Abstract
Viral fusogens merge viral and cell membranes during cell penetration. Their ectodomains drive fusion by undergoing large-scale refolding, but little is known about the functionally important regions located within or near the membrane. Here, we report the crystal structure of the full-length glycoprotein B, the fusogen from Herpes Simplex Virus, complemented by electron spin resonance measurements. The membrane-proximal (MPR), transmembrane (TMD), and cytoplasmic (CTD) domains form a uniquely folded trimeric pedestal beneath the ectodomain, which balances dynamic flexibility with extensive, stabilizing membrane interactions. Hyperfusogenic mutations within the CTD destabilize it, targeting trimeric interfaces, structural motifs, and membrane-interacting elements. Thus, we propose that the CTD trimer observed in the structure stabilizes gB in its prefusion state despite being appended to the postfusion ectodomain. Our data suggest a model for how this dynamic, membrane-dependent “clamp” controls the fusogenic refolding of gB.
Herpesviruses infect most of the world’s population for life by establishing latent infections from which they periodically reactivate. The three subfamilies of herpesviruses – α, β, and γ – have distinct replication strategies and pathogenesis. α-herpesviruses, which include Herpes Simplex viruses type 1 and 2 (HSV-1 and HSV-2), cause skin lesions, encephalitis1, and keratitis2. β-herpesviruses, including cytomegalovirus (CMV), are frequently asymptomatic but cause disseminated infections in immunocompromised patients, e.g., transplant recipients3, and developmental abnormalities in neonates4. γ-herpesviruses, which include Epstein-Barr virus5 (EBV), are linked causally to several cancers.
All herpesviruses share similar virion structures and penetrate cells by utilizing multiple viral and host proteins6 to catalyze the merger of their viral envelope with a host cell membrane. In HSV-1 and HSV-2, fusion requires four essential viral glycoproteins – gB, gD, gH, and gL – in addition to host receptors for gD and, potentially, other viral and host molecules6. Fusion by other herpesviruses also depends upon the core proteins gB, gH, and gL, paired with different tropism-determining partners7.
The conserved surface glycoprotein B (gB) is a fusogen. Viral fusogens are Type I membrane proteins anchored in the viral envelope that merge the viral and cell membranes during cell entry. They engage both membranes and are thought to provide energy to drive fusion by refolding from a high-energy prefusion to a low-energy postfusion conformation8. Structures of the soluble extracellular portions (or ectodomains) of many viral fusogens in both conformations revealed large-scale fusogenic rearrangements and pinpointed the location of the hydrophobic fusion peptides, or loops, that bind target cell membranes. Several crystal structures of gB ectodomains in the postfusion conformation9–12 and a cryoET reconstruction of an alternative conformation of HSV-1 gB on the surface of exosomes are available.
In addition to the ectodomain, gB contains three other regions: an external membrane-proximal region (MPR), single-pass transmembrane domain (TMD), and intraviral, or cytoplasmic, domain (CTD). These domains represent ~20% of the polypeptide and are essential for fusion13–16. The CTD also restrains the fusogenic activity of gB because point mutations, insertions, or truncations within this domain increase cell-cell fusion in the context of both infected cells and uninfected cells transfected with glycoproteins, and are referred to as hyperfusogenic14,17–19. But, in the absence of any structural information, the mechanistic contributions of the MPR-TMD-CTD to the gB-mediated fusion process remain unexplained.
Here, we report the crystal structure of full-length gB from HSV-1, in which the MPR-TMD-CTD forms a uniquely folded trimeric pedestal underneath the ectodomain. The structure and complementary electron spin resonance measurements reveal that the dynamic nature of this pedestal is offset by extensive, stabilizing membrane interactions. While the observed CTD trimer is appended to the postfusion ectodomain, hyperfusogenic mutations target either trimeric interfaces within the CTD or its membrane-interacting elements. Thus, the hyperfusogenic phenotype of these mutants can only be rationalized if this structure exists in the prefusion conformation. We propose that the ordered, membrane-bound CTD acts as a clamp that restrains the fusogenic activity of gB by stabilizing the ectodomain in prefusion conformation.
RESULTS
gB structure determination
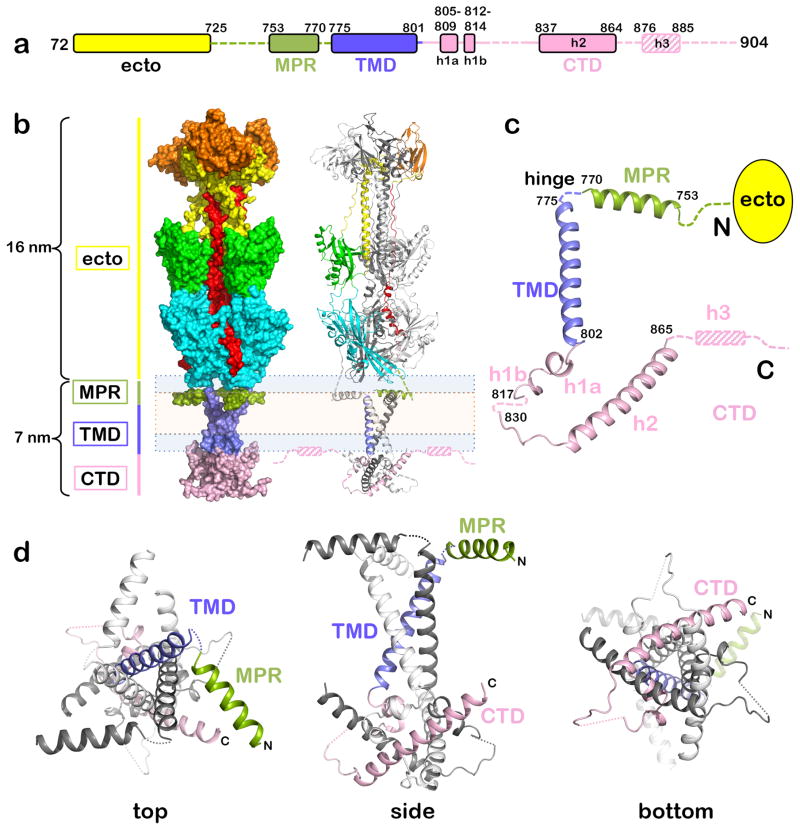
Construct gBΔ71, which lacks the signal peptide and proteolytically sensitive N terminus (Fig. 1a), was crystallized in a mixture of detergent and short amphipathic polymers (amphipols)20, yielding two crystal forms (Supplementary Fig. 1, Table 1). The 3.6-Å P321 structure and the 4.1-Å H32 structure were determined by molecular replacement using the ectodomain structure9 as a search model. The MPR and most of the TMD were resolved in both crystal forms likely due to the restriction of TMD movement by multiple interactions with the pendant hydrophobic groups of the amphipol, the “Gulliver effect” phenomenon20. However, the TMD C terminus and the CTD were resolved only in the P321 structure, presumably, due to favorable crystal contacts (Supplementary Fig. 1). The more complete P321 structure was thus used in all subsequent analyses.
Figure 1.
The structure of gBΔ71. (a) Bar diagram of the crystallized portion of gBΔ71. Unresolved regions are shown as dashed lines. Helix h3, unresolved in the crystals and identified from ESR experiments, is denoted by the hashed box. (b) The structure of gBΔ71 is shown in surface (left) and cartoon (right) representations. The ectodomain rests atop a pedestal formed by the MPR-TMD-CTD trimer. All three protomers are colored by domain in surface view, whereas in cartoon view, only one protomer is colored by domain. The inferred location of the membrane is shown schematically using peach (hydrophobic core) and light blue (polar head group) boxes. (c) Secondary structure elements of the MPR-TMD-CTD pedestal are shown in an isolated protomer. (d) Three views of the trimeric MPR-TMD-CTD pedestal, in which one protomer is colored by domain.
Table 1.
Data collection and refinement statistics (molecular replacement)
| FL-gBΔ71 (5V2S)a | FL-gBΔ71 (6BM8) | |
|---|---|---|
| Data collection | ||
| Space group | P321 | H32 |
| Cell dimensions | ||
| a, b, c (Å) | 118.66, 118.66, 216.46 | 118.60, 118.60, 800.58 |
| α, β, γ(°) | 90, 90, 120 | 90, 90, 120 |
| Resolution (Å) | 108.23–3.60 (3.73–3.60)b | 101.87–3.99 (4.37–3.99) |
| Rmerge | 0.3297 (1.833) | 0.109 (1.221) |
| Rmeas | 0.3256 (1.998) | 0.131 (1.46) |
| Rpim | 0.1196 (0.6654) | 0.072 (0.790) |
| I/σ(I) | 6.5 (1.15) | 7.00 (1.04) |
| CC1/2 | 0.98 (0.61) | 0.999 (0.506) |
| Completeness (%) | 99.70 (99.76) | 99.00 (99.10) |
| Redundancy | 8.4 (8.5) | 3.3 (3.3) |
| Refinement | ||
| Resolution (Å) | 108.23–3.60 | 101.87–4.10 |
| No. reflectionsc | 21048 (1673) | 17450 (1742) |
| Rwork / Rfree | 0.2444/0.2732 | 0.2686/0.2855 |
| No. atoms | ||
| Protein | 5646 | 5257 |
| Ligandd | 70 | 95 |
| B factors | ||
| Protein | 119.10 | 181.30 |
| Ligand | 150.39 | 221.81 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.003 | 0.005 |
| Bond angles (°) | 0.65 | 0.99 |
FL-gBΔ71-P321, 1 crystal; FL-gBΔ71-H32, 1 crystal.
Values in parentheses are for highest-resolution shell.
Values in parentheses are for the test set.
Ligands are 3 glycans (5 N-acetyl-glucosamine moieties) in P321 structure or 4 glycans (6 N-acetyl-glucosamine moieties and 1 beta-mannose) in H32 structure.
The nearly full-length gBΔ71 is a ~23-nm-long trimer, in which the ectodomain spike rests upon a pedestal composed of the MPR, TMD, and CTD (Fig. 1b). While the ectodomain adopted the postfusion conformation9, the uniquely folded trimeric pedestal is new.
