Abstract
The α-actinin proteins are a highly conserved family of actin crosslinkers that mediate interactions between several cytoskeletal and sarcomeric proteins. Non-sarcomeric α-actinin-1 and α-actinin-4 crosslink actin filaments in the cytoskeleton, while sarcomeric α-actinin-2 and α-actinin-3 serve a crucial role in anchoring actin filaments to the muscle Z-line. To assess the difference in turnover dynamics and structure/function properties between the α-actinin isoforms at the sarcomeric Z-line, we used Fluorescence Recovery After Photobleaching (FRAP) in primary myofiber cultures. We found that the recovery kinetics of these proteins followed three distinct patterns: α-actinin-2/α-actinin-3 had the slowest turn over, α-actinin-1 recovered to an intermediate degree, and α-actinin-4 had the fastest recovery. Interestingly, the isoforms’ patterns of recovery were reversed at adhesion plaques in fibroblasts. This disparity suggests that the different α-actinin isoforms have unique association kinetics in myofibers and that non-muscle isoform interactions are more dynamic at the sarcomeric Z-line. Protein domain-specific investigations using α-actinin-2/4 chimeric proteins showed that differential dynamics between sarcomeric and non-muscle isoforms are regulated by cooperative interactions between the N-terminal actin-binding domain, the spectrin-like linker region and the C-terminal calmodulin-like EF hand domain. Together, these findings demonstrate that α-actinin isoforms are unique in binding dynamics at the Z-line and suggest differentially evolved interactive and Z-line association capabilities of each functional domain.
Keywords: Fluorescence Recovery After Photobleaching (FRAP), actin, actin-binding, chimeric protein, EF hand
Introduction
α-Actinin, the first actin crosslinking protein discovered by Ebashi in 1964 [Ebashi and Ebashi, 1964; Maruyama and Ebashi, 1965], is now known to be a family of different isoforms expressed by four genes [Beggs et al., 1992; Honda et al., 1998; Youssoufian et al., 1990]. Skeletal muscle expresses two isoforms, α-actinin-2 and α-actinin-3, which are evolutionarily related but functionally distinct [Suzuki et al., 1973]. Two additional isoforms, α-actinin-1 and α-actinin-4, are expressed in non-muscle cells [Burridge and Feramisco, 1981; Duhaiman and Bamburg, 1984]. Studies in the past decade have revealed that in addition to F-actin binding and crosslinking, α-actinins are platforms allowing interactions with a plethora of proteins and thus contribute to the organization and stability of actin-based cell structures and participate in biosensing and the regulation of cellular functions.
The four mammalian α-actinin genes resulted from two gene duplication events [Dixson et al., 2003]. All four isoforms exhibit high levels of identity and similarity in their sequence and structure and play an important role in force generation, however, they have evolved to regulate actin bundles and networks differently depending on tissue-specific modifications and protein-protein interactions in the cells. Mammalian α-actinin-2 and α-actinin-3 evolved to be sarcomeric-specific isoforms that localize to the Z-lines of striated muscle and crosslink F-actin across neighboring sarcomeres, whereas α-actinin-1 and α-actinin-4 became non-muscle isoforms that crosslink F-actin in stress fibers and tether them at the focal adhesions of non-muscle cells. Additional splicing of α-actinin-1 also produces smooth muscle and brain-specific isoforms [Kremerskothen et al., 2002; Waites et al., 1992]. In all cases, the α-actinin proteins are crucial in maintaining the architecture and stability of the cytoskeleton, and compromises in their structure can lead to impaired cell function and various pathologies [Kaplan et al., 2000; Mohapatra et al., 2003; Yao et al., 2004; Chiu et al., 2010; Girolami et al., 2014; Bagnall et al., 2014; Haywood et al., 2016; Kunishima et al., 2013; Gueguen et al., 2013; Yasutomi et al., 2016].
Although the mammalian α-actinins have evolved into four separate isoforms, they are still very similar in structure. All α-actinin isoforms are composed of an N-terminal actin-binding domain (ABD), a C-terminal calmodulin-like EF hand domain, and a linker region with four spectrin-like repeats (SLR). Sequence identity amongst the isoforms ranges from 87–94% in the ABD, 68–83% in the SLR, and 68–85% in the EF hand domains. Functional α-actinin proteins exist as dimers through strong interactions between opposing SLRs, leading to dumbbell-shaped structures with an actin-binding motif on either end. Most molecules are thought to be homodimers, although there is evidence that herterodimers between skeletal muscle isoforms α-actinin-2 and α-actinin-3 may exist [Chan et al., 1998]. This structural design allows the resulting functional ~208 kD unit to crosslink F-actin and also to serve as a platform for additional protein-protein interactions [Otey and Carpen, 2004; Sjöblom et al., 2008].
The N-terminal ABD consists of two tandem calponin homology repeats (i.e., CH1 and CH2), which work synergistically to interact with F-actin [Korenbaum and Rivero, 2002]. The C-terminal CaM-like domain consists of two EF hands, which are helix-loop-helix motifs that bind calcium [Sjöblom et al., 2008]. The most significant divergence between sarcomeric and non-muscle isoforms is found in the EF hand domain. In non-muscle isoforms, calcium binding to this domain regulates the conformation of the ABD and at calcium concentrations greater than 10−7 M the affinity of these isoforms for F-actin is decreased [Burridge and Feramisco, 1981; Duhaiman and Bamburg, 1984; Tang et al., 2001]. In contrast, the sarcomeric isoforms have lost their calcium sensitivity due to several mutated residues predicted to abolish their calcium-binding ability [Moncrief et al., 1990; Beggs et al., 1992; Blanchard et al., 1989].
The central rod domain consists of four tandem SLRs and serves multiple roles, such as a mediator of protein self-dimerization, a linker and ruler for the functional ends of the dimer, and a platform for a wide variety of protein-protein interactions, as well as a source for elasticity along the protein [Djinovic-Carugo et al., 2002]. These characteristics allow for greater structural flexibility and mechanical stability, which is crucial for α-actinin function at focal adhesion points and sarcomeric Z-lines. In addition, the spectrin repeat region has been shown to interact with a variety of proteins and molecules, ranging from adhesion proteins (i.e., integrins, ICAMs, and L-selectin) [Carpen et al., 1992; Heiska et al., 1996; Otey et al., 1990; Pavalko and LaRoche, 1993; Pavalko et al., 1995] to muscle contractile apparatus components (i.e., nebulin, CapZ, and calsarcins) [Faulkner et al., 1999; Frey et al., 2000; Nave et al., 1990; Papa et al., 1999; Takada et al., 2001] to ion channels and neurotransmission proteins (i.e., Kv1.5, Nav1.4/5, NMDA receptor, densin-180, and rabphilin-3A) [Kato et al., 1996; Maruoka et al., 2000; Walikonis et al., 2001; Wyszynski et al., 1997; Ziane et al., 2010]. However, most of the partner proteins interaction sites on α-actinin have not been identified yet and further studies are required to understand the functional properties imparted by each interaction.
Studies have demonstrated that α-actinin isoforms are functionally and structurally similar [Chan et al., 1998] and that expression of only the highly conserved ABD domain is sufficient to correctly localize avian sarcomeric α-actinin in muscle [Wang et al., 2005]. Given that the localizations and interactions of the different isoforms differ based on tissue specificity, if they are artificially expressed in the same environmental conditions, the different isoforms could either function distinctly from each other, show complete redundancy, or exhibit partial redundancy. Despite their similarities, studies have started to parse out whether the α-actinin isoforms are functionally redundant. For example, previous findings indicate that complete redundancy is unlikely among the sarcomeric isoforms as a shift in muscle metabolic profiles was observed in mice where α-actinin-2 was substituted for α-actinin-3 [MacArthur et al., 2008]. Conversely, α-actinin-3 is not able to substitute for or rescue α-actinin-2 defects in Danio rerio [Gupta et al., 2012]. Likewise, studies have shown α-actinin-1 and α-actinin-4 to be similarly non-redundant since α-actinin-1 is not able to rescue the loss of α-actinin-4 [Shao et al., 2010b]. Determining whether the different α-actinins exhibit different properties and which structural features contribute to those differences will help to understand their evolution and specific roles in human disease.
To better understand the differences between the four mammalian α-actinin isoforms, we investigated their functional properties with regard to their ability to target to the skeletal muscle Z-lines and their dynamics in myofibers. More specifically, we examined whether there are differences in turnover dynamics between the sarcomeric isoforms (α-actinin-2 vs. α-actinin-3) as well as between the non-muscle and sarcomeric isoforms. To do this, we used fluorescence recovery after photobleaching (FRAP) in mouse myofibers to compare the rate and extent of protein recovery at the Z-line between α-actinin-1, α-actinin-2, α-actinin-3, and α-actinin-4. Domain-specific investigations between sarcomeric and non-muscle isoforms were done by using chimeric proteins with interchanged ABD, SLR, or EF domains.
Results
α-actinin proteins have differential turnover dynamics at the skeletal muscle Z-line
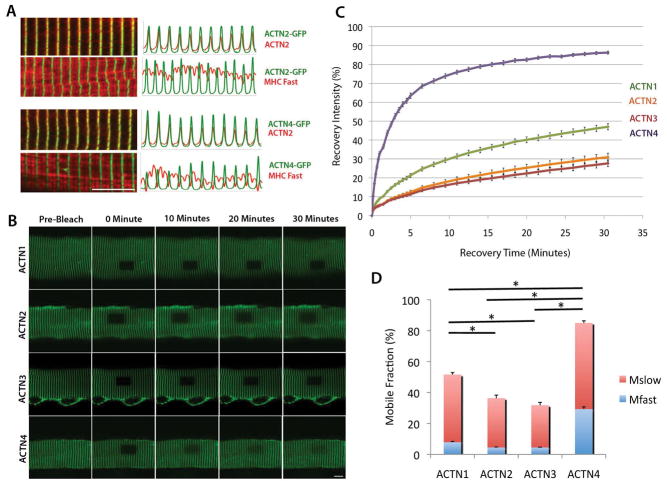
As an initial step in determining the unique properties of muscle α-actinin proteins at the Z-line, their dynamics were compared with those of the non-muscle isoforms. To assess the differential turnover dynamics at the Z-line by each of the four α-actinin isoforms, each human α-actinin gene was cloned and tagged with AcGFP (an altered form of GFP that is less likely to aggregate) and expressed in adult mouse dissociated flexor digitorum brevis (FDB) myofibers via adenoviral infection. Because the fiber type makeup of muscle can alter with age, only mice that were between 1 to 3 months old were used for sample consistency. Immunocytochemistry of infected fibers comparing the relative localization of the GFP-tagged α-actinins with myosin heavy chain (MHC) and endogenous α-actinin-2 revealed that the exogenously expressed GFP-tagged α-actinins localize to the Z-line and its expression pattern matches perfectly the endogenous α-actinin-2 (Figure 1A). This result indicates that the α-actinin proteins, regardless of isoform, are similar enough to the skeletal muscle isoforms to localize to the Z-line and allow for the use of FRAP as a method to assess their differential dynamics.
