Abstract
A cascade of alternative sigma factors directs developmental gene expression during spore formation by the bacterium Bacillus subtilis. As the spore develops, a tightly regulated switch occurs in which the early-acting sigma factor σF is replaced by the late-acting sigma factor σG. The gene encoding σG (sigG) is transcribed by σF and by σG itself in an autoregulatory loop; yet σG activity is not detected until σF-dependent gene expression is complete. This separation in σF and σG activities has been suggested to be due at least in part to a poorly understood intercellular checkpoint pathway that delays sigG expression by σF. Here we report the results of a careful examination of sigG expression during sporulation. Unexpectedly, our findings argue against the existence of a regulatory mechanism to delay sigG transcription by σF and instead support a model in which sigG is transcribed by σF with normal timing, but at levels that are very low. This low-level expression of sigG is the consequence of several intrinsic features of the sigG regulatory and coding sequence—promoter spacing, secondary structure potential of the mRNA, and start codon identity—that dampen its transcription and translation. Especially notable is the presence of a conserved hairpin in the 5’ leader sequence of the sigG mRNA that occludes the ribosome-binding site, reducing translation by up to 4-fold. Finally, we demonstrate that misexpression of sigG from regulatory and coding sequences lacking these features triggers premature σG activity in the forespore during sporulation, as well as inappropriate σG activity during vegetative growth. Altogether, these data indicate that transcription and translation of the sigG gene is tuned to prevent vegetative expression of σG and to ensure the precise timing of the switch from σF to σG in the developing spore.
Author summary
Global changes in gene expression occur during normal cellular growth and development, as well as during cancer cell transformation and bacterial pathogenesis. In this study we have investigated the molecular mechanisms that drive the switch from early to late developmental gene expression during spore formation by the model bacterium Bacillus subtilis. At early times, gene expression in the developing spore is directed by the transcription factor σF; at later times σF is replaced by σG. An important, yet poorly understood aspect of this σF-to-σG transition is how σG activation is delayed until the early, σF-directed phase of gene expression is complete. Here we have carefully examined expression of the gene encoding σG, sigG, and found that its transcription and translation are ordinarily dampened by several features of its regulatory and coding sequences. Moreover, we have found that this “tuning” of sigG expression is required for proper timing of the switch to σG. These results reframe our understanding of how sigG is regulated during B. subtilis sporulation and, more broadly, advance our understanding of how global changes in gene expression can be precisely executed at the molecular/genetic level.
Introduction
Cells across all domains of life alter their phenotypes through global changes in gene expression. In bacteria, global changes in gene expression drive phenotypic changes critical for growth, development, and pathogenesis. For example, the ability of the human pathogen Chlamydia trachomatis to progress through its infectious cycle requires sequential transitions between three stage-specific networks of gene regulation [1]. Here, we study a switch in gene expression that occurs during the developmental process of spore formation by the soil bacterium Bacillus subtilis, a premier model system for studies of regulation [2,3]. At the onset of B. subtilis sporulation, which is triggered by nutrient depletion, the rod shaped bacterial cell divides asymmetrically, resulting in two daughter cells of unequal size and fate. The smaller “forespore” becomes the mature, dormant spore, while the larger “mother cell” aids the development of the forespore but ultimately dies. Initially these two cells lie side-by-side; subsequently, the mother cell membranes migrate around the forespore in a process called engulfment, resulting in the forespore being pinched off as a free protoplast within the mother cell cytoplasm (Fig 1A). After engulfment, the forespore is encased in a protective peptidoglycan cortex and protein coat, and is then released into the environment as a mature spore upon lysis of the mother cell.
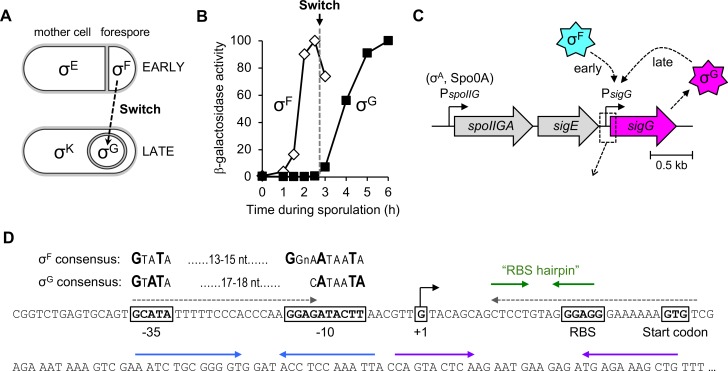
Fig 1. Expression of the sigG gene and the switch from σF to σG during B. subtilis sporulation.
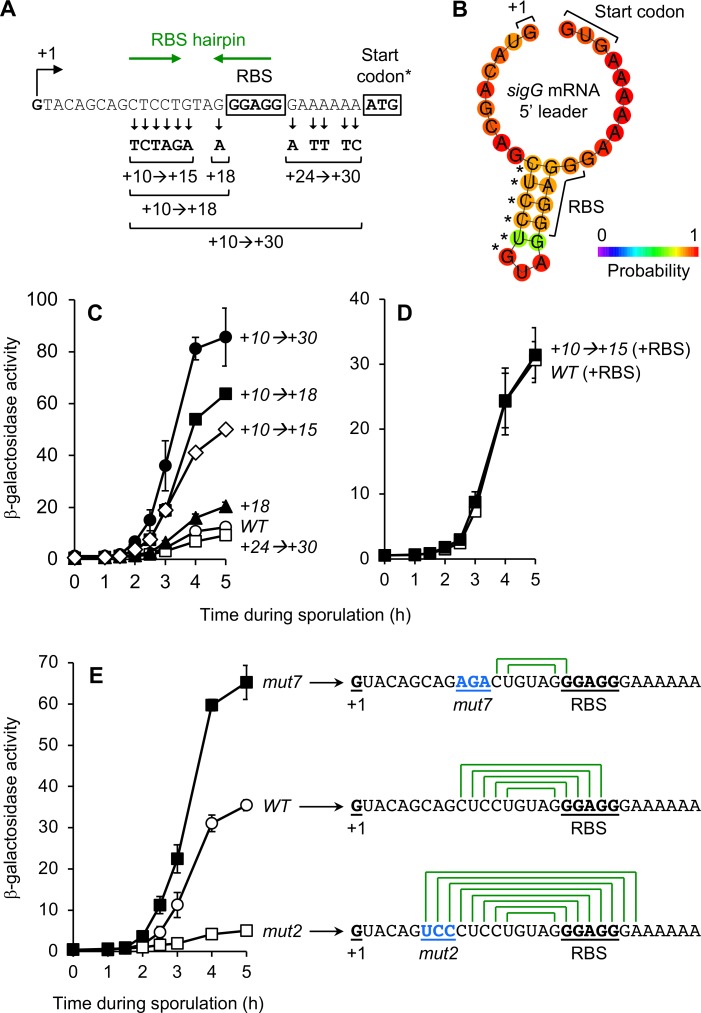
(A) Cartoon depicting the sigma factors that govern gene expression during early (top) and late (bottom) stages of B. subtilis sporulation. At early times, the sigma factors σF and σE control gene expression in the forespore and mother cell, respectively. At later times, after forespore engulfment by the mother cell, σF is replaced by σG and σE is replaced by σK. The switch from σF to σG is indicated by a dashed arrow. (B) The switch from σF to σG. The σF-dependent expression of a PspoIIQ-lacZ reporter (open diamonds) and σG-dependent expression of a PsspB-lacZ reporter (closed squares) were monitored during sporulation of wild type cells. (Strains AHB881 and AHB317, respectively.) All lacZ reporters used in this study, unless otherwise noted, were at the non-essential amyE locus. β-Galactosidase activity is reported as the percent maximum for each. The timing of the σF-to-σG switch, between sporulation hours 2.5 and 3, is indicated by a dashed gray line. (C) Cartoon depicting the sigG gene in its native chromosomal location downstream of the spoIIGA-sigE operon, and our new, simplified model for its transcriptional regulation by σF at early times and σG itself at late times. Dashed box/arrow indicates region depicted in (D). (D) Sequence of the sigG upstream regulatory region and 5’ coding region (codons 1–28). The -35 and -10 sigG promoter elements [9] are boxed, with the consensus σF and σG-recognized sequences shown above [38]. Also boxed are the sigG +1 transcription start site, RBS, and GTG start codon. Gray dashed arrows indicate complementary sequences predicted to form a hairpin structure that blocks translation of read-through transcripts originating from PspoIIG [52]; note that this hairpin cannot form in transcripts originating from PsigG. Green, blue, and purple solid arrows indicate complementary sequences predicted to form hairpin secondary structures in mRNA transcripts originating from PsigG.
The morphological events of sporulation are orchestrated by a complex gene regulatory network that coordinates the expression of hundreds of sporulation genes at the right time and in the right cell [3,4]. This gene regulatory network operates primarily to ensure the sequential and compartment-specific appearance of four sporulation sigma (σ) factors—σF, σE, σG, and σK—that bind and impart promoter specificity upon core RNA polymerase (RNAP). Early in sporulation, following asymmetric division, σF and σE direct gene expression in the forespore and mother cell, respectively. Later, after the completion of engulfment, σG takes over for σF in the forespore and σK replaces σE in the mother cell (Fig 1A). These two transitions, from σF- to σG-directed gene expression in the forespore and from σE- to σK-directed gene expression in the mother cell, are tightly regulated such that no temporal overlap between the activities of the early and late sigma factors can be detected [5]. However, the molecular mechanisms that control these global transitions in gene expression with such precision are not well understood.
In this study, we sought to identify molecular mechanisms that help to orchestrate the switch from σF to σG in the B. subtilis forespore (Fig 1A and 1B). The current model for the σF-to-σG switch, summarized here, is based on literature spanning several decades. To begin, σF is activated in the forespore soon after asymmetric cell division via a complex, but relatively well-characterized regulatory circuit [6]. In turn, σF directs the transcription of genes required for early forespore development as well as the gene sigG (previously spoIIIG) that encodes σG [7–9]. Any σG produced at these early times remains inactive; only after forespore engulfment is complete does σG become active and replace σF. The deactivation of σF is poorly understood, but involves the small protein Fin (previously YabK) as well as other unidentified mechanisms [10,11]. The subsequent activity of σG requires an intercellular channel apparatus comprised of the mother cell proteins SpoIIIAA-AH and forespore protein SpoIIQ (reviewed in [12]). Interestingly, this SpoIIIAA-AH•SpoIIQ channel does not specifically regulate σG, but instead is required more generally to maintain forespore physiology and to support any forespore gene expression at late times, even that engineered to be directed by a heterologous phage RNAP [13,14]. Finally, once active, σG directs the transcription of genes required for late forespore development, as well as its own gene in an autoregulatory loop [9], thus locking in the transition to the late program of developmental gene expression in the forespore.
A major unanswered question regarding the switch from early to late gene expression in the forespore is how σG activity is delayed until the early, σF-directed phase of gene expression is complete. One protein that has been implicated in controlling early σG activity is the anti-sigma factor CsfB (also called Gin), which is expressed under the control of σF at early times and is a potent antagonist of σG [15–18]. Deletion of csfB has been reported to cause premature activation of σG in a subset of sporulating cells [16]. But other studies have concluded that ΔcsfB does not alter the level nor timing of peak σG activity [19], and ΔcsfB cells display no defect in spore formation [15]. These results suggest that CsfB-independent mechanisms must also be in place to restrict σG activity at early times.
A second possible explanation for the delay in σG activation comes from reports that the transcription of sigG by σF is delayed by up to an hour relative to other σF target genes [20,21]. In addition, σF-dependent transcription of sigG, unlike that of other σF target genes, has been reported to require the forespore membrane protein SpoIIQ and the early-acting mother cell sigma factor σE [20,22,23]. These findings have led to speculation that a signaling pathway, perhaps involving SpoIIQ, specifically couples σF-dependent sigG transcription to the activation of σE in the mother cell [2,24]. Satisfyingly, a checkpoint mechanism such as this would account for the observed delay in sigG expression, given that both spoIIQ expression and σE activation require the earlier activity of σF [25–27]. To test this idea, several groups have monitored the timing of σG activation during sporulation of strains engineered to express sigG under the control of a more typical “early” σF-target promoter. The majority found no evidence of premature σG activity in such engineered strains [17,19,28], although one study reported that if sigG expression was boosted (by inserting three copies of the engineered sigG construct), inappropriate σG activity could be detected at early times [14]. This may hint at the importance of sigG expression levels, in addition to timing, in dictating the onset of σG activity. Overall, however, the regulation of sigG expression and its impact upon the timing of σG activity remains an open question.
