Abstract
Here we report a gain in function for mutant (mt) superoxide dismutase I (SOD1), a cause of familial amyotrophic lateral sclerosis (FALS), wherein small soluble oligomers of mtSOD1 acquire a membrane toxicity. Phosphatidylglycerol (PG) lipid domains are selectively targeted, which could result in membrane damage or “toxic channels” becoming active in the bilayer. This PG-selective SOD1-mediated membrane toxicity is largely reversible in vitro by a widely-available FDA-approved surfactant and membrane-stabilizer P188. Treatment of G93ASOD1 transgenic mice with P188 significantly delayed symptoms onset, extended survival and decreased motoneuron death. The use of P188 or an analogue, which targets mtSOD1 misfolding-induced membrane toxicity, may provide a new direction for ALS treatment.
Keywords: ALS, P188, F68, poloxamer, protein misfolding, superoxide dismutase, SOD1, G93A, atomic force microscopy, AFM, electrophysiology, membrane toxicity, lipid peroxidation
Approximately 5–10% of cases of amyotrophic lateral sclerosis (ALS) are inherited via more than 40 associated genes, and ~20% of these familial A LS (FALS) cases are caused by over 160 different mutations of superoxide dismutase 1 (SOD1) (Peters, Ghasemi, & Brown, 2015; Renton, Chiò, & Traynor, 2014; Taylor, Brown Jr, & Cleveland, 2016). Mutant (mt) SOD1 is thought to cause FALS because of a gain in toxicity rather than a loss in enzyme activity (Borchelt et al., 1994). Although a specific molecular mechanism underlying mtSOD1 toxicity has remained elusive, misfolding of the mutant protein has been hypothesized as being critical. A number of factors lead to the misfolding of mtSOD1, including certain mutations, metal deficiency (Bourassa, Brown, Borchelt, Vogt, & Miller, 2014; Galaleldeen et al., 2009; Hayward et al., 2002) and oxidation (Barber, Mead, & Shaw, 2006; Kabashi, Valdmanis, Dion, & Rouleau, 2007); these factors may also lead to misfolding of wild-type SOD1, which has been described in sporadic ALS (Ezzi, Urushitani, & Julien, 2007; Rotunno & Bosco, 2015).
We previously reported that mtSOD1 misfolding enables the protein to form a tetrameric channel-like structure in lipid bilayers that can have “toxic channel” activity, leading to motoneuronal death (Allen et al., 2012). We now show that misfolded SOD1 specifically targets lipid bilayer domains composed of phosphatidylglycerol (PG). We also find that the amphiphilic tri-block copolymer P188 alleviates this membrane toxicity in vitro and ameliorates disease in G93ASOD1 transgenic mice, a mouse model of ALS. Consistent with our findings, P188 has been shown to loosely interact with certain membrane surfaces while forming an apposed molecular adlayer capable of protecting membranes from physicochemical damage in a variety of cell systems (Lee, 2002; Lee, Canaday, & Hammer, 1993; Shelat, Plant, Wang, Lee, & Marks, 2013). These findings raise the possibility of a new therapeutic direction in which motoneurons in ALS are protected via stabilization of cellular membranes that are vulnerable to toxic SOD1 oligomers.
Materials and Methods
Materials
Expression plasmids harboring human wild-type (WT)-, A4V-, and G93A-SOD1, and the Src homology 3 (SH3) domain of c-Src kinase were employed in the present studies. Rabbit anti-SOD1 antibody was obtained from Enzo Life Sciences (Farmingdale, NY). A conformation-specific anti- G93ASOD1 antibody (C4F6) was a gift from Jean-Pierre Julien (Laval University, Québec). 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (POPG), 1,2 dioleoyl-sn-glycero-3-phosphoglycerol (DOPG); 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) 1,2 dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE); 1,2 dioleoly-sn-glycerol-3-phosphocholine (DOPC); 1,2 dioleoly-sn-glycerol-3-phosphoserine (DOPS); 1,2 dioleoly-sn-glycerol-3-phosphatidic acid (DOPA); 1,2 dioleoyl-3-tri-methyl ammonium propane (DOTAP) were obtained from Avanti Polar Lipids (Alabaster, AL). NSC-34 mouse cells were a gift from Dr. Neil Cashman (University of British Columbia). Highly purified P188 was obtained from Maroon Biotech (Chicago, IL). Male B6SJL-Tg(SOD1*G93A)1Gur/J mice were purchased from The Jackson Laboratory (Bar Harbor, ME, USA).
Expression, purification and characterization of human SOD1 proteins
Human wild-type (WT)-, A4V-, and G93A-SOD1 and a control protein, the Src homology 3 (SH3) domain of c-Src kinase, were expressed in E. coli BL21 (DE3), and purified on a Ni-column (Qiagen, Valencia, CA) (Allen et al., 2012; Scholle, Collart, & Kay, 2004). Proteins were desalted using 9K protein concentrators (Pierce, Rockford, IL) and stored in HBS (20 mM Hepes, 100 mM NaCl, pH 7.4) with 2.5% glycerol at −80° C. HBS and all solutions were made with BPC grade water (Sigma Aldrich, St. Louis, MO) unless stated otherwise.
Protein purity was determined by SDS-PAGE followed by Coomassie blue and silver staining, as well as by Western blots immunostained with antibody against WT- or mt-SOD1. The Western blots were developed with horseradish peroxidase (HRP)-linked anti-rabbit IgG (1:2000 dilution) followed by detection with an ECL-Plus detection kit (Amersham, NJ). SOD1 metalation was measured by Perkin Elmer ELAN dynamic reactive cell inductively-coupled mass spectrometry performed using EPA-approved guidelines (method 1638; Brooks Rand Labs, Seattle).
Superoxide dismutase activity of the purified human SOD1 proteins was detected (Supplementary Figure 1). Briefly, 500 or 1000 ng of WT, A4V, and G93A SOD1s and bovine SOD1 were electrophoresed on native 11% acrylamide gels containing nitroblue, followed by tetrazonium/riboflavin incubation in the dark. Activity bands were developed using a standard light box.
AFM
The atomic force microscope (Binnig, Quate, & Gerber, 1986) used was a Multimode AFM equipped with a Nanoscope IIIa controller with extender module or Nanoscope IV controller and E- and J-type scanners. The AFM system was isolated from external vibration by bungee cords with a 0.5–0.75 Hz vertical resonance. AFM images were obtained in fluid tapping mode with NP-STT and NPS silicon nitride (Bruker Nano Surfaces, Santa Barbara, CA) and OTR4 cantilevers (Olympus, Japan). The oxide sharpened probe/cantilevers used had a force constant of ~0.08 N/m and were oscillated between 8.8–9.8 kHz at low amplitude (i.e., several nanometers) to achieve a high resolution fluid tapping mode (HRF-AFM) with resolutions of 1.1 nm – 1. 5 nm lateral and 0.1 nm vertical (Möller, Allen, Elings, Engel, & Müller, 1999). Force curves were generated in fluid contact mode using modified NPG probes (Bruker, Santa Barbara, CA).
Preparation of SOD1 for HRF-AFM
Freshly prepared or thawed 10–100 nM purified SOD1 in HBS was incubated 3 min on freshly cleaved mica at RT, washed, and adsorbate scanned immediately under HBS. In some experiments, 10 μM purified SOD1 protein in HBS was boiled 15 min or incubated in 1 μM hydrogen peroxide for 1 hour at RT and analyzed immediately. Excess unbound protein was removed with 3 washes of HBS, and HRF-AFM scanning performed on fully hydrated, never-dried samples. The diameters of the protein structures were measured at full width half maximum (FWHM) using Nanoscope AFM software (Bruker, Santa Barbara, CA).
SOD1 and P188 adsorption to supported lipid bilayer
Individual phospholipids (POPG, DOPG, POPE, DOPE, DOPC, DOPS, DOTAP) or PG:PE blends mixed at ratios of 1:1, 3:1 and 9:1 (w/w) in chloroform were dried to a lipid cake against the wall of a glass vial under dry argon for 15 minutes. The lipid cake was resuspended at 1 mg/mL in HBS, diluted to 0.1 mg/mL, sonicated for 10–30 minutes and/or put through an extruder 11–15 times to form 400 nm diameter single-lamellar liposomes. The liposomes were adsorbed to a freshly cleaved mica surface in HBS in the presence of 2 mM CaCl2 for 45–80 minutes at 37°C, and via vesicle fusion formed a planar single lipid bilayer against the mica’s basal plane. Excess lipid was removed bythree washes of HBS with 2mM CaCl2. Supported lipid bilayer coverage was determined by AFM before exposing the lipid bilayer to purified SOD1 and/or 10 μM P188 poloxamer in HBS. Following HRF-AFM scanning to locate a bilayer area that was defect-free, several nanograms of the SOD1 proteins were allowed to incubate on the bilayer in a 50 μl total volume for 15 minutes at RT and re-scanned immediately.
