Abstract
Emerging evidence about the human microbiome, a collective term for all the microorganisms living in and on the human body, consistently demonstrates the critical influence it has on host physiology and disease risk. The microbiota in the gastrointestinal (GI) tract has the most significant and far-reaching effect on human physiology. The maternal GI microbiota can decrease the risk of adverse pregnancy outcomes by modulating energy extraction, glucose metabolism, vitamin production, and host immunity essential for optimal maternal and neonatal health. Moreover, the maternal GI microbiota is thought to influence colonization of the fetus and neonate that may predispose them to different health trajectories. This article provides a basic understanding about the influence of the structure of the maternal GI microbiota, the fundamental role it plays during pregnancy, and the factors that influence the structure, and subsequently function, of the GI microbiota in the general and pregnant population. While only a small number of studies have examined this topic during pregnancy, the preponderance of the evidence supports the need to clarify baseline structure and function of GI microbiota and its associations with pregnancy outcomes. In addition, the results from the studies conducted in the general population can be extrapolated to pregnancy in many cases. This knowledge is essential for clinicians who need to understand the implications of the microbiota for disease and wellness in order to address the care factors that may adversely influence the GI microbiota during pregnancy.
Keywords: microbiome, pregnancy, gastrointestinal microbiota
Emerging evidence about the human microbiome, a collective term for the microorganisms living in and on the human body, consistently demonstrates the critical influence the microbial communities have on host physiology and disease risk. Traditionally, microbes were thought to be pathogenic microorganisms associated with disease. However, current research is exposing the essential symbiotic nature of the relationship between the microbiota and the host. The change in our understanding is largely related to the development of culture-independent molecular techniques and computational tools, including polymerase chain reaction amplification, 16S ribosomal RNA sequencing techniques, and the Ribosomal Database Project Bayesian classifier used for taxonomic assignment. These new technologies allow scientists to characterize the microbes and isolate the physiologic functions they initiate in the human host (Caporaso et al., 2010; Cho & Blaser, 2012; Cole et al., 2009; Fadrosh et al., 2014; Kozich, Westcott, Baxter, Highlander, & Schloss, 2013; Kuczynski et al., 2012; Lasken & McLean, 2014; Q. Wang, Garrity, Tiedje, & Cole, 2007). The Human Microbiome Project (HMP), initiated by the National Institutes of Health in 2008, extended the Human Genome Project and focused on characterization of the microbial communities throughout the human body and analysis of their functional role in human health (Gevers, Pop, Schloss, & Huttenhower, 2012; Gevers, Knight, et al., 2012; HMP Consortium, 2012; Methé et al., 2012; Sellitto et al., 2012; Turnbaugh et al., 2007). Findings from the HMP have provided a baseline for understanding the structure and potential function of the unique microbial communities that reside in the various parts of the human body.
Of all the human microbial communities, the microbiota in the gastrointestinal (GI) tract has the most significant and far-reaching effect on human physiology. This is because the GI microbiota modulates host gene expression and significantly influences the development and maintenance of the immune system. Recent research about the brain–gut axis has also demonstrated the effect the GI microbiota has on cognitive function (Borre et al., 2014; Foster & McVey Neufeld, 2013; Ridaura & Belkaid, 2015; Sommer & Bäckhed, 2013; Xu & Gordon, 2003). The GI microbiota is a complex ecosystem comprised of tens to hundreds of trillions of microbes (1013–1014) that are distributed in unique clusters throughout the gut (Dethlefsen, McFall-Ngai, & Relman, 2007; National Institutes of Health Human Microbiome Project Working Group, 2009; Savage, 1977). Within this structure, the majority of the GI microbiota are bacteria, but archaea, eukaryote, and viruses are also present. Among them, they contain over 5 million genes—subunits of deoxyribonucleic acid (DNA) that encode instructions to produce specific proteins or enzymes that initiate particular physiologic functions in the host. The influence the organisms that comprise the microbiota have on human physiology is so profound that the GI microbiota is widely considered a metabolic organ in its own right, and the authors have referred to it as the “forgotten organ” (O’Hara & Shanahan, 2006). This viewpoint originated in recognition of the contribution these unique colonies of microbes make to immunologic and metabolic homeostasis in the host (Wells, Rossi, Meijerink, & van Baarlen, 2011).
The GI microbiota influences host physiology in a variety of ways, primarily by producing metabolites that modulate cell-signaling pathways, and changes in the structure of the communities such as low diversity or altered composition are associated with multiple disease processes including inflammatory bowel, cardiovascular, and neoplastic diseases and neurobehavioral symptoms (Borre et al., 2014; Foster & McVey Neufeld, 2013; Frank et al., 2007; Hope, Hold, Kain, & El-Omar, 2005; Louis, Hold, & Flint, 2014; Methé et al., 2012; Peterson, Frank, Pace, & Gordon, 2008; Ridaura & Belkaid, 2015; Sartor, 2008; Schwabe & Jobin, 2013; Wong, 2014; W. K. Wu et al., 2015; Zeng, Lazarova, & Bordonaro, 2014). Disruptions of GI microbiota are also linked with a wide range of health disorders including obesity, hypertension, and diabetes (Larsen et al., 2010; Ley, Turnbaugh, Klein, & Gordon, 2006; Qin et al., 2012; Ridaura et al., 2013; Turnbaugh et al., 2006; Turnbaugh, Hamady, et al., 2009; Vijay-Kumar et al., 2010; Vrieze et al., 2012). Alternately, the structure of the microbial communities can positively modulate energy extraction, glucose metabolism, vitamin production, and host immunity (Belizário & Napolitano, 2015; Qin et al., 2010).
The association of the GI microbiota with host physiologic function has particular relevance during pregnancy because of the high physiologic burden placed on the female, requiring metabolic alterations to meet the demands of the developing fetus. Inadequate adjustments of the maternal system are directly correlated to adverse pregnancy outcomes including gestational diabetes, preeclampsia, and preterm birth. Moreover, evidence suggests that the maternal GI microbiota influences immune development and colonization of the fetus and neonate that may predispose them to particular health trajectories (de Agüero et al., 2016; Pendse & Hooper, 2016).
The purpose of this article is to present the current state of the knowledge about the maternal GI microbiota, the fundamental role it plays during pregnancy, and the factors associated with the structure and function of the GI microbiota. This information is essential for practitioners, so that they are able to provide care that accounts for changes in the GI microbiota that could influence health outcomes.
Overview of the GI Microbiota
Organization of the GI Microbiota by Taxonomic Rank or Enterotypes
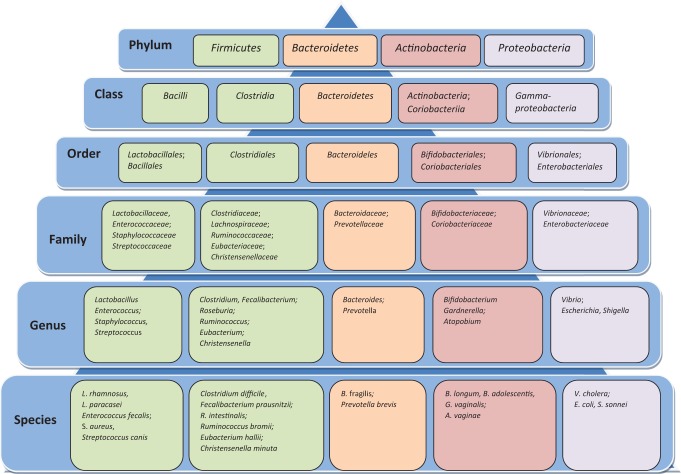
Microbes in the GI tract comprise the most abundant, diverse, and complex microbial ecosystem compared to those in other regions of the body. The inherited traits or features of the GI microbes have been classified into taxonomic ranks that define the relative levels of the groups within the specific kingdom. Phylum is the highest rank in terms of the taxonomic hierarchy followed by class, order, family, genus, and species (see Figure 1). The vast majority of the GI microbiota in adults belong to two bacterial phyla, Firmicutes and Bacteroidetes, with lower proportions classified into other phyla including Proteobacteria, Actinobacteria, and Verrucomicrobia (Tremaroli & Bäckhed, 2012).
Figure 1.
Limited subset of microbes found in the human gastrointestinal microbiome outlined by their taxonomic ranks, limited to species level. Strains within each species that are differentiated by a set of variable genes with distinct functional potentials are not included.
Phylum-level taxa are classified by genetic composition unrelated to specific functions. As knowledge has increased, more commonly these taxa have been clustered into enterotypes, which are ecosystems within the gut microbiome. This approach allows scientists to identify associations between enterotypes and disease processes or health outcomes. Some scientists have proposed that the GI microbiota cluster into three primary enterotypes, dominated by the Bacteroides, Prevotella, or Ruminococcus genera, each representing different functions in terms of the mechanisms used to generate energy from the available substrates in the colon (Table 1). The cluster dominated by the Bacteroides genus in the Bacteroidetes phylum derives energy primarily from carbohydrates and proteins. The cluster rich in the Prevotella genus, also in the Bacteroidetes phylum, degrades mucin glycoproteins in the mucosal layer of the gut for energy. The third cluster, rich in the Ruminococcus genus in the Firmicutes phylum, like Prevotella, degrades mucin. Enterotype clusters are unique, and their proportional dominance within the GI microbiota varies based on specialized functions, suggesting that specific microbial clusters are privileged based on the available substrates within the GI tract (Arumugam et al., 2011).
Table 1.
Enterotypes of Gastrointestinal (GI) Microbiota Representing Different Functions in Terms of the Mechanisms Used to Generate Energy From the Available Substrates in the Colon.
| Classification Model and Dominating Genus | Respective Phylum | Functions—Source of Energy |
|---|---|---|
| Three clusters (Arumugam et al., 2011) | ||
| Prevotella | Bacteroidetes | Mucin glycoproteins in mucosal layer of the gut |
| Bacteroides | Bacteroidetes | Carbohydrates and proteins |
| Ruminococcus | Firmicutes | Mucin glycoproteins in mucosal layer of the gut |
| Two clusters (G. D. Wu et al., 2011) | Functions—Associated Diet | |
| Prevotella | Bacteroidetes | Diet high in carbohydrates typically seen in people in Africa |
| Bacteroidetes + Ruminococcus | Bacteroidetes + Firmicutes | Diet high in protein and animal fats that is typically seen in people in Europe |
An alternative view is that there are only two primary enterotypes within the GI microbiota, which are influenced by long-term diet. In this perspective, the cluster rich in organisms of the Ruminococcus genus is fused with the cluster rich in organisms of the Bacteroides genus (G. D. Wu et al., 2011). The fused enterotype rich in both Ruminococcus and Bacteroides is associated with the diets high in protein and animal fats typically seen in people in Europe, whereas the Prevotella-rich enterotype is associated with the diets high in carbohydrates typically seen in people in Africa (De Filippo et al., 2010; G. D. Wu et al., 2011).
Despite the discrepancy over whether there are two or three enterotypes, there is agreement about the fact that the GI microbiota produce metabolites; for instance, Faecalibacterium prausnitzii, Roseburia intestinalis, and Bacteroides uniformis produce butyrate, among others (Qin et al., 2010). This finding is important because butyrate, among other metabolites, serves as an energy source for colonocytes, which maintain the integrity of the mucosal lining of the GI tract, thus limiting the ability of toxins and microbes to breach the protective intestinal barrier.
Structure and Function of GI Microbiota in the General Population
The structure of the GI microbiota comprises the composition, relative abundance, and diversity of microbes that is established over time beginning before birth. Composition describes the types of microbes that are present, while relative abundance describes the proportion of each type of microbe relative to the others. Diversity has two dimensions, namely, richness (the number of different phylotypes) and evenness (the proportionality of the taxa within the sample; Turnbaugh, Hamady, et al., 2009; Y. Wang et al., 2009). Diversity within samples or subjects is termed α diversity, whereas diversity between samples or subjects is called β diversity (Turnbaugh et al., 2007). The notion that the fetal sac is a sterile environment has been replaced by evidence generated from an increasing number of studies that shows traces of the maternal GI microbiota in the placenta, amniotic fluid, and fetal membranes in healthy term pregnancies without signs of infection, rupture of membrane, or labor. These findings suggest that colonization of the fetal GI tract starts in utero (Aagaard et al., 2014; Rautava, Collado, Salminen, & Isolauri, 2012; Satokari, Grönroos, Laitinen, Salminen, & Isolauri, 2009; Steel et al., 2005). This proposition is further supported by studies that have demonstrated that meconium contains microbes isolated from the amniotic fluid (Ardissone et al., 2014).
