Abstract
Hydrogel particles are versatile materials that provide exquisite, tunable control over the sequestration and delivery of materials in pharmaceutics, tissue engineering, and photonics. The favorable properties of hydrogel particles depend largely on their size, and particles ranging from nanometers to micrometers are used in different applications. Previous studies have only successfully fabricated these particles in one specific size regime and required a variety of materials and fabrication methods. We have developed a simple yet powerful system to easily tune the size of polypeptide-based, thermoresponsive hydrogel particles, from the nano to microscale, using a single starting material. We take advantage of the self-assembly and unique phase transition behavior of elastin-like polypeptides (ELPs) in bulk and within microfluidic-generated droplets to control particle size. These particles are then stabilized through ultraviolet irradiation of a photocrosslinkable unnatural amino acid (UAA) cotranslationally incorporated into the parent polypeptide. The thermoresponsive property of these particles provides an active mechanism for actuation and a dynamic responsive to the environment. This work represents a fundamental advance in the generation of crosslinked biomaterials, especially in the form of soft matter colloids, and is the one of the first demonstrations of successful use of UAAs in generating a novel material.
Keywords: unnatural amino acids, bioinspired materials, hydrogel particles, unstructured polypeptides, recombinant proteins
Graphical abstract
Thermoresponsive hydrogel particles that span four orders of magnitude in size, from the nano- to microscale, are developed using photocrosslinkable unnatural amino acids. These particles are fabricated from a single starting material, an intrinsically disordered polypeptide, using self-assembly and microfluidic templating. The particles are highly stable following crosslinking with ultraviolet light and exhibit a tunable, dynamic response to temperature.
Thermoresponsive hydrogel particles are versatile materials used in a variety of applications, ranging from photonics to tissue engineering and drug delivery.[1–6] These particles, made of crosslinked networks of polymers, proteins, or polypeptides, offer the ability to respond to local changes in temperature, which is manifested as reversible shrinking and swelling in solution due to changes in network solubility. These volumetric changes can be exploited for diverse applications in optics, actuation, cell capture, and drug release.[7–13] In addition to their thermal responsiveness, particle size governs many properties of interest (e.g., refractive index, cargo loading capacity, release kinetics, deformability). Despite the importance of this parameter, an overarching challenge in gel particle engineering is the ability to precisely design and synthesize particles of the same material with uniform size spanning multiple length scales. While techniques such as layer-by-layer deposition, molecular self-assembly, and microfluidic templating provide control over particle size,[14–20] these approaches limit size to a specific regime, such as either the nanoscale by self-assembly or the microscale by fluidic templating.[5, 20]
Here, we present a facile two-step method for fabricating crosslinked gel particles with programmable size across four orders of magnitude (nano-to-meso-to-microscale) that are composed of a thermoresponsive, artificial, repetitive polypeptide designed de novo for this application. Elastin-like polypeptides (ELPs), the building blocks of these particles, are stimuli responsive, intrinsically disordered polypeptides that exhibit lower critical solution temperature (LCST) phase behavior in aqueous solution.[21–28] At low temperatures, ELPs exist as soluble, disordered chains while at high temperatures above their concentration-dependent transition temperature (Tt) —also known as the cloud point— the chains desolvate to form polypeptide-rich globules known as coacervates.[29, 30] The consensus amino acid sequence of ELPs is (Val-Pro-Gly-Xaa-Gly)N, where N is the number of repeat units and Xaa is any amino acid except proline.[29] The influence of both these variables —N and Xaa— on the temperature-dependent phase behavior of ELPs is well-characterized[23, 28, 31, 32] and can be genetically encoded and independently controlled at the gene level. ELPs are an ideal starting material to fabricate thermoresponsive gel particles because they are genetically encodable, can be synthesized with perfect sequence control, have precisely specified molecular weight, are monodisperse, and can be recombinantly produced in high yield.[33, 34]
A major challenge in producing crosslinked, stimuli-responsive gel particles lies in the introduction of the crosslinking sites. ELPs can be crosslinked via lysine, cysteine, or tyrosine residues through the addition of an extrinsic crosslinker.[35–37] However, these methods are only moderately efficient and often cumbersome, requiring multiple additional processing steps and the use of cytotoxic crosslinking agents.[38–40] Further, because a useful application of these particles is as carriers of biological drugs for controlled release, this approach also limits the incorporation of biological cargo via attachment to these chemically reactive amino acids. To develop a bio-orthogonal crosslinking approach, and to eliminate the need for an extrinsic chemical crosslinker, we genetically encoded a photoreactive unnatural amino acid (UAA), para-azidophenylalanine (pAzF),[41] into the ELP backbone. The bioorthogonal aryl azide group on this residue is activated by irradiation with ultraviolet (UV) light to form a reactive nitrene, which readily inserts into nearby N-H or C-H bonds.[42, 43] Due to recent advancements in genomically recoded organisms[44–46] and multi-site UAA[47] incorporation by Isaacs and coworkers, multiple copies of a UAA can be site-specifically incorporated into proteins and polypeptides with high yield and >95% accuracy.[47] This renders the monodisperse biopolymers crosslinkable immediately after purification. Photocrosslinkable UAAs have previously been used successfully for probing protein interactions,[39, 48] photo-patterning,[49] and immobilization of proteins onto surfaces.[50, 51] This work is the first demonstration of incorporating multiple copies of a photocrosslinkable UAA into a polypeptide to create thermally responsive hydrogel particles whose size can be tuned across multiple orders of magnitude, ranging from tens of nanometers to several microns.
