Abstract
We used a temperature differential assay with the opportunistic fungal pathogen Cryptococcus neoformans as a simple screening platform to detect small molecules with antifungal activity in natural product extracts. By screening of a collection extracts from two different strains of the coprophilous fungus, Amphichorda felina, we detected strong, temperature-dependent antifungal activity using a two-plate agar zone of inhibition assay at 25 and 37 °C. Bioassay-guided fractionation of the crude extract followed by liquid chromatography–mass spectrometry (LC-MS) and nuclear magnetic resonance spectroscopy (NMR) identified cyclosporin C (CsC) as the main component of the crude extract responsible for growth inhibition of C. neoformans at 37 °C. The presence of CsC was confirmed by comparison with a commercial standard. We sequenced the genome of A. felina to identify and annotate the CsC biosynthetic gene cluster. The only previously characterized gene cluster for the biosynthesis of similar compounds is that of the related immunosuppressant drug cyclosporine A (CsA). The CsA and CsC gene clusters share a high degree of synteny and sequence similarity. Amino acid changes in the adenylation domain of the CsC nonribosomal peptide synthase’s sixth module may be responsible for the substitution of L-threonine compared to L-α-aminobutyric acid in the CsA peptide core. This screening strategy promises to yield additional antifungal natural products with a focused spectrum of antimicrobial activity.
Keywords: Adenylation domain, Antifungal, Ascomycota, Coprophilous fungi, Genome, Hypocreales, Nonribosomal peptide synthetase, Secondary metabolites, Thermal adaption
Introduction
Human infections by fungal pathogens affect vulnerable groups of patients, especially those with immune compromising conditions such AIDS, various cancers, and organ transplantation. Among these infections, cryptococcosis is due to Cryptococcus neoformans and C. gattii is the most common cause of fungal infections of the central nervous system (Park et al. 2009). Current therapies for this disease include a limited number of antifungal agents (amphotericin B, flucytosine, and fluconazole). Despite the emergence of drug-resistant strains (Smith et al. 2015), no new therapies for this condition have been introduced in the past few decades (Coelho and Casadevall 2016; Perfect 2017).
Core human body temperature (37 °C) represents an important innate barrier against infection by restricting the growth of most environmental fungi (Garcia-Solache and Casadevall 2010; Perfect 2006). Thus, a vulnerable facet of C. neoformans virulence is its obligate shift to growth at elevated temperatures upon entering and infecting a host (Chen et al. 2013b; Perfect 2006). In fact, experimental selection for strains adapted to grow at low temperature can negate virulence. Multiple C. neoformans signaling networks, including the calcineurin pathway, are necessary to cope with elevated host temperatures (Chen et al. 2013b). Calcineurin, a target of the immunosuppressive drugs FK506 and cyclosporin A, orchestrates growth, virulence, and drug resistance in a variety of fungal pathogens, including C. neoformans. Moreover, several lines of evidence have suggested that this pathway can be exploited for novel antifungal drug development (Juvvadi et al. 2014; Leach and Cowen 2013; Mody et al. 1988). Cryptococcus neoformans calcineurin A (cna1) mutants exhibit temperature-restricted growth in vitro and are avirulent in animal models of infection. Also, the C. neoformans cyclophilin A proteins, Cpa1 and Cpa2, which are cyclosporin A binding partners, are together required for growth at 37 °C. Additionally, the C. neoformans cpa1 mutants are attenuated for virulence (Wang et al. 2001).
CsA (Fig. 1) is a cyclic peptide produced by Tolypocladium inflatum and other fungi. This compound has potent immuno-suppressive activity and is used to prevent graft rejection in organ transplant recipients. CsA targets the highly conserved cyclophilins, and the CsA–cyclophilin complex in turn inhibits calcineurin activity (Cruz et al. 2000). This enzyme inhibition leads to impaired NF-κB function in mammalian immune cells, resulting in immunosuppression, as well as growth impairment of fungi.
Fig. 1.
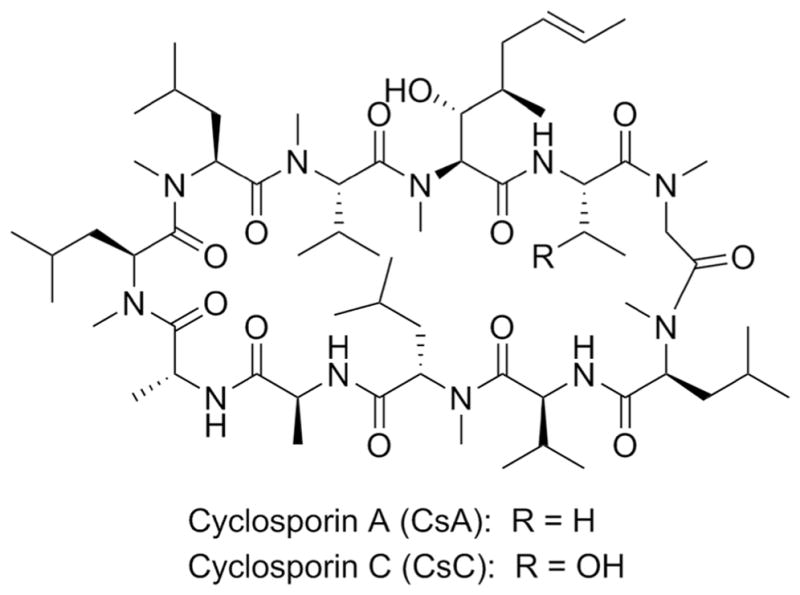
Structures of cyclosporins A and C
One challenge in identifying novel antifungal agents is the frequent presence of known compound classes, including polyenes, e.g., nystatin, peptaibols, e.g., antiamoebins, and potent mycotoxins, e.g., trichothecenes, in natural product extracts (Roemer et al. 2011). These agents have broad and temperature-independent antifungal activity. Although used clinically, polyenes are quite toxic to the host, limiting their potential applications, while most peptaibols and mycotoxins are generally considered a nuisance during natural product screening. We have therefore implemented a two-plate (25 and 37 °C) thermal sensitivity assay designed to detect natural products that interfere with thermal adaptation in C. neoformans. During early screening efforts with a set of about 750 uncharacterized crude fungal extracts, the two-plate assay recognized temperature-dependent zones of inhibition at 37 °C from extracts prepared from two different strains of the coprophilous fungus Amphichorda felina (Fig. S1). Coprophilous fungi represent a particularly rich source of natural products given the inherent microbial competition in short-lived animal excrement (Bills et al. 2013). Here, we describe the bioassay-guided fraction of one of these crude extracts followed by LC-MS and NMR studies, identifying the cyclic undecapeptide cyclosporin C (CsC, Fig. 1) as the main component responsible for antifungal activity at 37 °C. CsC differs from the immunosuppressant drug CsA by a substitution of L-threonine in the position of L-α-aminobutyric acid at the peptide’s sixth amino acid (AA). We sequenced the genome of A. felina, and we have identified and annotated the CsC nonribosomal peptide synthetase (NRPS) and biosynthetic gene cluster (BGC), comparing it to that of the CsA BGC from T. inflatum (Bushley et al. 2013). The discovery that strains of A. felina can produce CsC afforded us the opportunity to examine how CsC biosynthesis differs from that of CsA and thereby probe the origins of chemical variation in the cyclosporin family.
Materials and methods
Origin of strains
Strain TTI-0347 was isolated from synnemata formed on deer dung incubated in a humid chamber (Fig. S1), likely from white-tailed deer (Odocoileus virginianus), collected in October 2015 along U.S. Route 90, Amistad Village, Val Verde County, Texas. TTI-0469 was isolated from synnemata formed on dung of raccoon (Procyon lotor) collected in October 2016 in the Big Thicket National Preserve, near Saratoga, Hardin Co., Texas. Dung samples were rehydrated with deionized water and incubated at room temperature in deep-dish Petri plates lined with moist filter paper until synnemata formed on the dung surface. Conidia were dissected from synnemata and transferred to corn meal–dextrose agar supplemented with 50 μg/mL chlortetracycline and streptomycin sulfate. Germinated conidia were then transferred to YMA medium (1% malt extract, 0.2% yeast extract, and 2% agar) to establish axenic cultures. The isolates are maintained in the Texas Therapeutics Institute’s culture collection at the Brown Foundation Institute of Molecular Medicine, University of Texas Health Science Center at Houston, Houston, TX, USA. Subcultures were deposited in the Agriculture Research Service Culture Collection, Peoria, Illinois, USA, as NRRL 66746 (TTI-0347) and NRRL 66747 (TTI-0469).