Colonization continues during birth, although the composition varies based on the type of delivery. Babies born vaginally come into direct contact with, and are colonized by, the maternal vaginal microbial communities. In contrast, babies born by cesarean section are colonized by microbes common to the maternal skin surfaces (Dominguez-Bello et al., 2010; Penders et al., 2006). After birth, the method of feeding also influences the process of colonization. Breast milk harbors more than 200 species of bacteria, including those belonging to Bifidobacterium, Lactobacillus, Enterococcus, and Staphylococcus genera, which confer proven health benefits (Fukuda et al., 2011; LeBlanc et al., 2013). Breast milk is also rich in human milk oligosaccharides, which serve as prebiotics, substrates that promote the growth of beneficial microbes, specifically microbes such as Bifidobacteria (Jost, Lacroix, Braegger, & Chassard, 2015). Although the structure of the GI microbiota varies widely across neonates in general, in breastfed neonates, it is more stable and dominated by beneficial Bifidobacterium spp. than in formula-fed babies. Conversely, in those who are formula-fed, the microbiota is less diverse and has a lower density of lactic acid–producing Bifidobacterium spp. and Lactobacillus spp., which protect against pathogenic overgrowth (Jost et al., 2015; Martín et al., 2007; Penders et al., 2006; Sela et al., 2008). The structure of the GI microbiota in neonates is considerably less complex than in adults; however, as infants’ diet evolves to include complex solid foods, the microbiota stabilizes over time and comes to resemble the structure and functionality that of adults (Koenig et al., 2011; C. Palmer, Bik, DiGiulio, Relman, & Brown, 2007; Vaishampayan et al., 2010; Yatsunenko et al., 2012).
Understanding what an optimal structure for the maternal GI microbiota looks like is crucial because the microbes are hypothesized to pass through the permeable mucosal surface of the maternal GI tract into the blood stream and translocate to the placenta and intra-amniotic fluid where they are ingested by the fetus and colonize the fetal GI tract (Jiménez et al., 2005, 2008; Rautava, Collado, Salminen, & Isolauri, 2012; Rautava, Kalliomäki, & Isolauri, 2002). A similar mechanism is thought to transport maternal GI microbiota into the mammary gland, termed the enteromammary pathway (Jost et al., 2015). Likewise, findings from research on probiotic supplementation suggest that the maternal vaginal microbiota that colonizes the neonate during vaginal delivery are influenced by GI microbiota (Reid, 2012; Reid et al., 2003; Strus et al., 2012; Vitali et al., 2012; Vujic, Knez, Stefanovic, & Vrbanovic, 2013). This growing body of evidence provides insights that support the critical role that the structure of the maternal GI microbiota plays in the initial microbial colonization of the fetus and neonate, which may predispose the child to different health trajectories (Ajslev, Andersen, Gamborg, Sørensen, & Jess, 2011; Johansson, Sjögren, Persson, Nilsson, & Sverremark-Ekström, 2011; Kalliomäki, Collado, Salminen, & Isolauri, 2008; Rautava, Luoto, Salminen, & Isolauri, 2012; Renz, Brandtzaeg, & Hornef, 2012).
The GI Microbiota During Pregnancy
Very few studies have examined the structure of the GI microbiota during pregnancy. Much of the available evidence has been gathered via animal models and therefore may have limited application to humans. In addition, the findings are not consistent across studies, suggesting that more research is necessary to clarify the issues. This gap in the literature is particularly important in relation to defining the optimal structure of the GI microbiota during pregnancy due to the growing body of evidence suggesting that it is critical to both maternal and fetal health. Knowledge about the structure of the GI microbiota more generally will serve as a foundation to understanding its functions as they relate to pregnancy.
Composition and Relative Abundance
Despite the paucity of studies on the structure of the GI microbiota during pregnancy, investigators have studied the composition and relative abundance within the GI microbiota before and during pregnancy and in the early postpartum period (Table 2). In one study utilizing an animal model that examined the composition of the GI microbiota prior to and throughout pregnancy, researchers found significantly increased relative abundance of mucin-degrading microbes including Akkermansia (Verrucomicrobia phylum) and Bifidobacterium (Actinobacteria phylum) in pregnant versus nonpregnant mice (Gohir, Whelan, Surette, Moore, & Jonathan, 2015). The findings also showed that Akkermansia remained elevated throughout pregnancy. This finding replicated that of an earlier human study that examined the composition of GI microbiota throughout pregnancy (Collado, Isolauri, Laitinen, & Salminen, 2008). The finding is highly relevant because an increased abundance of mucin-degrading microbiota during pregnancy suggests increased capacity to extract energy in the absence of adequate dietary substrate. Mucin is essential to maintaining the mucosal barrier along the gut, and thus degradation of mucin may breach the integrity of the barrier. However, during pregnancy, the energy extracted is needed to accommodate increased maternal requirements to meet the needs of the growing fetus. In the same study (Collado et al., 2008), genera and species belonging to the phylum Firmicutes (e.g., Staphylococcus aureus), which are positively associated with increased storage of energy, increased from the first (T1) to the third trimester (T3). An increased abundance of Firmicutes may suggest an alteration in the physiologic capacity to extract additional energy necessary during pregnancy.
Table 2.
Structure of the Gastrointestinal (GI) Microbiota During Pregnancy.
| Structural Component | Pregnancy | Reference | ||
|---|---|---|---|---|
| Early | Late | Early Postpartum | ||
| Relative abundance | Increased mucin-degrading microbiota, including Akkermansia and Bifidobacterium | — | — | Gohir et al. (2015) |
| Akkermansia remained elevated throughout pregnancy | — | Gohir et al. (2015) | ||
| Genera and species belonging to phyla Firmicutes (e.g., Staphylococcus aureus) increased from T1 to T3 | — | Collado et al. (2008) | ||
| Members of pro-inflammatory Proteobacteria and Actinobacteria phyla increased from T1 to T3, while anti-inflammatory Faecalibacterium genus decreased from T1 to T3 | The composition during T3 is preserved in early postpartum period | Koren et al. (2012) | ||
| — | GI microbial community is relatively stable and does not change during period from T3 to 1-month postpartum | Jost et al. (2014) | ||
| Diversity | ||||
| α diversity (within individual) | High | Low | — | Koren et al. (2012) |
| β diversity (between individuals) | Low | High | — | Koren et al. (2012) |
| α- and β-diversity | Remained stable throughout pregnancy, unaffected by the progression of pregnancy | — | DiGiulio et al. (2015) | |
Note. T1 = first trimester; T3 = third trimester.
The profound changes in the microbial composition from T1 to T3 were validated in a different study that transplanted the GI microbiota from a subgroup of 5 out of 91 pregnant women into germ-free (gnotobiotic) mice (Koren et al., 2012). Regardless of diet or health status in the mice, the relative abundance of pro-inflammatory microbes in the Proteobacteria (e.g., Enterobacteria) and Actinobacteria phyla increased from T1 to T3, while that of microbes in the anti-inflammatory Faecalibacterium genus (Firmicutes phylum, Clostridiales order) decreased over the same period. The Proteobacteria phylum includes a wide variety of pathogenic genera, such as Escherichia, Salmonella, and Helicobacter, and increases in microbes from this phylum have been repeatedly associated with inflammation-related dysbiosis. In addition, microbes from the Actinobacteria phylum are associated with obesity, inflammatory bowel diseases, and respiratory diseases (Kassinen et al., 2007; Rintala, 2011; Turnbaugh, Hamady, et al., 2009). Not surprisingly, researchers noted metabolic changes similar to metabolic syndrome, characterized by increased weight, excess adiposity, insulin resistance, hyperglycemia, and low-grade inflammation, when they transplanted the human T3 GI microbiota, with increased abundance of members of both phyla, to the gnotobiotic mice (Koren et al., 2012; Vijay-Kumar et al., 2010). Such changes can be beneficial during late pregnancy because they promote fetal growth and facilitate the maternal energy storage necessary for lactation (Koren et al., 2012). In the neonate, the T3 GI microbiota is needed to extract additional energy and promote gluconeogenesis during the first few days of life when the volume of colostrum produced is low (Konstantinov, van der Woude, & Peppelenbosch, 2013).
In the study mentioned above that examined the GI microbial composition from T1 to T3, researchers reported that the composition of the GI microbiota during T3 is preserved into the early postpartum period in a subset of women (Koren et al., 2012). In another study that examined the relative abundance of GI microbiota in seven healthy pregnant women from T3 to 1-month postpartum, researchers showed that the relative abundance of the GI microbial community is relatively stable during that period (Jost, Lacroix, Braegger, & Chassard, 2014). Retention of a pro-inflammatory GI microbial composition into the postpartum period is likely related to the additional energy needs of the lactating mother since it promotes increased energy extraction (Koren et al., 2012).
Diversity
The diversity of the GI microbiota also changes significantly from T1 to T3. During T1, the richness (number of different species) and evenness (proportions of the species across the community) are similar to those of nonpregnant healthy adults. The α diversity (within individuals) is high, and the β diversity (between individuals) is low. The inverse is true, however, as pregnancy progresses; and by T3, the β diversity across women is significantly higher (Koren et al., 2012). However, this finding was not replicated in another study that showed that the α- and β diversities as well as the week-to-week stability of the microbial community were unaffected by the progression of pregnancy (DiGiulio et al., 2015). The differences in the results may be attributable to the use of rectal swabs in the second study (DiGiulio et al., 2015) as a means to estimate the GI microbiota as opposed to the stool samples used in the prior study (Koren et al., 2012). Rectal swabs are a convenient method of sampling, as they can be collected at will; however, the correlation of the microbial profiles between rectal swabs and stool samples is generally low. In addition, rectal swabs have only been used successfully to isolate specific GI microbial species, and their use increases the potential for contamination by skin bacteria (Arvelo et al., 2013; Budding et al., 2014; Duran et al., 2013; Kabayiza et al., 2013).
Factors Associated With the Structure and Function of the GI Microbiota
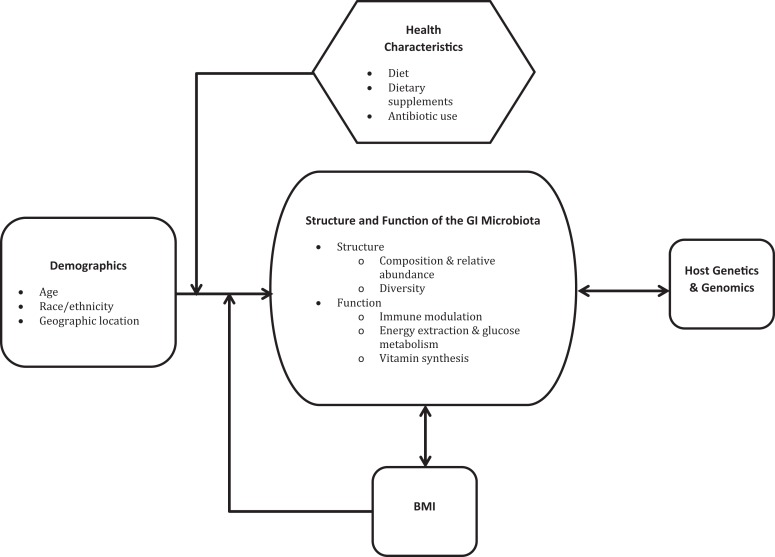
The factors associated with the structure and/or function of the GI microbiota include demographic and health characteristics such as chronological age, race, ethnicity, geographic location, dietary intake, use of supplements, antibiotic use, and body mass index (BMI) as well as host genetics (G. D. Wu et al., 2011; Figure 2). To this point, studies in pregnant women have been limited to exploring associations between the GI microbiota and diet, dietary supplements, antibiotics use, and BMI (Collado et al., 2008; Gohir et al., 2015). However, some of the results from studies conducted in the general population are applicable to pregnant women.
Figure 2.
Demographic characteristics, body mass index (BMI), and host genetics and genomics are associated with the structure of function of the gastrointestinal (GI) microbiota. The association between demographic characteristics and the GI microbiota is likely mediated by health characteristics and BMI. For simplicity’s sake, this figure does not depict the complex associations among these factors that are not directly related to the GI microbiota. BMI = body mass index.
Chronological Age
Chronological age of the host is a significant factor associated with the structure of the GI microbiota (Koenig et al., 2011; Yatsunenko et al., 2012). The α diversity increases with age and is significantly higher in adults than in children (Yatsunenko et al., 2012). On the other hand, β diversity decreases with age, with the composition of the GI microbiota being fairly similar among adults, whereas among children it is very different. In the elderly population, however, the structure of the GI microbiota once again becomes highly variable among individuals (Claesson et al., 2011). Differences in the structure of the microbial communities across ages may be attributable to changes in other factors dependent on aging such as diet. For example, the microbial structure changes when breastfeeding is stopped or when infants are weaned from exclusive milk feeding to solid food (Koenig et al., 2011). After weaning, the infant GI microbiota gradually changes until it closely resembles that of adults (Koenig et al., 2011; C. Palmer et al., 2007; Vaishampayan et al., 2010).
