCC susceptibility, as compared with non-drinking subjects with the same GG genotype. In the non-drinking individuals, mutant genotypes showed significant differences in HCC susceptibility, as compared with the wild genotype. These results indicate that mutant variants of ALDH2 and CYP2E1 genes may be protective factors for HCC susceptibility. Then, to verify these results, mRNA expression levels of ALDH2 and CYP2E1 after ethanol intervention were further studied. After intervention with different ethanol gradients, there were significant differences in ALDH2 mRNA expression levels in all G/A + A/A genotypes, as compared with no ethanol intervention, whereas some groups with the G/G genotype showed significant differences. Meanwhile, there were significant differences in CYP2E1 mRNA expression levels associated with the C1/C2 + C2/C2 genotype, as compared with no ethanol intervention, whereas there was no significant difference associated with the C1/C1 genotype, as compared with no ethanol intervention.
Many investigations of the ALDH2 rs671 polymorphism have reported its crucial role in disease, especially cancers, by alteration of ALDH2 expression levels. Mutation reduces ALDH2 expression owing to a dominant-negative effect and, importantly, leads to significant lowering of ALDH2 protein levels.19 By elevating the ALDH2 transcriptional level, ALDH2 activity increases with the restoration of arachidonic acid levels in hepatoma cell lines.20 Inhibition of ALDH2 expression may provide new insights into the associations between cancers and ALDH2.21 Low ALDH2 expression is associated with CCNE1 and SMAD3 expression, and possesses a potential prognostic value in upper tract urothelial carcinoma.22 It is well established that the activity of the ALDH2 enzyme, which is encoded by a mutant allele, was decreased by 30–50% in heterozygous individuals and to almost zero in individuals with the homozygous mutant.23 Decreased ALDH2 enzymatic activity, thus, causes AA accumulation as ALDH2 serves as the principal enzyme in AA metabolism. AA is highly toxic, mutagenic, and carcinogenic,24 and can induce ethanol-metabolizing cancers by its excessive accumulation, including esophageal squamous cell carcinoma,25,26 oropharyngolaryngeal cancer,27 gastric cancer,28–30 colorectal cancer,31–33 pancreatic cancer,34–36 and lung cancer.37–39
Nevertheless, the results of studies of HCC susceptibility remain controversial. In fact, the studies by Yokoyama et al,27 Yu et al,40 Zhou et al,41 and Koide et al42 indicated that the ALDH2*2 polymorphism had limited (not statistically significant) risk for HCC onset. Meanwhile, Tomoda et al43 revealed that ALDH2 rs671 alone was correlated with an increased risk for HCC development, and Liu et al44 and Munaka et al45 indicated that the ALDH2 gene had indirect significance on HCC, in combination with the alcohol dehydrogenase gene. Furthermore, Kato et al46 demonstrated that the combination of ALDH2 and CYP2E1 could modulate HCC development. The results of the present investigation indicated that mutant ALDH2 genotypes displayed significant difference in individuals with the C1/C1 genotype, as well as in the non-drinking populations. These findings are consistent with those of previous studies43,44,46,47 that genotypes, including mutant alleles, have more or less a direct or indirect impact on HCC development due to the ALDH2 gene alone or combined with other factors.
With respect to the CYP2E1 rs2031920 polymorphism, the role of its mutant allele (C2) on HCC susceptibility also remains controversial. Several studies42,48 have indicated that the mutant genotypes C1/C2 and C2/C2 had no impact on HCC susceptibility. Other studies,46,49–52 however, reported the unneglectable function on HCC vulnerability in either a direct or indirect manner. The results of the present study indicated that mutant genotypes of CYP2E1 had significant, but different, impacts on HCC susceptibility when combining ALDH2 and CYP2E1 for analysis. This finding is consistent with the above reports.
After various exposure times to different ethanol concentrations, ALDH2 mRNA expression levels in all groups with mutant genotypes increased significantly, while some groups with the wild genotype showed an increase. As a possible reason for this difference, the mutant genotypes of ALDH2 influence HCC susceptibility. In addition, there were significant differences among the mutant genotypes, while there was no significant difference in CYP2E1 gene expression associated with the wild genotype. Thus, the mutant genotypes of CYP2E1 may play a role in HCC susceptibility.