Cryptococcus neoformans H99 was purchased from the Fungal Genetics Stock Center (www.fgsc.net). The cna1 mutant strain (Cruz et al. 2000) was obtained from Joseph Heitman at Duke University.
Two-plate thermal sensitivity assay
Overnight cultures of C. neoformans H99 grown in YM broth (1% malt extract, 0.2%yeast extract in deionized H2O) at 25 °C were diluted with sterile H2O to an OD600 of 0.8. One-milliliter aliquots of the resulting mixture were combined with 35-mL aliquots of YMA at 45 °C and dispensed into Omnitray plates. A 24-well pattern of test wells (4 mm diameter) was aspirated from the solidified agar using the tip of a sterilized Luer-lock syringe. Sets of fungal fermentation extracts were tested at a density of 20 extracts/plate in paired plates. One plate of each pair was incubated at 25 and 37 °C. Zones of inhibition were photographed after 48 h.
CsA and CsC standards were purchased from Santa Cruz Biotechnology (SC-3503 and SC-203012), and rapamycin and FK506 from Sigma-Aldrich, and dissolved in DMSO. Amphotericin B solution (Sigma-Aldrich A2942) was the positive antifungal control.
Fermentation and purification of CsC
Agar plugs stored in frozen vials with 20% glycerol were inoculated into 250-mL Erlenmeyer flasks containing 50 mL of SMYA seed medium (1% bactoneopeptone, 4% maltose, 1% yeast extract, and 0.4% agar in deionized H2O). Seed media were incubated for 5 days with agitation at 220 rpm at 25 °C.
The strategy to generate small-scale extracts from fungi for antifungal screening has been detailed previously (Li et al. 2016, 2017). Briefly, each fungal strain from our collection was grown at a scale of 12 mL on a set of five media selected for their proven ability to stimulate production of secondary metabolites during 14 days. Each fermentation was extracted with an equal volume of methyl ethyl ketone (MEK). After thorough mixing of the solvent with the culture for at least 2 h, 5 mL of the organic phase was dried. The dried residue was re-suspended in 500 μL of DMSO. Aliquots (20 μL) of the crude DMSO extracts were applied to the wells of the assay plates as described above along with the corresponding controls.
A larger volume production fermentation was carried out in 10 Erlenmeyer flasks (500 mL), each containing 100 mL of MMK2 medium (4% mannitol, 0.5% yeast extract, and 0.43% Murashige and Skoog salts, Sigma Aldrich M5519, in deionized H2O). Five milliliters of seed cultures was inoculated into each flask. The production fermentation was carried out at 25 °C with agitation at 220 rpm for 14 days. The active compounds (CsC and minor cyclosporins) were separated by antifungal-guided fractionation. The total 1 L of fermented culture was extracted twice with an equal volume of MEK, and the organic phase was evaporated to dryness under vacuum afford the crude extract (4.0 g). One gram of crude extract was fractionated on a silica gel (20 g) and eluted with 0, 1, 2, 5, and 10% dichloromethane in methanol (v/v). The fourth fraction (5%) was subjected to a Sephadex LH-20 column and the fifth sub-fraction (80 mg) was further purified by semi-preparative reverse phase HPLC (Agilent Zorbax SB-C18 column; 5 μm; 9.4 × 250 mm; gradient elution 80–100% acetonitrile in H2O over 30 min; 2 mL/min), to afford the target compound CsC (8.2 mg, tR 13.5 min).
NMR analysis
The 1H and 13C NMR data were collected on a Bruker 500 MHz NMR equipped with a 5-mm triple resonance cryoprobe at 298 K. Residual solvent signals were used as reference (CD3Cl3—δH 7.26/δC 77.0).
HPLC-DAD-MS analysis
High-performance liquid chromatography–diode array detection–mass spectrometry (HPLC-DAD-MS) data were obtained on an Agilent 6120 single quadrupole LC-MS using positive electrospray ionization. A 10-μL aliquot of each dissolved extract was injected for HPLC-DAD-MS analysis on a linear gradient of 10–90% acetonitrile in water (with 0.1% formic acid) for 28 min at a flow rate of 1 mL/min on an Agilent Zorbax Eclipse Plus C18 re-verse phase column (4.6 × 150 mm, 5 μm). High-resolution mass spectra were acquired with an Orbitrap Elite (Thermo Scientific) instrument.
Determination of minimum inhibitory concentration values
In vitro antifungal activity was measured according to National Committee for Clinical Laboratory Standards (NCCLS) recommendations (Anonymous 2008). The minimum inhibitory concentration (MIC100) was determined by means of the serial dilution method in 96-well plates containing YM broth as the growth medium. Amphotericin B was used as the positive control. Test compounds were dissolved in DMSO and serially diluted in growth medium. Visual endpoint and optical density readings of microplate wells were measured relative to positive and negative controls. The strains were incubated at the indicated temperatures, and the MIC100 values were determined at 48 h for C. neoformans H99. Viability was determined with the aid of a plate reader using PrestoBlue® resazurin dye (Life Technologies) as the viability indicator and by direct inspection of the wells under a stereomicroscope. The spectrophotometric MIC100value was defined as the lowest concentration of a test compound that resulted in a culture with a density consistent with 100% inhibition and no detectable growth as observed by microscopy when compared to the growth of the untreated control.
Genome sequencing and annotation
Genomic DNA of A. felina TTI-0347 was prepared as previously described (Chen et al. 2013a). Genome sequencing and assembly services were provided by New York Genome Center (New York, New York, USA) and A2IDEA (Ann Arbor, Michigan, USA), respectively. A 1-μg aliquot of the genomic DNAwas prepared using the Illumina TruSeq PCR-free DNA HT sample preparation kit with 450 bp insert size. DNA library quality control included a measurement of the average size of library fragments using the BioAnalyzer (Agilent), estimation of the total concentration of DNA by PicoGreen, and a measurement of the yield and efficiency of the adaptor ligation process with a quantitative PCR assay (KAPA) using primers specific to the adaptor sequence. Sequencing was carried out on a HiSeq 2500 instrument (v4 chemistry) using 2 × 125 bp cycles, with libraries loaded onto the HiSeq 2500 flowcell for clustering on the cBot using the instrument-specific clustering protocol.
The de novo genome was assembled using velvet (Zerbino and Birney 2008), and assembly statistics were calculated with Assemblathon (Earl et al. 2011). Reads were assessed for standard quality with FastQC as provided by the Babrahma Bioinformatics Laboratory (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Paired end data with 315,701,801 total number of sequences were analyzed through FastQC. The sequence length was determined to be 125 nt (range 124–126 nt) with 49–50% GC content. The quality scores and both median and interquartile ranges were in acceptable ranges along each position in each sequence. The k-mer size for the paired end read data was calculated with Kmergenie (https://kmergenie.bx.psu.edu) (Chikhi and Medvedev 2014), resulting in an optimal k-mer of 91. The draft genome size was estimated at 32. 09 Mb, with a mean GC content of 54.19% (Table S1). Other quantitative details of assembly are summarized in Table S1. This draft genomic sequence was analyzed by the BGC-predictive algorithm antiSMASH (Blin et al. 2017) which recognized a cyclosporin-like gene cluster. Fgenesh and Augustus trained with Fusarium graminearum as a reference organism (Solovyev et al. 2006; Stanke and Morgenstern 2005) were used to refine gene predictions for the CsC BGC. Gene predictions were checked and adjusted with cDNA data from the CsA BGA (Bushley et al. 2013). The annotated gene cluster was deposited in GenBank (accession no. MF716954).
Adenylation domain analysis
To understand the underlying genetic changes encoding differences between the CsA and CsC peptides, we examined details of the key positions in A-domain binding pockets of the A-domain of the sixth module of the respective NRPSs. The A-domains of CsC NRPS were delimited based on the annotations from antiSMASH 4.0 (Blin et al. 2017). Then the predicted AA sequence of the sixth module’s A-domain was used to interrogate similar NRPS sequences in GenBank. Similar NRPS A-domains retrieved by the BLAST search that have been linked to known chemical structures were aligned to the CsC and CsA A-domains. The AAs were predicted from the A-domain by four different methods (Stachelhaus codes, NRPSpredictor2, HMM, and SEQL) (Khayatt et al. 2013; Knudsen et al. 2016; Röttig et al. 2011; Stachelhaus et al. 1999) and compared to the actual AAs in the CsA and CsC peptides.