Here we report the results of a careful examination of sigG expression that calls into question long-standing assumptions about its regulation. We argue here that sigG transcription by σF is not delayed, nor does it require a specific intercellular signaling pathway originating in the mother cell. Instead, we propose a simpler model (Fig 1C) in which sigG is first transcribed by σF with normal timing, but at levels that are very low and, as such, difficult to detect. Subsequently, the majority of sigG transcription occurs later under the control of σG itself. We present evidence that the low-level expression of sigG at early times is a consequence of four intrinsic features of the sigG regulatory and coding sequences that dampen its transcription and translation: suboptimal spacing between the -10 and -35 sigG promoter elements, a suboptimal translational start codon, a hairpin in the 5’ leader sequence of the sigG mRNA that occludes the ribosome-binding site (RBS), and a suboptimal 5’ coding sequence. Finally, we demonstrate that misexpression of sigG from regulatory sequences lacking these features results in inappropriate σG activity during vegetative growth and premature σG activity in the forespore during sporulation.
Results
sigG expression can be detected before and after the switch from σF to σG
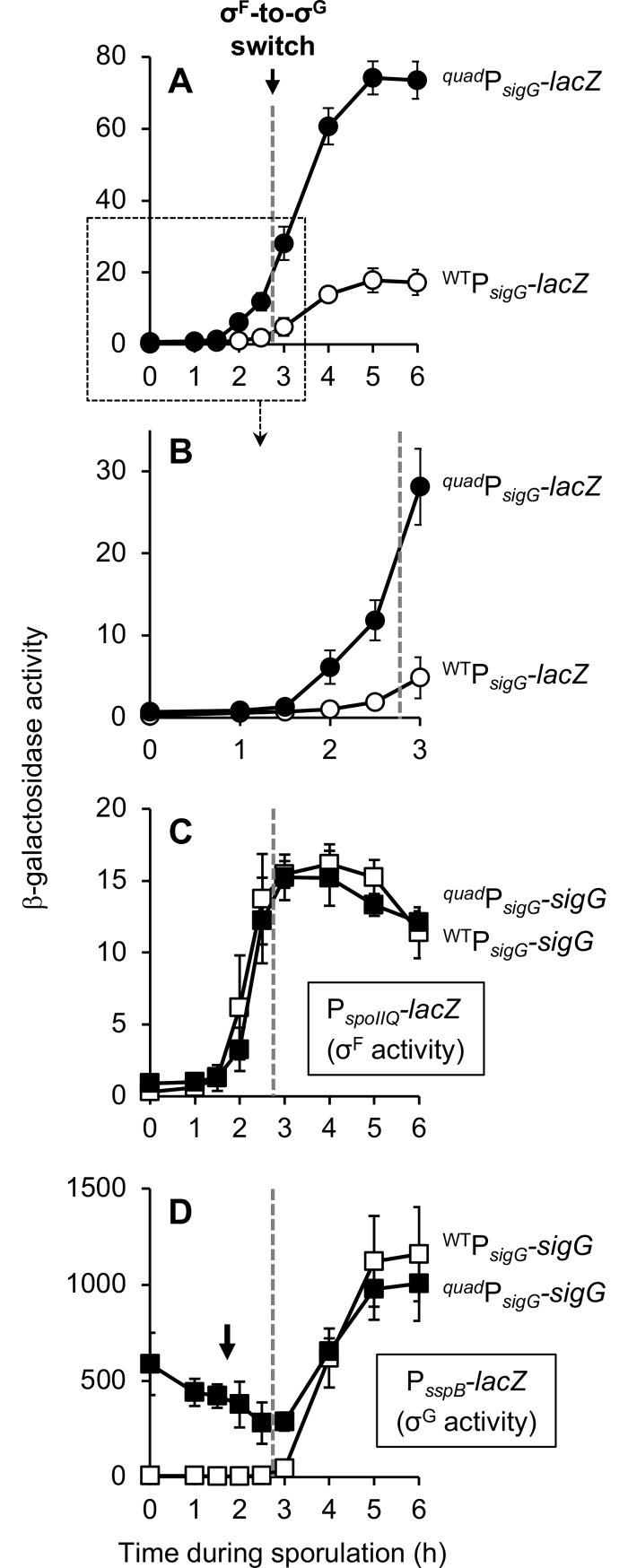
Before examining sigG expression during sporulation, we first determined the timing of the switch from σF to σG under our sporulation conditions using lacZ reporters fused to promoters under the exclusive control of each sigma factor. (All lacZ reporter genes in this study were integrated at the non-essential amyE locus.) As shown in Fig 1B, β-galactosidase production from a lacZ reporter fused to the σF-dependent spoIIQ promoter (PspoIIQ) [27] was first detected above background at approximately hour 1.5 of sporulation, peaked at hour 2.5, and declined thereafter. In contrast, a lacZ reporter fused to the σG-dependent sspB promoter (PsspB) [29] turned on at hour 3 of sporulation, after which time β-galactosidase production continued to rise through hour 5 (Fig 1B). Together, these findings indicate that under our sporulation conditions, σF is active from hours 1.5–2.5, σG is active from hours 3–6, and the switch from σF to σG occurs between hours 2.5–3.
To monitor sigG expression, we fused the entire sigG regulatory region (-203 to +114, in reference to a +1 transcription start site [9]), including the sigG promoter (PsigG), ribosome binding site (RBS), and first 28 codons, in-frame to lacZ (see Fig 1D). (In this and other experiments in this study, unless otherwise indicated, the native sigG gene was left intact, under the control of its wild-type regulatory sequences.) As shown in Fig 2A and 2B, expression of this PsigG-sigG1-28-lacZ reporter gene was first detectable above background at very low, but statistically significant levels at hour 2.5 of sporulation, after which time its expression increased substantially. This profile indicates that the majority of sigG expression occurs during times when σG is active (hours 3–6), but that very low levels of sigG expression can be detected at slightly earlier times (hour 2.5).
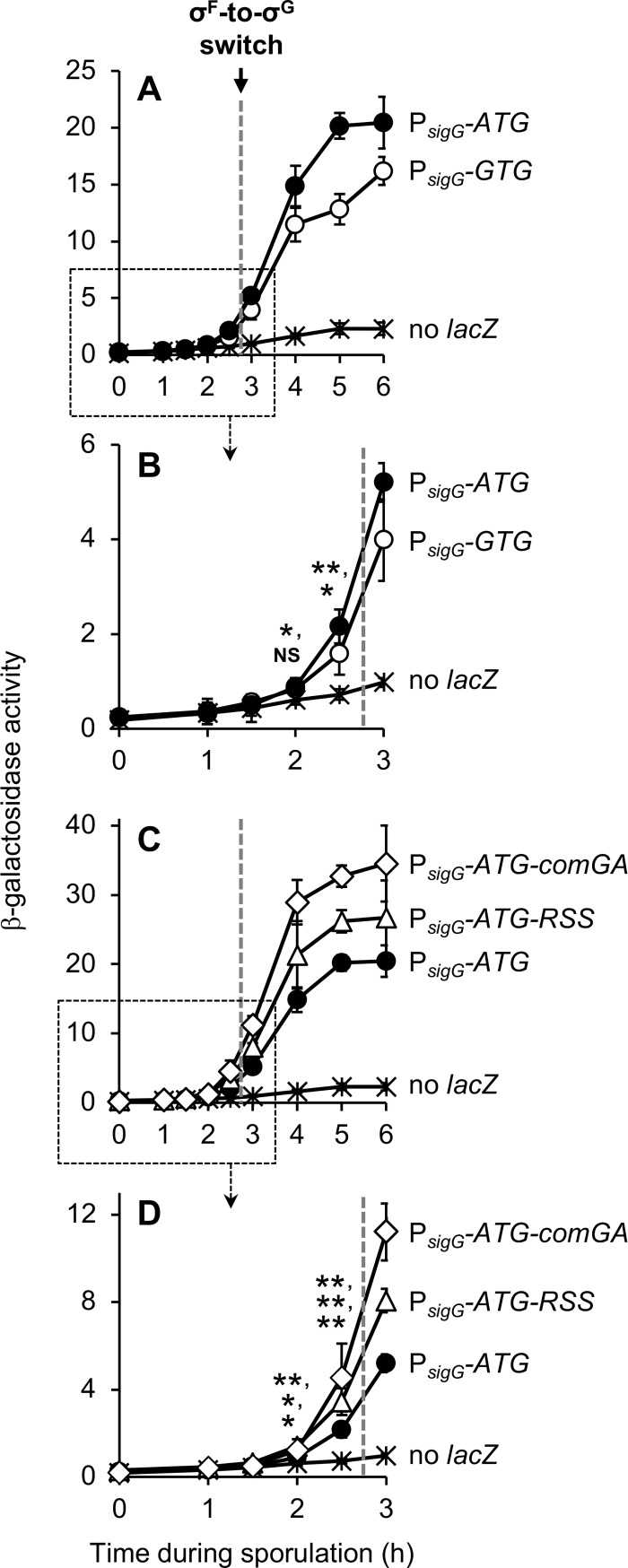
Fig 2. PsigG activity is detected both before and after the switch from σF to σG.
(A, B) The activities of PsigG reporters harboring the native GTG start codon, PsigG-sigG1-28-lacZ (PsigG-GTG; open circles) or engineered to harbor an ATG start codon, PsigG-ATG-sigG2-28-lacZ (PsigG-ATG; closed circles), were measured during a time course of sporulation. (Strains EBM177 and EBM175, respectively.) Background β-galactosidase activity was measured in a strain without a lacZ reporter, PY79 (no lacZ; asterisks). Note that (B) provides a zoomed view and statistical significance of early sporulation data points from the boxed area of (A). (C, D) Strains harboring PsigG reporters with altered sigG 5’ coding sequence, one engineered to reduce secondary structure (RSS) in sigG codons 2–28, PsigG-ATG-RSSsigG2-28-lacZ (PsigG-ATG-RSS; open triangles), and another harboring comGA codons 2–8 in place of sigG codons 2–28, PsigG-ATG-comGA2-8-lacZ (PsigG-ATG-comGA; open diamonds), were monitored for β-galactosidase production during sporulation. (Strains EBM237 and JJB31, respectively.) Data for the PsigG-ATG-sigG2-28-lacZ (PsigG-ATG; closed circles) strain (EBM175) and PY79 (no lacZ, asterisks), the same as shown in (A) and (B), are also included for reference. Note that (D) provides a zoomed view and statistical significance of early sporulation data points from the boxed area of (C). The timing of the σF-to-σG switch, between sporulation hours 2.5 and 3, is indicated in each panel by a dashed gray line. For all panels, error bars indicate ± standard deviations based on three independent experiments. *p < 0.05, **p < 0.01, NS not significant, Student’s t-test.
To improve our ability to detect low levels of PsigG transcription that may be occurring at early times of sporulation, we altered our lacZ reporter gene to optimize translation. First, we replaced the native sigG translation start codon, GUG (GTG), with the more efficiently utilized start codon AUG (ATG) [30]. Consistent with modestly improved translation, the modified reporter gene displayed a 1.3-fold increase in β-galactosidase activity at sporulation hours 2.5–6 relative to the original reporter gene harboring the native GTG start codon (Fig 2A). Notably, introduction of the ATG start codon also permitted the detection of weak, but statistically significant β-galactosidase activity at hour 2 of sporulation (Fig 2B). Given that the native sigG gene in this experiment was expressed from its unaltered wild-type regulatory sequences (including its suboptimal GTG start codon), any potential further amplification of sigG expression that might have resulted from enhanced σG autoregulation (i.e. if GTG had been replaced with ATG at the native site) was not measured in this assay.
To further optimize translation, we turned our attention to the remaining 27 sigG codons (codons 2–28) fused in frame to lacZ. The 5’ coding region can be a major determinant of translation efficiency, likely due to the presence of rare codons and/or the propensity of this region to form RNA secondary structures that impede translation initiation (reviewed in [31]). Notably, the sigG 5’ coding sequence has the potential to form several hairpin structures (Fig 1D, indicated by blue and purple arrows). As a first strategy to optimize the 5’ coding region of our PsigG reporter, we utilized the mRNA Optimizer tool [32] to redesign sigG codons 2–28 such that the potential for secondary structure was minimized without altering the encoded protein. (See S1 Fig for the optimized sequence and its secondary structure potential; also, we note that this tool does not optimize for species-specific biases in codon usage.) In a second approach, we altogether replaced sigG codons 2–28 with codons 2–8 of the highly expressed B. subtilis gene comGA, a strategy that has been demonstrated to significantly improve translation of another reporter gene [33]. As shown in Fig 2C and 2D, the resulting PsigG reporters with ATG start codons and optimized 5’ coding sequences were expressed more robustly (~1.5-fold and ~2-fold, respectively) at sporulation hours 2–6 relative to the reporter harboring only the ATG alteration. Together, this data indicates that sigG translation is ordinarily dampened by a suboptimal start codon and suboptimal 5’ coding sequence. Moreover, the improved reporter gene expression supports the conclusion that PsigG is transcriptionally active both before and after the switch from σF to σG.