The MPR structure
The top of the pedestal is formed by three symmetry-related MPR helices positioned underneath the ectodomain and parallel to the membrane bilayer. The N terminus of the MPR, which connects the MPR helix to the ectodomain, is unresolved in the structure (Fig. 1). The MPR helix makes no obvious contacts with the rest of the protein and is probably buttressed by the mixed micelle cushion. The poor side chain densities and the high B factors in this region are consistent with this absence of restraining contacts and suggest that the MPR helix is dynamic. Three residues within the MPR helix, V764, G766, and F770, are invariant among herpesviruses (Supplementary Fig. 2a) and may be functionally important. HSV-1 gB containing point mutations of residues G766 or F770 does not complement gB-null virus despite adequate surface expression15.
The TMD trimer has an inverted teepee shape
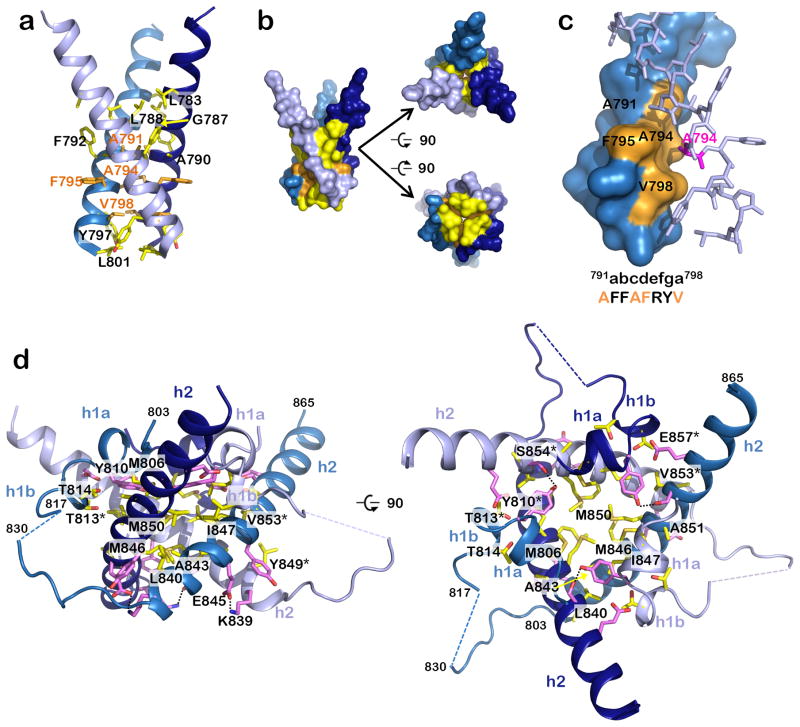
The TMD forms a nearly straight helix positioned perpendicular to the MPR helix (Fig. 1c). It includes a hydrophilic fragment previously assigned to the CTD that extends beyond the cytoplasmic face of the lipid bilayer (Fig. 1b). The three symmetry-related TMD helices cross at a ~46° angle to form a unique inverted teepee (Fig. 1, Fig. 2ab), in which each protomer buries ~770.5 Å2 of its accessible surface area (ASA). At the N terminus, the TMD helices are splayed and do not interact. Although the TMD trimer does not resemble a classic coiled-coil, its narrow C terminus (residues A791-V798) is stabilized by knob-in-hole packing (Fig. 2c). The close fit of the TMD helices is enabled by the small or absent side chains of G787, A790, A791, and A794 (Fig. 2a). These residues are completely conserved among α-herpesviruses implying a similar TMD arrangement (Supplementary Fig. 3). More distantly related β- and γ-herpesviruses (Supplementary Fig. 2a) have bulky residues at these positions, so their TMDs likely cross at different points and angles. Interactions between the hydrophilic C termini of the TMD helices and the CTD further stabilize this bundle. In the H32 structure, the TMD helices are more loosely packed, being unrestrained by the CTD (Supplementary Fig. 4).
Figure 2.
Residues involved in formation of the trimeric MPR-TMD-CTD pedestal. (a) The TMD helices interact exclusively through hydrophobic residues (yellow and orange side chains), which mediate both hydrophobic contacts (side chain atoms) and van der Waals contacts (main chain atoms). (b) Surface representation of “a” emphasizing contact between the helices, in three views. (c) A subset of the TMD interface residues participates in a stabilizing knob-in-hole interaction, where A791, A794, F795, and V798 (orange) form a hole into which the A794 (pink) knob of the neighboring protomer fits. (d) Side and top views of the CTD showing residues that form salt bridges and hydrogen bonds (pink) or hydrophobic interactions (yellow). Salt bridges and hydrogen bonds are indicated by dotted black lines.
The MPR-TMD hinge contains two invariant residues, P774 and G776 (Supplementary Fig. 2abc) but is unresolved and likely flexible. The greater MPR-TMD angle in the H32 structure (~100° vs. ~92°) further indicates that it is a dynamic region. This hinge is essential for proper gB folding15,21 and may help orient the MPR relative to the TMD.
The CTD forms an intertwined trimer
The resolved portion of the CTD – the CTD core – consists of two α helices (h1a and h2) and one 310 helix (h1b) (Fig. 1c). H1a and h1b form an elbow that juts outward towards the margin of the trimer while the long helix h2 crosses beneath the molecule and angles up toward the membrane. The unresolved linker connecting h1b and h2 is poorly conserved in sequence and length even among the closely related α-herpesvirus homologs and likely has a limited functional role because mutations within it, e.g., E830S/E831S, have no obvious effect on fusion efficiency22. The TMD, h1a, and h1b helices create a zigzag (Supplementary Fig. 2c). Residues P805 and P811 lie at the TMD/h1a and h1a/h1b junctions, respectively (Supplementary Fig. 2c), and likely stabilize this unusual structure, given the tendency of prolines to disrupt helices and create sharp bends23 in both soluble and membrane environments. These two prolines are the only invariant CTD core positions among all herpesviruses (Supplementary Fig. 2a), which suggests that the zigzag is an essential conserved structural element within the TMD-CTD module.
Below the proline zigzag, the remaining CTD core residues form a triangular base stabilized by multiple interactions (Fig. 2d, Fig. 3, Supplementary Table 1). Each protomer buries ~2,051 Å2 of ASA, with helix h2 embraced by helices h1a, h1b and h2 of one neighbor and contacting helices h1a, h1b, and h2 of the other neighbor (Fig. 1d, Fig. 2d). The trimeric CTD interface is highly conserved among α-herpesviruses (Supplementary Fig. 3, Supplementary Fig. 5) and is likely functionally important.
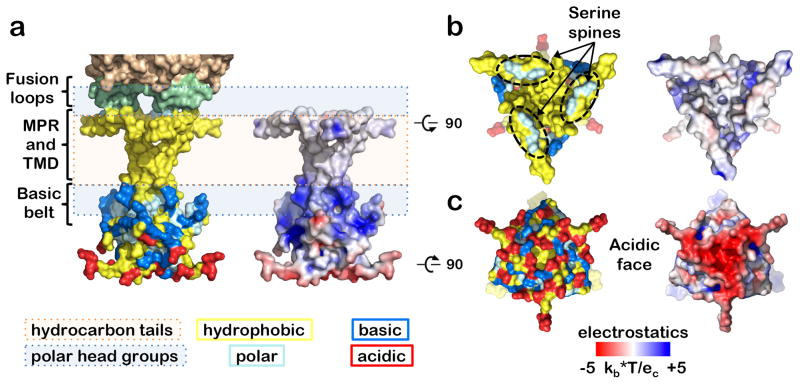
Figure 3.
Orientation of the MPR-TMD-CTD segment in the membrane. This orientation was inferred from residue distribution (left) and electrostatic surface potential (right) is shown in side (a), top (b), and bottom (c) views. Residues are shown as follows: hydrophobic (A, V, L, I, F, W, Y, G, P, M) in yellow, polar (S, T, N, Q, H) in cyan, acidic (D, E) in red, and basic (K, R) in marine. The inferred location of the membrane is shown schematically using peach (hydrophobic core) and light blue (polar head group) boxes. The hydrophobic MPR and TMD are buried in the nonpolar bilayer core except for the serine “spine” of the MPR, which protrudes into the polar headgroups of the outer leaflet. The basic belt in the TMD and the CTD interacts with the polar headgroups of the inner leaflet. The inner face of the CTD is largely acidic.
Charge distribution and orientation in the membrane
The placement of the MPR-TMD-CTD in the membrane was inferred from the surface distribution of hydrophobic, polar, and charged residues (Fig. 3a). Both the hydrophobic TMD and the hydrophobic MPR helix are embedded in the hydrocarbon core, with only the serine spine formed by the S762, S765, and S769 side chains protruding into the head groups of the outer leaflet (Fig. 3ab). This arrangement places the ectodomain fusion loops into the head groups of the outer leaflet, consistent with binding of the isolated gB ectodomain to membranes24. Such deep MPR placement within the membrane is expected in other homologs based on conservation of hydrophobic and polar residues (Supplementary Fig. 2a) and suggests that the MPR helices may promote fusion by facilitating lipid mixing.
On the opposite side of the bilayer, the side chains of R796 and R800 in the TMD and K807, R858, K862, K864, and K865 in the CTD form a positively charged “belt” (Fig. 3a). Most of these residues are highly conserved in α-herpesviruses, and even gB homologs from β- and γ-herpesviruses have at least one basic residue at the TMD C terminus and the h2 C terminus (Supplementary Fig. 2a). This “basic belt” could help brace the CTD against the membrane.
In contrast, the CTD base is negatively charged and rich in acidic residues (D834, D836, E837, E842, and E845) (Fig. 3c). Among 13 α-herpesvirus gB homologs (Supplementary Fig. 3), D836 is invariant and the other 4 residues are conserved. Substitutions that eliminate negative charge at several of these positions reduce the electrostatic potential of the CTD base. Yet, their distinct phenotypes imply that different sections of this “acidic face” have specific functional roles. The D836A mutant in HSV-2 gB is hyperfusogenic in virus-free cell-cell fusion assay25, and D836, located on the periphery of the CTD core, could participate in fusion by accepting fusion-triggering input from the gH cytotail22. Conversely, the E842S and E845S mutations in HSV-1 gB had no effect on fusion in the same assay22 and might instead provide a docking site for a positively charged binding partner, e.g., a tegument protein, during viral morphogenesis.