While our HRF-AFM studies utilized planar supported bilayers, similar levels of G93ASOD1 binding were observed to the curved lipid bilayer of intact PG-enriched liposomes in free suspension (Supplementary Figure 2). Briefly, 400 nm PG-enriched liposomes were loaded with G93ASOD1 protein over a 1 hr co-incubation in HBS at RT. Isolation of the G93ASOD1:DOPG proteoliposomes from the free G93ASOD1 protein was carried out using 300 kD MWCO dialysis media (Spectrum Labs, Rancho Dominguez, CA) against 2L HBS ON (protein gels were run to show the absence of free protein in the dialyzed fractions). The proteoliposomes were fused onto a AFM mica support forming large planar bilayers and scanned by AFM in HBS.
Nanoprobe fabrication
Gold-coated NPG probes (Bruker AXS, Santa Barbara, CA) were pretreated with UV-ozone for 35 min. Oxidized probes were washed three times with hot ethanol for 30 min then DMSO for 1 min. Probes were then incubated for 48 hours in a solution containing 2 mM biotinylated hexa(ethyleneglycol) undecane thiol (Biotin-PEG-Thiol) (Nanoscience Instruments, Phoenix, AZ) in DMSO under dry argon to form a self-assembled monolayer. Probes were washed 3x with DMSO for 3 min, then washed under a constant stream of ethanol for 20 sec, and dried by a stream of argon. Biotinylated probes were tested by force spectroscopy against an avidin-coated surface. Additionally, probes were characterized by fluorescence after incubating with 100 μg/ml FITC-avidin. Probes for experimentation were prepared as above, but using a 1:1 ratio of Biotin-PEG-Thiol and Hydroxy-PEG-Thiol. Biotinylated probes were rinsed with PBS and incubated in 10 μg/mL avidin for 5 min andwashed 4x in PBS. These probes were tested by force spectroscopy against a biotinylated-G93ASOD1 surface. Avidinylated probes were incubated with 1 μM G93ASOD1 for 5 min and washed 4x in PBS. Force curves of these probes were determined on a 10:1 POPG:POPE supported lipid bilayer in PBS.
Single bilayer electrophysiology (SB-EP)
A vertical single bilayer set-up with a Delrin pore 100–200 μm in diameter (Warner Instruments, Hamden, CT) was used as described previously (Allen et al., 2012). SOD1 samples were added to the cis side of the bilayers to a final concentration of 1 μM in HBS pH 7.4, and stirred. Recordings were made in “voltage clamp mode” with Ag/AgCl electrodes. For steady state recordings, membranes were held at −80 mV and for two-pulse recordings 0.5 sec pulses to +/−100 mV were delivered while the bilayer was held at 0 mV for 1.0 sec between pulse sets. Recording solutions were as follows: 150mM KCl in 10 mM Hepes pH 7.4 or SOS (10mM Hepes pH 7.4, 100 mM NaCl, 2mM KCl, 1mM MgCl2, 2mM CaCl2). A filter-cutoff of 3 kHz with sampling at 15 kHz was used. The raw SB-EP recordings are shown (no filtering was applied following acquisition). For the P188 recordings, 10 μM P188 was added to the chamber after bilayer formation, along with the protein, both to the cis side.
mtSOD1 transgenic mice
All animal protocols were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use of the University of Chicago. Forty 55 day old male B6SJL-Tg(SOD1*G93A)1Gur/J mice, which carry a high copy number of the G93ASOD1 transgene, were administered sterile filtered, purified P188 (10 mM in artificial cerebrospinal fluid [aCSF]) via mini-osmotic pumps (Alzet, model 1004, Cupertino, CA, USA) implanted in the lateral cerebral ventricle, starting at either age 57 days (n=9) or age 87 days (n=12) +/−3 days for both. A control group of G93ASOD1 transgenic received artificial cerebrospinal fluid (aCSF) infused via 200 μl mini-osmotic pumps at the same ages (early, n=6; late, n=7). Four additional control mice not receiving surgery showed no clinical and pathological differences with the 13 mice receiving aCSF (data not shown); 2 separate additional mice died on the same day of surgery due to trauma/anesthesia and were not included in the final analysis. The osmotic pump (delivering 0.15 μl/hr) provided 1.5 pM/hr of P188 solution or aCSF for 42 days.
Clinical assessment
The mice were weighed every 2 days and assessed as previously described (Wang et al., 2009). The onset of disease was defined as peak weight before a decline; early phase of disease was the period from peak weight until loss of 10% of maximal weight; and late phase of disease was the time from 10% loss in weight until death. Mortality was scored as the age when a mouse was unable to right itself within 20s after being put on its back in a supine position. In experiments monitoring disease progression and survival, P188-treated mice were compared with aCSF-treated control mice.
Pathology
For motoneuron counting, mice from both groups were sacrificed at end-stage and perfused with ice-cold PBS followed by 4% paraformaldehyde in PBS. The spinal cord was dissected and post-fixed for 4 hours. To compare the number of motoneurons in the spinal cord, Nissl-stained large cells on frozen sections of the lumbar spinal cord were counted from at least 20 sections from each of 4 mice in each group, as previously published (Wang et al., 2008). To assess SOD1 aggregation,4 similarly aged non-transgenic, aCSF-treated, and P188-treated end-stage mice were sacrificed and at least 20 spinal cord sections were assessed microscopically as previously described (Wang et al., 2009). Briefly, sacrificed mice were perfused with ice-cold PBS followed by 4% paraformaldehyde in PBS. The spinal cord was dissected and post-fixed for 4 hours. Paraffin frozen sections were stained immunohistochemically using a rabbit antibody that recognizes both the carboxyl end of mouse and human SOD1 (Deng et al., 2006).
Measurement of lipoperoxides
Lipoperoxides in frozen (−80° C) spinal cords harvested from mice treated with P188 or aCSF were measured directly using redox reactions with ferrous ions in chloroform, using a quantitative assay (Cayman Chemical, Ann Arbor, MI). Briefly, lipoperoxides were extracted from samples in ice cold chloroform and methanol. Triplicate samples and lipoperoxide standards were incubated in ferrous sulfate and ammonium thiocyanate solutions and transferred to a glass 96-well plate. Absorbance at 500 nm was read using a 96-well plate reader. Standard curves were constructed for each run, and hydroperoxide values of the samples for that run were calculated by linear regression.
Results
Metalation and ultrastructure of small soluble mtSOD1 oligomers
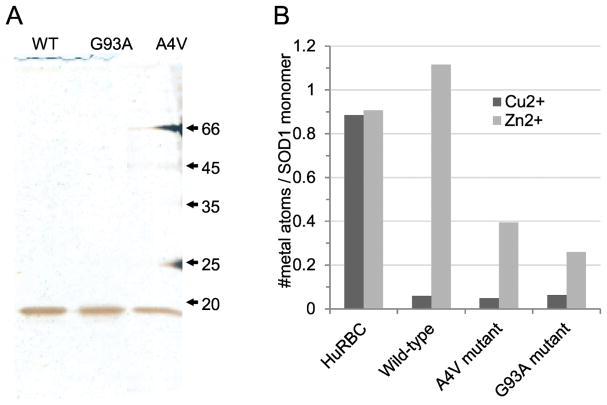
We compared the metalation of bacterially-expressed and purified human wild-type (WT), A4V and G93A mtSOD1 with control human erythrocyte SOD1 (HuRBC) (Figure 1). Consistent with proper folding, the control erythrocyte SOD1 monomer bound ~1 copper atom and ~1 atom of zinc. While WTSOD1 retained full zinc occupancy, the A4V- and G93ASOD1 exhibited binding deficiencies of ~60% and ~70%, respectively. Only 7% of the WT- and mt-SOD1 copper sites were occupied compared to control human erythrocyte SOD1 (Figure 1B), an undermetalated state not dissimilar to that found with mtSOD1s (Bourassa et al., 2014; Pratt et al., 2014; Shaw et al., 2008). All three bacterially-expressed SOD1s showed superoxide dismutase activity (Supplementary Figure 2), presumably because a portion of each of the SOD1s had enzymatic activity.
Figure 1.
(A) Silver-stained SDS-PAGE gel of the purified recombinant human SOD1s: WT, wild type SOD1; G93A, G93ASOD1; A4V, A4VSOD1 (mobility in kD). (B) Inductively-coupled plasma mass spectrometric copper and zinc metal analyses per SOD1 monomer of SOD1 and human erythrocyte SOD1 control, HuRBC.