The sigG promoter may be aberrantly activated by σF at late times in the absence of σG
In the generally accepted model for regulation of sigG expression, sigG is transcribed first under the control of σF (albeit with unique regulation, see Introduction and below) and then under the control of σG in an auto-regulatory loop. To determine the contribution of σF versus σG to the transcription of PsigG (using our strongest reporter, PsigG-ATG-comGA2-8-lacZ, henceforth simply PsigG-lacZ), we deleted the gene encoding σG (sigG) alone or in combination with sigF, which encodes σF. We predicted that ΔsigG would eliminate any late PsigG activity that depends upon σG, leaving intact the σF-dependent contribution to PsigG expression, which would then be eliminated upon introduction of ΔsigF. However, we found that PsigG-lacZ expression was not at all reduced at any time by ΔsigG, but in fact was slightly stimulated at later times; yet as predicted, the ΔsigF ΔsigG double mutant displayed no detectable β-galactosidase activity at any timepoint (Fig 3A). Close examination of the literature revealed that this effect of ΔsigG upon PsigG should not have been unexpected. Some of the earliest studies of sigG (then called spoIIIG) reported that PsigG activity was unaltered or even stimulated by sigG mutation [9,34,35], although other studies observed modest or even significant decreases [20,36].
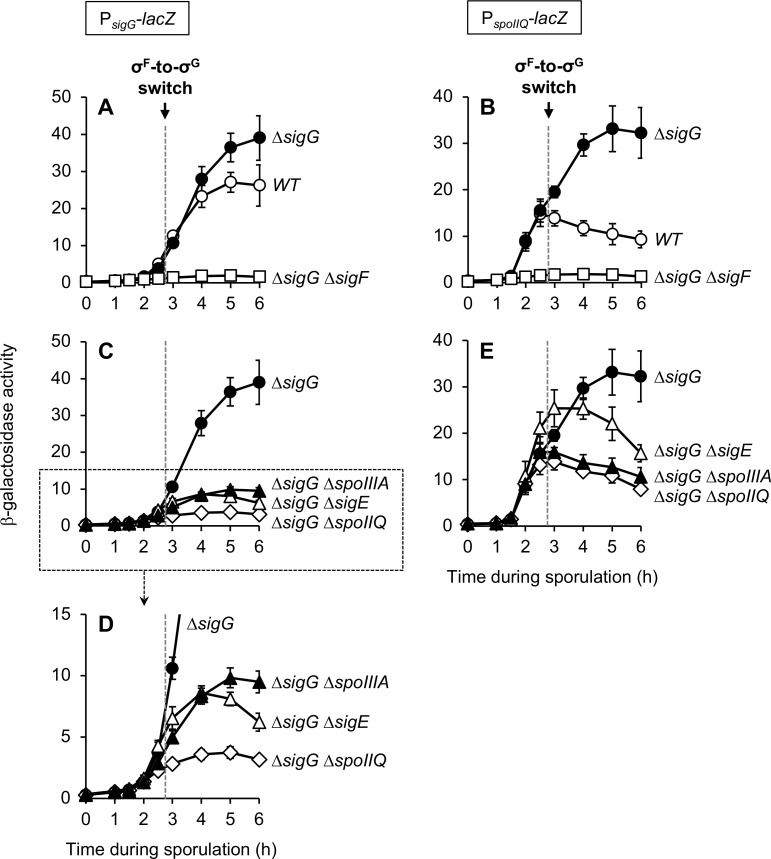
Fig 3. Late σF-dependent expression of both PsigG and PspoIIQ requires SpoIIQ, σE, and SpoIIIAA-AH.
The activity of PsigG-lacZ (A, C, D) or PspoIIQ-lacZ (B, E) was monitored during sporulation of strains with the following genotypes: wild type (WT; open circles), ΔsigG (closed circles), ΔsigG ΔsigF (open squares), ΔsigG ΔspoIIQ (open diamonds), ΔsigG ΔsigE (open triangles), and ΔsigG ΔspoIIIAA-AH (closed triangles). (PsigG-lacZ strains were JJB31, JJB73, JJB75, JJB79, JJB85, and JJB77, respectively. PspoIIQ-lacZ strains were AHB881, AHB882, AHB915, AHB916, AHB917, and AHB1017, respectively.) For clarity, only data from a subset of these strains are presented in each graph (as labeled) and, also for clarity, the data for the ΔsigG strain of each (closed circles) is presented in all graphs. Note that (D) provides a zoomed view of the data from the boxed area of (C). The timing of the σF-to-σG switch, between sporulation hours 2.5 and 3, is indicated in each panel by a dashed gray line. For all panels, error bars indicate ± standard deviations based on three independent experiments.
One interpretation of our results could be that all PsigG activity, at both early and late times during sporulation, is due exclusively to σF and not at all to σG. However, we find this to be an unsatisfactory interpretation given that σG has been shown to activate PsigG in vitro and in directed in vivo experiments [9,37,38]. Furthermore, there is no evidence that σF remains active at later times of sporulation during which σG is active; in fact, in one study, σF and σG activities could not be detected to overlap [5].
We instead suggest an alternative explanation for these data, namely that the late expression of PsigG-lacZ in ΔsigG cells is due to aberrant σF activity that is known to be unmasked in the absence of the late-acting sigma factor σG [13,15,39]. The cause of this aberrant activity is poorly understood but likely involves the σF inhibitor Fin, which is expressed partly under the control of σG, as well as other σG-dependent, Fin-independent mechanisms [10,11]. To demonstrate the likelihood that PsigG-lacZ is aberrantly activated by σF at late times in ΔsigG cells, we repeated the experiment presented for PsigG-lacZ (Fig 3A), but with strains harboring the exclusively σF-dependent PspoIIQ-lacZ reporter gene. As shown in Fig 3B and as we have previously reported [13], deletion of sigG unmasks a late phase of PspoIIQ-lacZ expression from hours 3–5 of sporulation. All PspoIIQ-lacZ expression was eliminated by the further introduction of ΔsigF (Fig 3B), indicating that the early (normal) and late (abnormal) transcription of PspoIIQ-lacZ in ΔsigG cells is driven by σF. It therefore seems plausible that the sigG promoter, like the spoIIQ promoter, is subject to aberrant late transcription by σF in the absence of σG. Importantly, this could make it appear as if σF—and not σG—ordinarily drives PsigG transcription at later times during sporulation. As such, we conclude that the contribution of σF versus σG to the transcription of PsigG in wild type cells cannot be determined with confidence through the use of a ΔsigG mutant.
PsigG is unlikely to be the target of a specific intercellular signaling pathway
Despite our inability to genetically dissect the contribution of σF versus σG to the activation of our PsigG-lacZ reporter, we reasoned that we could tentatively assign its early expression (hours 2–2.5) to σF and its late expression (hours 3–6) to σG, based on the timing of the switch from σF to σG under our conditions (between hours 2.5 and 3, Fig 1B). This simple interpretation is complicated, however, by the possibility that PsigG may not be a typical σF-dependent promoter. Unlike other σF-target promoters, σF-dependent transcription of PsigG has been reported to be delayed and to depend upon the mother cell sigma factor σE and the forespore protein SpoIIQ (itself under the control of σF) [20–23]. These data have been interpreted as evidence for an intercellular signaling pathway/checkpoint mechanism that specifically delays PsigG transcription by σF [2,24]. Upon closer examination of the original studies, however, we noted that the relevant experiments were performed in strains deleted for sigG to eliminate σG-dependent PsigG transcription. In light of our finding that PsigG may behave aberrantly in the absence of σG, as well as recent strides in our understanding of the function of SpoIIQ (see below), we reasoned that these data and their interpretation warranted re-evaluation.
To begin, we sought to reproduce these original findings with our PsigG-lacZ reporter. As shown in Fig 3C, PsigG-lacZ expression in ΔsigG cells was indeed significantly reduced at sporulation hour 3 and later when spoIIQ or sigE (the gene encoding σE) were deleted. Interestingly, however, β-galactosidase production at earlier times (hours 2–2.5), albeit relatively weak, was unaffected by deletion of spoIIQ or sigE (Fig 3D). These data indicate that in ΔsigG cells, σF-dependent expression of PsigG at late times (but not early times) requires σE and SpoIIQ, a finding that is mostly consistent with previous studies [20,22,23].
In the years since these previous studies were performed, SpoIIQ has been determined to assemble with the eight mother cell proteins SpoIIIAA-AH into a channel apparatus that connects the forespore and mother cell at intermediate stages of sporulation (reviewed in [12]). This SpoIIIAA-AH•SpoIIQ channel is generally required for any late gene expression in the forespore, including that normally directed by σG, abnormally directed by σF (as in a ΔsigG mutant), or engineered to be directed by a heterologous RNAP [13]. We reasoned, therefore, that the σE- and SpoIIQ-dependence of late σF-dependent PsigG expression might simply be another example of late forespore gene expression requiring the SpoIIIAA-AH•SpoIIQ channel (note that the spoIIIAA-AH operon is expressed under σE control). To investigate this possibility, we tested whether late PsigG activity in ΔsigG cells also depended upon the channel proteins SpoIIIAA-AH. As shown in Fig 3C, introduction of ΔspoIIIAA-AH caused PsigG-lacZ expression to be significantly reduced at late times (hour 3 and later) in a manner that was comparable to that observed with ΔsigE and ΔspoIIQ. And like ΔsigE and ΔspoIIQ, ΔspoIIIAA-AH did not alter β-galactosidase production at early times (hours 2–2.5) (Fig 3D). These findings therefore indicate that PsigG expression is dependent at late (but not early) times upon SpoIIQ, σE, and SpoIIIAA-AH. Although we cannot exclude the possibility of pleiotropic effects of these deletion mutants, we believe that the simplest interpretation of these data is that PsigG expression at late times is dependent—as appears to be any late forespore gene expression—upon the SpoIIIAA-AH•SpoIIQ channel.
To further demonstrate the likelihood that PsigG is not subject to unique regulation by SpoIIQ, σE, and SpoIIIAA-AH, we repeated the experiment presented for PsigG-lacZ (Fig 3C and 3D) with strains harboring the σF-dependent PspoIIQ-lacZ reporter gene. As shown in Fig 3E and (in part) as we have previously reported [13], the aberrant, late σF-directed expression of PspoIIQ-lacZ in ΔsigG cells was similarly dependent upon spoIIQ, sigE, and spoIIIAA-AH. (The relatively higher residual expression in the ΔsigE mutant is likely due to the abnormal formation of two forespore compartments, each with active σF [40].) Also similar to our findings for PsigG-lacZ, these mutations did not alter PspoIIQ-lacZ expression at early times (hours 2–2.5) (Fig 3E). We therefore conclude that PsigG and PspoIIQ both require spoIIQ, sigE, and spoIIIAA-AH for σF-dependent expression at late (but not early) times in ΔsigG cells. Overall, these findings are consistent with the general dependence of late forespore gene expression upon the SpoIIIAA-AH•SpoIIQ channel and, conversely, cast significant doubt upon the conclusion that these proteins comprise a regulatory pathway that specifically delays sigG transcription.
Suboptimal spacing between the PsigG -10 and -35 elements partially accounts for low-level expression of PsigG
If PsigG transcription is not delayed by an intercellular regulatory pathway, then why is sigG not more robustly expressed at early times? Bioinformatically the -10 and -35 elements of PsigG are recognized as a good match to the σF consensus (Figs 1D and 4A) [41], and PsigG is readily transcribed by σF•RNAP in vitro [9]. The sigG RBS and its spacing from the start codon (Fig 1D) appear to be ideal for translation initiation [30]. And, as we have shown, the less common GTG start codon of sigG, as well as its native 5’ coding sequence, only modestly reduce its translation efficiency (Fig 2B and 2D). We therefore reasoned that PsigG may be subject to a currently unknown, additional mode of negative regulation in vivo that weakens its expression at early times and, perhaps in turn, helps to properly time the switch to late, σG-directed gene expression in the developing spore.
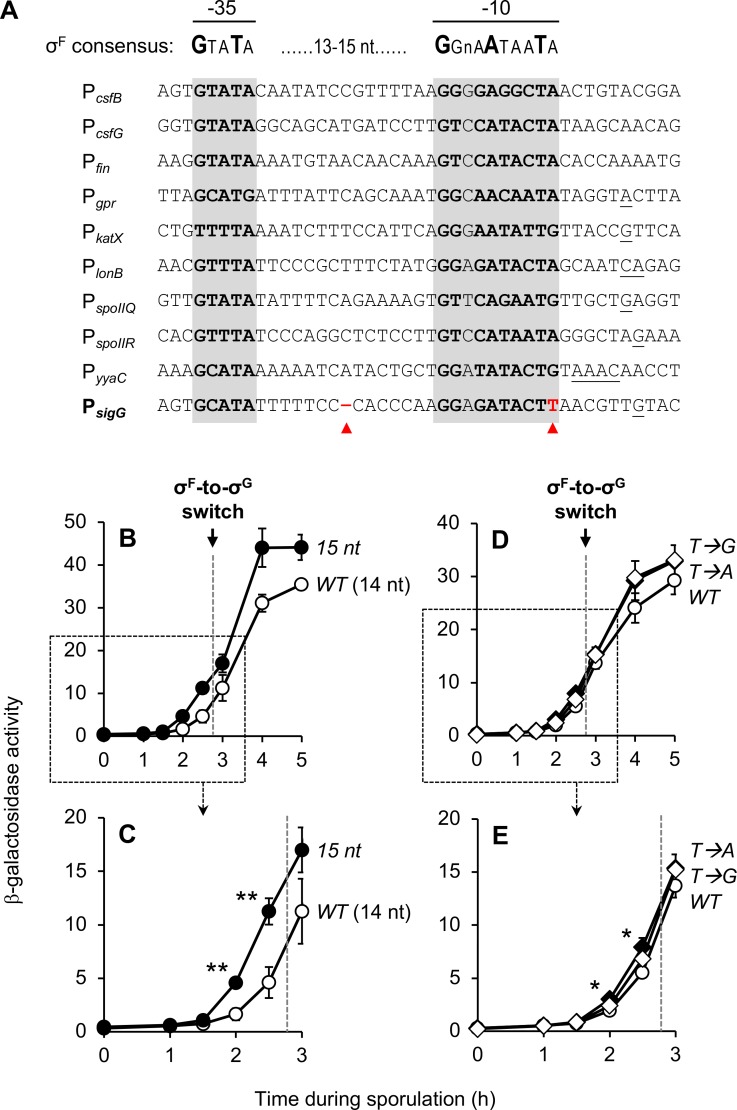
Fig 4. Suboptimal spacing of the PsigG -10 and -35 elements diminishes sigG expression.