Figure 1.
Exogenously expressed AcGFP-tagged α-actinin isoforms localize specifically to the myofiber Z-lines but have different turnover dynamics. (A) α-actinin-2-AcGFP and α-actinin-4-AcGFP were expressed in myofibers and samples were co-immunostained with either the α-actinin-2 or myosin heavy chain (MHC, fast-twitch; red) antibodies. Both α-actinin-2 and α-actinin-4 are analyzed for localization in relation to skeletal muscle markers. Confocal images (left) and fluorescence intensity plots (right) indicate that GFP-tagged isoforms co-localized with endogenous α-actinin-2 but alternated with MHC. Scale bar = 10μm (B) Fluorescence recovery of wild type isoforms α-actinin-1, α-actinin-2, α-actinin-3, and α-actinin-4 in myofibers. Representative recovery images show (left to right) pre-bleach and 0, 10, 20, and 30 minutes after photobleaching. Scale bar = 10 μm (C) Average FRAP recovery curves for α-actinin-1 (green), α-actinin-2 (orange), α-actinin-3 (red) and α-actinin-4 (purple) over 30 minutes. Values are mean +/− standard error at each time point based on between 8 and 22 replicate measurements in different myofibers. (D) Comparison of fast (blue) vs. slow (red) mobile fractions of α-actinin-1, 2, 3, and 4. Values are means +/− standard error. * p<0.05 in total mobile fraction (Mfast + Mslow) difference.
FRAP results of the four α-actinin isoforms showed differential turnover dynamics at the Z-line (Figure 1B–1D, Table 1). All isoforms were assessed for their fluorescence recovery over 30 minutes (Figure 1C). Muscle proteins have been found to have long half-lives, ranging from 3 to 9 days [Sanger et al., 2004]. Similar FRAP experiments done with protein synthesis inhibition by cycloheximide showed no effect on Z-line protein recovery on the scale of minutes to hours [Wang et al., 2005]; therefore, any observed recovery of over 30 minutes can be attributed to protein exchange that is independent of new protein synthesis.
Table 1.
Differential recovery phases, rates, and half-times of wild type α-actinin isoforms at myofiber Z-lines.
| M1 (%) | k1 (min−1) | t1/21 (min) | M2 (%) | k2 (min−1) | t1/22 (min) | |
|---|---|---|---|---|---|---|
| ACTN1 | 7.88 +/− 0.47 | 1.83 +/− 0.12 | 0.38 +/− 0.02 | 43.71 +/−1.29 | 0.07 +/− 0.00 | 9.96 +/− 0.37 |
| ACTN2 | 4.45 +/− 0.33 | 3.86 +/− 0.36 | 0.18 +/− 0.02 | 31.91 +/− 1.98 | 0.05 +/− 0.00 | 12.74 +/− 0.36 |
| ACTN3 | 4.49 +/− 0.22 | 4.78 +/− 0.71 | 0.15 +/− 0.02 | 27.33 +/− 1.86 | 0.06 +/− 0.00 | 12.47 +/− 0.46 |
| ACTN4 | 29.30 +/−1.45 | 1.69 +/− 0.08 | 0.41 +/− 0.02 | 55.73+/− 1.34 | 0.18 +/− 0.01 | 3.93 +/− 0.15 |
Values were derived by fitting fluorescence recovery of photobleached wild type α-actinin molecules in myofibers to R = M1(1-e^(-k1*t)) + M2(1-e^(-k2*t)). Data are shown as means +/− SE of 13, 22, 17 and 8 replicates respectively of ACTN1, ACTN2, ACTN3 and ACTN4.
Each isoform exhibited largely unique recovery kinetics. The skeletal muscle isoforms, α-actinin-2 and α-actinin-3, had the least amount of recovery at the Z-line (both at 30%, and were not significantly different from each other), followed by α-actinin-1 (at 50%), and finally α-actinin-4 (at 90%) (Figure 1C). These data indicate that of the four exogenous isoforms α-actinin-2 and α-actinin-3 are the most stable at the Z-line, whereas α-actinin-4 is highly dynamic and α-actinin-1 is intermediary in its turnover.
By applying the formula R = M1(1 − e^(-k1*t)) + M2(1-e^(-k2*t)) (Wang et al., 2005), the recovery dynamics can be separated into two mobile fractions: a fast phase (M1) and a slow phase (M2) (Figure 1D), where the M value describes the portion of recovery attributed to each phase and the k value describes the rate of recovery in each phase. The breakdown of α-actinin recovery dynamics into separate mobile fractions shows that while the amount of turnover differs between the isoforms, the ratios of fast to slow mobility are similar to each other and the majority of recovery for all four isoforms occur in the slow binding-turnover phase (60–90%) (Figure 1D, Table 1). These finding are concordant with the recovery profile for the avian sarcomeric isoform (α-actinin-2), where Wang et al. found that different Z-line proteins can recover to differing fast:slow phase ratios [Wang et al., 2005].
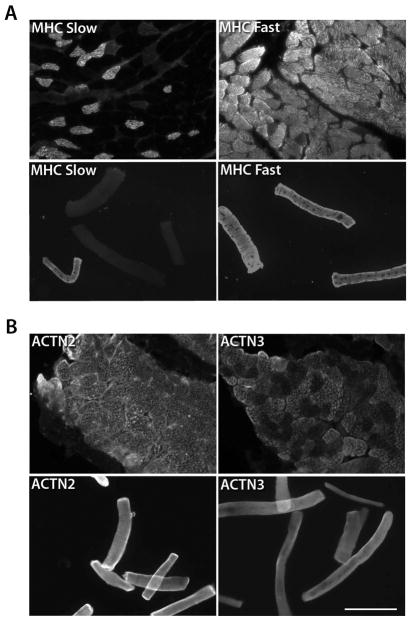
One factor that might contribute to variability of FRAP results might be heterogeneity of fiber types. To assess the fiber type composition of our experimental preparations, immunohistochemistry and immunocytochemistry were used to determine the composition of fast- versus slow-twitch fiber types as well as α-actinin-2- versus α-actinin-3-expressing fibers (Figure 2). The analysis for slow- versus fast-type fibers was done by myosin heavy chain (MHC) staining of frozen, sectioned muscle and cultured primary myofibers. The results showed that a large majority of fibers in the FDB are fast type II fibers (>90%), whereas a much smaller percentage are slow type I fibers (up to 20%) (Figure 2A). These numbers exceed 100% because some fibers may express both types of MHC. The analysis of α-actinin-2 versus α-actinin-3 expression revealed that all of the FDB fibers are α-actinin-2-positive, and a subset of these (66%) also express α-actinin-3 (Figure 2B), although the culturing process may have increased α-actinin-3 protein expression.
Figure 2.
The mouse FDB muscle consists of a heterogeneous mix of myofibers. Immunological analysis of FDB sections and dissociated, cultured FDB myofibers indicated heterogeneity in myofiber composition. (A) Analysis of fiber type using MHC Fast vs. Slow immunostaining showed 20% slow type fibers vs. 93% fast type fibers in sectioned tissue (upper panels) and 16% slow type fibers and 98% fast type fibers in cultured samples (lower panels). A total of 369 fibers were counted. (B) α-actinin-2-positive vs. α-actinin-3-positive immunological analysis of the FDB showed 100% of fibers to be α-actinin-2-expressing in sectioned tissue with 66% co-expressing α-actinin-3 (upper panels). Cultured myofiber samples (lower panels) were 100% positive for both α-actinin-2 and α-actinin-3. 190 fibers were counted. Scale bar = 25μm
α-actinin 2/4 chimeric isoforms are able to dimerize and form stable structures similar to native isoforms
The wild type α-actinin protein turnover dynamics suggested that the skeletal muscle α-actinin isoforms are more stable at the Z-lines when directly compared to the non-muscle isoforms. To investigate which domains of these proteins are responsible for the observed differences on Z-line turnover dynamics, we generated chimeric proteins with interchanged domains between muscle (α-actinin-2) and non-muscle (α-actinin-4) isoforms to characterize their effects on Z-line turnover dynamics (Figure 3A). α-actinin-2 and α-actinin-4 were chosen for this experiment since α-actinin-2 is a consistent component in all the samples, and α-actinin-4 Z-line dynamics are the least similar to α-actinin-2. Because the recovery of α-actinin-2 and α-actinin-4 are so dissimilar (p < .001, taking into account Bonferroni correction) from each other, changes in the recovery of α-actinin-2/α-actinin-4 chimeras were predicted to reflect whether the domain changes caused the proteins to shift toward α-actinin-2 or α-actinin-4 turnover rates.
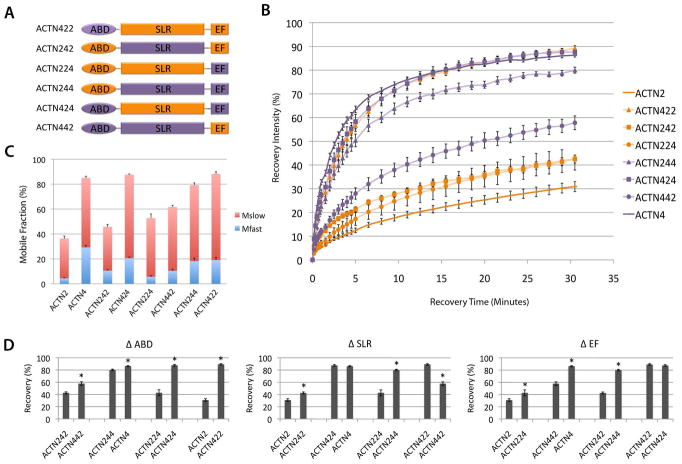
Figure 3.
