Rabbits have had a stronger impact on the landscape and plant communities of a remote island than one century of climate change.
Abstract
What are the long-term consequences of invasive species? After invasion, how long do ecosystems require to reach a new equilibrium? Answering these questions requires long-term, high-resolution data that are vanishingly rare. We combined the analysis of environmental DNA extracted from a lake sediment core, coprophilous fungi, and sedimentological analyses to reconstruct 600 years of ecosystem dynamics on a sub-Antarctic island and to identify the impact of invasive rabbits. Plant communities remained stable from AD 1400 until the 1940s, when the DNA of invasive rabbits was detected in sediments. Rabbit detection corresponded to abrupt changes of plant communities, with a continuous decline of a dominant plant species. Furthermore, erosion rate abruptly increased with rabbit abundance. Rabbit impacts were very fast and were stronger than the effects of climate change during the 20th century. Lake sediments can allow an integrated temporal analysis of ecosystems, revealing the impact of invasive species over time and improving our understanding of underlying mechanisms.
INTRODUCTION
Invasive alien species (IAS) are a major driver of biodiversity loss, with impacts that can include the modification of ecosystem functioning and the services ecosystems provide (1, 2). Impacts often are particularly evident on small islands, where IAS have determined multiple extinctions and complete modifications of biogeographical patterns (2–4). The spread and the impacts of IAS can be extremely rapid. In a spectacular example, American cane toads have invaded nearly 20% of Australia in less than 80 years and have caused the decline of multiple species of native predators, with cascading effects on the whole trophic web (5).
In some cases, the full impact of IAS does not occur immediately after their arrival in new ecosystems, but only after a so-called lag phase, during which IAS have limited abundances and are difficult to detect (6, 7). Understanding how quickly IAS spread and interact with native ecosystems is pivotal. Control actions performed during early invasion phases, when IAS have limited abundance, may be more successful, less costly, and can effectively prevent the consequences. On the other hand, even for the IAS with the fastest spread, it may take a long time before the full range of effects actually occur and the ecosystems shift to a new state (8, 9). Measuring the full range of the impact would require decades or centuries of monitoring, including periods before invasion; thus, in most cases, we do not have a full picture of changes of invaded ecosystems through time (8, 10).
European rabbits have been released on more than 800 islands all over the world, with heavy impacts on native communities, such as overgrazing, landscape degradation, and decline of multiple native plants (11). In the Kerguelen Islands, a remote sub-Antarctic archipelago (Fig. 1), rabbits were introduced in 1864. Recent analyses suggest that rabbit overgrazing is causing the decline of multiple endemic plants and a complete modification of soil communities (12, 13), but, as in most archipelagos, long-term information on how landscapes actually changed after rabbit introduction is limited (appendix S1).
Fig. 1. Lake La Poule.
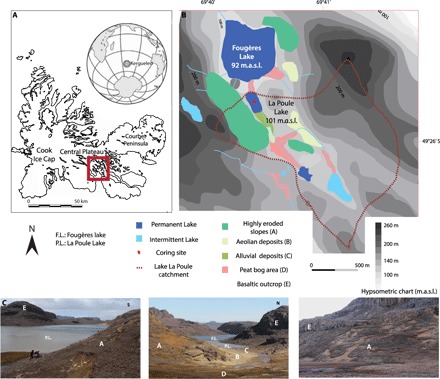
Location (A), map (B), and pictures (C) of the main landscape features of the watershed. F.L., Fougères Lake; P.L., La Poule Lake ; m.a.s.l., meters above sea level.
Retro-observational approaches, for instance, analyzing lake sediment cores, provide high-resolution information on multiple ecosystem features, thus allowing reconstruction of long-term environmental trajectories [for example, the study by Sabatier et al. (14)]. The recent observation that the DNA of organisms living in a lake watershed can be successfully amplified from lake sediments (sedDNA) has allowed exceptional reconstructions of community changes through time (15–17). SedDNA can be combined with pollen, spore, and non-pollen palynomorph (NPP) analysis to reconstruct paleoenvironments (17) and can provide unprecedented information on the tempo and magnitude of IAS impacts on the whole ecosystems, particularly if coped with additional sedimentological proxies.
Here, we combine sedDNA, analysis of coprophilous fungi, and reconstruction of erosion rate to evaluate the impact of introduced rabbits on a remote archipelago. Our analyses reveal the rapidity of rabbit effects on multiple ecosystem features and show that impacts on different ecosystem properties (native plant communities and modification of erosion patterns) can be highly heterogeneous if analyzed in a long-term perspective.
RESULTS
The lake sediments were characterized using grain-size distribution, loss on ignition (LOI), and sedimentary structures. The sediment consisted of olive-brown silt with (i) coarser (silt to fin sand) and (ii) dark-brown sediment interbedded thin layers. The grain-size distribution was rather homogeneous (median ± SD = 43 ± 10 μm; Fig. 2A and fig. S1). LOI presented relatively high organic content [mean of LOI at 550°C (LOI550) = 12.2 ± 3%], low to null carbonate content (<2%, detection limit of the method), and a mean NCIR (noncarbonate igneous residue) content of 87.7 ± 3% (fig. S1); these fractions significantly varied over the last 26 cm (Fig. 2B).
Fig. 2. Data from core POU14P1.
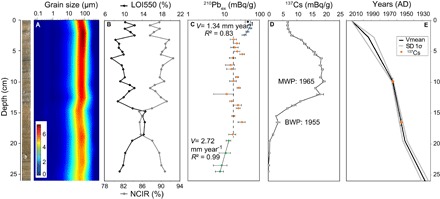
(A) Photograph and grain size contour plot. The color scale refers to the abundance in percentage of each grain-size class, with the less abundant in blue and the most abundant in red. (B) LOI550 (organic matter) and NCIR, (C) 210Pbex activity, (D) 137Cs activity, and (E) the age model for the upper 26 cm. MWP and BWP correspond, respectively, to maximum and beginning of nuclear weapon peak in the southern hemisphere.