Using our previous work on controlling the size of ELP coacervates within aqueous microdroplets as a guideline,[52] we designed two ELPs containing photocrosslinkable pAzF residues[53] to serve as the constituent polypeptides for the gel particles (Figure 1A and Table S1, Supporting Information). One polypeptide, termed photocrosslinkable ELP (PCE), is composed of eighty pentameric repeats of the VPGVG motif with four regularly-spaced pAzF residues, a sufficient number to form a crosslinked hydrogel network.[36] The second ELP in our system is a self-assembling photocrosslinkable diblock (PCD) comprised of two distinct repetitive blocks — an N-terminal hydrophobic PCE block and a C-terminal hydrophilic ELP block. We successfully expressed these polypeptides in a genomically recoded Escherichia coli strain[44–46] and purified them in high quantities (>50 mg L−1 culture; see Figure S1, Supporting Information) using inverse transition cycling, a non-chromatographic method for purifying ELPs.[29] To confirm the successful incorporation of functional pAzF, we labeled PCE and PCD with fluorophore by a “click” reaction[54] —a strain-promoted alkyne-azide cycloaddition with a dibenzocyclooctyne (DBCO)-conjugated Cy5 fluorophore— and visualized the labeled peptide polymers by fluorescent imaging of a sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel (Figure 1B). We further confirmed >99% incorporation efficiency of pAzF by electron spray ionization liquid chromatography mass spectrometry (ESI-LC/MS) (Figure S2, Supporting Information), which is consistent with previous reports.[47]
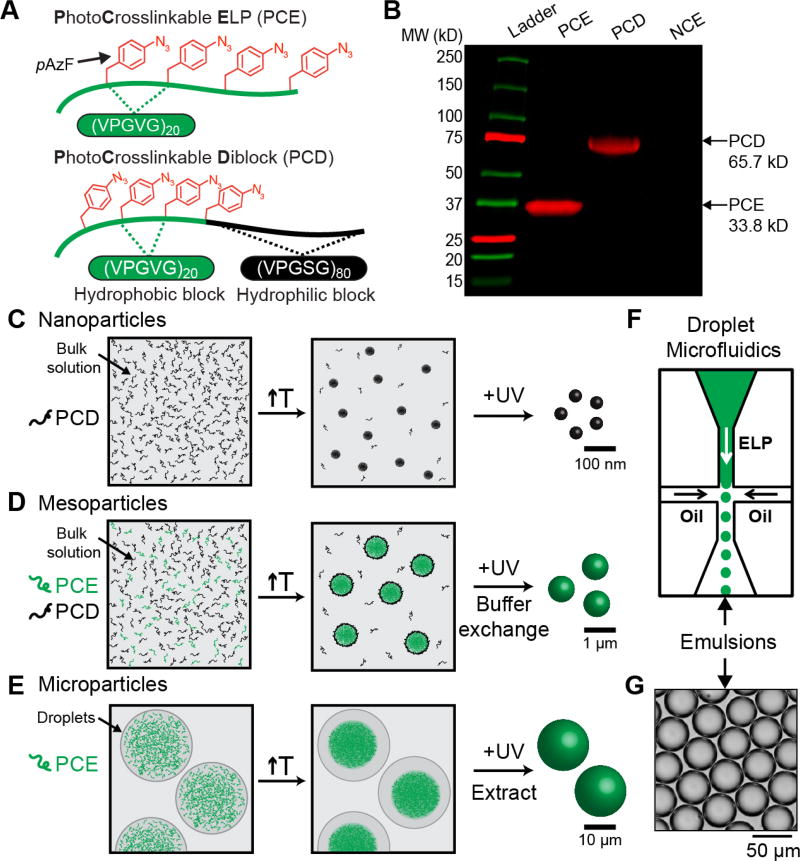
Figure 1. Design of size-controlled, thermoresponsive gel particles composed of disordered polypeptides containing photocrosslinkable unnatural amino acids.
(A) Schematic of the two disordered polypeptides used in this study — photocrosslinkable ELP (PCE) and photocrosslinkable diblock (PCD). (B) Fluorescent SDS-PAGE gel of PCE, PCD, and NCE (non-crosslinkable ELP, (VPGVG)80) labeled with DBCO-Cy5. PCE and PCD contain pAzF residues which react with DBCO-Cy5, while the negative control NCE is not fluorescently labeled via click. (C) Strategy for generating nanoparticles by heating and crosslinking PCD in bulk solution. (D) Strategy for generating mesoparticles by heating and crosslinking mixtures of PCE and PCD in bulk solution. The PCD acts as a surfactant to stabilize PCE particles after heating, and buffer exchanging removes free PCD following crosslinking. Mesoscale particles can also be generated in microfluidic droplets for fluorescent spatial tracking. (E) Strategy for generating microparticles by heating and crosslinking solutions of PCE in microfluidic droplets, followed by particle extraction from droplets. (F) Schematic of a chip-based microfluidic droplet generator for generating monodisperse ELP-containing water droplet templates. (G) Brightfield images of monodisperse water droplets containing photocrosslinkable ELPs.
After synthesizing pAzF-containing ELPs, we employed a two-step strategy to fabricate thermoresponsive gel particles of uniform size spanning multiple length scales (Figure 1C–E). We first tuned the temperature of solutions containing PCD, PCE, or a mixture of both to form particles, and then stabilized the resulting particles by crosslinking with UV light. For the smallest, nanoscale size regime we use only PCD, a thermally responsive amphiphile. Raising the temperature above its critical micellization temperature (CMT) in bulk solution triggers its self-assembly into spherical micelles (Figure 1C and Figure S3, Supporting Information) and subsequent UV-irradiation crosslinks the micelle cores, leading to the formation of nano-gels.
To create hydrogel particles that span the mesoscale, we combine diblock PCD and monoblock PCE in solution (Figure 1D). By tuning the temperature above both the CMT of the PCD and the Tt of PCE, but below the Tt of PCD, we can generate mesoscopic coacervates of PCE whose size is limited by the PCD that serves as a surfactant and limits coarsening of the PCE coacervates. By tuning the ratio of PCE to PCD, we can generate particles of different sizes spanning the mesoscale. UV-irradiation then stabilizes these hydrogel particles, which can be subsequently buffer exchanged to remove excess PCD for downstream applications.
At the largest, microscale size regime, we utilized microfluidic droplet-generating chips to create monodisperse water-in-oil emulsion droplets containing PCE (Figure 1E–G). Tuning the temperature either above or below the Tt of PCE and subsequent UV-irradiation crosslinks the PCE coacervates into microscopic particles. Gel particle size can be easily controlled by modulating the starting concentration of PCE and the temperature at which crosslinking occurs.
We first synthesized crosslinked nano-gels by thermally triggering the self-assembly of PCD in bulk solution into micelles and then crosslinking with UV light.[19, 55, 56] We use the terms “nxPCD” to denote the non-crosslinked micelles, and “xPCD” for these micelles after they are crosslinked into nano-gels by UV irradiation. We characterized the nanoscale assemblies of PCD before and after crosslinking with UV light in bulk solution. Thermal ramp dynamic light scattering (DLS) of dilute xPCD in solution confirmed the stabilization of self-assembled nano-sized gel particles (Figure 2A and Figure S4, Supporting Information). These xPCD particles demonstrate reversible thermal responsiveness by shrinking upon heating as the hydrophobic core desolvates and swelling upon cooling (Figure 2B). In contrast, the nxPCD particles assemble upon heating into nano-scale micelles but disassemble completely into single chains upon cooling (Figure 2A). Static light scattering (SLS), performed on both nxPCD and xPCD performed above the CMT corroborated the particle sizes measured by DLS (Table 1 and Figure S5, S6, Supporting Information).
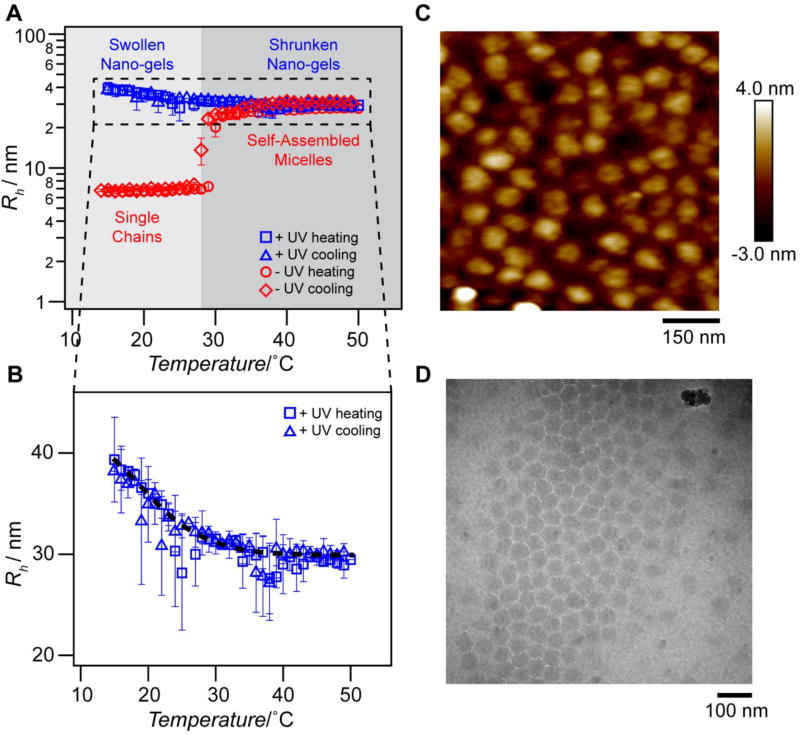
Figure 2. PCD self-assembles into micelles upon heating and can be photocrosslinked to form thermoresponsive nano-gels.
