Abstract
Unlike most other plant microsomal desaturases, the Δ6-fatty acid desaturase from borage (Borago officinalis) contains an N-terminal extension that shows homology to the small hemoprotein cytochrome (Cyt) b5. To determine if this domain serves as a functional electron donor for the Δ6-fatty acid desaturase, mutagenesis and functional analysis by expression in transgenic Arabidopsis was carried out. Although expression of the wild-type borage Δ6-fatty acid desaturase resulted in the synthesis and accumulation of Δ6-unsaturated fatty acids, this was not observed in plants transformed with N-terminally deleted forms of the desaturase. Site-directed mutagenesis was used to disrupt one of the axial heme-binding residues (histidine-41) of the Cyt b5 domain; expression of this mutant form of the Δ6-desaturase in transgenic plants failed to produce Δ6-unsaturated fatty acids. These data indicate that the Cyt b5 domain of the borage Δ6-fatty acid desaturase is essential for enzymatic activity.
The synthesis of unsaturated fatty acids is catalyzed by acyl-lipid-desaturases that introduce double bonds into preformed acyl chains by oxygen- and electron-donor-dependent desaturation (Heinz, 1993; Shanklin and Cahoon, 1998). The immediate electron donor for many microsomal desaturases is Cyt b5 (Lederer, 1994), a small hemoprotein that functions in a number of oxidation/reduction reactions in plants, including NADH-dependent acyl-group desaturation (Smith et al., 1990, 1998). Cyt b5 is also involved in fatty acid hydroxylation in higher plants (Kearns et al., 1991; Smith et al., 1992). Apart from the “free” microsomal form of the protein, Cyt b5 domains or folds have been found in a number of unrelated proteins, such as nitrate reductase, sulfite oxidase, and l-lactate dehydrogenase (Lederer, 1994; Sperling et al., 1995; Napier et al., 1997). These domains include the characteristic Cyt b5 motif His-Pro-Gly-Gly (HPGG), which forms the core of the heme-binding domain.
Recently, Cyt b5 domains have been identified in various positions in desaturases and hydroxylases. Thus, a fused C-terminal Cyt b5 domain has been detected in the Saccharomyces cerevisiae OLE1 microsomal Δ9-desaturase (Mitchell and Martin, 1995) and in OLE1 homologs from fungi (Meesters et al., 1997) and red alga (Itoh et al., 1998). Amino-terminal Cyt b5 domain have recently been identified in Δ6-fatty acid desaturases from plants (Sayanova et al., 1997), the nematode worm Caenorhabditis elegans (Napier et al., 1998), and mammals (Cho et al., 1999), and in Δ5-fatty acid desaturases from animals and fungi (Knutzon et al., 1998; Michaelson et al., 1998a, 1998b). These domains have also been detected in Δ8-sphingolipid desaturases from plants (Sperling et al., 1995, 1998) and in the FAH1 sphingolipid α-hydroxylase of yeast (Mitchell and Martin, 1997; Napier et al., 1997). The Δ6-fatty acid desaturase from the moss Physcomitrella patents also contains an additional N-terminal extension of 100 amino acids preceding its Cyt b5 domain, thus resembling fusion proteins such as nitrate reductase (Girke et al., 1998).
Although these Cyt b5-domain sequences have significantly diverged, this has occurred without modification of the HPGG motif, which forms the heme-binding core of the protein (Napier et al., 1997, 1999). The heme-binding nature of these domains has been demonstrated in sunflower sphingolipid desaturase by expression of the first 122 residues (essentially the entire Cyt b5 domain) in Escherichia coli (Sperling et al., 1995). The resulting recombinant protein exhibited redox absorbance spectra similar to those of plant microsomal Cyt b5 (Smith et al., 1994).