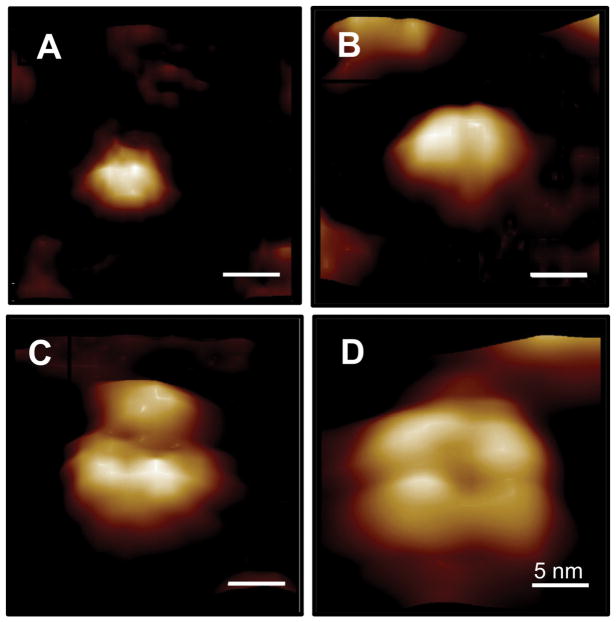
We previously showed that A4VSOD1, but not WTSOD1, formed tetrameric structures that functioned as ”toxic channels” when reconstituted in lipid membrane and also facilitated calcium uptake in motoneuron-like cells (Allen et al., 2012). Here, using high resolution fluid (HRF)-AFM, we first resolved 4 different size classes representing the small soluble oligomeric species of lipid-free G93ASOD1 (Figure 2): 4 nm monomer, 6–7 nm dimer, 8 nm tri-lobe, and 12–15 nm tetramer (Figures 2A–2D, respectively). Under native conditions, for all recombinant and control SOD1s, the dimer was the predominant species observed in HRF-AFM scans, approaching 100%. Monomer and tri-lobed structures were observed as the dominant molecular species, approaching 100%, following boiling and peroxide treatments, respectively. Tetramers were identified in approximately 1 in 50,000 structures scanned in spreads of peroxide-treated G93SOD1, and, in approximately 1 in 100,000 structures of untreated G93ASOD1. The non-native tri-lobed and tetrameric structuring likely result from large conformational changes due to peroxide-induced further decrease in zinc occupancy to below 20%, which is known to destabilize structural integrity (Strange et al., 2003). The determination of these oligomeric size classes for lipid-free G93ASOD1 was only used to assist later identification of membrane-bound G93ASOD1 species.
Figure 2.
HRF-AFM molecular diameters and topographies of the small soluble oligomeric species of lipid-free G93ASOD1 in HBS buffer pH 7.4. (A) monomer (4.1 ± 0.14 nm, n=20), (B) dimer (6.8 ± 0.15 nm, n=20), (C) tri-lobe (8.1 ± 0.32 nm, n=20), (D) tetramer (13.6 ± 0.31 nm, n=7). Full scale z-axis, 4 nm. Bar, 5 nm.
HRF-AFM scans of free G93ASOD1 occasionally revealed assemblies of 10 nm wide curving filaments 4 or more microns in contour length, similar to a previous report (Ivanova et al., 2014). Additionally, large diameter insoluble amorphous aggregates of G93ASOD1 (typically ranging from 50 nm in diameter to micron-sized clumps) were observed after incubation of the free protein for 24–72 hr in buffer at either RT or 37 °C.
Misfolded tetramer-sized G93ASOD1 oligomers form preferentially in PG bilayers
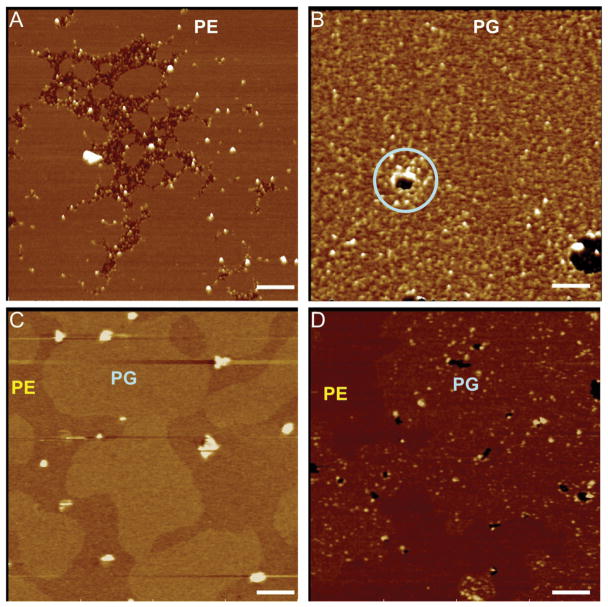
Using supported bilayers as binding targets, we asked whether untreated G93ASOD1 possessed the ability to bind physiologically relevant phospholipids. Starting with a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) bilayer, G93ASOD1 particles were observed only between unfused bilayers (Figure 3A), indicating that G93ASOD1 did not bind to POPE. In marked contrast, we observed dense binding of G93ASOD1 particles on 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (POPG) bilayers, demonstrating a specific binding of G93ASOD1 to POPG (Figure 3B). Notably, G93ASOD1 binding had also induced point defects (holes 10 nm – 200 nm in diameter) not present prior to exposure to G93ASOD1 (lt. blue circle in Figure 3B, also apparent within PG domains of 3D). Similar binding to PG bilayers occurred upon A4VSOD1 exposure, but not with WTSOD1 (Supplementary Figure 3).
Figure 3.
HRF-AFM shows small soluble oligomers of G93ASOD1 bind and damage bilayers composed of PG head groups, but not those of PE. Scans were performed before and after a 100 nM solution of G93ASOD1 protein was incubated 15 min atop the bilayers: (A) PE, where G93ASOD1 binding is seen only between unfused PE bilayers; (B) PG, where G93ASOD1 binds readily and locally damages the membrane (lt. blue circle); (C) 9:1 (w/w) PG:PE mixed bilayer, where no protein has been added, a 5 Å height difference exists between the two bilayers; (D) 9:1 (w/w) PG:PE mixed bilayer, where 100 nM G93ASOD1 protein is added and selectively binds PG subdomains. Full scale z-axis, 10 nm. Bar, 200 nm.
To obtain further evidence that G93ASOD1 binds selectively to PG, we exploited the 0.5 nm height difference between phosphoglycerol (PG) and phosphoethanolamine (PE) head groups in mixed bilayers (Picas et al., 2009). Thus, in mixed bilayers of POPG:POPE with a 9:1 (w/w) blend, the 0.5 nm height difference between these lipids clearly distinguishes POPG from POPE domains (Figure 3C). After exposing both subdomains to G93ASOD1, binding was only seen to the PG subdomain surface (Figure 3D).
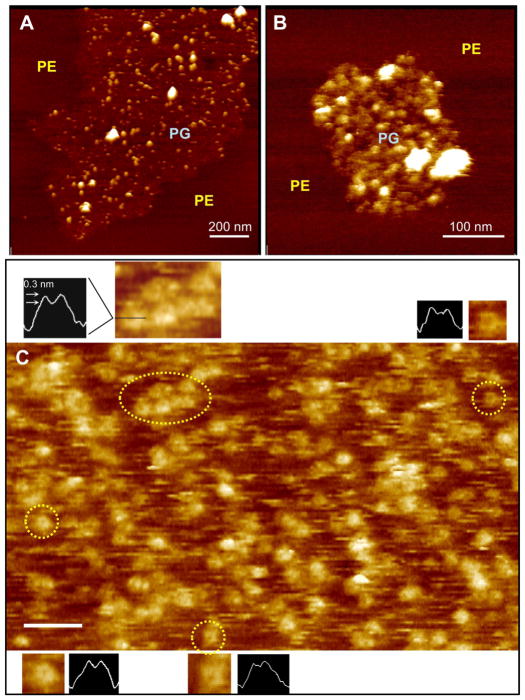
G93ASOD1 binding was then studied on small lipid raft-like PG subdomains surrounded by PE. On this bilayer, individual G93ASOD1 molecules dot the PG subdomain surface (Figures 4A, 4B). The diameters of these smaller G93ASOD1 structures were consistently 12–15 nm, corresponding most closely to the size of tetramers seen in Figure 2. Using higher magnification HRF-AFM scans, a number of “open” channel-like structures, each bearing a small central depression or pore, could be resolved dotting the DOPG bilayer surface following adsorption of G93ASOD1 (Figure 4C). These “open” channel-like structures exhibited diminished protrusion heights compared to the brighter tetramer-sized structures or lipid-free G93ASOD1 (1.3 ± 0.05 nm, n= 15 vs.2.6 ± 0.08 nm, n= 15, respectively) consistent with insertion of portions of the protein into the PG bilayer.
Figure 4.
HRF-AFM of untreated G93ASOD1 bound to PG membrane reveals tetramer-sized assemblies, in some cases, assuming “open” channel-like conformations. (A,B) Lipid raft-like PG subdomains surrounded by PE with selectively bound untreated G93ASOD1 molecules of a size consistent with tetrameric structuring (12–15 nm). The membrane-bound G93ASOD1 oligomers (diameter of smaller white bumps 13.5 nm ± 0.4, n=20) co-adsorbed with larger aggregates (larger brighter structures) bound only to the PG subdomains. (C) High magnification HRF-AFM scan reveals tetramer-sized G93ASOD1 bound to DOPG bilayer in aqueous buffer. The locations representing four pairs of figure insets enlarged to show more detail, including cross sectional line traces through the centers of each structure (dotted yellow circles, insets, and black horizontal line in top left inset). Note the small circular darkened areas or depressions at the centers of many of the tetramer-sized structures (insets; approximately 5–8 nm across, 0.3 nm deep) indicating “open” channel-like structures that have inserted into the membrane. Full scale z-axis, 5 nm. Bar, 50 nm.