α-actinin-2 and α-actinin-4 isoform Z-line dynamics are determined by cooperation between the ABD, SLR, and EF domains. (A) Schematic showing cloning of chimeric α-actinin-2/4 proteins: α-actinin-2 domains, orange; α-actinin-4 domains, purple. (B) Fluorescence recovery of chimeric isoforms: α-actinin-422 (orange triangle), α-actinin-242 (orange square), α-actinin-224 (orange circle), α-actinin-244 (purple triangle), α-actinin-424 (purple square), and α-actinin-442 (purple circle) compared to α-actinin-2 (dark orange) and α-actinin-4 (dark purple). Values are mean +/− standard error at each time point based on between 7 and 22 replicate measurements in different myofibers. (C) Comparison of fast (blue) vs. slow (red) mobile fractions of chimeric isoforms to α-actinin-2 and α-actinin-4. Values are means +/− standard error. (D) Recovery comparisons of α-actinin-2/4 wild type and chimeric isoforms. Panels (left to right) show pairwise comparisons of percentage fluorescent protein recovery at 30min when the ABD is changed (Δ ABD), SLR is changed (Δ SLR) and EF hands are changed (Δ EF). Values are mean +/− standard error at each time point. * p < .05 using Bonferroni correction.
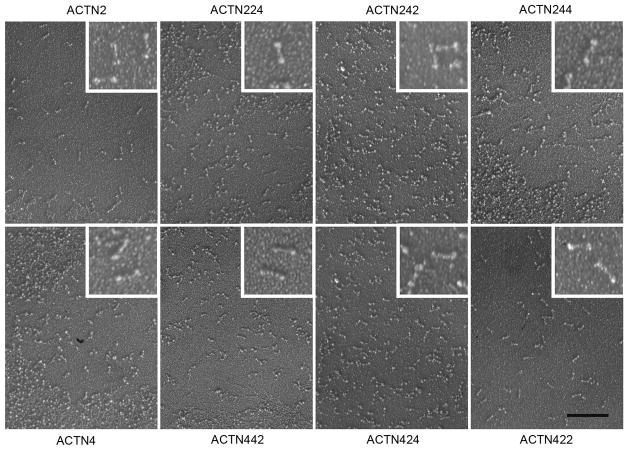
The ABD, the SLR, and the C-terminal EF hands of α-actinin-2 and α-actinin-4, were swapped at junctions within conserved portions of the linker regions between these domains. This lessened the possibility of altering the properties of any functional domain. To confirm that the resultant proteins dimerized with appropriate morphology, each isoform was examined by rotary metal-shadowing transmission electron microscopy (Figure 4). The wild type α-actinin-2 and α-actinin-4, and each of the chimeric isoforms, exhibited the distinct α-actinin protein morphology: 40 nm long rods with slightly enlarged ends [Meyer and Aebi, 1990]. Although there could still be unobservable anomalies, these results indicate that the chimeric proteins seem to have the expected tertiary structures and are able to dimerize.
Figure 4.
Electron microscopy of rotary shadowed α-actinin-2/4 wild type and chimeric isoforms. α-actinin proteins self-assemble into rod-shaped dimers with 40×5 nm dimensions. All chimeric proteins assembled into identical rods. Scale bar = 200 nm.
α-actinin protein turnover dynamics at the skeletal muscle Z-line is determined by cooperative composition of all three domains
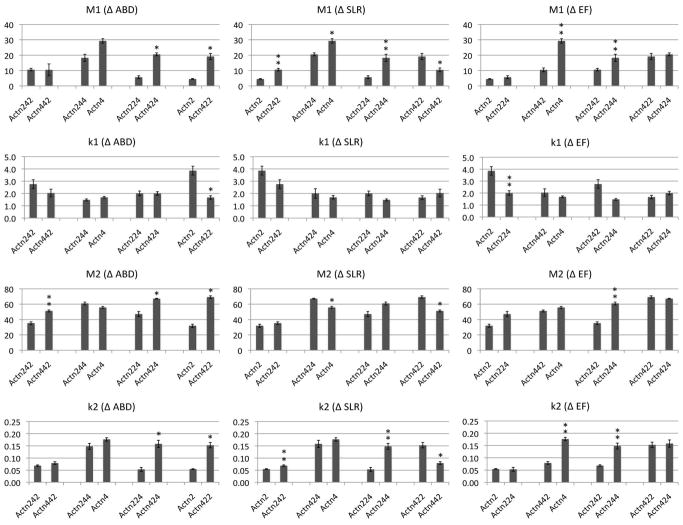
We hypothesized that there existed two possibilities in the Z-line dynamics of chimeric α-actinin-2/α-actinin-4 proteins: 1) recovery could be tightly correlated with the change of a single domain or 2) recovery could reflect cooperative contribution from multiple domains. Each chimeric isoform localized to Z-lines with negligible fluorescence signal in other sarcomeric regions, indicating retention of specificity for Z-line structures. The FRAP results of the chimeric α-actinin-2/α-actinin-4 isoforms showed that the Z-line dynamics of the proteins followed a trend, where each isoform’s recovery was reflective of the proportion of α-actinin-2 or α-actinin-4 it contained rather than dictated by one single domain (Figure 3B–D). As the α-actinin-2 protein increases in homology to α-actinin-4 in sequence, it becomes more dynamic at the Z-line and recovers more like α-actinin-4 (Figure 3B). For example, α-actinin-224 chimeric protein which contains α-actinin-2’s ABD and SLR domains and α-actinin-4’s EF domain, showed greater turnover than α-actinin-2, and α-actinin-244 chimeric protein which contains both SLR and EF domains coming from α-actinin-4 showed further dynamic turnover. By comparing the total percentage of protein turnover of each isoform after 30 minutes, we were able to do pairwise analyses of each functional domain’s effect on Z-line dynamics (Figure 3D). In addition to confirming the contributions of the SLR and EF domains on protein turnover at the Z-line, these comparisons indicated that changing the ABD also had an effect on recovery, despite its high conservation between the isoforms. These contributions were observed in total protein turnover after 30 minutes, as well as in analyses of fast and slow phase constants M1, k1, M2, and k2 (Figure 5). The breakdown of the chimeric α-actinin-2/α-actinin-4 proteins’ recoveries showed similar ratios of slow to fast phases as before, which reflects the previously observed dynamics of α-actinin protein, in comparison to other Z-line proteins [Wang et al., 2005] (Figure 3C) i.e. despite interchanging the domains, the chimeric proteins’ turnover profiles remained characteristic of that of previously established α-actinin. Further analysis of the half-times of both the slow and fast phases revealed that the chimeras that are mostly composed of α-actinin-2 tended to have faster half-times for fast phases and slower half-times for slow phases (Table 2). This trend suggests that the sarcomeric and non-muscle isoforms have different dynamics through Z-line and non-Z-line interactions.
Figure 5.
Pairwise comparisons of FRAP phase and rate constants of α-actinin-2/4 chimeric proteins at myofiber Z-lines. Values were derived by fitting recovery data to R = M1(1-e^(-k1*t)) + M2(1-e^(-k2*t)), and shown as mean +/− standard error. Panels, top to bottom, show comparisons of M1, k1, M2, and k2; panels, left to right show comparisons of each value when the ABD, SLR, or EF domain is changed from that of α-actinin-2 to α-actinin-4. Note the higher number of pairwise significance when the SLR or EF domain is changed, versus the ABD. * p<0.05 using Bonferroni correction. ** p<0.05 using Bonferroni correction when sample does not include α-actinin-422 or α-actinin-424.
Table 2.
Differential recovery phases, rates, and half-times of wild type and chimeric α-actinin2/4 isoforms at myofiber Z-lines.
| M1 (%) | k1 (min−1) | t1/21 (min) | M2 (%) | k2 (min−1) | t1/22 (min) | |
|---|---|---|---|---|---|---|
| ACTN2 | 4.45 +/− 0.33 | 3.86 +/− 0.36 | 0.18 +/− 0.02 | 31.91 +/− 1.98 | 0.05 +/− 0.00 | 12.74 +/− 0.36 |
| ACTN4 | 29.30 +/−1.45 | 1.69 +/− 0.08 | 0.41 +/− 0.02 | 55.73+/− 1.34 | 0.18 +/− 0.01 | 3.93 +/− 0.15 |
| ACTN242 | 10.59 +/− 0.84 | 2.76 +/− 0.36 | 0.26 +/− 0.03 | 35.23 +/− 1.78 | 0.07 +/− 0.00 | 10.10 +/− 0.43 |
| ACTN424 | 20.54 +/− 0.97 | 2.00 +/− 0.15 | 0.35 +/− 0.03 | 67.16 +/− 0.48 | 0.16 +/− 0.01 | 4.42 +/− 0.41 |
| ACTN224 | 5.63 +/− 0.93 | 2.01 +/− 0.19 | 0.35 +/− 0.03 | 47.11 +/− 3.34 | 0.05 +/− 0.01 | 13.29 +/− 1.99 |
| ACTN442 | 10.45 +/− 1.22 | 2.04 +/− 0.31 | 0.35 +/− 0.05 | 51.22 +/− 1.27 | 0.08 +/− 0.01 | 8.80 +/− 0.63 |
| ACTN244 | 18.29 +/− 2.34 | 1.48 +/− 0.08 | 0.47 +/− 0.02 | 60.95 +/− 1.86 | 0.15 +/− 0.01 | 4.71 +/− 0.39 |
| ACTN422 | 19.16 +/− 2.00 | 1.67 +/− 0.14 | 0.42 +/− 0.04 | 69.18 +/− 1.65 | 0.15 +/− 0.01 | 4.57 +/− 0.33 |
Values were derived by fitting fluorescence recovery of photobleached wild type and chimeric α-actinin molecules in myofibers to R = M1(1-e^(-k1*t)) + M2(1-e^(-k2*t)). Data are shown as means +/− SE of 22, 8, 10, 7, 8, 10, 9, and 11 replicates respectively of ACTN2, ACTN4, ACTN242, ACTN424, ACTN224, ACTN442, ACTN244 and ACTN422.
