Abstract
Purpose
Robust preclinical models of pediatric acute lymphoblastic leukemia (ALL) are essential in prioritizing promising therapies for clinical assessment in high-risk patients. Patient-derived xenograft (PDX) models of ALL provide a clinically relevant platform for assessing novel drugs, with efficacy generally assessed by enumerating circulating human lymphoblasts in mouse peripheral blood (PB) as an indicator of disease burden. While allowing indirect measurement of disease burden in real-time, this technique cannot assess treatment effects on internal reservoirs of disease. We explore benefits of bioluminescence imaging (BLI) to evaluate drug responses in ALL PDXs, compared to PB monitoring. BLI-based thresholds of drug response are also explored.
Experimental Design
ALL PDXs were lentivirally transduced to stably express luciferase and green fluorescent protein. In vivo PDX responses to an induction-type regimen of vincristine, dexamethasone and L-asparaginase were assessed by BLI and PB. Residual disease at Day 28 post-treatment initiation was assessed by flow cytometric analysis of major organs. BLI and PB were subsequently used to evaluate efficacy of the Bcl-2 inhibitor venetoclax.
Results
BLI considerably accelerated and enhanced detection of leukemia burden compared to PB and identified sites of residual disease during treatment in a quantitative manner, highlighting limitations in current PB-based scoring criteria. Using BLI alongside enumeration of human lymphoblasts in PB and bone marrow, we were able to redefine response criteria analogous to the clinical setting.
Conclusions
BLI substantially improves the stringency of preclinical drug testing in pediatric ALL PDXs, which will likely be important in prioritizing effective agents for clinical assessment.
Keywords: Leukemia, pediatric, bioluminescence, preclinical, venetoclax
Introduction
Continuing improvements to treatment strategies for childhood ALL have led to 5-year survival rates of approximately 90% (1). However, 10–20% of patients will relapse from or prove refractory to current treatments, with the majority succumbing to their disease (2–5). Large scale genomic profiling of childhood ALL cases has provided important insights into genetic lesions implicated in high-risk ALL, proffering novel targeting strategies for this disease (6–8). However despite ongoing identification of targetable aberrations within pediatric ALL, and the availability of a large number of potentially efficacious drugs developed for adult malignancies, the rarity of cases representing distinct ‘high-risk’ pediatric ALL subtypes restricts patient availability for clinical assessment of novel therapies.
Using immune-deficient mice as hosts for ALL patient-derived xenografts (PDXs) has the potential to accurately predict clinical efficacy of investigational drugs to prioritize them for clinical assessment. These PDXs accurately recapitulate fundamental characteristics of the original disease including immunophenotype, chromosomal translocations, gene mutations, gene expression profiles, DNA methylation patterns and drug sensitivity (9–13). Pediatric ALL PDXs used as part of the NCI-funded Pediatric Preclinical Testing Program (PPTP) have shown concordance between preclinical results and corresponding available clinical data for established and novel drugs (14).
Treatment response in the ALL PDX model is routinely assessed by flow cytometric enumeration of the proportion of circulating human lymphocytes (huCD45+ or huCD19+) in mouse peripheral blood (PB). However, disease monitoring in these models may benefit significantly from in vivo imaging techniques. Multiple groups have successfully introduced a bioluminescence imaging (BLI) capability into patient-derived ALL cells, using protocols including cell sorting and in vivo expansion of lentivirally transduced cells (15–17). These studies reported that BLI revealed engraftment considerably earlier than PB monitoring, suggesting migration of leukemic blasts to PB occurs relatively late during disease development. Drug efficacy experiments have highlighted enhanced sensitivity of BLI in measuring treatment response compared to PB monitoring, suggesting ablation of huCD45+ cells from PB may not equate to elimination of residual leukemia within sanctuary sites such as bone marrow (BM) (15, 16, 18, 19).
The sensitivity of BLI, alongside its ability to longitudinally monitor individual mice, may also provide a preclinical measure of minimal residual disease (MRD), a critical prognostic indicator in pediatric ALL (4, 20). MRD is assessed clinically by determining the percentage of blasts within a BM aspirate, by either flow cytometry or qPCR-based approaches (21–23), at multiple stages within the first 4–6 weeks of treatment (24). Equivalent BM sampling from mice, while possible (25), is technically challenging and may compromise the integrity of the bone sampled. Moreover, time-point harvesting of mice is ethically challenging and resource draining, and assumes that individuals represent the entire treatment group (26). BLI-based measurements of MRD may provide an alternative to these methods, facilitating real-time monitoring of residual disease in discrete anatomical sites (15).
Despite the potential benefits, significant limitations exist in establishing luciferase-expressing sub-lines of ALL PDXs. These include the inefficiency of transduction of continuous xenograft and primary cells, multiple rounds of expansion of cell numbers by passaging in mice, and sorting of transduced cells. Moreover, some ALL PDXs are refractory to lentiviral transduction, limiting the number and choice of samples of specific subtypes for testing targeted agents. Therefore, a detailed comparison of BLI and PB-based assessment of leukemic burden is warranted, to develop BLI-based criteria of response and to utilize BLI to inform improvements to existing PB-based criteria.
The present study used representative BLI-enabled ALL PDXs to evaluate potential benefits of BLI in enhancing the sensitivity of disease detection compared to PB monitoring. To assess drug efficacy over a range of responses, titrated doses of an established, induction-type drug regimen of vincristine, dexamethasone and L-asparaginase (VXL) were used (27). Responses were monitored via PB and BLI in parallel and subsequently scored using clinically relevant PPTP criteria (28). MRD was assessed 28 days post-treatment initiation to determine the ability of BLI to reflect flow cytometry results, and consequently the potential of BLI to measure MRD. Results from VXL treatments were used to inform testing of the Bcl-2 inhibitor venetoclax. Both venetoclax and its predecessor navitoclax have shown promise against chronic lymphocytic leukemia (CLL) (29), and although positive results have been reported against ALL PDXs (30), venetoclax is yet to be evaluated in the clinic against pediatric ALL.
Overall this study aimed to compare BLI and PB monitoring in preclinical drug testing for pediatric ALL. In doing so we were able to highlight the additional sensitivity of BLI in measuring drug response compared to PB monitoring, while using BLI to allow development of parameters to increase the stringency of testing in the existing ALL PDX model to ensure that only agents with the greatest likelihood of success will be advanced to clinical trials.
METHODS
Development of BLI-enabled PDXs
All experimental work was performed with approvals from the Human Research Ethics Committee and the Animal Care and Ethics Committee of UNSW Australia (Sydney, Australia). Experiments used continuous PDXs established previously in 20–25 g female non-obese diabetic/severe combined immuno-deficient (NOD.CB17-Prkdcscid/SzJ, NOD/SCID) or NOD/SCID/interleukin-2 receptor γ–negative (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, NSG) mice, as described elsewhere (31, 32). PDXs were propagated in mice through collection of human leukemia cells from spleen or BM of engrafted mice and re-inoculation into secondary recipient mice. The PDX lentiviral transduction protocol has been described elsewhere (33). ALL-11 and ALL-57 B-cell precursor ALL cells were transduced at a multiplicity of infection of 30. Following transduction for 24 h in virus containing medium cells were washed twice with PBS and inoculated into NSG mice for expansion. At high levels of leukemia burden, cells were harvested from spleens of engrafted mice and sorted to >99% purity based on GFP expression (FACSJazz or BD Influx, BD Biosciences, San Jose, CA). Pure GFP+ (>99%+) cell populations were then inoculated into secondary recipient NSG mice for expansion. The spleen-derived cells from this second expansion, named ALL-11-GL and ALL-57-GL denoting GFP and luciferase expression, were then used in all experiments described in this study.
PDX samples used were at tertiary passage for the initial transduction, therefore due to the additional expansion steps described, the samples used herein have undergone 5 in vivo passages at the beginning of the experiments described. Transduced cells were then validated against parental PDXs using single nucleotide polymorphism (SNP) validation as described elsewhere (34).
Bioluminescence imaging
Imaging data were acquired using the IVIS® SpectrumCT Preclinical In Vivo Imaging System (PerkinElmer). Groups of 2–5 mice at a time were administered D-luciferin (150 mg/kg) via intraperitoneal (IP) injection and anesthetized with isoflurane (2.5–3% induction, 1–1.5% maintenance). Previous experiments indicated peak photon emission at between 8–12 mins post-luciferin injection, therefore all images were acquired within this timeframe. Data were collected between 600–60,000 counts (photons/second, p/s) as per manufacturer’s instructions, then converted to photons/second/cm2/steradian (p/s/cm2/sr) to normalize variable parameters (exposure time, f-stop, binning) used between groups and weeks. Signal intensity was determined by drawing regions of interest (ROIs) around the whole animal, as well as areas encompassing the hind-legs (representing femoral/tibial BM), spleen, liver, spine and brain.