Results
Implementation of a two-plate zone of inhibition assay and detection of selective antifungal activity
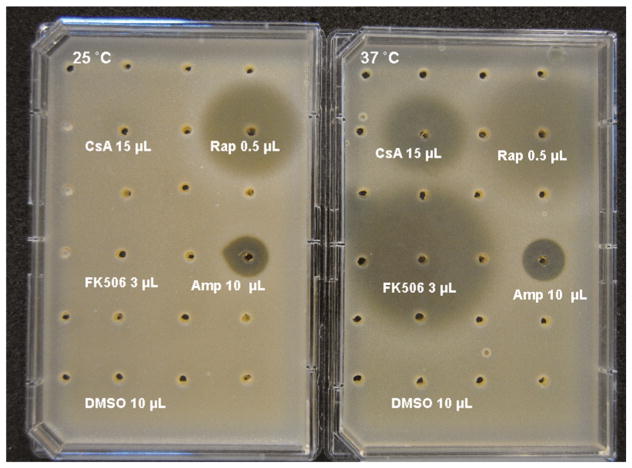
As previously reported (Cruz et al. 2000), CsA and FK506 both display strong differential antifungal activity toward C. neoformans when grown at 37 °C, with an MIC80 of 0.39 μg/mL and negligible activity at 24 °C. We implemented an agar diffusion assay in paired Omnitray plates in order to screen collections of crude natural product extracts and appropriate controls in a 24-wells-per-plate configuration. Pure samples of CsA and FK506 caused no zone of inhibition at 25 °C, but each compound potently inhibited growth at 37 °C (Fig. 2). In contrast, the immunosuppressant drug rapamycin which affects the TOR kinase via its complex with FKBP12 was potently active at both temperatures (Cruz et al. 1999) (Fig. 2). Likewise, the polyene antifungal drug, amphotericin B, inhibited growth equally at both temperatures.
Fig. 2.
Paired-plate temperature sensitivity assay with C. neoformans strain H99 after 48 h incubation at 25 and 37 °C. Note the temperature-sensitive effect of cyclosporin A (CsA) and FK506 versus rapamycin (Rap) and amphotericin B (Amp)
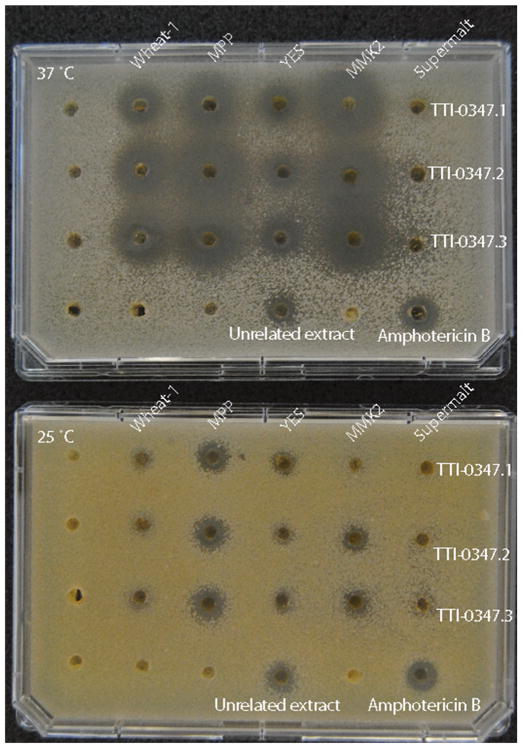
Two different strains of A. felina (TTI-0347, TTI-0469) were opportunistically included among a larger collection of fungi that were grown to produce a set of natural product extracts. All these strains were grown on a set of five different media selected for their proven ability to stimulate secondary metabolite production. In the case of TTI-0347, extracts from four of the five tested media exhibited substantially larger zones of inhibition at 37 °C (Fig. 3). In the case of TTI-0469, extracts of all the five media caused a zone of inhibition at 37 °C, but not at 25 °C (Fig. S2). None of the other approximately 750 extracts tested resulted in a strong difference in growth inhibition between 25 and 37 °C, even though about 15% of the extracts (about 110) caused zones of inhibition in C. neoformans seed plates at both temperatures.
Fig. 3.
Cryptococcus neoformans paired-plate temperature assay with extracts of A. felina TTI-0347 grown on five media. Extracts were concentrated 10-fold relative to original fermentation and suspended in DMSO. Extracts (10 μL of each of three replicates) were applied to wells for each of five media. Amphotericin B (10 μL of 256 μg/mL) was the positive control. An unrelated antifungal fermentation extract was also tested in the center of the bottom row. DMSO does not affect growth of C. neoformans
LC-MS bioassay-guided identification of the activity component for TTI-0347 and TTI-0469
The MEK extracts from scaled-up fermentations of two strains of A. felina (TTI-0347 and TTI-0469) were separated by HPLC, and their eluates were collected in 96-well plates at 300 μL/well using an integrated fraction collector. The solvent in each well was evaporated by vacuum centrifugation, and the residue in each well was assayed for growth inhibition of C. neoformans. The 96-well fractionation assay results showed that both extracts of TTI-0347 and TTI-0469 possessed the same activity component eluting at the same retention time range (23–26 min, Fig. S3A) and with the same molecular weight of 1217 (Fig. S3B).
Isolation and purification of cyclosporins from A. felina
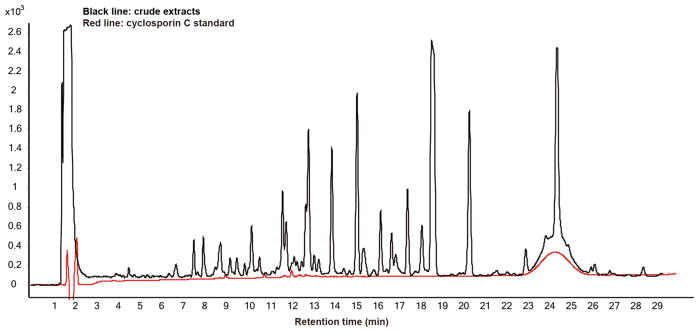
In order to isolate enough of the antifungal agent for structure confirmation, a 1-L scaled-up fermentation of A. felina TTI-0347 in MMK2 medium was extracted with MEK solvent. Upon evaporation of the solvent, fractionation of the resulting extract by preparative reversed-phase HPLC yielded 8.2 mg of pure CsC (Fig. 1). The structure of CsC was identified by comparison of its 1H, 13C NMR, and HRESIMS spectroscopic data (Fig. S4) with those described in the literature (Dreyfuss et al. 1976; Traber et al. 1977) and with the 1H NMR spectroscopic data of a commercial CsC standard (Fig. S5). Furthermore, the commercial standard co-eluted with the active peak identified as CsC in an HPLC-MS system (Fig. 4). As the data coincided with the values published and those measured from the commercial standard, the structure of CsC was identified as 6-L-threonine-cyclosporin A (Fig. 1).
Fig. 4.
HPLC-MS comparison of the crude extract from A. felina TTI-0347 grown on MMK2 medium with cyclosporin C standard (red). Note cyclosporin C co-elutes with an unknown compound with the same tR = 24.6 min
Potency of cyclosporins against C. neoformans in liquid dilution series assay
The potency of CsA and CsC was measured in a twofold liquid dilution series (Table 1). CsC showed a significant inhibitory effect against the C. neoformans clinical strain H99 with a similar potency as CsA (MIC100 values of 3.1 μg/mL at 37 °C, Table 1). Similar to the temperature-dependent antifungal effect of CsA, both CsA and CsC were much less potent at 30 °C (MIC100 12.5 μg/mL), and both compounds were essentially inactive as antifungal agents at 25 °C (MIC100 > 100 μg/mL). Amphotericin B, the positive control, showed a temperature-independent MIC100 value of 0.8 μg/mL (Table 1).
Table 1.
MIC100 (μg/mL) of cyclosporins A and C against C. neoformans H99 in twofold liquid dilution assaya
| Temperature | Cyclosporin A | Cyclosporin C | Amphotericin B |
|---|---|---|---|
| 25 °C | > 100 | 50–100 | 0.8 |
| 30 °C | 12.5 | 12.5 | Not tested |
| 37 °C | 3.1 | 3.1 | 0.8 |
The twofold serial dilution of cyclosporins from 100 to 0.098 μg/mL was tested in 96-well plates. The positive control was amphotericin B at a twofold serial dilution from 12.8 to 0.0125 μg/mL. The solvent control was 5% DMSO. The negative controls were medium and inoculum
Interestingly, both CsA and CsC displayed less potent antifungal activity against the cna1 calcineurin mutant strain compared to the isogenic wild type (Cruz et al. 2000). Since the cna1 strain grows poorly at 37 °C, the antifungal effect of CsA and CsC was tested at 30 °C for these two strains (Table 1). Both compounds failed to inhibit the growth of the cna1 mutant at high drug concentrations (MIC100 > 100 μg/mL), suggesting that both CsA and CsC function in a similar manner to inhibit cryptococcal calcineurin activity.