ESR analysis identifies a membrane-anchoring helix within the CTD C terminus
The CTD C terminus, residues 866-904, is unresolved in the crystal structure. This region restrains the fusogenic activity of gB because its truncation leads to a syncytial phenotype26. It contains a putative amphipathic helix h3 that forms in the presence of membranes and enables stable attachment of the isolated CTD to membranes19. To determine the structure of the CTD C terminus and its orientation within the membrane, we employed continuous-wave electron spin resonance spectroscopy (CW-ESR) on the isolated CTD, residues 801–904, bound to synthetic membrane vesicles. Single-cysteine CTD mutants from H861C to K885C were expressed in E. coli, purified, and spin-labeled. Mutations preserved gB fusogenicity, as assessed by virus-free cell-cell fusion assay (Supplementary Note 1, Supplementary Fig. 6), and likely did not globally perturb membrane interactions.
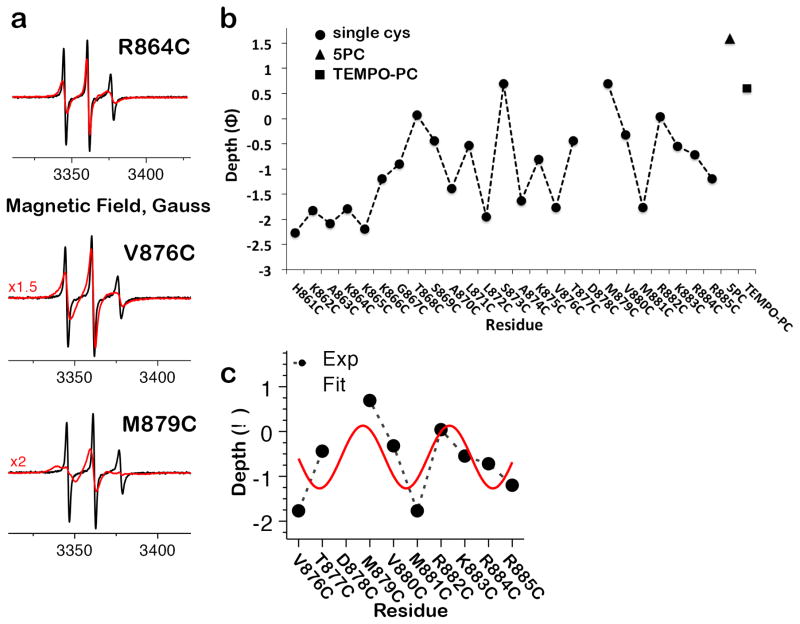
All CW-ESR spectra in solution displayed narrow, sharp peaks characteristic of a highly mobile spin label attached to unstructured protein (Fig. 4a). In contrast, the peaks of CW-ESR spectra collected in the presence of acidic liposomes were broadened to varying degrees, indicating restricted spin label mobility27 (Fig. 4a) and suggesting that residues H861-K885 become less dynamic upon membrane binding. The depth of spin label insertion (Fig. 4b) into the membrane was determined from CW-ESR microwave power saturation analysis of each position’s NiEDDA and O2 accessibility (Supplementary Fig. 7a). It follows a periodic pattern for V876-R884 with a period of ~3.65 residues, similar to the ideal α helix periodicity of 3.6, and we conclude that this span forms an amphipathic helix straddling the boundary of the head group and aqueous interface (Fig. 4c). The ESR-derived boundaries of h3 are consistent with secondary structure predictions and proteolytic sensitivity of the isolated CTD26, whereas the variable insertion depth along h3 suggests that it is kinked or otherwise distorted. The deepest residue, M879C, approaches the bilayer core location of the spin-labeled lipid 5PC) while the rest lie near the head group and aqueous interface. Structural features identified through the depth analysis are confirmed by the mobility pattern (Supplementary Fig. 7b), with the mobility of residues in h3 reflecting both their containment in this structural element and the extent to which membrane interactions confine them. Thus, binding of the unstructured CTD C terminus to the bilayer transforms it into a shallow membrane anchor composed of an amphipathic helix, one face of which interacts with the polar head groups. Amphipathic helices are also found in the HIV Env CTD28 and the fusion peptide of Influenza HA29, and may be employed by fusogens to grip the membranes they remodel.
Figure 4.
Membrane interactions of the CTD C terminus. (a) Representative CW-ESR spectra pairs for positions that display small (R864C), moderate (V876C) and large (M879C) mobility differences in the absence (black) and presence (red) of liposomes. Spectral broadening in the liposome-containing traces reflects secondary structure formation and the strength of membrane interactions at each position. All spectra were normalized to the same number of spins. In the plots, some of the spectra in lipid were scaled up by a factor of 1.5–2 for better visibility. (b) Dependence of the depth parameter (Φ) on spin-label position for CTD residues 861-885 and two spin-labeled lipid controls, 5PC and TEMPO-PC. (c) Periodic arrangement of residues 876-884 reflects their formation of the amphipathic helix h3, with one membrane-embedded and one solvent-exposed face.
DEER analysis of the free and membrane-bound CTD
Formation of helix h3 in the presence of membranes is accompanied by a global organization of the CTD, as determined by the double electron-electron resonance (DEER) spectroscopy on the isolated CTD. The CTD is a trimer in solution19, which allowed us to obtain inter-protomer distance distributions between symmetry-related spin-labels introduced by single cysteine mutations S803C, E816C, or E830C (Fig. 5a). Distance distributions for all mutants in solution were broad and centered at ~40 Å but narrowed in the presence of anionic liposomes (Fig. 5b), indicating that S803, E816 and E830 lie in highly dynamic regions that become more ordered upon membrane binding. This concurs with the helicity increase and proteolytic susceptibility decrease of the CTD in the presence of anionic liposomes26.
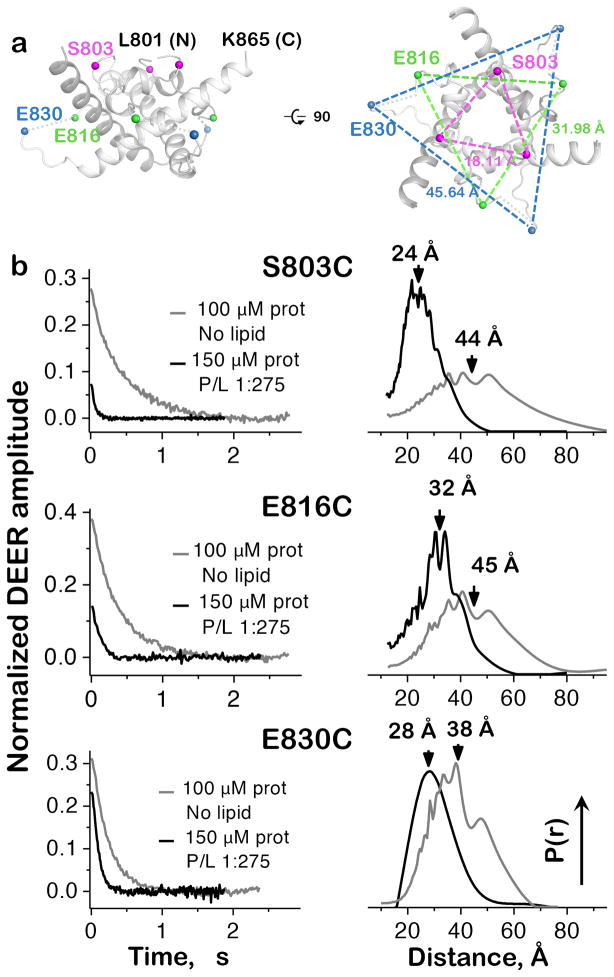
Figure 5.
Inter-protomer distances in the isolated CTD. (a) Positions of DEER labeling sites and their distances in the crystal structure of gBΔ71. Only residues 801-865 of the CTD are shown for clarity. (b) Experimental time-domain DEER data (left) and reconstructed inter-protomer distance distributions (right) for the CTD mutants S803C, E816C, and E830C. Protein in buffer alone is disordered, with broad, widely varying separation between equivalent residues on different protomers. Conversely, distance distributions for samples with a protein:PC/PA liposome ratio of 1:275 coalesce around shorter separation distances, indicating global organization of the CTD. Mutants S803C and E816C were tested once, while two biological replicates of E830C produced superimposable distance distributions.
Of the three CTD mutants, E816C displayed the closest agreement between the center of its membrane-bound DEER distance distribution and the E816(Cα)-E816′(Cα) crystal structure distance (where ′ denotes a neighboring protomer), with measurements of ~32 Å (Fig. 5b) and 31.98 Å (Fig. 5a), respectively. The distance distribution for membrane-bound S803C mutant was centered at ~24 Å (Fig. 5b), which is longer than the S803(Cβ)-S803′(Cα) distance of 18.11 Å in the crystal structure (Fig. 5a). Similar differences between inter-spin and backbone measurements have been reported previously30,31 and typically reflect contributions from the length and relative positions of spin label tethers. The lower than expected signal intensity observed with both the membrane-bound S803C and E816C mutants (Fig. 5b) may indicate that the spin label at these positions destabilizes the folded CTD structure.
In the third mutant, E830C, the strong signal in the presence of membrane and the relatively broad distance distribution are consistent with its position in a flexible loop. Yet, while this distribution is centered at ~28 Å (Fig. 5b), the E830(Cα)-E830′(Cα) distance in the crystal structure is 45.64 Å (Fig. 5a). We hypothesize that when the CTD binds the membrane bilayer, the E830-containing loop becomes more ordered and compact. Additionally, crystal packing of gBΔ71 or the absence of the TMD in the ESR constructs could affect the conformation of this loop.
Fusion regulation by the CTD
The gBΔ71 crystal structure together with ESR measurements provides an excellent model for understanding fusion regulation by the CTD. Certain mutations in the CTD and the TMD C terminus result in enhanced or impaired cell-cell fusion. We classified all known fusion-altering mutations in HSV-1 or HSV-2 gB based on their location in and potential effect on the structure (Fig. 6, Supplementary Table 2), regardless of whether they were identified in clinical HSV isolates or engineered.