We identified two exceptions to the observed trend, which are the chimeric isoforms α-actinin-424 and α-actinin-422. These proteins have one and two domains, respectively, from α-actinin-2, but show no characteristics of the binding dynamics of α-actinin-2. Instead, their dynamics mimic that of α-actinin-4. At first sight this may suggest that the ABD domain is the most important determinant of the binding dynamics at the Z-line. However, this is contradicted by the fact that α-actinin-442 presents a slower recovery than α-actinin-422. Therefore, we looked at shared aspects of the two chimeras, α-actinin-422 and α-actinin-424, that exhibited the fastest recoveries. Since these two chimeric proteins have the 4ABD-2SLR neck region in common, we hypothesized that the chimeric neck region might cause unexpected tertiary changes. This anomaly is further investigated through gelation assays.
Structural integrity of the ABD-SLR linker region is critical for actin crosslinking function
Despite the trend we observed in our myofiber FRAP analyses, where an increase in the number of α-actinin-2 functional domains led to increased stability at the FDB Z-line, there were 2 notable outliers: α-actinin-424 and α-actinin-422. These chimeric proteins have one and two α-actinin-2 domains, respectively, but instead of increased stability at the Z-line, they recover almost exactly like α-actinin-4 (almost 90% recovery after 30 minutes, p = 11.28 and 1.18, respectively).
To investigate the potential changes in protein interaction of α-actinin-422 and α-actinin-424, we tested their actin crosslinking ability by using a miniature falling ball viscometry assay, which assesses how well each α-actinin isoform gels F-actin [Weins et al., 2007]. This assay would reveal whether each chimeric isoform is able to crosslink F-actin and how efficiently it does so compared to wild-type isoforms, as well as whether each isoform is calcium-sensitive in its actin-binding ability.
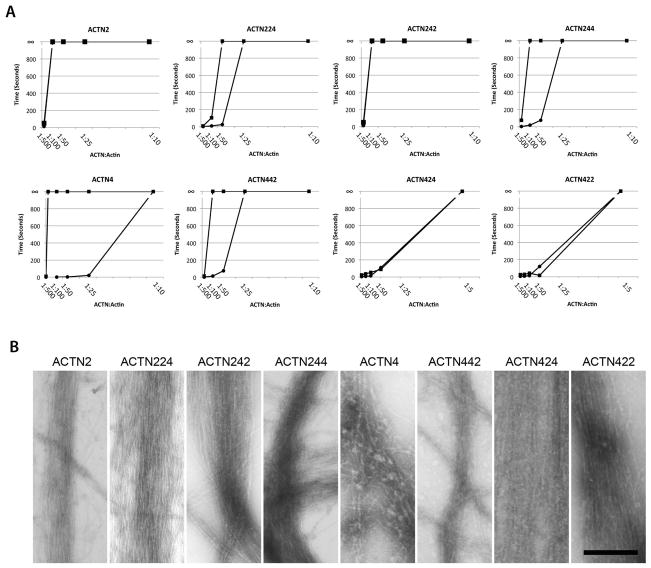
A viscometric analysis of actin-crosslinking at various α-actinin:actin ratios under calcium and calcium-free conditions confirmed that the wild-type isoforms of α-actinin-2 and α-actinin-4 retained their respective calcium-insensitive and sensitive properties (Figure 6A). In the absence of calcium, α-actinin-2 and α-actinin-4 isoforms were able to fully gel F-actin at an α-actinin:actin ratio of 1:100 and 1:500, respectively. With the addition of 0.5 mM calcium, α-actinin-2 retained its gelation activity at the 1:100 ratio, however, viscosity of α-actinin-4-bundled actin dropped significantly (Figure 6A), requiring a >50-fold molar increase in α-actinin-4 to regain gelation. These results were comparable to previously published observations regarding the calcium sensitivity of non-muscle isoforms and the calcium insensitivity of muscle isoforms [Burridge and Feramisco, 1981].
Figure 6.
Native structure of the α-actinin ABD-SLR neck region is essential for actin crosslinking and calcium sensitivity. (A) Viscometric assay comparing gel points of chimeric α-actinin-2/4 isoforms compared to α-actinin-2 and α-actinin-4 in the presence (circle) and absence (square) of 0.5mM calcium at varying α-actinin:actin molar ratios with actin fixed at 12.8 μM. Gel-to-solid transition is shown by ∞. Gel points of α-actinin-224 (upper row, second from left), α-actinin-242 (upper row, third from left), α-actinin-244 (upper row, right), and α-actinin-442 (bottom row, second from left) were comparable to those of α-actinin-2 and α-actinin-4 under calcium-free conditions with α-actinin-224, 244, and 442 exhibiting calcium sensitivity. α-actinin-424 (bottom row, third from left) and α-actinin-422 (bottom row, right) required a higher molar concentration of α-actinin protein to reach the gel point. (B) Electron microscopy of negatively stained actin filaments crosslinked by wild type and chimeric α-actinin-2/4 isoforms under calcium-free conditions. Note that parallel bundles were formed by all isoforms, indicating actin-crosslinking ability. Scale bar = 200 nm.
The analysis of chimeric α-actinin-2/α-actinin-4 proteins revealed that although most isoforms at an α-actinin:actin ratio of 1:25 formed gels with F-actin, α-actinin-422 and α-actinin-424 were unable to do so (Figure 6A). However, these isoforms were able to form gels at higher α-actinin concentrations (α-actinin:actin ratio of 1:5). These results suggest that chimeric isoforms with the α-actinin-4ABD-α-actinin-2SLR linker had compromised actin-crosslinking ability but did not lose all functionality, which correlates with the FRAP findings (Figure 3B). Negative staining of α-actinin/F-actin gels at high α-actinin:actin ratios showed similarly formed actin bundles for both native and chimeric α-actinin proteins (Figure 6B), establishing that all these chimeric isoforms are able to dimerize and crosslink F-actin.
α-actinin protein domains are similarly cooperative in turnover dynamics at fibroblast adhesion plaques
To investigate whether the dynamics observed in the native and chimeric protein FRAP experiments were resulting from differences in interactive stability with Z-line proteins via each isoforms’ functional domains, the same analysis was performed in non-muscle cells. In fibroblasts, α-actinin-4 is endogenously expressed and localizes to focal adhesion plaques, which are analogous to, but distinctly different from, the highly structured muscle Z-lines. As with the FDB myofibers, the wild-type and chimeric isoforms of α-actinin-2 are similar enough to α-actinin-4 to be able to localize to the focal adhesions of the 3T3 fibroblast cell line (Figure 7A), allowing investigation of their turnover dynamics. We hypothesized that chimeric α-actinin protein dynamics at fibroblast adhesion plaques would be converse to those from myofibers as α-actinin-4 should have more stable endogenous binding partners.
Figure 7.
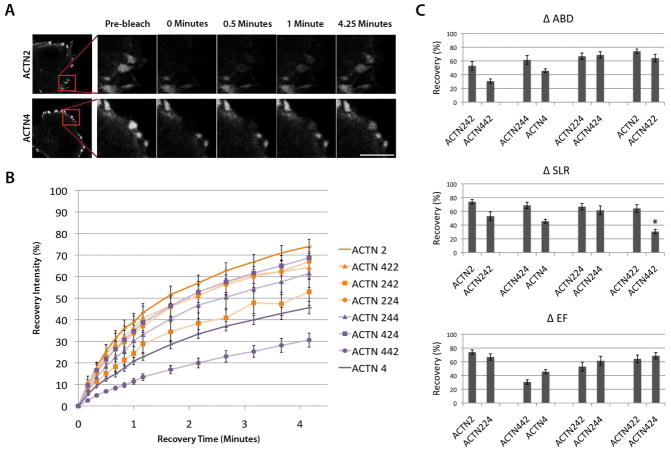
Sarcomeric α-actinin-2 isoform recovers faster at fibroblast focal adhesions than the non-muscle isoform. (A) Fluorescence recovery of α-actinin-2 and α-actinin-4 in fibroblasts. Representative images of recovery show (left to right) pre-bleach and 0, 0.5, 1, and 4.25 minutes after photobleaching at focal adhesions (magnified from inset of image at far left). Scale bar = 10μm (B) Fluorescence recovery at focal adhesions of chimeric isoforms α-actinin-422 (orange triangle), α-actinin-242 (orange square), α-actinin-224 (orange circle), α-actinin-244 (purple triangle), α-actinin-424 (purple square), and α-actinin-442 (purple circle) compared to α-actinin-2 (dark orange) and α-actinin-4 (dark purple). Values are mean +/− standard error at each time point based on between 4 and 7 replicate measurements in different cells. (C) Recovery comparisons of α-actinin-2/4 wild type and chimeric isoforms. Panels (top to bottom) show pairwise comparisons of isoforms when the ABD is changed (Δ ABD), SLR is changed (Δ SLR) and EF hands are changed (Δ EF). Values are means +/− standard error. No significance was observed between pairs except ACTN422 and ACTN442 as indicated by “*” (p = .02 following Bonferroni correction).
The fibroblast FRAP findings showed the opposite trend of that observed in the myofibers (Figure 7B, C). α-actinin-4 is more stable than α-actinin-2 at the focal adhesions (p = .004), recovering to 50% in just over 4 minutes, whereas α-actinin-2 is more dynamic in fibroblasts and recovers to 80% in the same time interval (Figure 7B). In addition, as α-actinin-4 domains are interchanged for α-actinin-2 domains, the chimeric protein becomes more dynamic at the cellular focal adhesions, although not all comparisons reached statistical significance at these sample sizes. Similar to FRAP findings in the myofibers (Figure 1), these recovery shifts seem to be due to cooperative contribution by all three functional domains rather than the effect of a single isolated domain. Analysis of the contribution of each domain and its effect on FRAP recovery points to a trend where as any single domain is changed from that of α-actinin-2 to α-actinin-4, it leads to a slight decrease in dynamic turnover at the focal adhesion (Figure 7C). A notable outlier in the fibroblast FRAP results is α-actinin-442, which has more stable dynamics at focal adhesions compared to α-actinin-4. This finding is possibly due to the loss of calcium sensitivity in the EF domain, which is known to be a calcium-dependent source for destabilizing α-actinin interaction with actin [Burridge and Feramisco, 1981].