Identification of CsC-producing strains and their relationships to other fungi
Strains were identified as A. felina on the basis of their conspicuous white, feathery synnemata formed on dung pellets (Fig. S1), the classic habitat for this species (de Hoog 1972; Seifert et al. 2011). We retrieved the internal transcribed spacer rDNA sequence (ITS) from the genome sequence and added it to a set of sequences recently used to delimit a new species of Amphichorda, A. guana, from bat dung in Guizhou province, China (Zhang et al. 2017b). Consistent with the morphological identification, ITS sequences from TTI-0347 grouped tightly with other strains of A. felina (Fig. S6), and as indicated previously Amphichorda appears to be a sister lineage to Tolypocladium species in the Ophiocordycipitaceae, Hypocreales (Zhang et al. 2017b).
Features of the CsC gene cluster and comparison to the CsA biosynthetic gene clusters
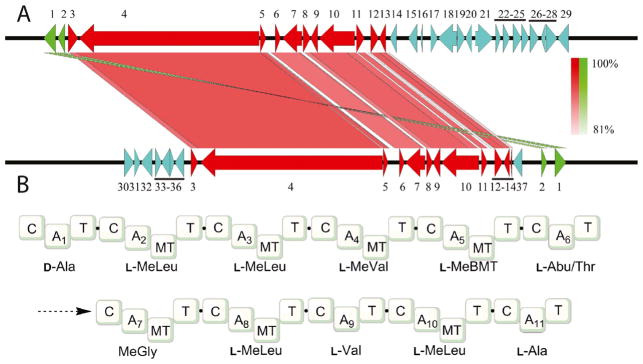
Because no previous data was available regarding the biosynthesis of CsC or its BGC, we searched for putative BGCs in the TTI-0347 genome. AntiSMASH analysis of the draft genome sequence of TTI-0347recognized 14 gene clusters with an NRPS core catalytic gene, including type 1 polyketide-NPRS hybrid type gene clusters. Surprisingly, AntiSMASH analysis identified the 11-module NRPS (15,273 AAs) on contig 1015 as a polyketide-NRPS hybrid gene cluster related to the leucinostatin A BGC instead of a close relative of the CsA BCG. The CsA and CsC BGCs (Fig. 5a) show a conservation of the overall genetic organization (genes 3–13, in red), with the positioning of orfs 1 and 2 transposed on opposite ends of each cluster (in green).
Fig. 5.
a Comparison of gene clusters responsible for biosynthesis of cyclosporin A from T. inflatum NRRL 8044 (lower) and cyclosporin C from A. felina TTI-0347 (NRRL 66746) (upper). Numbered genes in are orthologs. Intensity of shading between orthologs is proportional to identity of DNA sequences. Genes in blue indicate shared gene orthologs not involved in cyclosporin biosynthesis. Genes in green are transposed. Genes numbered 1–37 are: 1, hypothetical protein; 2, bZIP transcription factor; 3, F-box domain protein; 4, cyclosporin synthetase; 5, alanine racemase; 6, cyclophilin; 7, ABC transporter; 8, esterase-like protein; 9, cytochrome b2-like protein; 10, PKS; 11, hypothetical protein; 12, cytochrome P450; 13, aminotransferase; 14, hypothetical protein; 15–29, hypothetical proteins; 30, phosphotransferase family; 31, hypothetical protein; 32, ribonuclease P complex subunit (Pop1); 33, hypothetical protein; 34, Hsp70-like protein; 35, hypothetical protein; 36, C2H2 Zn-finger transcription factor; 37, dehydrogenase. b Predicted NRPS domain architecture and adenylation domain selectivity for the 11-amino-acid-encoding cyclosporin synthetase enzyme (gene 4). Abbreviations: C, condensation domain; A, adenylation domain; T, peptidyl carrier domain; MT, methyltransferase domain; BMT, butenyl-methyl-L-threonine; Abu, 2-aminobutyric acid
Direct comparison of this putative BGC in A. felina strain TTI-0347 with the gene sequence of the known CsABGC (Bushley et al. 2013) revealed that CsA biosynthesis gene orthologs were found at a contiguous locus on contig 1015 (Fig. 5a). Thirteen genes were highly similar (identities and similarities > 80%) with the BGC of CsA from T. inflatum NRRL 8004 (GenBank accession no. CAA8227, Table S2). The 13-gene BCG responsible for the production of CsC was defined based on previous transcriptional analysis (Bushley et al. 2013). Their functions are labeled in Fig. 5a and in Table S2. Besides simA, the core NRPS, the two gene clusters share nearly identical gene orthologs (Fig. 5a, Table S2) encoding for an alanine racemase necessary for conversion of L-alanine to D-alanine (Hoffmann et al. 1994), a polyketide synthase thought to be responsible for 3(R)-hydroxy-4(R)-methyl-6(E)-octenoic acid, the C9-backbone of the unusual amino acid (4R)-4-[(E)-2-butenyl]-4-methyl-l-threonine (Offenzeller et al. 1996); an aminotransferase likely involved in amination of branched chain precursors of L-valine and L-leucine; a cytochrome P450; a cyclophilin protein; an ABC transporter protein; a bZIP transcription factor; an F-Box domain protein; a esterase-like protein; and a cytochrome b2-like protein. Genes absent from the CsC BGC that were closely associated with the previously characterized CsA BGC were a dehydrogenase gene (Fig. 5, gene 37) and a C2H2 Zn-finger transcription factor (Fig. 5, gene 35). The predicted CsC NRPS encodes for 11 cyclosporin AAs with a modular organization that is identical to the T. inflatum CsA/CsC NRPS, including all seven N-methylated residues (Fig. 5b).
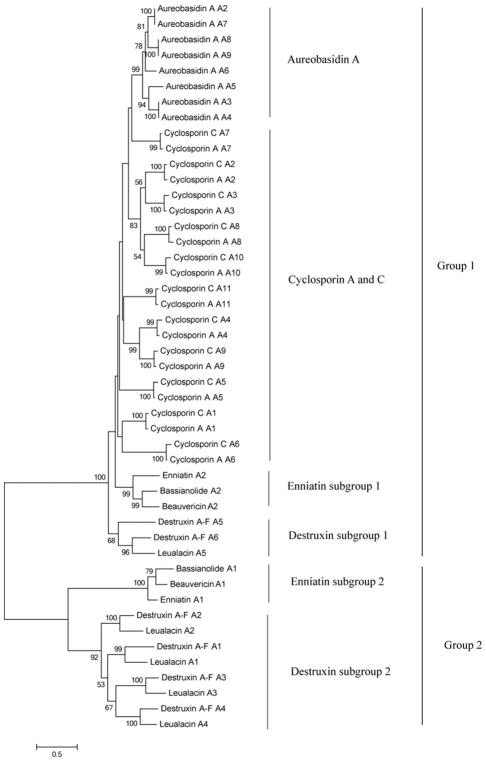
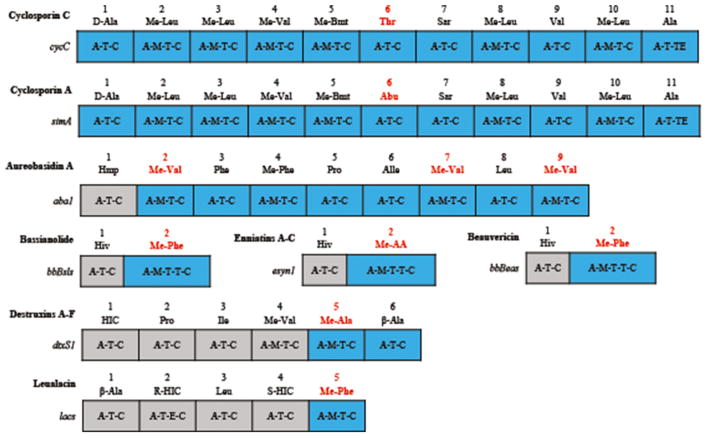
AntiSMASH and similarity searches with the 11 adenylation (A)-domains from the 11 modules of the CsC NRPS recognized 36 other highly similar A-domains from seven NRPSs (CsA, aureobasidin A, bassianolide, enniatin A–C, beauvericin, leualacin, and destruxin A–F). Their AA sequences were used to construct a phylogenetic maximum likelihood tree (Fig. 6). The analysis partitioned the set of 47 A-domains into two groups (Figs. 6, 7); group 1 included all the A-domains of CsC, CsA, and aureobasidin A, and group 2 consisted of the A-domains of bassianolide, enniatin A–C, and beauvericin (Figs. 6, 7). The pattern was consistent with a previous classification of NRPS A-domains (Bushley et al. 2013; Bushley and Turgeon 2010), except that the inclusion of fifth A-domain a leualacin NRPS (Zhang et al. 2017a) was aligned with the A-domains of group 1. The other four A-domains from the leualacin NRPS were close orthologs of the destruxin NRPS (Fig. 7, Table 2), even though their substrate AAs and final products differed.