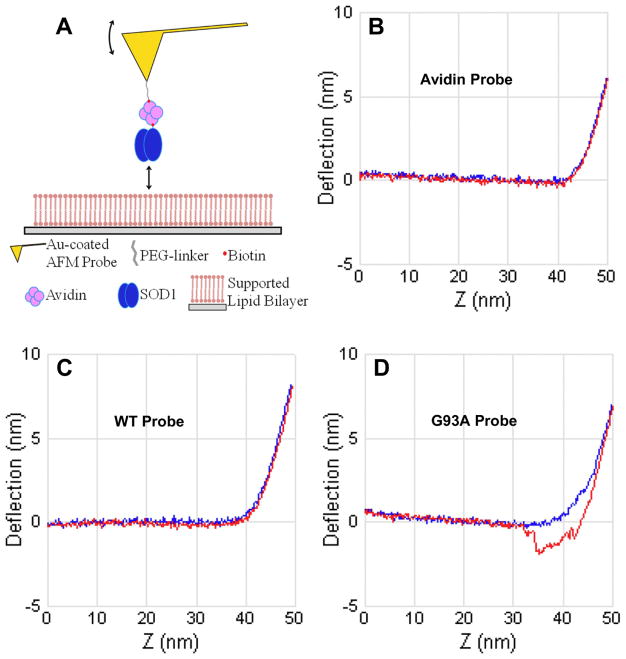
To further characterize the nature of the binding interaction between G93ASOD1 and PG bilayer, single molecule interfacial force spectroscopy was performed using gold nanoprobes coated with WT-, G93A-SOD1, or avidin molecules to compare the adherence of these different molecules to the PG bilayer (Figure 5A). Nanoprobes coated with WTSOD1 consistently showed little to no adhesion force when pulled from the PG membrane surface (Figures 5B, 5C). Similar results were observed when we coated the nanoprobes with avidin as a negative control. In contrast, nanoprobes coated with G93ASOD1 demonstrated an adhesion force of ~100 pN when pulled from the PG bilayer (Figure 5D). These observations independently confirm the specificity of G93ASOD1 binding to PG-bilayer compared to WTSOD1, consistent with our HRF-AFM results.
Figure 5.
Single-molecule interfacial force spectroscopy measures the nanoscale unbinding force from a PG membrane surface of G93ASOD1 compared to WTSOD1. (A) Schematic of the fabricated nanoprobe. To measure unbinding, the nanoprobe is extended into controlled soft contact with the PG bilayer surface and the bending of the integrated microcantilever is monitored during controlled withdrawal. Forces down to ~50 pN can be measured with the HRF-AFM readout used. (B–D) Deflection (nm) vs Z-travel (nm) plots show the nanoprobe extension (from left to right) to the PG bilayer surface (horizontal blue line). When the nanoprobe contacts the bilayer surface the cantilever deflects upward (inflection point in blue line moves upward at right in plots). The nanoprobe is then withdrawn from the bilayer and the adhesion force, if any, is recorded (red line). Only the G93ASOD1 nanoprobe showed significant adhesion to the PG bilayer surface (as seen in D), corresponding to a force of ~100 pN. Data representative of three separate experiments and >1000 force scans for each plot shown.
P188 rescues PG-membrane from G93A- and A4V-SOD1 selective targeting in vitro
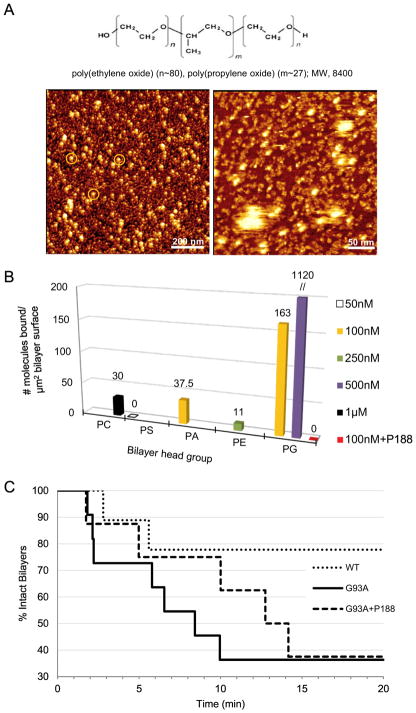
The triblock co-polymer P188 interacts with lipid bilayers and cellular membranes and restores membrane integrity following induced damage. We therefore tested whether P188 could partially rescue PG-bilayer from G93ASOD1 targeting (Figures 6, 7). We preliminarily characterized P188 using HRF-AFM. Figure 6A shows scans of protein- and lipid-free preparations of P188 molecules in micellar and worm-like assemblies (see Supplementary Figure 4). In addition, we performed a series of HRF-AFM measurements showing the binding of untreated G93ASOD1 to a panel of phospholipid bilayers differing in head group: phosphocholine (PC), phosphoserine (PS), phosphatidic acid (PA), (PE), or (PG), following a 15 min incubation period. Topographic scans taken on 5 different planar lipid bilayers before and after timed incubation with various concentrations of G93ASOD1 protein allowed a count of the protein molecules bound per unit surface area for each bilayer type (Figure 6B; see Supplementary Figure 5). Here, G93ASOD1 displayed a distinct binding pattern with strong targeting of PG membrane. However, G93ASOD1 binding to PG was completely blocked in the presence of P188 (Figure 6B).
Figure 6.
P188 suppresses G93ASOD1 membrane toxicity. (A, upper) Chemical structure of tri-block copolymer P188. (A, lower) P188 molecules visualized directly using HRF-AFM. (Left image) Micelles each show a compacted hydrophobic core of ~20 nm in diameter (white spots) with a surrounding hydrophilic corona with a halo of ~5 nm (orange circles, left image). (Right image) Close inspection of a smaller area scan reveals a sub-monolayer coating of lumpy 10 nm aggregates, some assembled into higher ordered 10 nm wide twisted worm-like structures (right image) situated between the ~20 nm particles seen in A. P188 was adsorbed from a solution of 1 mM P188 in water and then scanned at RT. Full scale z-axis, 10 nm. (B) Binding assays as measured by HRF-AFM. Solutions of G93ASOD1 were incubated against each of 5 planar lipid bilayers composed of differing head groups. Binding of G93ASOD1 to PG is seen, but relatively little to PC, PS, PA or PE membranes. When 10 μM P188 is added to the assay, G93ASOD1 is no longer able to bind PG membrane (red square). Multiple areas across the bilayer surfaces were sampled to confirm representative binding. WTSOD1, the control Src homology 3 (SH3) domain of c-Src kinase, and avidin proteins showed little to no binding against the bilayers. (C) Single bilayer electrophysiology (SB-EP) shows that P188 suppresses PG-selective G93ASOD1 membrane toxicity. After ~10 min, G93ASOD1 reached its maximum toxicity on the PG-enriched reporting membrane (7 of 11 reporting membranes were ruptured, solid line). In contrast, membrane rupture was suppressed by a factor of ~2 (only 3 of 11 reporting membranes were ruptured, dashed line) in the presence of P188. In separate control experiments, 100% of the PG:PE reporting membranes that were stable for 2–3 min after formation maintained their stability over a total of ~20 min in buffer or P188 solution.
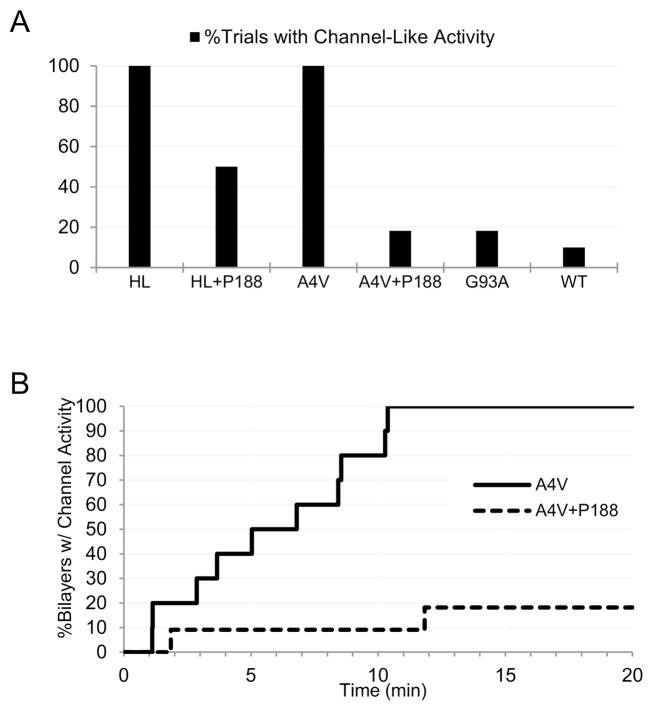
Figure 7.
P188 suppresses A4VSOD1 membrane toxicity. (A) SB-EP recordings of highly purified A4VSOD1 consistently exhibit “toxic channel” activity (12/12 trials), while the addition of 10 μM P188 reduces this channel activity by ~80% (2/11 trials). Channel activity for hemolysin (HL) was observed in 100% of trials (9/9), and reduced to 50% (3/6) in the presence of P188. (B) By ~10 min post-addition of A4VSOD1, 100% of bilayers under test exhibit “toxic channel” activity (12/12), while only 9% of this activity (1/11) was observed in the presence of P188.