(A) Alignment of ten B. subtilis promoters activated by σF, including PsigG. Nucleotides comprising the -10 and -35 elements of each promoter are in bold and shaded gray. Transcription start sites, if known, are underlined [41]. The consensus promoter sequence for σF is shown above [38]. Red arrowheads indicate two notable features of PsigG that differ from the other σF-target promoters: shorter spacing of the -10 and -35 promoter elements (14 nt as opposed to the more common 15 nt; left arrowhead), and a T at position -7 (more typically an A or G; right arrowhead). (B, C) PsigG activation is significantly stimulated by increasing the spacing between the -10 and -35 elements to 15 nt. β-Galactosidase production was monitored during sporulation of strains harboring PsigG-lacZ (WT [14 nt]; open circles) and 15ntPsigG-lacZ (15 nt; closed circles), a variant in which a single nucleotide was inserted between the PsigG -10 and -35 elements to increase their spacing to 15 nt. (Strains JJB31 and JJB51, respectively.) Note that (C) provides a zoomed view of the data from the boxed area of (B). (D, E) The T at position -7 at most only modestly influences PsigG activation. The activity of PsigG-lacZ (WT; closed circles),T→APsigG-lacZ (T→A; closed diamonds), and T→GPsigG-lacZ (T→G; open diamonds) was measured during sporulation. (Strains JJB31, JJB87, and JJB89, respectively.) The T→A and T→G variants were engineered to harbor an A or G, respectively, at position -7 in place of T. Note that (E) provides a zoomed view of the data from the boxed area of (D). The timing of the σF-to-σG switch, between sporulation hours 2.5 and 3, is indicated in each panel by a dashed gray line. For all relevant panels, error bars indicate ± standard deviations based on three independent experiments. *p < 0.05, **p < 0.01 Student’s t-test.
To identify novel mechanisms of sigG regulation, we first turned our attention to the PsigG -10 and -35 promoter elements. As shown in Fig 4A, there were two notable differences when we compared PsigG to several other promoters also recognized by σF in B. subtilis. First was that the spacing between the PsigG -10 and -35 elements is 14 nts, whereas the majority of promoters had a spacing of 15 nts. Second, the nucleotide at position -7 in PsigG is a T, whereas in the majority of promoters the equivalent position is an A or G.
To test whether one or both of these unique features could explain the low level expression of PsigG, we constructed variants of our PsigG-lacZ reporter in which the spacing between the -10 and -35 elements was increased to 15 nt by insertion of single base pair (15ntPsigG-lacZ), or in which the T at position -7 was switched to A or G (T→APsigG-lacZ or T→GPsigG-lacZ, respectively). As shown in Fig 4B, the 15ntPsigG-lacZ reporter displayed a notable increase in expression relative to the corresponding wild type reporter at both early and late times of sporulation. Interestingly, this stimulation was most pronounced at times corresponding to σF activity: at hours 2 and 2.5, the 15ntPsigG-lacZ reporter displayed a nearly 3-fold increase in expression (Fig 4C). In contrast, switching the T at position -7 to either A or G had very little effect on the extent of PsigG expression, although a slight, statistically-significant increase for the T→APsigG-lacZ reporter (~1.5-fold relative to the wild type PsigG-lacZ reporter) was detected at hours 2 and 2.5 (Fig 4D and 4E). Finally, we combined the 15nt alteration with the T→A or T→G mutations to test for a synergistic effect on PsigG activity, but no additional stimulation was observed (S2 Fig). Together these findings suggest that the ability of σF to activate PsigG at early times is considerably diminished by the shorter spacing between the PsigG -10 and -35 elements, and is only modestly (if at all) affected by the identity of the nucleotide at position -7 (T vs. A vs. G).
sigG translation is reduced by formation of an mRNA stem-loop structure that occludes the sigG RBS
Next, our attention was drawn to the possibility that nucleotides adjacent to the sigG RBS might also contribute to the low levels of sigG expression. This idea came from characterization of a PsigG-lacZ variant, originally constructed for another line of investigation, in which 12 nucleotides ranging from positions +10 to +30, both upstream and downstream of the sigG core RBS, were randomly mutated (Fig 5A). (Note that the PsigG-lacZ reporter utilized here also harbored an ATG start codon in place of the native GTG start codon.) As shown in Fig 5C, this PsigG+10→+30-lacZ reporter was expressed ~5-8-times more robustly than the corresponding wild type reporter at both early and late times of sporulation. To identify the nucleotides responsible for this effect, we constructed PsigG-lacZ variants harboring subsets of the original 12 mutations (Fig 5A). Through this analysis, we discovered that the 6 alterations introduced at positions +10 through +15 accounted for the majority of the observed stimulation. β-Galactosidase production from PsigG+10→+15-lacZ was increased ~3-4-fold at both early and late times of sporulation compared to the corresponding wild type PsigG-lacZ, while all other mutations either had no effect or only modestly affected expression (Fig 5C).
Fig 5. An mRNA hairpin formed between the sigG leader sequence and RBS significantly decreases PsigG-lacZ translation.
(A) Depiction of the mutations in the PsigG+10→+30-lacZ reporter variant, as well as variants designed to harbor only a subset of these mutations. The sigG transcription start site (+1), sigG ribosome-binding site (RBS), and ATG start codon are indicated. (*Note that the PsigG-lacZ reporter gene used here harbored the non-native ATG start codon in place of the native sigG GTG start codon.) Also indicated, with green arrows, are complementary sequences predicted to form the RBS hairpin structure shown in (B). (B) Depiction of the sigG RBS-hairpin structure predicted to form in the sigG mRNA leader sequence. The 5’ end of the sigG mRNA (+1), the sigG core RBS, and native GTG (GUG) translation start codon are labeled. Asterisks indicate the six nucleotide positions altered in the +10→+15 mutant. The prediction and graphic were generated by ViennaRNA, with bases color-coded according to their partition function probabilities [68]. (C) Alteration to nucleotides upstream of the sigG RBS stimulates expression from a PsigG-lacZ reporter gene. The activities of PsigG+10→+30-lacZ (+10→+30; closed circles), PsigG+24→+30-lacZ (+24→+30; open squares), PsigG+10→+18-lacZ (+10→+18; closed squares), PsigG+18-lacZ (+18; closed triangles), PsigG+10→+15-lacZ (+10→+15; open diamonds), and the corresponding wild type PsigG-lacZ (WT; open circles) were measured during sporulation. (Strains AHB883, AHB2124, AHB2126, JC68, JC70, and AHB1274, respectively.) (D) The +10→+15 alterations do not impact β-galactosidase production from a transcriptional lacZ reporter. β-Galactosidase production was monitored during sporulation of strains harboring PsigG-RBS-lacZ (WT [+RBS]; open squares) and PsigG+10→+15-RBS-lacZ (+10→+15 [+RBS]; closed squares). (Strains AM3 and AM4, respectively.) In these transcriptional reporters, lacZ is separated from the sigG RBS by a spacer and provided with an engineered RBS. (E) Mutations that weaken/eliminate or strengthen the sigG RBS-hairpin increase or decrease PsigG-lacZ expression, respectively. The activities of PsigGmut7-lacZ (mut7; closed squares), PsigGmut2-lacZ (mut2; open squares), and the corresponding wild type PsigG-lacZ (WT; open circles) were measured during sporulation. (Strains JJB55, JJB37, and JJB31, respectively.) The mRNA leader sequence for each PsigG-lacZ variant is indicated to the right, with the 5’ end of the mRNA (+1), core RBS, and the mut7 or mut2 mutations (in blue) labeled and underlined. Also depicted is the capacity for formation of a hairpin structure (green lines connecting complementary base-pairs). For all relevant panels, error bars indicate ± standard deviations based on three independent experiments.
Positions +10 through +15 are located downstream of the sigG transcription start site (+1) and upstream of the core sigG RBS (positions +19 to +23) (Fig 5A). These nucleotides therefore might exert their inhibitory effect upon sigG expression at the level of transcription, mRNA stability, and/or mRNA translation. To distinguish these modes of regulation, we introduced the +10→+15 mutations into a transcriptional PsigG-lacZ reporter (referred to here as PsigG-RBS-lacZ), in which lacZ was separated from the sigG RBS by a spacer and provided with an engineered, optimal RBS. As shown in Fig 5D, β-galactosidase production from the PsigG-RBS-lacZ reporter was unaffected by the +10→+15 mutations, arguing strongly for a role of these nucleotides specifically in the regulation of translation initiation at the sigG RBS.
One mechanism for regulation of translation initiation involves the occlusion of the RBS by complementary base-pairing with adjacent nucleotides in the 5’ mRNA leader sequence (reviewed in [42]). To determine whether this may be the case for sigG, we analyzed the sigG 5’ mRNA leader sequence for potential secondary structures. Strikingly, we found that a stem-loop structure comprised of five sequential base-pairs was predicted to form between the majority of the sigG RBS and upstream nucleotides (Fig 5A and 5B), with a calculated free energy of -6.2 kcal/mol [43]. Interestingly (and serendipitously), five of the six nucleotides altered in the +10→+15 mutant correspond exactly to the five nucleotides that participate in formation of this sigG “RBS hairpin” (asterisks in Fig 5B). Re-analysis of the sigG+10→+15 5’ mRNA leader sequence for potential secondary structure confirmed that this mutant was no longer predicted to form this hairpin [43]. Together, these findings are suggestive of a model in which sigG translation is ordinarily dampened by a stem-loop structure that occludes the sigG RBS.
To further confirm the role of the identified RBS hairpin in regulating sigG translation, we introduced mutations into our PsigG-lacZ reporter that were specifically designed to weaken/eliminate (“mut7”) or strengthen (“mut2”) the sigG RBS hairpin (Fig 5E, right). As expected, the PsigGmut7-lacZ reporter, which retained the potential for only two complementary base pairs, was expressed more robustly, producing ~2-fold more β-galactosidase than the corresponding wild type reporter at both early and late times of sporulation (Fig 5E). We do note that this effect was not as robust as the 3-4-fold stimulation we observed for the +10→+15 variant (see Fig 5C). Although we cannot be certain, we speculate that this difference is not due to residual hairpin formation, but rather secondary consequences of these mutations (such as decreased mRNA stability). In contrast, the “strengthened hairpin” PsigGmut2-lacZ reporter displayed very low levels of expression (Fig 5E). Altogether, these findings indicate that the sigG RBS hairpin ordinarily dampens sigG translation by 2-4-fold and, when strengthened, can almost entirely block translation from the sigG mRNA.
Together, the identified transcriptional and translational regulation of sigG diminishes expression by 4-6-fold
Our data reveal that expression of sigG is diminished by at least four mechanisms: (i) suboptimal spacing of the PsigG -10 and -35 elements, (ii) a hairpin in the sigG mRNA 5’ leader sequence predicted to occlude the RBS, (iii) a suboptimal GTG start codon, and (iv) secondary structure in the sigG 5’ coding sequence. To visualize the full extent of sigG inhibition by these mechanisms, we built a PsigG-lacZ reporter construct (referred to as quadPsigG-lacZ) that simultaneously removed or “repaired” all of these features. Strikingly, quadPsigG-lacZ was expressed ~4-6-fold more robustly than the original PsigG-sigG1-28-lacZ reporter (Fig 6A and 6B). This effect is in line with what would be predicted by the individual effects of each alteration (4-13-fold), suggesting that these four features operate independently to modulate sigG expression during sporulation. Importantly, this 4-6-fold increase is almost certainly an underestimate of the actual increase that would result if the sigG gene itself (as opposed to a lacZ reporter gene) were misexpressed, given the amplification of sigG expression that would occur via σG autoregulation.
Fig 6. The identified transcriptional and translational regulation of sigG diminishes sigG expression by 4-6-fold and is required to prevent aberrant activity of σG.