Short-lived radionuclides allowed the establishment of a chronological framework (18). A logarithmic plot of 210Pbex activity (Fig. 2C) showed a general decrease with two distinct linear trends separating a 15-cm-long period with almost constant 210Pbex, suggesting a higher sedimentation rate (dashed line in Fig. 2C). According to the constant flux constant sedimentation rate (CFCS) model (19), as applied to the lower and upper parts of the profile, the levels of 210Pbex indicate mean accumulation rates of 2.72 ± 0.15 mm year−1 between depths of 26 and 19 cm and 1.34 ± 0.33 mm year−1 in the upper 2.8 cm of the core (Fig. 2C); the most superficial sample showed very low 210Pbex and was not taken into account. No sedimentation rate could be determined from 210Pbex in the central part of this core. The plot of 137Cs activities (Fig. 2D) displays an increase at 16.5 cm and a maximum peak at 9.85 cm. According to previous studies in the southern hemisphere, these two depths correspond to the first appearance of 137Cs dated to AD 1955 and the maximum peak at AD 1965, respectively (20–22). These two dates allow us to calculate mean sedimentation rates for the period with constant 210Pbex activities of 2.5 mm year−1 (between 2.8 and 9.85 cm) and 6.6 mm year−1 (between 9.85 and 19 cm). The age model of this sediment core (Fig. 2E) derived from the two 210Pbex-CFCS model, constrained by the artificial radionuclide data, provides a continuous age-depth relationship with a major sedimentation rate increase in ~1950 ± 4 years and then a progressive decrease to the current rate, lower than before this change. Finally, radiocarbon dating was performed on a terrestrial macroremain and provided a calibrated 14C age range of 329 to 509 years cal BP (AD 1621–1441) at 2σ uncertainty, allowing to calculate an age-depth relationship for the lower part of this sediment core (fig. S2) (23).
Mammal occurrence chronology
Among the NPP, spores of coprophilous fungi (Sporormiella) have been proposed as a proxy for the density of wild and domestic herbivores (24–27) and are well recorded in the last 21 cm of the sequence. Sporormiella was completely absent from the sediments until AD 1941, and the first occurrence of this coprophilous fungus was recorded in sediments deposited between 1941 and 1948. The amount (influx) of Sporormiella sharply increased in subsequent years, with a rise in 1952–1955 (Fig. 3 and table S1). The abundance of Sporormiella remained high through the last 50 years, except in the last two decades, when it tended to decrease. Between 1941 and 1959, we also detected Podospora ascospores and multicellular ascospores of Sporormiella. Podospora provides additional evidence of herbivore presence (25, 26); multicellular ascospores reflect a low residence time in the watershed and thus fast transport to the lake system, because these fungal ascospores are generally rapidly separated into individual spores.
Fig. 3. Temporal variation of biological and sedimentological proxies.
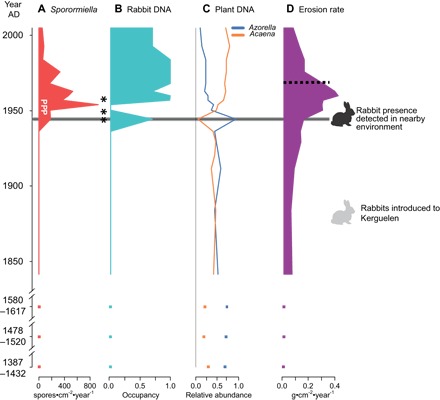
(A) accumulation rate of spores of coprophilous fungi (Sporormiella spp.; influx), occurrence of Podospora spp. (P), and multicellular ascospores of Sporormiella (*). (B) Rabbit sedDNA. (C) sedDNA of plants (proportion). (D) Erosion rate. Dashed lines represent the abrupt changes detected by regression tree and breakpoint analysis.
We did not detect mammal DNA before AD 1941 (Fig. 3). SedDNA sequences assigned to Oryctolagus cuniculus (rabbit) were first detected in 1941–1948 [1/8 polymerase chain reaction (PCR) replicates] and were then present in most of the subsequent samples. Rabbit sedDNA was generally detected in 2/8 to 4/8 of PCRs in samples from the period 1950–1988 and remained present in recent samples, but with a lower frequency of detections (Fig. 3 and table S2). Occupancy modeling (28) indicated that, when present, the detection probability of rabbit sedDNA was 31.5%, and the estimated rate of false detection was low (<1%); thus, detection of even a single instance of sedDNA per sample provides a good indication of the presence of rabbits (probability of presence = 0.72; Fig. 3). The presence of rabbit sedDNA was strongly correlated with the influx of Sporormiella spores (r = 0.74, n = 20, P = 0.0002), although the temporal pattern of these two proxies of rabbit occurrence was not identical (Fig. 3).
Variation of plant communities
We detected sedDNA of eight plant taxa: Azorella selago, Acaena magellanica, Ranunculus biternatus, Agrostis magellanica, Taraxacum officinale, Pringlea antiscorbutica, Festuca contracta, and Deschampsia antarctica (table S2). All are native to Kuerguelen, except T. officinale, which is an alien species native to Europe and was only detected after 1948 at low abundance. Until 1941, A. selago was the dominant plant species, accounting for 44 to 72% of plant sedDNA reads, whereas Acaena magellanica was the second most abundant species. The abundance of A. selago sedDNA peaked in 1941–1948; during this time period, A. selago accounted for 95% of DNA reads, but then sharply declined in subsequent years. A multivariate regression tree (MRT) (cross-validation error = 0.461) indicated that the composition of plant communities abruptly changed once during the study period (after 1948), with a shift from communities dominated by A. selago to communities dominated by Acaena magellanica (Fig. 3A). The analysis of similarities confirmed the significant differences between the communities observed until 1948 and the most recent ones (permutation test: P < 0.0001, r = 0.73).
Abundance of Sporormiella and rabbit DNA was a strong predictor of changes in plant communities (Fig. 4B) and explained most community variation [redundancy analysis (RDA): P < 0.0001, R2 = 0.72). This finding confirms that the shift from Azorella-dominated to Acaena-dominated communities is strongly related to rabbit invasion.
Fig. 4. Ecological variation of plant communities through time.
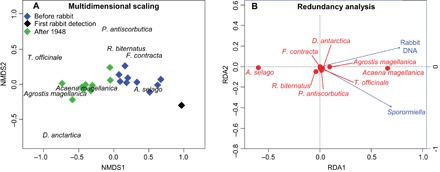
(A) Nonmetric multidimensional scaling (NMDS) representing the similarity of communities from sediment samples from different periods. (B) Constrained RDA showing the relationship between the proxies of rabbit abundance (sedDNA and Sporormiella) and the abundance of different plant taxa. Taxa with negative scores along the first RDA axis have negative association with rabbit abundance.