The essential role of the Cyt b5-like domain in these fusion-desaturases has been demonstrated for yeast OLE1p Δ9-acyl-CoA desaturase (Mitchell and Martin, 1995). Expression of the OLE1 gene rescued yeast double mutants that lacked both OLE1 and microsomal Cyt b5 genes, whereas rescue of ole1 mutants by a rat microsomal Δ9-desaturase required the additional presence of a functional Cyt b5 gene. Truncation or disruption of the Cyt b5 domain of the OLE1 desaturase, even in cells with wild-type levels of “free” Cyt b5, resulted in unsaturated fatty acid auxotrophy (Mitchell and Martin, 1995). Since OLE1 complements ole1 mutants in a Cyt b5-deletion yeast strain (unlike the rat Δ9-desaturase, which lacks the C-terminal Cyt b5 domain), this implies that the 100-residue extension present in OLE1 functions as the electron donor (Mitchell and Martin, 1995). However, in that study, the disruption of the Cyt b5 domain was achieved via an internal 100-bp deletion, which may have resulted in a perturbation of the secondary structure of the enzyme and rendered it nonfunctional.
Although it is possible that the presence of a fused Cyt b5 domain allows for a more efficient reaction mechanism, it is interesting that none of the acyl-desaturases with Δ12- and Δ15-regio-selectivities (Arondel et al., 1992; Okuley et al., 1994) or the related oleate hydroxylase (van de Loo et al., 1995) have a Cyt b5 domain, even though the Δ12- and Δ15-desaturases are much more prevalent in the plant kingdom. However, all of the enzymes that contain N-terminal Cyt b5 domains are involved in “proximal” or “front-end” modifications of lipid substrates, and it is possible that this fusion is an obligate requirement for correct function of this distinct class of proteins (Sperling et al., 1998; Napier et al., 1999).
Recently, we isolated a cDNA clone (pBdes6) encoding the microsomal fatty acid Δ6-desaturase from developing seeds of borage (Borago officinalis). This cDNA encoded a protein with a N-terminal Cyt b5 fusion domain and was functionally characterized by expression in transgenic tobacco plants, resulting in the accumulation of γ-linolenic acid (GLA; 18:36,9,12) and octadecatetraenoic acid (OTA; 18:46,9,12,15) (Sayanova et al., 1997). As part of our continuing studies on this enzyme, we sought to determine the functional role of the Cyt b5 domain. We used two complementary approaches, N-terminal deletion and site-directed mutagenesis, to define this domain. Thus, two N-terminally truncated forms of the borage Δ6-desaturase were generated, as well as a point-mutated form in which the conserved His (His-41) of the heme-binding HPGG motif was replaced with an Ala. These three mutant forms of the desaturase were expressed in transgenic Arabidopsis and functionally compared with lines expressing the full-length wild-type borage Δ6-desaturase.
MATERIALS AND METHODS
Plasmid Construction for Plant Expression
PCR products encoding N-terminally deleted forms of the Δ6-desaturase were amplified by removing 112 and 146 residues from the N terminus of the borage Δ6-fatty acid desaturase. Two oligonucleotides were synthesized based on the pBdes6 coding sequence: primer M2, 5′GCGTCGACATGTTTGCAACTTTGTGC 3′ (annealing to the Met-113, indicated in boldface type) and primer M3, 5′-GCGTCGACATGGGGTTTCTTTGGATTC-3′ (annealing to the Met-147, indicated in boldface type). The SalI restriction site is underlined. Reverse primer D, containing a SmaI site, was as described in Sayanova et al. (1997) and these primers were used for PCR amplification with pBdes6 as described previously. Reactions were run on a DNA thermal cycler (Perkin-Elmer Cetus, Foster City, CA) using a program of 1 min at 94°C, 1 min at 55°C, and 2 min at 72°C for 32 cycles, followed by extension for 10 min at 72°C. PCR amplification products were gel-purified and subcloned into vector pJD330. Digestion of the two resulting plasmids with XbaI released fragments containing ORFs of different lengths (starting either with Met-113 or Met-147 and designated Δ112 and Δ146, respectively), together with the CaMV 35S promoter containing an Ω-translational enhancer and the nopaline synthase (nos) termination sequence. These XbaI-fragments were gel-purified and cloned into pBIN19 to generate plasmids pBdesM2 and pBdesM3, which were transformed into Agrobacterium tumefaciens strain GV 3101 + VirG+) by electroporation.
Plant Transformation
The Columbia ecotype of Arabidopsis was transformed by A. tumefaciens-mediated transformation according to the in planta vacuum infiltration method (Bechtold et al., 1993) Transformed seeds were selected on plates containing selective medium: Murashige and Skoog basal salts (4.3 g/L), 10 g/L Suc, and 50 mg/L kanamycin. Kanamycin-resistant seedlings (the T1 generation) were transferred to soil for production of T2 seeds.