Assessment of in vivo drug efficacy
All in vivo treatment groups (6–10 mice/group) were monitored using PB sampling, as described elsewhere (31), as well as BLI, measured on the same day to allow comparison. To standardize bioluminescence measurements between PDXs, treatment response was measured by log10 fold change, calculated using the ratio of values from the group median of the lowest BLI output in response to treatment (p/s/cm2/sr) or signal at the end of the treatment period divided by the cohort’s median Day 0 value. Treatment initiation, endpoint and overall response were measured using PPTP criteria (28). Briefly, treatment began when the median of the groups reached >1% huCD45+ cells in PB, with event considered as 25% huCD45+ cells in PB. Response to treatment was evaluated using two methods: (1) leukemia growth delay (LGD), representing the difference between the median event-free survival (EFS) of the drug-treated cohort and that of the vehicle-treated control cohort (T-C); and, (2) objective response measures (ORMs) developed by the PPTP to mimic stringent clinical criteria, assessed at Day 42 post-treatment initiation as described elsewhere (28) and in Supplementary Fig. S1. For VXL treatments all drugs were purchased from Clifford Hallam Healthcare (NSW, Australia) and administered by IP injection. Dexamethasone (X, 5 mg/kg) and L-asparaginase (L, 1000 U/kg) were administered once daily, 5 days on, 2 days off, with the addition of vincristine (V, 0.15 mg/kg) on the first day of treatment each week. VXL treatments were for 2 or 4 weeks, and at 1x, 0.5x, 0.25x and 0.125x the above doses. Venetoclax (obtained from AbbVie under a standard Material Transfer Agreement) was administered daily for 21 days via oral gavage at 100 mg/kg.
Statistical analysis
The exact log-rank test, implemented using Proc StatXact for SAS®, was used to compare event-free survival distributions between treatment and control groups (two-tailed; P ≤ 0.05 considered significant). Correlations were analyzed using Pearson correlation analysis. Group analysis with >2 groups and a normal distribution were analyzed using a one-way ANOVA, while multiple comparisons applied Tukey’s multiple comparisons test.
RESULTS
BLI augments assessment of drug response by PB monitoring
To comprehensively compare PB and BLI in measuring leukemia burden in response to chemotherapy, a treatment strategy to achieve a spectrum of responses was chosen, allowing comparison to response categories established by the PPTP (Supplementary Fig. S1) (28). Consequently, the VXL combination was used against two BLI-enabled BCP-ALL PDXs, ALL-11-GL and ALL-57-GL, and administered at the dose established previously (27) (1x), as well as attenuated doses of half (0.5x), a quarter (0.25x) and an eighth (0.125x) of the full dose for 2 weeks, with the exception of the ALL-11-GL 1x group, which was for 4 weeks.
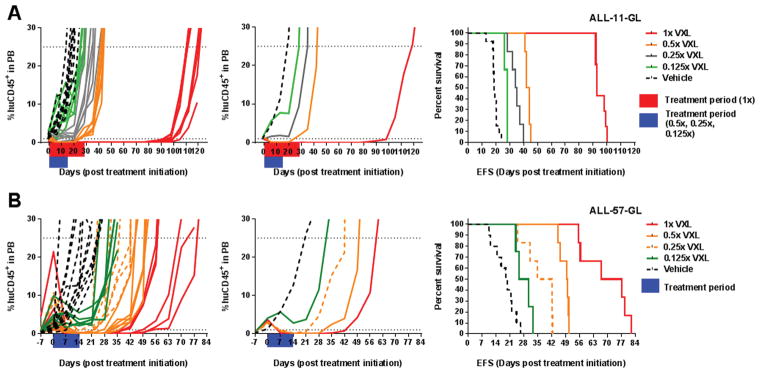
The titrated VXL doses successfully separated the responses of treatment groups in both PDXs. For ALL-11-GL, the 1x, 0.5x, 0.25x and 0.125x doses elicited LGDs of 75, 25.4, 16.6 and 10.2 days, respectively (Fig. 1A and Supplementary Table S1). Similarly, in ALL-57-GL the 1x, 0.5x, 0.25x and 0.125x doses achieved LGDs of 47.7, 30.5, 19.2 and 8.8 days, respectively (Fig. 1B and Supplementary Table S1). When scored using PPTP criteria, the 1x and 0.5x groups achieved objective responses against ALL-11-GL, scoring a maintained complete response (MCR) and a complete response (CR) respectively (Fig. 1A and Supplementary Table S1). Against ALL-57-GL, the 1x group achieved an MCR, while the 0.5x and 0.25x groups scored CRs (Fig. 1B and Supplementary Table S1).
Figure 1. In vivo efficacy of titrated VXL doses against ALL-11-GL and ALL-57-GL as measured by the percentage of human CD45+ cells in mouse PB.
The responses of ALL-11-GL (A) and ALL-57-GL (B) to the combination treatment VXL at a range of doses; 1x, 0.5x, 0.25x and 0.125x the doses of vincristine (0.15 mg/kg), dexamethasone (5 mg/kg) and L-asparaginase (1000 U/kg). Treatment periods are shown in red and blue on the individual graphs. Cohorts are labeled on the legend for each graph set and are colored to match the ORM score according to PPTP criteria. Response to each treatment is visualized as the %huCD45+ cells in mouse PB for individual mice (left panels), the median values for each cohort (middle panels) and the EFS for individual mice, represented by Kaplan-Meier plots (right panels).
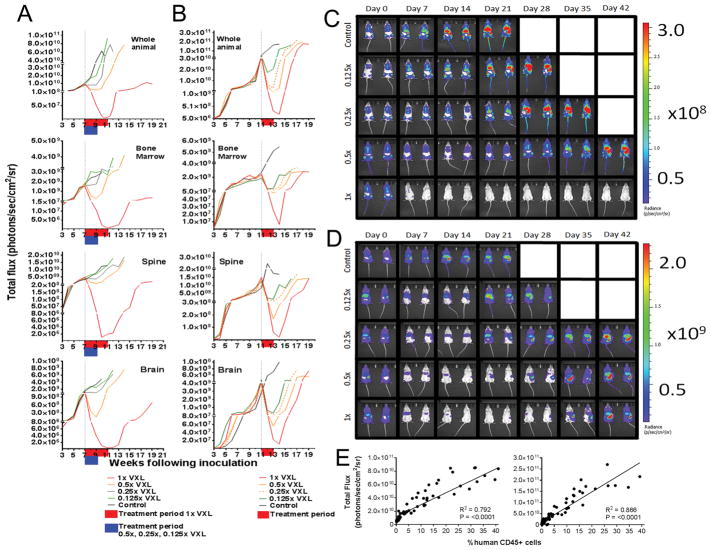
Corresponding whole-animal imaging data showed a similar dose response to PB measurements for all cohorts (Fig. 2A–B). However imaging showed a considerably greater range and sensitivity of detection compared to PB, highlighting persistent disease at time-points where circulating lymphocytes were undetectable in PB (Fig. 1 and Fig. 2A–D). To assess differences in the dynamic range of both techniques, median Day 0 values (%huCD45+ for PB, p/s/cm2/sr for BLI) for the 1x treatment groups for both PDXs were compared to their respective values at the lowest measurable level of leukemia burden. A lower threshold of reliability of 0.1% (10 events in 10,000 acquired) was used for PB values, as below this detection of huCD45+ cells in PB becomes unreliable. ALL-11-GL and ALL-57-GL exhibited a 9-fold and a 36-fold decrease from Day 0 values of 0.9% and 3.6%, respectively, before leukemia became undetectable. In comparison, ALL-11-GL and ALL-57-GL exhibited a 2,260-fold and a 243-fold decrease in bioluminescence, respectively.
Figure 2. In vivo efficacy of titrated VXL doses against ALL-11-GL and ALL-57-GL measured by bioluminescence imaging (BLI).