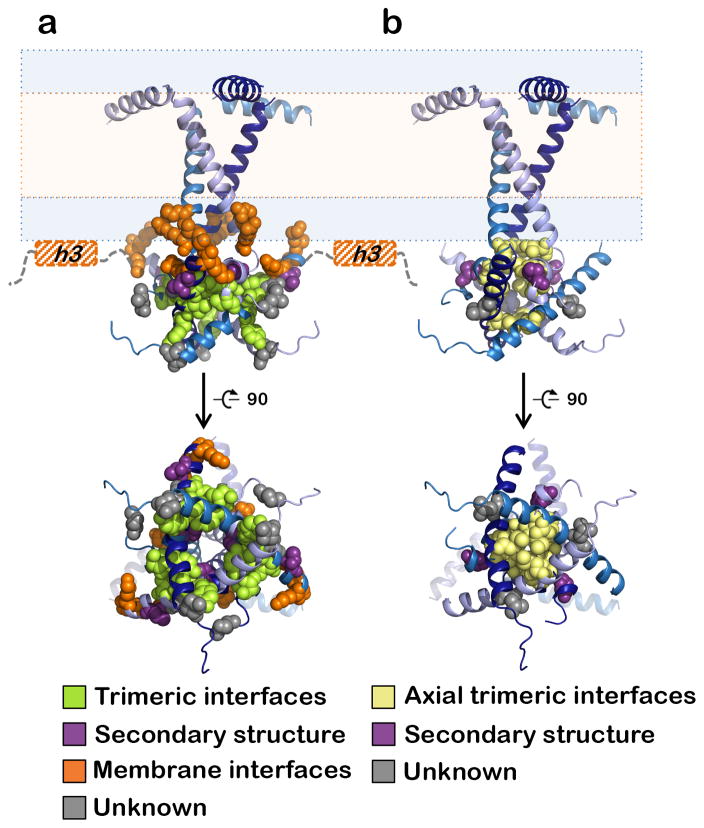
Figure 6.
Locations of fusion altering mutations within the MPR-TMD-CTD structure. Mutated residues are shown as spheres and colored according to the scheme below. (a) Hyperfusogenic mutations are grouped into 4 groups depending on their location and effect on the CTD. Truncations of the amphipathic h3 helix (hashed orange boxes) disrupt membrane interactions and are assigned to Group 3. (b) Surface expression of gB is eliminated by alanine substitutions at several positions, notably, hydrophobic residues lining the central CTD axis (yellow).
The largest group of hyperfusogenic mutations 14,17,22,25,32–36, group 1, targets residues located at the trimeric CTD interfaces. These mutations are predicted to disrupt interprotomer bonds including hydrogen bonds (Y810A25, Y849A14, S854F32, E857A25, E857D33), van der Waals contacts (T813I17) or hydrophobic contacts (V853A25) due to side chain substitutions. Group 2 contains mutations P805A25 and T859P37 that could indirectly destabilize trimeric interfaces by disrupting important structural elements, the zigzag structure and helix h2, respectively. Group 3 contains mutations that target membrane binding elements like the “basic belt”: R796A17 and R800A17 in the TMD and R858H34,38/R858C17, K864S22, and K865S22 in the CTD. These mutations probably weaken membrane interactions of the CTD by reducing the positive charge in the vicinity of the membrane. Group 3 also contains truncations of helix h318,26,39. Eliminating this important membrane anchor reduces the kinetic energy barrier that gB must overcome during pre-to-postfusion refolding22,40 in HSV-118,26,39 and EBV40–42 gB and may account for enhanced fusion in HIV Env truncation mutants43. Finally, group 4 contains mutations the hyperfusogenic phenotype of which cannot yet be readily explained. These include D836A25 and A855V35,44 in h2 as well as L817P36, L817H35, or insertions between E816 and L81734 in the region immediately preceding the unresolved CTD loop. All of these residues are located at the periphery of the CTD core and could accept fusion-triggering input from the gH cytotail22. Nevertheless, most hyperfusogenic mutations map to either the trimeric or the membrane interfaces and would be expected to disrupt either the CTD trimer or its membrane interactions.
Unlike hyperfusogenic mutants, CTD mutants that fuse poorly are uncommon and are typically expressed on cell surface at very low levels25 likely due to proteins misfolding. Structure analysis shows that most of these mutations shorten the side chains of hydrophobic residues along the trimeric CTD axis and likely eliminate interactions critical for basal CTD trimer stability (Fig. 6b). The only known CTD mutant that reduces the rate of viral entry despite adequate surface expression, A851V17, may impede fusion by increasing the number of hydrophobic contacts at the trimeric interface.
The clustering of hyperfusogenic mutations in the CTD indicates that one of its central functions is the stabilization of gB in its prefusion form. Given that these mutations map to key interfaces and structural motifs of the observed CTD structure, their phenotype can only be rationalized if this structure exists in the prefusion gB. We propose that the membrane-bound, trimeric CTD structure, observed in the crystals and inferred from the ESR data, is essential for restraining the fusogenic activity of gB.
DISCUSSION
Many viral fusogens require regions lying within or near the membrane for activity. In HIV Env, the MPER houses epitopes of several broadly-neutralizing antibodies45 whereas the TMD influences its conformation, stability, and antigenicity46. In Influenza HA, the TMD is required to bring fusion to completion47. Additionally, CTDs of herpesvirus gB17,41,48, HIV Env45, and paramyxovirus F49 can control the fusogenic activity of their ectodomains across the membrane. However, structural information on these juxta-membrane regions is limited. Structures of the isolated MPER and TMD of HIV Env have been determined by NMR46,50, but no structures for large, lipid-associated CTDs have been described. Moreover, the only available high-resolution structures of full-length fusogens are of class II fusogens from dengue51 and Zika52 viruses, which lack CTDs and have “hook-shaped” TMDs that do not span the membrane.
Here we presented the most complete structure of a fusogen with a membrane-spanning TMD and a large CTD. The gB structure shows that the MPR-TMD-CTD forms a uniquely folded trimeric pedestal beneath the ectodomain that interacts extensively with the membrane. The structure defines the TMD boundaries more precisely and identifies regions of potential functional importance throughout the pedestal, including the MPR serine spine and the basic belt and acidic face of the CTD. Membrane binding organizes the CTD core and is accompanied by the formation of the stabilizing C-terminal amphipathic helix bilayer anchor.
Collectively, structure-based analysis of a large panel of fusion altering CTD mutations17,25,34–36,38 supports two important conclusions. First, CTD trimerization is essential for gB folding and surface expression, and proper regulation of fusogenic activity requires membrane-binding of this trimer. Second, because hyperfusogenic mutations both destabilize prefusion gB and disrupt elements that reinforce the observed CTD trimer structure, this CTD structure must stabilize the prefusion gB. Given that the gB ectodomain in our structure has adopted the postfusion conformation, we hypothesize that the CTD took on the prefusion conformation in the structure due under the influence of amphipol and favorable crystal packing. Nevertheless, we further hypothesize that the CTD adopts similar conformation in the prefusion and the postfusion state. Alternatively, the gBΔ71 structure is a hybrid of a postfusion ectodomain and the prefusion CTD.
We propose that the CTD – including the core observed in the crystals and the C-terminal amphipathic helix – constitutes an inhibitory membrane-dependent “clamp” that stabilizes the gB ectodomain in its prefusion conformation. Lacking this clamp, the isolated gB ectodomain adopts only the postfusion conformation9,53. Moreover, the fully folded CTD clamp requires intact membrane, which explains why even full-length gB adopts the postfusion conformation upon detergent solubilization (this work and 54). Solubilization disrupts the bilayer scaffold surrounding gB and destabilizes the CTD clamp, enabling the ectodomain to refold into the postfusion conformation. In agreement with this hypothesis, non-postfusion gB conformations have thus far only been observed in gB-containing exosomes55,56. Our results illustrate the need to study complete fusogens in a bilayer environment, especially those containing large membrane-anchoring segments.
The gB structure presented here may help illuminate its unusual activation mechanism. All viral fusogens face the challenge of deploying at the correct moment. Class I and class II fusogens rely upon priming systems that involve proteolytic cleavage of an inactive precursor or chaperone to prevent premature activation, while VSV G and gp64 from class III circumvent the issue through reversible conformational rearrangements8. Lacking both a priming “safety system” and the ability to regain its prefusion state, gB is instead stabilized in its prefusion conformation by its CTD and activated by other glycoproteins including the gH-gL heterodimer.
Although the ectodomain of the gH-gL is thought to activate gB by directly binding its ectodomain6, the 14-residue gH cytotail is also essential for gB activation 57,58. Cell-cell fusion is reduced by truncations of the gH cytotail, and fusion levels are proportionate to the length of the remaining gH cytotail22, such that an 8-residue tail maintains efficient fusion, a 5-residue tail reduces it 2-fold, and further truncations yield progressively lower fusion until 1-residue tail achieves only background fusion levels. Although the precise role of the gH cytotail remains unclear, we hypothesize that the gH cytotail releases the gB clamp by acting as a wedge that disrupts either the CTD core or membrane interactions, and because its effectiveness is vastly reduced by truncation, gH cytotail must be able to reach a certain point on the gB CTD to do this22. Its potential targets could thus be any CTD feature that stabilizes the gB clamp; for example, interprotomer contacts in the CTD core or h3/membrane interactions. An extended 8-residue peptide would nearly span the height of the gB CTD and could interact with the CTD at multiple sites. Hyperfusogenic HSV-1 gB constructs still require activation by gH-gL and are similarly sensitive to gH tail length22 but, their mutated clamps are on a “hair-trigger” that is more easily released.
Combining our structure analysis and the published data, we propose a “clamp-and-wedge” model of HSV fusion. (Fig. 7). The CTD clamp initially restrains the gB ectodomain in its high-energy prefusion form by an unknown mechanism. Binding of gD to one of its receptors results in activation of the gH-gL heterodimer, prompting the gH cytotail wedge to interact with the CTD clamp and destabilize it in some manner. Freed from the CTD’s restraining influence, the prefusion ectodomain then refolds irreversibly into its thermodynamically favored postfusion conformation while the CTD returns to its initial conformation. This mechanism for releasing the CTD clamp may explain the participation of multiple glycoproteins in gB activation in both HSV-1 and other herpesvirus homologs. Although the CTD trimer must be destabilized, it does not need to come apart completely. Major ectodomain rearrangements coupled to restricted CTD motion could be accommodated by asymmetric refolding of the ectodomain8 and extensive rearrangement of the flexible MPR. However, if the observed CTD trimer represents solely its prefusion form, this domain could undergo permanent refolding or unfolding during fusion.