Fatty Acid Analysis
Fatty acids from seeds, roots, siliques, and leaves were extracted and transmethylated according to the method of Miquel and Browse (1992). Analyses of fatty acid methyl esters (FAMEs) were carried out using a GC (model 5880A, Hewlett-Packard, Wilmington, DE) equipped with a 25- × 0.32-mm bonded capillary column (RSL-500BP, Hewlett-Packard) with a flame ionization detector. Fatty acids were identified by comparison of retention times with FAME standards (Sigma, St. Louis) separated on the same GC.
Site-Directed Mutageneses
Mutagenesis of His-41 to Ala was performed using a site-directed mutagensis kit (QuikChange, Stratagene, La Jolla, CA) according to the manufacturer's protocol. Two mutagenic oligonucleotide primers, each complementary to the opposite strands of the p35Bdes6 construct, were synthesized as shown.
Forward primer A, 5′GATTGGGTGAAAGACGCTCCAGGTGGCAGC 3′, and reverse primer B, 5′GCTGCCACCTGGAGCGTCTTTCACCCAATC 3′. The bold letters indicate altered nucleotides. Mutation of His-41 to Ala was performed on the plant expression construct containing the borage Δ6-desaturase coding sequence. The sequence of the mutated fragment was checked for the presence of the correct alteration and was used to ensure that other mutations had not occurred. This plasmid was used for Arabidopsis transformation via A. tumefaciens (see above).
Nothern-Blot Analyses
RNA from expanding Arabidopsis leaves was extracted using a kit (RNAeasy, Qiagen, Valencia, CA). About 10 μg of total RNA was run on 1% (v/v) formaldehyde gel and transferred to a nylon membrane (Hybond N+, Amersham, Uppsala) according to the manufacturer's protocol. Probes were made from the truncated Δ146 form of the pBdes6 cDNA by random priming using a kit (Prime-it II, Stratagene) according to the supplier's instructions.
RESULTS AND DISCUSSION
Expression of a Δ6 Fatty Acid Desaturase Gene in Arabidopsis
The full-length (wild-type) borage Δ6-desaturase cDNA (Sayanova et al., 1997) was initially expressed in Arabidopsis plants and the distributions of fatty acids in total lipid fractions prepared from leaves, roots, siliques, and seeds compared with those in control transformants (Table I). GLA and OTA accumulated to levels of 4.8% and 9.4% of the total fatty acids, respectively (all fatty acid percentages are expressed as a mole percentage of the total). This bias toward the accumulation of OTA may be have been due to the preference of the Δ6-desaturase for α-linolenic acid as a substrate or to high levels of endogenous leaf Δ15-desaturase activity, reducing the levels of linoleic acid available for conversion to GLA. The endogenous levels of both 18:2 and α-18:3 in roots were reduced by expression of the borage Δ6-desaturase, with a significant increase in GLA. In mid-stage (S2) developing seeds, the accumulation of GLA and OTA was low (Table I) and declined to undetectable levels as the seeds matured (data not shown). This may have been due to reduced levels of the Δ6-desaturase enzyme (as a result of reduced promoter activity) or to the exclusion of Δ6-desaturated fatty acids from seed lipids by endogenous acyltransferases.
Table I.
Fatty acid composition (moles percent of total) of seed and non-seed tissues of transgenic Arabidopsis transformed with either full-length borage Δ6-desaturase gene (AT6) or a control transformation (WT)
| Fatty Acid | Leaves
|
Roots
|
S2 Seeds
|
|||
|---|---|---|---|---|---|---|
| WT | AT6 | WT | AT6 | WT | AT6 | |
| 16:0 | 15.5 | 15.5 | 25.0 | 27.0 | 20.1 | 14.0 |
| 16:1 | 3.2 | 4.4 | 1.6 | 1.0 | – | – |
| 16:3 | 16.0 | 22.0 | – | – | – | – |
| 18:0 | 0.5 | 0.7 | 2.0 | 2.5 | 2.7 | 2.5 |
| 18:1 | 1.3 | 2.1 | 2.7 | 3.0 | 12.4 | 10.4 |
| 18:2 | 11.0 | 10.1 | 40.0 | 30.1 | 35.1 | 34.0 |
| 18:3 | 46.0 | 35.0 | 26.2 | 15.2 | 26.0 | 28.7 |
| GLA | – | 4.90 | – | 15.1 | – | 0.5 |
| OTA | – | 9.30 | – | 7.0 | – | 1.3 |
Measurements were determined from a number of runs; se for all measurements was <3%. S2 seeds are defined as being at the midstage of seed development.