As shown by the results of bioinformatic analysis, the ALDH2 rs671 mutation was correlated with splicing events (ESE or ESS). An ESE consisting of six bases within an exon is a DNA sequence motif that directs or enhances accurate splicing of pre-mRNA into mRNA. An ESS is a short region, usually 4–18 nucleotides, of an exon, and serves as silencer or inhibitor of splicing of the pre-mRNA and, thus, results in alternate and constitutive splicing.53 To elicit its silencing or inhibiting effects, ESS recruits many proteins that negatively affect the splicing mechanism.53
Prediction results of changing splicing regulation were also determined with four tools: ESEfinder, ESRSearchi, PESX, and RESCUE_ESE. Abnormal splicing changes are thought to contribute to cancer onset and progression.54–57 These splicing changes induce a nonsynonymous mutation at the rs671 site. The protein coding tools PolyPhen, SIFT, and SNPs3D predicted probable damaging effects to the rs671 site. Therefore, we hypothesized that mutation to the rs671 site would induce changes in splicing and protein coding that may serve as potential influencing factors in HCC susceptibility.
The CYP2E1 rs2031920 polymorphism is associated with TFBS, according to the SNP function prediction tool. TFBS is a type of non-DNA molecular binding site that is bound by transcriptional factors and participates in the regulation of transcription of DNA to RNA. With varying degrees of affinity for different transcriptional factors, some binding sites have been reported to undergo rapid evolutionary change.58 However, the results of GoldenPath, a SNP function prediction tool, indicated no transcriptional regulation of the rs2031920 polymorphism. Therefore, we speculated that the low affinity of some transcriptional factors to the rs2031920 genetic polymorphism site could explain these discrepancies. So, further studies should explore the mechanisms of both the ALDH2 rs671 and CYP2E1 polymorphisms. Moreover, further studies concentrating on ALDH2 and CYP2E1 expression levels with HCC prognosis are also warranted.
The findings of this study could serve as a reference to investigate the causes of the high prevalence of HCC in certain areas in China. These findings can also be utilized to distinguish individuals with ALDH2 gene deficiency from the normal ALDH2 genotype population in order to diminish exposure to factors associated with HCC susceptibility. Furthermore, once the roles of these two genes in HCC susceptibility are validated, therapeutic targets and serum biomarkers can be explored to improve early diagnosis and prognosis of HCC.
There were several limitations to this study that should be addressed. First, the samples size was relatively small, thus additional studies with larger sample sizes should be conducted to further validate the roles of these two genes in HCC susceptibility. Second, to eliminate potential confounding factors, more patient data should be collected for analysis of HCC susceptibility and prognosis. Third, multi-center, multi-racial, clinical studies are required to generalize these findings and clarify the underlying mechanisms.
In conclusion, this study was the first to investigate whether the ALDH2 rs671 and CYP2E1 rs2031920 polymorphisms are associated with HCC susceptibility in Guangxi Zhuang Autonomous Region, a region in China with a high prevalence of HCC. In addition, we found that mutant variants of ALDH2-rs671 and CYP2E1-rs2031920 genes may serve as protective factors for HCC susceptibility. These findings provide reference to identify new biomarkers for early diagnosis and treatment of HCC.
Acknowledgments
The authors thank Professors Bin Chen, Kaiyin Xiao, Minhao Peng, Lequn Li, Xiao Qin, Dinghua Yang, Xigang Chen, Ya Guo, Gang Chen, Qiming Feng, Shan Li, and directord Min He, Jiaquan Li, Zhixiong Su, Ning Peng, Xiwen Liao, Yayun Liu, Tingdong Yu, Chengkun Yang, Ketuan Huang, and Liying Huang for their invaluable help to our study. The GTEx Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by National Cancer Institute, National Human Genome Research Institute, National Heart, Lung and Blood Institute, National Institute on Drug Abuse, National Institute of Mental Health and National Institute of Neurological Disorders and Stroke. The data used for the analyses described in this manuscript were obtained from https://gtexportal.org/home/the GTEx Portal on July 1, 2017.
This work was supported in part by the National Nature Science Foundation of China (Nos.: 81560535, 81072321, 30760243, 30460143 and 30560133), 2009 Program for New Century Excellent Talents in University (NCET), Guangxi Nature Sciences Foundation (No.: GuiKeGong 1104003A-7), and Guangxi Health Ministry Medicine Grant (Key-Scientific Research-Grant Z201018), Innovation Project of Guangxi Graduate Education (JGY2018037), and Self-raised Scientific Research Fund of the Health and Family Planning Commission of Guangxi Zhuang Autonomous Region (Z2016318).