To further characterize this protection of PG membrane by P188, we used single bilayer electrophysiology (SB-EP). Figure 6C shows the summary of a series of SB-EP recordings on ~30 separate PG-enriched reporter bilayers following their exposure to WTSOD1 or G93ASOD1 alone or in the presence of P188. By 10 min, P188 had rescued more than half of the PG membranes that G93SOD1 alone was able to rupture over the same period (presumably via point defects formed in the bilayers). In sum, these results show that a G93ASOD1-mediated membrane toxicity selectively damaging to PG-membranes is reduced in the presence of P188.
Similar to the SB-EP results we published previously (Allen et al., 2012), the ion channel-like activity of A4VSOD1 in PG-enriched bilayer was observed as frequently as that of the positive control, alpha-hemolysin (Figure 7A, see Supplementary Figure 6). When performing the same measurement in the presence of P188, the frequency of A4VSOD1’s channel-like activity was reduced by 80% (Figure 7A). Channel-like activities for G93ASOD1 and WTSOD1 were also observed by SB-EP, however, at frequencies too low to assess the inhibitory effects of P188. Moreover, channel-like activity for A4VSOD1 was observed in SB-EP trials 82% more often than for G93ASOD1, and approximately 200% more often than WTSOD1 (Figure 7A). Using SB-EP we monitored the time that had elapsed from the point of initial exposure of the bilayer to A4VSOD1 to the point at which channel-like events began to occur. All of the bilayers exposed to A4VSOD1 alone exhibited channel-like activity by 10 minutes (Figure 7B). In contrast, the presence of P188 prevented 91% of the bilayers from demonstrating A4VSOD1’s channel-like activity within 10 minutes.
These results indicate a G93ASOD1 channel-like activity only slightly higher than that of WTSOD1. However, as seen in Figure 6C, membrane rupture events occur at a significantly higher rate for G93ASOD1 than WTSOD1. This suggests that G93ASOD1 is significantly more damaging to PG-membrane than WTSOD1. We speculate that rupture events may be occurring to the SB-EP reporter membrane before G93ASOD1 channel-like activity can be observed.
P188 ameliorates G93ASOD1-induced pathology
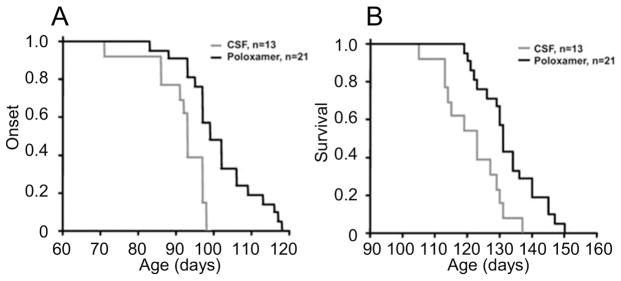
Having found that P188 prevents alterations to PG membrane due to the activity of mtSOD1, we next tested whether P188 treatment could ameliorate the pathology induced in G93ASOD1 transgenic mice (Figure 8). Accordingly, we implanted an osmotic mini-pump to continuously deliver P188 or control aCSF into the cerebral lateral ventricle of G93ASOD1 transgenic mice for 42 days beginning at 57 or 87 days. Disease progression was monitored every two days. P188-treated mice had a mean onset of disease of 101.4 ± 9.4 days (range: 83–118 days) compared with a mean of 91.7 ± 7.4 days (range: 71–98 days) in aCSF-treated mice (P < 0.004) (Figure 8A). Of note, the 4 mice that registered the latest disease onsets in the P188-treated group, these onsets occurred at 113–118 days; while the four mice that registered the latest onsets in the aCSF-treated group, these onsets occurred at 97–98 days. Mean survival was significantly longer in the P188-treated mice (P188: 132.5 ± 9.2 days vs. aCSF: 121.5 ± 9.2 days; P < 0.002) (Figure 8B). Again, of note, the 4 mice that registered the longest survival in the P188-treated group had a survival of 145–150 days, while the same metric in the aCSF-treated group was 129–137 days. Body weight showed no statistical differences at any time point between the aCSF-treated mice and the P188-treated mice (see Supplementary Figure 8).
Figure 8.
Plots of disease onset (A) and survival (B) of P188- vs. aCSF-treated G93ASOD1 mice.
P188 provides protection of G93ASOD1 mouse motoneurons and neuronal membranes
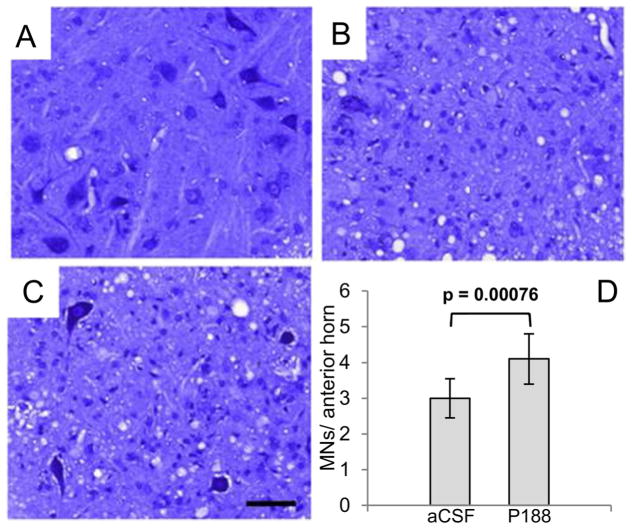
To determine whether P188 treatment altered spinal motoneuron death, motoneurons from the lumbar spinal cord were stained from end-stage G93ASOD1 mice treated with P188, end-stage aCSF-treated mice, and untreated non-transgenic mice (Figures 9A–C). Nissl-stained sections showed the number of motoneurons in aCSF-treated mice was significantly lower than for the P188 treated mice (P < 0.001) indicating P188 protection of motoneurons in the ALS model (Figure 9D). Of note, we observed no differences in SOD1 aggregation levels between aCSF- and P188-treated mice at end-stage (Supplementary Figure 7).
Figure 9.
Motoneuron pathology in aCSF control vs. P188-treated G93ASOD1 end-stage mice. Bar, 50 μm. Representative Nissl-stained sections of lumbar spinal cord of (A) Non-transgenic mice, (B) aCSF-treated G93ASOD1 mice, and (C) P188-treated G93ASOD1 mice. Note extensive loss of motoneurons in B relative to A and C. (D) Mean motoneuron numbers (MNs) from 4 different mice from both the aCSF and P188-treated G93A mice. Mice receiving P188 showed significantly higher numbers of motoneurons at end-stage (P < 0.001). Nissl-sections on healthy adult non-TG mice showed counts of 24 ±3 MNs/anterior horn.
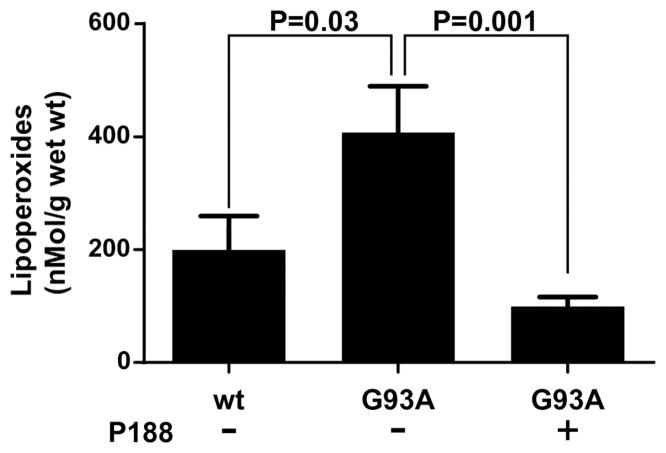
Finally, we investigated whether P188 treatment had stabilized mouse neuronal membranes against SOD1 toxicity via inhibition of lipid peroxidation, which is caused by oxidative stress. The anterior horns of cervical and lumbar spinal cords were isolated from non-transgenic mice, aCSF-treated G93ASOD1 mice, and P188-treated G93ASOD1 mice. Total lipoperoxides were measured in chloroform from deproteinated samples using a highly specific assay utilizing redox reactions with ferrous ions (Figure 10). Mean lipoperoxide content in anterior horns of spinal cords from eight non-transgenic mice was 199.5 ± 60 nMol/g, significantly less than mean lipoperoxide content in spinal cords from aCSF-treated G93ASOD1 mice (407.3 ± 83 nMol/g, P < 0.05). Mean lipoperoxide content in spinal cords from P188-treated G93ASOD1 mice was significantly decreased compared with aCSF-treated G93ASOD1 mice as well as non-transgenic mice (Figure 10). These data indicate that, as expected, aCSF-treated G93ASOD1 mice have increased lipoperoxide accumulation compared to non-transgenic mice. Importantly, however, P188 treatment decreased lipoperoxide accumulation in G93ASOD1 mice.
Figure 10.
Total lipid peroxides measured from spinal cords of non-transgenic mice (wt), a control group of aCSF-treated G93ASOD1 mice (G93A-), and P188-treated G93ASOD1 mice (G93A+).
Discussion
Identified over two decades ago as a central cause of FALS, the nature of the gain-in-function toxicity of mtSOD1 remains unclear. An attractive hypothesis that fits in with proposals concerning toxicity in other neurodegenerative diseases is that misfolding of a cognate protein leads to motoneuron death. In the case of mtSOD1-induced FALS, missense/truncation mutations, undermetalation, and oxidative stress have been implicated in playing a role in motoneuronal toxicity. Our study investigates the toxicity of mtSOD1, and finds evidence that it targets a certain minor lipid membrane moiety.