(A, B) Expression of a PsigG reporter is increased by 4-6-fold in the absence of the four regulatory features identified in this study. β-Galactosidase production was monitored during sporulation of strains harboring PsigG-sigG1-28-lacZ (WTPsigG-lacZ; open circles) or a variant in which all four features of sigG that dampen expression were simultaneously removed or repaired, 15ntPsigGmut7-ATG-RSSsigG2-28-lacZ (quadPsigG-lacZ; closed circles) (strains EBM177 and EBM262, respectively.) Note that (B) provides a zoomed view of the data from the boxed area of (A). (C, D) Expression of sigG from regulatory sequences lacking the four regulatory features identified in this study causes aberrant σG activity during a time course of sporulation. β-Galactosidase production from (C) the σF-dependent PspoIIQ-lacZ reporter or (D) the σG-dependent PsspB-lacZ reporter was monitored during sporulation of strains in which sigG was expressed from its wild type regulatory sequences (WTPsigG-sigG; open squares) or from regulatory sequences modified to remove or repair the four features identified in this study to dampen sigG expression (quadPsigG-sigG; closed squares). (PspoIIQ-lacZ strains were CFB429 and CFB431, respectively. PsspB-lacZ strains were CFB435 and CFB437, respectively.) The black arrow in (D) indicates aberrant σG activity at early times of sporulation. The timing of the σF-to-σG switch, between sporulation hours 2.5 and 3, is indicated in each panel by a dashed gray line. For all panels, error bars indicate ± standard deviations based on three independent experiments.
Misregulation of sigG causes inappropriate σG activity during vegetative growth and disrupts the timing of σG activation during sporulation
We hypothesized that the four identified mechanisms of sigG negative regulation help to ensure the proper execution of the switch from σF to σG in the developing forespore, most likely by preventing premature σG activation. To test this prediction, we constructed a strain in which sigG itself (i.e. not a lacZ reporter gene) was expressed from regulatory sequences altered to remove or repair the four features that ordinarily reduce sigG expression (these alterations were identical to those in the quadPsigG-lacZ reporter, see above). This engineered sigG gene, referred to as quadPsigG-sigG, was inserted at an ectopic locus in a strain deleted for the native sigG gene; a strain harboring sigG under the control of wild type regulatory sequences (PsigG-sigG) was constructed in an identical manner as a control. The resulting quadPsigG-sigG strain displayed no detectable defect in heat resistant spore formation relative to the wild type PsigG-sigG control strain (S3 Fig), indicating that misregulation of sigG does not drastically compromise sporulation.
To determine whether sigG misregulation interferes with the switch from σF to σG, we measured the activities of these two sigma factors during sporulation of the quadPsigG-sigG and wild type PsigG-sigG control strains using lacZ reporter genes. As shown in Fig 6C, we observed no detectable difference in the timing or extent of σF-dependent β-galactosidase production from a PspoIIQ-lacZ reporter. In contrast, σG-dependent PsspB-lacZ expression was significantly altered such that the quadPsigG-sigG strain displayed up to 100-fold more β-galactosidase activity at sporulation hours 0–3, but appeared to be relatively normal thereafter (Fig 6D). These results could be consistent with inappropriate early activation of σG in the forespores of quadPsigG-sigG cells during sporulation, but the presence of significant β-galactosidase activity at hour 0 (i.e. prior to the onset of sporulation) also suggests that σG may be inappropriately active in vegetative cells.
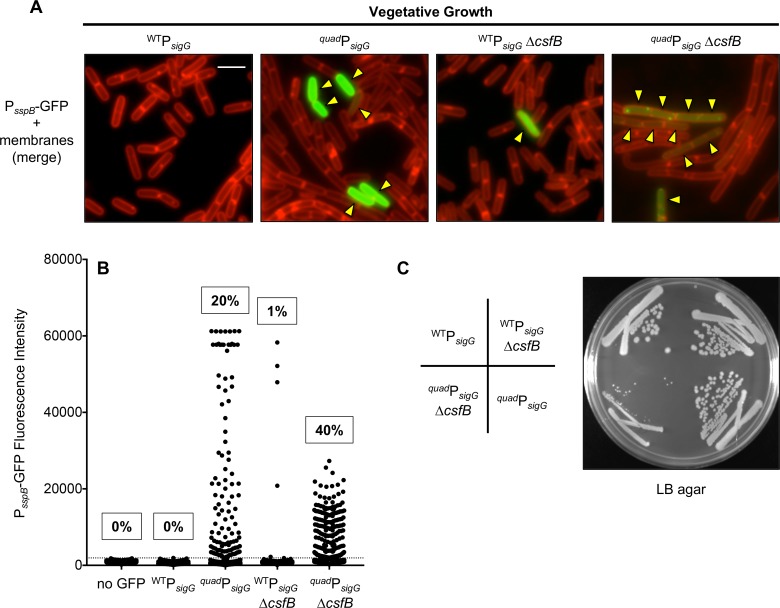
To identify the sub-population(s) of quadPsigG-sigG cells exhibiting inappropriate σG activity, we monitored and quantified σG-dependent expression of a PsspB-gfp reporter gene in single cells by fluorescence microscopy during both vegetative growth and sporulation. As shown in Fig 7A and 7B, we detected GFP fluorescence significantly above background in 20% of vegetative quadPsigG-sigG cells, as compared to 0% of the wild type PsigG-sigG control cells. Inappropriate σG activity in a subset of vegetative cells has also been reported for cells lacking the σG inhibitor CsfB [16,44]. Consistent with these reports, introduction of a ΔcsfB deletion into our wild type PsigG-sigG control strain caused 1% of vegetative cells to display σG-dependent GFP expression (Fig 7A and 7B). Interestingly, when the quadPsigG-sigG and ΔcsfB mutations were combined, the resulting double mutant exhibited aberrant σG activity in 40% of vegetative cells (Fig 7A and 7B). The double mutant also appeared sickly: the intensity of GFP fluorescence per cell was lower than in the respective single mutants (Fig 7A and 7B), and GFP protein aggregates accumulated in most cells (Fig 7A). Last, it is worth noting that the quadPsigG-sigG ΔcsfB double mutant was difficult to construct and gave rise to very small colonies when grown on LB plates, unlike the two single mutants (Fig 7C). Growth curve analysis in liquid LB media revealed that the double mutant had a significantly longer doubling time during log phase (~32 min vs. ~25 min for the corresponding wild type strain), and failed to reach the same maximal cell density during stationary phase (S4 Fig). In contrast, growth of the two single mutants was indistinguishable from wild type with the exception of the quadPsigG-sigG strain, which had a slight but statistically significant increase in doubling time during exponential growth (S4 Fig). Together, these data indicate that the sigG regulatory features identified in this study act synergistically with CsfB to prevent inappropriate σG activity during vegetative growth and that, in the absence of this regulation, vegetative cells are at a severe fitness disadvantage during both exponential growth and stationary phase.
Fig 7. Misexpression of sigG causes ectopic σG activity in a subset of vegetative cells.
(A, B) GFP production from a σG-dependent PsspB-gfp reporter was monitored by fluorescence microscopy of vegetatively growing cells in which sigG was expressed from its wild type regulatory sequences (WTPsigG) or from regulatory sequences modified to remove or repair the four features identified in this study to dampen sigG expression (quadPsigG). Additionally, cells either harbored wild type csfB, or were deleted for the gene (ΔcsfB). (Strains EBM192 [WTPsigG], EBM276 [quadPsigG], EBM282 [WTPsigG ΔcsfB], and EBM287 [quadPsigG-sigG ΔcsfB].) In (A), representative microscopy images are shown with GFP fluorescence (false-colored green) merged with membrane fluorescence from the dye FM 4–64 (false-colored red). Yellow arrowheads indicate vegetative cells with visible GFP fluorescence. Scale bar 5 μm. In (B), GFP fluorescence intensity (with background subtracted) for more than 500 cells of each strain, including a “no GFP” control strain (PY79) lacking the PsspB-gfp reporter, is shown in column scatter graph format, with each cell represented by a black dot. Cells exhibiting fluorescence intensity above the cut off value (three standard deviations above mean auto-fluorescence of the “no GFP” strain; gray dashed line) were determined to have detectable σG activity. The percentage of cells with this activity is indicated above the respective strain. Note that ~60000 fluorescence units is the upper limit of detection under our microscopy settings. (C) Simultaneous deletion of csfB and misexpression of sigG from quadPsigG causes synthetic toxicity to vegetatively growing B. subtilis. The same strains described in (A) were struck onto LB agar and were photographed after ~18 hours of growth at 37°C.
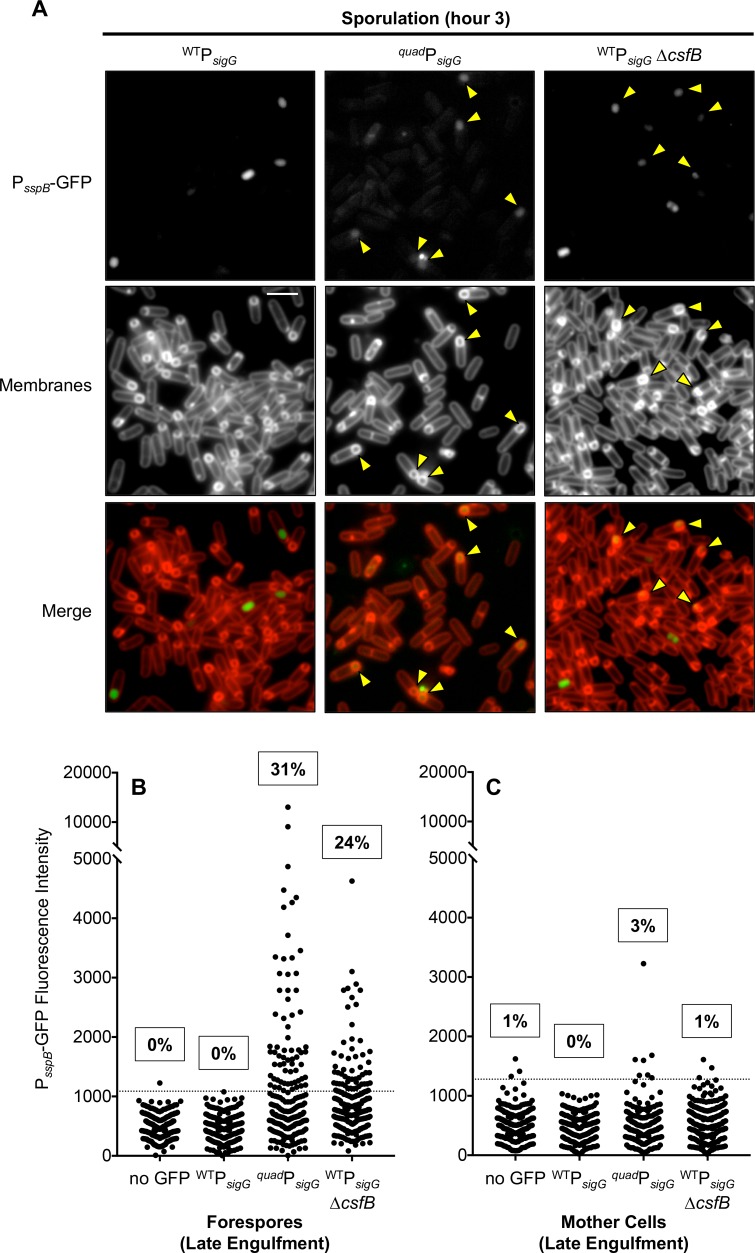
Next, we observed cells during sporulation to determine whether misexpression of sigG might also cause σG to become prematurely active in the forespore. As expected, σG-dependent expression of the PsspB-gfp reporter gene in wild type PsigG-sigG forespores was observed only after the process of forespore engulfment was complete, and was not detected in any mother cells (Fig 8A–8C). In contrast, when sigG expression was misregulated in the quadPsigG-sigG mutant, 31% of forespores displayed aberrant σG activity prior to engulfment (Fig 8A and 8B). Based on a previous report [16], we anticipated that deletion of csfB in the wild type PsigG-sigG control strain might give rise to a similar effect. Indeed, we detected significant σG-dependent GFP fluorescence in 24% of ΔcsfB forespores that had not yet completed engulfment (Fig 8B). In both cases, aberrant σG activity was specific to forespores, given that only 3% of quadPsigG-sigG mother cells and 1% of ΔcsfB mother cells displayed σG-dependent fluorescence (Fig 8C). Unfortunately, due to the poor vegetative growth of the double quadPsigG-sigG ΔcsfB mutant (Fig 7C), as well as the pre-existing σG activity in 40% of vegetative cells (Fig 7A and 7B), we were unable to determine with confidence the combined influence of these mutations on premature σG activation during sporulation. Nevertheless, our findings reveal that proper regulation of sigG expression by the features identified in this study are required to prevent premature σG activation in the developing forespore.
Fig 8. Misexpression of sigG causes premature σG activity in the forespore during sporulation.