We recently determined the lipid classes present in the vegetative tissues of homozygous lines of transgenic tobacco plants expressing the borage Δ6-desaturase (Sayanova et al., 1997) and also in seed tissue (O. Sayanova, P.R. Shewry, and J.A. Napier, unpublished data). GLA accumulated in the leaves of the transgenic tobacco plants at levels between 12.9% of total fatty acids in young leaves and 20.1% in mature leaves, with OTA levels of 8.5% and 14.9%, respectively. However, there was only a low level of accumulation of GLA (2.6%) in mature seeds with insignificant levels of OTA. Thus, there is a difference in the proportions of the GLA and OTA in Arabidopsis and tobacco leaves, with GLA being the predominant Δ6-fatty acid in tobacco and OTA being predominant in Arabidopsis. This difference may reflect the relative levels of desaturation occurring in the chloroplasts of the two species, as some 50% of cellular glycerolipids are synthesized in the plastids of 16:3 plants such as spinach and Arabidopsis, compared with only about 20% in 18:3 plants such as tobacco. Such observations confirm the contribution of the plastid in glycerolipid synthesis (Somerville and Browse, 1991), especially in 16:3 species such as Arabidopsis.
Truncation of the Cyt b5-Domain of the Borage Δ6 Fatty Acid Desaturase
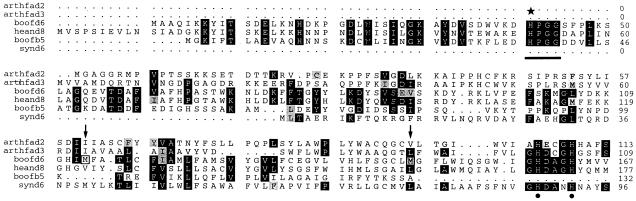
Comparison of the deduced amino acid sequences of the borage Δ6-desaturase with other microsomal desaturases indicated that borage protein contained an N-terminal Cyt b5 domain, with residues 1 to 112 corresponding to the hydrophilic portion of microsomal Cyt b5 (see Fig. 1.). If this N-terminal Cyt b5 domain was the result of a gene fusion event, then Met-113 in the borage Δ6-desaturase could correspond to the initiating Met of the ancestral desaturase domain. Alternatively, the ancestral desaturase ORF could start at Met-147, which corresponds to the end of plant microsomal Cyt b5 (Fig. 1). To determine whether the desaturase domain of the borage Δ6-enzyme could function with other (independent) electron donors instead of its own (linked) Cyt b5 domain, two truncations of the N-terminal domain were generated by deleting either the first 112 or 146 residues. These deletions, designated Δ112 and Δ146, respectively, were expressed in transgenic Arabidopsis plants.
Figure 1.
Sequence alignments of the first 167 residues of the borage Δ6-desaturase (boofd6) with the Arabidopsis FAD2 (arthfad2) and FAD3 (arthfad3) microsomal desaturases, sunflower Δ8-sphingolipid desaturase (heand8), cyanobacteria Δ6 desaturase (synd6), and borage Cyt b5 (boofb5). The conserved HPGG motif is underlined, with the essential His (H41) indicated by a star. The position of the internal Met residues (Met-113 and Met-147) are boxed and arrowed. The conserved His residues of the first “His box” of the microsomal desaturases are also indicated by black dots.
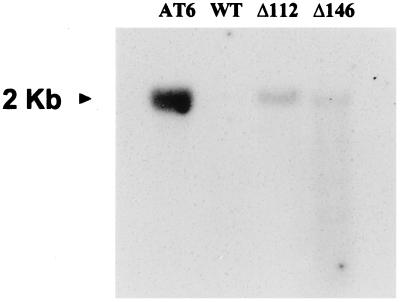
GC of FAMEs prepared from leaf lipids showed no evidence of GLA or OTA in the control transformants or in plants transformed with Δ112 and Δ146, in contrast to plants transformed with the full-length Cyt b5 Δ6-fatty acid desaturase fusion. Northern analysis was therefore performed to confirm that the truncated forms of the borage Δ6-desaturase were expressed in the transgenic Arabidopsis lines (Fig. 2). This analysis showed that the full-length and truncated forms of the borage desaturase had reduced levels of transcript accumulation, although all constructs were driven by the same promoter (CaMV 35S). In particular, transcripts of the more severe N-terminal deletion (Δ146) appeared to be less stable, as judged by the signal from the northern blot.