Race, Ethnicity, and Geographic Location
Race and ethnicity are strongly associated with significant differences in the microbial structure (Aagaard et al., 2013; HMP Consortium, 2012; Zhou et al., 2007). Early studies showed that, in the U.S. population, strains of microbiota are distributed differentially between individuals of Asian, Black, Mexican, Puerto Rican, and White races and ethnicities (HMP Consortium, 2012). Similarly, the composition of microbial communities differs across populations based on geographic location. For example, adults in Columbia, the United States, Europe, and Asia have distinctly divergent GI microbial compositions (Escobar, Klotz, Valdes, & Agudelo, 2014). In Columbia, researchers found that the GI microbiota of adults was composed primarily of Firmicutes and Bacteroidetes, with widely varying relative proportions. In the U.S., European, and Asian populations, the GI microbiota varied equally but had lower proportions of Firmicutes to Bacteroidetes. In another study, the structure and functional representation of the GI microbiota differed significantly between adults and breastfed infants living in the United States and those living in Venezuela and Malawi (Yatsunenko et al., 2012). Adults in non-U.S. populations were found to have higher abundance of the genus Prevotella compared to those living in the United States. That study also found that the β diversity was lower in U.S. adults compared to those in other geographic locations. In addition, the microbial genes encoding enzymes that degrade amino acids and simple sugars were higher in adults in the United States, while genes encoding enzymes that break down starch were higher in the other populations. Babies born in Venezuela and Malawi had a higher abundance of microbes endowed with the genetic capacities needed to synthesize specific vitamins and metabolize carbohydrates compared to babies born in the United States (Yatsunenko et al., 2012).
These findings suggest that differences in the structure and function of the GI microbiota are shaped by external factors, such nutrient availability, that are unique to the demands of living in a particular social or geographic location. A major factor contributing to differences in nutrient availability is, of course, food availability in different geographical regions. For example, the GI microbial structure in African children, who typically have a diet high in polysaccharides and low in fat and protein, is characterized by a higher abundance of Bacteroidetes, which break down polysaccharides. Conversely, the abundance of Bacteroidetes found in European children, who typically consume a Western diet high in fat and calories, is significantly lower. In European children, the GI microbial structure was, instead, characterized by an overabundance of Firmicutes and Enterobacteriaceae (Proteobacteria phylum), which are typically associated with obesity (De Filippo et al., 2010). These observations demonstrate that the structure of GI microbial communities is tailored to provide a mechanism to digest the available nutrients in specific regions, implying that diet might potentially be used as a means to modulate the structure and function of the GI microbiota to support host physiology.
Dietary Intake
Individual dietary patterns shape the structure of the GI microbiota (Chen, He, & Huang, 2014; Saha & Reimer, 2014; Trompette et al., 2014; Vieira, Pagovich, & Kriegel, 2014; W. K. Wu et al., 2015). For example, the gut composition in people who eat meat is characterized by a higher abundance of the Roseburia–Eubacterium rectale group within Clostridium cluster XIV in the Firmicutes phylum (Kabeerdoss, Devi, Mary, & Ramakrishna, 2012). Alternatively, vegans or vegetarians have reduced densities of Escherichia coli (Proteobacteria phylum), the Enterobacteriaceae family (Proteobacteria phylum), Bacteroides spp. (Bacteroidetes phylum), and Bifidobacterium spp. (Actinobacteria phylum; Zimmer et al., 2012). In addition, the ratio of protein to carbohydrates in diet influences the composition of the asaccharolytic species that is typically found in the distal colon and harvests energy from protein (Neis, Dejong, & Rensen, 2015; Scott, Gratz, Sheridan, Flint, & Duncan, 2013). Diets high in fat are associated with reductions in Bifidobacteria (Actinobacteria phylum), which produce vitamins and inhibit pathogens (Brinkworth, Noakes, Clifton, & Bird, 2009; LeBlanc et al., 2013), while high-fiber diets are associated with a lower ratio of Firmicutes to Bacteroidetes, which decrease risk of obesity (Saha & Reimer, 2014). These findings suggest that the microbes comprising the GI microbiota require particular types of food to survive, and differences in structure across individuals are likely associated with unique characteristics of the diet.
Long-term dietary patterns are also associated with two distinct enterotypes that are grouped in relation to metabolic function, one rich in Bacteroides genus and the other rich in Prevotella. The Bacteroides-rich enterotype has been associated with animal protein and saturated fats, and the Prevotella-rich enterotype has been found with diets high in carbohydrates and simple sugars (G. D. Wu et al., 2011). Although both the Bacteroides and Prevotella genera belong to the Bacteroidetes phylum, the two enterotypes are associated with different long-term dietary patterns. This observation suggests that functional ability is not associated with membership in a particular phylum; rather, even within a single phylum dysbiosis can lead to adverse changes in the function. Once again, these findings illustrate the therapeutic potential of diet for targeting particular metabolic functions.
This idea is further supported by studies showing the influence of dietary interventions on changes in the structure and function of the GI microbiota. In mice, switching from a diet low in fat and high in plant polysaccharide to a diet high in fat and sugars resembling the “Western” diet increased the density of members of the Firmicutes phylum and decreased the proportions of members of the Bacteroidetes phylum within 20 hr. Functionally, the host’s genetic capacities associated with carbohydrate metabolism were upregulated in the mice fed the Western diet (Turnbaugh, Ridaura, et al., 2009).
Similarly, in humans fed either a high-fat or high-fiber diet over a 10-day period, the GI microbiota underwent changes within 24 hr (G. D. Wu et al., 2011). The compositional changes that occurred also reflected differences in function that suggest that species within each phylum have a specific purpose or role in host metabolism and gene expression. Likewise, changes that occur when infants transition from milk to solid food provide further evidence that components within the diet stimulate shifts in the GI microbiota (Koenig et al., 2011; C. Palmer et al., 2007; Vaishampayan et al., 2010). This phenomenon is likely due to an upsurge of microbes that are genetically endowed to utilize a particular energy source found in specific types of food.
Investigators have reported similar findings in pregnant mice, where fat content in the maternal diet prior to and during pregnancy modulated the structure of the GI microbiota (Gohir et al., 2015). The differences between the two groups of mice, one fed a high- and one a low-fat diet, were significant for microbes in 26 genera, the majority belonging to the Firmicutes phylum, particularly the Clostridiales order. The changes were functionally related with metabolism of energy and vitamin production regardless of diet grouping, with the shift in the microbial community over time in the mice fed a high-fat diet favoring lipid and glucose metabolism compared to the group fed a low-fat diet. These findings illustrate the critical importance of diet during pregnancy in relation to optimal metabolic processing.
Dietary Supplements
Dietary supplements confer benefits that can distinctly influence health and well-being. Prebiotics, for example, are nondigestible polysaccharides such as fiber, plant cell wall polysaccharides, resistant starch, and oligosaccharides that serve as nutrition for microbes in the human body (Lenoir-Wijnkoop et al., 2007). Previous research found that adding fermentable (oligofructose) or nonfermentable dietary fiber (microcrystalline cellulose) to a normal diet strengthened the integrity of the intestinal barrier and reduced inflammation associated with intestinal permeability (Cani et al., 2009). This function is thought to be mediated by specific microbes, particularly the Bifidobacteria and Lactobacillus species that increase with prebiotic supplementation. Although the exact mechanisms are unclear, the study authors proposed that Bifidobacteria and Lactobacillus protect the intestinal epithelial barrier by not degrading the glycoproteins in mucus, thereby leaving the mucosal barrier intact, which in turn prevents translocation of pathogens or pathogenic substances to the host compartment.
Alternatively, probiotic supplementation is the introduction of live microbes, such as the Bifidobacteria and Lactobacillus species, typically found in the gut of healthy humans (Walker, 2008). Probiotic supplementation during the perinatal period has distinct health benefits including reduced rates of preterm birth and gestational diabetes (Barrett, Dekker Nitert, Conwell, & Callaway, 2014; Gjessing et al., 2011; Yang et al., 2015). The causal mechanism the authors have proposed to explain the relationship between probiotic supplementation and preterm birth is optimization of the structures of the vaginal microbiota that corrects dysbiotic states thought to initiate labor (Krauss-Silva, Almada-Horta, Alves, Camacho, & Moreira, 2014; Reid, 2012; Vitali et al., 2012; Yang et al., 2015). The strains of microbes within probiotic supplements vary widely, but they frequently contain species whose functions reestablish a symbiotic state within microbial communities and subsequently between the microbes and the host (Macklaim et al., 2013; Vitali et al., 2012). In relation to the GI microbiota and its potential to initiate labor, there is a growing body of evidence demonstrating the crucial role that it plays in moderating systemic inflammatory pathways that are key in the onset of labor (Gracie et al., 2011; MacIntyre, Sykes, Teoh, & Bennett, 2012; Romero, Dey, & Fisher, 2014; Romero et al., 2006).
In gestational diabetes mellitus, several well-controlled randomized trials reported improved blood glucose metabolism, insulin regulation, and homeostatic model assessment-insulin resistance levels in pregnant women receiving probiotics supplementation (Asemi et al., 2013; Laitinen, Poussa, Isolauri, & the Nutrition, Allergy, Mucosal Immunology and Intestinal Microbiota Group, 2009; Luoto, Laitinen, Nermes, & Isolauri, 2010). There have been, however, inconsistencies in this finding across studies, which may be due in part to differences in the strains of microbes included in the probiotic supplement as well as the point in pregnancy at which they are introduced (Lindsay et al., 2014, 2015). These discrepancies strongly suggest the need for additional research to clarify the optimal type and timing of probiotic supplementation during pregnancy.
Probiotic supplementation during pregnancy also has implications for the health of the child. In mothers with a history of environmental allergies, researchers found that supplementation with a combination of Lactobacillus rhamnosus LPR and Bifidobacterium longum BL999 or Lactobacillus paracasei ST11 and B. longum BL999, started 2 months before the expected date of birth and continued until 2 months postpartum reduced the risk of the infant developing eczema during the first 2 years of life (Rautava, Kainonen, Salminen, & Isolauri, 2012). However, while probiotic supplementation was effective in reducing the prevalence of eczema, it did not prevent asthma or wheezing in the infants (Azad et al., 2013). This finding is surprising because eczema and asthma share similar physiologic triggers (Saunes et al., 2012), suggesting that additional research is needed to more clearly delineate the mechanisms involved and identify specific strains of microbiota that target those functions. These findings are important, as they suggest an alternative for treating common disease processes without the risk of iatrogenic complications associated with pharmacotherapeutic agents (Floch, 2014).
Conversely, certain supplements compromise commensal microbes and act to benefit pathogens. For example, iron (Fe) is essential for metabolic processes associated with microbial replication, such as DNA synthesis and defense against reactive oxygen species. Supplementation with Fe benefits many anaerobic microbes because they have a very high affinity for it (Andrews, Robinson, & Rodríguez-Quiñones, 2003; Schaible & Kaufmann, 2004). In infants and children, Fe supplementation for 3 and 6 months, respectively, was associated with decreased Lactobacillus and increased Bacteroides spp. and Enterobacteriaceae (e.g., Escherichia, Shigella) in both age groups (Krebs et al., 2013; Zimmermann et al., 2010). More recently, researchers in a randomized controlled trial reported that Fe supplementation in infants was associated with increased levels of inflammatory markers and pathogenic microbes belonging to the Escherichia, Shigella, and Clostridium genera (Jaeggi et al., 2015). Similarly, in a study that used a rat model, Fe supplementation following a diet deficient in Fe was associated with an increased abundance of species belonging to Bacteroidetes phylum and Clostridium genera as well as higher level of metabolites such as butyrate (Dostal, Lacroix, et al., 2014). Conversely, Fe deficiency was associated with decreased Bacteroides species (Bacteroidetes phylum), Roseburia spp., and Eubacterium rectale within the Clostridiales order as well as lower levels of butyrate and propionate (Dostal et al., 2012). In a third trial among three groups of children—iron-deficient children who received placebo treatment, iron-deficient children who received Fe supplementation, and iron-sufficient children—researchers found no association between Fe supplementation and changes in microbes and metabolites (Dostal, Baumgartner, et al., 2014). Despite the single study that showed no effect, the preponderance of evidence suggests that Fe supplementation may adversely alter the abundance of specific species in the gut, thus changing their functional activities.
Of particular interest is the effect of Fe supplementation during pregnancy since it is recommended for all women to reduce the risk of iron-deficiency anemia and low infant birth weight (Haider et al., 2013). In one recent study, however, researchers found that maternal iron supplementation increased the risk of celiac disease in the child (Størdal, Haugen, Brantsæter, Lundin, & Stene, 2014). Celiac disease is an autoimmune enteropathy triggered by gluten ingestion that manifests symptoms that include diarrhea. Studies of this disease process have shown that it is associated with a decreased abundance of beneficial microbes such as Lactobacillus and Bifidobacteria (de Sousa Moraes, Grzeskowiak, de Sales Teixeira, & do Carmo Gouveia Peluzio, 2014). More specifically, researchers found that in individuals diagnosed with celiac disease, the composition of GI microbiota was characterized by a decreased abundance of members of the Bacteroidetes and Firmicutes phyla (e.g., Clostridiaceae, Eubacteriaceae) and increased levels of members of the Proteobacteria phylum (e.g., Enterobacteriaceae; Wacklin et al., 2014). These findings suggest that health-care providers should carefully consider the risks and benefits of Fe supplementation because it may be implicated in iatrogenic complications as a response to changes in the GI microbial composition.