Our single molecule experiments with misfolded mtSOD1 demonstrate a membrane toxicity, involving non-native oligomers 12–15 nm in diameter, that specifically targets phosphatidylglycerol (PG) domains and disrupts normal membrane integrity. PG is an important target because although it is a trace, negatively-charged phospholipid that comprises ~1% of all membranes in mammals, it exists primarily within mitochondrial membranes as a substrate for cardiolipin synthesis – and is therefore key to subsequent ATP production (Acehan et al., 2011). Furthermore, precursors to PG, such as diacylglycerolphosphate and phosphatidylglycerol phosphate, are known to be present in ER membranes. Of note, ER membranes are physically and functionally networked with mitochondria via mitochondrial-associated membranes which facilitate exchange of lipids (Flis & Daum., 2013; Lahiri et al., 2014; Wu et al., 2017). Therefore, the ER, in addition to mitochondria, would be expected to harbor membrane-binding sites for toxic SOD1. Moreover, pertinent to our finding of a PG-selective membrane toxicity, the ER and mitochondria are sites that have been emphasized as primary targets underlying ALS pathophysiology (Beal, 2000; Borthwick, Johnson, Ince, Shaw, & Turnbull, 1999; Higgins, Jung, & Xu, 2003; Jaarsma et al., 2000; Kong & Xu, 1998; Liu et al., 2004; Manfredi & Kawamata, 2016; Paillusson et al., 2016; Tan, Pasinelli, & Trotti, 2014; Velde, Miller, Cashman, & Cleveland, 2008; Watanabe et al., 2016). Furthermore, binding interactions of toxic SOD1 with outer mitochondrial membrane (Velde, Miller, Cashman, & Cleveland, 2008) and the ER membrane (Sun et al., 2015) have been reported. In addition, integral insertion of undermetalated SOD1 into phosphatidylcholine micelles has been reported (Lim et al., 2015; Lim et al., 2016) as well as transmembrane insertion into PG-enriched bilayers forming “toxic channels” (Allen et al., 2012). Additionally pertinent to our finding of membrane toxicity, alteration to motoneuron electrophysiology has been reported in the ALS literature (Choi et al., 2011; Kuwabara & Kanai, 2007; Pieri et al., 2003; Ruffoli, Bartalucci, Frati, & Fornai, 2015). Regarding our in vitro observations of small “islands” of toxic G93ASOD1 complexed with PG-membrane resembling lipid rafts; in vivo such structures could diffuse laterally across intracellular membrane systems and also via vesicles extracellularly, leading to spread of membrane abnormalities to the cell membrane and also surrounding cells.
Generally, mtSOD1s exhibit increased hydrophobicity compared with native SOD1 (Hayward et al., 2002). While this observation may explain the propensity of mtSOD1 to aggregate, it may also underlie the acquired ability to interact with lipids, as we have observed. Regarding initial mtSOD1 interaction with the bilayer surface, this likely involves bonding between mtSOD1’s positively-charged N-terminal domain and negatively-charged PG headgroups. As to events that follow surface binding, the NMR-based results of Lim et al., 2015 depicting SOD1 insertion into micelles provide an intriguing support for our observations of mtSOD1 insertion into PG bilayer. Wherein, the adjacent N-terminal domain of SOD1 containing β-barrel strands 1–4 “flips” into the bilayer’s hydrophobic core and is stabilized by a conformational change to α-helical domains that expose hydrophobic residues now spanning the core environment. Importantly, the long unstructured middle loop, containing the unoccupied zinc binding domain, would protrude up and away from the bilayer surface into the aqueous milieu. As for the C-terminus, β-strands 6,7 undergo the same “flip”/conformational change to α-helices stable within the hydrophobic core. The result of these insertion events is an extramembraneous disordered middle loop flanked by two anchoring α-helical domains. Our results suggest that cooperative interactions between monomers within a tetrameric assembly further stabilize insertion and provide for the passage of ions.
The finding of membrane toxicity by mtSOD1 led us to consider a membrane-stabilizing agent as a new therapeutic approach to ALS. We tested whether such an intervention could decrease mtSOD1 membrane toxicity in vitro and ameliorate disease in the G93ASOD1 mouse. We focused attention on the tri-block copolymer P188 since it had been shown to possess membrane-stabilizing and -resealing properties (Lee, 2002; Lee et al., 1993; Shelat et al., 2013). By HRF-AFM and SB-EP, we found that P188 was indeed able to block the binding of G93ASOD1 to PG membrane, and to also inhibit membrane rupture and “toxic channel” activity of G93ASOD1 and A4VSOD1. Furthermore, we administered P188 to G93ASOD1 transgenic mice and found significant disease amelioration and motoneuron protection. Given the cellular uptake of P188 (Gigout, Buschmann, & Jolicoeur, 2008) and the large number of poloxameric structures that can be precisely synthesized, a structural analogue to P188 may significantly improve treatment efficacy.
Our finding that mtSOD1 acts as a small diffusible membrane toxin may partly explain why motoneurons and oligodendrocytes are preferentially killed in FALS and ALS. The long narrow tubular shapes of motoneuronal axons and dendrites and long myelin membranes of oligodendrocytes contrast dramatically with other less complex neural cell types that possess far less membrane. These shape differences alone increase the probability of a small soluble toxin coming into contact with a cellular membrane. Although additional factors are likely involved, simple mass action could represent an important driver of selective motoneuronal loss and oligodendrocyte death. For example, the distance from the center of the motoneuronal axon along its short axis to the outer cell membrane (i.e., its cross-sectional radius) is 6–7 times shorter than that of a typical spheroid-shaped mammalian cell. Since diffusion time scales quadratically with distance, the axonal cell membrane would experience ~42 times more toxin collisions than the typical cell membrane over unit time. As such, increased vulnerability to membrane toxicity could contribute to the selectivity of motoneuronal loss.
Our results of lipoperoxide measures in the G93ASOD1 mice are consistent with the finding that P188 protects membranes against attack by reactive-oxygen species (ROS) (Marks, Pan et al. 2001). Although our in vitro studies showed that P188 reduces G93ASOD1 membrane toxicity, we were unable to determine the effect of controlled levels of lipid oxidation on this toxicity. However, since lipid oxidation results in the degradation of lipid bilayer structural integrity (Wong-Ekkabut et al., 2007), it is quite possible that both oxidative stress and SOD1-mediated membrane toxicity act in concert in the G93ASOD1 mouse.
We and others have shown that P188 enters neurons and protects PG-membranes from mtSOD1 toxicity in vitro and neuronal membranes from lipid peroxidation in vivo. Therefore, it is not unlikely that these same P188 actions provide protection to mitochondrial, ER and other cellular membranes in G93ASOD1 mouse motoneurons. Moreover, absent intervention by P188, the mtSOD1-mediated membrane toxicity reported here would be expected to increase superoxide release into the cytosol with resultant oxidative stress and lipid peroxidation. Based primarily on (1) our previous results showing a rise of intracellular calcium ions in live motoneuron-like cells exposed to mtSOD1 (Allen et al., 2012) (2) evidence that ALS mutations in SOD1 can increase peroxynitrite production (Parakh, Spencer et al. 2013) and (3) our current results showing high levels of lipid peroxidation in the spinal cords of G93ASOD1 mice; we propose a disease mechanism where mtSOD1-mediated PG-selective membrane toxicity- potentially 40 times more active in motoneurons- results in a calcium-dependent increase in ROS and nitric oxide to produce ONOO- a highly potent ROS. Although we cannot directly link disease amelioration and a decrease in lipoperoxides in the P188-treated G93ASOD1 mouse to the P188-mediated decrease in G93ASOD1 membrane toxicity observed in vitro, it is a striking parallel observation and, at minimum, serves to generate further investigation into how P188 alters mtSOD1 gain-in-function toxicity in models of FALS.
Conclusions
In summary, we report that an FDA-approved non-ionic surfactant, tri-block copolymer P188, reverses SOD1 toxicity in vitro and significantly ameliorates disease in the most commonly used ALS model. Our findings raise the possibility that P188 or a close analogue may be effective in FALS patients bearing SOD1 mutations. An attractive feature of P188 is that it is known to be safe when delivered systemically and can efficiently cross the blood-brain-barrier (Gelperina et al., 2010). Since there is evidence that WTSOD1 in sporadic ALS can misfold in a way similar to that seen in mtSOD1-induced FALS (Bosco et al., 2010; Rotunno & Bosco, 2015), a membrane toxicity may also be present in sporadic ALS - and the use of tri-block copolymer treatment may be effective more broadly in ALS.
Supplementary Material
Highlights.