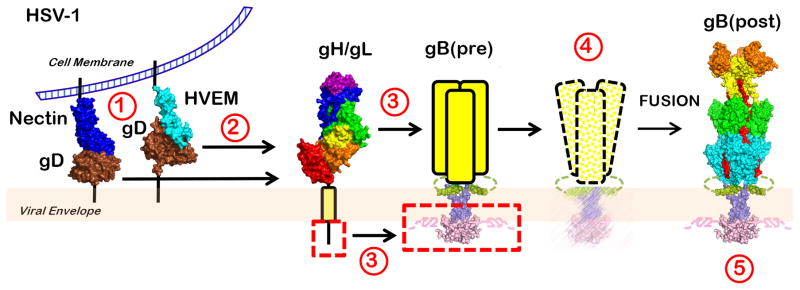
Figure 7.
The clamp-and-wedge model of HSV fusion. The CTD clamp restrains the gB ectodomain in its high-energy prefusion form. Binding of gD to one of its receptors (1) relieves it from an autoinhibited dimeric state (not shown) and results in activation of the gH/gL heterodimer (2). The ectodomain of activated gH/gL interacts with the gB ectodomain, while the gH cytotail releases the clamp by wedging between the intertwined CTD protomers (3). The destabilized clamp loses its hold on the ectodomain, and gB proceeds to refold into its thermodynamically favored postfusion conformation, drawing the virion and cell membranes together (4). The clamp then returns into its initial conformation (5).
To influence the ectodomain conformation, the CTD must convey its own status through the intervening MPR and TMD. Extension of the TMD helices beyond the inner leaflet and into the CTD “space” could enable them to serve as levers to efficiently couple the CTD movements to the TMD. Indeed, when the CTD core is ordered, the TMD helices are fully assembled into an inverted teepee. However, a disordered CTD core enforces no such constraints on the TMD. This contrast is an extreme example of how rearrangements in the regulatory CTD might propagate across the membrane. Nevertheless, the same pathway could transmit smaller, functionally relevant conformational changes within the CTD due to syncytial mutations or in response to a fusion triggering signal.
How these changes are transmitted to the ectodomain through the MPR is less clear. The N terminus of the MPR is unresolved in both of our structures and, since the visible C terminus (the MPR helix) does not appear to contact the postfusion ectodomain extensively enough to influence either its stability or conformation, this domain likely looks very different in prefusion gB. Furthermore, if the prefusion and postfusion CTD conformations are similar, major rearrangement of the MPR (and possibly the TMD) might be necessary to couple it to the vastly different ectodomain conformations that are predicted.
Our model of full-length HSV-1 gB provides a first look at its fusion-restraining clamp as well as the most comprehensive picture of the membrane interacting domains of any fusogen containing a membrane-spanning TMD and a large CTD. Given the many ways in which the structure of this clamp is dependent on the surrounding viral envelope, we hypothesize that the structure and function of the herpesvirus fusogen gB is uniquely controlled by the membrane. This new paradigm provides a starting point for exploring the persistent mysteries that surround gB, such as the nature of its interaction with gH-gL and the structure of its prefusion conformation.
Online methods
Construction of gB CTD Mutants
Single cysteine CTD mutants were cloned in pET24b. Many mutants were subcloned into pKH52 (WT CTD with a C-terminal His6 tag) from equivalent FL-gB single cysteine plasmids with PstI and XhoI. Others were prepared using a Quickchange PCR strategy using the primers listed in Supplementary Table 3. Finally, some constructs were prepared using “splicing overlap extension” PCR (SOE PCR) to modify pKH52.
Construction of Full Length gB (FL-gB) for functional assays
Full-length gB (FL-gB) constructs with single-cysteine mutations were cloned in pCAGGS background. Mutants K862C, K864C, K865C, K866C, V876C, T877C, D878C, M879C, V880C, M881C, R882C, K883C, R884C, and R885C were generated in pPEP98 (WT FL-gB) by SOE PCR. Other FL-gB single-cysteine mutants, namely S803C, E830C, H861C, A863C, G867C, T868C, S869C, A870C, L871C, L872C, S873C, A874C, and K875C, were subcloned from their equivalent gB CTD single-cysteine mutant expression construct with SacI and XhoI. To facilitate this process, a modified pPEP98 construct called pRC30 was created in which an XhoI site was introduced after residue 904 while other SacI and XhoI sites were destroyed. This resulted in amino acids LE between residue 904 and the stop codon. All primers are listed in Supplementary Table 3.
Construction of gBΔ71
The crystallization construct gBΔ71 encodes residues 72-904 of full-length gB, an N-terminal honeybee melittin signal sequence and a C-terminal His6 tag in a pFastBac1 vector (pRC4). This construct lacks the signal peptide, residues 1-29, which is cleaved during protein maturation and is absent from the mature gB, and the proteolytically sensitive N terminus, residues 30–71, that causes heterogeneity in gB samples. To make pRC4, a segment of the gB CTD with a C-terminal His6 tag was excised from pKH11 using NheI and MfeI and subcloned into full-length, untagged gB (pEV34) generating FL-gB (pRC2). Next, SOE PCR using flanking primers 5′-CGCGGAATTCAAAGGCCTAC-3′ and 5′-CTCGACGATGCAGTTTACCG-3′ along with mutagenesis primers 5′-ATCTATGCGAACAAAAAACCGAAAAACCCAACG-3′ and 5′-GGTTTTTTGTTCGCATAGATGTAAGAAATGTACAC-3′ was used to remove codons 30–71.
Expression and Purification of gBΔ71
Recombinant baculovirus encoding C-terminally His6-tagged HSV-1 gBΔ71 was generated using Bac-to-Bac technology (ThermoFisher). Spodoptera frugiperda (Sf9) cells (ThermoFisher) were grown in Sf900-SFM media (ThermoFisher) to the density of 2X106 cells/mL and infected with recombinant baculovirus by adding 14 ml of the viral stock from the third passage (P3) to 1.4 L cells. 60 to 72 hrs post-infection, cells were harvested by centrifugation at 4,000 g and resuspended in 50 mM Tris pH 8.0, 150 mM NaCl, and 10% glycerol (solubilization buffer, SB) plus 0.1 mM PMSF. After an additional centrifugation, cell pellets were frozen at −80 °C.
Prior to purification, cell pellets were defrosted at 4 °C and resuspended in SB with 1 mM PMSF and 1x Roche protease inhibitor cocktail. The suspension was lysed by passing it three times through a M-110S microfluidizer, per manufacturer’s instructions, and clarified by centrifugation at 4,000 g for 25 minutes. Crude membrane fraction was isolated from the clarified lysate by ultracentrifugation at 150,000 g for 90 minutes and resuspended in 50 ml SB plus 0.1 mM PMSF using a dounce homogenizer. Membrane proteins were extracted by adding 1.2% w/v n-Dodecyl-β-D-Maltopyranoside (DDM, D310LA, Anatrace) and incubated with 1 ml Ni superflow resin (GE Healthcare) in the presence of 10 mM imidazole.
After 16-hr incubation, the resin was washed with 15 CV of 20 mM Tris pH 8, 150 mM NaCl, 5% glycerol, 0.05% DDM and 20 mM imidazole (WB1) and 25 CV of 20 mM Tris pH 8.0, 150 mM NaCl, 5% glycerol, 0.05% DDM and 35 mM imidazole (WB2). GBΔ71 was eluted with 20 mM Tris pH 7.5, 150 mM NaCl, 5% glycerol, 0.05% DDM and 300 mM imidazole (EB) and concentrated in Ultra-15 (MW cutoff 100 kDa) concentrator (Millipore EMD). gBΔ71 was further purified by size-exclusion chromatography using a Superdex 200 column (GE Healthcare) equilibrated with 20mM Tris-HCl pH 7.5, 150mM NaCl, 5% glycerol, and 0.05% DDM (GF Buffer).
Crystallization and structure determination
Initially, gBΔ71 crystallized in several maltosides, yet the MPR-TMD-CTD portion was largely unresolved in these crystals. To stabilize the membrane regions, we obtained crystals in the presence of amphipols. Protein in GF buffer was concentrated to 3.6 mg/ml using an Ultra-4 (MW cutoff 100 kDa) concentrator (Millipore EMD) and passed through a 0.1 μm Ultrafree centrifugal filter (Millipore EMD). The protein was supplemented with A8-35 (Anatrace), mixed gently, and incubated at room temperature for 30 minutes. Crystals were grown by vapor diffusion in hanging drops containing 1 μl of protein and 1 μl of reservoir solution. Crystals appeared after 3–4 weeks and grew for an additional week. Higher PEG 3350 concentrations (16–18%) favored the formation of crystals in the P321 space group, whereas 12–14% PEG 3350 yielded crystals in the H32 space group. Some drops in the middle of this range contained H32 crystals at their periphery and P321 crystals in their center, potentially due to uneven distribution of precipitant, detergent, and/or amphipol. While we hypothesize that association with A8-35 is necessary to stabilize the TMD and enable P321 packing of gB, this highly anionic amphipol is thought to hinder crystallization thereby requiring increased precipitant60. The P321 crystal was grown using gBΔ71 in 0.05% DDM and 0.01% A8-35 and reservoir solution containing 16% PEG 3350, 0.1 M HEPES pH 7.2, and 0.15 M sodium formate. The H32 crystal was grown using gBΔ71 in 0.075% UM and 0.0075% A8-35 and reservoir solution containing 11% w/v PEG 4000, 0.1 M Tris-HCl pH 7.5, and 0.1 M NaCl.
Cryoprotectant solution containing 20% glycerol was prepared by mixing 2 parts 50% glycerol with 3 parts reservoir solution. Crystals were cryoprotected by brief incubation in cryoprotectant followed by plunging into liquid N2. X-ray data was collected at 100K on beamline 24ID-E and 24ID-C at the Advanced Photon Source at the Argonne National Laboratories. Both the P321 and H32 data sets were processed using XDS61 as implemented in the Northeastern Collaborative Access Team (NE-CAT) software pipeline RAPD (https://rapd.nec.aps.anl.gov/rapd/).