(A) Hydrodynamic radius of nxPCD (red; non-crosslinked) and xPCD (blue; crosslinked) as a function of temperature. nxPCD self-assembles from single chains into monodisperse micelles above the critical micellization temperature (TCMT ≈ 27 °C at 100 µm), whereas xPCD does not disassemble into single chains when the temperature is lowered below the CMT. (B) Hydrodynamic radius (Rh) of xPCD as a function of temperature demonstrates that xPCD nano-gels exhibit thermal responsiveness as seen by the decrease in Rh with increasing temperature. Black dashed line represents the best sigmoidal curve fit for visualization. Error bars represent the polydispersity percentages (n = 5). (C) Tapping mode atomic force microscopy (AFM) images of xPCD below the CMT (T = 22 °C) indicate highly monodisperse, crosslinked nano-gels with a diameter of 60.4 ± 11.7 nm. (D) Cryo-transmission electron microscopy (TEM) images of xPCD above the CMT (T = 37 °C).
Table 1.
Static light scattering characterization of nxPCD and xPCD at 37 °C.
| Rg [nm] |
Rh [nm] |
ρ | MWmicelle [g mol−1] |
Nagg | |
|---|---|---|---|---|---|
| nxPCD | 16.4 | 27.4 | 0.60 | 1.1 × 107 | 162 |
| xPCD | 19.2 | 29.1 | 0.66 | 1.3 × 107 | 200 |
We visualized the xPCD particles by tapping mode atomic force microscopy (AFM) performed below the CMT, which showed highly monodisperse nano-gels and confirmed their spherical shape (Figure 2C and Figure S7, S8, Supporting Information). At the same temperature, nxPCD does not form any visible nanoscale assemblies in the absence of crosslinks to stabilize the chains against dissociation (Figure S9, Supporting Information). We further confirmed, using cryo-transmission electron microscopy (cryo-TEM), that xPCD exists as monodisperse particles above the CMT (Figure 2D and Figure S10, Supporting Information). These uniform, spherical xPCD nano-gels are ideally suited as delivery vehicles as they can be generated in solution and exist as stable micelles across a range of temperatures. Their size and solvation, both of which decrease with increasing temperature, are useful tunable attributes of these thermally responsive nano-gels, as they may provide an “active” mechanism to release encapsulated cargo as compared to the typical “passive” diffusive mechanism of release from most hydrogels.[57–60]
We next developed crosslinked particles with sizes spanning the mesoscopic range from 100 nm to 1 µm in both bulk solution and microdroplets utilizing insights from our previous study. In that work, we showed that mesoscopic coacervates of uniform size could be obtained by emulsifying mixtures of ELP diblocks and monoblocks, and thermally triggering the phase separation of both species.[52] Upon heating these mixtures to a temperature, T, such that the CMTdiblock < T < Tt-diblock and T > Tt-monoblock we observed two concurrent events: (i) the assembly of diblocks into nanoscale micelles and (ii) the assembly of monoblocks into stable mesoscopic coacervate droplets where the diblock micelles prevent coalescence of the coacervate droplets. Importantly, we showed that the size of the mesoscale coacervates can be easily tuned by adjusting the relative ratio of mono-to-diblock ELP.
To determine the appropriate temperature for mesoscale particle formation, we first investigated the bulk LCST phase separation of PCE (Figure S11, Supporting Information). To visualize the phase separation within microdroplets, we fluorescently labeled the N-termini of the polypeptides for spatial tracking with fluorescence microscopy. When emulsified and heated, PCE undergoes the canonical liquid-liquid phase separation of monoblock ELPs and coarsening to form uniform liquid spheres (Figure S12 and Movie S1, Supporting Information), indicating that the pAzF residues do not have an adverse effect on PCE phase behavior. Next, we carried out light scattering of PCD to determine the CMT of PCD and its Tt – the temperature at which micelles of PCD undergo phase separation into a coacervate (Figure S3A and S11, Supporting Information). These experiments allowed us to identify a range of temperatures, T, such that CMTPCD < T < Tt-PCD and T > Tt-PCE for a given concentration in order to take advantage of these distinct phase behaviors and form mesoscale particles. For example, at 100 µm, CMTPCD is ≈27 °C, Tt-PCD is ≈65 °C, and Tt-PCE is ≈23 °C; therefore, heating to any temperature in the range of 27 °C < T < 65 °C will result in mesoscale particle formation. Having verified that PCE and PCD satisfied these conditions, we next created microdroplets containing equimolar concentrations of PCE and PCD and heated to T = 30 °C, to induce the phase separation of PCE and self-assembly of PCD (Movie S2, Supporting Information). Heating PCE in the presence of PCD results in visible PCE coacervates that do not coarsen over time but rather exhibit Brownian motion due to the surfactant-like PCD chains at the interface between the PCE-rich and PCE-poor phases (Movie S2, Supporting Information). Exposing the mesoscopic PCE-PCD coacervates to UV light efficiently crosslinks them into particles that do not re-solubilize upon cooling below the cloud point temperature of PCE (Figure 3A and Movie S3, Supporting Information). By varying the ratio of PCE: PCD, it is possible to create meso-gels whose size spans the entire mesoscale regime; increasing the concentration of PCE relative to PCD creates coacervate droplets with increasing size which then can be photocrosslinked into hydrogel particles (Figure S13, Supporting Information). Scanning electron microscopy (SEM) imaging of these particles confirms the ability to easily tune their size within the mesoscale range (Figure 3B and Figure S14, S15, Supporting Information).
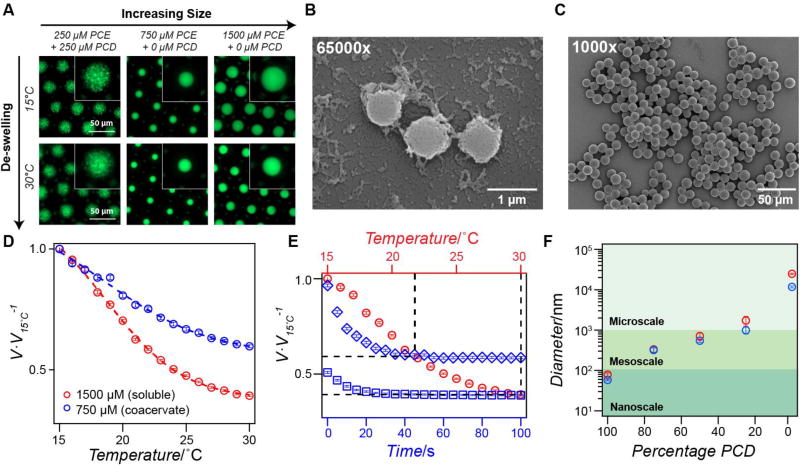
Figure 3. Thermoresponsive meso- and micro-gels with tunable swelling and de-swelling behavior.