Erosion rate
Assuming that the flux of biogenic silica (diatom) is constant over time and negligible with regard to terrigenous flux in the lake system, we observed two changes in the sedimentation rate (Fig. 2) related to variations in the terrigenous sediment supply (mass accumulation) from the watershed and that may be attributed to soil erosion (Fig. 3) (14, 29). The existence of two sharp changes in erosion rate was confirmed by breakpoint analysis (table S3): Erosion rate was generally <0.15 g cm−2 year−1 until 1941, then abruptly increased at values >0.3 after 1948, and decreased again after 1964 (Fig. 3). The relationship between the variation in erosion rate and rabbit abundance was very strong (autoregressive model: t18 = 6.45, P < 0.0001, likelihood ratio R2 = 0.70).
Potential role of climate change
In the Kerguelen archipelago, average precipitation tended to decrease, whereas mean temperature tended to increase through the 20th century, with the strongest changes occurring after 1970 (table S4). An RDA relating plant communities during the 20th century to rabbit occurrence and climate explained 86.3% of variation. Both rabbits and climate explained a significant amount of the variation of plant communities, but rabbit abundance explained much more of the variation than climate. Rabbit occurrence explained 33.6% of variation in plant communities (P < 0.001), climatic variables explained 19.5% of variation (P = 0.015), and 30.5% of variation was explained by the joint effects of rabbits and climate.
When we included climatic variables in the analysis of erosion rate, rabbit abundance remained the variable best explaining the variation of erosion (t11 = 3.70, P = 0.004), whereas relationships between erosion and climate were weaker and nonsignificant (precipitation: t11 = 1.06, P = 0.32; temperature: t11 = −2.15, P = 0.06).
DISCUSSION
Using present-day observations alone, we are often incapable of reconstructing invasion dynamics or of understanding whether the ecosystem state many decades after the introduction of IAS still corresponds to a postintroduction crisis, or to new ecological equilibria (8, 10). Although the effects of rabbit introduction on the southern islands have been described multiple times (11), the actual dynamics of ecosystem changes remain poorly understood. In which order (rabbit population growth, change of native plants, increase of soil erosion, and introduction of alien plant species) and with which kinetics do the changes occur? The combination of sedDNA, spore, and lake sediment analyses is a powerful approach for the study of IAS through time because it allows a high-resolution reconstruction of landscape changes across centuries, providing a clear idea of the velocity, kinetics, and breadth of IAS impact on multiple properties of this isolated ecosystem. Rabbits were introduced in eastern Kerguelen in 1874. However, the few available historical records suggest that rabbits were absent from the study area in 1932, but probably present in the 1960s (appendix S1) (30, 31). The excellent match between sedimentary and historical records suggests that our reconstructions of invasion dynamics are reliable, allowing accurate timing of past ecosystem change in the landscape surrounding the lake.
In principle, Sporormiella spores can originate from other introduced herbivorous mammalian species that are present in Kerguelen, such as reindeer and mouflons (32). However, Sporormiella spores and additional NPPs (multicellular ascospores and Podospora spores) have been continuously detected from the 1941–1948 sample onward (Fig. 3), whereas reindeer and mouflons were first introduced only in 1955–1957 (32). Furthermore, we never detected reindeer or mouflon sedDNA, despite the fact that these species should have been detectable with the primers used here (table S5) (15). Therefore, rabbits are the most likely explanation for the observed pattern, which combines multiple palaeoecological proxies. Nevertheless, the detection probability of environmental DNA in sediments is often low, and existing species may remain undetected even if multiple samples are analyzed (33); thus, we cannot exclude a contribution from other mammals to Sporormiella inputs for recent samples. Despite a good correlation between Sporormiella abundance and rabbit sedDNA, the match between these two proxies was not perfect. Both Sporormiella and sedDNA are generally transported only limited distances; therefore, they probably provide a signal of ecological processes occurring at the local or landscape scale (17, 25, 34). However, spores of coprophilous fungi are generally carried by surface runoff either from the shores or from rivers that are hydrologic inputs to the lake (24, 25). SedDNA transport to sediments is also linked to surface run-off, but appears to be additionally favored by erosion, as DNA can be protected and transported more easily when adsorbed on clay particles (17, 35). Finally, during periods of high mortality, such as after the introduction of myxomatosis in 1955–1956, additional DNA transport could have occurred through dead animals (32), whereas Sporormiella flux might be weakened by such an event. All these processes, combined with the low detection probability of environmental DNA (33), can contribute to the differences between proxies.
A. selago currently is among the most abundant plant species in the areas of Kerguelen without herbivores (12, 13, 36). SedDNA confirmed that A. selago was extremely abundant over the last 600 years in the La Poule Lake watershed, and until AD 1941 only minor oscillations in plant abundance occurred (Fig. 3 and table S1). The relative stability of plant communities during this period was confirmed by MRT, which did not detect community changes until the 1940s. However, the situation abruptly changed after 1941, when in the same sample rabbit sedDNA was detected together with spores of the coprophilous fungus Sporormiella, indicating the arrival of hitherto absent herbivores. The first detection of rabbits corresponds to an immediate peak in the abundance of A. selago sedDNA, which in this sample accounted for 95% of plant reads. The peak of A. selago sedDNA observed in 1941–1948 probably corresponds to heavy grazing and/or burrowing, which determines the transport of plant DNA from the watershed to the lake, and does not represent a true increase in the abundance of this plant in the ecosystem. Direct consumption of A. selago by rabbits has been mentioned by previous reports (31, 37); this plant is particularly sensitive to rabbit burrowing and erosion, which are additional processes transforming vegetation (12, 32). However, after the first wave of invaders has heavily reduced the most sensitive species, this peak disappears, sedDNA clearly detects a shift of plant communities, and the continuous drop of A. selago DNA represents the dramatic decline of this plant, which currently is nearly extinct in the watershed (Fig. 1). A. selago is a keystone species in the sub-Antarctic islands. Its cushions provide shelter and increase resources for other plants and invertebrates; thus, this plant improves microhabitat and influences the distribution of multiple species (38, 39). Therefore, the effects of rabbit invasion are likely not limited to its impact on specific plant species, and cascading effects may occur throughout the whole soil community (13).
Nonnative plant species and climate change are additional threats to the Kerguelen flora (12). During the last century, Kerguelen experienced a significant warming and a decrease of precipitation (table S4), and these environmental changes probably had major impacts on communities (12). Nevertheless, climate change has mostly occurred since the 1970s (table S4), whereas in our study site, the strongest community shifts coincided with the arrival of rabbits in the 1940s (Fig. 3). Furthermore, although multivariate analyses detected a significant role of climate, rabbit occurrence explains much more of the variation than climate. This finding suggests that in some ecosystems, the impact of invasive herbivores can be particularly profound, even stronger than the ones of climate change.