Footnotes
Author contributions
Xinping Ye, Xiangkun Wang, Liming Shang, and Tao Peng designed this manuscript; Xinping Ye, Xiangkun Wang, Liming Shang, Guangzhi Zhu, Hao Su, Chuangye Han, Guanghui Li, and Tao Peng conducted the study and collected HCC tissues and blood samples as well as corresponding baseline data of HCC and control groups; Xinping Ye and Xiangkun Wang analyzed the data and wrote this manuscript; Tao Peng guided the writing. All authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379(9822):1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 3.Fan H, Zhang Q, Zhao X, Lv P, Liu M, Tang H. Transcriptomic profiling of long non-coding RNAs in hepatitis B virus-related hepatocellular carcinoma. Oncotarget. 2017;8(39):65421–65434. doi: 10.18632/oncotarget.18897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bierut LJ, Agrawal A, Bucholz KK, et al. A genome-wide association study of alcohol dependence. Proc Natl Acad Sci USA. 2010;107(11):5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seitz HK, Meier P. The role of acetaldehyde in upper digestive tract cancer in alcoholics. Transl Res. 2007;149(6):293–297. doi: 10.1016/j.trsl.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Bellentani S, Saccoccio G, Costa G, et al. Drinking habits as cofactors of risk for alcohol induced liver damage. Gut. 1997;41(6):845–850. doi: 10.1136/gut.41.6.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang B, Zan RY, Wang SY, et al. Radiofrequency ablation versus percutaneous ethanol injection for hepatocellular carcinoma: a meta-analysis of randomized controlled trials. World J Surg Oncol. 2015;13:96. doi: 10.1186/s12957-015-0516-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu W, Qian J, Ma L, Ma P, Yang F, Shu Y. MiR-346 suppresses cell proliferation through SMYD3 dependent approach in hepatocellular carcinoma. Oncotarget. 2017;8(39):65218–65229. doi: 10.18632/oncotarget.18060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altekruse SF, McGlynn KA, Dickie LA, Kleiner DE. Hepatocellular carcinoma confirmation, treatment, and survival in surveillance, epidemiology, and end results registries, 1992–2008. Hepatology. 2012;55(2):476–482. doi: 10.1002/hep.24710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song BJ. Ethanol-inducible cytochrome P450 (CYP2E1): biochemistry molecular biology and clinical relevance: 1996 update. Alcohol Clin Exp Res. 1996;20(8 Suppl):138a–146a. doi: 10.1111/j.1530-0277.1996.tb01764.x. [DOI] [PubMed] [Google Scholar]
- 11.Lieber CS. Alcohol and the liver: 1994 update. Gastroenterology. 1994;106(4):1085–1105. doi: 10.1016/0016-5085(94)90772-2. [DOI] [PubMed] [Google Scholar]
- 12.Lucas D, Menez C, Girre C, et al. Cytochrome P450 2E1 genotype and chlorzoxazone metabolism in healthy and alcoholic Caucasian subjects. Pharmacogenetics. 1995;5(5):298–304. doi: 10.1097/00008571-199510000-00005. [DOI] [PubMed] [Google Scholar]
- 13.Li YF, Sung FC, Tsai MH, et al. Interactions between cigarette smoking and polymorphisms of xenobiotic-metabolizing genes: the risk of oral leukoplakia. Dis Markers. 2013;34(4):247–255. doi: 10.3233/DMA-130967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarty CA, Reding DJ, Commins J, et al. Alcohol, genetics and risk of breast cancer in the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. Breast Cancer Res Treat. 2012;133(2):785–792. doi: 10.1007/s10549-012-1972-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee H, Kim SS, You KS, et al. Asian flushing: genetic and sociocultural factors of alcoholism among East Asians. Gastroenterol Nurs. 2014;37(5):327–336. doi: 10.1097/SGA.0000000000000062. [DOI] [PubMed] [Google Scholar]