Antibiotics
Pharmacotherapeutic agents, in particular antibiotics, significantly alter the structure of the GI microbiota (Dethlefsen & Relman, 2011). Antibiotics are designed to eradicate microbes, and they cannot selectively target a particular pathogen. Therefore, they eliminate commensal microbes in equal numbers, disrupting the microbial composition. Although the magnitude of the changes differs across individuals, the type of antibiotic and the duration of the treatment are associated with decreases in the number (richness) and proportionality of the strains (evenness) within the GI microbiota (Fouhy et al., 2012). In general, these changes occur within 3–7 days of initiation of the antibiotic, and the microbial structure only partially returns to its prior state 1–4 weeks after the end of the treatment, although in some cases the effects can persist for years (Dethlefsen, Huse, Sogin, & Relman, 2008; Dethlefsen & Relman, 2011; Ferrer, dos Santos, Ott, & Moya, 2013; Fouhy et al., 2012; Jakobsson et al., 2010; Jernberg, Löfmark, Edlund, & Jansson, 2007). In relation to particular antibiotics, by day 3–4 of a 5-day course of Ciprofloxacin, adults exhibited decreased diversity of the GI microbiota. The decreases in diversity stabilized and the microbiota partially returned to the normalized state, 1 week after the end of the treatment (Dethlefsen & Relman, 2011). A 7-day course of clindamycin caused profound alterations in the GI microbial structure that persisted for 2 years (Jernberg et al., 2007). Changes in microbial communities associated with treatment with antibiotics include an increased abundance of Clostridium difficile (C. difficile), which is associated with diarrhea (Barbato et al., 2014; Bartlett, 2002). These changes are a result of disruptions in the normal gut flora that reduce protection from pathogen overgrowth, thereby permitting organisms like C. difficile to bloom (Britton & Young, 2012). Although narrow-spectrum antibiotics can be used to target C. difficile, use of those antibiotics can also promote microbial resistance (Jernberg et al., 2007; A. C. Palmer & Kishony, 2013).
Host colonization by antibiotic-resistant microbes such as C. difficile has forced scientists to develop alternative treatment modalities. In the case of C. difficile, specifically, enteric fecal microbial transplants from healthy individuals (usually those who live in close proximity to the infected patient) have proven to be remarkably effective, especially in cases of recurrent C. difficile colonization (Kelly et al., 2015). In many cases, fecal microbial transplantation successfully treated C. difficile-related symptoms including frequent watery stools and protected against future recurrent infections (Jorup-Rönström et al., 2012; Kassam, Hundal, Marshall, & Lee, 2012). Although the precise mechanism is not clear, fecal microbial transplants appear to restore a “normal” flora. Despite these findings, the U.S. Food and Drug Administration has yet to approve fecal microbial transplantation. Clinicians must obtain regulatory authorization to use this treatment modality, which promotes standardized implementation but also hinders its use. Larger controlled clinical trials and regulatory support are needed to facilitate the broader use of fecal microbial transplantation.
Maternal use of antibiotics during pregnancy is associated with adverse health outcomes in the child, including increased risks of asthma, celiac disease, and obesity (Lapin et al., 2015; Mårild, Ludvigsson, Sanz, & Ludvigsson, 2014; Mueller et al., 2015; Stensballe, Simonsen, Jensen, Bønnelykke, & Bisgaard, 2013). The demonstrated effect that antibiotics have on the GI microbiota has particular relevance in neonates, as parenteral ampicillin and gentamicin are commonly given within the first 2 days of life to treat sepsis and pneumonia. In one study, this practice was associated with a higher proportion of microbes in the Proteobacteria phylum and a reduced microbial diversity in neonates 1-month posttreatment that persisted for at least another month. It was also associated with lower proportions of microbes in the Actinobacteria phylum, Bifidobacteria, and Lactobacillus genera at 1-month posttreatment that had partially recovered by the second month (Fouhy et al., 2012). Use of antibiotics in early life is also associated with a higher prevalence of necrotizing enterocolitis (NEC), an inflammatory intestinal disorder that can lead to mortality in extremely low birth weight babies (Cotton, 2010). Researchers found that infants who developed NEC had received antibiotics for a longer duration than those who did not develop the disorder. In babies with NEC, the gut microbiota is characterized by a low diversity and an increased abundance of Gammaproteobacteria (Proteobacteria phylum; Y. Wang et al., 2009). Antibiotic use alters the composition of the GI microbiota to one that initiates inflammation in the mucosal lining, leading to development of NEC. Furthermore, exposure to antibiotics during infancy is strongly associated with the onset of allergic diseases such as childhood asthma and eczema (Marra et al., 2006; Tsakok, McKeever, Yeo, & Flohr, 2013). The authors hypothesize that this relationship is mediated by perturbations in the GI microbiota resulting from antibiotic use (Noverr & Huffnagle, 2005).
Host Genetics
There is limited research about the direct effect that host genetics may have on the GI microbiota or vice versa. There is a small body of research conducted in the general population that shows that microbes do interact with the host genetic makeup, thereby influencing gene transcription and expression. For example, when microbes isolated in the stool of lean and obese donors were transplanted into germ-free mice, the mice expressed the same patterns of weight gain as their donors, and microbial genes expressed in mice were discordant between the two groups (Ridaura et al., 2013).
Conversely, the GI microbiota is also influenced by host genetics (Benson et al., 2010; Goodrich et al., 2014; Kovacs et al., 2011; McKnite et al., 2012; Murphy et al., 2015; Spor, Koren, & Ley, 2011). In a recent study comparing the GI microbiota in 416 mono- and dizygotic twins, the findings demonstrated that the structure of the GI microbiota is more similar in monozygotic than dizygotic twin pairs. The finding suggests that similarities in the host genotype define the types and abundance of the microbes within the GI communities (Goodrich et al., 2014). This conclusion is supported by another finding from the same study showing that certain types of microbes, such as those in the Christensenellaceae family (Clostridiales order), are heritable and that the heritability is driven by host genetics (Goodrich et al., 2014). The effect of host genetics on microbiota has also been demonstrated with the genes involved with innate and adaptive immunity, such as those encoding toll-like receptors (TLRs; Vijay-Kumar et al., 2010) and myeloid differentiation primary response 88 (Myd88; Wen et al., 2008). In a mouse-model study, mice without TLR-5 had a lower abundance of microbes from the Bacteroidetes phylum and Lachnospiraceae family compared to mice that encoded TLR-5. Similarly, mice without Myd88 had a higher abundance of microbes from the Porphyromonadaceae, Lactobacillaceae, and Rikenellaceae families compared to mice with Myd88. These studies support the linkage between the expression of genes involved in maintaining host immunity and the GI microbiota.
Similarly, the composition of the GI microbiota is associated with gene expression related to host metabolism. Specifically, the gene that codes for leptin, a hormone derived from adipocytes that regulates appetite, energy expenditure, and metabolism and acts as a cytokine by influencing immune cells, is associated with a lower abundance and altered composition of fecal bacteria (Waldram et al., 2009). These findings support the symbiotic and highly interactive nature of the relationship between the microbes in the gut and the host genotype that defines the phenotypic expression and in turn differentiates the structure of the microbiota.
BMI
The structure and function of the GI microbiota are also associated with BMI and obesity (Dominianni et al., 2015; Ley et al., 2005; Turnbaugh, Hamady, et al., 2009; Turnbaugh, Ridaura, et al., 2009). In both humans and mice, increased BMI was associated with reduced diversity and proportions of particular types of GI microbiota at the phylum level. In mice, obesity correlates with decreased diversity and a significantly lower abundance of Bacteroidetes and increased levels of Firmicutes (Ley et al., 2005). In addition, when the microbiota from obese donors were transplanted into lean germ-free mice, the phenotypes of the donors were expressed and the recipients became obese (Ridaura et al., 2013; Turnbaugh, Ridaura, et al., 2009). In obese compared to lean individuals, researchers observed the same changes in the microbial diversity and density of Bacteroidetes, though they found no significant difference in the levels of Firmicutes, and there was a greater proportion of Actinobacteria (Turnbaugh, Hamady, et al., 2009). These studies effectively demonstrate the functional role that the GI microbiota plays in obesity.
Functionally, microbial genes in obese individuals are associated with the metabolism of carbohydrates, lipids, and amino acids (Turnbaugh, Hamady, et al., 2009). Short-chain fatty acids (SCFA: acetate, propionate, and butyrate), significant metabolites of GI microbiota, are known to have a profound effect on glucose and lipid metabolic processes such as hepatic gluconeogenesis and hepatic and cholesterol synthesis and therefore could play a significant role in regulating the process of fat storage and weight gain (den Besten et al., 2013).
The associations among weight gain, the GI microbiota, and BMI have implications for pregnancy because women are physiologically predisposed to weight gain during pregnancy due to increased insulin resistance, which is likely the mechanism to ensure adequate energy for the fetus (Hadden & McLaughlin, 2009). GI microbes influence physiologic functions that alter homeostasis and can adversely affect health outcomes. For instance, the composition of the GI microbiota is linked to the production of hormones and enzymes, such as estrogen and insulin, that facilitate weight gain during pregnancy (Bäckhed et al., 2004; Barbour et al., 2007; Fuhrman et al., 2014; Picciano, 2003; Santacruz et al., 2010). Moreover, the GI microbiota also regulates appetite by increasing circulating leptin, which is reduced in the obese population (Bäckhed et al., 2004; Clément et al., 1998; Ravussin et al., 2012). Furthermore, probiotic supplementation is associated with reduced body weight and fat mass in obese women (Sanchez et al., 2014).
Prepregnancy weight and BMI correlate with differences in the density of Bacteroides (Bacteroidetes phylum) and Staphylococcus and Clostridium (Firmicutes phylum; Collado et al., 2008). Overall, concentrations of Bacteroides are significantly higher in overweight women. In addition, during the first trimester, Staphylococcus is positively correlated with increased BMI, while the microbes in the Clostridium group tended to correlate negatively with BMI (Collado et al., 2008). During the third trimester, the GI microbial composition functionally manifests physiologic responses associated with metabolic syndrome (Koren et al., 2012). Although research in this area is still at an early stage, the findings provide support for the close relationship between BMI and the GI microbiota. As much as it is important to consider the role of BMI in shaping the composition of the GI microbiota, it is also essential to account for the role that the GI microbiota plays in host energy metabolism (Kimura et al., 2013; Neuman, Debelius, Knight, & Koren, 2015; Tremaroli & Bäckhed, 2012), not only in the general population but also in pregnant women. This body of knowledge lends credibility to the claim that the GI microbial composition prior to and during pregnancy is key to understanding and managing energy metabolism.
Discussion
The GI microbiota has coevolved with mammals to adapt to the unique ecosystems in the host. Microbes harvest energy from nondigestible polysaccharides and produce substrates and metabolites including vitamins and SCFAs that are involved with the initiation of metabolic and immunologic pathways essential to host physiologic function. A growing body of knowledge about the structure and function of the GI microbiota in health and disease suggests that it plays an augmented role during pregnancy to meet the metabolic needs associated with the developing fetus.
Previous studies have identified significant reductions in the relative abundance and phylogenetic diversity of the GI microbiota associated with pregnancy as well as the role of the microbiota in maintaining physiologic homeostasis. The evidence linking alterations in the GI microbiota with changes in physiologic pathways during pregnancy demonstrates the crucial need for increased understanding about the temporal dynamics of the microbiota during pregnancy. This need is more pressing because many of the extant studies have important limitations, and a number of studies examined changes in the GI microbiota composition during the first and the third trimesters only (Collado et al., 2008; Koren et al., 2012). The maintenance of maternal homeostasis is essential during the second trimester, however, to allow the body to build physiologic reserves that promote fetal growth and development (Bystron, Blakemore, & Rakic, 2008; Zhang et al., 2013). In another large trial, investigators used rectal swabs as opposed to stool samples to estimate the structure of the GI microbiota (DiGiulio et al., 2015). Yet as we discussed above, anal swabs are not a proxy for stool samples and are capable of measuring only a limited number of microbes (Arvelo et al., 2013; Budding et al., 2014; Duran et al., 2013; Kabayiza et al., 2013). Finally, the majority of the studies have been conducted in animal models (Gohir et al., 2015). Although animal models contribute to understanding phenomena, they do not mirror human conditions in pregnancy and therefore have limited application (Bolker, 2012; McCarthy, Kingdom, Kenny, & Walsh, 2011).
To address these limitations, future research should examine the GI microbiota during the second trimester of pregnancy using stool samples. New information about the temporal dynamics of the GI microbiota during pregnancy could provide scientists with the ability to understand key functions of the GI microbiota. In addition, knowledge about the baseline structure will demonstrate how deviations are associated with adverse pregnancy outcomes. That knowledge is also necessary to capitalize on the capacity of the GI microbiota to maintain physiologic homeostasis, which has significant application in the clinical management of pregnancy. Strengthening the research community’s understanding about the factors associated with the GI microbiota has the potential to lead to an enhanced ability to monitor and manipulate that ecosystem, which, in turn, holds the promise of possible therapeutic strategies designed to enhance the beneficial effects of the GI microbiota and, subsequently, larger health outcomes for both mothers and infants.
Conclusion
As the growing evidence suggests, health and disease processes in humans cannot be adequately explained without accounting for the symbiotic relationship between host genetics and the microbiome. The human microbiome is a potentially powerful therapeutic target that can be directly modulated with low-risk interventions such as supplemental prebiotics, probiotics, or even fecal transplants. Biopsychosocial factors, including diet, that influence the microbial structure can serve as secondary therapeutic targets.