Figure 2.
Northern-blot analysis of transgenic Arabidopsis plants expressing the full-length borage Δ6-desaturase (AT6) compared with lines expressing two N-terminally truncated forms (Δ112 and Δ146). The approximate size of the detected transcript is indicated. A control transformed Arabidopsis (WT) sample was also probed. Ten micrograms of RNA was loaded in each lane.
It is possible that the failure to detect Δ6-fatty acids in plants expressing the deleted forms of the desaturase could be due to alterations in mRNA processing or translation of the synthetic transgene, rather than from any loss of enzyme activity. Alternatively, deletion of the Cyt b5 domain could result in aberrant folding of the modified desaturase, rendering the enzyme nonfunctional or reducing stability. Unfortunately, the lack of a suitable antibody prevented us from comparing the levels of wild-type and N-terminally deleted forms of the Δ6-desaturase. The role of the C-terminal Cyt b5 domain of the yeast OLE1 Δ9-desaturase was similarly studied via internal deletion of 34 amino acid residues by restriction digestion and re-ligation (Mitchell and Martin, 1995). In that study, the mutated form of OLE1 was nonfunctional. Moreover, in gene-replacement yeast strains, steady-state transcripts for the mutant OLE1 were at a reduced level compared with that observed for the wild-type OLE1 gene. An equally important observation was that even in the functional OLE1 enzyme, the protein displayed a very short half-life (<2 mins) (Mitchell and Martin, 1995); therefore, altered forms of the protein may be even less stable.
Site-Directed Mutagenesis
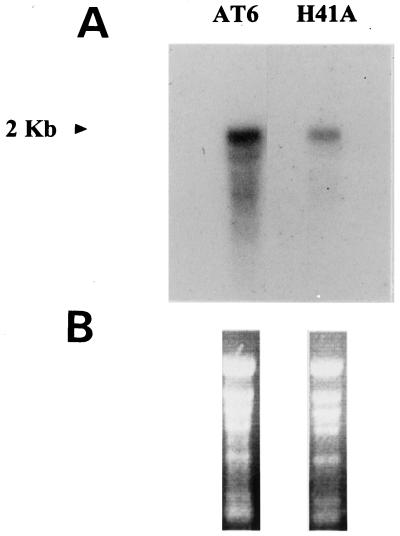
In view of our results for the truncated Δ6-desaturase and the above-mentioned studies of Michell and Martin (1995) on the OLE1 Δ9 desaturase, it was clear that a different approach was required to study the role of the Cyt b5 domain. Site-directed mutagenesis is a more precise tool for the definition of important amino acid determinants in a protein, and such mutations would not be expected to affect mRNA stability and translational efficiency. We therefore mutated the highly conserved His of the HPGG heme-binding domain in the N-terminal Cyt b5 extension. This residue is considered essential for the function of Cyt b5 by acting as a heme ligand (Lederer, 1994). His-41 was converted to Ala by a 3-bp mutation in the desaturase coding sequence. Ala was chosen as the replacement residue because it is uncharged and should therefore not be able to act as a heme-binding residue. The mutated Δ6-desaturase (named H41A) was introduced into Arabidopsis, and a number of transgenic lines were regenerated. GC analysis of the leaf lipids extracted from these transgenic plants again showed no accumulation of GLA and OTA (data not shown; compared with approximately 15% Δ6 desaturate fatty acids in line AT6 expressing the wild-type Δ6-desaturase; compare with Table I), although northern blots showed mRNA levels equivalent to that in plants expressing the wild-type borage enzyme (Fig. 3). This suggests that the conserved residue His-41 of the HPGG-binding domain is likely to be essential for enzymatic function of the Δ6-fatty acid desaturase and is most likely to coordinate the axial heme-binding center of that domain.
Figure 3.