The responses of ALL-11-GL (A, C) and ALL-57-GL (B, D) to the combination treatment VXL at a range of doses; 1x, 0.5x, 0.25x and 0.125x the doses of vincristine (0.15 mg/kg), dexamethasone (5 mg/kg) and L-asparaginase (1000 U/kg). Treatment began once a median of 1% huCD45+ cells in PB was reached for each cohort. Weekly imaging data representing whole animal and organ-specific bioluminescence are presented graphically as the median bioluminescence intensity for each treatment group (p/s/cm2/sr) (A, B), with treatment periods shown in red and blue on the individual graphs and Day 0 represented by a vertical dotted line. Cohorts are labeled on the legend for each graph set, with the cohorts colored to match the objective response measure (ORM score according to PPTP criteria). Images of representative mice for each cohort are shown at weekly intervals throughout the evaluation period of 42 days, acquired using the Spectrum CT in vivo imaging system (C, D). Images were analyzed using the LivingImage v4.4 software. (E) Where available (>0.1% huCD45+ cells), PB data were plotted against matched whole animal BLI values for ALL-11-GL (left panel) and ALL-57-GL (right panel) and subjected to Pearson correlation. R, Pearson correlation coefficient; P-value, two-tailed P-value of the Pearson correlation.
To standardize BLI values between PDXs, a log10 fold change value was calculated for each treatment group, comparing Day 0 values with the lowest BLI output after drug treatment (Supplementary Table S2). The ALL-11-GL 1x VXL group achieved a log10 fold change of −3.35, while the 0.5x group showed a more marginal decrease of −0.88, despite scoring a CR. The 0.25x and 0.125x groups failed to achieve remission in ALL-11-GL, as observed by PB (Fig. 1A) and BLI (Fig. 2A), and therefore scored a positive log10 fold change (Supplementary Table S2). Leukemia regression was observed for all ALL-57-GL VXL treatment groups, with log10 fold changes of −2.39, −1.57, −0.99 and −0.35 for the 1x, 0.5x, 0.25x and 0.125x groups, respectively (Fig. 2B and Supplementary Table S2).
Quantification of bioluminescence and log10 fold change was also calculated for individual sites of leukemia infiltration (Fig. 2A–B, Supplementary Table S2). A similar dose response to whole animal data was observed for individual organ measurements for both PDXs across regions representative of BM, spine and brain (Supplementary Table S2). Interestingly, BLI data representing spine burden in ALL-11-GL 1x treated mice identified relapse during the last week of treatment while for all other sites relapse occurred post-treatment cessation (Fig. 2A). The ALL-57-GL 0.125x treatment group showed an increase in BM signal during treatment, indicating leukemia re-growth following initial regression (Fig. 2B).
In order to test the sensitivity of BLI in identifying low levels of leukemia in discrete sites, previously determined values of total flux/cell were used to estimate the number of leukemia cells present in the femurs of representative ALL-11-GL mice following 4 weeks of 1x VXL treatment. Using an algorithm provided in the LivingImage analysis software, based on 3D reconstruction of BLI and computed tomography images, treated mice exhibited a median of 5.5 × 104 cells in femoral BM (data not shown). To provide a reference for highly engrafted BM, this calculation was also performed on control mice, which exhibited a median of 3.1 × 107 cells in femoral BM (data not shown). These values strongly reflect cell counts performed using trypan blue exclusion following femoral BM harvest of highly engrafted ALL-11-GL mice (median = 2.2 × 107, data not shown), as well as cell numbers observed via isolation of murine femoral BM by Mahajan et al. (2016) (35).
While BLI enhanced the sensitivity of disease monitoring compared to PB, there was a strong concordance between these techniques in determining overall treatment response. BLI values were plotted against PB values where engraftment was measurable in PB, showing a significant correlation for both ALL-11-GL (R2 = 0.792, P < 0.0001) and ALL-57-GL (R2 = 0.866, P < 0.0001) (Fig. 2E).
BLI allows longitudinal measurement of residual disease during and post-treatment
Given the prognostic importance of early treatment response in ALL (24, 36), the utility of a BLI-based MRD measurement was assessed. First, BLI and flow cytometry were compared in measuring organ infiltration following treatment. At Day 28 post-treatment initiation, representative mice from each treatment group were imaged and immediately humanely killed, with blood, BM, spleen and liver harvested to measure %huCD45+ cells. Consistent with treatment response observed for the ALL-11-GL 1x VXL-treated group, these mice showed low levels of leukemia in all compartments, with a median 0.8% and 0.3% huCD45+ cells in BM and spleen, respectively (Fig. 3A). Also consistent with PB responses, the 0.25x, 0.125x and vehicle-treated ALL-11-GL groups showed high levels of engraftment in all organs (Fig. 3A).
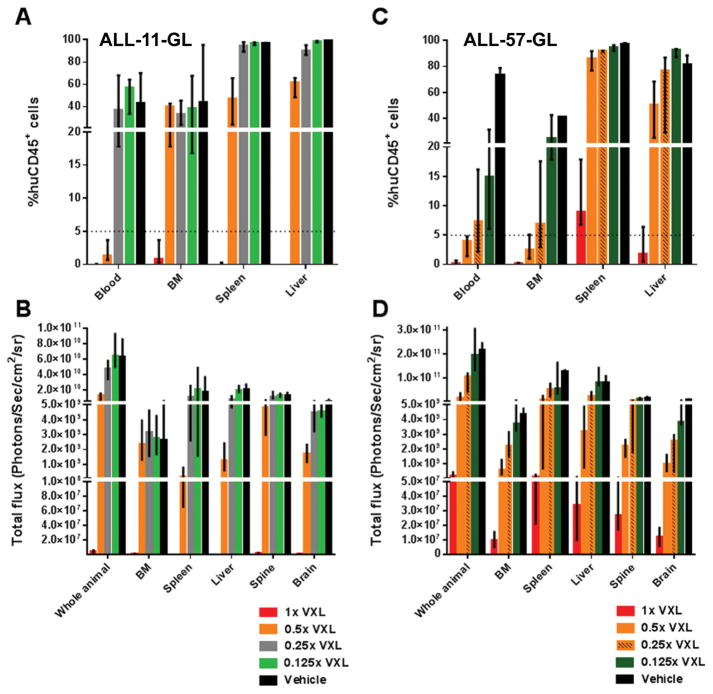
Figure 3. Residual leukemia burden at Day 28 post-treatment initiation for ALL-11-GL- and ALL-57-GL-bearing mice, as measured by flow cytometry and BLI.
Leukemia burden for ALL-11-GL (A, B) and ALL-57-GL (C, D) mice in all VXL treatment groups (1x, 0.5x, 0.25x and 0.125x) 28 days post-treatment initiation was measured first by BLI (B, D), then mice were humanely killed and blood, femoral BM and spleen samples taken for determination of %huCD45+ cells by flow cytometry (A, C). Cohorts are as labeled, colored to match the ORM score according to PPTP criteria. Values are shown as median + range.
Interestingly, the 0.5x group, which scored a CR by PPTP response criteria, exhibited high leukemic burden in BM (median 40.5%) and spleen (median 46.9%), despite a median 1.4% huCD45+ cells in PB. Supporting the flow cytometry data, BLI BM values for the 0.5x group (median = 2.29 × 109 p/s/cm2/sr) resembled more those of control mice (median = 2.65 × 109 p/s/cm2/sr) than the 1x group (median = 7.51 × 105 p/s/cm2/sr) (Fig. 3B). This was reflected in a one-way ANOVA of BM values, including a multiple comparisons analysis, whereby no significant difference in either %huCD45+ or BLI values were observed between the 0.5x, 0.25x, 0.125x and control groups (P < 0.05). Applying a clinical criteria of complete response (<5% blasts in the BM following 2–4 weeks of treatment) (37) to this dataset highlights that only the 1x treatment was sufficiently effective to reflect a clinical CR. While brain and spine data were not measured by flow cytometry, BLI data indicated persistent disease in regions representative of CNS disease in all groups, albeit significantly less so in the 1x cohort. These data suggest that a CR by PB measurements, according to PPTP criteria, does not reflect the clinical definition of a CR for ALL-11-GL.
Unlike ALL-11-GL, flow cytometry values for BM burden in ALL-57-GL reflected PB values. Median BM values of 0.2%, 2.5%, 6.9%, and 24.8% for the 1x, 0.5x, 0.25x and 0.125x groups were similar to their median PB percentages of 0.2%, 4%, 7.3% and 15%, respectively (Fig. 3C). A similar dose-response was observed in BLI BM values, however BLI data showed a >60-fold difference between the 0.5x group (median = 5.97 × 108 p/s/cm2/sr) and the 1x group (median = 9.89 × 106 p/s/cm2/sr), compared to the 12.5-fold difference observed by %huCD45+ values (Fig. 3D).