- 16.Lai CL, Yao CT, Chau GY, et al. Dominance of the inactive Asian variant over activity and protein contents of mitochondrial aldehyde dehydrogenase 2 in human liver. Alcohol Clin Exp Res. 2014;38(1):44–50. doi: 10.1111/acer.12215. [DOI] [PubMed] [Google Scholar]
- 17.Chen YC, Peng GS, Tsao TP, Wang MF, Lu RB, Yin SJ. Pharmacokinetic and pharmacodynamic basis for overcoming acetaldehyde-induced adverse reaction in Asian alcoholics, heterozygous for the variant ALDH2*2 gene allele. Pharmacogenet Genomics. 2009;19(8):588–599. doi: 10.1097/FPC.0b013e32832ecf2e. [DOI] [PubMed] [Google Scholar]
- 18.Harada S, Agarwal DP, Goedde HW. Aldehyde dehydrogenase deficiency as cause of facial flushing reaction to alcohol in Japanese. Lancet. 1981;2(8253):982. doi: 10.1016/s0140-6736(81)91172-7. [DOI] [PubMed] [Google Scholar]
- 19.Jin S, Chen J, Chen L, et al. ALDH2(E487K) mutation increases protein turnover and promotes murine hepatocarcinogenesis. Proc Natl Acad Sci USA. 2015;112(29):9088–9093. doi: 10.1073/pnas.1510757112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Canuto RA, Ferro M, Salvo RA, et al. Increase in class 2 aldehyde dehydrogenase expression by arachidonic acid in rat hepatoma cells. Biochem J. 2001;357(Pt 3):811–818. doi: 10.1042/0264-6021:3570811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amanuma Y, Ohashi S, Itatani Y, et al. Protective role of ALDH2 against acetaldehyde-derived DNA damage in oesophageal squamous epithelium. Sci Rep. 2015;5:14142. doi: 10.1038/srep14142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu S, Chen J, Dong P, et al. Global gene expression profiling identifies ALDH2, CCNE1 and SMAD3 as potential prognostic markers in upper tract urothelial carcinoma. BMC Cancer. 2014;14:836. doi: 10.1186/1471-2407-14-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crabb DW, Edenberg HJ, Bosron WF, Li TK. Genotypes for aldehyde dehydrogenase deficiency and alcohol sensitivity. The inactive ALDH2 (2) allele is dominant. J Clin Invest. 1989;83(1):314–316. doi: 10.1172/JCI113875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salaspuro MP. Acetaldehyde, microbes, and cancer of the digestive tract. Crit Rev Clin Lab Sci. 2003;40(2):183–208. doi: 10.1080/713609333. [DOI] [PubMed] [Google Scholar]
- 25.Yokoyama A, Kato H, Yokoyama T, et al. Genetic polymorphisms of alcohol and aldehyde dehydrogenases and glutathione S-transferase M1 and drinking, smoking, and diet in Japanese men with esophageal squamous cell carcinoma. Carcinogenesis. 2002;23(11):1851–1859. doi: 10.1093/carcin/23.11.1851. [DOI] [PubMed] [Google Scholar]
- 26.Yokoyama A, Muramatsu T, Omori T, et al. Alcohol and aldehyde dehydrogenase gene polymorphisms and oropharyngolaryngeal, esophageal and stomach cancers in Japanese alcoholics. Carcinogenesis. 2001;22(3):433–439. doi: 10.1093/carcin/22.3.433. [DOI] [PubMed] [Google Scholar]
- 27.Yokoyama A, Muramatsu T, Ohmori T, et al. Alcohol-related cancers and aldehyde dehydrogenase-2 in Japanese alcoholics. Carcinogenesis. 1998;19(8):1383–1387. doi: 10.1093/carcin/19.8.1383. [DOI] [PubMed] [Google Scholar]
- 28.Cao HX, Li SP, Wu JZ, et al. Alcohol dehydrogenase-2 and aldehyde dehydrogenase-2 genotypes, alcohol drinking and the risk for stomach cancer in Chinese males. Asian Pac J Cancer Prev. 2010;11:1073–1077. [PubMed] [Google Scholar]
- 29.Yokoyama A, Yokoyama T, Omori T, et al. Helicobacter pylori, chronic atrophic gastritis, inactive aldehyde dehydrogenase−2, macrocytosis and multiple upper aerodigestive tract cancers and the risk for gastric cancer in alcoholic Japanese men. J Gastroenterol Hepatol. 2007;22(2):210–217. doi: 10.1111/j.1440-1746.2006.04377.x. [DOI] [PubMed] [Google Scholar]
- 30.Matsuo K, Oze I, Hosono S, et al. The aldehyde dehydrogenase 2 (ALDH2) Glu504Lys polymorphism interacts with alcohol drinking in the risk of stomach cancer. Carcinogenesis. 2013;34(7):1510–1515. doi: 10.1093/carcin/bgt080. [DOI] [PubMed] [Google Scholar]