A 3.6-Å P321 data set was used to determine the structure by molecular replacement in Phaser62 using the HSV-1 gB ectodomain (2GUM)9 as a search model. There is a single gB polypeptide in the asymmetric unit, and a gB trimer is generated by a 3-fold crystallographic symmetry operation. The initial electron density revealed the locations of the MPR, TMD, and the CTD, in addition to the ectodomain. The ectodomain adopted the postfusion conformation, presumably during membrane disruption caused by detergent extraction. The overall fold of the ectodomain within the gBΔ71 structure is very similar to that of the isolated gB ectodomain9, with RMSD of 1.00 over 576 residues. An initial MPR-TMD-CTD model was built manually into the experimental density using Coot63 and refined against data truncated to 3.6 Å. Prior to refinement, 8% of reflections were set aside for cross-validation. Model refinement included gradient minimization refinement of XYZ coordinates and individual thermal parameters with optimization of X-ray/stereochemistry and X-ray/ADP weights, as implemented in phenix.refine64. Secondary structure restraints were used throughout refinement. Rotamer restraints were imposed until the last stage of refinement. Iterative rounds of model rebuilding were carried out in Coot. The final model encompasses residues 104-865 (unresolved 71-103, 477-491, 726-752, 771-775, 818-829, 866-904, and the C-terminal His6 tag) and 4 N-acetyl-glucosamine moieties at residues N141, N398, and N674. Sequence 754-770 was assigned to a helical density within the MPR region in the vicinity of the TMD, but due to poor side chain density, its precise register is unclear. The unresolved loop following residue 817 was also paired with the nearest residue 830, rather than this position in more distant symmetry mates. This path is suggested by the weak electron density, and of the three possible pairings, it is the only one that does not require the polypeptide chain to make a long detour. According to Molprobity as implemented in phenix.refine64, 95.7% residues lie in the most favored regions of the Ramachandran plot and 4.3% lie in the additionally allowed regions. Relevant crystallographic statistics are listed in Table 1.
A 4.0-Å H32 data set was used to determine the structure by molecular replacement in Phaser62 using the HSV-1 gB ectodomain structure (2GUM) 9 as a search model. As in P321 crystals, there is a single gB polypeptide in the asymmetric unit, and a gB trimer is generated by a 3-fold crystallographic symmetry operation. While the MPR and most of the TMD were visible in the initial electron density, in addition to the ectodomain, the CTD was unresolved. The model of MPR-TMD was built manually into the experimental density using Coot and refined against 4.1-Å data. Prior to refinement, 10% of reflections were set aside for cross-validation. Model refinement included gradient minimization refinement of XYZ coordinates and individual thermal parameters, all as implemented in phenix.refine. Secondary structure restraints were used throughout refinement. Rotamer restraints were imposed until the last stage of refinement. Iterative rounds of model rebuilding were carried out in Coot. The final model encompasses residues 103-794 (unresolved 71-101, 483-489, 731-752, 796-904, and the C-terminal His6 tag) and 7 N-acetyl-glucosamine moieties at residues N141, N398, N430, and N674. As in the P321 structure, the precise location of residues 753-770 cannot be ascertained. But, residues 726-730 and the hinge between the MPR and the TMD, residues 771-774, were resolved in the H32 structure, in contrast to the P321 structure. According to Molprobity, 92.56% residues lie in the most favored regions and 7.13% lie in the additionally allowed regions of the Ramachandran plot. Two residues, P130 and P774, are Ramachandran outliers. Relevant crystallographic statistics are listed in Table 1.
Structure analysis
The sequence alignments were generated using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) and ESPRIPT65 (http://espript.ibcp.fr/ESPript/ESPript/). Interfaces were analyzed using PISA (http://www.ebi.ac.uk/msd-srv/prot_int/cgi-bin/piserver) and CCP4 Contact66. All structure figures were made in PyMOL (http:www.pymol.org). Electrostatic surface potential was calculated using the PyMOL ABPS Tools v. 2.1 plugin (pH 7, monovalent ion concentration 0.15 M, and dielectric constant of 2.0 for protein and 78.0 for solvent). Helix packing was analyzed using Socket (http://coiledcoils.chm.bris.ac.uk/socket/server.html).
CTD Expression and Purification
Soluble CTD proteins were expressed in E. coli Rosetta pLysS cells (Novagen). Freshly transformed Rosetta cells were grown at 37°C in LB supplemented with 50 μg/ml kanamycin and 34 μg/ml chloramphenicol to an optical density at 600 nm (OD600) of approximately 0.6. Protein production was induced with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG). After 3 h, cells were harvested and lysed in 50 mM Tris-HCl (pH 8.0), 250 mM NaCl, 3mM β-mercaptoethanol (BME), and 0.2 mM phenylmethylsulfonyl fluoride (PMSF) (lysis buffer) using a fluidizer. The cell lysate was cleared by centrifugation at 13,000g and nutated with 2-ml Ni-Sepharose 6B fast-flow resin (GE Healthcare) for 2 hrs. The resin was washed twice with 7.5 CV each of 50 mM Tris-HCl (pH 8.0), 250 mM NaCl, 3mM β-mercaptoethanol (BME), and 0.1 mM phenylmethylsulfonyl fluoride (PMSF) (rinse buffer) and rinse buffer supplemented with 6 mM imidazole (WB1). It was then loaded into a gravity column and washed with 17.5 CV of rinse buffer containing 20 mM imidazole (WB2). Protein was eluted with rinse buffer containing 300mM imidazole (EB). The eluate was concentrated in an Ultra-15 30-kDa-cutoff concentrator (Millipore). Imidazole removal and further purification of the eluate were done by size-exclusion chromatography (SEC) using a Superdex 200 column (GE Healthcare) equilibrated with 20mM Tris-HCl (pH 8.0), 150mM NaCl, 1 mM EDTA, and 200 mM TCEP (TNET). Peak fractions were combined, supplemented with 0.1 mM PMSF, and flash frozen in liquid N2 for storage at −80 °C. Mutant D878C precipitated and was excluded from subsequent analysis.
Liposome preparation
Single lamellar vesicles (SUVs) were prepared by mixing a 1:1 molar ratio of POPC/POPA lipids in chloroform, drying the lipids under N2 gas stream and then in a vacuum chamber overnight. Lipid mixture was then dissolved in 25 mM Tris/HCl pH 7.2, 150 mM NaCl, 1 mM EDTA (EDTA was absent from samples with NiEDDA) and allowed to fully hydrate for 1 hr at 4 °C. SUVs were formed by sonication and used within the same day.
ESR sample preparation and spin labeling
To reduce disulfide bonds that had formed during storage, the CTD cysteine mutants were incubated with 600 μM free TCEP for 2–3 hrs. Thereafter, the buffer with high TCEP concentration was exchanged by passing the protein solution through a NAP-5 size exclusion column (GE Healthcare) equilibrated with 20 mM Tris/HCl pH 8.0, 150 mM NaCl, 50 μM TCEP and 0.5–1 mM EDTA. 100 μM reduced protein samples were spin-labeled with 3-(2-iodoacetamido)-PROXYL spin label (ISL) at 1:10 protein-to-ISL molar ratio. Spin labeling proceeded for 7–8 h at 4 °C. Thereafter, the unreacted spin label was removed by a combination of NAP-10 size-exclusion column (GE Healthcare) filtration and extensive washing with buffer of 25 mM Tris/HCl pH 7.2 (at room temperature, RT), 150 mM NaCl, and 40 μM TCEP in microconcentrators at 4 °C and 4000×g. Whenever the ESR spectra indicated trace amounts of free spin label, samples were further purified by passing through Zeba spin desalting columns (Thermo-Fisher). Finally, 200 μM spin-labeled protein stock solutions were prepared and used for final dilutions to measure ESR spectra.
CW-ESR measurements
CW-ESR spectra were recorded on multiple spin-labeled protein variants in helix h2 and helix h3 in buffer solution. CW-ESR spectra were recorded for all spin-labeled cysteine mutants in gB CTD at a protein monomer-to-lipid molar ratio (P/L) of 1:660. The final protein concentration for both samples in buffer solution and in liposomes was 53 μM. A control sample with the same POPC and POPA molar proportions, plus spin-labeled lipid 1-palmitoyl-2-stearoyl-(5-doxyl)-sn-glycero-3-phosphocholine (16:0–5 Doxyl PC or 5PC) or 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho(tempo)choline (TEMPO-PC), added to a 1:500 molar ratio of labeled-to-unlabeled lipids, were also prepared and measured.
In the case of lipid-bound protein samples, complete binding of the protein to liposomes was verified by recording additional CW-ESR spectra at P/L’s of 1:730 (58 μM protein) and 1:980 (43 μM protein) for residue 879C. No difference in the spectral lineshape was observed. Therefore, the highest protein concentration was selected for the experiments. All samples for CW-ESR measurements were placed into 50 μL precision micro-capillary glass pipettes (Kimble Glass, Vineland, NJ) after closing their bottoms; sample lengths were 10 mm (i.e., the volume of 6.7 μl). All ESR measurements were performed at 25 °C with the ELEXIS E500 ESR spectrometer (Bruker, GmbH) using ER 4122-SHQE super-high-Q resonator and VT-31 variable-temperature accessory. The full-width nitroxide ESR spectra were recorded under the following conditions: incident microwave (MW) power was 1.26 mW; the field modulation amplitude was 1.1 G for samples in buffer and 1.6–2.1 G for liposome samples depending on the spectral lines width for different mutants. All spectra were normalized to the same number of spins.
The MW power saturation experiments were performed on the central line of the nitroxide CW-ESR spectrum67, to measure the accessibility of the spin labels to the commonly used fast-relaxing agents, vis. oxygen (O2) and Ni(II)-diammine-2,2′-(ethane-1,2-diyldiimino) diacetic acid complex (NiEDDA). O2 has high solubility in the hydrophobic lipid membrane core, whereas NiEDDA is water-soluble. Measurements were performed on regular gB CTD samples and 5PC, and TEMPO-PC in the presence of O2, deoxygenated and argon-filled samples, and deoxygenated and argon-filled samples containing NiEDDA. NiEDDA was added to the POPC/POPA lipid suspension in buffer, which was then sonicated to form liposomes with a uniform distribution of this fast-relaxing agent. Two final concentrations of NiEDDA were tried, 4.5 μM and 7 μM. However, some CW-ESR spectral broadening was observed with the 7 μM, particularly in the case of spin-label attached to highly solvent-exposed residue positions. Therefore 4.5 μM NiEDDA was used throughout the accessibility scanning experiments. Sample deoxygenation was performed on a vacuum line by repeatedly evacuating the capillary tube with the sample to soft vacuum and filling it with argon gas. Finally, the capillary tube, filled with argon to ~0.9 bar, was flame sealed.