Because nurses are on the front line of care, it is important that they are equipped with a basic knowledge about the structure and function of the GI microbiota, the factors that influence it, and the implications it has for health and disease. Caregivers can employ modifiable factors such as diet and prebiotics to restore disruptions in the microbial community and help reduce adverse pregnancy outcomes. Clinicians should also consider the influence of routine medical treatments such as the use of iron supplementation or antibiotics on the structure of the GI microbiota in pregnant women. Over the longer term, research findings will provide nurses with evidence-based practices aimed at improving health outcomes in the mother and child by optimizing the structure and function of the GI microbiota.
Glossary
Composition of microbiome: types of microbiota that reside in the gut.
Diversity of the microbiome: diversity has two dimensions, namely, richness, which is the number of different strains, and evenness, which is the proportionality of the strains within a sample. Diversity within a sample or a subject is termed α diversity, whereas diversity between samples or subjects is called β diversity.
Microbiome: a collective term for all the microorganisms living in and on the human body.
Relative abundance: Proportions of each type of microbe among all.
Perturbations: disruptions in the structure of the microbial community.
Phylotypes: a classification of groups of microbes with similar genetic sequences that may reflect evolutionary relationships.
Structure of the microbiome: the composition, relative abundance, and diversity of the microbial community.
Taxonomic ranks: relative levels of groups of organisms, for example, the microbes. Under the kingdom categorizing the microbiota, phylum is the highest rank in terms of the taxonomic hierarchy followed by class, order, family, genus, and species.
Footnotes
Author Contribution: Seon-Yoon Chung contributed to conception, design, data acquisition, data analysis, and interpretation; drafted the manuscript; gave final approval; and agrees to be held accountable for all aspects of work, ensuring integrity and accuracy. Jacques Ravel contributed to conception, critically revised the manuscript, gave final approval, and agrees to be held accountable for all aspects of work, ensuring integrity and accuracy. Mary Regan contributed to conception, design, data acquisition, data analysis, and interpretation; critically revised the manuscript; gave final approval; and agrees to be held accountable for all aspects of work, ensuring integrity and accuracy
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was funded by the National Institute of Nursing Research (1R01NR014826).
References
- Aagaard K., Ma J., Antony K. M., Ganu R., Petrosino J., Versalovic J. (2014). The placenta harbors a unique microbiome. Science Translational Medicine, 6, 237ra65 doi:10.1126/scitranslmed.3008599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aagaard K., Petrosino J., Keitel W., Watson M., Katancik J., Garcia N.…Versalovic J. (2013). The Human Microbiome Project strategy for comprehensive sampling of the human microbiome and why it matters. FASEB Journal, 27, 1012–1022. doi:10.1096/fj.12-220806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajslev T. A., Andersen C. S., Gamborg M., Sørensen T. I. A., Jess T. (2011). Childhood overweight after establishment of the gut microbiota: The role of delivery mode, pre-pregnancy weight and early administration of antibiotics. International Journal of Obesity, 35, 522–529. doi:10.1038/ijo.2011.27 [DOI] [PubMed] [Google Scholar]
- Andrews S. C., Robinson A. K., Rodríguez-Quiñones F. (2003). Bacterial iron homeostasis. FEMS Microbiology Reviews, 27, 215–237. doi:10.1016/S0168-6445(03)00055-X [DOI] [PubMed] [Google Scholar]
- Ardissone A. N., De La Cruz D. M., Davis-Richardson A. G., Rechcigl K. T., Li N., Drew J. C.…Neu J. (2014). Meconium microbiome analysis identifies bacteria correlated with premature birth. PLoS One, 9, 1–8. doi:10.1371/journal.pone.0090784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam M., Raes J., Pelletier E., Paslier D. L., Yamada T., Mende D. R.…Bork P. (2011). Enterotypes of the human gut microbiome. Nature, 473, 174–180. doi:10.1038/nature09944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvelo W., Hall A. J., Estevez A., Lopez B., Gregoricus N., Vinjé J.…Lindblade K. A. (2013). Diagnostic performance of rectal swab versus bulk stool specimens for the detection of rotavirus and norovirus: Implications for outbreak investigations. Journal of Clinical Virology, 58, 678–682. doi:10.1016/j.jcv.2013.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asemi Z., Samimi M., Tabassi Z., Naghibi Rad M., Rahimi Foroushani A., Khorammian H., Esmaillzadeh A. (2013). Effect of daily consumption of probiotic yoghurt on insulin resistance in pregnant women: A randomized controlled trial. European Journal of Clinical Nutrition, 67, 71–74. doi:10.1038/ejcn.2012.189 [DOI] [PubMed] [Google Scholar]
- Azad M. B., Coneys J. G., Kozyrskyj A. L., Field C. J., Ramsey C. D., Becker A. B.…Zarychanski R. (2013). Probiotic supplementation during pregnancy or infancy for the prevention of asthma and wheeze: Systematic review and meta-analysis. British Medical Journal, 347, f6471 doi:10.1136/bmj.f6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F., Ding H., Wang T., Hooper L. V., Koh G. Y., Nagy A.…Gordon J. I. (2004). The gut microbiota as an environmental factor that regulates fat storage. Proceedings of the National Academy of Sciences, 101, 15718–15723. doi:10.1073/pnas.0407076101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbato V. A., Hasty R., Fonarov I., Valdes P., Jabbar Q., Klein J., Reed J. (2014). Clostridium difficile colitis: Epidemiology, diagnosis, treatment and modalities. Osteopathic Family Physician, 6, 13–18. [Google Scholar]
- Barbour L. A., McCurdy C. E., Hernandez T. L., Kirwan J. P., Catalano P. M., Friedman J. E. (2007). Cellular mechanisms for insulin resistance in normal pregnancy and gestational diabetes. Diabetes Care, 30, S112–S119. doi:10.2337/dc07-s202 [DOI] [PubMed] [Google Scholar]
- Barrett H. L., Dekker Nitert M., Conwell L. S., Callaway L. K. (2014). Probiotics for preventing gestational diabetes. Cochrane Database of Systematic Reviews, 2, 1–31. doi:10.1002/14651858.CD009951.pub2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett J. G. (2002). Antibiotic-associated diarrhea. New England Journal of Medicine, 346, 334–339. doi:10.1056/NEJMcp011603 [DOI] [PubMed] [Google Scholar]
- Belizário J. E., Napolitano M. (2015). Human microbiomes and their roles in dysbiosis, common diseases, and novel therapeutic approaches. Frontiers in Microbiology, 6, 1050 doi:10.3389/fmicb.2015.01050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson A. K., Kelly S. A., Legge R., Ma F., Low S. J., Kim J.…Pomp D. (2010). Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proceedings of the National Academy of Sciences, 107, 18933–18938. doi:10.1073/pnas.1007028107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolker J. (2012). Model organisms: There’s more to life than rats and flies. Nature, 491, 31–33. doi:10.1038/491031a [DOI] [PubMed] [Google Scholar]
- Borre Y. E., O’Keeffe G. W., Clarke G., Stanton C., Dinan T. G., Cryan J. F. (2014). Microbiota and neurodevelopmental windows: Implications for brain disorders. Trends in Molecular Medicine, 20, 509–518. doi:10.1016/j.molmed.2014.05.002 [DOI] [PubMed] [Google Scholar]
- Brinkworth G. D., Noakes M., Clifton P. M., Bird A. R. (2009). Comparative effects of very low-carbohydrate, high-fat and high-carbohydrate, low-fat weight-loss diets on bowel habit and faecal short-chain fatty acids and bacterial populations. British Journal of Nutrition, 101, 1493–1502. doi:10.1017/S0007114508094658 [DOI] [PubMed] [Google Scholar]
- Britton R. A., Young V. B. (2012). Interaction between the intestinal microbiota and host in Clostridium difficile colonization resistance. Trends in Microbiology, 20, 313–319. doi:10.1016/j.tim.2012.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budding A. E., Grasman M. E., Eck A., Bogaards J. A., Vandenbroucke-Grauls C. M. J. E., van Bodegraven A. A., Savelkoul P. H. M. (2014). Rectal swabs for analysis of the intestinal microbiota. PLoS One, 9, e101344 doi:10.1371/journal.pone.0101344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystron I., Blakemore C., Rakic P. (2008). Development of the human cerebral cortex: Boulder Committee revisited. Nature Reviews Neuroscience, 9, 110–122. doi:10.1038/nrn2252 [DOI] [PubMed] [Google Scholar]
- Cani P. D., Possemiers S., Van de Wiele T., Guiot Y., Everard A., Rottier O.…Delzenne N. M. (2009). Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut, 58, 1091–1103. doi:10.1136/gut.2008.165886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K.…Knight R. (2010). QIIME allows analysis of high- throughput community sequencing data. Nature Methods, 7, 335–336. doi:10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., He X., Huang J. (2014). Diet effects in gut microbiome and obesity. Journal of Food Science, 79, 442–451. doi:10.1111/1750-3841.12397 [DOI] [PubMed] [Google Scholar]
- Cho I., Blaser M. J. (2012). The human microbiome: At the interface of health and disease. Nature Reviews Genetics, 13, 260–270. doi:10.1038/nrg3182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson M. J., Cusack S., O’Sullivan O., Greene-Diniz R., de Weerd H., Flannery E.…O’Toole P. W. (2011). Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proceedings of the National Academy of Sciences, 108, 4586–4591. doi:10.1073/pnas.1000097107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clément K., Vaisse C., Lahlou N., Cabrol S., Pelloux V., Cassuto D.…Guy-Grand B. (1998). A mutation in the human leptin receptor gene causes obesity and pituitary dysfunction. Nature, 392, 398–401. doi:10.1038/32911 [DOI] [PubMed] [Google Scholar]
- Cole J. R., Wang Q., Cardenas E., Fish J., Chai B., Farris R. J.…Tiedje J. M. (2009). The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Research, 37, D141–D145. doi:10.1093/nar/gkn879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M. C., Isolauri E., Laitinen K., Salminen S. (2008). Distinct composition of gut microbiota during pregnancy in overweight and normal-weight women. American Journal of Clinical Nutrition, 88, 894–899. [DOI] [PubMed] [Google Scholar]
- Cotton C. M. (2010). Early, prolonged use of postnatal antibiotics increased the risk of necrotising enterocolitis. Archives of Disease in Childhood. Education and Practice Edition, 95, 94 doi:10.1136/adc.2010.187732 [DOI] [PubMed] [Google Scholar]
- de Agüero M. G., Ganal-Vonarburg S. C., Fuhrer T., Rupp S., Uchimura Y., Li H.…Macpherson A. J. (2016). The maternal microbiota drives early postnatal innate immune development. Science, 351, 1296–1302. doi:10.1017/CBO9781107415324.004 [DOI] [PubMed] [Google Scholar]
- De Filippo C., Cavalieri D., Di Paola M., Ramazzotti M., Poullet J. B., Massart S.…Lionetti P. (2010). Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proceedings of the National Academy of Sciences, 107, 14691–14696. doi:10.1073/pnas.1005963107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Besten G., van Eunen K., Groen A. K., Venema K., Reijngoud D.-J., Bakker B. M. (2013). The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. Journal of Lipid Research, 54, 2325–2340. doi:10.1194/jlr.R036012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa Moraes L. F., Grzeskowiak L. M., de Sales Teixeira T. F., & do Carmo Gouveia Peluzio M. (2014). Intestinal microbiota and probiotics in celiac disease. Clinical Microbiology Reviews, 27, 482–489. doi:10.1128/CMR.00106-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L., Huse S. M., Sogin M. L., Relman D. A. (2008). The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16 S rRNA sequencing. PLoS Biology, 6, e280 doi:10.1371/journal.pgen.1000255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L., McFall-Ngai M., Relman D. A. (2007). An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature, 449, 811–818. doi:10.1038/nature06245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L., Relman D. A. (2011). Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proceedings of the National Academy of Sciences, 108, 4554–4561. doi:10.1073/pnas.1000087107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiulio D. B., Callahan B. J., McMurdie P. J., Costello E. K., Lyell D. J., Robaczewska A.…Relman D. A. (2015). Temporal and spatial variation of the human microbiota during pregnancy. Proceedings of the National Academy of Sciences, 112, 11060–11065. doi:10.1073/pnas.1502875112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bello M. G., Costello E. K., Contreras M., Magris M., Hidalgo G., Fierer N., Knight R. (2010). Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proceedings of the National Academy of Sciences, 107, 11971–11975. doi:10.1073/pnas.1002601107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominianni C., Sinha R., Goedert J. J., Pei Z., Yang L., Hayes R. B., Ahn J. (2015). Sex, body mass index, and dietary fiber intake influence the human gut microbiome. PLoS One, 10, e0124599 doi:10.1371/journal.pone.0124599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostal A., Baumgartner J., Riesen N., Chassard C., Smuts C. M., Zimmermann M. B., Lacroix C. (2014). Effects of iron supplementation on dominant bacterial groups in the gut, faecal SCFA and gut inflammation: A randomised, placebo-controlled intervention trial in South African children. British Journal of Nutrition, 112, 547–556. doi:10.1017/S0007114514001160 [DOI] [PubMed] [Google Scholar]