In contrast, ALL-57-GL spleen samples exhibited >80% huCD45+ cells for all but the 1x cohort, which showed a median of 8.9% huCD45+ cells (Fig. 3C). A one-way ANOVA comparison of %huCD45+ values showed that the means of all groups were statistically different (P = <0.0001), however a multiple comparisons test showed that while the 1x group was significantly different to all other groups, and the 0.5x group was significantly different from the 0.125x and control groups, there was no significant difference between all other comparisons (P = >0.05). This suggests that spleen infiltration at Day 28 for the 0.25x and 0.125x resembled that of control mice, while the 0.5x group was more similar to the 0.25x group than the 1x group.
Median liver infiltration values for all groups except the 1x cohort were >50% huCD45+ (Fig. 3C), reflecting the comparably high bioluminescent output observed in these groups (Fig. 3D). Despite lower liver engraftment by both flow cytometry and BLI when compared to other groups, persistent disease remained in the livers of the 1x VXL treated mice (Fig. 3C–D). Residual disease in the CNS, measured by BLI, was also present in all groups (Fig. 3D). These data suggest that relapse in these mice was driven primarily by residual disease in hematolymphoid organs, and that Day 28 BM measurements may not be a significant enough improvement to existing scoring criteria.
BLI evaluation of the efficacy of the novel Bcl-2 inhibitor venetoclax
The ability of BLI to increase the stringency of novel drug testing was assessed using the Bcl-2 inhibitor venetoclax. This drug was chosen based on previous testing of venetoclax and its predecessor navitoclax against a large number of ALL PDXs, with both drugs eliciting a broad range of responses (30, 32).
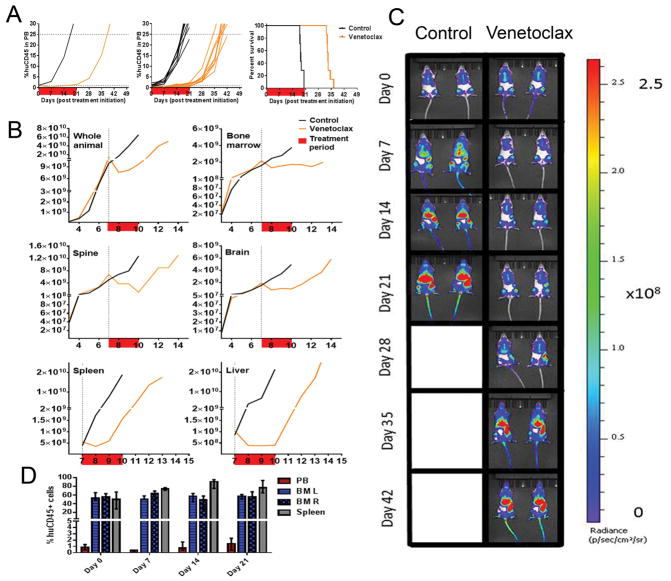
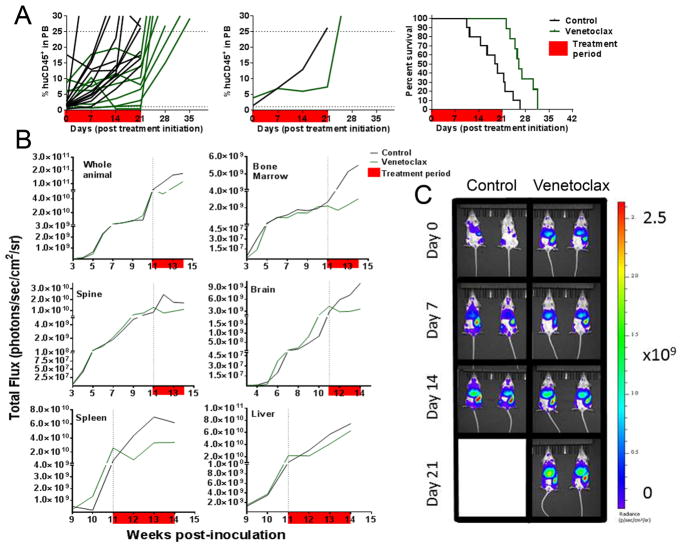
Venetoclax elicited significant regression of ALL-11-GL using PB measures, achieving a CR and an LGD of 14.5 days (Fig. 4A and Supplementary Table S1). However, despite treatment maintaining <1% huCD45+ cells in the PB for 2 consecutive weeks, the median %huCD45+ value increased almost two-fold from Day 7 (0.4%) to Day 14 (0.7%), and continued to increase thereafter. Whole animal BLI showed a similar profile to PB measurements, highlighting regression in the first week of treatment (log10 fold change = −0.26), before the signal began to increase at Day 14 (log10 fold change = −0.23), and continued to increase throughout the treatment and monitoring periods (Fig. 4B).
Figure 4. In vivo efficacy of venetoclax against ALL-11-GL.
The response of ALL-11-GL to venetoclax (100 mg/kg) as measured by %huCD45+ cells in mouse PB (A), showing median group PB values (left), individual mouse PB values (center) and Kaplan-Meier survival plot (right). (B) Efficacy of venetoclax against ALL-11-GL as measured by BLI for the whole animal and individual organs (as labeled). Response to each treatment is visualized as the median bioluminescence intensity (p/s/cm2/sr) for each cohort. The treatment period is shown in red. (C) Images of representative mice from both venetoclax-treated and control cohorts are shown at weekly intervals throughout the evaluation period of 42 days. (D) Representative ALL-11-GL venetoclax treated mice were also harvested weekly throughout the treatment period to assess leukemia burden in the PB, BM and spleen. All imaging data were acquired using the Spectrum CT in vivo imaging system. Images were analyzed with the LivingImage v4.4 software. Values in (D) are shown as median + range.
Interestingly, BLI values for BM, brain and liver of treated mice were more characteristic of disease stabilization following the initial regression at Day 7, while spine burden continued to decrease until Day 21 (log10 fold change = −0.85) (Fig. 4B). The pattern of relapse following transient regression observed in whole animal imaging was strongly reflected by BLI values from the spleen, suggesting its role in persistent disease through treatment (Fig. 4B). This was also observed in representative raw bioluminescence images (Fig. 4C). Representative mice were humanely killed at weekly intervals throughout treatment to assess disease burden in organs using flow cytometry. These results support imaging data, showing stabilization of leukemia in the BM and a steady increase in %huCD45+ cells in the spleen (Fig. 4D).
Venetoclax showed minimal efficacy against ALL-57-GL, eliciting a score of progressive disease 1 (PD1) and an LGD of 6.8 days (Fig. 5A and Supplementary Table S1). While median PB values showed regression at Day 14 compared to Day 7, this was not as clear based on values for individual mice. Imaging data showed disease regression at Day 7 by whole animal imaging (log10 fold decrease of −0.12). This was reflected in imaging values for individual organs (Fig. 5B). Persistent disease was also observed throughout treatment in raw images of representative mice (Fig. 5C). Importantly, bioluminescent signal from the liver appeared to plateau from Day 0 to Day 7 (log10 fold change = 0.01), and thereafter increase similarly to controls, suggesting this organ as a reservoir of persistent disease (Fig. 5B).
Figure 5. In vivo efficacy of venetoclax against ALL-57-GL.
The response of ALL-57-GL to venetoclax (100 mg/kg) as measured by %huCD45+ cells in mouse PB (A), showing individual mouse PB values (left), median group PB values (center) and Kaplan-Meier survival plot (right). (B) Efficacy of venetoclax against ALL-57-GL as measured by BLI for the whole animal and individual organs (as labeled). Response to each treatment is visualized as the median bioluminescence intensity (p/s/cm2/sr) for each cohort. The treatment period is shown in red. (C) Images of representative mice from both venetoclax-treated and control cohorts are shown at weekly intervals throughout the evaluation period of 21 days (to event). All imaging data were acquired using the Spectrum CT in vivo imaging system. Images were analyzed with the LivingImage v4.4 software.