Northern-blot analysis of transgenic Arabidopsis plants expressing either the full-length borage Δ6-desaturase (AT6) or a mutated form in which His-41 was converted to an Ala (H41A). Approximately 10 μg of RNA was loaded for both samples, as judged by ethidium bromide staining (B), and the approximate size of the detected transcript is indicated (A).
The above results also suggest that the presence of an N-terminal Cyt b5 domain in the so-called front-end desaturases (i.e. enzymes that desaturate between pre-existing double bonds and the carboxyl-group) has arisen as an integral part of the reaction mechanisms of this class of enzymes. The situation for other desaturase-like enzymes involved in proximal lipid modification is less clear and is summarized in Table II. The presence of an N-terminal Cyt b5 domain has been observed in higher plant Δ8-sphingolipid desaturase (Sperling et al., 1998) and in yeast FAH1 sphingolipid α-hydroxylase (Mitchell and Martin, 1997). However, in FAH1, an Arabidopsis homolog of this enzyme did not contain any domain with homology to Cyt b5. When this plant FAH1 homolog was expressed in yeast fah1 deletion mutants, it was capable of α-hydroxylation of substrate long-chain fatty acids. However, the plant enzyme did not fully restore hydroxylated fatty acids to wild-type levels even though it was overexpressed via a GAL1 promoter on a multicopy plasmid (Mitchell and Martin, 1997). It may be that the efficiency/activity of the (Cyt b5 domain-lacking) plant enzyme is reduced, though obviously other factors (such as expression in a heterologous system) could also be influential. The authors of that study also noted the presence of an N-terminal Cyt b5 domain in a putative FAH1 homolog from C. elegans, but this domain was missing in a Schizosaccharomyces pombe FAH1 homolog. This would indicate that, at least in the α-hydroxylation of the sphingolipid long-chain fatty acids, the Cyt b5 domain is not an obligate requirement for enzyme function. This is in contrast to our observations with the Δ6-fatty acid desaturase and to that reported for the OLE1 Δ9-desaturase (Mitchell and Martin, 1995). It will be of interest to determine if further examples of front-end fatty acid-modifying enzymes exist as fusions with their electron donor.
Table II.
Examples of “front-end” desaturases and related proximal lipid modifying enzymes
| Enzyme | Organism | Accession No. | b5 | |
|---|---|---|---|---|
| Δ5-Fatty acid desaturases | ||||
| Mortierella alpina | Fungus | AF054824 | ✓ | |
| Caenorhabditis elegans | Nematode | AF078796 | ✓ | |
| Dictyostelium discoideum | Slug | AB022097 | ✓ | |
| Δ6-Fatty acid desaturases | ||||
| B. officinalis | Plant | U79010 | ✓ | |
| Physcomitrella patens | Moss | AJ980222 | ✓a | |
| C. elegans | Nematode | AF031477 | ✓ | |
| Homo sapiens | Human | Z44979 | ✓ | |
| Synechocystis | Cyanobacteria | L11421 | ✗b | |
| Δ8-Sphingolipid desaturases | ||||
| Arabidopsis | Plant | AJ224161 | ✓ | |
| Helianthus annuus | Plant | X87143 | ✓ | |
| Brassica napus | Plant | AJ224160 | ✓ | |
| Ricinus communis | Plant | AF005096 | ✓ | |
| FAH1 sphingolipid α-hydroxylase | ||||
| S. cerevisiae | Yeast | Z49260 | ✓ | |
| C. elegans | Nematode | Z81038 | ✓ | |
| Arabidopsis | Plant | AF021804 | ✗ | |
| Schizosaccharomyces pombe | Yeast | Z97209 | ✗ |
The presence of a N-terminal Cyt b5 domain is indicated by a tick. The GenBank accession numbers for all sequences are given.
The Cyt b5 domain present in this Δ6-desaturase is internal.
The electron donor for cyanobacterial desaturases is ferredoxin.
CONCLUSIONS
We have shown that His-41 of the Cyt b5 domain is likely essential for the enzymatic activity of the borage Δ6-fatty acid desaturase, as determined by site-directed mutagenesis and expression in transgenic plants. These results support the hypothesis that the presence of a N-terminal Cyt b5 domain in this front-end desaturase is an obligate requirement for enzymatic activity.