(A) Fluorescence microscopy images of AlexaFluor 488-labeled meso-gels (left), 12 µm xPCE micro-gels (middle), and 25 µm xPCE micro-gels (right) within water-in-oil emulsion droplets. The concentration of PCE and PCD is shown above each panel. At T < Tcloud point PCE the gels are swollen (top panels) and T > Tcloud point PCE the gels are shrunken (bottom panels). (B) SEM images of mesoscale particles with mean particle diameter 865 nm ± 199 nm (n ≥ 200). (C) SEM images of microscale particles with mean particle diameter 12.1 µm ± 0.8 µm (n ≥ 200). (D) Gel volume (normalized to the volume at 15 °C) as a function of temperature for 750 µm xPCE particles (blue points) and 1500 µm xPCE particles (red points). The dashed lines represent the best sigmoidal fit and the error bars represent the SEM (n = 150 droplets). (E) Normalized gel volume as a function of time for xPCE micro-gels (1500 µm) quench heated from 15 °C to 22 °C (blue diamonds) or 22 °C to 30 °C (blue boxes). The red circles indicate the equilibrium volume of xPCE gels as a function of temperature; the black dashed lines are visual aids that intersect at the equilibrium volume at 22 °C and 30 °C. The error bars represent the SEM (n = 150). (F) Gel particle diameter as a function of PCD content (PCE percentage = 100 – PCD) demonstrating the ability to fabricate uniform gel particles spanning multiple length scales. The two data points shown represent the largest (red) and smallest (blue) particles observed via DLS (100% – 25% PCD) illustrating the size range that can be achieved by modulating temperature, with error bars representing the polydispersity percentage (n ≥ 5). The red and blue points at 0% PCD illustrate the particle size obtained with different starting concentrations of PCE, and error bars represent the standard deviation (n ≥ 100).
To extend the size of the particles further into the microscale, and expand their utility for drug delivery, optics, cosmetics, and other applications, we created microdroplets containing only PCE. Without PCD chains present in solution, PCE phase separates to form spherical, microscale coacervates with dimensions dictated by the template droplet size (Movie S1, Supporting Information). We can tune the size of these particles by modulating the initial concentration of PCE and then crosslinking at a temperature either above or below its Tt. We used this approach to generate microparticles of 12 µm and 25 µm in diameter, respectively. To generate 25 µm micro-gels, we crosslinked droplets containing soluble PCE (T < Tcloud point PCE) at a concentration above chain overlap to enable network formation. After crosslinking, these micro-gels rapidly reach their equilibrium concentration and upon heating expel water and shrink due to enhanced chain-chain interactions and desolvation (Figure 3A). To create the 12 µm microparticles, we heated PCE droplets (T > Tcloud point PCE) to form uniform micro-coacervates (one coacervate per droplet) and then irradiated the droplets with UV light to crosslink the coacervates into micro-gels. These micro-gel particles were extracted from their template droplets and imaged by SEM, revealing highly monodisperse, spherical particles (Figure 3C and Figure S14, S15 Supporting Information). These particles are also highly stable; they can be lyophilized and reconstituted in solution, and stored at temperatures ranging from −80 °C to 37 °C for at least one week without loss of structural integrity or change in size (data not shown).
We next investigated the thermal responsivity of xPCE micro-gels by monitoring their size as a function of temperature using temperature-controlled fluorescence microscopy. We observed that the 12 µm and 25 µm xPCE micro-gels exhibit well-controlled de-swelling upon slow heating from 15 °C to 30 °C (Figure 3D and Movie S4, S5, Supporting Information). In both micro-gels, the temperature-dependent de-swelling follows a sigmoidal relationship similar to that observed for xPCD nano-gels, though this effect is more prominent for the 25 µm xPCE micro-gels due to a less densely packed network. This steady decrease in micro-gel size with increasing temperature is due to the decreased excluded volume of each monomer comprising the network, which in turn directly affects the swelling behavior of the gel.[61] These results demonstrate that by simply changing the temperature at which crosslinking is performed (i.e., upon solutions of soluble or insoluble PCE), it is possible to generate micro-gels that controllably shrink down to 40% of their original volume (Figure 3D).
Finally, we evaluated the de-swelling kinetics of the xPCE micro-gels to better understand their response rate to changes in temperature. We performed two step-wise heating ramps (from 15 °C to 22 °C and 22 °C to 30 °C) and monitored the size of the 25 µm xPCE micro-gels as a function of time after reaching the target temperature (Figure 3E and Movie S6, S7, Supporting Information). In both instances, the micro-gels respond similarly and reach their equilibrium size in ~40 s, indicating a rapid response to the temperature change of the environment, which is ideal for applications requiring on-demand feedback. The de-swelling dynamics we observed are consistent with those reported for poly(N-isopropylacrylamide) micro-gels of similar size.[62] In thermoresponsive particles, the swelling/de-swelling response rate is proportional to the rate of diffusion of water into/out of the particle, which is governed by the overall size and density of the polymer network. Given ELPs’ finely tuned and well-characterized response to temperature, these particles are well-positioned to serve as ideal carrier vehicles, actuators, and sensors.
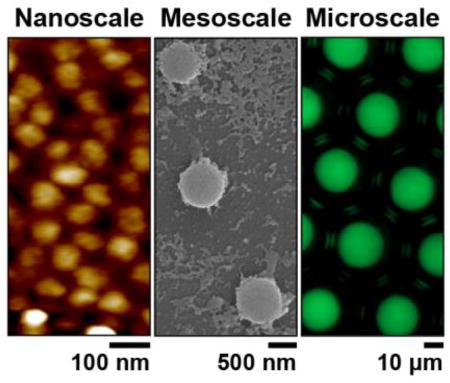
In conclusion, we have developed a simple and convenient method to fabricate thermoresponsive gel particles from genetically encoded polypeptides that are tunable from the nanoscale to microscale in size (Figure 3F). Particle size can be easily adjusted by tuning the relative concentrations of the pAzF-containing ELP diblock and monoblock. Particle size is increased from the nano- to mesoscale by increasing the ratio of monoblock (PCE) to diblock ELP (PCD). To extend into the microscale regime, microfluidic droplets provide a necessary level of control for templating PCE coacervates, with droplet dimensions directly dictating particle size. The reversible swelling/de-swelling behavior of the particles is rapid (i.e., on the order of seconds) and easily tuned by specifying the phase —soluble versus coacervate— of the polypeptides prior to crosslinking. Future work with this material system will allow further fine-tuning of gel network density at any given temperature. For example, changing the ELP sequence (i.e. guest residue, length) will modulate the functional dependence of monomer-excluded volume on temperature,[61] and altering the concentration of pAzF residues can also easily tune the swelling properties. The simplicity and flexibility of this new method for creating thermoresponsive nano-, meso-, and micro-gels enables the fabrication of particles with potential utility in a wide variety of applications in biomaterials, optics, and pharmaceutics.