Development of novel BLI-based criteria for assessment of drug efficacy
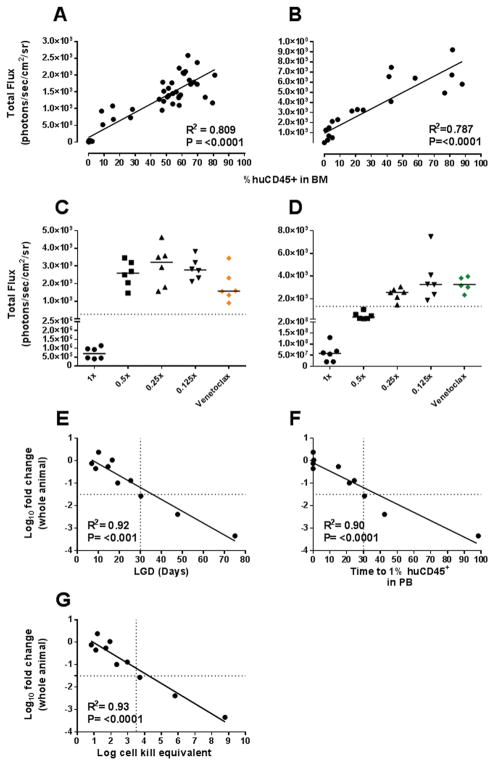
The spectrum of responses achieved using VXL and venetoclax allowed investigation of BLI-based criteria to increase the sensitivity and predictive power of drug efficacy testing in the ALL PDX model. First, establishment of a BLI surrogate for a clinical CR measurement (<5% blasts in BM at Day 28) was investigated, using a BLI-based threshold representative of 5% huCD45+ cells in the BM. Imaging values and %huCD45+ data for BM samples compiled from VXL experiments were used to model a standard curve, from which a BLI value at 5% huCD45+ cells in the BM was obtained (2.67 × 108 p/s/cm2/sr for ALL-11-GL and 1.33 × 109 p/s/cm2/sr for ALL-57-GL) (Fig. 6A–D).
Figure 6. Development of BLI criteria representative of a clinical complete response.
The correlation between BM BLI values and respective %huCD45+ values for harvested BM samples was assessed for both ALL-11-GL (A, C) and ALL-57-GL (B, D). Subsequently, the BLI value representing 5% huCD45+ cells in the BM (dashed lines in C and D) was applied as a cut-off to BLI data for all treatment groups for both ALL-11-GL and ALL-57-GL. Each data point represents an individual mouse, with the median of the group shown as a black line. (E) The median log10 fold change value for each cohort was plotted against each group’s respective leukemia growth delay (LGD) value. (F) The median log10 fold change value for each cohort was plotted against each group’s respective TT1% value. (G) The median log10 fold change value for each cohort was plotted against each group’s respective log cell kill equivalent value. R, Pearson correlation coefficient; P-value, two-tailed P-value of the Pearson correlation.
The 1x VXL cohorts for both PDXs were considerably below the cut-off, confirming their score of MCR (Fig. 6C–D). For ALL-11-GL, all other VXL treatment groups failed to meet this criterion (Fig. 6C), thus the 0.5x group did not achieve the clinical definition of CR, despite having scored a CR by PPTP criteria. This distinction is less clear for ALL-57-GL samples, as while the cut-off excluded the 0.25x (CR) and 0.125x (PD1) groups, the 0.5x group met the criteria despite showing considerably higher BM signal compared to the 1x group (Fig. 6D) and persistent disease in the spleen and liver at Day 28 (Fig. 3B).
The BLI-based CR threshold was also applied to venetoclax data, with BM values recorded at Day 28 for ALL-11-GL or Day 21 for ALL-57-GL (due to morbidity prior to Day 28). Venetoclax treated ALL-57-GL mice showed similar BM signals to the 0.125x VXL group (Fig. 6D), consistent with both treatments scoring a PD (PD2 and PD1, respectively) (Supplementary Table S1). Interestingly, despite scoring a CR by PB measures, the BM values for ALL-11-GL failed to meet the cut-off, with values approaching those of the 0.5x VXL group (Fig. 6C). Therefore the application of this additional threshold would preclude venetoclax from achieving a CR against ALL-11-GL.
In addition to BLI-based criteria, this study aimed to reassess PB-based criteria to increase the stringency of drug testing where BLI is not available. The median log10 fold change value for each group was therefore compared to parameters measurable by PB; LGD, time to 1% huCD45+ in PB (TT1%) and an approximation of log cell kill (log cell kill equivalent, LCKe). First, comparison of log10 fold change and LGD showed significant correlations for ALL-11-GL (R2 = 0.98, P = 0.001) and ALL-57-GL (R2 = 0.99, P = <0.001) (Supplementary Fig. S2A–B) and both PDXs together (R2 = 0.92, P = <0.001) (Fig. 6E). No correlation was evident when median %huCD45+ values were plotted against LGD values for each treatment group (R2 = 0.29, P = 0.164) (Supplementary Fig. S2E). Second, log10 fold change data were plotted against each group’s TT1% values, as Day 28 data suggested values >1% huCD45+ cells in PB were indicative of considerable leukemia burden in other sites. A strong inverse correlation was observed for ALL-11-GL (R2 = 0.99, P = 0.0005) and ALL-57-GL (R2 = 0.95, P = 0.026) separately (Supplementary Fig. S2C–D), as well as together (R2 = 0.90, P = <0.0001) (Fig. 6F), indicating that the depth of response by BLI is predictive of reappearance of leukemia in the PB.
Determination of LCKe values used the equation LCKe = T-C/(3.32 × TVDT) (38), where T-C is equivalent to LGD (Supplementary Table S1) and TVDT represents tumor volume doubling time. TVDT values were determined via interpolation of a standard curve created using PB values of control mice (Supplementary Fig. S3A–B). LCKe values ranged from 0.8–8.8 (Supplementary Table S3), with the values for each treatment group correlating strongly with respective log10 fold change values for ALL-11-GL (R2 = 0.98, P = 0.0012) and ALL-57-GL (R2 = 0.99, P = 0.0003) (Supplementary Fig. S3C–D), as well as for both PDXs combined (R2 = 0.93, P = <0.0001) (Fig. 6G). Combining these data with previous clinical CR classifications, each of the three groups scoring a clinical CR achieved a log10 fold change >1.5, an LGD of >30 days, a TT1% of >30 days and an LCKe of >3.5. These parameters may therefore be used alongside existing scoring criteria to indirectly assess a drug’s ability to induce a clinical CR.
DISCUSSION
In this study we report that BLI enhances the sensitivity of treatment monitoring in pediatric ALL PDXs, allowing more robust assessment of drug efficacy compared to PB monitoring. These data support reports outlining BLI’s ability to sensitively monitor the treatment response of leukemia models (15, 16, 39). However, treatments used in those studies were relatively ineffective, achieving a maximum reduction of one order of magnitude by BLI, and no direct comparisons between BLI data and %huCD45+ values in PB or individual organs were provided (15, 16).
In monitoring the efficacy of the VXL regimen, BLI was able to monitor leukemia regression over a range of >3 orders of magnitude, while PB measures were restricted by the threshold of detection for flow cytometry and the reliance on measuring only circulating lymphocytes. This discrepancy was epitomized by BLI enabling measurement of changes in disease burden in the ALL-11-GL 1x VXL group during 10 weeks where leukemia was undetectable by PB. Moreover, BLI enabled real-time visualization of residual disease, allowing determination of timing and anatomical location of relapse.
While identification of sites of residual disease using BLI has been reported previously (15, 16, 39), there have been no attempts to correlate organ burden measured by BLI with traditional methods, such as flow cytometry, across diverse treatment responses. Real-time measurements of organ burden by BLI will allow for more comprehensive predictions of clinical efficacy, as assessing MRD levels during early treatment is one of the primary clinical indicators of outcome for pediatric ALL (4, 20). The significant correlation between BLI and flow cytometry allowed the establishment of a BLI-based measurement of 5% blasts in the BM, approximating a clinical measurement of CR (37). When applied to VXL treatments, the number of objective responses evaluated through PB was reduced from 5 to 3. Moreover, the threshold also excluded the venetoclax-treated ALL-11-GL group, which achieved a CR by PB measurements. It is notable that 3 of 4 groups scoring a CR by PB measurements did not meet the 5% cut-off. The venetoclax-treated ALL-11-GL group failing to meet BLI-based criteria highlights the benefits of BLI in assessing the efficacy of novel agents, and the need for additional stringency in drug efficacy scoring criteria.