GFP production from a σG-dependent PsspB-gfp reporter was monitored by fluorescence microscopy of sporulating cells (hour 3.5) in which sigG was expressed from its wild type regulatory sequences (WTPsigG) or from regulatory sequences modified to remove or repair the four features identified in this study to dampen sigG expression (quadPsigG). Additionally, cells either harbored wild type csfB, or were deleted for the gene (ΔcsfB). (Strains EBM192 [WTPsigG], EBM276 [quadPsigG] and EBM282 [WTPsigG ΔcsfB].) (A) Representative microscopy images for each strain. GFP fluorescence is depicted in grayscale (PsspB-GFP) or false-colored green (Merge). Membrane fluorescence from the dye FM 4–64 is shown in grayscale (Membranes) or false-colored red (Merge). Following asymmetric division and during engulfment, the FM 4–64 dye labels all membranes including the double membranes separating the mother cell and forespore; after the completion of engulfment, the membranes surrounding the forespore are no longer accessible to the dye and therefore remain unlabeled. Yellow arrowheads indicate late engulfment forespores, identifiable by their FM 4-64-labeled engulfing membranes, with visible GFP fluorescence. Scale bar 5 μm. (B) GFP fluorescence intensity (with background subtracted) for more than 200 late-engulfment forespores or (C) their corresponding mother cells for the three strains described in (A) as well as a “no GFP” control strain (PY79) lacking the PsspB-gfp reporter are shown in column scatter graph format, with each cell represented by a black dot. Cells exhibiting fluorescence intensity above the cut off value (three standard deviations above mean auto-fluorescence of the “no GFP” strain; gray dashed line) were determined to have detectable σG activity. The percentage of cells with this activity is noted above the respective strain.
Discussion
In this study, we sought to unravel a decades old puzzle in the B. subtilis sporulation pathway; namely, how does the late acting transcription factor σG become active at the proper time during forespore development? At the outset, we hypothesized that precise regulation of the transcription and/or translation of sigG plays a key role in ensuring that σG does not become active prematurely. Here we report a careful re-evaluation of sigG expression that calls into question long-standing notions regarding its regulation, including models in which sigG expression is delayed or dependent upon an intercellular “checkpoint”. Instead, our data support a new working model in which sigG expression is neither delayed nor subject to a specific intercellular regulatory pathway, but rather is dampened at the transcriptional and translational levels by at least four features intrinsic to its regulatory and coding sequence. As we hypothesized, these features are required to prevent premature σG activity in the forespore. We also found that they are required to prevent inappropriate σG activity in vegetatively growing cells, suggesting that these controls serve not only to fine-tune the timing of σG activation during sporulation, but also to prevent its inappropriate activation under non-sporulating conditions.
A new working model for regulation of sigG expression
The long-standing working model for regulation of sigG expression (see, for example, [2,24]) posits that an intercellular checkpoint mechanism, perhaps involving SpoIIQ, delays σF-dependent sigG expression until the early phase of σE-dependent, mother cell gene expression is underway. Here we present evidence that the data supporting this original working model (see Introduction and Results) can be reinterpreted in a manner that does not invoke the existence of an enigmatic, sigG-specific intercellular checkpoint. First, the apparently delayed activation of PsigG by σF can be ascribed to aberrant, late σF activity that is unmasked in the absence of σG (note that the original experiments were performed in a ΔsigG genetic background) [13,15,39]. Second, the dependence upon sigE and spoIIQ can be explained by the failure of these mutants to assemble the SpoIIIAA-AH•SpoIIQ channel, which is required for any late gene expression in the forespore, including that directed by aberrantly active σF [13].
As such, we propose here a new, simpler working model for sigG regulation (Fig 1C). At early times, we posit that sigG is transcribed under the control of σF without delay, but at levels that are very low and therefore difficult to detect. Then, at later times, σG drives its own transcription in an autoregulatory loop, during which time the majority of sigG/σG is produced. We have established here that at least four features of the sigG regulatory and coding sequences, affecting both transcription and translation, account for its low-level expression: reduced spacing between the -35 and -10 promoter elements, a GTG start codon, a suboptimal 5′ coding sequence, and an RBS-sequestering hairpin located in the 5’ leader sequence.
Promoter spacer
The influence of the spacing between the -35 and -10 promoter elements upon promoter activity reflects the requirement that the sigma factor subunit of RNAP simultaneously bind to both sites. Even single base-pair increases or decreases in the spacer length can significantly affect promoter activity (for example, [45–48]). Here, we determined that sub-optimal, reduced spacing between the sigG promoter elements does contribute to the weak expression of sigG. Interestingly, lengthening the promoter spacer most significantly increased expression during early sporulation, perhaps suggesting that σF is more sensitive to promoter element spacing than σG. Furthermore, this transcriptional mechanism for dampening sigG expression appears to be conserved among endospore formers from the class Bacilli. In a survey of 15 representative species from this class (including B. subtilis), we found that 14 display a reduced (14 bp) spacing between their sigG promoter elements (S5 Fig). In contrast, endospore formers from the class Clostridia more often space their sigG promoter elements by 15 bp; only 3 out of 13 species from this class display the reduced 14 bp spacing (S5 Fig). These observations suggest that the spacing between the sigG promoter elements is important in the regulation and possibly optimization of the sporulation program in many Bacilli species.
GTG start codon
In B. subtilis, the start codon GTG is weak in terms of translational efficiency compared to TTG and ATG, which have been reported in some cases to produce up to 3- and 7-fold more translational product, respectively [30]. We found that replacing the sigG GTG start codon with ATG improved translational efficiency modestly (~1.3-fold). Nevertheless, it is interesting to note that 11 out of 15 sigG genes from representative Bacilli utilize a non-ATG start codon (6 GTG, 5 TTG). In contrast, only 1 out of 13 representative Clostridia utilize a non-ATG start codon (GTG) (S5 Fig). This suggests that, despite the modest impact on translational efficiency, initiating sigG translation with non-ATG start codons is advantageous for members of the Bacilli.
5’ coding sequence
In addition to the identity of the start codon, the 5’ coding sequence of a gene can also influence translational efficiency, in some cases by orders of magnitude (see for example [33,49–51]). This influence is thought to be mediated by the presence (or lack thereof) of RNA secondary structures and/or rare codons that impede translation initiation (reviewed in [31]). When we replaced the native sigG 5’ coding sequence with synonymous codons selected to minimize secondary structure, or with the 5’ coding sequence of the highly expressed gene comGA, expression increased by ~1.5- or 2-fold, respectively. We interpret this data to indicate that the sigG 5’ coding sequence ordinarily dampens translation efficiency, at least in part through the formation of secondary structures such as those noted in Fig 1D. However, we cannot exclude the possibility that one or more rare codons and/or the abundance of certain tRNAs in the forespore during sporulation, contribute to this effect.
RBS hairpin
Finally, we found that sigG expression is dampened ~2-4-fold via a hairpin structure that forms between the RBS and nucleotides in the adjacent 5’ mRNA leader sequence. This small “RBS hairpin”, which is comprised of five complementary base pairs, is distinct from the very large hairpin (comprised of 20+ complementary base pairs) proposed to block translation of a read-through transcript originating from the upstream spoIIGA-sigE operon (Fig 1D) [9,52]. (This read-through transcript is not required for sigG expression during sporulation, nor does the large hairpin form in the transcript originating from the sigG promoter [9,52].) Through targeted mutagenesis we were able to weaken/eliminate or strengthen the RBS hairpin with predictable consequences for sigG expression. Interestingly, RBS-sequestering hairpins of similar free energies (ranging from -6.0 to -8.9 kcal/mol) are predicted to form in the sigG leader sequences of many endospore-forming Bacilli species (found in all 15 of the representatives listed in S5 Fig), but are less common or weaker in Clostridia (hairpins with free energies of -6.0 kcal/mol or lower were only found in 4 of the 13 representatives listed in S5 Fig) [43]. A subset of these predicted sigG RBS-sequestering hairpins are shown in S6 Fig.
In the simplest scenario, the RBS hairpin dampens translation of the sigG transcript at all times, with the ribosome gaining access only during transient moments of hairpin unzipping. This is consistent with the fact that weakening and strengthening the hairpin increases and decreases expression, respectively, both before and after the σF-to-σG switch. However, this doesn’t exclude a second possible scenario in which the sigG RBS hairpin changes conformation dynamically during sporulation, such that the RBS is exposed only at later stages of sporulation (i.e. after the switch from σF to σG). This regulation could occur through the binding of a small molecule, protein, or small RNA (reviewed in [42]). Given the hypothesis that the forespore becomes dependent on nutrients from the mother cell, delivered through the SpoIIIAA-AH•SpoIIQ channel at late times [13], it is tempting to speculate that dynamic control of sigG translation could be tuned to a specific metabolite through a riboswitch-like mechanism. However, the sigG 5’ leader is relatively short (30 nt) and does not match any known class of riboswitch according to several web-based bioinformatics tools [53–55]. Nevertheless, exploring the possibility that the sigG RBS hairpin is regulated, perhaps by a novel mechanism, is an exciting challenge for future work.
Do sigG/σG regulatory mechanisms serve primarily to protect from σG toxicity in non-sporulating cells?
A collective take-home message from this and many other studies of sigG/σG is that this gene/protein is subject to layer upon layer of negative regulation, at the transcriptional, translational, and post-translational levels. While it is tempting to imagine that these layers of regulation evolved to ensure the precise timing of σG activation in the forespore during sporulation, it has become increasingly evident that they function in addition, or in some cases instead, to prevent aberrant σG activity in the mother cell and/or in non-sporulating, vegetative cells. Given that sigG is autoregulated, even very low levels of “leaky” expression could trigger a positive feedback loop of σG synthesis and activity. The anti-sigma factor SpoIIAB and the serine protease LonA, which inhibit σG post-translationally, counteract/eliminate σG produced in the mother cell and in non-sporulating cells, with no apparent role in σG regulation in the forespore [28,44,56–59]. In contrast, the anti-sigma factor CsfB post-translationally inhibits σG in the forespore (at early times), mother cell, and in non-sporulating cells, an impressive feat that is accomplished via csfB regulatory sequences that permit transcription by σF (at early times in the forespore), σK (at late times in the mother cell), and σG itself (in vegetative cells, but not in the forespore) (this study and [15,16,44,59,60]).
Here we have found that the regulatory features that dampen sigG transcription and translation, like the aforementioned σG inhibitors, are not solely dedicated to preventing premature σG activity in the forespore. While misexpression of sigG indeed caused nearly one third of forespores to display premature σG activity, thus supporting our original hypothesis, it also led to aberrant σG activity in up to 20% of vegetative cells. (We found no evidence for any effect in the mother cell.) We do not currently know why the sigG/σG autoregulatory loop was triggered in this particular subpopulation of vegetative cells, but we speculate that it could be due to activation of PsigG by σF (itself produced by leaky expression) or spurious transcription through the sigG coding sequence. We do note that read-through transcription from the spoIIGA-sigE operon [9,52], which resides upstream of the native sigG gene, could not have been a contributing factor given that sigG was located at an ectopic locus in our strain background.
Strikingly, misexpression of sigG in cells also lacking CsfB led to a synergistic and severe effect on vegetative cells: nearly half of the population displayed aberrant σG activity and the strain was sickly and exhibited a small colony morphology, consistent with the documented toxicity of σG to vegetative cells [44,61]. Unfortunately, this toxicity was so severe that we were unable to assess synergistic effects in the forespore during sporulation. These results, along with the fact that early σG activity in the forespore (i.e. in quadPsigG-sigG or ΔcsfB cells) does not appear to be detrimental to the sporulation program (this study and [15]), raise the speculative idea that the majority of sigG/σG regulation has evolved primarily to prevent ectopic σG activity in vegetative cells, and not to precisely regulate the switch from early to late gene expression in the developing spore. In this light, a key challenge for future studies will be to understand how σG escapes these many layers of inhibition in order to drive late gene expression in the developing spore.
Materials and methods
General methods
All B. subtilis strains were derived from the laboratory strain PY79 [62]. Tables of strains, plasmids, primers, and synthetic DNA fragments used in this study, as well as details of strain and plasmid construction, are provided as supplementary material (S1, S2, S3, S4 Tables, S1 Text). Bacterial strains were propagated in lysogeny broth (LB), either in liquid culture or on solid plates with 1.6% agar. When appropriate, antibiotics were included as follows: chloramphenicol (5 μg/mL), erythromycin plus lincomycin (1 μg/mL and 25 μg/mL, respectively), spectinomycin (100 μg/mL), kanamycin (5 μg/mL), and ampicillin (100 μg/mL). For growth curves, colonies grown overnight on LB agar from a frozen stock were inoculated directly (to minimize selection of suppressors) into 1.5 mL liquid LB in clear 12-well flat-bottom plates. Plates were incubated at 37°C with continuous shaking in a Synergy H1M plate reader (BioTek, Inc.), and optical density at 600 nm (OD600) was measured every 5 min. To quantify spore formation, cells were induced to sporulate by nutrient exhaustion in Difco sporulation medium (DSM) [63,64]. After 24 h at 37°C, the number of colony-forming units that survived heat treatment (20 min at 80°C) was calculated. For all other experiments, sporulation was induced by the resuspension method [64,65]. β-Galactosidase activity was measured as previously described [13] and is reported in arbitrary units (rate of substrate conversion normalized to cell density), unless otherwise noted.