The rapidity of rabbit impacts was also visible in patterns of erosion. Erosion rate remained low from AD 1400 to AD 1940, but again the situation markedly changed immediately after rabbit arrival. Rabbits can increase erosion by burrowing and reducing vegetation cover (11, 32). Similar to effects on plants, the kinetics of rabbit impact were very rapid: The highest erosion rate was observed only 10 years after the first detection of rabbits and matched well the period of highest abundance of coprophilous spores. The abundance of rabbit proxies has decreased in recent samples, suggesting rabbit decline in the recent decades, perhaps because of the myxomatosis virus introduced in 1955–1956 for biological control (appendix S1) (32) or because of landscape degradation and disappearance of plant species with the highest food value. The recent rabbit decline was followed by a reduction of erosion at rates comparable to the preinvasion values, suggesting that rabbit control can be effective in reestablishing this major ecosystem property. The reduction of erosion might also have occurred because the system shifted to a new state, for instance, because the most easily mobilized materials have already been eroded. Conversely, plant communities did not recover during this period, perhaps because plant and soil communities require longer periods for a full recovery (12, 13), or because rabbits continue to exert their impact through browsing, even if they have a limited abundance.
High-resolution information on IAS presence and ecosystem features through time provides a means of answering major questions on invasions and their consequences on environments. Rabbit impact was rapid, because less than 10 years from the first rabbit detection was enough for an abrupt shift in plant communities and erosion patterns. In practice, instead of a lag phase, rabbits exerted their strongest impacts immediately. In these situations, only prompt management actions can prevent the negative consequences on ecosystems. The eradication and control of invasive mammals from islands are possible and have been successfully performed multiple times (11, 40). Unfortunately, although the eradication of IAS allows substantial recovery of native species and enhancement of ecosystems (40), the velocity and effectiveness of recovery can be heterogeneous among ecosystem properties. Some physical features (for example, erosion, as shown here) and the abundance of some native vertebrates can quickly switch back to their original values (40). However, for other biological features, the recovery can be more difficult or even impossible, for instance, if the system is evolving toward a new state. Plant and soil communities may require many decades to attain a new equilibrium or to recover after eradications, and interactions among species and with other changing environmental features (for example, climate) can make ecosystem trajectories even more complex (12, 13). Long-term observations are needed to ascertain the actual effectiveness of management practices; emerging approaches that allow an integrated temporal analysis of ecosystems on the basis of lake sediment paleoenvironmental data (15, 17) are opening new avenues for obtaining such hitherto unavailable information.
MATERIALS AND METHODS
Our study focused on Lake La Poule (49°25′5″S, 69°40′25″E, 100 m.a.s.l.), located on the main Island of Kerguelen archipelago. Lake La Poule is a 220 × 120–m low-altitude lake in the central plateau of Kerguelen (Fig. 1). The lake watershed (surface area = 1.5 km2) is dominated by basaltic rocks without soils and vegetation. On the south of the lake and along the main river, a peaty area occurs. On the west of the catchment area and above the banks of the lake, volcanic soils that developed on pyroclastic deposits were observed. Acaena magellanica is the dominant plant species on these slopes, also characterized by the presence of numerous burrows of rabbits and intense erosion marks (Fig. 1). A 90-cm-long core (POU14P1) was collected from the deepest part of the lake in December 2014 using an Uwitec gravity corer. In small lakes, collecting one core from the deepest part of the lake is the standard in sedimentological, spore, and sedDNA studies, and multiple cores analyzed from the same lake generally provide reproducible signal of environmental changes (41). For instance, the reconstruction of pastoral pressure using spores and NPPs, using two cores from the deepest parts of two basins of the same lake, provided highly consistent reconstructions of human activities (24).
Logging
In the laboratory, the core was split, photographed, and logged in detail, noting all physical sedimentary structures and the vertical succession of facies. The grain-size distributions were determined using a Malvern Mastersizer S at a continuous interval of 2 cm. After inserting the bulk sediment into the fluid module of the granulometer, ultrasound was applied to minimize particle flocculation. The core was also sampled at 0.5-cm steps adapted to lithological variations for the upper 15 cm and at 1 cm below this depth and dried at 60°C over the course of 4 days to obtain its dry bulk density (DBD), and then the LOI of each 1-cm interval was measured following the study of Heiri et al. (42). LOI550 and LOI950 (LOI at 950°C) correspond to the organic and carbonate components of the sediment, respectively. NCIR was obtained by removing LOI550 and LOI950 from the initial dry weight. The mass accumulation was calculated as DBD × sedimentation rate, expressed in g cm−2 year−1.
Dating
The same sampling method as with LOI was applied to determine 210Pb, 226Ra, and 137Cs activities using well-type, germanium detectors placed at the Laboratoire Souterrain de Modane following the study by Reyss et al. (43). Counting times of 24 to 48 hours were required to reach a statistical error of less than 10% for excess 210Pb in the deepest samples and for the 137Cs peak. In each sample, the 210Pb excess activities (10Pbex) were calculated by subtracting the 226Ra-supported activity from the total 210Pb activity. A 14C measurement was carried out by an accelerator mass spectrometer at the Poznan Radiocarbon Laboratory. The calibration curve SHCal04 (44) was used for the 14C age calibration. Then, we used a smooth spline interpolation with the R package clam to generate the age model (23).
Sedimentary DNA analyses
From the sediment core, we sampled 22 slices of 2-cm thickness covering the whole core, following strict laboratory precautions (15). For the first 30 cm, the core was continuously sampled, that is, we collected one sample each 2 cm of the core. For each sediment slice, we mixed ~15 g of sediment with 15 ml of saturated phosphate buffer (Na2HPO4; 0.12 M, pH ≈ 8) during 15 min. The mixture was then centrifuged (10 min at 10,000g). Twelve milliliters of the resulting supernatant was transferred to Amicon Ultra-15 10K Centrifugal Filter Devices (Millipore) and centrifuged (20 min at 4000g) to concentrate sedDNA. Four hundred microliters of the resulting concentrate was kept as starting material for DNA extraction using the NucleoSpin Soil Kit (Macherey-Nagel) (45). Four extraction controls were performed. Mammal and vascular plant sedDNA was amplified with the primer pairs P007 and g-h, respectively (15, 46). These primers have a very limited mismatch with sequences of mammals and plants living in Kerguelen (table S5). In addition to extraction controls, we also amplified two PCR controls containing PCR mix but no DNA template, and two PCR positive controls, each containing a mix of 0.18 ng of DNA extracted from taxa absent both in Europe and Kerguelen [the marsupial Dideplhis marsupialis and three tropical plants: Passiflora edulis (Passifloraceae), Hylocereus undatus (Cactaceae), and Carica papaya (Caricaceae)] (17). All samples and controls underwent eight PCR replicates (33). Sequencing was performed by 2 × 125–base pair pair-end sequencing on Illumina HiSeq 2500 platform. DNA sequences were filtered using the OBITools software (47), as described in the study by Pansu et al. (16). Using the ecotag program, mammal sequences were assigned to the relevant taxa by comparing them with a global mammal database, generated from EMBL release 117; plant sequences were assigned using an exhaustive database of the vascular plants found on Kerguelen (13). We only kept sequences with a match >97% with a mammal or a plant genus. Vertical migration (leaching) of sedDNA has not been observed in lake sediments (48), suggesting that it allows accurate temporal reconstruction.