It is important to note that novel response criteria such as the threshold of 5% huCD45+ cells in the BM of treated mice should be used in concert with other parameters measuring the extent of treatment response. The approximation of a clinical CR in the PDX model provides a strong indication of drug activity, however the clinical measurement is both an assessment of leukemia reduction as well as the regeneration of normal hematopoiesis, which is impossible to reflect in PDX models. Future studies may benefit from the adoption of a more directly clinically applicable measurement, such as MRD analysis by flow cytometry or PCR methods, which may be used to increase the clinical relevance of the Day 28 measurement.
The use of BLI also has the potential to allow investigators to test novel compounds in a more clinically relevant setting - against residual disease following eradication of bulk disease by an induction-type regimen. The use of targeted agents against a lower disease burden during this remission period may assist in ensuring that an appropriate amount of drug reaches and eradicates all leukemia cells, thereby reducing the chance of cells surviving drug exposure and developing drug resistance. Further, BLI may also enable sensitive testing of drugs targeting leukemia stem cell (LSC) populations, however these experiments would require thorough characterization of such cells with respect to stem cell properties and the inherent differences between the proposed LSCs and the bulk leukemia cells.
In addition to enabling quantitation of residual disease, BLI also highlighted differential efficacy of venetoclax in different anatomical compartments. Interestingly, disease stabilization in the brain and regression in the spine for ALL-11-GL-bearing mice in response to venetoclax is consistent with reports this drug is able to infiltrate the CNS (40). Spleen involvement observed during treatment is contrary to clinical findings in CLL, where venetoclax significantly reduced splenomegaly (41). However, CLL is largely a Bcl-2 dependent disease and is therefore likely to be more sensitive to Bcl-2 inhibitors compared to ALL (42, 43). The ability of BLI to longitudinally monitor organ-specific disease burden may prove crucial for testing agents targeting leukemic niches and leukemic stem cells, as well as identifying novel drug combinations based on a drug’s ability to target specific leukemic niches (44–46). Site-specific drug efficacy has been observed previously against an ALL PDX using VXL combined with a CD19-directed antibody drug conjugate, SAR3419. Despite clearance of leukemia from PB, BM and spleen, mice succumbed to disease harbored within the CNS, purportedly due to the inability of the ADC to cross the blood-brain barrier (47). CNS-borne leukemia can be monitored using BLI, as shown herein and elsewhere (48).
While BLI would ideally become routine in preclinical efficacy testing for pediatric ALL, considerable limitations exist with this technique. Lentiviral transduction of PDX cells is inefficient, requiring the time consuming protocols used herein and elsewhere (15, 16), while some PDXs remain completely refractory to transduction. Developing PDX panels representing defined ALL subtypes is therefore limited, as is using BLI for personalized medicine approaches whereby mice are used as avatars for a patient’s cancer to inform treatment decisions in a clinically relevant time-frame (49, 50). Potential changes to PDXs introduced by transduction procedures are also an important consideration. However, a number of groups have now reported the development of GFP/Luciferase+ leukemia PDXs without significant alterations, as measured by various techniques including targeted sequencing of common leukemia-associated mutations, short tandem repeat (STR) and SNP analyses and comparative drug sensitivity (15, 16, 18).
In establishing the GFP/Luciferase+ PDXs used herein, ALL-11-GL and ALL-57-GL cells were engrafted concomitantly with their respective parental PDXs as passage-matched controls, with no significant difference in engraftment rates observed between the transduced and parental cells (Supplementary Fig. S4). Further, comparison of gene expression profiles between ALL-11-GL cells and ALL-11 cells showed no significantly differentially expressed genes using a false discovery rate of 0.1 (L Jones, unpublished data). A comparison of the mean expression values between ALL-11-GL and ALL-11 across all 34,602 genes assessed by the array was also performed, with no significant differences observed (R2 = 0.99, P = <0.0001) (Supplementary Fig. S5). While not performed for ALL-57, this comparison was performed for another GFP/luciferase+ PDX, ALL-4-GL, also showing no significant differences between transduced and non-transduced cells (R2 = 0.99, P = <0.0001) (L Jones, unpublished data).
Additionally, the sensitivity of ALL-11-GL to VXL treatment strongly reflected historical data of VXL response for ALL-11 (LGD = 99.7 days) (27). Though it is important to note that the comparison using VXL response was for a non-targeted treatment regimen, and it has been reported that the production of eGFP may influence, either positively or negatively, response to certain targeted therapies (51). The use of targeted agents using these BLI-enabled PDXs should therefore include either direct comparison to non-transduced cells or to historical data of the parental PDX where available.
The limitations in developing BLI-enabled ALL PDXs supports development of stringent response criteria based on the BLI data and organ infiltration measurements herein, which can be indirectly evaluated using PB measurements. Importantly, determination of a clinical CR in this study could be translated to four additional parameters; a log10 BLI fold change >-1.5, an LGD >30 days, an LCKe >3.5 and TT1% >30 days, three of which are evaluable without BLI. Interestingly, the LCKe threshold is potentially relatable to clinical MRD measurements, as a >3.5 log reduction in leukemia burden approximates an MRD value of ≤ 0.01%, which is indicative of positive outcome (52). Of the criteria proposed herein, however, TT1% was perhaps the most important parameter in grouping the ALL-57-GL 0.5x group (CR) with the two MCRs, as this measure highlighted that %huCD45+ levels in the PB were maintained below 1% for two consecutive weeks following treatment cessation. Existing CR criteria require two consecutive weeks <1% huCD45+ cells in PB, however this is highly likely to include the treatment period and therefore may not reflect a durable response. Therefore the stringency of existing objective response criteria could be significantly improved by incorporating the measurement of a clinical CR, requiring a log10 fold change >-1.5 (where BLI is possible), an LGD >30 days, an LCKe of >3.5 and <1% huCD45+ cells in PB for two consecutive weeks following treatment cessation. The definition of a CR would then become similar to that of an MCR, which requires PB levels <1% huCD45+ cells for the last 3 weeks of the 42-day monitoring period. This additional stringency will likely discount the majority of novel agents as, using previous scores of MCR for comparison, only 4/57 drugs tested against the PPTP ALL PDX panel have achieved an MCR in >50% of PDXs (14). However this may be improved with more biomarker-driven testing, as 16/57 drugs induced at least one MCR (14). It is noteworthy that the ALL-57-GL 0.5x VXL group achieved a clinical CR despite the presence of considerable extramedullary disease, however clinically the prognosis of BM relapse is significantly poorer than that of extramedullary relapse (53, 54), supporting the use of the BM-based threshold in this study.
In summary, the present study highlights that BLI-enabled PDX models of pediatric ALL can significantly enhance the predictive capacity of preclinical assessment of novel drugs, and can be used to re-define stringent response criteria based on measurements of leukemic burden in the BM and PB. The more widespread adoption of a minimum log10 fold change by BLI (>-1.5), a minimum LGD (>30 days) and LCKe (>3.5), as well as the requirement of <1% huCD45+ cells in PB for two consecutive weeks following treatment cessation may allow for more confident predictions of clinical efficacy of novel agents, thereby helping to ensure that only drugs with the greatest likelihood of success are advanced to clinical trials.
Supplementary Material
Statement of translational relevance.
Childhood acute lymphoblastic leukemia (ALL) patient-derived xenografts (PDXs) are widely used for preclinical assessment of novel agents to inform new agent prioritization decisions. Despite the widespread use of these PDXs, uniform criteria for assessing drug responses relevant to the clinical setting are yet to be adopted. Methods of assessment include enumerating leukemic blasts in the murine peripheral blood over time or assessing leukemic infiltration of hematolymphoid organs at specific times post treatment initiation and, more recently, bioluminescence imaging (BLI) of lentivirally transduced PDXs. This article describes establishing stable luciferase expressing childhood ALL PDXs, and provides a detailed analysis of how assessing leukemic burden through BLI can enhance surrogate and traditional methods of disease monitoring. The more widespread application of these findings in other leukemia models will likely increase the stringency with which new drug candidates are evaluated in the preclinical setting for advancement into clinical trials.
Acknowledgments
GRANT SUPPORT
This work was partially supported by grants from the National Institutes of Health and National Cancer Institute (NOI-CM-42216 and NOI-CM-91001-03), and the National Health and Medical Research Council of Australia (NHMRC). R.B.L. is supported by a fellowship from the NHMRC. L.J. was supported by an Australian Postgraduate Award from the Australian Government Department of Education and Training.