Small soluble oligomers of mutant SOD1 exhibit a selective membrane toxicity
Toxicity involves physical damage to bilayers and “toxic channel” activity
Membrane stabilization with P188 significantly reduces disease in ALS SOD1 mouse
Pathology results show P188 protects motor neurons and reduces lipid peroxides
Acknowledgments
Support: Research reported in this publication was supported by NIH U54 GM087519: Membrane Protein Structural Dynamics Consortium to AMC, National Institute of Neurological Disorders and Stroke of the National Institutes of Health under award numbers R01NS056313 to JDM, and R01NS067247 to MJA. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We are grateful to D. Pfaffinger and V. Bindokas for their technical support, R. Lee for advice regarding the use of tri-block copolymers, and F. Bezanilla for guidance and feedback regarding the single bilayer electrophysiology. Research reported in this publication was supported by NIH U54 GM087519: Membrane Protein Structural Dynamics Consortium to AMC; National Institute of Neurological Disorders and Stroke of the National Institutes of Health under award numbers R01NS056313 to JDM, and R01NS067247 to MJA. This work is dedicated to the memory of Dr. R. S. Allen, D.V.M.
Footnotes
Conflict of Interest: There are no conflicts for the authors regarding any aspect of the work represented in this manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acehan D, Malhotra A, Xu Y, Ren M, Stokes DL, Schlame M. Cardiolipin affects the supramolecular organization of ATP synthase in mitochondria. Biophysical Journal. 2011;100(9):2184–2192. doi: 10.1016/j.bpj.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen MJ, Lacroix JJ, Ramachandran S, Capone R, Whitlock JL, Ghadge GD, … Lal R. Mutant SOD1 forms ion channel: implications for ALS pathophysiology. Neurobiology of Disease. 2012;45(3):831–838. doi: 10.1016/j.nbd.2011.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banquy X, Lee DW, Kristiansen K, Gebbie MA, Israelachvili JN. Interaction forces between supported lipid bilayers in the presence of PEGylated polymers. Biomacromolecules. 2015;17(1):88–97. doi: 10.1021/acs.biomac.5b01216. [DOI] [PubMed] [Google Scholar]
- Barber SC, Mead RJ, Shaw PJ. Oxidative stress in ALS: a mechanism of neurodegeneration and a therapeutic target. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease. 2006;1762(11):1051–1067. doi: 10.1016/j.bbadis.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Beal MF. Mitochondria and the pathogenesis of ALS. Brain. 2000;123(7):1291–1292. doi: 10.1093/brain/123.7.1291. [DOI] [PubMed] [Google Scholar]
- Binnig G, Quate CF, Gerber C. Atomic force microscope. Physical Review Letters. 1986;56(9):930. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- Borchelt DR, Lee MK, Slunt HS, Guarnieri M, Xu ZS, Wong PC, … Cleveland DW. Superoxide dismutase 1 with mutations linked to familial amyotrophic lateral sclerosis possesses significant activity. Proceedings of the National Academy of Sciences. 1994;91(17):8292–8296. doi: 10.1073/pnas.91.17.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borthwick GM, Johnson MA, Ince PG, Shaw PJ, Turnbull DM. Mitochondrial enzyme activity in amyotrophic lateral sclerosis: implications for the role of mitochondria in neuronal cell death. Annals of Neurology. 1999;46(5):787–790. doi: 10.1002/1531-8249(199911)46:5<787::aid-ana17>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Bosco DA, Morfini G, Karabacak NM, Song Y, Gros-Louis F, Pasinelli P, … McKenna-Yasek D. Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nature Neuroscience. 2010;13(11):1396–1403. doi: 10.1038/nn.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourassa MW, Brown HH, Borchelt DR, Vogt S, Miller LM. Metal-deficient aggregates and diminished copper found in cells expressing SOD1 mutations that cause ALS. Frontiers in Aging Neuroscience. 2014;6:110. doi: 10.3389/fnagi.2014.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotherton TE, Li Y, Cooper D, Gearing M, Julien JP, Rothstein JD, … Glass JD. Localization of a toxic form of superoxide dismutase 1 protein to pathologically affected tissues in familial ALS. Proceedings of the National Academy of Sciences. 2012;109(14):5505–5510. doi: 10.1073/pnas.1115009109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chng CP, Strange RW. Lipid-associated aggregate formation of superoxide dismutase-1 is initiated by membrane-targeting loops. Proteins: Structure, Function, and Bioinformatics. 2014;82(11):3194–3209. doi: 10.1002/prot.24688. [DOI] [PubMed] [Google Scholar]
- Choi I, Song HD, Lee S, Yang YI, Nam JH, Kim SJ, … Yi J. Direct observation of defects and increased ion permeability of a membrane induced by structurally disordered Cu/Zn-superoxide dismutase aggregates. PloS One. 2011;6(12):e28982. doi: 10.1371/journal.pone.0028982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng HX, Shi Y, Furukawa Y, Zhai H, Fu R, Liu E, … Bigio EH. Conversionto the amyotrophic lateral sclerosis phenotype is associated with intermolecular linked insoluble aggregates of SOD1 in mitochondria. Proceedings of the National Academy of Sciences. 2006;103(18):7142–7147. doi: 10.1073/pnas.0602046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzi SA, Urushitani M, Julien JP. Wild-type superoxide dismutase acquires binding and toxic properties of ALS-linked mutant forms through oxidation. Journal of Neurochemistry. 2007;102(1):170–178. doi: 10.1111/j.1471-4159.2007.04531.x. [DOI] [PubMed] [Google Scholar]
- Flis VV, Daum G. Lipid transport between the endoplasmic reticulum and mitochondria. Cold Spring Harbor Perspectives in Biology. 2013;5(6):a013235. doi: 10.1101/cshperspect.a013235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa Y, Anzai I, Akiyama S, Imai M, Cruz FJC, Saio T, … Ishimori K. Conformational disorder of the most immature Cu, Zn-superoxide dismutase leading to amyotrophic lateral sclerosis. Journal of Biological Chemistry. 2016;291(8):4144–4155. doi: 10.1074/jbc.M115.683763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galaleldeen A, Strange RW, Whitson LJ, Antonyuk SV, Narayana N, Taylor AB, … Hart PJ. Structural and biophysical properties of metal-free pathogenic SOD1 mutants A4V and G93A. Archives of Biochemistry and Biophysics. 2009;492(1):40–47. doi: 10.1016/j.abb.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelperina S, Maksimenko O, Khalansky A, Vanchugova L, Shipulo E, Abbasova K, … Kreuter J. Drug delivery to the brain using surfactant-coated poly (lactide-co-glycolide) nanoparticles: influence of the formulation parameters. European Journal of Pharmaceutics and Biopharmaceutics. 2010;74(2):157–163. doi: 10.1016/j.ejpb.2009.09.003. [DOI] [PubMed] [Google Scholar]
- Gigout A, Buschmann MD, Jolicoeur M. The fate of Pluronic F-68 in chondrocytes and CHO cells. Biotechnology and Bioengineering. 2008;100(5):975–987. doi: 10.1002/bit.21840. [DOI] [PubMed] [Google Scholar]
- Hayward LJ, Rodriguez JA, Kim JW, Tiwari A, Goto JJ, Cabelli DE, … Brown RH. Decreased metallation and activity in subsets of mutant superoxide dismutases associated with familial amyotrophic lateral sclerosis. Journal of Biological Chemistry. 2002;277(18):15923–15931. doi: 10.1074/jbc.M112087200. [DOI] [PubMed] [Google Scholar]
- Higgins CM, Jung C, Xu Z. ALS-associated mutant SOD1 G93A causes mitochondrial vacuolation by expansion of the intermembrane space and by involvement of SOD1 aggregation and peroxisomes. BMC Neuroscience. 2003;4(1):16. doi: 10.1186/1471-2202-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova MI, Sievers SA, Guenther EL, Johnson LM, Winkler DD, Galaleldeen A, … Eisenberg DS. Aggregation-triggering segments of SOD1 fibril formation support a common pathway for familial and sporadic ALS. Proceedings of the National Academy of Sciences. 2014;111(1):197–201. doi: 10.1073/pnas.1320786110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaarsma D, Haasdijk ED, Grashorn JAC, Hawkins R, van Duijn W, Verspaget HW, … Holstege JC. Human Cu/Zn superoxide dismutase (SOD1) overexpression in mice causes mitochondrial vacuolization, axonal degeneration, and premature motoneuron death and accelerates motoneuron disease in mice expressing a familial amyotrophic lateral sclerosis mutant SOD1. Neurobiology of Disease. 2000;7(6):623–643. doi: 10.1006/nbdi.2000.0299. [DOI] [PubMed] [Google Scholar]
- Kabashi E, Valdmanis PN, Dion P, Rouleau GA. Oxidized/misfolded superoxide dismutase-1: the cause of all amyotrophic lateral sclerosis? Annals of Neurology. 2007;62(6):553–559. doi: 10.1002/ana.21319. [DOI] [PubMed] [Google Scholar]