Footnotes
IACR-Long Ashton Research Station receives granted-aided support from the Biotechnology and Biological Sciences Research Council, UK. This work was partially funded by Scotia Pharmaceuticals (Stirling, UK).
LITERATURE CITED
- Arondel V, Lemieux B, Hwang I, Gibson S, Goodman HM, Somerville CR. Map-based cloning of a gene controlling omega-3 fatty acid desaturation in Arabidopsis. Science. 1992;258:1353–1355. doi: 10.1126/science.1455229. [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Ser III Life Sci. 1993;316:1194–1199. [Google Scholar]
- Cho HP, Nakamura MT, Clarke SD. Cloning, expression and nutritional regulation of the mammalian Δ6 desaturase. J Biol Chem. 1999;274:4711–4717. doi: 10.1074/jbc.274.1.471. [DOI] [PubMed] [Google Scholar]
- Girke T, Schmidt H, Zahringer U, Reski R, Heinz E. Identification of a novel Δ6-acyl-group desaturase by targeted gene disruption in Physcomitrella patens. Plant J. 1998;15:39–48. doi: 10.1046/j.1365-313x.1998.00178.x. [DOI] [PubMed] [Google Scholar]
- Heinz E. Biosynthesis of polyunsaturated fatty acids. In: Moore TS Jr, editor. Lipid Metabolism in Plants. Boca Raton, FL: CRC Press; 1993. pp. 33–89. [Google Scholar]
- Itoh R, Toda K, Takahashi H, Takano H, Kuroiwa T. Delta 9 fatty acid desaturase gene containing a carboxyl-terminal cytochrome b5 domain from the red alga Cyanidioschyzon merolae. Curr Genet. 1998;33:165–170. doi: 10.1007/s002940050323. [DOI] [PubMed] [Google Scholar]
- Kearns EV, Hugly S, Somerville CR. The role of cytochrome b5 in Δ12 desaturation of oleic acid by microsomes of safflower (Carthamus tinctorius L.) Arch Biochem Biophys. 1991;284:431–436. doi: 10.1016/0003-9861(91)90319-e. [DOI] [PubMed] [Google Scholar]
- Knutzon DS, Thurmond JM, Huang Y-S, Chaudhary S, Bobik EG, Chan GM, Kirchner SJ, Mukerji P. Identification of Δ5-desaturase from Mortierella alpina by heterologous expression in Baker's yeast and canola. J Biol Chem. 1998;273:29360–29366. doi: 10.1074/jbc.273.45.29360. [DOI] [PubMed] [Google Scholar]
- Lederer F. The cytochrome b5-fold: an adaptable molecule. Biochimie. 1994;76:974–992. doi: 10.1016/0300-9084(94)90144-9. [DOI] [PubMed] [Google Scholar]
- Meesters PAEP, Springer J, Eggink G. Cloning and expression of the Δ9 fatty acid desaturase gene from Cryptococcus curvatus ATCC 20509 containing histidine boxes and cytochrome b5 domain. Appl Microbiol Biotechnol. 1997;47:663–667. doi: 10.1007/s002530050992. [DOI] [PubMed] [Google Scholar]
- Michaelson LV, Lazarus CM, Griffiths G, Napier JA, Stobart AK. Isolation of a Δ5-fatty acid desaturase gene from Mortierella alpina. J Biol Chem. 1998a;273:19055–19059. doi: 10.1074/jbc.273.30.19055. [DOI] [PubMed] [Google Scholar]
- Michaelson LV, Napier JA, Lazarus CM, Griffiths G, Stobart AK. Isolation of a Δ5-desaturase gene from Caenorhabditis elegans. FEBS Lett. 1998b;439:215–218. doi: 10.1016/s0014-5793(98)01385-4. [DOI] [PubMed] [Google Scholar]
- Miquel M, Browse J. Arabidopsis mutants deficient in polyunsaturated fatty acid synthesis. J Biol Chem. 1992;267:1502–1509. [PubMed] [Google Scholar]
- Mitchell AG, Martin CE. A novel cytochrome b5-like domain is linked to the carboxyl terminus of the Saccharomyces cerevisiae Δ9 fatty acid desaturase. J Biol Chem. 1995;270:29766–29772. doi: 10.1074/jbc.270.50.29766. [DOI] [PubMed] [Google Scholar]
- Mitchell AG, Martin CE. Fah1p, a Saccharomyces cerevisiae cytochrome b5 fusion protein, and its Arabidopsis thaliana homolog that lacks the cytochrome b5 domain both function in the α-hydroxylation of sphingolipid-associated very long chain fatty acids. J Biol Chem. 1997;272:28281–28288. doi: 10.1074/jbc.272.45.28281. [DOI] [PubMed] [Google Scholar]