Experimental Section
Materials
pET24+ vectors were purchased from Novagen (Madison, WI). Oligonucleotides and gBlocks encoding sequences of interest were purchased from Integrated DNA Technologies (Coralville, IA). Ligation enzymes, restriction enzymes and calf intestinal alkaline phosphatase were purchased from New England Biolabs (Ipswich, MA). EB5α chemically competent Escherichia coli cells were purchased from Bioline (Taunton, MA). Genomically recoded E. coli cells were generously provided by Professor Farren Isaacs (Yale University). All E. coli cultures were grown in 2×YT media comprised of sodium chloride (5 g L−1; Alfa Aesar, Ward Hill, MA), tryptone (16 g L−1, Becton, Dickinson and Co., Franklin Lakes, NJ, and yeast extract (10 g L−1, Becton, Dickinson and Co., Franklin Lakes, NJ). p-Azidophenylalanine hydrochloride was purchased from Synchem, Inc (Elk Grove Village, IL). Kanamycin sulfate was purchased from EMD Millipore (Billerica, MA) and chloramphenicol was purchased from Sigma-Aldrich (St. Louis, MO). Protein expression was induced with isopropyl β-D-1-thiogalactopyranoside (IPTG) from Gold Biotechnology (St. Louis, MO) and L-(+)-arabinose from Sigma-Aldrich (St. Louis, MO). All salts were purchased from Alfa Aesar (Ward Hill, MA). 1× phosphate buffered saline (PBS) tablets (10 mm phosphate buffer, 140 mm NaCl, 3 mm KCl, pH 7.4 at 25 °C) were purchased from EMD Millipore (Billerica, MA). Molecular organic fluorophores (NHS-Alexa Fluor® 488, dibenzocyclooctyne-Cy5) were purchased from Life Technologies (Grand Island, NY) and Sigma-Aldrich (St. Louis, MO), respectively. DNA extraction kits, DNA gel purification kits and PCR purification kits were purchased from Qiagen Inc. (Germantown, MD). Mineral oil was purchased from Sigma-Aldrich (St. Louis, MO). ABIL® EM 90 and TEGOSOFT® DEC surfactants were purchased from Evonik Industries (Essen, Germany). A glass single emulsion droplet-generating chip was purchased from Dolomite Microfluidics (Royston, United Kingdom). Syringe pumps were purchased from Chemyx Inc. (Stafford, TX). Isobutanol was purchased from IBI Scientific (Peosta, IA). All solvents for liquid chromatography/mass spectroscopy (LC/MS) analysis were LC/MS grade, purchased from Thermo Fisher Scientific (Waltham, MA). Scanning electron microscopy (SEM) samples were deposited on silicon wafers on aluminum specimen mounts, purchased from Electron Microscopy Sciences (Hatfield, PA). Quantifoil grids (Quantifoil Micro Tools GmbH, Großlöbichau, Germany) or Lacey carbon grids (Ted Pella, Redding, CA) were used for TEM imaging.
Gene Synthesis
Genes encoding PCE and PCD were first synthesized using recursive directional ligation by plasmid reconstruction, as described elsewhere.[34] In brief, we modified a pET-24+ cloning vector (m-pET-24+) to contain endonuclease recognition sites for AcuI, BseRI, and BglI. We digested the modified vector with BseRI and ligated the desired ELP repeat sequence into the vector. The ELP sequence was created by annealing together complementary ssDNA strands that encode for the desired amino acid sequence along with “sticky end” overhangs. An amber stop codon (DNA sequence “TAG”) was introduced as an oligonucleotide into a second population of m-pET-24+ between the start and stop codons by digesting with BseRI. The vector containing the ELP sequence was then digested with AcuI and BglI to create an “A” population and the TAG-containing vector was digested with BseRI and BglI to create a “B” population. The two populations of cut vectors are complementary to one another such that when ligated together the ELP sequence and TAG codon are seamlessly encoded between the start and stop codon of m-pet-24+ vector. This process was repeated to form populations of an “A” cut of ELP-TAG and a “B” cut of ELP-TAG that were subsequently ligated together to double the length of the base repeat sequences. This method was continued until the desired length of the gene was obtained. The genes encoding the final sequences of PCE and PCD (see Table S1, S2) were confirmed by DNA sequencing. The gene encoding NCE (non-crosslinkable ELP) was constructed with the same protocol and confirmed by DNA sequencing (Table S1, S2).
We replaced the T7 promoter and terminator sequences of m-pET-24+ with a pTac promoter and rrnB terminator, with vector re-construction performed by GenScript USA Inc. (Piscataway, NJ). This vector, m-pTac, is compatible with expression in the genomically recoded E. coli. We transferred the genes of interest by digesting the PCE- and PCD-containing m-pET-24+ vectors with BseRI/BamHI, extracting the insert using agarose gel separation and purification, and ligating with similarly digested m-pTac. These vectors were then co-transformed with the modified pEvol tRNA/aaRS vector that contained two copies of the pAcFRS.1.t1 synthetase into the genomically recoded E. coli (both generously provided by F. J. Isaacs).
Protein Expression, Purification and Characterization
Liquid cell cultures (50 mL) of strains harboring pEvol and ELP plasmids were inoculated from frozen glycerol stocks and grown to confluence overnight. Cultures were then inoculated at 1:20 dilution in 2×YT media (1 L) supplemented with kanamycin (45 µg ml−1) and chloramphenicol (25 µg ml−1). aaRS expression was simultaneously induced by the addition of arabinose (0.2%) and pAzF (1 mm). Cells were grown at 34 °C in a shaking incubator at 200 r.p.m. for 6 h, at which time ELP expression was induced by the addition of IPTG (1 mm), and the cultures incubated at 34 °C for an additional 18 h. Cell pellets were harvested by centrifugation at 3500×g and resuspended in 1×PBS (20 mL). Cells were lysed by sonication for a total of 3 min (Misonix; Farmingdale, NY) and DNA was precipitated by addition of polyethylenimine (10%; MP Biomedicals, Santa Ana, CA). Precipitated DNA and cellular debris were removed by centrifugation at 20,000×g at 4 °C. Proteins were then purified using four rounds of inverse transition cycling, as described elsewhere.[63, 64] Briefly, solutions of proteins were heated and salt (NaCl) was added to induce the phase transition of the ELP, centrifuged to collect all insoluble material at 35 °C, 20,000×g (“hot spin”), and re-suspended in cold 1×PBS. Upon cooling, the ELP resolubilizes, while contaminants remain insoluble and can be removed by centrifugation at 35 °C, 20,000×g (“cold spin”). Protein purity was characterized by 4–20% gradient Tris-HCl (Biorad, Hercules, CA) sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and staining with copper chloride (0.5 m; Fisher Scientific, Hampton, NH). Protein yield was determined gravimetrically after dialysis into Nanopure water and lyophilization. Incorporation of pAzF was qualitatively confirmed by fluorescent labeling with excess dibenzocyclooctyne-Cy5 (DBCO-Cy5) followed by separation on SDS-PAGE and imaging on a Typhoon 9410 Variable Mode Imager (GE Healthcare, Pittsburgh, PA). All protein samples were kept in the dark to protect pAzF from ambient light.