- Dostal A., Chassard C., Hilty F. M., Zimmermann M. B., Jaeggi T., Rossi S., Lacroix C. (2012). Iron depletion and repletion with ferrous sulfate or electrolytic iron modifies the composition and metabolic activity of the gut microbiota in rats. Journal of Nutrition, 142, 271–277. doi:10.3945/jn.111.148643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostal A., Lacroix C., Pham V. T., Zimmermann M. B., Del’homme C., Bernalier-Donadille A., Chassard C. (2014). Iron supplementation promotes gut microbiota metabolic activity but not colitis markers in human gut microbiota-associated rats. British Journal of Nutrition, 111, 2135–2145. doi:10.1017/S000711451400021X [DOI] [PubMed] [Google Scholar]
- Duran C., Nato F., Dartevelle S., Thi Phuong L. N., Taneja N., Ungeheuer M. N.…Germani Y. (2013). Rapid diagnosis of diarrhea caused by Shigella sonnei using dipsticks; comparison of rectal swabs, direct stool and stool culture. PLoS One, 8, e80267 doi:10.1371/journal.pone.0080267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar J. S., Klotz B., Valdes B. E., Agudelo G. M. (2014). The gut microbiota of Colombians differs from that of Americans, Europeans and Asians. BMC Microbiology, 14, 311 doi:10.1186/s12866-014-0311-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadrosh D. W., Ma B., Gajer P., Sengamalay N., Ott S., Brotman R. M., Ravel J. (2014). An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome, 2, 6 doi:10.1186/2049-2618-2-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer M., Martins dos Santos V. A., Ott S. J., Moya A. (2013). Gut microbiota disturbance during antibiotic therapy: A multi-omic approach. Gut Microbes, 62, 1591–1601. doi:10.1136/gutjnl-2012-303184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floch M. (2014). Recommendations for probiotic use in humans—A 2014 update. Pharmaceuticals, 7, 999–1007. doi:10.3390/ph7100999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J. A., McVey Neufeld K. A. (2013). Gut-brain axis: How the microbiome influences anxiety and depression. Trends in Neurosciences, 36, 305–312. doi:10.1016/j.tins.2013.01.005 [DOI] [PubMed] [Google Scholar]
- Fouhy F., Guinane C. M., Hussey S., Wall R., Ryan C. A., Dempsey E. M.…Cotter P. D. (2012). High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrobial Agents and Chemotherapy, 56, 5811–5820. doi:10.1128/AAC.00789-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D. N., St Amand A. L., Feldman R. A., Boedeker E. C., Harpaz N., Pace N. R. (2007). Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proceedings of the National Academy of Sciences, 104, 13780–13785. doi:10.1073/pnas.0706625104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman B. J., Feigelson H. S., Flores R., Gail M. H., Xu X., Ravel J., Goedert J. J. (2014). Associations of the fecal microbiome with urinary estrogens and estrogen metabolites in postmenopausal women. Journal of Clinical Endocrinology and Metabolism, 99, 4632–4640. doi:10.1210/jc.2014-2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda S., Toh H., Hase K., Oshima K., Nakanishi Y., Yoshimura K.…Ohno H. (2011). Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature, 469, 543–547. doi:10.1038/nature09646 [DOI] [PubMed] [Google Scholar]
- Gevers D., Knight R., Petrosino J. F., Huang K., McGuire A. L., Birren B. W.…Huttenhower C. (2012). The Human Microbiome Project: A community resource for the healthy human microbiome. PLoS Biology, 10, e1001377 doi:10.1371/journal.pbio.1001377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D., Pop M., Schloss P. D., Huttenhower C. (2012). Bioinformatics for the Human Microbiome Project. PLoS Computational Biology, 8, e1002779 doi:10.1371/journal.pcbi.1002779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjessing K., Sengpiel V., Meltzer H. M., Myhre R., Brantsæter A. L., Myking S.…Jacobsson B. (2011). Intake of probiotic food and risk of spontaneous preterm delivery. American Journal of Clinical Nutrition, 93, 151–157. doi:10.3945/ajcn.110.004085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohir W., Whelan F. J., Surette M. G., Moore C., Jonathan D. (2015). Pregnancy-related changes in the maternal gut microbiota are dependent upon the mother’s periconceptional diet. Gut Microbes, 6, 310–320. doi:10.1080/19490976.2015.1086056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J. K., Waters J. L., Poole A. C., Sutter J. L., Koren O., Blekhman R.…Ley R. E. (2014). Human genetics shape the gut microbiome. Cell, 159, 789–799. doi:10.1016/j.cell.2014.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracie S., Pennell C., Ekman-Ordeberg G., Lye S., McManaman J., Williams S.…Gravett M. (2011). An integrated systems biology approach to the study of preterm birth using “-omic” technology—A guideline for research. BMC Pregnancy and Childbirth, 11, 71 doi:10.1186/1471-2393-11-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadden D. R., McLaughlin C. (2009). Normal and abnormal maternal metabolism during pregnancy. Seminars in Fetal and Neonatal Medicine, 14, 66–71. doi:10.1016/j.siny.2008.09.004 [DOI] [PubMed] [Google Scholar]
- Haider B. A., Olofin I., Wang M., Spiegelman D., Ezzati M., Fawzi W. W. (2013). Anaemia, prenatal iron use, and risk of adverse pregnancy outcomes: Systematic review and meta-analysis. British Medical Journal, 346, f3443 doi:10.1136/bmj.f3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope M. E., Hold G. L., Kain R., El-Omar E. M. (2005). Sporadic colorectal cancer—Role of the commensal microbiota. FEMS Microbiology Letters, 244, 1–7. doi:10.1016/j.femsle.2005.01.029 [DOI] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium. (2012). Structure, function and diversity of the healthy human microbiome. Nature, 486, 207–214. doi:10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeggi T., Kortman G. A. M., Moretti D., Chassard C., Holding P., Dostal A.…Zimmermann M. B. (2015). Iron fortification adversely affects the gut microbiome, increases pathogen abundance and induces intestinal inflammation in Kenyan infants. Gut, 64, 731–742. doi:10.1136/gutjnl-2014-307720 [DOI] [PubMed] [Google Scholar]
- Jakobsson H. E., Jernberg C., Andersson A. F., Sjölund-Karlsson M., Jansson J. K., Engstrand L. (2010). Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One, 5, e9836 doi:10.1371/journal.pone.0009836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernberg C., Löfmark S., Edlund C., Jansson J. K. (2007). Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME Journal, 1, 56–66. doi:10.1038/ismej.2007.3 [DOI] [PubMed] [Google Scholar]
- Jiménez E., Fernández L., Marín M. L., Martín R., Odriozola J. M., Nueno-Palop C.…Rodríguez J. M. (2005). Isolation of commensal bacteria from umbilical cord blood of healthy neonates born by cesarean section. Current Microbiology, 51, 270–274. doi:10.1007/s00284-005-0020-3 [DOI] [PubMed] [Google Scholar]
- Jiménez E., Marín M. L., Martín R., Odriozola J. M., Olivares M., Xaus J.…Rodríguez J. M. (2008). Is meconium from healthy newborns actually sterile? Research in Microbiology, 159, 187–193. doi:10.1016/j.resmic.2007.12.007 [DOI] [PubMed] [Google Scholar]
- Johansson M. A., Sjögren Y. M., Persson J. O., Nilsson C., Sverremark-Ekström E. (2011). Early colonization with a group of Lactobacilli decreases the risk for allergy at five years of age despite allergic heredity. PLoS One, 6, e23031 doi:10.1371/journal.pone.0023031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorup-Rönström C., Håkanson A., Sandell S., Edvinsson O., Midtvedt T., Persson A.-K., Norin E. (2012). Fecal transplant against relapsing Clostridium difficile-associated diarrhea in 32 patients. Scandinavian Journal of Gastroenterology, 47, 548–552. doi:10.3109/00365521.2012.672587 [DOI] [PubMed] [Google Scholar]
- Jost T., Lacroix C., Braegger C., Chassard C. (2014). Stability of maternal GIMB during late and early lactation. Current Microbiology, 68, 419–427. doi:10.1007/s00284-013-0491-6 [DOI] [PubMed] [Google Scholar]
- Jost T., Lacroix C., Braegger C., Chassard C. (2015). Impact of human milk bacteria and oligosaccharides on neonatal gut microbiota establishment and gut health. Nutrition Reviews, 73, 426–437. doi:10.1093/nutrit/nuu016 [DOI] [PubMed] [Google Scholar]
- Kabayiza J.-C., Andersson M. E., Welinder-Olsson C., Bergström T., Muhirwa G., Lindh M. (2013). Comparison of rectal swabs and faeces for real-time PCR detection of enteric agents in Rwandan children with gastroenteritis. BMC Infectious Diseases, 13, 447 doi:10.1186/1471-2334-13-447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeerdoss J., Devi R. S., Mary R. R., Ramakrishna B. S. (2012). Faecal microbiota composition in vegetarians: Comparison with omnivores in a cohort of young women in southern India. British Journal of Nutrition, 108, 953–957. doi:10.1017/S0007114511006362 [DOI] [PubMed] [Google Scholar]
- Kalliomäki M., Collad M. C, Salmine S, Isolauri E. (2008). Early differences in fecal microbiota composition in children may predict overweight. American Journal of Clinical Nutrition, 87, 534–538. [DOI] [PubMed] [Google Scholar]
- Kassam Z., Hundal R., Marshall J. K., Lee C. H. (2012). Fecal transplant via retention enema for refractory or recurrent Clostridium difficile infection. Archives of Internal Medicine, 172, 191–193. doi:10.1001/archinte.172.2.191 [DOI] [PubMed] [Google Scholar]
- Kassinen A., Krogius-Kurikka L., Mäkivuokko H., Rinttilä T., Paulin L., Corander J.…Palva A. (2007). The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology, 133, 24–33. doi:10.1053/j.gastro.2007.04.005 [DOI] [PubMed] [Google Scholar]
- Kelly C. R., Kahn S., Kashyap P., Laine L., Rubin D., Atreja A.…Wu G. (2015). Update on fecal microbiota transplantation 2015: Indications, methodologies, mechanisms, and outlook. Gastroenterology, 149, 223–237. doi:10.1053/j.gastro.2015.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura I., Ozawa K., Inoue D., Imamura T., Kimura K., Maeda T.…Tsujimoto G. (2013). The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nature Communications, 4, 1829 doi:10.1038/ncomms2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig J. E., Spor A., Scalfone N., Fricker A. D., Stombaugh J., Knight R.…Ley R. E. (2011). Succession of microbial consortia in the developing infant gut microbiome. Proceedings of the National Academy of Sciences, 108, 4578–4585. doi:10.1073/pnas.1000081107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinov S. R., van der Woude C. J., Peppelenbosch M. P. (2013). Do pregnancy-related changes in the microbiome stimulate innate immunity? Trends in Molecular Medicine, 19, 454–459. doi:10.1016/j.molmed.2013.06.002 [DOI] [PubMed] [Google Scholar]
- Koren O., Goodrich J. K., Cullender T. C., Spor A., Laitinen K., Bäckhed H. K.…Ley R. E. (2012). Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell, 150, 470–480. doi:10.1016/j.cell.2012.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs A., Ben-Jacob N., Tayem H., Halperin E., Iraqi F. A., Gophna U. (2011). Genotype is a stronger determinant than sex of the mouse gut microbiota. Microbial Ecology, 61, 423–428. doi:10.1007/s00248-0 [DOI] [PubMed] [Google Scholar]