- Napier JA, Hey SJ, Lacey DJ, Shewry PR. Identification of a Caenorhabditis elegans Δ6-fatty-acid-desaturase by heterologous expression in Saccharomyces cerevisiae. Biochem J. 1998;330:611–614. doi: 10.1042/bj3300611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napier JA, Sayanova O, Sperling P, Heinz E. A growing family of cytochrome b5 fusion desaturases. Trends Plant Sci. 1999;4:2–5. [Google Scholar]
- Napier JA, Sayanova O, Stobart AK, Shewry PR. A new class of cytochrome b5 fusion proteins. Biochem J. 1997;328:717–720. [PMC free article] [PubMed] [Google Scholar]
- Okuley J, Lightner J, Feldmann K, Yadav N, Lark E, Browse J. Arabidopsis FAD2 gene encodes the enzyme this is essential for polyunsaturated lipid synthesis. Plant Cell. 1994;6:147–158. doi: 10.1105/tpc.6.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayanova O, Smith MA, Lapinskas P, Stobart AK, Dobson G, Christie WW, Shewry PR, Napier JA. Expression of a borage desaturase cDNA containing an N-terminal cytochrome b5 domain results in the accumulation of high levels of Δ6-desaturated fatty acids in transgenic tobacco. Proc Natl Acad Sci USA. 1997;94:4211–4216. doi: 10.1073/pnas.94.8.4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanklin J, Cahoon EB. Desaturation and related modifications of fatty acids. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:611–641. doi: 10.1146/annurev.arplant.49.1.611. [DOI] [PubMed] [Google Scholar]
- Smith MA, Cross AR, Jones OTG, Griffiths WT, Stymne S, Stobart AK. Electron-transport components of the 1-acyl-2-oleoyl-sn-glycero-3-phosphocholine Δ12-desaturase (Δ12-desaturase) in microsomal preparations from developing safflower (Carthamus tinctorius L.) cotyledons. Biochem J. 1990;272:23–29. doi: 10.1042/bj2720023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Jonsson L, Stymne S, Stobart AK. Evidence for cytochrome b5 as electron donor in ricinoleic acid biosynthesis in microsomal preparations from developing castor bean (Ricinus communis L.) Biochem J. 1992;287:141–144. doi: 10.1042/bj2870141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Napier JA, Stymne S, Tatham AS, Shewry PR, Stobart AK. Expression of a biologically active plant cytochrome b5 in Escherichia coli. Biochem J. 1994;303:73–79. doi: 10.1042/bj3030073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Stobart AK, Shewry PR, Napier JA. Cytochrome b5 and polyunsaturated fatty acid biosynthesis. In: Shewry PR, Napier JA, Davis PJ, editors. Engineering Crop Plants for Industrial End Uses. Portland Press London; 1998. pp. 181–188. [Google Scholar]
- Somerville CR, Browse J. Plant lipids: metabolism, mutants and membranes. Science. 1991;252:80–87. doi: 10.1126/science.252.5002.80. [DOI] [PubMed] [Google Scholar]
- Sperling P, Schmidt H, Heinz E. A cytochrome-b5-containing fusion protein similar to acyl lipid desaturases. Eur J Biochem. 1995;232:798–805. [PubMed] [Google Scholar]
- Sperling P, Zähringer U, Heinz E. A sphingolipid desaturase from higher plants: identification of a new cytochrome b5 fusion protein. J Biol Chem. 1998;273:28590–28596. doi: 10.1074/jbc.273.44.28590. [DOI] [PubMed] [Google Scholar]
- van de Loo FJ, Broun P, Turner S, Somerville C. An oleate 12-hydroxylase from Ricinus communis L. is a fatty acyl desaturase homolog. Proc Natl Acad Sci USA. 1995;92:6743–6747. doi: 10.1073/pnas.92.15.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]