Temperature-controlled Spectrophotometry
Cloud point transition temperatures (Tt) were determined by temperature-controlled spectrophotometry using a Cary 300 (Agilent Technologies). Samples containing various concentrations of the polypeptide in 1×PBS were heated at 1 °C min−1 and the optical turbidity at 650 nm was recorded every 0.1 °C. The cloud point was determined as the maximum of the first derivative of the turbidity as a function of temperature.
Droplet Formation
To create water-in-oil emulsion droplets, two liquid phases – a dispersed, aqueous phase of ELPs in 1×PBS and an organic, continuous phase comprised of TEGOSOFT® DEC/ABIL® EM 90/mineral oil (75%/5%/20% vol/vol) – were injected into the microfluidic droplet generator at constant flow rates using precision syringe pumps. The flow rates of the dispersed and continuous fluids were tuned to ensure droplet formation in the dripping regime; in these experiments, the dripping regime was achieved using a constant flow rate of 250 µL hr−1 for the organic continuous phase and 75–100 µL hr−1 for the aqueous, dispersed phase. The production of droplets within the microfluidic device was monitored using a 5× objective on an inverted microscope (Leica) equipped with a digital microscopy camera (Lumenera Infinity 3-1 CCD).
Particle Extraction from Droplets
Microscale particles were extracted from droplets using isobutanol to disrupt the water/oil phase boundary and create two continuous phases. Isobutanol was added at a ratio of 10:1 with droplets in solution and vortexed gently to mix. Droplet rupture and therefore particle extraction was evident by a change in the solution from turbid and yellow-white to clear and colorless. This solution was then centrifuged for 1 min at 20,000×g and the supernatant removed. The remaining pellet contained the stable, crosslinked particles and was resuspended in PBS or water for SEM imaging. Particle integrity, shape, and size were confirmed to be intact and unchanged before and after droplet extraction via light microscopy.
Mass Spectrometry
Solutions of PCE and PCD (20 µm) were prepared in 5% acetonitrile/0.2% formic acid/water and mass spectra acquired on an Agilent 1100 LC/MSD Trap SL (Agilent Technologies, Santa Clara, CA). Samples were injected into a Phenomenex Luna C18 column (50 × 1 mm, 3 µm; 0.2% formic acid in water as buffer A, 0.2% formic acid in acetonitrile as buffer B) and then into the mass spectrometer using a fully automated system. Spectra were acquired in positive mode followed by analysis and deconvolution using LC/MSD Trap Data Analysis software (Agilent Technologies, Santa Clara, CA). Mass spectra were acquired at the Mass Spectrometry shared facility at Duke University.
Ultraviolet-initiated Crosslinking
PCE and PCD were UV-crosslinked using an Omnicure® Series 1000 lamp (Ontario, Canada) equipped with a liquid light guide and a 250 – 450 nm filter set. PCD micelles were crosslinked to form nano-gels by heating a solution of PCD in 1×PBS to 40 °C for 5 min to ensure micellization, and then exposed to 50% intensity UV for 10 s. Similarly, emulsions containing PCE or mixtures of PCE + PCD were brought to a desired temperature (T = 40 °C for photocrosslinking PCD + PCE mixtures, T = 15 °C for photocrosslinking soluble PCE, T = 30 °C for photocrosslinking PCE coacervates) on the Linkam LTS120 precision Peltier heating and cooling microscope stage and subsequently exposed to 50% intensity UV for 10 s.
Light Scattering
Dynamic light scattering (DLS) measurements were performed over a temperature range of 15−50 °C using a Wyatt DynaPro temperature-controlled instrument (Wyatt Technology, Santa Barbara, CA). Samples for the DLS system were prepared in 1×PBS and filtered through Whatman Anotop sterile syringe filters (0.2 µm; GE Healthcare Life Sciences, Pittsburgh, PA), into a quartz crystal cuvette (12 µL; Wyatt Technology, Santa Barbara, CA). Five acquisitions were taken at each temperature, and the results presented represent the mean Rh or Dh of the sample at each temperature. The error bars represent the polydispersity percentage determined from the five data points taken at each temperature. It should be noted that the sizing measurements on all PCD + PCE combination mixtures demonstrated a small percentage of PCD nanoscale micelles and, at some temperatures, larger populations that accounted for small percentages of the overall composition. These data points are not presented in Figure 3D.
Static light scattering (SLS) measurements were performed using an ALV/CGS-3 goniometer system (Langen, Germany). Samples for the ALV/CGS-3 goniometer system were prepared in 1×PBS and filtered through Whatman Anotop sterile syringe filters (0.2 µm) into a disposable borosilicate glass tube (10 mm; Fischer Scientific, Pittsburgh, PA). Simultaneous SLS measurements were obtained at angles between 30°–150° at 5° increments, with each angle consisting of 3 runs for 15 s, at 37 °C. The differential refractive index (dn dc−1) was determined by measuring refractive index at five different concentrations at 37 °C using an Abbemat 500 refractometer (Anton Paar, Graz, Austria). Static light scattering data were analyzed by partial Zimm plots using the ALVSTAT software to determine the radius of gyration (Rg) and molecular weight. Nagg was determined by dividing the particle molecular weight by molecular weight of an individual PCD chain, as calculated from the amino acid sequence.
Atomic Force Microscopy
Samples for atomic force microscopy (AFM) imaging were prepared by placing a drop (~20 µl) of sample solution (6 mg ml−1 in water) onto a freshly cleaved mica surface. The samples were then incubated for 15 min and subsequently gently rinsed with Milli-Q H2O and dried with N2 gas. AFM images were acquired in Tapping Mode under ambient conditions using a Nanoscope MultiMode AFM (Bruker). TappingMode silicon cantilevers were used for all the AFM images (kF = 40 N mol−1, fres = 300 kHz). The particle sizes were determined using ImageJ open source software.
Cryogenic Transmission Electron Microscopy (Cryo-TEM)
Cryo-EM imaging was performed on a Tecnai G2 Spirit BioTWIN (FEI-Company, Eindhoven, the Netherlands, and Hillsboro, OR) operated at 120 kV. xPCD samples were adsorbed onto holey carbon grids. Either 2.0 µm holes, 2.0 µm spacing, Quantifoil grids (Quantifoil Micro Tools GmbH, Großlöbichau, Germany) or Lacey carbon grids (Ted Pella, Redding, CA) covered with a thin continuous carbon film were used. Samples were vitrified using an FEI Vitrobot Mark IV device. xPCD samples were loaded onto the grid in the Vitrobot chamber at RT (22°C) with the relative humidity set to 100%. Samples were blotted for 2 s with a force set to 3, plunged directly into liquid ethane and transferred into liquid nitrogen. Grids were then transferred into a Gatan 626 cryo-holder (GATAN Inc. Pleasanton, CA) and inserted into the TEM. 2D micrographs were acquired using an FEI Eagle 4k×4K CCD camera in low-dose condition at varying magnifications with a dose not exceeding 15 e Å−2 s−1.
Scanning Electron Microscopy (SEM)
Microparticles were extracted from droplets and resuspended in water. An aliquot of sample (5 µL) was drop-cast onto a silicon wafer attached to an aluminum stub with double-sided adhesive carbon tape and allowed to air dry for at least 4 hours. The dried samples were sputter-coated with gold for 250 s (Denton Desk IV, Moorestown, NJ) and then imaged with an FEI XL30 SEM-FEG at 7 kV. Mesoparticles were crosslinked in bulk and diluted 1:100 in water prior to sample preparation.