To obtain the half-saturation parameter, P1/2, the central line of the nitroxide CW-ESR spectrum (~30 G width for lipid samples and ~15 G for protein without lipid) was recorded as a function of the MW power (varied from 0.5 mW to 200 mW in 20 steps). The measured intensity A of the central line was plotted as a function of the square root of the applied MW power; and the data were fitted to the equation:
| (1) |
where P is the applied MW power, ε is a line-homogeneity parameter, and I is a constant. We tested ε values between 1 and 1.5, and in general, for all samples, good fits were obtained using ε = 1.5 (which is typical of a highly homogeneous spectrum), resulting in a goodness of fit parameter R2 > 0.98. The accessibilities to O2 and NiEDDA were calculated using the following expression:
| (2) |
where ΔP1/2 is the offset of P1/2 is samples with O2 or NiEDDA compared to sample in argon for the same cysteine mutant; ΔH (or ΔHref) is the line width and superscript “ref” indicates the reference. We used the data for spin-labeled residue 884C in deoxygenated solution as a reference, similarly to previous studies67,68.
The depth parameter Φ, which visualizes better the difference in Π(O2) and Π(NiEDDA) was calculated as:
| (3) |
The spin label mobility was estimated using the inverse width (1/ΔH) of the central line of the nitroxide CW-ESR spectrum 27. The Φ and 1/ΔH data for residues in h3 were fitted to the following periodic function:
| (4) |
where y is either Φ or 1/ΔH, n is residue number, N is the periodicity, b is offset, and y0 is a constant. The obtained N values for Φ and 1/ΔH were 3.65 and 3.75, respectively.
All fittings of CW-ESR power saturation data and spin label mobility were performed in OriginLab software (OriginLab Corporation, Northampton, MA, USA). The measurements were performed twice on most of the singly labeled residues in helix 3 and the 5PC-labeled sample.
Inter-spin distance measurements by DEER spectroscopy
Four-pulse DEER69 measurements at 17.3 GHz and 60 K were performed using a home-built Ku-band pulse ESR spectrometer70 under standard experimental conditions: The π/2-π-π pulse widths were 16 ns, 32 ns and 32 ns, respectively, and the π pump pulse was 32 ns. A frequency separation of 70 MHz between detection and pump pulses was used. The detection pulses were applied at the low-field edge and the pump pulse was positioned near the central maximum of the nitroxide spin-label spectrum. The inter-spin distanced were reconstructed from the time-domain DEER data using L-curve Tikhonov regularization71 and refined by the maximum entropy method72.
Data Availability
Atomic coordinates and structure factors for the HSV-1 gB structures have been deposited to the RCSB Protein Data Bank under accession number 5V2S (P321) and 6BM8 (H32). Source data for Supplementary Figure 6 are available with the paper online. Source data for Figures 4 and 5 and for Supplementary Figure 7 are available upon reasonable request.
Supplementary Material
Acknowledgments
We thank NE-CAT staff for help with x-ray data collection and Henry Rogalin for help with the cell-cell fusion assays. We also thank P.G. Spear, R. J. Eisenberg, and G. H. Cohen for the gift of plasmids, and J.M. Coffin for the gift of CHO cells. This work was funded by the NIH grant 1R21AI107171 (E.E.H.), the Burroughs Wellcome Fund (E.E.H.), and by the NIH Ruth L. Kirschstein NRSA postdoctoral fellowship 1F32GM115060 (R.S.C.). The research of E.E.H. was supported in part by a Faculty Scholar grant from Howard Hughes Medical Institute. ESR experiments were funded by NIH grants P41GM103521 (J.H.F, ACERT) and R01GM123779 (J.H.F. and E.R.G.). This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P41 GM103403). The Pilatus 6M detector on 24-ID-C beam line is funded by a NIH-ORIP HEI grant (S10 RR029205). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. All software was installed and maintained by SBGrid59.
References
- 1.Kennedy PG, Steiner I. Recent issues in herpes simplex encephalitis. J Neurovirol. 2013;19:346–50. doi: 10.1007/s13365-013-0178-6. [DOI] [PubMed] [Google Scholar]
- 2.Rowe AM, et al. Herpes keratitis. Prog Retin Eye Res. 2013;32:88–101. doi: 10.1016/j.preteyeres.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Santos CA. Cytomegalovirus and Other beta-Herpesviruses. Semin Nephrol. 2016;36:351–361. doi: 10.1016/j.semnephrol.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 4.Britt WJ. Congenital Human Cytomegalovirus Infection and the Enigma of Maternal Immunity. J Virol. 2017;91 doi: 10.1128/JVI.02392-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Young LS, Yap LF, Murray PG. Epstein-Barr virus: more than 50 years old and still providing surprises. Nat Rev Cancer. 2016;16:789–802. doi: 10.1038/nrc.2016.92. [DOI] [PubMed] [Google Scholar]
- 6.Eisenberg RJ, et al. Herpes virus fusion and entry: a story with many characters. Viruses. 2012;4:800–32. doi: 10.3390/v4050800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sathiyamoorthy K, Chen J, Longnecker R, Jardetzky TS. The COMPLEXity in herpesvirus entry. Curr Opin Virol. 2017;24:97–104. doi: 10.1016/j.coviro.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrison SC. Viral membrane fusion. Virology. 2015;479–480C:498–507. doi: 10.1016/j.virol.2015.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heldwein EE, et al. Crystal structure of glycoprotein B from herpes simplex virus 1. Science. 2006;313:217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- 10.Backovic M, Longnecker R, Jardetzky TS. Structure of a trimeric variant of the Epstein-Barr virus glycoprotein B. Proc Natl Acad Sci U S A. 2009;106:2880–5. doi: 10.1073/pnas.0810530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burke HG, Heldwein EE. Crystal Structure of the Human Cytomegalovirus Glycoprotein B. PLoS Pathog. 2015;11:e1005227. doi: 10.1371/journal.ppat.1005227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandramouli S, et al. Structure of HCMV glycoprotein B in the postfusion conformation bound to a neutralizing human antibody. Nat Commun. 2015;6:8176. doi: 10.1038/ncomms9176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones NA, Geraghty RJ. Fusion activity of lipid-anchored envelope glycoproteins of herpes simplex virus type 1. Virology. 2004;324:213–28. doi: 10.1016/j.virol.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 14.Fan Z, et al. Truncation of herpes simplex virus type 2 glycoprotein B increases its cell surface expression and activity in cell-cell fusion, but these properties are unrelated. J Virol. 2002;76:9271–83. doi: 10.1128/JVI.76.18.9271-9283.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wanas E, Efler S, Ghosh K, Ghosh HP. Mutations in the conserved carboxy-terminal hydrophobic region of glycoprotein gB affect infectivity of herpes simplex virus. J Gen Virol. 1999;80(Pt 12):3189–98. doi: 10.1099/0022-1317-80-12-3189. [DOI] [PubMed] [Google Scholar]
- 16.Lin E, Spear PG. Random linker-insertion mutagenesis to identify functional domains of herpes simplex virus type 1 glycoprotein B. Proc Natl Acad Sci U S A. 2007;104:13140–13145. doi: 10.1073/pnas.0705926104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gage PJ, Levine M, Glorioso JC. Syncytium-inducing mutations localize to two discrete regions within the cytoplasmic domain of herpes simplex virus type 1 glycoprotein B. J Virol. 1993;67:2191–201. doi: 10.1128/jvi.67.4.2191-2201.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baghian A, Huang L, Newman S, Jayachandra S, Kousoulas KG. Truncation of the carboxy-terminal 28 amino acids of glycoprotein B specified by herpes simplex virus type 1 mutant amb1511-7 causes extensive cell fusion. J Virol. 1993;67:2396–401. doi: 10.1128/jvi.67.4.2396-2401.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chowdary TK, Heldwein EE. Syncytial phenotype of C-terminally truncated herpes simplex virus type 1 gB is associated with diminished membrane interactions. Journal of virology. 2010;84:4923–35. doi: 10.1128/JVI.00206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zoonens M, Popot JL. Amphipols for each season. J Membr Biol. 2014;247:759–96. doi: 10.1007/s00232-014-9666-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng Z, Maidji E, Tugizov S, Pereira L. Mutations in the carboxyl-terminal hydrophobic sequence of human cytomegalovirus glycoprotein B alter transport and protein chaperone binding. J Virol. 1996;70:8029–40. doi: 10.1128/jvi.70.11.8029-8040.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogalin HB, Heldwein EE. The interplay between the HSV-1 gB cytodomains and the gH cytotail during cell-cell fusion. J Virol. 2015 doi: 10.1128/JVI.02391-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langelaan DN, Wieczorek M, Blouin C, Rainey JK. Improved helix and kink characterization in membrane proteins allows evaluation of kink sequence predictors. J Chem Inf Model. 2010;50:2213–20. doi: 10.1021/ci100324n. [DOI] [PubMed] [Google Scholar]
- 24.Hannah BP, et al. Herpes simplex virus glycoprotein B associates with target membranes via its fusion loops. J Virol. 2009;83:6825–36. doi: 10.1128/JVI.00301-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruel N, Zago A, Spear PG. Alanine substitution of conserved residues in the cytoplasmic tail of herpes simplex virus gB can enhance or abolish cell fusion activity and viral entry. Virology. 2006;346:229–37. doi: 10.1016/j.virol.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 26.Silverman JL, Greene NG, King DS, Heldwein EE. Membrane Requirement for Folding of the Herpes Simplex Virus 1 gB Cytodomain Suggests a Unique Mechanism of Fusion Regulation. Journal of virology. 2012;86:8171–84. doi: 10.1128/JVI.00932-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McHaourab HS, Lietzow MA, Hideg K, Hubbell WL. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 1996;35:7692–704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 28.Haffar OK, Dowbenko DJ, Berman PW. The cytoplasmic tail of HIV-1 gp160 contains regions that associate with cellular membranes. Virology. 1991;180:439–41. doi: 10.1016/0042-6822(91)90054-f. [DOI] [PubMed] [Google Scholar]