Fig. 6.
A-domain phylogeny from cyclosporin synthetases and related fungal NRPSs. The phylogenetic maximum likelihood (ML) tree was constructed with Mega 6.0. Amino acid sequences were aligned by Clustal W. Tree was calculated with the best fit models, LG amino acid model, and Gamma Distribution. 1000 bootstrap repeats were used to estimate statistical support for the branch nodes. Bootstrap values greater than 50% are shown above branches
Fig. 7.
Modular domain structure of cyclosporin synthetases and six most similar nonribosomal peptide synthetases. Codes above modules are A-domain specificities of NRPS. Blue modules belong to group 1 based on phylogenetic analysis in Fig. 6. Red amino acids are the final products of the sixth A-domain of cyclosporin synthetases and the corresponding nearest A-domains of six other synthetases. Me, N-methyl
Table 2.
Comparative analysis and prediction of the sixth A-domain of the cyclosporin C synthetase and closely related A-domains of other fungal nonribosomal peptides
| Compound name |
Position of A- domain |
Stachelhaus codesa |
Stachelhaus predictionb |
NRPSpredictor2 predictionc |
NRPSpredictor3 predictionb |
PrediCATb | SANDPUMAb | HMM prediction4d |
SEQL predictione |
Final productf |
|---|---|---|---|---|---|---|---|---|---|---|
| Cyclosporin C | 6th | DAWFHSIAY | ABU | VAL|LEU|ILE|ABU|IVA | N/A | N/A | N/A | LEU | ABU-IVA | THR |
| Cyclosporin A | 6th | DAWFHAVAY | ABU | VAL|LEU|ILE|ABU|IVA | N/A | ABU | ABU | LEU | ABU-IVA | ABU |
| Aureobasidin A | 2nd, 7th, and 9th | DAWMFAAIL | VAL | VAL|LEU|ILE|ABU|IVA | N/A | N/A | N/A | LEU | VAL | Me-VAL |
| Bassianolide | 2nd | DGYIIGGVF | N/A | VAL|LEU|ILE|ABU|IVA | TRP | N/A | nme-fhorn | LEU | LEU | Me-LEU |
| EnniatinA-C | 2nd | DGWFIGIII | VAL | VAL|LEU|ILE|ABU|IVA | N/A | N/A | N/A | LEU | VAL | Me-VALMe-LEUMe-ILE |
| Beauvericin | 2nd | DGYIMAAVM | N/A | VAL|LEU|ILE|ABU|IVA | N/A | N/A | N/A | LEU | VAL | Me-PHE |
| Leualacin | 5th | DPWTYGAVV | N/A | VAL|LEU|ILE|ABU|IVA | PHE | N/A | PHE | LEU | ALA | Me-PHE |
| DestruxinA-F | 5th | DAWFYGGTF | ILE|LEU|VAL | VAL|LEU|ILE|ABU|IVA | VAL | N/A | VAL | LEU | LEU | Me-ALA |
| Compound name | NRPSpredictor3 predictionb | SANDPUMAb | SEQL predictione | Identityg | Species, strain | Reference |
|---|---|---|---|---|---|---|
| Cyclosporin C | N/A | N/A | ABU-IVA | 100% | Amphichorda felinaTTI-0347 | This work |
| Cyclosporin A | N/A | ABU | ABU-IVA | 94% | Tolypocladium inflatum NRRL 8004 | (Bushley et al. 2013) |
| Aureobasidin A | N/A | N/A | VAL | 55% | Aureobasidium pullulans BP-1938 | (Slightom et al. 2009) |
| Bassianolide | TRP | nme-fhorn | LEU | 49% | Beauveria bassiana ATCC 7159 | (Xu et al. 2009) |
| EnniatinA-C | N/A | N/A | VAL | 53% | Fusarium equiseti | (Haese et al. 1993) |
| Beauvericin | N/A | N/A | VAL | 50% | Beauveria bassiana ATCC 7159 | (Xu et al. 2008) |
| Leualacin | PHE | PHE | ALA | 50% | Hapsidospora irregularis | (Zhang et al. 2017a) |
| DestruxinA-F | VAL | VAL | LEU | 49% | Metarhizium robertsii ARSEF 23 | (Wang et al. 2012) |
Me N-methyl
All the Stachelhaus codes were extracted by NRPSpredictor2 (except the 10th code of Lys514) (Blin et al. 2017; Röttig et al. 2011; Stachelhaus et al. 1999)
Stachelhaus, NRPSpredictor3, PrediCAT, and SANDPUMA predictions were from http://fungismash.secondarymetabolites.org
NRPS predictor2: http://nrps.informatik.uni-tuebingen.de/Controller?cmd=SubmitJob
HMM prediction: http://www.cmbi.ru.nl/NRPS-PKS-substrate-predictor/
SEQL prediction: https://services.birc.au.dk/seql-nrps/
Final product means the amino acid in the final products
Blast: https://blast.ncbi.nlm.nih.gov/Blast.cg, and all the coverage is 100%
Comparative analysis of the A-domain specificity between the CsC and Cs NRPSs
Similarity searches with the predicted AA sequence of the A-domain from the sixth module of the CsC NRPS indicated a high identity to NRPS modules from seven other fungal peptides (CsA, aureobasidin A, bassianolide, enniatin A–C, beauvericin, leualacin, and destruxin A–F) (Bushley et al. 2013; Zhang et al. 2017a). A comparison of the predicted and actual AAs from each peptide and the corresponding A-domain binding pocket sequence (Stachelhaus codes) was compiled (Table 2). As expected, the predicted binding pocket sequence of the sixth A-domain of the CsC NRPS (DAWFHSIAY) was most similar to that of the sixth A-domain from cyclosporin synthetase from T. inflatum (DAWFHAVAY). Based on the Stachelhaus codes for these two A-domain sequences, strain TTI-0347 (SI) differed from T. inflatum (AV) by two AAs. These subtle differences may be critical to why the CsC sixth module A-domain preferentially activates L-threonine, while in the CsA sixth module A-domain, L-α-aminobutyric acid is preferentially activated. Of the various AAs predicted from the five prediction algorithms, SEQL most reliably predicted the sixth AA of CsC, predicting either L-isovaline or L-α-aminobutyric acid (Table 2). However, SEQL was unable to discriminate between the CsC or CsA sixth A-domain (Table 1). Similarly, the other NRPS prediction algorithms were variably successful in predicting the correct AA from the other homologous fungal A-domains (Table 2).
Discussion
Construction of temperature-sensitive mutant phenotypes has been fundamental to gene identification and for dissecting functions of essential genes in microorganisms (Ben-Aroya et al. 2010). However, application of temperature sensitivity screening for antibiotics that interfere with thermal adaption is a relatively newly recognized possibility (Cordeiro et al. 2016; Cowen et al. 2009; Steinbach et al. 2007). We have shown that a simple two-plate temperature sensitivity assay detected cyclosporins in crude extracts from fungal fermentations. Calcineurin, the ultimate molecular target of cyclosporins, has been shown to be a promising target for antifungal drug development due to its requirement for virulence and the potential for differential function of the pathways in the mammalian host. Implementation of an economical screening method is critical for high-throughput discovery and lead optimization studies. Furthermore, the assay may detect compounds that inhibit alternative pathways and targets involved in thermal adaption in C. neoformans, including Ras-mediated signaling pathways and trehalose biosynthesis (Esher et al. 2016; Perfect 2017).
Our recognition of CsC-producing strains of A. felina afforded active extracts with a clear-cut assay readout, with little or no growth inhibition at 25 °C and a large zone of inhibition at 37 °C. However, we also foresee that continued screening may reveal extracts that exhibit more ambiguous contrasts in growth inhibition between ambient and human body temperatures. Identifying compounds with enhanced activity at host temperatures has several real and theoretical benefits. First, it will clearly distinguish compounds with novel antifungal activities from those of commonly identified anti-fungal compounds, such as polyenes, that typically inhibit fungal growth in a temperature-independent manner. In theory, because the assay is fungus based, we may be able to detect previously unrecognized calcineurin inhibitors with selective activity towards fungal cells, as opposed to mammalian cells. Additionally, this strategy has the potential to identify compounds that target other pathways responding to host stress signals, and thereby providing new insights into microbial processes required for pathogenesis.