The authors thank AbbVie Inc. for providing venetoclax (ABT-199), as well as Dr. Tzong-tyng Hung, Dr. Brendan Lee and Dr. Carl Power of the Biological Resources Imaging Laboratory at the University of New South Wales for their bioluminescence imaging support and equipment. Children’s Cancer Institute is affiliated with UNSW Australia and the Sydney Children’s Hospitals Network.
Footnotes
Conflicts of Interest
The authors have declared that no conflict of interest exists.
References
- 1.Pui CH, Yang JJ, Hunger SP, Pieters R, Schrappe M, Biondi A, et al. Childhood acute lymphoblastic leukemia: Progress through collaboration. J Clin Oncol. 2015;33:2938–48. doi: 10.1200/JCO.2014.59.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tallen G, Ratei R, Mann G, Kaspers G, Niggli F, Karachunsky A, et al. Long-term outcome in children with relapsed acute lymphoblastic leukemia after time-point and site-of-relapse stratification and intensified short-course multidrug chemotherapy: Results of trial ALL-REZ BFM 90. J Clin Oncol. 2010;28:2339–47. doi: 10.1200/JCO.2009.25.1983. [DOI] [PubMed] [Google Scholar]
- 3.Parker C, Waters R, Leighton C, Hancock J, Sutton R, Moorman AV, et al. Effect of mitoxantrone on outcome of children with first relapse of acute lymphoblastic leukaemia (ALL R3): an open-label randomised trial. The Lancet. 2010;376:2009–17. doi: 10.1016/S0140-6736(10)62002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Locatelli F, Schrappe M, Bernardo ME, Rutella S. How I treat relapsed childhood acute lymphoblastic leukemia. Blood. 2012;120:2807–16. doi: 10.1182/blood-2012-02-265884. [DOI] [PubMed] [Google Scholar]
- 5.Waanders E, Dobson SM, Ma XT, Payne-Turner D, Song GC, Fan YP, et al. Genomic landscape of relapsed acute lymphoblastic leukemia. Blood. 2015;126:692. [Google Scholar]
- 6.Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang Y-L, Pei D, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med. 2014;371:1005–15. doi: 10.1056/NEJMoa1403088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts Kathryn G, Morin Ryan D, Zhang J, Hirst M, Zhao Y, Su X, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22:153–66. doi: 10.1016/j.ccr.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts KG, Mullighan CG. Genomics in acute lymphoblastic leukaemia: insights and treatment implications. Nat Rev Clin Oncol. 2015;12:344–57. doi: 10.1038/nrclinonc.2015.38. [DOI] [PubMed] [Google Scholar]
- 9.Neale G, Su X, Morton CL, Phelps D, Gorlick R, Lock RB, et al. Molecular characterization of the pediatric preclinical testing panel. Clin Cancer Res. 2008;14:4572–83. doi: 10.1158/1078-0432.CCR-07-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liem NLM, Papa RA, Milross CG, Schmid MA, Tajbakhsh M, Choi S, et al. Characterization of childhood acute lymphoblastic leukemia xenograft models for the preclinical evaluation of new therapies. Blood. 2004;103:3905–14. doi: 10.1182/blood-2003-08-2911. [DOI] [PubMed] [Google Scholar]
- 11.Jing D, Bhadri VA, Beck D, Thoms JAI, Yakob NA, Wong JWH, et al. Opposing regulation of BIM and BCL2 controls glucocorticoid-induced apoptosis of pediatric acute lymphoblastic leukemia cells. Blood. 2015;125:273–83. doi: 10.1182/blood-2014-05-576470. [DOI] [PubMed] [Google Scholar]
- 12.Moradi Manesh D, El-Hoss J, Evans K, Richmond J, Toscan CE, Bracken LS, et al. AKR1C3 is a biomarker of sensitivity to PR-104 in preclinical models of T-cell acute lymphoblastic leukemia. Blood. 2015;126:1193–202. doi: 10.1182/blood-2014-12-618900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong NC, Bhadri VA, Maksimovic J, Parkinson-Bates M, Ng J, Craig JM, et al. Stability of gene expression and epigenetic profiles highlights the utility of patient-derived paediatric acute lymphoblastic leukaemia xenografts for investigating molecular mechanisms of drug resistance. BMC Genomics. 2014;15:416. doi: 10.1186/1471-2164-15-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones L, Carol H, Evans K, Richmond J, Houghton PJ, Smith MA, et al. A review of new agents evaluated against pediatric acute lymphoblastic leukemia by the pediatric preclinical testing program. Leukemia. 2016 doi: 10.1038/leu.2016.192. [DOI] [PubMed] [Google Scholar]
- 15.Barrett DM, Seif AE, Carpenito C, Teachey DT, Fish JD, June CH, et al. Noninvasive bioluminescent imaging of primary patient acute lymphoblastic leukemia: a strategy for preclinical modeling. Blood. 2011;118:e112–e7. doi: 10.1182/blood-2011-04-346528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terziyska N, Alves CC, Groiss V, Schneider K, Farkasova K, Ogris M, et al. In vivo imaging enables high resolution preclinical trials on patients leukemia cells growing in mice. PLoS ONE. 2012;7:e52798. doi: 10.1371/journal.pone.0052798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bomken S, Buechler L, Rehe K, Ponthan F, Elder A, Blair H, et al. Lentiviral marking of patient-derived acute lymphoblastic leukaemic cells allows in vivo tracking of disease progression. Leukemia. 2013;27:718–21. doi: 10.1038/leu.2012.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vick B, Rothenberg M, Sandhöfer N, Carlet M, Finkenzeller C, Krupka C, et al. An advanced preclinical mouse model for acute myeloid leukemia using patients’ cells of various genetic subgroups and in vivo bioluminescence imaging. PLoS ONE. 2015;10:e0120925. doi: 10.1371/journal.pone.0120925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christoph S, Schlegel J, Alvarez-Calderon F, Kim Y-M, Brandao LN, DeRyckere D, et al. Bioluminescence imaging of leukemia cell lines in vitro and in mouse xenografts: effects of monoclonal and polyclonal cell populations on intensity and kinetics of photon emission. J Hematol Oncol. 2013;6:10. doi: 10.1186/1756-8722-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eckert C, Biondi A, Seeger K, Cazzaniga G, Hartmann R, Beyermann B, et al. Prognostic value of minimal residual disease in relapsed childhood acute lymphoblastic leukaemia. The Lancet. 2001;358:1239–41. doi: 10.1016/S0140-6736(01)06355-3. [DOI] [PubMed] [Google Scholar]
- 21.Karawajew L, Dworzak M, Ratei R, Rhein P, Gaipa G, Buldini B, et al. Minimal residual disease analysis by eight-color flow cytometry in relapsed childhood acute lymphoblastic leukemia. Haematologica. 2015;100:935–44. doi: 10.3324/haematol.2014.116707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flohr T, Schrauder A, Cazzaniga G, Panzer-Grumayer R, van der Velden V, Fischer S, et al. Minimal residual disease-directed risk stratification using real-time quantitative PCR analysis of immunoglobulin and T-cell receptor gene rearrangements in the international multicenter trial AIEOP-BFM ALL 2000 for childhood acute lymphoblastic leukemia. Leukemia. 2008;22:771–82. doi: 10.1038/leu.2008.5. [DOI] [PubMed] [Google Scholar]
- 23.Morley AA, Latham S, Brisco MJ, Sykes PJ, Sutton R, Hughes E, et al. Sensitive and specific measurement of minimal residual disease in acute lymphoblastic leukemia. J Mol Diagn. 2009;11:201–10. doi: 10.2353/jmoldx.2009.080048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campana D. Minimal residual disease in acute lymphoblastic leukemia. ASH Education Program Book. 2010;2010:7–12. doi: 10.1182/asheducation-2010.1.7. [DOI] [PubMed] [Google Scholar]
- 25.Chung YR, Kim E, Abdel-Wahab O. Femoral bone marrow aspiration in live mice. J Vis Exp. 2014:e51660. doi: 10.3791/51660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Workman P, Aboagye EO, Balkwill F, Balmain A, Bruder G, Chaplin DJ, et al. Guidelines for the welfare and use of animals in cancer research. Br J Cancer. 2010;102:1555–77. doi: 10.1038/sj.bjc.6605642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szymanska B, Wilczynska-Kalak U, Kang MH, Liem NLM, Carol H, Boehm I, et al. Pharmacokinetic modeling of an induction regimen for in vivo combined testing of novel drugs against pediatric acute lymphoblastic leukemia xenografts. PLoS ONE. 2012;7:e33894. doi: 10.1371/journal.pone.0033894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Houghton PJ, Morton CL, Tucker C, Payne D, Favours E, Cole C, et al. The pediatric preclinical testing program: description of models and early testing results. Pediatr Blood Cancer. 2007;49:928–40. doi: 10.1002/pbc.21078. [DOI] [PubMed] [Google Scholar]