- Kozich J. J., Westcott S. L., Baxter N. T., Highlander S. K., Schloss P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Applied and Environmental Microbiology, 79, 5112–5120. doi:10.1128/AEM.01043-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss-Silva L., Almada-Horta A., Alves M. B., Camacho K. G., Moreira M. E. L. (2014). Basic vaginal pH, bacterial vaginosis and aerobic vaginitis: Prevalence in early pregnancy and risk of spontaneous preterm delivery, a prospective study in a low socioeconomic and multiethnic South American population. BMC Pregnancy and Childbirth, 14, 107 doi:10.1186/1471-2393-14-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs N. F., Sherlock L. G., Westcott J., Culbertson D., Hambidge K. M., Feazel L. M.…Frank D. N. (2013). Effects of different complementary feeding regimens on iron status and enteric microbiota in breastfed infants. Journal of Pediatrics, 163, 416–423. e4 doi:10.1016/j.jpeds.2013.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczynski J., Stombaugh J., Anton Walters W., González J., Caporaso G., Knight R. (2012). Using QIIME to analyze 16S rRNA gene sequences from Microbial Communities. Current Protocols in Bioinformatics, 1E-5, 1–30. doi:10.1002/9780471729259.mc01e05s27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laitinen K., Poussa T., Isolauri E. , & the Nutrition, Allergy, Mucosal Immunology and Intestinal Microbiota Group. (2009). Probiotics and dietary counselling contribute to glucose regulation during and after pregnancy: A randomised controlled trial. British Journal of Nutrition, 101, 1679–1687. doi:10.1017/S0007114508111461 [DOI] [PubMed] [Google Scholar]
- Lapin B., Piorkowski J., Ownby D., Freels S., Chavez N., Hernandez E.…Persky V. (2015). Relationship between prenatal antibiotic use and asthma in at-risk children. Annals of Allergy, Asthma and Immunology, 114, 203–207. doi:10.1016/j.anai.2014.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen N., Vogensen F. K., van den Berg F. W., Nielsen D. S., Andreasen A. S., Pedersen B. K.…Jakobsen M. (2010). Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One, 5, e9085 doi:10.1371/journal.pone.0009085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasken R. S., McLean J. S. (2014). Recent advances in genomic DNA sequencing of microbial species from single cells. Nature Reviews Genetics, 15, 577–584. doi:10.1038/nrg3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc J. G., Milani C., de Giori G. S., Sesma F., van Sinderen D., Ventura M. (2013). Bacteria as vitamin suppliers to their host: A gut microbiota perspective. Current Opinion in Biotechnology, 24, 160–168. doi:10.1016/j.copbio.2012.08.005 [DOI] [PubMed] [Google Scholar]
- Lenoir-Wijnkoop I., Sanders M. E., Cabana M. D., Caglar E., Corthier G., Rayes N.…Wolvers D. A. W. (2007). Probiotic and prebiotic influence beyond the intestinal tract. Nutrition Reviews, 65, 469–489. doi:10.1301/nr.2007.nov.469 [DOI] [PubMed] [Google Scholar]
- Ley R. E., Bäckhed F., Turnbaugh P., Lozupone C. A., Knight R. D., Gordon J. I. (2005). Obesity alters gut microbial ecology. Proceedings of the National Academy of Sciences, 102, 11070–11075. doi:10.1073/pnas.0504978102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R. E., Turnbaugh P. J., Klein S., Gordon J. I. (2006). Human gut microbes associated with obesity. Nature, 444, 1022–1023. doi:10.1038/nature4441021a [DOI] [PubMed] [Google Scholar]
- Lindsay K. L., Brennan L., Kennelly M. A., Maguire O. C., Smith T., Curran S.…McAuliffe F. M. (2015). Impact of probiotics in women with gestational diabetes mellitus on metabolic health: A randomized controlled trial. American Journal of Obstetrics & Gynecology, 212, 496.e1–496.e11. [DOI] [PubMed] [Google Scholar]
- Lindsay K. L., Kennelly M., Smith T., Maguire O., Shanahan F., Brennan L., McAuliffe F. (2014). Probiotics in obese pregnancy do not reduce maternal fasting glucose: A double-blind, placebo-controlled, randomized trial (Probiotics in Pregnancy Study). American Journal of Clinical Nutrition, 99, 1432–1439. doi:10.3945/ajcn.113.079723. [DOI] [PubMed] [Google Scholar]
- Louis P., Hold G. L., Flint H. J. (2014). The gut microbiota, bacterial metabolites and colorectal cancer. Nature Reviews Microbiology, 12, 661–672. doi:10.1038/nrmicro3344 [DOI] [PubMed] [Google Scholar]
- Luoto R., Laitinen K., Nermes M., Isolauri E. (2010). Impact of maternal probiotic-supplemented dietary counselling on pregnancy outcome and prenatal and postnatal growth: A double-blind, placebo-controlled study. British Journal of Nutrition, 103, 1792–1799. doi:10.1017/S0007114509993898 [DOI] [PubMed] [Google Scholar]
- MacIntyre D. A., Sykes L., Teoh T. G., Bennett P. R. (2012). Prevention of preterm labour via the modulation of inflammatory pathways. Journal of Maternal-Fetal and Neonatal Medicine, 25, 17–20. doi:10.3109/14767058.2012.666114 [DOI] [PubMed] [Google Scholar]
- Macklaim J. M., Fernandes A. D., Di Bella J. M., Hammond J.-A., Reid G., Gloor G. B. (2013). Comparative meta-RNA-seq of the vaginal microbiota and differential expression by Lactobacillus iners in health and dysbiosis. Microbiome, 1, 12 doi:10.1186/2049-2618-1-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mårild K., Ludvigsson J., Sanz Y., Ludvigsson J. F. (2014). Antibiotic exposure in pregnancy and risk of coeliac disease in offspring: A cohort study. BMC Gastroenterology, 14, 75 doi:10.1186/1471-230X-14-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra F., Lynd L., Coombes M., Richardson K., Legal M., FitzGerald J. M., Marra C. A. (2006). Does antibiotic exposure during infancy lead to development of asthma? A systematic review and metaanalysis. Chest, 129, 610–618. doi:10.1378/chest.129.3.610 [DOI] [PubMed] [Google Scholar]
- Martín R., Heilig H. G. H. J., Zoetendal E. G., Jiménez E., Fernández L., Smidt H., Rodríguez J. M. (2007). Cultivation-independent assessment of the bacterial diversity of breast milk among healthy women. Research in Microbiology, 158, 31–37. doi:10.1016/j.resmic.2006.11.004 [DOI] [PubMed] [Google Scholar]
- McCarthy F. P., Kingdom J. C., Kenny L. C., Walsh S. K. (2011). Animal models of preeclampsia; Uses and limitations. Placenta, 32, 413–419. doi:10.1016/j.placenta.2011.03.010 [DOI] [PubMed] [Google Scholar]
- McKnite A. M., Perez-Munoz M. E., Lu L., Williams E. G., Brewer S., Andreux P. A.…Ciobanu D. C. (2012). Murine gut microbiota is defined by host genetics and modulates variation of metabolic traits. PLoS One, 7, e39191 doi:10.1371/journal.pone.0039191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methé B. A., Nelson K. E., Pop M., Creasy H. H., Giglio M. G., Huttenhower C.…White O. (2012). A framework for human microbiome research. Nature, 486, 215–221. doi:10.1038/nature11209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller N. T., Whyatt R., Hoepner L., Oberfield S., Dominguez-Bello M. G., Widen E. M.…Rundle A. (2015). Prenatal exposure to antibiotics, cesarean section and risk of childhood obesity. International Journal of Obesity, 39, 665–670. doi:10.1038/ijo.2014.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K., O’ Shea C. A., Ryan C. A., Dempsey E. M., O’ Toole P. W., Stanton C., Ross R. P. (2015). The gut microbiota composition in dichorionic triplet sets suggests a role for host genetic factors. PLoS One, 10, e0122561 doi:10.1371/journal.pone.0122561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institutes of Health Human Microbiome Project Working Group. (2009). The NIH Human Microbiome Project. Genome Research, 19, 2317–2323. doi:10.1101/gr.096651.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neis E., Dejong C., Rensen S. (2015). The role of microbial amino acid metabolism in host metabolism. Nutrients, 7, 2930–2946. doi:10.3390/nu7042930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman H., Debelius J. W., Knight R., Koren O. (2015). Microbial endocrinology: The interplay between the microbiota and the endocrine system. FEMS Microbiology Reviews, 39, 509–521. doi:10.1093/femsre/fuu010 [DOI] [PubMed] [Google Scholar]
- Noverr M. C., Huffnagle G. B. (2005). The “microflora hypothesis” of allergic diseases. Clinical Experimental Allergy, 35, 1511–1520. doi:10.1111/j.1365-2222.2005.02379.x [DOI] [PubMed] [Google Scholar]
- O’Hara A. M., Shanahan F. (2006). The gut flora as a forgotten organ. EMBO Reports, 7, 688–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer A. C., Kishony R. (2013). Understanding, predicting and manipulating the genotypic evolution of antibiotic resistance. Nature Reviews. Genetics, 14, 243–248. doi:10.1038/nrg3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C., Bik E. M., DiGiulio D. B., Relman D. A., Brown P. O. (2007). Development of the human infant intestinal microbiota. PLoS Biology, 5, e177 doi:10.1371/journal.pbio.0050177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penders J., Thijs C., Vink C., Stelma F. F., Snijders B., Kummeling I.…Stobberingh E. E. (2006). Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics, 118, 511–521. doi:10.1542/peds.2005-2824 [DOI] [PubMed] [Google Scholar]
- Pendse M., Hooper L. V. (2016). Mum’s microbes boost baby’s immunity. Nature, 533, 42–43. [DOI] [PubMed] [Google Scholar]
- Peterson D. A., Frank D. N., Pace N. R., Gordon J. I. (2008). Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host and Microbe, 3, 417–427. doi:10.1016/j.chom.2008.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picciano M. F. (2003). Pregnancy and lactation: Physiological adjustments, nutritional requirements and the role of dietary supplements. Journal of Nutrition, 133, 1997S–2002S. [DOI] [PubMed] [Google Scholar]
- Qin J., Li R., Raes J., Arumugam M., Burgdorf K. S., Manichanh C.…Wang J. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature, 464, 59–65. doi:10.1038/nature08821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Li Y., Cai Z., Li S. S., Zhu J., Zhang F.…Wang J. J. (2012). A metagenome-wide association study of gut microbiota in Type 2 diabetes. Nature, 490, 55–60. doi:10.1038/nature11450 [DOI] [PubMed] [Google Scholar]
- Rautava S., Collado M. C., Salminen S., Isolauri E. (2012). Probiotics modulate host-microbe interaction in the placenta and fetal gut: A randomized, double-blind, placebo-controlled trial. Neonatology, 102, 178–184. doi:10.1159/000339182 [DOI] [PubMed] [Google Scholar]
- Rautava S., Kainonen E., Salminen S., Isolauri E. (2012). Maternal probiotic supplementation during pregnancy and breast-feeding reduces the risk of eczema in the infant. Journal of Allergy and Clinical Immunology, 130, 1355–1360. doi:10.1016/j.jaci.2012.09.003 [DOI] [PubMed] [Google Scholar]
- Rautava S., Kalliomäki M., Isolauri E. (2002). Probiotics during pregnancy and breast-feeding might confer immunomodulatory protection against atopic disease in the infant. Journal of Allergy and Clinical Immunology, 109, 119–121. doi:10.1067/mai.2002.120273 [DOI] [PubMed] [Google Scholar]
- Rautava S., Luoto R., Salminen S., Isolauri E. (2012). Microbial contact during pregnancy, intestinal colonization and human disease. Nature Reviews Gastroenterology & Hepatology, 9, 565–576. doi:10.1038/nrgastro.2012.144 [DOI] [PubMed] [Google Scholar]
- Ravussin Y., Koren O., Spor A., LeDuc C., Gutman R., Stombaugh J.…Leibel R. L. (2012). Responses of gut microbiota to diet composition and weight loss in lean and obese mice. Obesity, 20, 738–747. doi:10.1038/oby.2011.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid G. (2012). Probiotic and prebiotic applications for vaginal health. Journal of AOAC International, 95, 31–34. doi:http://doi.org/http://dx.doi.org/10.5740/jaoacint [DOI] [PubMed] [Google Scholar]
- Reid G., Charbonneau D., Erb J., Kochanowski B., Beuerman D., Poehner R., Bruce A. W. (2003). Oral use of Lactobacillus rhamnosus GR-1 and L. fermentum RC-14 significantly alters vaginal flora: Randomized, placebo-controlled trial in 64 healthy women. FEMS Immunology and Medical Microbiology, 35, 131–134. doi:10.1016/S0928-8244(02)00465-0 [DOI] [PubMed] [Google Scholar]
- Renz H., Brandtzaeg P., Hornef M. (2012). The impact of perinatal immune development on mucosal homeostasis and chronic inflammation. Nature Reviews Immunology, 12, 9–23. doi:10.1038/nri3112 [DOI] [PubMed] [Google Scholar]
- Ridaura V. K., Belkaid Y. (2015). Gut microbiota: The link to your second brain. Cell, 161, 193–194. doi:10.1016/j.cell.2015.03.033 [DOI] [PubMed] [Google Scholar]
- Ridaura V. K., Faith J. J., Rey F. E., Cheng J., Alexis E., Kau A. L.…Gordon J. I. (2013). Cultured gut microbiota from twins discordant for obesity modulate adiposity and metabolic phenotypes in mice. Science, 341, 1–22. doi:10.1126/science.1241214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rintala H. (2011). Actinobacteria in indoor environments: Exposures and respiratory health effects. Frontiers in Bioscience, 3, 1273–1284. doi:10.2741/225 [DOI] [PubMed] [Google Scholar]