Discussion
α-Actinin proteins are highly conserved across species and isoforms. However, despite their high levels of similarity in structure and function, previous studies have shown that their isoforms are not able to fully compensate for one another. Studies of the non-muscle isoforms found that α-actinin-1 can functionally compensate for α-actinin-4 in some instances but not others [Quick and Skalli, 2010; Shao et al., 2010a] and studies of the muscle isoforms found that α-actinin-2 can partially compensate for loss of α-actinin-3 but not vice versa [Gupta et al., 2012; MacArthur et al., 2007]. These findings indicate that each isoform has evolved to have specific and optimal interactions within their endogenous environments. To investigate the extent of muscle specificity of isoforms, we undertook FRAP studies to analyze their differential dynamics at the skeletal muscle Z-line.
Previous studies using FRAP to analyze the turnover rates of α-actinin under various conditions, such as injecting in vitro generated proteins into cells and utilizing myofibers differentiated in vitro from myoblasts, showed very variable turnover rates [Hasebe-Kishi and Shimada, 2000; McKenna et al., 1986; Wang et al., 2005]. To better model endogenous conditions in mature skeletal muscle, in the present study we expressed proteins in cultured adult myofibers differentiated in vivo.
Our findings showed that the four human α-actinin isoforms have widely varying dynamics at muscle Z-lines, ranging from FRAP recoveries of 30% to 90% over a 30 minute time period. α-Actinin-2 and α-actinin-3 were least dynamic at the Z-line (30% recovery), followed by α-actinin-1 (60%) and α-actinin-4 (90%). These results showed that muscle-specific isoforms are most stably associated at the Z-lines, suggesting that muscle isoforms have evolved specific interactions in striated muscle despite similarities in their sequence and function with non-muscle isoforms. The differences observed indicate that as the muscle isoforms branched off from the ancestral gene [Dixson et al., 2003], they evolved interactions with muscle-specific proteins and lost certain properties of the ancestral isoform, such as calcium sensitivity (retained by the non-muscle splice isoform of α-actinin-1 and α-actinin-4). These developments may have led α-actinin-2 and α-actinin-3 to be more strongly associated at the Z-line.
Although α-actinin proteins are commonly thought to form primarily homodimers, Chan et al [Chan et al., 1998] showed that α-actinin-2 and α-actinin-3 are capable of forming heterodimers at low concentrations in vitro and in vivo. In our study, it was unclear to what extent α-actinin-2 and α-actinin-3 formed heterodimers in cultured myofibers due to their similarity in recovery. Since dimers are formed via anti-parallel interactions through the SLR domain, greater sequence variability between the muscle and non-muscle isoforms may have prevented heterodimer formation. If heterodimer formation between α-actinin-2 and α-actinin-4 was common and affecting recovery, then we might expect to see greater variability in recovery kinetics of α-actinin-4 in myofibers, which express high levels of endogenous α-actinin-2, as the ratio of homodimers to heterodimers would be expected to vary between myofibers expressing different levels of transduced α-actinin-4. The standard errors of the means were proportionally similar for ACTN2 and ACTN4 constructs, suggesting this was not a major confounding factor (Figure 1, Table 1).
According to the FRAP models previously presented [Sprague and McNally, 2005], the recovery curves we observed for α-actinin isoforms indicated dynamics of diffusion-uncoupled recovery (i.e. diffusion happens immediately after photobleaching, followed by a much longer recovery phase reflective of the exchange of binding partners). Therefore, a vast majority of the recovery curve is due to binding and unbinding events. A breakdown of the dynamics into fast versus slow phases revealed that, at the Z-line, muscle isoforms had faster half-times for fast phases and slower half-times for slow phases. As the fast phases are indicative of turnover with non-specific partners and the slow phases with specific interactors, our results suggest that muscle isoforms have faster turnover with non-specific interactors, and slower turnover with specific binding partners, thus suggesting more numerous and/or stable binding with Z-line proteins.
The calcium sensitivity of non-muscle isoforms may account for part of their decreased stability at the Z-line, as it was found that calcium concentrations >100 nM lead to a decrease in non-muscle isoforms’ actin-binding ability [Burridge and Feramisco, 1981], and the resting calcium concentration of FDB was found to be at 106 +/− 2 nM [Williams et al., 1990]. Although calcium sensitivity is one main difference between muscle and non-muscle isoforms, the FRAP results indicate protein interactions as an additional source for the observed differences. Although α-actinin-1 is calcium-sensitive, it was over 30% more stable than its fellow non-muscle isoform α-actinin-4. A probable reason is that it is able to better interact with skeletal muscle proteins despite having calcium-sensitive actin binding, as a calcium-insensitive splice isoform of it is normally expressed in smooth muscle [Youssoufian et al., 1990].
The goal of this study was to delineate which portion of the α-actinin protein dictated the observed recovery differences. The analysis of the turnover of chimeric α-actinin-2/α-actinin-4 proteins showed that the ABD, SLR, and EF domains all contributed significantly to the differential dynamics of the muscle and non-muscle isoforms. The presence of the calcium-insensitive α-actinin-2 EF hands appeared to be a general predictor of stability at Z-lines (Figure 3), while the α-actinin-4 SLR region seems important for stability at focal adhesions (Figure 7), perhaps explaining the peculiar dynamics of ACTN242, which exhibited recovery dynamics similar to α-actinin-2 in myofibers, and similar to α-actinin-4 in fibroblasts.
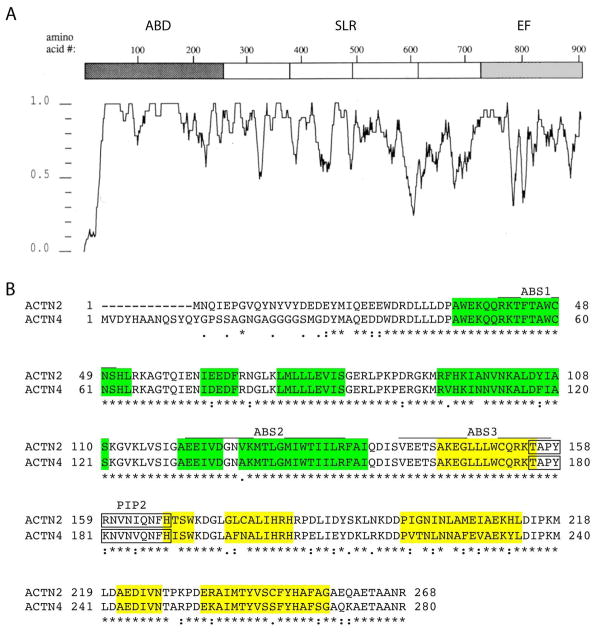
Although the calcium sensitivity of the EF hands and the sequence diversity of the SLR domain suggested that these domains might contribute to differences in turnover dynamics, the nature of the ABD (the most highly conserved domain) surprisingly also significantly affected Z-line dynamics. This was unexpected as the function of this region is well-studied: it is the most highly conserved amongst all four isoforms, and there are no known muscle-specific interactions at the ABD. Sequence comparison of the ABDs of α-actinin-2 and α-actinin-4 reveals that the three actin binding sites are virtually identical, varying only at a single residue in actin binding site 2 (Figure 8). The greatest degree of divergence is within the carboxy-terminal calponin homology domain 1 (CH1), particularly in the E′ alpha helix of the closed confirmation molecule [Franzot et al., 2005], inviting speculation that this structure may play differential roles in context-dependent interactions at Z-lines and focal adhesions. Overall, these observations suggest that interactions between α-actinins and F-actin may be altered in an isoform-dependent manner, and that as an α-actinin-2 isoform’s domains are replaced by those of α-actinin-4, molecules incrementally become more dynamic at the Z-line.
Figure 8.
Sequence comparisons among α-actinin isoforms. (A) Graphical representation of average sequence identity among α-actinin-1, α-actinin-2, α-actinin-3 and α-actinin-4 illustrating co-linear nature of the four isoforms with exception of sequences at the amino terminus proximal to the ABDs and within the EF hands. The highest degree of conservation is within the ABDs. (B) Amino acid sequence alignment of the ABDs of α-actinin-2 and α-actinin-4. ABS = actin binding site, PIP2 indicates the PtdIns(4,5)P2 binding site. The six major α-helical regions in each of the two calponin homology domains are highlighted in yellow (CH1) and green (CH2) after [Franzot et al., 2005].
Breakdown of the dynamics into fast and slow phases showed that overall, chimeric proteins that are predominantly α-actinin-2 had faster half-times for fast phases and slower half-times for slow phases compared to those that are predominantly α-actinin-4. The faster half-times for fast phases indicate that as an α-actinin-4 isoform gains α-actinin-2 domains, it loses interactions with non-Z-line binding partners. Alternatively, its slower half-times for slow phases suggest that as α-actinin-4 domains are swapped for those of α-actinin-2, the resulting isoform becomes less dynamic at the Z-line due to more numerous and/or stable interactions with α-actinin-2-specific binding partners. Together, these results suggest that muscle isoforms are different from non-muscle isoforms because as they are transported through the myoplasm, each of the three domains help direct them to more efficiently bypass non-specific interactions and more stably bind at the Z-line.
Analysis of the chimeric isoforms at fibroblast focal adhesions showed the converse in turnover dynamics. In the non-muscle environment, the α-actinin-2 isoforms attained more stable dynamics at focal adhesions as its domains were swapped for those of α-actinin-4. This observed difference is unlikely to be due to differences in actin protein structure, as skeletal muscle alpha actin and fibroblast beta actin are 94% similar. Rather, this change indicates that not only did the muscle α-actinin isoforms evolved to gain specific interactions at muscle Z-lines, but also that they may have lost properties of the ancestral α-actinin protein that allow non-muscle isoforms to maintain stability in non-muscle cells. An additional possibility is that α-actinin-1 and α-actinin-4 isoforms gained non-muscle specific properties after the α-actinin-2 and α-actinin-3 divergence.