Heating and Optical Imaging
The PCE or PCD + PCE emulsion samples were collected on a glass microscope slide and heated using a precision Peltier heating and cooling stage (Linkam LTS120) equipped with a Linkam PE95 digital temperature control unit. The spatial distribution of Alexa Fluor 488-labeled (25% molar fraction N-terminal labeled) PCE was characterized via fluorescence microscopy using an upright Zeiss Axio Imager A2 microscope with a 20× objective and the appropriate filter set (ex 470/40, em 525/50). Particle sizing of visible microscale particles was done using MATLAB.
Supplementary Material
Acknowledgments
S.A.C. and J.R.S. contributed equally to this work. The authors would like to acknowledge Cedric Bouchet-Marquis (FEI-Company, Hillsboro, OR) for assistance with Cryo-TEM imaging. We are grateful for support from the National Institutes of Health (NIH) (R01 GM61232, A.C.), from the National Science Foundation (NSF) Research Triangle MRSEC (DMR-1121107), from DuPont, Inc. (F.J.I), from The Arnold and Mabel Beckman Foundation (F.J.I.), and the NSF Graduate Research Fellowship Program (DGF1106401, S.A.C., J.R.S.).
Footnotes
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Simone A. Costa, NSF Research Triangle Materials Research Science and Engineering Center, Department of Biomedical Engineering, Duke University, Durham, NC 27708, USA
Dr. Joseph R. Simon, NSF Research Triangle Materials Research Science and Engineering Center, Department of Biomedical Engineering, Duke University, Durham, NC 27708, USA
Prof. Miriam Amiram, Department of Biotechnology Engineering, Ben-Gurion University, Beer-Sheva, 8410501, P.O 653, Israel Department of Molecular, Cellular, and Developmental Biology, Yale University, New Haven, CT 06520, USA.
Dr. Lei Tang, NSF Research Triangle Materials Research Science and Engineering Center, Department of Mechanical Engineering and Materials Science, Duke University, Durham, NC 27708, USA
Prof. Stefan Zauscher, NSF Research Triangle Materials Research Science and Engineering Center, Department of Mechanical Engineering and Materials Science, Duke University, Durham, NC 27708, USA
Prof. Eric M. Brustad, NSF Research Triangle Materials Research Science and Engineering Center, Department of Chemistry, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA
Prof. Farren J. Isaacs, Department of Molecular, Cellular, and Developmental Biology, Yale University, New Haven, CT 06520, USA
Prof. Ashutosh Chilkoti, NSF Research Triangle Materials Research Science and Engineering Center, Department of Biomedical Engineering, Duke University, Durham, NC 27708, USA
References
- 1.Brett WG, Tong C, Santaneel G, Zhibing H, Arup N. Appl. Phys. Express. 2009;2:057001. [Google Scholar]
- 2.Debord JD, Lyon LA. J Phys. Chem. B. 2000;104:6327. [Google Scholar]
- 3.Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM. Nature. 2009;460:894. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McDaniel JR, MacEwan SR, Li X, Radford DC, Landon CD, Dewhirst M, Chilkoti A. Nano Lett. 2014;14:2890. doi: 10.1021/nl5009376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oh JK, Drumright R, Siegwart DJ, Matyjaszewski K. Prog. Polym. Sci. 2008;33:448. doi: 10.1016/j.progpolymsci.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan X, Zhu L, Wang K, Wang B, Wu Y, Xie W, Huang C, Chan BP, Du Y. Adv. Healthcare Mater. 2017;6:1601152. doi: 10.1002/adhm.201601152. [DOI] [PubMed] [Google Scholar]
- 7.Toma M, Jonas U, Mateescu A, Knoll W, Dostalek J. J Phys. Chem. C. 2013;117:11705. doi: 10.1021/jp400255u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamidi M, Azadi A, Rafiei P. Adv. Drug Delivery Rev. 2008;60:1638. doi: 10.1016/j.addr.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 9.Sharma N, Petri C, Jonas U, Dostalek J. Opt. Express. 2016;24:2457. doi: 10.1364/OE.24.002457. [DOI] [PubMed] [Google Scholar]
- 10.Liang L, Feng XD, Martin PFC, Peurrung LM. J Appl. Polym. Sci. 2000;75:1735. [Google Scholar]
- 11.Hoffmann J, Plotner M, Kuckling D, Fischer WJ. Sens. Actuators, A. 1999;77:139. [Google Scholar]
- 12.van den Brom CR, Anac I, Roskamp RF, Retsch M, Jonas U, Menges B, Preece JA. J Mater. Chem. 2010;20:4827. [Google Scholar]
- 13.Gan D, Lyon LA. J Am. Chem. Soc. 2001;123:7511. doi: 10.1021/ja010609f. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Angelatos AS, Caruso F. Chem. Mater. 2008;20:848. [Google Scholar]
- 15.Hartgerink JD, Beniash E, Stupp SI. Proc. Natl. Acad. Sci. U.S.A. 2002;99:5133. doi: 10.1073/pnas.072699999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Utada AS, Lorenceau E, Link DR, Kaplan PD, Stone HA, Weitz DA. Science. 2005;308:537. doi: 10.1126/science.1109164. [DOI] [PubMed] [Google Scholar]
- 17.a) Chu LY, Utada AS, Shah RK, Kim JW, Weitz DA. Angew. Chem. Int. Ed. Engl. 2007;46:8970. doi: 10.1002/anie.200701358. [DOI] [PubMed] [Google Scholar]; b) Chu LY, Utada AS, Shah RK, Kim JW, Weitz DA. Angew. Chem. 2007;47:9128. doi: 10.1002/anie.200701358. [DOI] [PubMed] [Google Scholar]
- 18.Nie Z, Li W, Seo M, Xu S, Kumacheva E. J Am. Chem. Soc. 2006;128:9408. doi: 10.1021/ja060882n. [DOI] [PubMed] [Google Scholar]
- 19.Dreher MR, Simnick AJ, Fischer K, Smith RJ, Patel A, Schmidt M, Chilkoti A. J Am. Chem. Soc. 2008;130:687. doi: 10.1021/ja0764862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shah RK, Kim JW, Agresti JJ, Weitz DA, Chu LY. Soft Matter. 2008;4:2303. [Google Scholar]
- 21.Roberts S, Dzuricky M, Chilkoti A. FEBS Lett. 2015;589:2477. doi: 10.1016/j.febslet.2015.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quiroz FG, Chilkoti A. Nat. Mater. 2015;14:1164. doi: 10.1038/nmat4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer DE, Chilkoti A. Biomacromolecules. 2004;5:846. doi: 10.1021/bm034215n. [DOI] [PubMed] [Google Scholar]