The CsA-producing T. inflatum is an insect parasite; however, the trophic relationships of A. felina remain enigmatic. It is an asexual species typically recognized because of its conspicuous synnemata that form on dung and its mode of conidiogenesis (de Candolle 1815; de Hoog 1972; Seifert et al. 2011). However, strains of this species have also been recovered from soils, marine sediments, and prepared yeast (Anonymous 2017; de Hoog 1972; Langenfeld et al. 2011; Yurchenko et al. 2014). Various secondary metabolites have been reported from A. felina, including the isariins (Deffieux et al. 1981), isarfelins (Guo et al. 2005), oxirapentyns (Yurchenko et al. 2014), isariketides (Smetanina et al. 2017), and felinones (Du et al. 2014). Thus, its genome sequence will offer the raw data needed to link this rich array of secondary metabolites to their cognate gene clusters. Dreyfuss (1986) listed several kinds of fungi that produced cyclosporins and mentioned that a strain designated as A. felina S249/F (as Isaria felina) was a low-titer producer of cyclosporin, but did not specify which cyclosporin. Thus, we report here that the primary cyclosporin from A. felina is CsC and that possibly CsP and other cyclosporins are produced at low titers (data not shown).
Rather than producing a microheterogeneous mixture of up to 25 cyclosporin variants as in fermentations of T. inflatum (Traber and Dreyfuss 1996; Traber et al. 1987), A. felina produces primarily CsC, suggesting a high fidelity of the corresponding NRPS for incorporation of L-threonine during biosynthesis. A similar pattern of CsC production has also been reported for Gliomastix luzulae (as Acremonium luzulae) (Moussaïf et al. 1997). To understand why this fungus differs in incorporation of AAs into the cyclosporine peptide, we sequenced its genome and annotated the CsC gene cluster, including the mega gene simA responsible for encoding the 11-module NRPS. Accurate substrate prediction from the A-domain sequences for fungal NRPSs remains computationally elusive. In part, poor predictability is the result of the unavailability of computational methods trained on well-curated data sets linking the genetic code of nonribosomal peptides of A-domains to their cognate substrates, and perhaps equally important in the case of fungi, due to the presence of novel and uncharacterized substrate-recognizing motifs in A-domains (Kalb et al. 2013). Suites of predictive rules eventually will need to be tailored to decipher the nonribosomal code for the major subfamilies of fungal NRPSs (Kalb et al. 2013), as exemplified by the cyclosporin family of NRPSs (Bushley and Turgeon 2010) (Table 2, Fig. 6). The predicted binding pocket AA sequence in the sixth module of CsC synthetase differed by at least two AAs compared to the same A-domain in the CsA synthetase. Thus, conformational changes in the A-domain binding pocket are a plausible explanation for the alternative sixth position AAs.
In conclusion, a two-plate, temperature-differential assay was designed to detect small molecules that selectively prevent the growth of the fungal meningitis pathogen, Cryptococcus neoformans, at human body temperature. Implementation of the assay by screening a collection of crude fermentation extracts of filamentous fungi led to identification of the calcineurin inhibitor CsC from the fungus Amphichorda felina. Therefore, the two-plate temperature assay affords a general method to detect fungal-specific calcineurin pathway inhibitors, including cyclosporins, from complex natural product mixtures. The assay has the potential to discover additional small molecules that interrupt other pathways that enable the pathogen to grow at human body temperature. Recognition of a CsC-producing fungus offered an opportunity to identify the CsC biosynthetic gene cluster and to understand why this fungus preferentially produces this particular cyclosporin variant. Differences in the amino acid sequence of the cyclosporin NRPSs at their sixth adenylation domain binding pocket are likely responsible for substitution of L-threonine and L-α-aminobutyric acid between cyclosporins A and C.
Supplementary Material
Acknowledgments
Funding This work was supported by University of Texas Health Science Center at Houston new faculty start-up funds, the Kay and Ben Fortson Endowment (to G.B.), the Chinese Scholarship Council (to L.X.), and a grant from the NIH (R01 GM121458). Genome sequencing and assembly services were provided by New York Genome Center (New York, New York, USA) and A2IDEA (Ann Arbor, Michigan, USA), respectively.
Footnotes
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s00253-018-8792-0) contains supplementary material, which is available to authorized users.
Compliance with ethical standards
Conflict of interest J.B.B., B.R.B., and G.L.V. have financial interests in Lifemine Therapeutics. None of the other authors declare any potential conflicts of interest.
Ethical approval No work appearing in this article involved studies with human participants or animals.
References
- Anonymous. Approved standard M27-A3. 3. Clinical and Laboratory Standards Institute; Wayne, PA: 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts. [Google Scholar]
- Anonymous. Westerdijk Institute Culture Collection. Utrecht, The Netherlands: 2017. [Google Scholar]
- Ben-Aroya S, Pan X, Boeke JD, Hieter P. Making temperature-sensitive mutants. Meth Enzymol. 2010;470:181–204. doi: 10.1016/S0076-6879(10)70008-2. https://doi.org/10.1016/S0076-6879(10)70008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bills GF, Gloer JB, An Z. Coprophilous fungi: antibiotic discovery and functions in an underexplored arena of microbial defensive mutualism. Curr Opin Microbiol. 2013;16(5):549–565. doi: 10.1016/j.mib.2013.08.001. https://doi.org/10.1016/j.mib.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Blin K, Wolf T, Chevrette MG, Lu XW, Schwalen CJ, Kautsar SA, Duran HGS, Santos E, Kim HU, Nave M, Dickschat JS, Mitchell DA, Shelest E, Breitling R, Takano E, Lee SY, Weber T, Medema MH. AntiSMASH 4.0—improvements in chemistry prediction and gene cluster boundary identification. Nucl Acids Res. 2017;45(W1):W36–W41. doi: 10.1093/nar/gkx319. https://doi.org/10.1093/nar/gkx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushley KE, Raja R, Jaiswal P, Cumbie JS, Nonogaki M, Boyd AE, Owensby CA, Knaus BJ, Elser J, Miller D, Di Y, McPhail KL, Spatafora JW. The genome of Tolypocladium inflatum: evolution, organization, and expression of the cyclosporin biosynthetic gene cluster. PLoS Gen. 2013;9(6):e1003496. doi: 10.1371/journal.pgen.1003496. https://doi.org/10.1371/journal.pgen.1003496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushley KE, Turgeon BG. Phylogenomics reveals subfamilies of fungal nonribosomal peptide synthetases and their evolutionary relationships. BMC Evol Biol. 2010;10(1):26. doi: 10.1186/1471-2148-10-26. https://doi.org/10.1186/1471-2148-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Yue Q, Zhang X, Xiang M, Wang C, Li S, Che Y, Ortiz-Lopez FJ, Bills GF, Liu X, An Z. Genomics-driven discovery of the pneumocandin biosynthetic gene cluster in the fungus Glarea lozoyensis. BMC Genomics. 2013a;14(1):339. doi: 10.1186/1471-2164-14-339. https://doi.org/10.1186/1471-2164-14-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-L, Lehman VN, Lewit Y, Averette AF, Heitman J. Calcineurin governs thermotolerance and virulence of Cryptococcus gattii. G3. 2013b;3(3):527–539. doi: 10.1534/g3.112.004242. https://doi.org/10.1534/g3.112.004242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikhi R, Medvedev P. Informed and automated k-mer size selection for genome assembly. Bioinformatics. 2014;30(1):31–37. doi: 10.1093/bioinformatics/btt310. https://doi.org/10.1093/bioinformatics/btt310. [DOI] [PubMed] [Google Scholar]