Although there was an apparent domain-dependent trend in our FRAP results, the chimeric proteins α-actinin-422 and α-actinin-424 both turned over more dynamically than expected. Viscometry assessment showed that, compared to other isoforms, they have compromised gelation ability and required higher concentrations of α-actinin protein to form gels with F-actin, indicating a decreased ability to crosslink F-actin. As these isoforms have in common the α-actinin-4ABD/α-actinin-2SLR linker, changes in properties of this linker is a possible cause for the gelation defect. Rotary shadowing indicated that their dimerization ability is unaltered and their α-actinin-4-like dynamics suggest they are functional proteins, albeit with less stable binding at the Z-line. Although the linker is not part of a conserved domain, it closely neighbors both the ABD and SLR and interacts with the opposing dimer’s EF domain. The hybrid linker region may have disrupted tertiary structure at and around the junction and affected interactions that may have lowered the isoforms’ ability to efficiently crosslink F-actin.
The viscometry assessment revealed unexpected calcium sensitivity of the chimeric isoform α-actinin-442, despite having a calcium-insensitive EF domain from α-actinin-2. The reason for this result is unclear, especially since its dynamics in fibroblast FRAP experiments indicated that the α-actinin-2 EF domain in this isoform may have contributed to calcium-related stability. In conjunction with the gelation results of α-actinin-422 and α-actinin-424, this indicates that the ABD domain is susceptible to functional changes as a result of changes in its neighboring linker region and/or the other two domains, perhaps through steric interactions.
Mutations of α-actinin-2 and α-actinin-4 have both been associated with human genetic diseases: α-actinin-2 with hypertrophic cardiomyopathy and dilated cardiomyopathy [Chiu et al., 2010; Mohapatra et al., 2003] and α-actinin-4 with focal segmental glomerulosclerosis (FSGS) [Kaplan et al., 2000]. Rather than localizing α-actinin-2-associated disease to a specific functional domain, mutations observed in patients were found in all three domains. However, mutations for FSGS have been found to localize to the distal end of the α-actinin-4 ABD. These findings coincide with our results in supporting the sensitivity of the α-actinin-4 ABD-SLR regions and the cooperative contribution of all three domains in α-actinin function.
In this study, we determined that despite the conservation of α-actinin isoforms, they have highly variable dynamics in skeletal muscle. This variability could be attributed to muscle isoforms evolving muscle-specific interactions and losing properties of non-muscle isoforms that allow them to be optimally stable at the Z-line. Moreover, we found that turnover differences result from cooperative contribution of all three domains and that the ABD-SLR linker region is essential for α-actinin protein function. Further studies would be necessary to determine the specific protein interactions that account for differences found in α-actinin dynamics. In addition, although we have found the ABD, SLR, and EF domains to cooperatively contribute to dynamics, it is still unclear whether α-actinin domains functionally regulate each other or their dimer partners. Together, our findings indicate that there are functional differences between isoforms of the α-actinin family of proteins, which warrant isoform-specificity in future studies.
Materials and Methods
FDB myofiber culture
FDB muscles from 1–3 month old wild type C57BL/6J mice were isolated and cultured based on a technique previously described [Ravenscroft et al., 2007]. Myofibers were seeded on 20μg/mL laminin-coated Mattek P35G-1.5-20-C glass-bottom dishes for FRAP studies or Thermo Scientific Lab-Tek 8-well glass chamber slides for immuno-studies.
Protein expression in myofibers
Human wild type α-actinin 1–4 (NM_001102.3, NM_001103.3, NM_001104.2, NM_004924.4) were tagged with AcGFP and cloned into pAd/CMV/V5-DEST using Gateway cloning. Chimeric α-actinin 2/4 were made by swapping individual domains in modules consisting of the following nucleotides (based on reference sequences above) where the adenine of the initiating methionine is numbered as base 1: ABDs were ACTN2 c.1–806, ACTN4 c.1–861. SLRs were ACTN2 c.786–2240 and ACTN4 c.839–2294. EF hand domains were ACTN2 c.2222–2683 and ACTN4 c.2275–2734. The different modules were then assembled using GENEART Seamless Cloning. The final listed bases were followed immediately by a linker (CGGTACCGCGGGCCCGGGATCCACCGGTC), then AcGFP, to create a C-terminal fusion. Vectors were transfected into HEK293A cells using Lipofectamine 2000, (Invitrogen). Adenoviral production was performed as previously described [Untergasser et al., 2008]. Viral particles were purified and used to infect FDB myofibers at 1 day in vitro (DIV) in DMEM with 10% FBS and 5μg/mL polybrene. Media was changed to DMEM with 20% FBS at 2 DIV and samples were used at 4 DIV. Viral transductions were performed with limiting doses such that roughly 80% of myofibers were transduced with 1–3 infections particles as evidenced by foci of GFP expression at 24 hours post infection. Cultures were maintained for three more days at which time a majority of myofibers exhibited uniform GFP expression throughout their length.
Immuno-chemistry assays for fiber typing
Myofibers and frozen sections were fixed using 100% methanol. Myosin heavy chain, slow type (NOQ7.5.4D, Sigma) and fast type (MY-32, Sigma) monoclonal antibodies were used at 1:100 in 10% FBS in PBS. α-Actinin-2 (4B3) and α-actinin-3 (5B4) rabbit polyclonal antibodies (described by Beggs et al) [Beggs et al., 1992] were used at 1:100. Alexa Fluor 488 Goat anti-Mouse IgG and Alexa Fluor 488 Goat anti-Rabbit IgG antibodies were used at 1:200 for visualization. Samples were imaged at 25× using Nikon Elements on a Nikon Eclipse TE2000-S.
Immuno-cytochemistry for co-localization
Myofibers were infected for α-actinin-2-AcGFP and α-actinin-4-AcGFP expression and fixed at 4 DIV in 4% PFA and perforated using 3% Triton X-100. Myosin heavy chain, fast type (MY-32) and α-actinin-2 (4B3) antibodies were used at 1:100. All confocal images were collected with Zeiss LSM 700 using a 60× oil immersion objective. Protein co-localization analysis was done using ImageJ.
Fluorescence Recovery After Photobleaching
Myofibers expressing α-actinin-AcGFP were imaged for FRAP at 4 DIV in DMEM without phenol red and with 20% FBS. Samples were kept in a Tokai-Hit stage-top incubator set at 37°C with 5% CO2. All images were collected using a Nikon A1R with the Perfect Focus System using a 60× water immersion objective with Nikon-Elements. Areas containing 5–6 Z-lines were bleached with pinhole set to 1.5 AU (1.0μm). Post-bleach images were collected every 10 seconds for the first minute, every 30 seconds for the subsequent 3 minutes, and every 1.5 minutes for the remaining 25.5 minutes. Fluorescence recovery was measured for areas of each image restricted to the Z-lines to exclude any recovery due to non-specific diffusion or binding in non Z-line areas. For each construct, between 7 and 22 replicate areas were bleached in different myofibers and values were averaged to determine recovery curves and kinetic parameters. Fibroblasts expressing α-actinin-AcGFP were similarly imaged and analyzed at 2 DIV in DMEM without phenol red and with 10% FBS. Samples were kept at 29°C during image collection to minimize cell movement. Bleach areas contained 1–2 focal adhesions and between 4 and 7 replicate cells were bleached and analyzed for each construct. Image stacks of each series were aligned and fluorescence recovery was measured by averaging changes in fluorescence intensity of each Z-line in the bleached region. Time points were chosen to ensure adequate data collection during the dynamic phases of fluorescence recovery out to a point where recovery curves approached a plateau to permit accurate calculation of recovery kinetics. An unbleached area was recorded for each time point and background intensity was subtracted from both bleached and unbleached intensity values. The resulting values were corrected as a bleached:unbleached ratio. The initial post-bleached ratio (t=0) was subtracted from each ratio, and all values were divided by the pre-bleach value to set the pre-bleach value to 1 and initial post-bleach value to 0. Resultant values from 5 Z-lines or 1–2 focal adhesions from each replicate were then averaged and analyzed using ImageJ and processed to fit R = M1(1 − e^(-k1*t)) + M2(1-e^(-k2*t)) using KaleidaGraph (Synergy Software) as previously described [Wang et al., 2005]. Half-time of each recovery phase (t1/2) was derived from (ln2)/k.
Generation of recombinant proteins
Ac-GFP-tagged human wild type and chimeric α-actinin-2/4 were cloned into pFastBac HT C. Bacmids were produced and transfected into Sf9 cells for baculovirus production according to the Bac-to-Bac System (Invitrogen). Cells were grown at 25°C and 5% CO2 in spinner flasks and lysed using 1% Triton X-100. Proteins were purified using Ni-NTA columns as described in Nakamura et al [Nakamura et al., 2007]. Purified proteins were concentrated using Amicon Ultra-15 100k filter units. Recombinant proteins molecular weights were confirmed by SDS-PAGE and Coomassie Blue staining.
Rotary shadowing
Recombinant α-actinin, 0.5 μM, in 5 mM sodium phosphate and 50% glycerol, were sprayed onto freshly cleaved mica [Hartwig et al., 1980], dried under vacuum, and metal-coated with 1 nm of platinum at 6° with rotation and 3.0 nm of carbon at 90° without rotation. Replicas were floated in water, picked up on copper grids cleaned with 10% formic acid, and photographed in a JEOL 1200-EX electron microscope at 80 kV.
Viscometry assay
Gel-filtered rabbit skeletal muscle actin, stored at −70° C in 0.2 mM CaCl2, 0.5 mM ATP, 0.2 mM MgCl2, and 2 mM Tris, pH 7.0 was rapidly thawed, diluted to 40 μM in the same buffer, stored at 4° C for 12 hr, and clarified by centrifugation at 200,000 × g for 30 min. A final actin concentration of 12.8 μM was mixed with increasing amounts of the different recombinant α-actinin preparations and polymerization initiated with 0.1 M KCl and 2 mM MgCl2 in the presence of a final concentration of either 0.5 mM EGTA or 0.2 mM CaCl2. The solutions were loaded into 100 μl glass capillary tubes, the bottom sealed with hematocrit sealant, and incubated for 30 min at room temperature. The time required for small stainless-steel balls to fall through the solution was measured at 90°.