- Romero R., Dey S. K., Fisher S. J. (2014). Preterm labor: One syndrome, many causes. Science, 345, 760–765. doi:10.1126/science.1251816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero R., Espinoza J., Goncalves L. F., Kusanovic J. P., Friel L. A., Nien J. K. (2006). Inflammation in preterm and term labour and delivery. Seminars in Fetal & Neonatal Medicine, 11, 317–326. doi:10.1016/j.siny.2006.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha D. C., Reimer R. A. (2014). Long-term intake of a high prebiotic fiber diet but not high protein reduces metabolic risk after a high fat challenge and uniquely alters gut microbiota and hepatic gene expression. Nutrition Research, 34, 789–796. doi:10.1016/j.nutres.2014.08.004 [DOI] [PubMed] [Google Scholar]
- Sanchez M., Darimont C., Drapeau V., Emady-Azar S., Lepage M., Rezzonico E.…Tremblay A. (2014). Effect of Lactobacillus rhamnosus CGMCC1.3724 supplementation on weight loss and maintenance in obese men and women. British Journal of Nutrition, 111, 1507–1519. doi:10.1017/S0007114513003875 [DOI] [PubMed] [Google Scholar]
- Santacruz A., Collado M. C., García-Valdés L., Segura M. T., Martín-Lagos J. A., Anjos T.…Sanz Y. (2010). Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. British Journal of Nutrition, 104, 83–92. doi:10.1017/S0007114510000176 [DOI] [PubMed] [Google Scholar]
- Sartor R. B. (2008). Microbial influences in inflammatory bowel diseases. Gastroenterology, 134, 577–594. doi:10.1053/j.gastro.2007.11.059 [DOI] [PubMed] [Google Scholar]
- Satokari R., Grönroos T., Laitinen K., Salminen S., Isolauri E. (2009). Bifidobacterium and Lactobacillus DNA in the human placenta. Letters in Applied Microbiology, 48, 8–12. doi:10.1111/j.1472-765X.2008.02475.x [DOI] [PubMed] [Google Scholar]
- Saunes M., Oien T., Dotterud C. K., Romundstad P. R., Storro O., Holmen T. L., Johnsen R. (2012). Early eczema and the risk of childhood asthma: A prospective, population-based study. BMC Pediatrics, 12, 168 doi:10.1186/1471-2431-12-168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage D. C. (1977). Microbial ecology of the gastrointestinal tract. Annual Review of Microbiology, 31, 107–133. [DOI] [PubMed] [Google Scholar]
- Schaible U. E., Kaufmann S. H. (2004). Iron and microbial infection. Nature Reviews Microbiology, 2, 946–953. doi:10.1038/nrmicro1116 [DOI] [PubMed] [Google Scholar]
- Schwabe R., Jobin C. (2013). The microbiome and cancer. Nature Reviews Cancer, 13, 800–812. doi:10.1038/nrc3610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K. P., Gratz S. W., Sheridan P. O., Flint H. J., Duncan S. H. (2013). The influence of diet on the gut microbiota. Pharmacological Research, 69, 52–60. doi:10.1016/j.phrs.2012.10.020 [DOI] [PubMed] [Google Scholar]
- Sela D. A., Chapman J., Adeuya A., Kim J. H., Chen F., Whitehead T. R.…Mills D. A. (2008). The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proceedings of the National Academy of Sciences, 105, 18964–18969. doi:10.1073/pnas.0809584105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellitto M., Bai G., Serena G., Fricke W. F., Sturgeon C., Gajer P.…Fasano A. (2012). Proof of concept of microbiome-metabolome analysis and delayed gluten exposure on celiac disease autoimmunity in genetically at-risk infants. PLoS One, 7, e33387 doi:10.1371/journal.pone.0033387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer F., Bäckhed F. (2013). The gut microbiota—Masters of host development and physiology. Nature Reviews Microbiology, 11, 227–238. doi:10.1038/nrmicro2974 [DOI] [PubMed] [Google Scholar]
- Spor A., Koren O., Ley R. (2011). Unravelling the effects of the environment and host genotype on the gut microbiome. Nature Reviews Microbiology, 9, 279–290. doi:10.1038/nrmicro2540 [DOI] [PubMed] [Google Scholar]
- Steel J. H., Malatos S., Kennea N., Edwards a. D., Miles L., Duggan P.…Sullivan M. H. F. (2005). Bacteria and inflammatory cells in fetal membranes do not always cause preterm labor. Pediatric Research, 57, 404–411. doi:10.1203/01.PDR.0000153869.96337.90 [DOI] [PubMed] [Google Scholar]
- Stensballe L. G., Simonsen J., Jensen S. M., Bønnelykke K., Bisgaard H. (2013). Use of antibiotics during pregnancy increases the risk of asthma in early childhood. Journal of Pediatrics, 162, 832.e3–838.e3. doi:10.1016/j.jpeds.2012.09.049 [DOI] [PubMed] [Google Scholar]
- Størdal K., Haugen M., Brantsæter A. L., Lundin K. E. A., Stene L. C. (2014). Maternal iron supplement intake during pregnancy and risk of celiac disease in children. Clinical Gastroenterology and Hepatology, 12, 624.e2–631.e2. doi:10.1016/j.cgh.2013.09.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strus M., Chmielarczyk A., Kochan P., Adamski P., Chełmicki Z., Chełmicki A.…Heczko P. B. (2012). Studies on the effects of probiotic Lactobacillus mixture given orally on vaginal and rectal colonization and on parameters of vaginal health in women with intermediate vaginal flora. European Journal of Obstetrics Gynecology and Reproductive Biology, 163, 210–215. doi:10.1016/j.ejogrb.2012.05.001 [DOI] [PubMed] [Google Scholar]
- Tremaroli V., Bäckhed F. (2012). Functional interactions between the gut microbiota and host metabolism. Nature, 489, 242–249. doi:10.1038/nature11552 [DOI] [PubMed] [Google Scholar]
- Trompette A., Gollwitzer E. S., Yadava K., Sichelstiel A. K., Sprenger N., Ngom-Bru C.…Marsland B. J. (2014). Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nature Medicine, 20, 159–168. doi:10.1038/nm.3444 [DOI] [PubMed] [Google Scholar]
- Tsakok T., McKeever T. M., Yeo L., Flohr C. (2013). Does early life exposure to antibiotics increase the risk of eczema? A systematic review. British Journal of Dermatology, 169, 983–991. doi:10.1111/bjd.12476 [DOI] [PubMed] [Google Scholar]
- Turnbaugh P. J., Hamady M., Yatsunenko T., Cantarel B. L., Duncan A., Ley R. E.…Gordon J. I. (2009). A core gut microbiome in obese and lean twins. Nature, 457, 480–484. doi:10.1038/nature07540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J., Ley R. E., Hamady M., Fraser-liggett C., Knight R., Gordon J. I. (2007). The Human Microbiome Project: Exploring the microbial part of ourselves in a changing world. Nature, 449, 804–810. doi:10.1038/nature06244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J., Ley R. E., Mahowald M. A., Magrini V., Mardis E. R., Gordon J. I. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature, 444, 1027–1031. 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- Turnbaugh P. J., Ridaura V. K., Faith J. J., Rey F. E., Knight R., Gordon J. I. (2009). The effect of diet on the human gut microbiome: A metagenomic analysis in humanized gnotobiotic mice. Science Translational Medicine, 1, 6ra14 doi:10.1126/scitranslmed.3000322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishampayan P. A., Kuehl J. V., Froula J. L., Morgan J. L., Ochman H., Francino M. P. (2010). Comparative metagenomics and population dynamics of the gut microbiota in mother and infant. Genome Biology and Evolution, 2, 53–66. doi:10.1093/gbe/evp057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira S. M., Pagovich O. E., Kriegel M. A. (2014). Diet, microbiota and autoimmune diseases. Lupus, 23, 518–526. doi:10.1177/0961203313501401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar M., Aitken J. D., Carvalho F. A., Cullender T. C., Mwangi S., Srinivasan S.…Gewirtz A. T. (2010). Metabolic syndrome and altered gut microbiota in mice lacking toll-like receptor 5. Science, 328, 228–232. doi:10.1126/science.1179721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali B., Cruciani F., Baldassarre M. E., Capursi T., Spisni E., Valerii M. C.…Brigidi P. (2012). Dietary supplementation with probiotics during late pregnancy: Outcome on vaginal microbiota and cytokine secretion. BMC Microbiology, 12, 236 doi:10.1186/1471-2180-12-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze A., Van Nood E., Holleman F., Salojärvi J., Kootte R. S., Bartelsman J. F. W. M.…Nieuwdorp M. (2012). Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology, 143, 913.e7–916.e7. doi:10.1053/j.gastro.2012.06.031 [DOI] [PubMed] [Google Scholar]
- Vujic G., Knez A. J., Stefanovic V. D., Vrbanovic V. K. (2013). Efficacy of orally applied probiotic capsules for bacterial vaginosis and other vaginal infections: A double-blind, randomized, placebo-controlled study. European Journal of Obstetrics Gynecology and Reproductive Biology, 168, 75–79. doi:10.1016/j.ejogrb.2012.12.031 [DOI] [PubMed] [Google Scholar]
- Wacklin P., Laurikka P., Lindfors K., Collin P., Salmi T., Lähdeaho M.-L.…Kaukinen K. (2014). Altered duodenal microbiota composition in celiac disease patients suffering from persistent symptoms on a long-term gluten-free diet. American Journal of Gastroenterology, 109, 1933–1941. doi:10.1038/ajg.2014.355 [DOI] [PubMed] [Google Scholar]
- Waldram a., Holmes E., Wang Y., Rantalainen M., Wilson I. D., Tuohy K. M.…Nicholson J. K. (2009). Top-down systems biology modeling of host metabotype-microbiome associations in obese rodents. Journal of Proteome Research, 8, 2361–2375. doi:10.1021/pr8009885 [DOI] [PubMed] [Google Scholar]
- Walker W. A. (2008). Mechanisms of action of probiotics. Clinical Infectious Diseases, 46, 87–91. doi:10.1086/523335 [DOI] [PubMed] [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology, 73, 5261–5267. doi:10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Hoenig J. D., Malin K. J., Qamar S., Petrof E. O., Sun J.…Claud E. C. (2009). 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME Journal, 3, 944–954. doi:10.1038/ismej.2009.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J. M., Rossi O., Meijerink M., van Baarlen P. (2011). Epithelial crosstalk at the microbiota-mucosal interface. Proceedings of the National Academy of Sciences, 108, 4607–4614. doi:10.1073/pnas.1000092107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L., Ley R. E., Volchkov P. Y., Stranges P. B., Avanesyan L., Stonebraker A. C.…Chervonsky A. V. (2008). Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature, 455, 1109–1113. doi:10.1038/nature07336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J. M. W. (2014). Gut microbiota and cardiometabolic outcomes: Influence of dietary patterns and their associated components. American Journal of Clinical Nutrition, 100, 369S–377S. doi:10.3945/ajcn.113.071639 [DOI] [PubMed] [Google Scholar]
- Wu G. D., Chen J., Hoffmann C., Bittinger K., Chen Y., Keilbaugh S. A.…Lewis J. D. (2011). Linking long-term dietary patterns with gut microbial enterotypes. Science, 334, 105–108. doi:10.1126/science.1208344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W. K., Panyod S., Ho C.-T., Kuo C.-H., Wu M.-S., Sheen L.-Y. (2015). Dietary allicin reduces transformation of L-carnitine to TMAO through impact on gut microbiota. Journal of Functional Foods, 15, 408–417. doi:10.1016/j.jff.2015.04.001 [Google Scholar]
- Xu J., Gordon J. I. (2003). Honor thy symbionts. Proceedings of the National Academy of Sciences, 100, 10452–10459. doi:10.1073/pnas.1734063100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Reid G., Challis J. R. G., Kim S. O., Gloor G. B., Bocking A. D. (2015). Is there a role for probiotics in the prevention of preterm birth? Frontiers in Immunology, 6, 1–8. doi:10.3389/fimmu.2015.00062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T., Rey F. E., Manary M. J., Trehan I., Dominguez-Bello M. G., Contreras M.…Gordon J. I. (2012). Human gut microbiome viewed across age and geography. Nature, 486, 222–227. doi:10.1038/nature11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H., Lazarova D. L., Bordonaro M. (2014). Mechanisms linking dietary fiber, gut microbiota and colon cancer prevention. World Journal of Gastrointestinal Oncology, 6, 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Hou Z., Lin X., Teng G., Meng H., Zang F.…Liu S. (2013). Development of the fetal cerebral cortex in the second trimester: Assessment with 7T postmortem MR imaging. American Journal of Neuroradiology, 34, 1462–1467. doi:10.3174/ajnr.A3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Brown C. J., Abdo Z., Davis C. C., Hansmann M. A., Joyce P.…Forney L. J. (2007). Differences in the composition of vaginal microbial communities found in healthy Caucasian and Black women. ISME Journal, 1, 121–133. doi:10.1038/ismej.2007.12 [DOI] [PubMed] [Google Scholar]
- Zimmer J., Lange B., Frick J.-S., Sauer H., Zimmermann K., Schwiertz a.…Enck P. (2012). A vegan or vegetarian diet substantially alters the human colonic faecal microbiota. European Journal of Clinical Nutrition, 66, 53–60. doi:10.1038/ejcn.2011.141 [DOI] [PubMed] [Google Scholar]
- Zimmermann M. B., Chassard C., Rohner F., Goran K. N. E., Nindjin C., Dostal A.…Hurrell R. F. (2010). The effects of iron fortification on the gut microbiota in African children: A randomized controlled trial in Côte d’Ivoire. American Journal of Clinical Nutrition, 92, 1406–1415. doi:10.3945/ajcn.110.004564.1 [DOI] [PubMed] [Google Scholar]