- Coelho C, Casadevall A. Cryptococcal therapies and drug targets: the old, the new and the promising. Cell Microbiol. 2016;18(6):792–799. doi: 10.1111/cmi.12590. https://doi.org/10.1111/cmi.12590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordeiro RA, Evangelista AJJ, Serpa R, Marques FJF, Melo CVS, Oliveira JS, Franco JS, Alencar LP, Bandeira TJPG, Brilhante RSN, Sidrim JJC, Rocha MFG. Inhibition of heat-shock protein 90 enhances the susceptibility to antifungals and reduces the virulence of Cryptococcus neoformans/Cryptococcus gattii species complex. Microbiol. 2016;162(2):309–317. doi: 10.1099/mic.0.000222. https://doi.org/10.1099/mic.0.000222. [DOI] [PubMed] [Google Scholar]
- Cowen LE, Singh SD, Kohler JR, Collins C, Zaas AK, Schell WA, Aziz H, Mylonakis E, Perfect JR, Whitesell L, Lindquist S. Harnessing Hsp90 function as a powerful, broadly effective therapeutic strategy for fungal infectious disease. Proc Nat Acad Sci USA. 2009;106(8):2818–2823. doi: 10.1073/pnas.0813394106. https://doi.org/10.1073/pnas.0813394106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz MC, Cavallo LM, Görlach JM, Cox G, Perfect JR, Cardenas ME, Heitman J. Rapamycin antifungal action is mediated via conserved complexes with FKBP12 and TOR kinase homologs in Cryptococcus neoformans. Mol Cell Biol. 1999;19(6):4101–4112. doi: 10.1128/mcb.19.6.4101. https://doi.org/10.1128/MCB.19.6.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz MC, Del Poeta M, Wang P, Wenger R, Zenke G, Quesniaux VFJ, Movva NR, Perfect JR, Cardenas ME, Heitman J. Immunosuppressive and nonimmunosuppressive cyclosporine analogs are toxic to the opportunistic fungal pathogen Cryptococcus neoformans via cyclophilin-dependent inhibition of calcineurin. Antimicrob Agents Chemotherap. 2000;44(1):143–149. doi: 10.1128/aac.44.1.143-149.2000. https://doi.org/10.1128/AAC.44.1.143-149.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Candolle AP. Flore Française. 1815;6:1–662. [Google Scholar]
- de Hoog GS. The genera Beauveria, Isaria, Tritirachium and Acrodontium gen. nov. Stud Mycol. 1972;1:1–41. [Google Scholar]
- Deffieux G, Merlet D, Baute R, Bourgeois G, Baute MA, Neveu A. New insecticidal cyclopeptides from the fungus Isaria felina 2. Structure elucidation of isariin B, isariin C and isariin D. J Antibiot. 1981;34(10):1266–1270. doi: 10.7164/antibiotics.34.1266. https://doi.org/10.7164/antibiotics.34.1266. [DOI] [PubMed] [Google Scholar]
- Dreyfuss M, Härri E, Hofmann H, Kobel H, Pache W, Tscherter H. Cyclosporin A and C, new metabolites from Trichoderma polysporum (Link ex Pers.) Rifai. Eur J Appl Microbiol Biotech. 1976;3(2):125–133. https://doi.org/10.1007/BF00928431. [Google Scholar]
- Dreyfuss MM. Neue Erkenntnisse aus einem pharmakologischen Pilz-screening. Sydowia. 1986;39:22–36. [Google Scholar]
- Du F-Y, Li X-M, Zhang P, Li C-S, Wang B-G. Cyclodepsipeptides and other O-containing heterocyclic metabolites from Beauveria felina EN-135, a marine-derived entomopathogenic fungus. Marine Drugs. 2014;12(5):2816–2826. doi: 10.3390/md12052816. https://doi.org/10.3390/md12052816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl D, Bradnam K, St John J, Darling A, Lin D, Fass J, Yu HOK, Buffalo V, Zerbino DR, Diekhans M, Nguyen N, Ariyaratne PN, Sung W-K, Ning Z, Haimel M, Simpson JT, Fonseca NA, Birol İ, Docking TR, Ho IY, Rokhsar DS, Chikhi R, Lavenier D, Chapuis G, Naquin D, Maillet N, Schatz MC, Kelley DR, Phillippy AM, Koren S, Yang S-P, Wu W, Chou W-C, Srivastava A, Shaw TI, Ruby JG, Skewes-Cox P, Betegon M, Dimon MT, Solovyev V, Seledtsov I, Kosarev P, Vorobyev D, Ramirez-Gonzalez R, Leggett R, MacLean D, Xia F, Luo R, Li Z, Xie Y, Liu B, Gnerre S, MacCallum I, Przybylski D, Ribeiro FJ, Yin S, Sharpe T, Hall G, Kersey PJ, Durbin R, Jackman SD, Chapman JA, Huang X, DeRisi JL, Caccamo M, Li Y, Jaffe DB, Green RE, Haussler D, Korf I, Paten B. Assemblathon 1: a competitive assessment of de novo short read assembly methods. Genome Res. 2011;21(12):2224–2241. doi: 10.1101/gr.126599.111. https://doi.org/10.1101/gr.126599.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esher SK, Ost KS, Kozubowski L, Yang D-H, Kim MS, Bahn Y-S, Alspaugh JA, Nichols CB. Relative contributions of prenylation and postprenylation processing in Cryptococcus neoformans pathogenesis. mSphere. 2016;1(2):e00084–e00015. doi: 10.1128/mSphere.00084-15. https://doi.org/10.1128/mSphere.00084-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Solache MA, Casadevall A. Global warming will bring new fungal diseases for mammals. MBio. 2010;1:e00061–e00010. doi: 10.1128/mBio.00061-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YX, Liu QH, Ng TB, Wang HX. Isarfelin, a peptide with antifungal and insecticidal activities from Isaria felina. Peptides. 2005;26(12):2384–2391. doi: 10.1016/j.peptides.2005.05.020. https://doi.org/10.1016/j.peptides.2005.05.020. [DOI] [PubMed] [Google Scholar]
- Haese A, Schubert M, Herrmann M, Zocher R. Molecular characterization of the enniatin synthetase gene encoding a multifunctional enzyme catalysing N-methyldepsipeptide formation in Fusarium scirpi. Mol Microbiol. 1993;7(6):905–914. doi: 10.1111/j.1365-2958.1993.tb01181.x. https://doi.org/10.1111/j.1365-2958.1993.tb01181.x. [DOI] [PubMed] [Google Scholar]
- Hoffmann K, Schneider-Scherzer E, Kleinkauf H, Zocher R. Purification and characterization of eucaryotic alanine racemase acting as key enzyme in cyclosporin biosynthesis. J Biol Chem. 1994;269(17):12710–12714. [PubMed] [Google Scholar]
- Juvvadi PR, Lamoth F, Steinbach WJ. Calcineurin as a multifunctional regulator: unraveling novel functions in fungal stress responses, hyphal growth, drug resistance, and pathogenesis. Fungal Biol Rev. 2014;28(2–3):56–69. doi: 10.1016/j.fbr.2014.02.004. https://doi.org/10.1016/j.fbr.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalb D, Lackner G, Hoffmeister D. Fungal peptide synthetases: an update on functions and specificity signatures. Fungal Biol Rev. 2013;27(2):43–50. https://doi.org/10.1016/j.fbr.2013.05.002. [Google Scholar]
- Khayatt BI, Overmars L, Siezen RJ, Francke C. Classification of the adenylation and acyl-transferase activity of NRPS and PKS systems using ensembles of substrate specific hidden Markov models. PLoS One. 2013;8(4):e62136. doi: 10.1371/journal.pone.0062136. https://doi.org/10.1371/journal.pone.0062136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen M, Søndergaard D, Tofting-Olesen C, Hansen FT, Brodersen DE, Pedersen CNS. Computational discovery of specificity-conferring sites in non-ribosomal peptide synthetases. Bioinformatics. 2016;32(3):325–329. doi: 10.1093/bioinformatics/btv600. https://doi.org/10.1093/bioinformatics/btv600. [DOI] [PubMed] [Google Scholar]
- Langenfeld A, Blond A, Gueye S, Herson P, Nay B, Dupont J, Prado S. Insecticidal cyclodepsipeptides from Beauveria felina. J Nat Prod. 2011;74(4):825–830. doi: 10.1021/np100890n. https://doi.org/10.1021/np100890n. [DOI] [PubMed] [Google Scholar]
- Leach MD, Cowen LE. Surviving the heat of the moment: a fungal pathogens perspective. PLoS Path. 2013;9(3):e1003163. doi: 10.1371/journal.ppat.1003163. https://doi.org/10.1371/journal.ppat.1003163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Yue Q, Jayanetti DR, Swenson DC, Bartholomeusz GA, An Z, Gloer JB, Bills GF. Anti-Cryptococcus phenalenones and cyclic tetrapeptides from Auxarthron pseudauxarthron. J Nat Prod. 2017;80(7):2101–2109. doi: 10.1021/acs.jnatprod.7b00341. https://doi.org/10.1021/acs.jnatprod.7b00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Yue Q, Krausert NM, An Z, Gloer JB, Bills GF. Emestrins: anti-Cryptococcus epipolythiodioxopiperazines from Podospora australis. J Nat Prod. 2016;79(9):2357–2363. doi: 10.1021/acs.jnatprod.6b00498. https://doi.org/10.1021/acs.jnatprod.6b00498. [DOI] [PubMed] [Google Scholar]