- 31.Matsuo K, Hamajima N, Hirai T, et al. Aldehyde dehydrogenase 2 (ALDH2) genotype affects rectal cancer susceptibility due to alcohol consumption. J Epidemiol. 2002;12(2):70–76. doi: 10.2188/jea.12.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiang CP, Wu CW, Lee SP, et al. Expression pattern, ethanol-metabolizing activities, and cellular localization of alcohol and aldehyde dehydrogenases in human small intestine. Alcohol Clin Exp Res. 2012;36(12):2047–2058. doi: 10.1111/j.1530-0277.2012.01836.x. [DOI] [PubMed] [Google Scholar]
- 33.Murata M, Tagawa M, Watanabe S, Kimura H, Takeshita T, Morimoto K. Genotype difference of aldehyde dehydrogenase 2 gene in alcohol drinkers influences the incidence of Japanese colorectal cancer patients. Jpn J Cancer Res. 1999;90(7):711–719. doi: 10.1111/j.1349-7006.1999.tb00805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyasaka K, Hosoya H, Tanaka Y, et al. Association of aldehyde dehydrogenase 2 gene polymorphism with pancreatic cancer but not colon cancer. Geriatr Gerontol Int. 2010;10(Suppl 1):S120–S126. doi: 10.1111/j.1447-0594.2010.00616.x. [DOI] [PubMed] [Google Scholar]
- 35.Miyasaka K, Kawanami T, Shimokata H, Ohta S, Funakoshi A. Inactive aldehyde dehydrogenase-2 increased the risk of pancreatic cancer among smokers in a Japanese male population. Pancreas. 2005;30(2):95–98. doi: 10.1097/01.mpa.0000147084.70125.41. [DOI] [PubMed] [Google Scholar]
- 36.Kanda J, Matsuo K, Suzuki T, et al. Impact of alcohol consumption with polymorphisms in alcohol-metabolizing enzymes on pancreatic cancer risk in Japanese. Cancer Sci. 2009;100(2):296–302. doi: 10.1111/j.1349-7006.2008.01044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eriksson CJ. Genetic-epidemiological evidence for the role of acetaldehyde in cancers related to alcohol drinking. Adv Exp Med Biol. 2015;815:41–58. doi: 10.1007/978-3-319-09614-8_3. [DOI] [PubMed] [Google Scholar]
- 38.Minegishi Y, Tsukino H, Muto M, et al. Susceptibility to lung cancer and genetic polymorphisms in the alcohol metabolite-related enzymes alcohol dehydrogenase 3, aldehyde dehydrogenase 2, and cytochrome P450 2E1 in the Japanese population. Cancer. 2007;110(2):353–362. doi: 10.1002/cncr.22795. [DOI] [PubMed] [Google Scholar]
- 39.Eom SY, Zhang YW, Kim SH, et al. Influence of NQO1, ALDH2, and CYP2E1 genetic polymorphisms, smoking, and alcohol drinking on the risk of lung cancer in Koreans. Cancer Causes Control. 2009;20(2):137–145. doi: 10.1007/s10552-008-9225-7. [DOI] [PubMed] [Google Scholar]
- 40.Yu SZ, Huang XE, Koide T, et al. Hepatitis B and C viruses infection, lifestyle and genetic polymorphisms as risk factors for hepatocellular carcinoma in Haimen, China. Jpn J Cancer Res. 2002;93(12):1287–1292. doi: 10.1111/j.1349-7006.2002.tb01236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou D, Xiao L, Zhang Y, et al. Genetic polymorphisms of ALDH2 and ADH2 are not associated with risk of hepatocellular carcinoma among East Asians. Tumour Biol. 2012;33(3):841–846. doi: 10.1007/s13277-011-0309-8. [DOI] [PubMed] [Google Scholar]
- 42.Koide T, Ohno T, Huang XE, et al. HBV/HCV infection, alcohol, tobacco and genetic polymorphisms for hepatocellular carcinoma in Nagoya, Japan. Asian Pac J Cancer Prev. 2000;1(3):237–243. [PubMed] [Google Scholar]
- 43.Tomoda T, Nouso K, Sakai A, et al. Genetic risk of hepatocellular carcinoma in patients with hepatitis C virus: a case control study. J Gastroenterol Hepatol. 2012;27(4):797–804. doi: 10.1111/j.1440-1746.2011.06948.x. [DOI] [PubMed] [Google Scholar]
- 44.Liu J, Yang HI, Lee MH, et al. Alcohol drinking mediates the association between polymorphisms of ADH1B and ALDH2 and hepatitis B-related hepatocellular carcinoma. Cancer Epidemiol Biomarkers Prev. 2016;25(4):693–699. doi: 10.1158/1055-9965.EPI-15-0961. [DOI] [PubMed] [Google Scholar]