- Kong J, Xu Z. Massive mitochondrial degeneration in motor neurons triggers the onset of amyotrophic lateral sclerosis in mice expressing a mutant SOD1. Journal of Neuroscience. 1998;18(9):3241–3250. doi: 10.1523/JNEUROSCI.18-09-03241.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara S, Kanai K. Altered axonal ion channel function in amyotrophic lateral sclerosis. Brain and Nerve= Shinkei Kenkyu No Shinpo. 2007;59(10):1109–1115. [PubMed] [Google Scholar]
- Lahiri S, Chao JT, Tavassoli S, Wong AK, Choudhary V, Young BP, … Prinz WA. A conserved endoplasmic reticulum membrane protein complex (EMC) facilitates phospholipid transfer from the ER to mitochondria. PLoS biology. 2014;12(10):e1001969. doi: 10.1371/journal.pbio.1001969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC. Cytoprotection by stabilization of cell membranes. Annals of the New York Academy of Sciences. 2002;961(1):271–275. doi: 10.1111/j.1749-6632.2002.tb03100.x. [DOI] [PubMed] [Google Scholar]
- Lee RC, Canaday DJ, Hammer SM. Transient and stable ionic permeabilization of isolated skeletal muscle cells after electrical shock. Journal of Burn Care & Research. 1993;14(5):528–540. doi: 10.1097/00004630-199309000-00007. [DOI] [PubMed] [Google Scholar]
- Lim L, Lee X, Song J. Mechanism for transforming cytosolic SOD1 into integral membrane proteins of organelles by ALS-causing mutations. Biochimica et Biophysica Acta (BBA)-Biomembranes. 2015;1848(1):1–7. doi: 10.1016/j.bbamem.2014.10.002. [DOI] [PubMed] [Google Scholar]
- Lim L, Song J. SALS-linked WT-SOD1 adopts a highly similar helical conformation as FALS-causing L126Z-SOD1 in a membrane environment. Biochimica et Biophysica Acta (BBA)-Biomembranes. 2016;1858(9):2223–2230. doi: 10.1016/j.bbamem.2016.06.027. [DOI] [PubMed] [Google Scholar]
- Liu J, Lillo C, Jonsson PA, Velde CV, Ward CM, Miller TM, … Andersen PM. Toxicity of familial ALS-linked SOD1 mutants from selective recruitment to spinal mitochondria. Neuron. 2004;43(1):5–17. doi: 10.1016/j.neuron.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Manfredi G, Kawamata H. Mitochondria and endoplasmic reticulum crosstalk in amyotrophic lateral sclerosis. Neurobiology of Disease. 2016;90:35–42. doi: 10.1016/j.nbd.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks JD, Pan CY, Bushell T, Cromie C, Lee RC. Amphiphilic, tri-block copolymers provide potent, membrane-targeted neuroprotection. FASEB Journal Express Article. 2001 doi: 10.1096/fj.00-0547fje. [DOI] [PubMed] [Google Scholar]
- Möller C, Allen M, Elings V, Engel A, Müller DJ. Tapping-mode atomic force microscopy produces faithful high-resolution images of protein surfaces. Biophysical Journal. 1999;77(2):1150–1158. doi: 10.1016/S0006-3495(99)76966-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillusson S, Stoica R, Gomez-Suaga P, Lau DH, Mueller S, Miller T, Miller CC. There’s Something Wrong with my MAM; the ER–Mitochondria Axis and Neurodegenerative Diseases. Trends in Neurosciences. 2016;39(3):146–157. doi: 10.1016/j.tins.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parakh S, Spencer DM, Halloran MA, Soo KY, Atkin JD. Redox Regulation in Amyotrophic Lateral Sclerosis. Oxidative Medicine and Cellular Longevity. 2013;2013:12. doi: 10.1155/2013/408681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters OM, Ghasemi M, Brown RH. Emerging mechanisms of molecular pathology in ALS. The Journal of Clinical Investigation. 2015;125(5):1767–1779. doi: 10.1172/JCI71601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picas L, Montero MT, Morros A, Cabanas ME, Seantier B, Milhiet PE, Hernández-Borrell J. Calcium-induced formation of subdomains in phosphatidylethanolamine phosphatidylglycerol bilayers: a combined DSC, 31P NMR, and AFM study. The Journal of Physical Chemistry B. 2009;113(14):4648–4655. doi: 10.1021/jp8102468. [DOI] [PubMed] [Google Scholar]
- Pieri M, Albo F, Gaetti C, Spalloni A, Bengtson CP, Longone P, … Zona C. Altered excitability of motor neurons in a transgenic mouse model of familial amyotrophic lateral sclerosis. Neuroscience Letters. 2003;351(3):153–156. doi: 10.1016/j.neulet.2003.07.010. [DOI] [PubMed] [Google Scholar]
- Pratt AJ, Shin DS, Merz GE, Rambo RP, Lancaster WA, Dyer KN, … Crane BR. Aggregation propensities of superoxide dismutase G93 hotspot mutants mirror ALS clinical phenotypes. Proceedings of the National Academy of Sciences. 2014;111(43):E4568–E4576. doi: 10.1073/pnas.1308531111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratovitski T, Corson LB, Strain J, Wong P, Cleveland DW, Culotta VC, Borchelt DR. Variation in the biochemical/biophysical properties of mutant superoxide dismutase 1 enzymes and the rate of disease progression in familial amyotrophic lateral sclerosis kindreds. Human Molecular Genetics. 1999;8(8):1451–1460. doi: 10.1093/hmg/8.8.1451. [DOI] [PubMed] [Google Scholar]
- Renton AE, Chiò A, Traynor BJ. State of play in amyotrophic lateral sclerosis genetics. Nature Neuroscience. 2014;17(1):17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotunno MS, Bosco DA. An emerging role for misfolded wild-type SOD1 in sporadic ALS pathogenesis. Cellular and Molecular Mechanisms of Motor Neuron Death in Amyotrophic Lateral Sclerosis. 2015:110. [Google Scholar]
- Ruffoli R, Bartalucci A, Frati A, Fornai F. Ultrastructural studies of ALS mitochondria connect altered function and permeability with defects of mitophagy and mitochondriogenesis. Frontiers in Cellular Neuroscience. 2015:9. doi: 10.3389/fncel.2015.00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholle MD, Collart FR, Kay BK. In vivo biotinylated proteins as targets for phage-display selection experiments. Protein Expression and Purification. 2004;37(1):243–252. doi: 10.1016/j.pep.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Shaw BF, Lelie HL, Durazo A, Nersissian AM, Xu G, Chan PK, … Borchelt DR. Detergent-insoluble aggregates associated with amyotrophic lateral sclerosis in transgenic mice contain primarily full-length, unmodified superoxide dismutase-1. Journal of Biological Chemistry. 2008;283(13):8340–8350. doi: 10.1074/jbc.M707751200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelat PB, Plant LD, Wang JC, Lee E, Marks JD. The membrane-active tri-block copolymer pluronic F-68 profoundly rescues rat hippocampal neurons from oxygen–glucose deprivation-induced death through early inhibition of apoptosis. The Journal of Neuroscience. 2013;33(30):12287–12299. doi: 10.1523/JNEUROSCI.5731-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange RW, Antonyuk S, Hough MA, Doucette PA, Rodriguez JA, Hart PJ, … Hasnain SS. The structure of holo and metal-deficient wild-type human Cu, Zn superoxide dismutase and its relevance to familial amyotrophic lateral sclerosis. Journal of Molecular Biology. 2003;328(4):877–891. doi: 10.1016/s0022-2836(03)00355-3. [DOI] [PubMed] [Google Scholar]
- Tan W, Pasinelli P, Trotti D. Role of mitochondria in mutant SOD1 linked amyotrophic lateral sclerosis. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease. 2014;1842(8):1295–1301. doi: 10.1016/j.bbadis.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JP, Brown RH, Jr, Cleveland DW. Decoding ALS: from genes to mechanism. Nature. 2016;539(7628):197–206. doi: 10.1038/nature20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velde CV, Miller TM, Cashman NR, Cleveland DW. Selective association of misfolded ALS-linked mutant SOD1 with the cytoplasmic face of mitochondria. Proceedings of the National Academy of Sciences. 2008;105(10):4022–4027. doi: 10.1073/pnas.0712209105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Deng HX, Grisotti G, Zhai H, Siddique T, Roos RP. Wild-type SOD1 overexpression accelerates disease onset of a G85R SOD1 mouse. Human Molecular Genetics. 2009;18(9):1642–1651. doi: 10.1093/hmg/ddp085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Sharma K, Deng HX, Siddique T, Grisotti G, Liu E, Roos RP. Restricted expression of mutant SOD1 in spinal motor neurons and interneurons induces motor neuron pathology. Neurobiology of disease. 2008;29(3):400–408. doi: 10.1016/j.nbd.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Ilieva H, Tamada H, Nomura H, Komine O, Endo F, … Yamanaka K. Mitochondria-associated membrane collapse is a common pathomechanism in SIGMAR1-and SOD1-linked ALS. EMBO Molecular Medicine. 2016b;8(12):1421–1437. doi: 10.15252/emmm.201606403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong-Ekkabut J, Xu Z, Triampo W, Tang IM, Tieleman DP, Monticelli L. Effect of lipid peroxidation on the properties of lipid bilayers: a molecular dynamics study. Biophysical Journal. 2007;93(12):4225–4236. doi: 10.1529/biophysj.107.112565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Whiteus C, Xu CS, Hayworth KJ, Weinberg RJ, Hess HF, De Camilli P. Contacts between the endoplasmic reticulum and other membranes in neurons. Proceedings of the National Academy of Sciences. 2017;114(24):E4859–E4867. doi: 10.1073/pnas.1701078114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.