- Mody CH, Toews GB, Lipscomb MF. Cyclosporin A inhibits the growth of Cryptococcus neoformans in a murine model. Infect Immun. 1988;56(1):7–12. doi: 10.1128/iai.56.1.7-12.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussaïf M, Jacques P, Schaarwächter P, Budzikiewicz H, Thonart P. Cyclosporin C is the main antifungal compound produced by Acremonium luzulae. App Environ Microbiol. 1997;63:1739–1743. doi: 10.1128/aem.63.5.1739-1743.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offenzeller M, Santer G, Totschnig K, Su Z, Moser H, Traber R, Schneider-Scherzer E. Biosynthesis of the unusual amino acid (4R)-4-[(E)-2-Butenyl]-4-methyl-L-threonine of cyclosporin A: enzymatic analysis of the reaction sequence including identification of the methylation precursor in a polyketide pathway. Biochemistry. 1996;35(25):8401–8412. doi: 10.1021/bi960224n. https://doi.org/10.1021/bi960224n. [DOI] [PubMed] [Google Scholar]
- Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23(4):525–530. doi: 10.1097/QAD.0b013e328322ffac. https://doi.org/10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- Perfect JR. Cryptococcus neoformans: the yeast that likes it hot. FEMS Yeast Res. 2006;6(4):463–468. doi: 10.1111/j.1567-1364.2006.00051.x. https://doi.org/10.1111/j.1567-1364.2006.00051.x. [DOI] [PubMed] [Google Scholar]
- Perfect JR. The antifungal pipeline: a reality check. Nat Rev Drug Discov. 2017;16(9):603–616. doi: 10.1038/nrd.2017.46. https://doi.org/10.1038/nrd.2017.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemer T, Xu D, Singh SB, Parish CA, Harris G, Wang H, Davies JE, Bills GF. Confronting the challenges of natural product-based antifungal discovery. Chem Biol. 2011;18(2):148–164. doi: 10.1016/j.chembiol.2011.01.009. https://doi.org/10.1016/j.chembiol.2011.01.009. [DOI] [PubMed] [Google Scholar]
- Röttig M, Medema MH, Blin K, Weber T, Rausch C, Kohlbacher O. NRPSpredictor2 - a web server for predicting NRPS adenylation domain specificity. Nucl Acids Res. 2011;39(suppl_2):W362–W367. doi: 10.1093/nar/gkr323. https://doi.org/10.1093/nar/gkr323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert KA, Morgan-Jones G, Gams W, Kendrick B. The genera of Hyphomycetes. CBS-KNAW Fungal Biodiversity Centre; Utrect, The Netherlands: 2011. [Google Scholar]
- Slightom JL, Metzger BP, Luu HT, Elhammer AP. Cloning and molecular characterization of the gene encoding the aureobasidin a biosynthesis complex in Aureobasidium pullulans BP-1938. Gene. 2009;431(1–2):67–79. doi: 10.1016/j.gene.2008.11.011. https://doi.org/10.1016/j.gene.2008.11.011. [DOI] [PubMed] [Google Scholar]
- Smetanina OF, Yurchenko AN, Ivanets EV, Kalinovsky AI, Khudyakova YV, Dyshlovoy SA, von Amsberg G, Yurchenko EA, Afiyatullov SS. Unique prostate cancer-toxic polyketides from marine sediment-derived fungus Isaria felina. J Antibiot. 2017;70(7):856–858. doi: 10.1038/ja.2017.53. https://doi.org/10.1038/ja.2017.53. [DOI] [PubMed] [Google Scholar]
- Smith KD, Achan B, Huppler Hullsiek K, McDonald T, Okagaki LH, Akampurira A, Rhein JR, Meya DB, Boulware DR, Nielsen K. Increased antifungal drug resistance in Ugandan clinical isolates of Cryptococcus neoformans. Antimicrob Agents Chemotherap. 2015;59(12):7197–7204. doi: 10.1128/AAC.01299-15. https://doi.org/10.1128/AAC.01299-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovyev V, Kosarev P, Seledsov I, Vorobyev D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 2006;7(Suppl 1):S10. doi: 10.1186/gb-2006-7-s1-s10. https://doi.org/10.1186/gb-2006-7-s1-s10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachelhaus T, Mootz HD, Marahiel MA. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol. 1999;6(8):493–505. doi: 10.1016/S1074-5521(99)80082-9. https://doi.org/10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- Stanke M, Morgenstern B. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucl Acids Res. 2005;33(Web Server):W465–W467. doi: 10.1093/nar/gki458. https://doi.org/10.1093/nar/gki458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbach WJ, Reedy JL, Cramer RA, Jr, Perfect JR, Heitman J. Harnessing calcineurin as a novel anti-infective agent against invasive fungal infections. Nat Rev Microbiol. 2007;5(6):418–430. doi: 10.1038/nrmicro1680. https://doi.org/10.1038/nrmicro1680. [DOI] [PubMed] [Google Scholar]
- Traber R, Dreyfuss MM. Occurrence of cyclosporins and cyclosporin-like peptolides in fungi. J Indust Microbiol. 1996;17:397–401. [Google Scholar]
- Traber R, Hofmann H, Loosli H-R, Ponelle M, von Wartburg A. Neue cyclosporine aus Tolypocladium inflatum. Die cyclosporine K–Z. Helvet Chim Acta. 1987;70(1):13–36. https://doi.org/10.1002/hlca.19870700103. [Google Scholar]
- Traber R, Kuhn M, Rüegger A, Lichti H, Loosli H-R, von Wartburg A. Die Struktur von Cyclosporin C. Helvet Chim Acta. 1977;60(4):1247–1255. doi: 10.1002/hlca.19770600414. https://doi.org/10.1002/hlca.19770600414. [DOI] [PubMed] [Google Scholar]
- Wang B, Kang Q, Lu Y, Bai L, Wang C. Unveiling the biosynthetic puzzle of destruxins in Metarhizium species. Proc Nat Acad Sci USA. 2012;109(4):1287–1292. doi: 10.1073/pnas.1115983109. https://doi.org/10.1073/pnas.1115983109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Cardenas ME, Cox GM, Perfect JR, Heitman J. Two cyclophilin A homologs with shared and distinct functions important for growth and virulence of Cryptococcus neoformans. EMBO Rep. 2001;2(6):511–518. doi: 10.1093/embo-reports/kve109. https://doi.org/10.1093/embo-reports/kve109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Orozco R, Kithsiri Wijeratne EM, Espinosa-Artiles P, Leslie Gunatilaka AA, Patricia Stock S, Molnár I. Biosynthesis of the cyclooligomer depsipeptide bassianolide, an insecticidal virulence factor of Beauveria bassiana. Fungal Gen Biol. 2009;46(5):353–364. doi: 10.1016/j.fgb.2009.03.001. https://doi.org/10.1016/j.fgb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Xu Y, Orozco R, Wijeratne EMK, Gunatilaka AAL, Stock SP, Molnár I. Biosynthesis of the cyclooligomer depsipeptide beauvericin, a virulence factor of the entomopathogenic fungus Beauveria bassiana. Chem Biol. 2008;15(9):898–907. doi: 10.1016/j.chembiol.2008.07.011. https://doi.org/10.1016/j.chembiol.2008.07.011. [DOI] [PubMed] [Google Scholar]
- Yurchenko AN, Smetanina OF, Kalinovsky AI, Pushilin MA, Glazunov VP, Khudyakova YV, Kirichuk NN, Ermakova SP, Dyshlovoy SA, Yurchenko EA, Afiyatullov SS. Oxirapentyns F-K from the marine-sediment-derived fungus Isaria felina KMM 4639. J Nat Prod. 2014;77(6):1321–1328. doi: 10.1021/np500014m. https://doi.org/10.1021/np500014m. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18(5):821–829. doi: 10.1101/gr.074492.107. https://doi.org/10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Qiu Y, Kakule TB, Lu Z, Xu F, Lamb JG, Reilly CA, Zheng Y, Sham SWS, Wang W, Xuan L, Schmidt EW, Zhan J. Identification of cyclic depsipeptides and their dedicated synthetase from Hapsidospora irregularis. J Nat Prod. 2017a;80(2):363–370. doi: 10.1021/acs.jnatprod.6b00808. https://doi.org/10.1021/acs.jnatprod.6b00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZF, Liu F, Zhou X, Liu XZ, Liu SJ, Cai L. Culturable mycobiota from Karst caves in China, with descriptions of 20 new species. Persoonia. 2017b;39:1–31. doi: 10.3767/persoonia.2017.39.01. https://doi.org/10.3767/persoonia.2017.39.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.