In ancient DNA studies, the pattern of DNA postmortem damage has often been used to authenticate DNA molecules (17). This approach is particularly efficient with metagenomics, but it is not suitable with metabarcoding studies; thus, we used extensive controls and replicated analyses to ensure the quality of the sedDNA results (17, 49). We removed all sequences with <100 reads over the whole sample and all sequences detected in only one PCR replicate from all the sediment samples. To remove potential contaminants, we removed all sequences with ≥10 reads in ≥2% of extraction and PCR-negative controls, and a taxon was considered present in a PCR replicate only if the number of reads was greater than that detected in any of the controls (50). Two plant taxa (Agrostis magellanica and the invasive T. officinale) were detected with a large number of reads in one single control out of the 64 analyzed for plants, suggesting sporadic contamination. Because the frequency of these plants in sediment samples was much higher than in controls (28 and 7% of PCR replicates in samples, respectively), we included them in the analyses (17, 49). Repeating the analyses without these additional samples led to identical results (appendix S2). In two sediment samples (dating back to approximately 1771 and 1682) PCR yielded no sequences; therefore, we excluded these samples from the analyses.
For mammal sedDNA, we used site occupancy detection modeling to estimate the detection probability of species, and the frequency of false detections (33) using Bayesian models (28). Estimates of occupancy, detection probability, and probability of false detection were then used to calculate the probability of species presence in a given sample. This approach allows successfully limiting false positives in sedDNA analyses; the probability of presence is a measure of the reliability of species presence in a given sample and may be considered as a measure of abundance of the target DNA (28, 33, 51). For Kerguelen plant communities, the relative abundance of sedDNA in a given sample (measured as the proportion of reads) is strongly and linearly related with plant cover estimated using traditional methods (13). For each species, we thus calculated the relative abundance of sedDNA in each sample as the proportion of reads assigned to that taxon. However, it should be remarked that the relationship between plant cover and sedDNA abundance might not hold in other systems, for instance, if the number of mismatches in the priming region is highly variable among species. Furthermore, the efficiency of DNA (and partly, spores of coprophilous fungi) transport to the lake may be affected by several factors beyond species abundance, such as the proximity of DNA sources (for example, plants, rabbit latrines) to the lake or to streams that transport sediments to the lake, and/or by the processes determining DNA transport (for example, erosion, grazing; see Discussion). Measuring relative sedDNA abundance was not possible with mammals, as rabbits were the only detected species.
Spores and NPP analysis
Samples from the sediment slices analyzed for sedDNA were also processed following standard methods in palynology using HCl, KOH baths (52). Four Lycopodium clavatum tablets were added to each sample to enable quantitative analysis of pollen and spore concentrations and influx. After treatment, the residue was suspended in glycerol, mounted onto microscope slides, and counted using a Leica DM 1000 microscope. Pollen grains and spores were identified using previous works performed in Kerguelen and South Georgia (53, 54), as well as reference microscopic slides. Among the NPPs, only two strict coprophilous fungal spore types (cells of Podospora sp./HdV-368 and Sporormiella sp./HdV-113) were counted following the study by Etienne and Jouffroy-Bapicot (55). Sporormiella has a very limited dispersal and provides robust quantitative measures of mammal abundance at fine spatial scale (25, 27). Sporormiella influx was expressed as accumulation rates (N cm−2 year−1). We also repeated the analyses by expressing Sporormiella as concentration (N cm−3) and obtained identical results (appendix S2). One has to mention the occurrence of complete multicellular ascospores of Sporormiella (large spore-bearing structure) in three samples (Fig. 2 and table S1). In this area, the pollen assemblages are not the best marker to reconstruct past vegetation changes, as it was demonstrated in previous works undertaken in sub-Antarctic territories (56); therefore, they were not analyzed here.
Statistical analyses
For plant communities, we used a series of multivariate analyses to describe variation of plant communities through time and to test the significance of rabbit abundance as a potential driver of community changes. We first used a time-constrained MRT to identify the existence of breakpoints in the plant community time series (57). In MRT, we built 1000 cross-validated trees. To obtain a parsimonious solution, we selected the most parsimonious tree with cross-validation performance within one standard error from the tree with the lowest cross-validation performance (58). Subsequently, we used the analysis of similarity (ANOSIM) to test whether plant communities before and after the breakpoint(s) detected by MRT were significantly different. We also used nonmetric multidimensional scaling to visualize differences in plant communities through time (58). We then assessed whether shifts in plant communities are significantly related to rabbit occurrence using RDA (58). In RDA, the relative abundance of plant communities was the independent matrix. As predictor, we used a matrix composed by two proxies of rabbit presence: accumulation rate of Sporormiella spores and detection of rabbit sedDNA. Plant abundance data of the sample corresponding to 1941–1948 were outliers (Figs. 3 and 4 and table S1), as in this sample almost only the DNA of A. selago was detected, and this can be explained by a different process determining plant DNA transport (rabbit feces; see Discussion). Therefore, the 1941–1948 sample was excluded from the RDA analyses, but we obtained identical results when we repeated the analyses including this sample (fig. S2). Temporal autocorrelation of the residuals was not significant (P > 0.05 for all the species). Significance of ANOSIM and RDA was tested through 30,000 permutations using the vegan package in R (www.r-project.org).
For analysis of erosion rate, we first used a dynamic programming algorithm to detect and date the occurrence of breakpoints in the erosion time series (59). The best number of breakpoints was assessed using Bayesian information criterion. Subsequently, we used generalized least squares to assess the relationship between rabbit abundance (measured as the abundance of Sporormiella spores) and erosion rate, including a first-order autocorrelation term to take into account potential temporal autocorrelation issues. Results were identical if rabbit sedDNA was used as a proxy of rabbit presence, instead of the Sporormiella influx (appendix S2).