Negative staining
F-actin and α-actinin solutions were removed from the 100 μl capillary tubes used for gel point determinations, diluted to 2 μM F-actin in actin polymerization buffer (100 mM KCl, 0.5 mM ATP, 2 mM MgCl2, 2 mM Tris-HCl, pH 7.0 containing either 0.2 mM CaCl2 or 0.5 mM EGTA), and adhered to the surface of carbon and formvar-coated copper grids. Samples were washed twice in polymerization buffer and negatively stained with 1% uranyl acetate.
Acknowledgments
The authors wish to thank Dr. J. Sanger for many helpful discussions and technical advice on FRAP studies of cytoskeletal proteins, Drs. J. Pan and F. Nakamura for their guidance and resources in generating recombinant proteins and members of the L.M. Kunkel and E. Gussoni laboratories for critically reading the manuscript. This work was supported by the Muscular Dystrophy Association (MDA383249), National Institute of Health grant from the National Institute of Arthritis and Musculoskeletal and Skin Diseases R01 AR044345; and NIH HL 104145 (JHH). This work was also made possible by expert technical assistance from the Nikon Imaging Center at Harvard Medical School and the Intellectual and Developmental Disability Research Center Imaging and Molecular Genetics Core Laboratories at Boston Children’s Hospital (U54 HD090255). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Footnotes
Conflict of Interest
Authors have no conflicts of interest to declare.
References
- Bagnall RD, Molloy LK, Kalman JM, Semsarian C. Exome sequencing identifies a mutation in the ACTN2 gene in a family with idiopathic ventricular fibrillation, left ventricular noncompaction, and sudden death. BMC Med Genet. 2014;15:99. doi: 10.1186/s12881-014-0099-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beggs AH, Byers TJ, Knoll JH, Boyce FM, Bruns GA, Kunkel LM. Cloning and characterization of two human skeletal muscle alpha-actinin genes located on chromosomes 1 and 11. J Biol Chem. 1992;267(13):9281–8. [PubMed] [Google Scholar]
- Blanchard A, Ohanian V, Critchley D. The structure and function of alpha-actinin. J Muscle Res Cell Motil. 1989;10(4):280–9. doi: 10.1007/BF01758424. [DOI] [PubMed] [Google Scholar]
- Burridge K, Feramisco JR. Non-muscle alpha actinins are calcium-sensitive actin-binding proteins. Nature. 1981;294(5841):565–7. doi: 10.1038/294565a0. [DOI] [PubMed] [Google Scholar]
- Carpen O, Pallai P, Staunton DE, Springer TA. Association of intercellular adhesion molecule-1 (ICAM-1) with actin-containing cytoskeleton and alpha-actinin. J Cell Biol. 1992;118(5):1223–34. doi: 10.1083/jcb.118.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y, Tong HQ, Beggs AH, Kunkel LM. Human skeletal muscle-specific alpha-actinin-2 and -3 isoforms form homodimers and heterodimers in vitro and in vivo. Biochem Biophys Res Commun. 1998;248(1):134–9. doi: 10.1006/bbrc.1998.8920. [DOI] [PubMed] [Google Scholar]
- Chiu C, Bagnall RD, Ingles J, Yeates L, Kennerson M, Donald JA, Jormakka M, Lind JM, Semsarian C. Mutations in alpha-actinin-2 cause hypertrophic cardiomyopathy: a genome-wide analysis. J Am Coll Cardiol. 2010;55(11):1127–35. doi: 10.1016/j.jacc.2009.11.016. [DOI] [PubMed] [Google Scholar]
- Dixson JD, Forstner MJ, Garcia DM. The alpha-actinin gene family: a revised classification. J Mol Evol. 2003;56(1):1–10. doi: 10.1007/s00239-002-2374-5. [DOI] [PubMed] [Google Scholar]
- Djinovic-Carugo K, Gautel M, Ylanne J, Young P. The spectrin repeat: a structural platform for cytoskeletal protein assemblies. FEBS Lett. 2002;513(1):119–23. doi: 10.1016/s0014-5793(01)03304-x. [DOI] [PubMed] [Google Scholar]
- Duhaiman AS, Bamburg JR. Isolation of brain alpha-actinin. Its characterization and a comparison of its properties with those of muscle alpha-actinins. Biochemistry. 1984;23(8):1600–8. doi: 10.1021/bi00303a003. [DOI] [PubMed] [Google Scholar]
- Ebashi S, Ebashi F. A New Protein Factor Promoting Contraction of Actomyosin. Nature. 1964;203:645–6. doi: 10.1038/203645a0. [DOI] [PubMed] [Google Scholar]
- Faulkner G, Pallavicini A, Formentin E, Comelli A, Ievolella C, Trevisan S, Bortoletto G, Scannapieco P, Salamon M, Mouly V, et al. ZASP: a new Z-band alternatively spliced PDZ-motif protein. J Cell Biol. 1999;146(2):465–75. doi: 10.1083/jcb.146.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzot G, Sjöblom B, Gautel M, Djinovic Carugo K. The crystal structure of the actin binding domain from alpha-actinin in its closed conformation: structural insight into phospholipid regulation of alpha-actinin. J Mol Biol. 2005;348(1):151–65. doi: 10.1016/j.jmb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Frey N, Richardson JA, Olson EN. Calsarcins, a novel family of sarcomeric calcineurin-binding proteins. Proc Natl Acad Sci U S A. 2000;97(26):14632–7. doi: 10.1073/pnas.260501097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girolami F, Iascone M, Tomberli B, Bardi S, Benelli M, Marseglia G, Pescucci C, Pezzoli L, Sana ME, Basso C, et al. Novel alpha-actinin 2 variant associated with familial hypertrophic cardiomyopathy and juvenile atrial arrhythmias: a massively parallel sequencing study. Circ Cardiovasc Genet. 2014;7(6):741–50. doi: 10.1161/CIRCGENETICS.113.000486. [DOI] [PubMed] [Google Scholar]
- Gueguen P, Rouault K, Chen JM, Raguenes O, Fichou Y, Hardy E, Gobin E, Pan-Petesch B, Kerbiriou M, Trouve P, et al. A missense mutation in the alpha-actinin 1 gene (ACTN1) is the cause of autosomal dominant macrothrombocytopenia in a large French family. PLoS One. 2013;8(9):e74728. doi: 10.1371/journal.pone.0074728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V, Discenza M, Guyon JR, Kunkel LM, Beggs AH. alpha-Actinin-2 deficiency results in sarcomeric defects in zebrafish that cannot be rescued by alpha-actinin-3 revealing functional differences between sarcomeric isoforms. FASEB J. 2012;26(5):1892–908. doi: 10.1096/fj.11-194548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig JH, Tyler J, Stossel TP. Actin-binding protein promotes the bipolar and perpendicular branching of actin filaments. J Cell Biol. 1980;87(3 Pt 1):841–8. doi: 10.1083/jcb.87.3.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasebe-Kishi F, Shimada Y. Dynamics of actin and alpha-actinin in nascent myofibrils and stress fibers. J Muscle Res Cell Motil. 2000;21(8):717–24. doi: 10.1023/a:1010374424143. [DOI] [PubMed] [Google Scholar]
- Haywood NJ, Wolny M, Rogers B, Trinh CH, Shuping Y, Edwards TA, Peckham M. Hypertrophic cardiomyopathy mutations in the calponin-homology domain of ACTN2 affect actin binding and cardiomyocyte Z-disc incorporation. Biochem J. 2016;473(16):2485–93. doi: 10.1042/BCJ20160421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiska L, Kantor C, Parr T, Critchley DR, Vilja P, Gahmberg CG, Carpen O. Binding of the cytoplasmic domain of intercellular adhesion molecule-2 (ICAM-2) to alpha-actinin. J Biol Chem. 1996;271(42):26214–9. doi: 10.1074/jbc.271.42.26214. [DOI] [PubMed] [Google Scholar]
- Honda K, Yamada T, Endo R, Ino Y, Gotoh M, Tsuda H, Yamada Y, Chiba H, Hirohashi S. Actinin-4, a novel actin-bundling protein associated with cell motility and cancer invasion. J Cell Biol. 1998;140(6):1383–93. doi: 10.1083/jcb.140.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan JM, Kim SH, North KN, Rennke H, Correia LA, Tong HQ, Mathis BJ, Rodriguez-Perez JC, Allen PG, Beggs AH, et al. Mutations in ACTN4, encoding alpha-actinin-4, cause familial focal segmental glomerulosclerosis. Nat Genet. 2000;24(3):251–6. doi: 10.1038/73456. [DOI] [PubMed] [Google Scholar]
- Kato M, Sasaki T, Ohya T, Nakanishi H, Nishioka H, Imamura M, Takai Y. Physical and functional interaction of rabphilin-3A with alpha-actinin. J Biol Chem. 1996;271(50):31775–8. doi: 10.1074/jbc.271.50.31775. [DOI] [PubMed] [Google Scholar]
- Korenbaum E, Rivero F. Calponin homology domains at a glance. J Cell Sci. 2002;115(Pt 18):3543–5. doi: 10.1242/jcs.00003. [DOI] [PubMed] [Google Scholar]
- Kremerskothen J, Teber I, Wendholt D, Liedtke T, Bockers TM, Barnekow A. Brain-specific splicing of alpha-actinin 1 (ACTN1) mRNA. Biochem Biophys Res Commun. 2002;295(3):678–81. doi: 10.1016/s0006-291x(02)00734-9. [DOI] [PubMed] [Google Scholar]
- Kunishima S, Okuno Y, Yoshida K, Shiraishi Y, Sanada M, Muramatsu H, Chiba K, Tanaka H, Miyazaki K, Sakai M, et al. ACTN1 mutations cause congenital macrothrombocytopenia. Am J Hum Genet. 2013;92(3):431–8. doi: 10.1016/j.ajhg.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur DG, Seto JT, Chan S, Quinlan KG, Raftery JM, Turner N, Nicholson MD, Kee AJ, Hardeman EC, Gunning PW, et al. An Actn3 knockout mouse provides mechanistic insights into the association between alpha-actinin-3 deficiency and human athletic performance. Hum Mol Genet. 2008;17(8):1076–86. doi: 10.1093/hmg/ddm380. [DOI] [PubMed] [Google Scholar]