- 24.Ribeiro A, Arias FJ, Reguera J, Alonso M, Rodriguez-Cabello JC. Biophys. J. 2009;97:312. doi: 10.1016/j.bpj.2009.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urry DW, Hugel T, Seitz M, Gaub HE, Sheiba L, Dea J, Xu J, Parker T. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2002;357:169. doi: 10.1098/rstb.2001.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Urry DW. Prog. Biophys. Mol. Biol. 1992;57:23. doi: 10.1016/0079-6107(92)90003-o. [DOI] [PubMed] [Google Scholar]
- 27.Urry DW, Haynes B, Harris RD. Biochem. Biophys. Res. Commun. 1986;141:749. doi: 10.1016/s0006-291x(86)80236-4. [DOI] [PubMed] [Google Scholar]
- 28.Urry DW, Luan CH, Parker TM, Gowda DC, Prasad KU, Reid MC, Safavy A. J Am. Chem. Soc. 1991;113:4346. [Google Scholar]
- 29.Meyer DE, Chilkoti A. Nat. Biotechnol. 1999;17:1112. doi: 10.1038/15100. [DOI] [PubMed] [Google Scholar]
- 30.Urry DW. J Phys. Chem. B. 1997;101:11007. [Google Scholar]
- 31.Fluegel S, Fischer K, McDaniel JR, Chilkoti A, Schmidt M. Biomacromolecules. 2010;11:3216. doi: 10.1021/bm100965y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDaniel JR, Radford DC, Chilkoti A. Biomacromolecules. 2013;14:2866. doi: 10.1021/bm4007166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacEwan SR, Chilkoti A. Biopolymers. 2010;94:60. doi: 10.1002/bip.21327. [DOI] [PubMed] [Google Scholar]
- 34.McDaniel JR, Mackay JA, Quiroz FG, Chilkoti A. Biomacromolecules. 2010;11:944. doi: 10.1021/bm901387t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chung C, Lampe KJ, Heilshorn SC. Biomacromolecules. 2012;13:3912. doi: 10.1021/bm3015279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raphel J, Parisi-Amon A, Heilshorn S. J Mater. Chem. 2012;22:19429. doi: 10.1039/C2JM31768K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim DW, Nettles DL, Setton LA, Chilkoti A. Biomacromolecules. 2007;8:1463. doi: 10.1021/bm061059m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leitner A, Walzthoeni T, Kahraman A, Herzog F, Rinner O, Beck M, Aebersold R. Mol. Cell Proteomics. 2010;9:1634. doi: 10.1074/mcp.R000001-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Preston GW, Wilson AJ. Chem. Soc. Rev. 2013;42:3289. doi: 10.1039/c3cs35459h. [DOI] [PubMed] [Google Scholar]
- 40.Sinz A. Mass Spectrom. Rev. 2006;25:663. doi: 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- 41.Chin JW, Santoro SW, Martin AB, King DS, Wang L, Schultz PG. J Am. Chem. Soc. 2002;124:9026. doi: 10.1021/ja027007w. [DOI] [PubMed] [Google Scholar]
- 42.Gritsan NP, Platz MS. Chem. Rev. 2006;106:3844. doi: 10.1021/cr040055+. [DOI] [PubMed] [Google Scholar]
- 43.Gritsan NP, Platz MS. Org. Azides. John Wiley & Sons, Ltd; 2009. p. 311. [Google Scholar]
- 44.Lajoie MJ, Rovner AJ, Goodman DB, Aerni HR, Haimovich AD, Kuznetsov G, Mercer JA, Wang HH, Carr PA, Mosberg JA, Rohland N, Schultz PG, Jacobson JM, Rinehart J, Church GM, Isaacs FJ. Science. 2013;342:357. doi: 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Isaacs FJ, Carr PA, Wang HH, Lajoie MJ, Sterling B, Kraal L, Tolonen AC, Gianoulis TA, Goodman DB, Reppas NB, Emig CJ, Bang D, Hwang SJ, Jewett MC, Jacobson JM, Church GM. Science. 2011;333:348. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gan R, Perez JG, Carlson ED, Ntai I, Isaacs FJ, Kelleher NL, Jewett MC. Biotechnol. Bioeng. 2017;114:1074. doi: 10.1002/bit.26239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amiram M, Haimovich AD, Fan C, Wang Y-S, Aerni H-R, Ntai I, Moonan DW, Ma NJ, Rovner AJ, Hong SH, Kelleher NL, Goodman AL, Jewett MC, Soll D, Rinehart J, Isaacs FJ. Nat. Biotechnol. 2015;33:1272. doi: 10.1038/nbt.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pham ND, Parker RB, Kohler JJ. Curr. Opin. Chem. Biol. 2013;17:90. doi: 10.1016/j.cbpa.2012.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeForest CA, Tirrell DA. Nat. Mater. 2015;14:523. doi: 10.1038/nmat4219. [DOI] [PubMed] [Google Scholar]
- 50.Van't Veer IL, Leloup NO, Egan AJ, Janssen BJ, Martin NI, Vollmer W, Breukink E. Chembiochem. 2016;17:2250. doi: 10.1002/cbic.201600461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang K, Diehl MR, Tirrell DA. J Am. Chem. Soc. 2005;127:10136. doi: 10.1021/ja051457h. [DOI] [PubMed] [Google Scholar]
- 52.Simon JR, Carroll NJ, Rubinstein M, Chilkoti A, Lopez GP. Nat. Chem. 2017;9:509. doi: 10.1038/nchem.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reddington S, Watson P, Rizkallah P, Tippmann E, Jones DD. Biochem. Soc. Trans. 2013;41:1177. doi: 10.1042/BST20130094. [DOI] [PubMed] [Google Scholar]
- 54.Sletten EM, Bertozzi CR. Acc. Chem. Res. 2011;44:666. doi: 10.1021/ar200148z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hassouneh W, Zhulina EB, Chilkoti A, Rubinstein M. Macromolecules. 2015;48:4183. doi: 10.1021/acs.macromol.5b00431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McDaniel JR, Weitzhandler I, Prevost S, Vargo KB, Appavou MS, Hammer DA, Gradzielski M, Chilkoti A. Nano Lett. 2014;14:6590. doi: 10.1021/nl503221p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patenaude M, Hoare T. ACS Macro Lett. 2012;1:409. doi: 10.1021/mz200121k. [DOI] [PubMed] [Google Scholar]
- 58.Yan Q, Hoffman AS. Polymer. 1995;36:887. [Google Scholar]
- 59.Nakayama M, Okano T, Miyazaki T, Kohori F, Sakai K, Yokoyama M. J Controlled Release. 2006;115:46. doi: 10.1016/j.jconrel.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 60.Coughlan DC, Quilty FP, Corrigan OI. J Controlled Release. 2004;98:97. doi: 10.1016/j.jconrel.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 61.Rubinstein M, Colby R. Polymer Physics. Oxford; 2003. [Google Scholar]
- 62.Chu LY, Kim JW, Shah RK, Weitz DA. Adv. Funct. Mater. 2007;17:3499. [Google Scholar]
- 63.Meyer DE, Chilkoti A. Protein-Protein Interact. Cold Spring Harbor Laboratory Press; 2002. p. 329. [Google Scholar]
- 64.MacEwan SR, Hassouneh W, Chilkoti A. J Visualized Exp. 2014 doi: 10.3791/51583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meyer DE, Chilkoti A. Biomacromolecules. 2002;3:357. doi: 10.1021/bm015630n. [DOI] [PubMed] [Google Scholar]
- 66.McPherson DT, Xu J, Urry DW. Protein Expression Purif. 1996;7:51. doi: 10.1006/prep.1996.0008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.