Climate change is an alternative mechanism that might have major impacts on communities and erosion. To test the potential role of climate change, we obtained data on mean precipitation and temperature from the 2014 version of the University of Delaware time series of monthly global gridded high-resolution station data (60). First, we calculated mean monthly precipitation and mean annual temperature for the 0.5° grid cell containing the La Poule Lake; because of missing data, we focused on the period 1912–2013. We then repeated RDA and generalized least squares analyses for this period, including both rabbit occurrence and climate as explanatory variables.
Supplementary Material
Acknowledgments
We thank the Laboratoire Souterrain de Modane facilities for the gamma spectrometry measurements and the Environnement Dynamique et Territoires de Montagne for the x-ray fluorescence analyses. Two reviewers provided useful comments on an early version of the manuscript. Funding: This study was partially funded by the ESKER project thanks to the Dispositif de Partenariat en Ecologie et Environnement Grenoble-Chambery. UDel_AirT_Precip data were provided by the NOAA/OAR/ESRL PSD (Boulder, CO) from their web site at www.esrl.noaa.gov/psd/. We are grateful to IPEV, the French Polar Institute, for providing necessary logistical and financial support for field expedition (programme 1094-PALAS). Author contributions: G.F.F., J.P., P.S., P.T., and F.A. conceived the study. J.P., F.A., J.B., and E.S. performed the fieldwork. G.F.F., J.P., P.S., E.M., L.G., A.L., D.E., E.M., B.F., J.-L.R., P.T., and F.A. performed the analyses. G.F.F. wrote the first draft of the manuscript, with subsequent contribution by all the authors. Competing interests: P.T. and L.G. are inventors on a U.S. patent application related to this work (application no. 20110143354). All the other authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/5/eaar4292/DC1
appendix S1. The Kerguelen Islands: Historical information on rabbit introduction.
appendix S2. Supplementary results.
appendix S3. Amplification of plant and mammal taxa using the P007 and g-h primers.
table S1. Detection of NPPs in the analyzed sediment samples.
table S2. Detection of sedDNA in the analyzed sediment samples.
table S3. Estimation of breakpoints in the erosion rate performed using a dynamic programming algorithm (59).
table S4. Average climatic variables during the period 1912–2013, calculated on the basis of the 2014 version of the University of Delaware time series of monthly global gridded high-resolution station data (60).
table S5. Match between primers and sequences from plants and mammals living in Kerguelen.
fig. S1. Data from core POU14P1 with photograph, grain size median, LOI550 (organic matter), and NCIR.
fig. S2. Age-depth model for the upper 65 cm of the sediment core POU14P1 based on radiocarbon (blue) and short-lived radionuclides derived ages (green).
fig. S3. Constrained RDA, showing the relationship between the proxies of rabbit abundance (sedDNA and Sporormiella) and the abundance of different plant taxa.
fig. S4. Temporal variation of the accumulation rate of spores of coprophilous fungi (Sporormiella sp.), measured as concentration (spores/cm3).
REFERENCES AND NOTES
- 1.Pejchar L., Mooney H. A., Invasive species, ecosystem services and human well-being. Trends Ecol. Evol. 24, 497–504 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Vilà M., Espinar J. L., Hejda M., Hulme P. E., Jarošík V., Maron J. L., Pergl J., Schaffner U., Sun Y., Pyšek P., Ecological impacts of invasive alien plants: A meta-analysis of their effects on species, communities and ecosystems. Ecol. Lett. 14, 702–708 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Helmus M. R., Mahler D. L., Losos J. B., Island biogeography of the Anthropocene. Nature 513, 543–546 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Reaser J. K., Meyerson L. A., Cronk Q., De Poorter M., Eldrege L. G., Green E., Kairo M., Latasi P., Mack R. N., Mauremootoo J., O’Dowd D., Orapa W., Sastroutomo S., Saunders A., Shine C., Thrainsson S., Vaiutu L., Ecological and socioeconomic impacts of invasive alien species in island ecosystems. Environ. Conserv. 34, 98–111 (2007). [Google Scholar]
- 5.Shine R., The ecological impact of invasive cane toads (Bufo marinus) in Australia. Q. Rev. Biol. 85, 253–291 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Aikio S., Duncan R. P., Hulme P. E., Lag-phases in alien plant invasions: Separating the facts from the artefacts. Oikos 119, 370–378 (2010). [Google Scholar]
- 7.Essl F., Dullinger S., Rabitsch W., Hulme P. E., Hülber K., Jarošík V., Kleinbauer I., Krausmann F., Kühn I., Nentwig W., Vilà M., Genovesi P., Gherardi F., Desprez-Loustau M.-L., Roques A., Pyšek P., Socioeconomic legacy yields an invasion debt. Proc. Natl. Acad. Sci. U.S.A. 108, 203–207 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strayer D. L., Eight questions about invasions and ecosystem functioning. Ecol. Lett. 15, 1199–1210 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Strayer D. L., Evinver V. T., Jeschke J. M., Pace M. L., Understanding the long-term effects of species invasions. Trends Ecol. Evol. 21, 645–651 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Delibes R., Delibes-Mateos M., Linking historical ecology and invasion biology: Some lessons from European rabbit introductions into the new world before the nineteenth century. Biol. Invasions 17, 2505–2515 (2015). [Google Scholar]
- 11.H. V. Thompson, C. M. King, Eds., The European Rabbit, The History and Biology of a Successful Colonizer (Oxford Univ. Press, 1994). [Google Scholar]
- 12.Chapuis J.-L., Frenot Y., Lebouvier M., Recovery of native plant communities after eradication of rabbits from the subantarctic Kerguelen Islands, and influence of climate change. Biol. Conserv. 117, 167–179 (2004). [Google Scholar]
