To the Editor
ATAC-seq (assay for transposase-accessible chromatin with high-throughput sequencing) enables rapid, genomewide analysis of chromatin accessibility in a variety of cell types1. However, follow-up and hypothesis-driven analyses typically focus on only a small subset of these DNA elements. To facilitate high-throughput, low-cost analysis of chromatin accessibility, we developed ATAC Primer Tool (APT) for primer design and the identification of accurate normalization controls for ATAC-qPCR (Fig. 1a, Supplementary Fig. 1). We demonstrate that with APT, ATAC-qPCR and allelic ATAC-PCR can accurately quantify differential and allele-specific chromatin accessibility at targeted loci for biomarker or functional studies, including the detection of functional somatic mutations in DNA regulatory elements in cancer cells.
Figure 1.
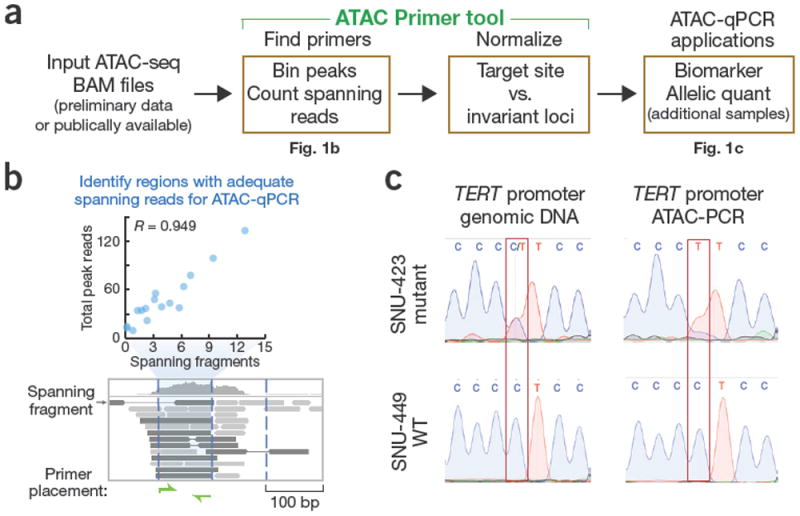
ATAC Primer Tool enables targeted and allele-specific quantification by ATAC-qPCR. (a) Workflow for primer design and identification of normalization controls using APT and applications of ATAC-qPCR. (b) Illustration of the APT algorithm for identifying optimal ATAC-qPCR primers. Peaks of interest are binned into overlapping windows, and the correlation between the number of spanning fragments and peak height is calculated for each window. Windows with adequate spanning fragments (blue) are selected for primer design. (c) ATAC-PCR of allele-specific accessibility at the TERT promoter. Sequencing of genomic DNA and ATAC-PCR is shown for SNU-423 (top; heterozygous for −124 C>T TERT promoter mutation) and SNU-449 (bottom; wild-type TERT promoter).
Many fragments generated by Tn5 transposition during ATACseq are less than 100 base pairs long and thus are too short for quantification by qPCR. To overcome this limitation, APT identifies optimal regions for ATAC-qPCR primers within peaks by comparing the number of spanning fragments in overlapping windows to the normalized peak height across samples (Fig. 1b, Supplementary Table 1).
Differences in background signal and transcription start site enrichment between ATAC libraries can produce misleading ATAC-qPCR results if data are not normalized correctly. Normalization to elements with low variability between samples is analogous to sequencing-depth correction during standard ATAC-seq analysis. We identified universal normalization controls based on low-variance peaks across a panel of diverse human and mouse tissues from ATAC-seq data in ENCODE (Supplementary Fig. 2). APT can also identify custom normalization controls based on user-supplied ATAC-seq data. We tested the correlation between ATAC-qPCR and ATAC-seq, using a panel of differentially accessible elements in human fibroblasts after topoisomerase II inhibition, as well as a panel of random monoallelically accessible elements in mouse neural progenitor cells2 (Supplementary Table 2 and 3). Systematic comparison of ATAC-qPCR with ATAC-seq showed that normalization to invariant peaks, as implemented in APT, greatly improved the correlation between ATAC-qPCR results and read counts from deeply sequenced ATAC-seq libraries (average change from R = 0.747 to R = 0.957); we obtained similar results with cell-type-specific and universal normalization controls (Supplementary Fig. 3).
A targeted allelic ATAC assay is highly desirable because only a small fraction of DNA elements (~1%) both possess informative sequence variants and demonstrate allelic regulation2. Inclusion of an informative SNP between primer-binding sites allows for targeted analysis of allele-specific accessibility by ATAC-PCR, for example, to assess the effect of disease-associated variants on allele-specific accessibility. For well-characterized TERT promoter mutations3,4, ATAC-PCR and Sanger sequencing demonstrated that only the mutant allele conferred TERT promoter chromatin accessibility (Fig. 1c, Supplementary Table 4). We further validated allelic ATAC-PCR on a panel of DNA elements with random monoallelic accessibility in hybrid mouse NPC cells2 (Supplementary Table 5). Allele-specific quantification by ATAC-PCR correlated strongly with the allelic accessibility determined by ATAC-seq (R = 0.962) (Supplementary Fig. 4), thus demonstrating accurate quantification of targeted and allelespecific accessibility by ATAC-PCR enabled by APT.
Further information on experimental design is available in the Supplementary Methods and the Life Sciences Reporting Summary.
Software availability
The current APT software is available from GitHub (https://github.com/ChangLab/ATACPrimerTool) and as part of the Galaxy Tool Shed (details at https://github.com/ChangLab/ATACPrimerTool/blob/master/Running_APT_with_Galaxy.md).
Data availability
ATAC-seq data from human and mouse tissues were obtained from ENCODE; accession numbers are available in Supplementary Tables 6 and 7. ATAC-seq data from hybrid mouse neural progenitor cells are available at the Gene Expression Omnibus (GSE84646).
Supplementary Material
Acknowledgments
We thank members of the Chang and Greenleaf labs for discussion. This work was supported by the NIH (grant P50-HG007735 to H.Y.C.), the National Science Foundation (Graduate Research Fellowship DGE-1656518 to K.E.Y.) and Stanford University (Graduate Fellowships to K.E.Y. and A.C.C.).
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
K.E.Y., A.C.C. and H.Y.C. conceived and designed the project. K.E.Y., A.C.C. and U.L. performed experiments. K.E.Y., A.C.C., U.L. and J.X. analyzed sequencing data. K.E.Y. wrote the software tool with input from J.X. and A.C.C. K.E.Y. and H.Y.C. wrote the manuscript with input from all other authors. H.Y.C. supervised the project.
COMPETING INTERESTS
H.Y.C. is a founder of Accent Therapeutics and Epinomics and is a consultant for Spring Discovery. Stanford University has filed a patent on ATAC-seq, on which H.Y.C. is named as an inventor.
References
- 1.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu J, et al. Nat Genet. 2017;49:377–386. doi: 10.1038/ng.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang FW, et al. Science. 2013;339:957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stern JL, Theodorescu D, Vogelstein B, Papadopoulos N, Cech TR. Genes Dev. 2015;29:2219–2224. doi: 10.1101/gad.269498.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ATAC-seq data from human and mouse tissues were obtained from ENCODE; accession numbers are available in Supplementary Tables 6 and 7. ATAC-seq data from hybrid mouse neural progenitor cells are available at the Gene Expression Omnibus (GSE84646).