Fluorescence microscopy and quantification of σG activity in single cells
Cells harboring the σG-dependent PsspB-gfp reporter were collected during mid-log phase to observe vegetative cells, or at hour 3 of sporulation to observe forespores and their corresponding mother cells during late-engulfment. Harvested cells were resuspended in 1x phosphate-buffered saline (PBS) with 1 μg/ml FM 4–64 (Molecular Probes) to stain membranes, and spotted on thin 1% agarose pads with poly-L-lysine (Sigma) treated coverslips. Fluorescence microscopy was performed with a Nikon Eclipse Ti-U inverted microscope fitted with filter sets 49002 (Chroma; excitation 470/40x, dichroic 495Ipxr, and emission 525/50m) and 49017 (Chroma; excitation 560/40x, dichroic 590Ipxr, and emission 590lp) for GFP and FM 4–64 detection, respectively. Images were captured with an ORCA-Flash4.0 digital CMOS camera (Hamamatsu Photonics K.K.) using NIS-Elements Advanced Research software (Nikon Instruments Inc.).
For each strain, GFP fluorescence intensity above background was quantified for more than 500 vegetative cells and more than 200 late-engulfment forespores (defined as cells that were more than halfway engulfed, but not fully engulfed as determined by FM 4–64 membrane staining) and their corresponding mother cells, using the ImageJ software distributed within the Fiji package [66,67]. Cells to be quantified were obtained from two or three biological replicates (experiments carried out on different days). For each replicate sample, at least three fields were selected at random from the same slide and images were acquired with identical settings. Cells with a net GFP fluorescence intensity more than three standard deviations higher than autofluorescence (as determined by images collected in parallel from PY79, the wild type parent strain lacking a gfp reporter), were considered to have detectable σG activity. Percentages were calculated based on the total number of cells or forespores quantified. Representative images had background subtraction applied, were false colored, and overlaid using the aforementioned ImageJ software.
Supporting information
An AUG (ATG) start codon followed by codons 2–28 of B. subtilis sigG are shown (top sequence), as is a sequence comprised of synonymous codons selected by the mRNA Optimizer tool [32] to minimize secondary structure (bottom sequence). Note that the latter is the sequence present in the PsigG-ATG-RSSsigG2-28-lacZ reporter construct used in Fig 2. Nucleotide differences are indicated in red and blue text. Both sequences were analyzed for secondary structure by the RNAstructure method [43]. Potential secondary structures are displayed above (red) or below (blue) using RNAbow diagrams [69], with base pairs indicated by arcs, the thickness and shading of which is proportional to their probability. The partition function free energy (G) for each is indicated.
(TIF)
β-Galactosidase production was monitored during sporulation of strains harboring PsigG-lacZ (WT; open circles), 15ntPsigG-lacZ (15 nt; closed circles), T→APsigG-lacZ (T→A; open diamonds), T→GPsigG-lacZ (T→G; closed diamonds) 15nt,T→APsigG-lacZ (15 nt, T→A; open triangles), and 15nt,T→GPsigG-lacZ (15 nt, T→G; open squares). (Strains JJB31, JJB51, JJB87, JJB89, JJB99 and JJB101, respectively.) For clarity, only data from a subset of these strains are presented in each graph (as labeled) and, also for clarity, the WT and 15 nt data is presented in all graphs. Note that (B) and (D) provide zoomed views of the data from the boxed areas of (A) and (C), respectively.
(TIF)
The total number of heat-resistant spores per mL was determined for a wild type strain harboring unaltered sigG at its native locus (WT; strain PY79), a strain deleted for sigG (ΔsigG; strain AHB98), or strains deleted for sigG and harboring at the non-essential ylnF locus a copy of the sigG gene either under the control of wild type regulatory sequences (+ylnF::WTPsigG-sigG; strain LMB39) or regulatory sequences modified to remove or repair the four features identified in this study to dampen sigG expression (+ylnF::quadPsigG-sigG; strain AHB2819). Error bars indicate standard deviation based on three or more independent experiments.
(TIF)
(A) Representative growth curves in liquid LB for cells expressing sigG from its wild type regulatory sequences (WTPsigG) or from regulatory sequences modified to remove or repair the four features identified in this study to dampen sigG expression (quadPsigG). Additionally, cells either harbored wild type csfB, or were deleted for the gene (ΔcsfB). (Strains EBM192 [WTPsigG; purple diamonds], EBM276 [quadPsigG; orange triangles], EBM282 [WTPsigG ΔcsfB; blue squares], and EBM287 [quadPsigG-sigG ΔcsfB; green circles].) (B) Doubling time during exponential growth and maximal OD600 during stationary phase for each strain are reported as the average ± standard deviation based on three independent experiments. **p < 0.01, ****p < 0.0001, ns not significant, Student’s t-test.
(TIF)
Upstream regulatory sequences for sigG were retrieved from the genome sequences of representative endospore forming bacteria from the classes Bacilli and Clostridia. The sequence for the model organism Bacillus subtilis is boxed. The -35 and -10 promoter elements are bolded and shaded with gray boxes, and the consensus sequences for σF and σG are shown above [38]. The known transcription start site for the B. subtilis sigG transcript is also bolded. Known or putative ribosome binding sites are underlined. Shortened spacing between the -35 and -10 promoter elements as well as non-ATG start codons are indicated in red.
(TIF)
Upstream regulatory sequences for sigG were retrieved from the genome sequences of representative endospore forming bacteria from the classes Bacilli and Clostridia. The 5’ mRNA leader sequences for each were queried for secondary structure by the RNAstructure method [43]. Potential secondary structures for a subset of the 5’ leader sequences are displayed using RNAbow diagrams [69], with base pairs are indicated by arcs, the thickness and shading of which is proportional to their probability. The partition function free energy (G) is indicated. The +1 transcription start site, RBS, and start codon for each are underlined. “+1*” indicates start sites selected based on alignment with the known B. subtilis sigG transcriptional start site.
(TIF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
We are grateful to Wade Winkler and Cordelia Weiss for helpful input regarding the sigG RBS-hairpin, and we thank Richard Losick as well as other members of the Camp Lab for valuable conversations and comments on the manuscript. This study is dedicated to the memory of co-author Laura Suzanne Murphy (1993–2016), Mount Holyoke College Class of 2015, whose undergraduate thesis research is included herein. Laura tragically lost her life at the hands of a drunk driver a month before she was set to begin graduate studies in Microbiology at the University of California, Berkeley. She had a bright future, and will be forever missed and remembered as a scientist, student-athlete, daughter, sister, niece, cousin, and friend.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by National Institutes of Health (https://www.nih.gov) grant R15 GM101559 to AHC. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Tan M. Temporal gene regulation during the Chlamydial developmental cycle. In: Tan M, Bavoil PM, editors. Intracellular Pathogens I: Chlamydiales. Washington, D.C; 2012. pp. 149–169. [Google Scholar]
- 2.Hilbert DW, Piggot PJ. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol Mol Biol Rev. 2004;68: 234–262. doi: 10.1128/MMBR.68.2.234-262.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Hoon MJL, Eichenberger P, Vitkup D. Hierarchical evolution of the bacterial sporulation network. Curr Biol. 2010;20: R735–R745. doi: 10.1016/j.cub.2010.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arrieta-Ortiz ML, Hafemeister C, Bate AR, Chu T, Greenfield A, Shuster B, et al. An experimentally supported model of the Bacillus subtilis global transcriptional regulatory network. Mol Syst Biol. 2015;11: 839–839. doi: 10.15252/msb.20156236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Z, Piggot PJ. Development of a two-part transcription probe to determine the completeness of temporal and spatial compartmentalization of gene expression during bacterial development. Proc Natl Acad Sci USA. 2001;98: 12538–12543. doi: 10.1073/pnas.221454798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barák I, Wilkinson AJ. Where asymmetry in gene expression originates. Mol Microbiol. 2005;57: 611–620. doi: 10.1111/j.1365-2958.2005.04687.x [DOI] [PubMed] [Google Scholar]
- 7.Schmidt R, Margolis P, Duncan L, Coppolecchia R, Moran CP, Losick R. Control of developmental transcription factor σF by sporulation regulatory proteins SpoIIAA and SpoIIAB in Bacillus subtilis. Proc Natl Acad Sci USA. 1990;87: 9221–9225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Partridge SR, Foulger D, Errington J. The role of σF in prespore-specific transcription in Bacillus subtilis. Mol Microbiol. 1991;5: 757–767. [DOI] [PubMed] [Google Scholar]
- 9.Sun DX, Cabrera-Martinez RM, Setlow P. Control of transcription of the Bacillus subtilis spoIIIG gene, which codes for the forespore-specific transcription factor σG. J Bacteriol. 1991;173: 2977–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camp AH, Wang AF, Losick R. A small protein required for the switch from σF to σG during sporulation in Bacillus subtilis. J Bacteriol. 2011;193: 116–124. doi: 10.1128/JB.00949-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Erickson AF, Deighan P, Chen S, Barrasso K, Garcia CP, Martínez-Lumbreras S, et al. A novel RNA polymerase-binding protein that interacts with a sigma-factor docking site. Mol Microbiol. 2017;105: 652–662. doi: 10.1111/mmi.13724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crawshaw AD, Serrano M, Stanley WA, Henriques AO, Salgado PS. A mother cell-to-forespore channel: current understanding and future challenges. FEMS Microbiol Lett. 2014;358: 129–136. doi: 10.1111/1574-6968.12554 [DOI] [PubMed] [Google Scholar]
- 13.Camp AH, Losick R. A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis. Genes Dev. 2009;23: 1014–1024. doi: 10.1101/gad.1781709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doan T, Morlot C, Meisner J, Serrano M, Henriques AO, Moran CP, et al. Novel secretion apparatus maintains spore integrity and developmental gene expression in Bacillus subtilis. PLoS Genet. 2009;5: e1000566 doi: 10.1371/journal.pgen.1000566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Decatur A, Losick R. Identification of additional genes under the control of the transcription factor σF of Bacillus subtilis. J Bacteriol. 1996;178: 5039–5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chary VK, Xenopoulos P, Piggot PJ. Expression of the σF-directed csfB locus prevents premature appearance of σG activity during sporulation of Bacillus subtilis. J Bacteriol. 2007;189: 8754–8757. doi: 10.1128/JB.01265-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karmazyn-Campelli C, Rhayat L, Carballido-López R, Duperrier S, Frandsen N, Stragier P. How the early sporulation sigma factor σF delays the switch to late development in Bacillus subtilis. Mol Microbiol. 2008;67: 1169–1180. doi: 10.1111/j.1365-2958.2008.06121.x [DOI] [PubMed] [Google Scholar]
- 18.Rhayat L, Duperrier S, Carballido-López R, Pellegrini O, Stragier P. Genetic dissection of an inhibitor of the sporulation sigma factor σG. J Mol Biol. 2009;390: 835–844. doi: 10.1016/j.jmb.2009.05.073 [DOI] [PubMed] [Google Scholar]
- 19.Camp AH, Losick R. A novel pathway of intercellular signalling in Bacillus subtilis involves a protein with similarity to a component of type III secretion channels. Mol Microbiol. 2008;69: 402–417. doi: 10.1111/j.1365-2958.2008.06289.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Partridge SR, Errington J. The importance of morphological events and intercellular interactions in the regulation of prespore-specific gene expression during sporulation in Bacillus subtilis. Mol Microbiol. 1993;8: 945–955. [DOI] [PubMed] [Google Scholar]
- 21.Karow ML, Piggot PJ. Construction of gusA transcriptional fusion vectors for Bacillus subtilis and their utilization for studies of spore formation. Gene. 1995;163: 69–74. [DOI] [PubMed] [Google Scholar]
- 22.Sun YL, Sharp MD, Pogliano K. A dispensable role for forespore-specific gene expression in engulfment of the forespore during sporulation of Bacillus subtilis. J Bacteriol. 2000;182: 2919–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evans L, Feucht A, Errington J. Genetic analysis of the Bacillus subtilis sigG promoter, which controls the sporulation-specific transcription factor σG. Microbiology. 2004;150: 2277–2287. doi: 10.1099/mic.0.26914-0 [DOI] [PubMed] [Google Scholar]
- 24.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30: 297–341. doi: 10.1146/annurev.genet.30.1.297 [DOI] [PubMed] [Google Scholar]
- 25.Karow ML, Glaser P, Piggot PJ. Identification of a gene, spoIIR, that links the activation of σE to the transcriptional activity of σF during sporulation in Bacillus subtilis. Proc Natl Acad Sci USA. 1995;92: 2012–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Londoño-Vallejo JA, Stragier P. Cell-cell signaling pathway activating a developmental transcription factor in Bacillus subtilis. Genes Dev. 1995;9: 503–508. [DOI] [PubMed] [Google Scholar]