- 13.Pansu J., Winkworth R. C., Hennion F., Gielly L., Taberlet P., Choler P., Long-lasting modification of soil fungal diversity associated with the introduction of rabbits to a remote sub-Antarctic archipelago. Biol. Lett. 11, 20150408 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sabatier P., Poulenard J., Fanget B., Reyss J.-L., Develle A.-L., Wilhelm B., Ployon E., Pignol C., Naffrechoux E., Dorioz J.-M., Montuelle B., Arnaud F., Long-term relationships among pesticide applications, mobility, and soil erosion in a vineyard watershed. Proc. Natl. Acad. Sci. U.S.A. 111, 15647–15652 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giguet-Covex C., Pansu J., Arnaud F., Rey P.-J., Griggo C., Gielly L., Domaizon I., Coissac E., David F., Choler P., Poulenard J., Taberlet P., Long livestock farming history and human landscape shaping revealed by lake sediment DNA. Nat. Commun. 5, 3211 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Pansu J., Giguet-Covex C., Ficetola G. F., Gielly L., Boyer F., Zinger L., Arnaud F., Poulenard J., Taberlet P., Choler P., Reconstructing long-term human impacts on plant communities: An ecological approach based on lake sediment DNA. Mol. Ecol. 24, 1485–1498 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Parducci L., Bennett K. D., Ficetola G. F., Alsos I. G., Suyama Y., Wood J. R., Pedersen M. W., Ancient plant DNA in lake sediments. New Phytol. 214, 924–942 (2017). [DOI] [PubMed] [Google Scholar]
- 18.E. Golberg, Geochronology with 210Pb, in Radioactive Dating (International Atomic Energy Agency, 1963), pp. 121–131. [Google Scholar]
- 19.Krishnaswamy S., Lal D., Martin J. M., Meybeck M., Geochronology of lake sediments. Earth Planet. Sci. Lett. 11, 407–414 (1971). [Google Scholar]
- 20.Graham I. J., Ditchburn R. G., Barry B. J., 210Pb–137Cs dating of glacial lake sediments. New Zealand Sci. Rev. 61, 45–47 (2004). [Google Scholar]
- 21.Pfitzner J., Brunskill G., Zagorskis I., 137Cs and excess 210Pb deposition patterns in estuarine and marine sediment in the central region of the Great Barrier Reef Lagoon, north-eastern Australia. J. Environ. Radioact. 76, 81–102 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Arnaud F., Magand O., Chapron E., Bertrand S., Boës X., Charlet F., Mélières M.-A., Radionuclide dating (210Pb, 137Cs, 241Am) of recent lake sediments in a highly active geodynamic setting (lakes Puyehue and Icalma—Chilean Lake District). Sci. Total Environ. 366, 837–850 (2006). [Google Scholar]
- 23.Blaauw M., Methods and code for ‘classical’ age-modelling of radiocarbon sequences. Quat. Geochronol. 5, 512–518 (2010). [Google Scholar]
- 24.Etienne D., Wilhelm B., Sabatier P., Reyss J.-L., Arnaud F., Influence of sample location and livestock numbers on Sporormiella concentrations and accumulation rates in surface sediments of Lake Allos, French Alps. J. Paleolimnol. 49, 117–127 (2013). [Google Scholar]
- 25.Baker A. G., Cornelissen P., Bhagwat S. A., Vera F. W. M., Willis K. J., Quantification of population sizes of large herbivores and their long-term functional role in ecosystems using dung fungal spores. Methods Ecol. Evol. 7, 1273–1281 (2016). [Google Scholar]
- 26.Doyen E., Etienne D., Ecological and human land-use indicator value of fungal spore morphotypes and assemblages. Veg. Hist. Archaeobot. 26, 357–367 (2017). [Google Scholar]
- 27.Gill J. L., McLauchlan K. K., Skibbe A. M., Goring S., Zirbel C. R., Williams J. W., Linking abundances of the dung fungus Sporormiella to the density of bison: Implications for assessing grazing by megaherbivores in palaeorecords. J. Ecol. 101, 1125–1136 (2013). [Google Scholar]
- 28.Lahoz-Monfort J. J., Guillera-Arroita G., Tingley R., Statistical approaches to account for false positive errors in environmental DNA samples. Mol. Ecol. Resour. 16, 673–685 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Bajard M., Sabatier P., David F., Develle A.-L., Reyss J.-L., Fanget B., Malet E., Arnaud D., Augustin L., Crouzet C., Poulenard J., Arnaud F., Erosion record in Lake La Thuile sediments (Prealps, France): Evidence of montane landscape dynamics throughout the Holocene. Holocene 26, 350–364 (2016). [Google Scholar]
- 30.Lésel L., Contribution à l’étude écologique de quelques mammifères importés aux îles Kerguelen. Terres Australes et Antarctiques Françaises 38, 3–40 (1967). [Google Scholar]
- 31.de La Rue A., Observations sur les caractères et la répartition de la végétation des lies Kerguelen. CNFRA 1, 1–60 (1964). [Google Scholar]
- 32.Chapuis J. L., Boussès P., Barnaud G., Alien mammals, impact and management in the French subantarctic islands. Biol. Conserv. 67, 97–104 (1994). [Google Scholar]
- 33.Ficetola G. F., Pansu J., Bonin A., Coissac E., Giguet-Covex C., De Barba M., Gielly L., Lopes C. M., Boyer F., Pompanon F., Rayé G., Taberlet P., Replication levels, false presences, and the estimation of the presence/absence from eDNA metabarcoding data. Mol. Ecol. Resour. 15, 543–556 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Boessenkool S., McGlynn G., Epp L. S., Taylor D., Pimentel M., Gizaw A., Nemomissa S., Brochmann C., Popp M., Use of ancient sedimentary DNA as a novel conservation tool for high-altitude tropical biodiversity. Conserv. Biol. 28, 446–455 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Levy-Booth D. J., Campbell R. G., Gulden R. H., Hart M. M., Powell J. R., Klironomos J. N., Pauls K. P., Swanton C. J., Trevors J. T., Dunfield K. E., Cycling of extracellular DNA in the soil environment. Soil Biol. Biochem. 39, 2977–2991 (2007). [Google Scholar]
- 36.Frenot Y., Gloaguen J. C., Massé L., Lebouvier M., Human activities, ecosystem disturbance and plant invasions in subantarctic Crozet, Kerguelen and Amsterdam Islands. Biol. Conserv. 101, 33–50 (2001). [Google Scholar]
- 37.M. R. Clark, P. R. Dingwall, Conservation of Islands in The Southern Ocean: A Review of the Protected Areas of Insulantarctica (International Union for Conservation of Nature, 1985), pp. 188. [Google Scholar]
- 38.Hugo E. A., McGeoch M. A., Marshall D. J., Chown S. L., Fine scale variation in microarthropod communities inhabiting the keystone species Azorella selago on Marion Island. Polar Biol. 27, 466–473 (2004). [Google Scholar]