- 45.Munaka M, Kohshi K, Kawamoto T, et al. Genetic polymorphisms of tobacco-and alcohol-related metabolizing enzymes and the risk of hepatocellular carcinoma. J Cancer Res Clin Oncol. 2003;129(6):355–360. doi: 10.1007/s00432-003-0439-5. [DOI] [PubMed] [Google Scholar]
- 46.Kato S, Tajiri T, Matsukura N, et al. Genetic polymorphisms of aldehyde dehydrogenase 2, cytochrome p450 2E1 for liver cancer risk in HCV antibody-positive japanese patients and the variations of CYP2E1 mRNA expression levels in the liver due to its polymorphism. Scand J Gastroenterol. 2003;38(8):886–893. doi: 10.1080/00365520310004489. [DOI] [PubMed] [Google Scholar]
- 47.Sakamoto T, Hara M, Higaki Y, et al. Influence of alcohol consumption and gene polymorphisms of ADH2 and ALDH2 on hepatocellular carcinoma in a Japanese population. Int J Cancer. 2006;118(6):1501–1507. doi: 10.1002/ijc.21505. [DOI] [PubMed] [Google Scholar]
- 48.Wong NA, Rae F, Simpson KJ, Murray GD, Harrison DJ. Genetic polymorphisms of cytochrome p4502E 1 and susceptibility to alcoholic liver disease and hepatocellular carcinoma in a white population: a study and literature review, including meta-analysis. Mol Pathol. 2000;53(2):88–93. doi: 10.1136/mp.53.2.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ladero JM, Agundez JA, Rodriguez-Lescure A, Diaz-Rubio M, Benitez J. RsaI polymorphism at the cytochrome P4502E1 locus and risk of hepatocellular carcinoma. Gut. 1996;39(2):330–333. doi: 10.1136/gut.39.2.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu MW, Gladek-Yarborough A, Chiamprasert S, Santella RM, Liaw YF, Chen CJ. Cytochrome P450 2E1 and glutathione S-transferase M1 polymorphisms and susceptibility to hepatocellular carcinoma. Gastroenterology. 1995;109(4):1266–1273. doi: 10.1016/0016-5085(95)90587-1. [DOI] [PubMed] [Google Scholar]
- 51.Zhou J, Wen Q, Li SF, et al. Significant change of cytochrome P450s activities in patients with hepatocellular carcinoma. Oncotarget. 2016;7(31):50612–50623. doi: 10.18632/oncotarget.9437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao J, Zhou J, He XP, et al. Changes in cytochrome P450s-mediated drug clearance in patients with hepatocellular carcinoma in vitro and in vivo: a bottom-up approach. Oncotarget. 2016;7(19):28612–28623. doi: 10.18632/oncotarget.8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goren A, Ram O, Amit M, et al. Comparative analysis identifies exonic splicing regulatory sequences—the complex definition of enhancers and silencers. Mol Cell. 2006;22(6):769–781. doi: 10.1016/j.molcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 54.Skotheim RI, Nees M. Alternative splicing in cancer: noise, functional, or systematic? Int J Biochem Cell Biol. 2007;39(7–8):1432–1449. doi: 10.1016/j.biocel.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 55.He C, Zhou F, Zuo Z, Cheng H, Zhou R. A global view of cancer-specific transcript variants by subtractive transcriptome-wide analysis. PloS One. 2009;4(3):e4732. doi: 10.1371/journal.pone.0004732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fackenthal JD, Godley LA. Aberrant RNA splicing and its functional consequences in cancer cells. Dis Model Mech. 2008;1(1):37–42. doi: 10.1242/dmm.000331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sveen A, Kilpinen S, Ruusulehto A, Lothe R, Skotheim R. Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene. 2016;35(19):2413–2427. doi: 10.1038/onc.2015.318. [DOI] [PubMed] [Google Scholar]
- 58.Borneman AR, Gianoulis TA, Zhang ZD, et al. Divergence of transcription factor binding sites across related yeast species. Science. 2007;317(5839):815–819. doi: 10.1126/science.1140748. [DOI] [PubMed] [Google Scholar]