- 27.Londoño-Vallejo JA, Fréhel C, Stragier P. SpoIIQ, a forespore-expressed gene required for engulfment in Bacillus subtilis. Mol Microbiol. 1997;24: 29–39. [DOI] [PubMed] [Google Scholar]
- 28.Serrano M, Neves A, Soares CM, Moran CP, Henriques AO. Role of the anti-sigma factor SpoIIAB in regulation of σG during Bacillus subtilis sporulation. J Bacteriol. 2004;186: 4000–4013. doi: 10.1128/JB.186.12.4000-4013.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun D, Fajardo-Cavazos P, Sussman MD, Tovar-Rojo F, Cabrera-Martinez RM, Setlow P. Effect of chromosome location of Bacillus subtilis forespore genes on their spo gene dependence and transcription by EσF: identification of features of good EσF-dependent promoters. J Bacteriol. 1991;173: 7867–7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vellanoweth RL, Rabinowitz JC. The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo. Mol Microbiol. 1992;6: 1105–1114. [DOI] [PubMed] [Google Scholar]
- 31.Tuller T, Zur H. Multiple roles of the coding sequence 5' end in gene expression regulation. Nucleic Acids Res. 2015;43: 13–28. doi: 10.1093/nar/gku1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaspar P, Moura G, Santos MAS, Oliveira JL. mRNA secondary structure optimization using a correlated stem-loop prediction. Nucleic Acids Res. 2013;41: e73 doi: 10.1093/nar/gks1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Veening J-W, Smits WK, Hamoen LW, Jongbloed JDH, Kuipers OP. Visualization of differential gene expression by improved cyan fluorescent protein and yellow fluorescent protein production in Bacillus subtilis. Appl Environ Microbiol. 2004;70: 6809–6815. doi: 10.1128/AEM.70.11.6809-6815.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foulger D, Errington J. The role of the sporulation gene spoIIIE in the regulation of prespore-specific gene expression in Bacillus subtilis. Mol Microbiol. 1989;3: 1247–1255. [DOI] [PubMed] [Google Scholar]
- 35.Smith K, Bayer ME, Youngman P. Physical and functional characterization of the Bacillus subtilis spoIIM gene. J Bacteriol. 1993;175: 3607–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karmazyn-Campelli C, Bonamy C, Savelli B, Stragier P. Tandem genes encoding σ-factors for consecutive steps of development in Bacillus subtilis. Genes Dev. 1989;3: 150–157. [DOI] [PubMed] [Google Scholar]
- 37.Steil L, Serrano M, Henriques AO, Völker U. Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis. Microbiology. 2005;151: 399–420. doi: 10.1099/mic.0.27493-0 [DOI] [PubMed] [Google Scholar]
- 38.Wang ST, Setlow B, Conlon EM, Lyon JL, Imamura D, Sato T, et al. The forespore line of gene expression in Bacillus subtilis. J Mol Biol. 2006;358: 16–37. doi: 10.1016/j.jmb.2006.01.059 [DOI] [PubMed] [Google Scholar]
- 39.Bagyan I, Casillas-Martinez L, Setlow P. The katX gene, which codes for the catalase in spores of Bacillus subtilis, is a forespore-specific gene controlled by σF, and KatX is essential for hydrogen peroxide resistance of the germinating spore. J Bacteriol. 1998;180: 2057–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewis PJ, Partridge SR, Errington J. σ factors, asymmetry, and the determination of cell fate in Bacillus subtilis. Proc Natl Acad Sci USA. 1994;91: 3849–3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sierro N, Makita Y, de Hoon M, Nakai K. DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Res. 2008;36: D93–D96. doi: 10.1093/nar/gkm910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyer MM. The role of mRNA structure in bacterial translational regulation. Wiley Interdiscip Rev RNA. 2017;8: e1370 doi: 10.1002/wrna.1370 [DOI] [PubMed] [Google Scholar]
- 43.Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11: 129 doi: 10.1186/1471-2105-11-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Serrano M, Real G, Santos J, Carneiro J, Moran CP, Henriques AO. A negative feedback loop that limits the ectopic activation of a cell type-specific sporulation sigma factor of Bacillus subtilis. PLoS Genet. 2011;7: e1002220 doi: 10.1371/journal.pgen.1002220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stefano JE, Gralla JD. Spacer mutations in the lac ps promoter. Proc Natl Acad Sci USA. 1982;79: 1069–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Russell DR, Bennett GN. Construction and analysis of in vivo activity of E. coli promoter hybrids and promoter mutants that alter the -35 to -10 spacing. Gene. 1982;20: 231–243. [DOI] [PubMed] [Google Scholar]
- 47.Aoyama T, Takanami M, Ohtsuka E, Taniyama Y, Marumoto R, Sato H, et al. Essential structure of E. coli promoter: effect of spacer length between the two consensus sequences on promoter function. Nucleic Acids Res. 1983;11: 5855–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mulligan ME, Brosius J, McClure WR. Characterization in vitro of the effect of spacer length on the activity of Escherichia coli RNA polymerase at the TAC promoter. J Biol Chem. 1985;260: 3529–3538. [PubMed] [Google Scholar]
- 49.Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324: 255–258. doi: 10.1126/science.1170160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goodman DB, Church GM, Kosuri S. Causes and effects of N-terminal codon bias in bacterial genes. Science. 2013;342: 475–479. doi: 10.1126/science.1241934 [DOI] [PubMed] [Google Scholar]
- 51.Allert M, Cox JC, Hellinga HW. Multifactorial determinants of protein expression in prokaryotic open reading frames. J Mol Biol. 2010;402: 905–918. doi: 10.1016/j.jmb.2010.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Masuda ES, Anaguchi H, Yamada K, Kobayashi Y. Two developmental genes encoding σ factor homologs are arranged in tandem in Bacillus subtilis. Proc Natl Acad Sci USA. 1988;85: 7637–7641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mukherjee S, Sengupta S. Riboswitch Scanner: an efficient pHMM-based web-server to detect riboswitches in genomic sequences. Bioinformatics. 2016;32: 776–778. doi: 10.1093/bioinformatics/btv640 [DOI] [PubMed] [Google Scholar]
- 54.Abreu-Goodger C, Merino E. RibEx: a web server for locating riboswitches and other conserved bacterial regulatory elements. Nucleic Acids Res. 2005;33: W690–W692. doi: 10.1093/nar/gki445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang T-H, Huang H-D, Wu L-C, Yeh C-T, Liu B-J, Horng J-T. Computational identification of riboswitches based on RNA conserved functional sequences and conformations. RNA. 2009;15: 1426–1430. doi: 10.1261/rna.1623809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmidt R, Decatur AL, Rather PN, Moran CP, Losick R. Bacillus subtilis Lon protease prevents inappropriate transcription of genes under the control of the sporulation transcription factor σG. J Bacteriol. 1994;176: 6528–6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Serrano M, Hövel S, Moran CP, Henriques AO, Völker U. Forespore-specific transcription of the lonB gene during sporulation in Bacillus subtilis. J Bacteriol. 2001;183: 2995–3003. doi: 10.1128/JB.183.10.2995-3003.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rather PN, Coppolecchia R, DeGrazia H, Moran CP. Negative regulator of σG-controlled gene expression in stationary-phase Bacillus subtilis. J Bacteriol. 1990;172: 709–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Serrano M, Gao J, Bota J, Bate AR, Meisner J, Eichenberger P, et al. Dual-specificity anti-sigma factor reinforces control of cell-type specific gene expression in Bacillus subtilis. PLoS Genet. 2015;11: e1005104 doi: 10.1371/journal.pgen.1005104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Flanagan KA, Comber JD, Mearls E, Fenton C, Wang Erickson AF, Camp AH. A membrane-embedded amino acid couples the SpoIIQ channel protein to anti-sigma factor transcriptional repression during Bacillus subtilis sporulation. J Bacteriol. 2016;198: 1451–1463. doi: 10.1128/JB.00958-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kirchman PA, DeGrazia H, Kellner EM, Moran CP. Forespore-specific disappearance of the sigma-factor antagonist SpoIIAB: implications for its role in determination of cell fate in Bacillus subtilis. Mol Microbiol. 1993;8: 663–671. [DOI] [PubMed] [Google Scholar]
- 62.Youngman P, Perkins JB, Losick R. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid. 1984;12: 1–9. [DOI] [PubMed] [Google Scholar]
- 63.Schaeffer P, Millet J, Aubert JP. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci USA. 1965;54: 704–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nicholson WL, Setlow P. Sporulation, germination and outgrowth. In: Harwood CR, Cutting SM, editors. Molecular Biology Methods for Bacillus. New York City, NY; 1990. pp. 391–450. [Google Scholar]
- 65.Sterlini JM, Mandelstam J. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem J. 1969;113: 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9: 676–682. doi: 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gruber AR, Bernhart SH, Lorenz R. The ViennaRNA web services. Methods Mol Biol. 2015;1269: 307–326. doi: 10.1007/978-1-4939-2291-8_19 [DOI] [PubMed] [Google Scholar]
- 69.Aalberts DP, Jannen WK. Visualizing RNA base-pairing probabilities with RNAbow diagrams. RNA. 2013;19: 475–478. doi: 10.1261/rna.033365.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
An AUG (ATG) start codon followed by codons 2–28 of B. subtilis sigG are shown (top sequence), as is a sequence comprised of synonymous codons selected by the mRNA Optimizer tool [32] to minimize secondary structure (bottom sequence). Note that the latter is the sequence present in the PsigG-ATG-RSSsigG2-28-lacZ reporter construct used in Fig 2. Nucleotide differences are indicated in red and blue text. Both sequences were analyzed for secondary structure by the RNAstructure method [43]. Potential secondary structures are displayed above (red) or below (blue) using RNAbow diagrams [69], with base pairs indicated by arcs, the thickness and shading of which is proportional to their probability. The partition function free energy (G) for each is indicated.
(TIF)
β-Galactosidase production was monitored during sporulation of strains harboring PsigG-lacZ (WT; open circles), 15ntPsigG-lacZ (15 nt; closed circles), T→APsigG-lacZ (T→A; open diamonds), T→GPsigG-lacZ (T→G; closed diamonds) 15nt,T→APsigG-lacZ (15 nt, T→A; open triangles), and 15nt,T→GPsigG-lacZ (15 nt, T→G; open squares). (Strains JJB31, JJB51, JJB87, JJB89, JJB99 and JJB101, respectively.) For clarity, only data from a subset of these strains are presented in each graph (as labeled) and, also for clarity, the WT and 15 nt data is presented in all graphs. Note that (B) and (D) provide zoomed views of the data from the boxed areas of (A) and (C), respectively.
(TIF)
The total number of heat-resistant spores per mL was determined for a wild type strain harboring unaltered sigG at its native locus (WT; strain PY79), a strain deleted for sigG (ΔsigG; strain AHB98), or strains deleted for sigG and harboring at the non-essential ylnF locus a copy of the sigG gene either under the control of wild type regulatory sequences (+ylnF::WTPsigG-sigG; strain LMB39) or regulatory sequences modified to remove or repair the four features identified in this study to dampen sigG expression (+ylnF::quadPsigG-sigG; strain AHB2819). Error bars indicate standard deviation based on three or more independent experiments.
(TIF)
(A) Representative growth curves in liquid LB for cells expressing sigG from its wild type regulatory sequences (WTPsigG) or from regulatory sequences modified to remove or repair the four features identified in this study to dampen sigG expression (quadPsigG). Additionally, cells either harbored wild type csfB, or were deleted for the gene (ΔcsfB). (Strains EBM192 [WTPsigG; purple diamonds], EBM276 [quadPsigG; orange triangles], EBM282 [WTPsigG ΔcsfB; blue squares], and EBM287 [quadPsigG-sigG ΔcsfB; green circles].) (B) Doubling time during exponential growth and maximal OD600 during stationary phase for each strain are reported as the average ± standard deviation based on three independent experiments. **p < 0.01, ****p < 0.0001, ns not significant, Student’s t-test.
(TIF)
Upstream regulatory sequences for sigG were retrieved from the genome sequences of representative endospore forming bacteria from the classes Bacilli and Clostridia. The sequence for the model organism Bacillus subtilis is boxed. The -35 and -10 promoter elements are bolded and shaded with gray boxes, and the consensus sequences for σF and σG are shown above [38]. The known transcription start site for the B. subtilis sigG transcript is also bolded. Known or putative ribosome binding sites are underlined. Shortened spacing between the -35 and -10 promoter elements as well as non-ATG start codons are indicated in red.
(TIF)
Upstream regulatory sequences for sigG were retrieved from the genome sequences of representative endospore forming bacteria from the classes Bacilli and Clostridia. The 5’ mRNA leader sequences for each were queried for secondary structure by the RNAstructure method [43]. Potential secondary structures for a subset of the 5’ leader sequences are displayed using RNAbow diagrams [69], with base pairs are indicated by arcs, the thickness and shading of which is proportional to their probability. The partition function free energy (G) is indicated. The +1 transcription start site, RBS, and start codon for each are underlined. “+1*” indicates start sites selected based on alignment with the known B. subtilis sigG transcriptional start site.
(TIF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.