- 39.Cerfonteyn M. E., Le Roux P. C., Van Vuuren B. J., Born C., Cryptic spatial aggregation of the cushion plant Azorella selago (Apiaceae) revealed by a multilocus molecular approach suggests frequent intraspecific facilitation under sub-Antarctic conditions. Am. J. Bot. 98, 909–914 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Jones H. P., Holmes N. D., Butchart S. H. M., Tershy B. R., Kappes P. J., Corkery I., Aguirre-Muñoz A., Armstrong D. P., Bonnaud E., Burbidge A. A., Campbell K., Courchamp F., Cowan P. E., Cuthbert R. J., Ebbert S., Genovesi P., Howald G. R., Keitt B. S., Kress S. W., Miskelly C. M., Oppel S., Poncet S., Rauzon M. J., Rocamora G., Russell J. C., Samaniego-Herrera A., Seddon P. J., Spatz D. R., Towns D. R., Croll D. A., Invasive mammal eradication on islands results in substantial conservation gains. Proc. Natl. Acad. Sci. U.S.A. 113, 4033–4038 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilhelm B., Sabatier P., Arnaud F., Is a regional flood signal reproducible from lake sediments? Sedimentology 62, 1103–1117 (2015). [Google Scholar]
- 42.Heiri O., Lotter A. F., Lemcke G., Loss on ignition as a method for estimating organic and carbonate content in sediments: Reproducibility and comparability of results. J. Paleolimnol. 25, 101–110 (2001). [Google Scholar]
- 43.Reyss J.-L., Schmidt S., Legeleux F., Bonté P., Large low background well-type detectors for measurements of environmental radioactivity. Nucl. Instrum. Methods Phys. Res. A 357, 391–397 (1995). [Google Scholar]
- 44.McCormac F. G., Hogg A. G., Blackwell P. G., Buck C. E., Higham T. F. G., Reimer P. J., SHCal04 southern hemisphere calibration, 0-11.0 cal kyr BP. Radiocarbon 46, 1087–1092 (2004). [Google Scholar]
- 45.Taberlet P., Prud’Homme S. M., Campione E., Roy J., Miquel C., Shehzad W., Gielly L., Rioux D., Choler P., Clément J.-C., Melodelima C., Pompanon F., Coissac E., Soil sampling and isolation of extracellular DNA from large amount of starting material suitable for metabarcoding studies. Mol. Ecol. 21, 1816–1820 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Taberlet P., Coissac E., Pompanon F., Gielly L., Miquel C., Valentini A., Vermat T., Corthier G., Brochmann C., Willerslev E., Power and limitations of the chloroplast trn L (UAA) intron for plant DNA barcoding. Nucleic Acids Res. 35, e14 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boyer F., Mercier C., Bonin A., Le Bras Y., Taberlet P., Coissac E., obitools: A unix-inspired software package for DNA metabarcoding. Mol. Ecol. Resour. 16, 176–182 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Sjögren P., Edwards M. E., Gielly L., Langdon C. T., Croudace I. W., Merkel M. K. F., Fonville T., Alsos I. G., Lake sedimentary DNA accurately records 20th century introductions of exotic conifers in Scotland. New Phytol. 213, 929–941 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Champlot S., Berthelot C., Pruvost M., Bennett E. A., Grange T., Geigl E.-M., An efficient multistrategy DNA decontamination procedure of PCR reagents for hypersensitive PCR applications. PLOS ONE 5, e13042 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Barba M., Miquel C., Boyer F., Mercier C., Rioux D., Coissac E., Taberlet P., DNA metabarcoding multiplexing and validation of data accuracy for diet assessment: Application to omnivorous diet. Mol. Ecol. Resour. 14, 306–323 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Furlan E. M., Gleeson D., Hardy C. M., Duncan R. P., A framework for estimating the sensitivity of eDNA surveys. Mol. Ecol. Resour. 16, 641–654 (2016). [DOI] [PubMed] [Google Scholar]
- 52.K. Fægri, J. Iversen, Textbook of Pollen Analysis (Wiley, 1989). [Google Scholar]
- 53.Barrow C. J., Palynological studies in South Georgia: 1. Pollen and spore morphology of the native vascular species. Brit. Antarct. Surv. Bull. 43, 63–75 (1976). [Google Scholar]
- 54.N. Bellair, Sédimentologie et palynologie d’une tourbière des Kerguelen (Diplôme d’études supérieures en sciences naturelles, Faculté des sciences de l’Université de Paris, 1967). [Google Scholar]
- 55.Etienne D., Jouffroy-Bapicot I., Optimal counting limit for fungal spore abundance estimation using Sporormiella as a case study. Veg. Hist. Archaeobot. 23, 743–749 (2014). [Google Scholar]
- 56.Van der Putten N., Verbruggen C., Björck S., de Beaulieu J.-L., Barrow C. J., Frenot Y., Is palynology a credible climate proxy in the Subantarctic? Holocene 22, 1113–1121 (2012). [Google Scholar]
- 57.Legendre P., Gauthier O., Statistical methods for temporal and space–time analysis of community composition data. Proc. Biol. Sci. 281, 20132728 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.P. Legendre, L. Legendre, Numerical Ecology (Elsevier, 2012), pp. 1006. [Google Scholar]
- 59.Zeileis A., Kleiber C., Krämer W., Hornik K., Testing and dating of structural changes in practice. Comput. Stat. Data Anal. 44, 109–123 (2003). [Google Scholar]
- 60.Nickl E., Willmott C. J., Matsuura K., Robeson S. M., Changes in annual land-surface precipitation over the twentieth and early twenty-first century. Ann. Assoc. Am. Geogr. 100, 729–739 (2010). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/5/eaar4292/DC1
appendix S1. The Kerguelen Islands: Historical information on rabbit introduction.
appendix S2. Supplementary results.
appendix S3. Amplification of plant and mammal taxa using the P007 and g-h primers.
table S1. Detection of NPPs in the analyzed sediment samples.
table S2. Detection of sedDNA in the analyzed sediment samples.
table S3. Estimation of breakpoints in the erosion rate performed using a dynamic programming algorithm (59).
table S4. Average climatic variables during the period 1912–2013, calculated on the basis of the 2014 version of the University of Delaware time series of monthly global gridded high-resolution station data (60).
table S5. Match between primers and sequences from plants and mammals living in Kerguelen.
fig. S1. Data from core POU14P1 with photograph, grain size median, LOI550 (organic matter), and NCIR.
fig. S2. Age-depth model for the upper 65 cm of the sediment core POU14P1 based on radiocarbon (blue) and short-lived radionuclides derived ages (green).
fig. S3. Constrained RDA, showing the relationship between the proxies of rabbit abundance (sedDNA and Sporormiella) and the abundance of different plant taxa.
fig. S4. Temporal variation of the accumulation rate of spores of coprophilous fungi (Sporormiella sp.), measured as concentration (spores/cm3).