- MacArthur DG, Seto JT, Raftery JM, Quinlan KG, Huttley GA, Hook JW, Lemckert FA, Kee AJ, Edwards MR, Berman Y, et al. Loss of ACTN3 gene function alters mouse muscle metabolism and shows evidence of positive selection in humans. Nat Genet. 2007;39(10):1261–5. doi: 10.1038/ng2122. [DOI] [PubMed] [Google Scholar]
- Maruoka ND, Steele DF, Au BP, Dan P, Zhang X, Moore ED, Fedida D. alpha-actinin-2 couples to cardiac Kv1.5 channels, regulating current density and channel localization in HEK cells. FEBS Lett. 2000;473(2):188–94. doi: 10.1016/s0014-5793(00)01521-0. [DOI] [PubMed] [Google Scholar]
- Maruyama K, Ebashi S. Alpha-actinin, a new structural protein from striated muscle. II. Action on actin. J Biochem. 1965;58(1):13–9. doi: 10.1093/oxfordjournals.jbchem.a128158. [DOI] [PubMed] [Google Scholar]
- McKenna NM, Johnson CS, Wang YL. Formation and alignment of Z lines in living chick myotubes microinjected with rhodamine-labeled alpha-actinin. J Cell Biol. 1986;103(6 Pt 1):2163–71. doi: 10.1083/jcb.103.6.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RK, Aebi U. Bundling of actin filaments by alpha-actinin depends on its molecular length. J Cell Biol. 1990;110(6):2013–24. doi: 10.1083/jcb.110.6.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra B, Jimenez S, Lin JH, Bowles KR, Coveler KJ, Marx JG, Chrisco MA, Murphy RT, Lurie PR, Schwartz RJ, et al. Mutations in the muscle LIM protein and alpha-actinin-2 genes in dilated cardiomyopathy and endocardial fibroelastosis. Mol Genet Metab. 2003;80(1–2):207–15. doi: 10.1016/s1096-7192(03)00142-2. [DOI] [PubMed] [Google Scholar]
- Moncrief ND, Kretsinger RH, Goodman M. Evolution of EF-hand calcium-modulated proteins. I. Relationships based on amino acid sequences. J Mol Evol. 1990;30(6):522–62. doi: 10.1007/BF02101108. [DOI] [PubMed] [Google Scholar]
- Nakamura F, Osborn TM, Hartemink CA, Hartwig JH, Stossel TP. Structural basis of filamin A functions. J Cell Biol. 2007;179(5):1011–25. doi: 10.1083/jcb.200707073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nave R, Furst DO, Weber K. Interaction of alpha-actinin and nebulin in vitro. Support for the existence of a fourth filament system in skeletal muscle. FEBS Lett. 1990;269(1):163–6. doi: 10.1016/0014-5793(90)81144-d. [DOI] [PubMed] [Google Scholar]
- Otey CA, Carpen O. Alpha-actinin revisited: a fresh look at an old player. Cell Motil Cytoskeleton. 2004;58(2):104–11. doi: 10.1002/cm.20007. [DOI] [PubMed] [Google Scholar]
- Otey CA, Pavalko FM, Burridge K. An interaction between alpha-actinin and the beta 1 integrin subunit in vitro. J Cell Biol. 1990;111(2):721–9. doi: 10.1083/jcb.111.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa I, Astier C, Kwiatek O, Raynaud F, Bonnal C, Lebart MC, Roustan C, Benyamin Y. Alpha actinin-CapZ, an anchoring complex for thin filaments in Z-line. J Muscle Res Cell Motil. 1999;20(2):187–97. doi: 10.1023/a:1005489319058. [DOI] [PubMed] [Google Scholar]
- Pavalko FM, LaRoche SM. Activation of human neutrophils induces an interaction between the integrin beta 2-subunit (CD18) and the actin binding protein alpha-actinin. J Immunol. 1993;151(7):3795–807. [PubMed] [Google Scholar]
- Pavalko FM, Walker DM, Graham L, Goheen M, Doerschuk CM, Kansas GS. The cytoplasmic domain of L-selectin interacts with cytoskeletal proteins via alpha-actinin: receptor positioning in microvilli does not require interaction with alpha-actinin. J Cell Biol. 1995;129(4):1155–64. doi: 10.1083/jcb.129.4.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick Q, Skalli O. Alpha-actinin 1 and alpha-actinin 4: contrasting roles in the survival, motility, and RhoA signaling of astrocytoma cells. Exp Cell Res. 2010;316(7):1137–47. doi: 10.1016/j.yexcr.2010.02.011. [DOI] [PubMed] [Google Scholar]
- Ravenscroft G, Nowak KJ, Jackaman C, Clement S, Lyons MA, Gallagher S, Bakker AJ, Laing NG. Dissociated flexor digitorum brevis myofiber culture system--a more mature muscle culture system. Cell Motil Cytoskeleton. 2007;64(10):727–38. doi: 10.1002/cm.20223. [DOI] [PubMed] [Google Scholar]
- Sanger JW, Sanger JM, Franzini-Armstrong C. In: Assembly of the skeletal muscle cell. Myology Engel AG, Franzini-Armstrong C., editors. New York: McGraw-Hill; 2004. pp. 45–65. [Google Scholar]
- Shao H, Wang JH, Pollak MR, Wells A. alpha-actinin-4 is essential for maintaining the spreading, motility and contractility of fibroblasts. PLoS One. 2010a;5(11):e13921. doi: 10.1371/journal.pone.0013921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, Wu C, Wells A. Phosphorylation of alpha-actinin 4 upon epidermal growth factor exposure regulates its interaction with actin. J Biol Chem. 2010b;285(4):2591–600. doi: 10.1074/jbc.M109.035790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöblom B, Salmazo A, Djinovic-Carugo K. Alpha-actinin structure and regulation. Cell Mol Life Sci. 2008;65(17):2688–701. doi: 10.1007/s00018-008-8080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague BL, McNally JG. FRAP analysis of binding: proper and fitting. Trends Cell Biol. 2005;15(2):84–91. doi: 10.1016/j.tcb.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Goll DE, Stromer MH, Temple J. -actinin from red and white porcine muscle. Biochim Biophys Acta. 1973;295(1):188–207. doi: 10.1016/0005-2795(73)90087-1. [DOI] [PubMed] [Google Scholar]
- Takada F, Vander Woude DL, Tong HQ, Thompson TG, Watkins SC, Kunkel LM, Beggs AH. Myozenin: an alpha-actinin- and gamma-filamin-binding protein of skeletal muscle Z lines. Proc Natl Acad Sci U S A. 2001;98(4):1595–600. doi: 10.1073/pnas.041609698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Taylor DW, Taylor KA. The three-dimensional structure of alpha-actinin obtained by cryoelectron microscopy suggests a model for Ca(2+)-dependent actin binding. J Mol Biol. 2001;310(4):845–58. doi: 10.1006/jmbi.2001.4789. [DOI] [PubMed] [Google Scholar]
- Untergasser A, Dumortier J, Oberwinkler H, Protzer U. Production of Adenoviral Vectors Application in Test Animals. Untergasser’s Lab; 2008. [Google Scholar]
- Waites GT, Graham IR, Jackson P, Millake DB, Patel B, Blanchard AD, Weller PA, Eperon IC, Critchley DR. Mutually exclusive splicing of calcium-binding domain exons in chick alpha-actinin. J Biol Chem. 1992;267(9):6263–71. [PubMed] [Google Scholar]
- Walikonis RS, Oguni A, Khorosheva EM, Jeng CJ, Asuncion FJ, Kennedy MB. Densin-180 forms a ternary complex with the (alpha)-subunit of Ca2+/calmodulin-dependent protein kinase II and (alpha)-actinin. J Neurosci. 2001;21(2):423–33. doi: 10.1523/JNEUROSCI.21-02-00423.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Shaner N, Mittal B, Zhou Q, Chen J, Sanger JM, Sanger JW. Dynamics of Z-band based proteins in developing skeletal muscle cells. Cell Motil Cytoskeleton. 2005;61(1):34–48. doi: 10.1002/cm.20063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weins A, Schlondorff JS, Nakamura F, Denker BM, Hartwig JH, Stossel TP, Pollak MR. Disease-associated mutant alpha-actinin-4 reveals a mechanism for regulating its F-actin-binding affinity. Proc Natl Acad Sci U S A. 2007;104(41):16080–5. doi: 10.1073/pnas.0702451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DA, Head SI, Bakker AJ, Stephenson DG. Resting calcium concentrations in isolated skeletal muscle fibres of dystrophic mice. J Physiol. 1990;428:243–56. doi: 10.1113/jphysiol.1990.sp018210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyszynski M, Lin J, Rao A, Nigh E, Beggs AH, Craig AM, Sheng M. Competitive binding of alpha-actinin and calmodulin to the NMDA receptor. Nature. 1997;385(6615):439–42. doi: 10.1038/385439a0. [DOI] [PubMed] [Google Scholar]
- Yao J, Le TC, Kos CH, Henderson JM, Allen PG, Denker BM, Pollak MR. Alpha-actinin-4-mediated FSGS: an inherited kidney disease caused by an aggregated and rapidly degraded cytoskeletal protein. PLoS Biol. 2004;2(6):e167. doi: 10.1371/journal.pbio.0020167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasutomi M, Kunishima S, Okazaki S, Tanizawa A, Tsuchida S, Ohshima Y. ACTN1 rod domain mutation associated with congenital macrothrombocytopenia. Ann Hematol. 2016;95(1):141–4. doi: 10.1007/s00277-015-2517-6. [DOI] [PubMed] [Google Scholar]
- Youssoufian H, McAfee M, Kwiatkowski DJ. Cloning and chromosomal localization of the human cytoskeletal alpha-actinin gene reveals linkage to the beta-spectrin gene. Am J Hum Genet. 1990;47(1):62–72. [PMC free article] [PubMed] [Google Scholar]
- Ziane R, Huang H, Moghadaszadeh B, Beggs AH, Levesque G, Chahine M. Cell membrane expression of cardiac sodium channel Na(v)1.5 is modulated by alpha-actinin-2 interaction. Biochemistry. 2010;49(1):166–78. doi: 10.1021/bi901086v. [DOI] [PMC free article] [PubMed] [Google Scholar]