- 29.Lai AL, Park H, White JM, Tamm LK. Fusion peptide of influenza hemagglutinin requires a fixed angle boomerang structure for activity. J Biol Chem. 2006;281:5760–70. doi: 10.1074/jbc.M512280200. [DOI] [PubMed] [Google Scholar]
- 30.Stoll S, et al. Double electron-electron resonance shows cytochrome P450cam undergoes a conformational change in solution upon binding substrate. Proc Natl Acad Sci U S A. 2012;109:12888–93. doi: 10.1073/pnas.1207123109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zou P, Bortolus M, McHaourab HS. Conformational cycle of the ABC transporter MsbA in liposomes: detailed analysis using double electron-electron resonance spectroscopy. J Mol Biol. 2009;393:586–97. doi: 10.1016/j.jmb.2009.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walev I, Lingen M, Lazzaro M, Weise K, Falke D. Cyclosporin A resistance of herpes simplex virus-induced “fusion from within” as a phenotypical marker of mutations in the Syn 3 locus of the glycoprotein B gene. Virus Genes. 1994;8:83–6. doi: 10.1007/BF01703606. [DOI] [PubMed] [Google Scholar]
- 33.Muggeridge MI. Characterization of cell-cell fusion mediated by herpes simplex virus 2 glycoproteins gB, gD, gH and gL in transfected cells. J Gen Virol. 2000:2017–2027. doi: 10.1099/0022-1317-81-8-2017. [DOI] [PubMed] [Google Scholar]
- 34.Cai WH, Gu B, Person S. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J Virol. 1988;62:2596–604. doi: 10.1128/jvi.62.8.2596-2604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engel JP, Boyer EP, Goodman JL. Two novel single amino acid syncytial mutations in the carboxy terminus of glycoprotein B of herpes simplex virus type 1 confer a unique pathogenic phenotype. Virology. 1993;192:112–20. doi: 10.1006/viro.1993.1013. [DOI] [PubMed] [Google Scholar]
- 36.Diakidi-Kosta A, Michailidou G, Kontogounis G, Sivropoulou A, Arsenakis M. A single amino acid substitution in the cytoplasmic tail of the glycoprotein B of herpes simplex virus 1 affects both syncytium formation and binding to intracellular heparan sulfate. Virus Res. 2003;93:99–108. doi: 10.1016/s0168-1702(03)00070-4. [DOI] [PubMed] [Google Scholar]
- 37.Muggeridge MI, Grantham ML, Johnson FB. Identification of syncytial mutations in a clinical isolate of herpes simplex virus 2. Virology. 2004;328:244–53. doi: 10.1016/j.virol.2004.07.027. [DOI] [PubMed] [Google Scholar]
- 38.Bzik DJ, Fox BA, DeLuca NA, Person S. Nucleotide sequence of a region of the herpes simplex virus type 1 gB glycoprotein gene: mutations affecting rate of virus entry and cell fusion. Virology. 1984;137:185–90. doi: 10.1016/0042-6822(84)90022-9. [DOI] [PubMed] [Google Scholar]
- 39.Foster TP, Melancon JM, Kousoulas KG. An alpha-helical domain within the carboxyl terminus of herpes simplex virus type 1 (HSV-1) glycoprotein B (gB) is associated with cell fusion and resistance to heparin inhibition of cell fusion. Virology. 2001;287:18–29. doi: 10.1006/viro.2001.1004. [DOI] [PubMed] [Google Scholar]
- 40.Chen J, Zhang X, Jardetzky TS, Longnecker R. The Epstein-Barr virus (EBV) glycoprotein B cytoplasmic C-terminal tail domain regulates the energy requirement for EBV-induced membrane fusion. J Virol. 2014;88:11686–95. doi: 10.1128/JVI.01349-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garcia NJ, Chen J, Longnecker R. Modulation of Epstein-Barr virus glycoprotein B (gB) fusion activity by the gB cytoplasmic tail domain. MBio. 2013;4:e00571–12. doi: 10.1128/mBio.00571-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haan KM, Lee SK, Longnecker R. Different functional domains in the cytoplasmic tail of glycoprotein B are involved in Epstein-Barr virus-induced membrane fusion. Virology. 2001;290:106–14. doi: 10.1006/viro.2001.1141. [DOI] [PubMed] [Google Scholar]
- 43.Postler TS, Desrosiers RC. The tale of the long tail: the cytoplasmic domain of HIV-1 gp41. J Virol. 2013;87:2–15. doi: 10.1128/JVI.02053-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haanes EJ, Nelson CM, Soule CL, Goodman JL. The UL45 gene product is required for herpes simplex virus type 1 glycoprotein B-induced fusion. J Virol. 1994;68:5825–34. doi: 10.1128/jvi.68.9.5825-5834.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen J, et al. HIV-1 ENVELOPE. Effect of the cytoplasmic domain on antigenic characteristics of HIV-1 envelope glycoprotein. Science. 2015;349:191–5. doi: 10.1126/science.aaa9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dev J, et al. Structural basis for membrane anchoring of HIV-1 envelope spike. Science. 2016;353:172–5. doi: 10.1126/science.aaf7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kemble GW, Danieli T, White JM. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell. 1994;76:383–91. doi: 10.1016/0092-8674(94)90344-1. [DOI] [PubMed] [Google Scholar]
- 48.Nixdorf R, Klupp BG, Karger A, Mettenleiter TC. Effects of truncation of the carboxy terminus of pseudorabies virus glycoprotein B on infectivity. J Virol. 2000;74:7137–7145. doi: 10.1128/jvi.74.15.7137-7145.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waning DL, Russell CJ, Jardetzky TS, Lamb RA. Activation of a paramyxovirus fusion protein is modulated by inside-out signaling from the cytoplasmic tail. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9217–22. doi: 10.1073/pnas.0403339101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun ZY, et al. Disruption of helix-capping residues 671 and 674 reveals a role in HIV-1 entry for a specialized hinge segment of the membrane proximal external region of gp41. J Mol Biol. 2014;426:1095–108. doi: 10.1016/j.jmb.2013.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang X, et al. Cryo-EM structure of the mature dengue virus at 3.5-A resolution. Nat Struct Mol Biol. 2013;20:105–10. doi: 10.1038/nsmb.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sirohi D, et al. The 3.8 A resolution cryo-EM structure of Zika virus. Science. 2016;352:467–70. doi: 10.1126/science.aaf5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vitu E, Sharma S, Stampfer SD, Heldwein EE. Extensive mutagenesis of the HSV-1 gB ectodomain reveals remarkable stability of its postfusion form. Journal of molecular biology. 2013;425:2056–71. doi: 10.1016/j.jmb.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patrone M, et al. Enhanced expression of full-length human cytomegalovirus fusion protein in non-swelling baculovirus-infected cells with a minimal fed-batch strategy. PLoS One. 2014;9:e90753. doi: 10.1371/journal.pone.0090753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zeev-Ben-Mordehai T, et al. Two distinct trimeric conformations of natively membrane-anchored full-length herpes simplex virus 1 glycoprotein B. Proc Natl Acad Sci U S A. 2016;113:4176–81. doi: 10.1073/pnas.1523234113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fontana J, et al. The Fusion Loops of the Initial Prefusion Conformation of Herpes Simplex Virus 1 Fusion Protein Point Toward the Membrane. MBio. 2017;8 doi: 10.1128/mBio.01268-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harman A, Browne H, Minson T. The transmembrane domain and cytoplasmic tail of herpes simplex virus type 1 glycoprotein H play a role in membrane fusion. J Virol. 2002;76:10708–16. doi: 10.1128/JVI.76.21.10708-10716.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Browne HM, Bruun BC, Minson AC. Characterization of herpes simplex virus type 1 recombinants with mutations in the cytoplasmic tail of glycoprotein H. J Gen Virol. 1996;77(Pt 10):2569–73. doi: 10.1099/0022-1317-77-10-2569. [DOI] [PubMed] [Google Scholar]
- 59.Morin A, et al. Collaboration gets the most out of software. Elife. 2013;2:e01456. doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Charvolin D, Picard M, Huang LS, Berry EA, Popot JL. Solution behavior and crystallization of cytochrome bc(1) in the presence of amphipols. J Membr Biol. 2014;247:981–96. doi: 10.1007/s00232-014-9694-4. [DOI] [PubMed] [Google Scholar]
- 61.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica. Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–8. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 66.Collaborative Computational Project N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 67.Georgieva ER, Xiao S, Borbat PP, Freed JH, Eliezer D. Tau binds to lipid membrane surfaces via short amphipathic helices located in its microtubule-binding repeats. Biophys J. 2014;107:1441–52. doi: 10.1016/j.bpj.2014.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zou P, McHaourab HS. Alternating access of the putative substrate-binding chamber in the ABC transporter MsbA. J Mol Biol. 2009;393:574–85. doi: 10.1016/j.jmb.2009.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pannier M, Veit S, Godt A, Jeschke G, Spiess HW. Dead-time free measurement of dipole-dipole interactions between electron spins. 2000. J Magn Reson. 2011;213:316–25. doi: 10.1016/j.jmr.2011.08.035. [DOI] [PubMed] [Google Scholar]
- 70.Borbat PP, Crepeau RH, Freed JH. Multifrequency two-dimensional Fourier transform ESR: an X/Ku-band spectrometer. J Magn Reson. 1997;127:155–67. doi: 10.1006/jmre.1997.1201. [DOI] [PubMed] [Google Scholar]
- 71.Chiang YW, Borbat PP, Freed JH. The determination of pair distance distributions by pulsed ESR using Tikhonov regularization. J Magn Reson. 2005;172:279–95. doi: 10.1016/j.jmr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 72.Chiang YW, Borbat PP, Freed JH. Maximum entropy: a complement to Tikhonov regularization for determination of pair distance distributions by pulsed ESR. J Magn Reson. 2005;177:184–96. doi: 10.1016/j.jmr.2005.07.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the HSV-1 gB structures have been deposited to the RCSB Protein Data Bank under accession number 5V2S (P321) and 6BM8 (H32). Source data for Supplementary Figure 6 are available with the paper online. Source data for Figures 4 and 5 and for Supplementary Figure 7 are available upon reasonable request.