- 29.Anderson MA, Huang D, Roberts A. Targeting BCL2 for the treatment of lymphoid malignancies. Semin Hematol. 2014;51:219–27. doi: 10.1053/j.seminhematol.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 30.Khaw SL, Suryani S, Evans K, Richmond J, Robbins A, Kurmasheva RT, et al. Venetoclax responses of pediatric ALL xenografts reveal MLLr ALL sensitivity, but overall requirement to target both BCL2 and BCLXL. Blood. 2016 doi: 10.1182/blood-2016-03-707414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lock RB, Liem N, Farnsworth ML, Milross CG, Xue C, Tajbakhsh M, et al. The nonobese diabetic/severe combined immunodeficient (NOD/SCID) mouse model of childhood acute lymphoblastic leukemia reveals intrinsic differences in biologic characteristics at diagnosis and relapse. Blood. 2002;99:4100–8. doi: 10.1182/blood.v99.11.4100. [DOI] [PubMed] [Google Scholar]
- 32.Suryani S, Carol H, Chonghaile TN, Frismantas V, Sarmah C, High L, et al. Cell and molecular determinants of in vivo efficacy of the BH3 mimetic ABT-263 against pediatric acute lymphoblastic leukemia xenografts. Clin Cancer Res. 2014;20:4520–31. doi: 10.1158/1078-0432.CCR-14-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jamieson SMF, Gu Y, Manesh DM, El-Hoss J, Jing D, MacKenzie KL, et al. A novel fluorometric assay for aldo-keto reductase 1C3 predicts metabolic activation of the nitrogen mustard prodrug PR-104A in human leukaemia cells. Biochem Pharmacol. 2014;88:36–45. doi: 10.1016/j.bcp.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 34.El-Hoss J, Jing D, Evans K, Toscan C, Xie J, Lee H, et al. A single nucleotide polymorphism genotyping platform for the authentication of patient derived xenografts. Oncotarget. 2016 doi: 10.18632/oncotarget.11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahajan MM, Cheng B, Beyer AI, Mulvaney US, Wilkinson MB, Fomin ME, et al. A quantitative assessment of the content of hematopoietic stem cells in mouse and human endosteal-bone marrow: a simple and rapid method for the isolation of mouse central bone marrow. BMC Hematology. 2015;15:9. doi: 10.1186/s12878-015-0031-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conter V, Bartram CR, Valsecchi MG, Schrauder A, Panzer-Grumayer R, Moricke A, et al. Molecular response to treatment redefines all prognostic factors in children and adolescents with B-cell precursor acute lymphoblastic leukemia: results in 3184 patients of the AIEOP-BFM ALL 2000 study. Blood. 2010;115:3206–14. doi: 10.1182/blood-2009-10-248146. [DOI] [PubMed] [Google Scholar]
- 37.Gaynon PS, Desai AA, Bostrom BC, Hutchinson RJ, Lange BJ, Nachman JB, et al. Early response to therapy and outcome in childhood acute lymphoblastic leukemia. Cancer. 1997;80:1717–26. doi: 10.1002/(sici)1097-0142(19971101)80:9<1717::aid-cncr4>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 38.Wild R, Fager K, Flefleh C, Kan D, Inigo I, Castaneda S, et al. Cetuximab preclinical antitumor activity (monotherapy and combination based) is not predicted by relative total or activated epidermal growth factor receptor tumor expression levels. Mol Cancer Ther. 2006;5:104–13. doi: 10.1158/1535-7163.MCT-05-0259. [DOI] [PubMed] [Google Scholar]
- 39.Duy C, Hurtz C, Shojaee S, Cerchietti L, Geng H, Swaminathan S, et al. BCL6 enables Ph+ acute lymphoblastic leukaemia cells to survive BCR-ABL1 kinase inhibition. Nature. 2011;473:384–8. doi: 10.1038/nature09883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ackler S, Oleksijew A, Chen J, Chyla BJ, Clarin J, Foster K, et al. Clearance of systemic hematologic tumors by venetoclax (ABT-199) and navitoclax. Pharmacol Res Perspect. 2015;3:e00178. doi: 10.1002/prp2.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roberts AW, Seymour JF, Brown JR, Wierda WG, Kipps TJ, Khaw SL, et al. Substantial susceptibility of chronic lymphocytic leukemia to BCL2 inhibition: results of a Phase I study of navitoclax in patients with relapsed or refractory disease. J Clin Oncol. 2012;30:488–96. doi: 10.1200/JCO.2011.34.7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Del Gaizo Moore V, Brown JR, Certo M, Love TM, Novina CD, Letai A. Chronic lymphocytic leukemia requires BCL2 to sequester prodeath BIM, explaining sensitivity to BCL2 antagonist ABT-737. J Clin Invest. 2007;117:112–21. doi: 10.1172/JCI28281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Letai A. A new face of BCL-2 inhibition in CLL. Blood. 2011;117:2750–1. doi: 10.1182/blood-2011-01-328658. [DOI] [PubMed] [Google Scholar]
- 44.Parameswaran R, Yu M, Lim M, Groffen J, Heisterkamp N. Combination of drug therapy in acute lymphoblastic leukemia with a CXCR4 antagonist. Leukemia. 2011;25:1314–23. doi: 10.1038/leu.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hartwell KA, Miller PG, Mukherjee S, Kahn AR, Stewart AL, Logan DJ, et al. Niche-based screening identifies small-molecule inhibitors of leukemia stem cells. Nat Chem Biol. 2013;9:840–8. doi: 10.1038/nchembio.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bakker E, Qattan M, Mutti L, Demonacos C, Krstic-Demonacos M. The role of microenvironment and immunity in drug response in leukemia. Biochim Biophys Acta. 2016;1863:414–26. doi: 10.1016/j.bbamcr.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 47.Carol H, Szymanska B, Evans K, Boehm I, Houghton PJ, Smith MA, et al. The anti-CD19 antibody-drug conjugate SAR3419 prevents hematolymphoid relapse postinduction therapy in preclinical models of pediatric acute lymphoblastic leukemia. Clin Cancer Res. 2013;19:1795–805. doi: 10.1158/1078-0432.CCR-12-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frishman-Levy L, Shemesh A, Bar-Sinai A, Ma C, Ni Z, Frenkel S, et al. Central nervous system acute lymphoblastic leukemia: role of natural killer cells. Blood. 2015;125:3420–31. doi: 10.1182/blood-2014-08-595108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malaney P, Nicosia SV, Davé V. One mouse, one patient paradigm: New avatars of personalized cancer therapy. Cancer Lett. 2014;344:1–12. doi: 10.1016/j.canlet.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trahair TN, Lock RB, Sutton R, Sia KC, Evans K, Richmond J, et al. Xenograft-directed personalized therapy for a patient with post-transplant relapse of ALL. Bone Marrow Transplant. 2016 doi: 10.1038/bmt.2016.122. [DOI] [PubMed] [Google Scholar]
- 51.Ansari AM, Ahmed AK, Matsangos AE, Lay F, Born LJ, Marti G, et al. Cellular GFP toxicity and immunogenicity: Potential confounders in in vivo cell tracking experiments. Stem Cell Rev. 2016;12:553–9. doi: 10.1007/s12015-016-9670-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borowitz MJ, Wood BL, Devidas M, Loh ML, Raetz EA, Salzer WL, et al. Prognostic significance of minimal residual disease in high risk B-ALL: a report from Children’s Oncology Group study AALL0232. Blood. 2015;126:964–71. doi: 10.1182/blood-2015-03-633685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nguyen K, Devidas M, Cheng SC, La M, Raetz EA, Carroll WL, et al. Factors influencing survival after relapse from acute lymphoblastic leukemia: a Children’s Oncology Group study. Leukemia. 2008;22:2142–50. doi: 10.1038/leu.2008.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bhojwani D, Pui C-H. Relapsed childhood acute lymphoblastic leukaemia. Lancet Oncol. 2013;14:e205–e17. doi: 10.1016/S1470-2045(12)70580-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.