Abstract
Tissue-resident memory CD8+ T cells (TRM) are localized in non-lymphoid tissues throughout the body where they mediate long-lived protective immunity at common sites of pathogen exposure. As the signals controlling TRM differentiation are uncovered, it is becoming apparent that the dynamic activities of numerous transcription factors are intricately involved in TRM formation. Here, we highlight known transcriptional regulators of TRM differentiation and discuss how understanding the transcriptional programming of CD8+ T cell residency in non-lymphoid tissues can be leveraged to prevent or treat disease.
Introduction: TRM as a distinct memory CD8+ T cell subset
CD8+ T cells are critical for rapid eradication of intracellular pathogens. Following viral or bacterial clearance, the majority of the effector CD8+ T cell population undergoes contraction while a small subset persists indefinitely as memory T cells, mediating durable protective immunity. Memory CD8+ T cells can be broadly segregated into three distinct subsets: central memory (TCM), effector memory (TEM), and tissue-resident memory (TRM) cells [1–4]. TCM and TEM populations are predominantly located in the blood and secondary lymphoid organs [4–7], although TEM and TCM can exhibit the capacity to survey non-lymphoid tissues (NLTs) [3]. TRM are generally defined as memory CD8+ T cells that permanently reside in NLTs without egress [3,8–10]; however, lymphoid tissues may also harbor non-circulating memory subsets as well [11–13]. While effective memory responses likely require the integrated activities of all three memory CD8+ T cell subsets [14,15], TRM exhibit sentinel immune surveillance activity and are critical in the earliest phases of secondary immune responses [10,15,16]. TRM recognizing cognate antigen rapidly induce inflammatory responses, proliferate in situ [17,18], and trigger the trafficking of diverse immune cell types to the site of infection [15,16]. Infection-induced-TRM have been identified in both barrier (e.g. skin, lung, intestine) and non-barrier (e.g. kidney, liver, brain) sites, and the protective activities of TRM have been validated in numerous infection models including HSV [19–21], vaccinia [9,22–24], LCMV [25], influenza [26–28], Listeria [29], malaria [30], as well as in models of malignancy [14,23,31,32]. Given these protective attributes, targeting TRM to enhance vaccine responses represents a powerful approach to combatting infections and cancer.
Transcriptional regulation of CD8+ T cell memory differentiation
Differential expression of CD127 and KLRG1 can be used to distinguish splenic effector CD8+ T cell populations with differing memory potential [33,34]. KLRG1hiCD127lo cells comprise a shorter-lived, terminally-differentiated population of cells referred to as “terminal-effector” cells (TE), whereas KLRG1loCD127hi cells are long-lived with the capacity for proliferation and self renewal, referred to as “memory-precursor” cells (MP) [35]. While circulating memory cells are primarily derived from the latter population, TE can persist for extended periods of time into the memory phase of infection [36]. A number of signals ranging from antigen exposure to inflammatory signals impact this bifurcation in CD8+ T cell differentiation [35]. Further, it is apparent that multiple transcription factors (TFs) operate in concert to instruct MP vs. TE differentiation and circulating memory CD8+ T cell fates. MP cells and TCM are dependent on key TFs such as Id3 [37,38], TCF1 [39], Eomes [40,41], and Bcl6 [42] whereas optimal TE and TEM differentiation requires Id2 [38,43], Blimp1 [44,45], T-bet [33], and Zeb2 [46,47] (Figure 1). As TRM have emerged as a distinct memory subset, understanding how these canonical effector/memory TF relationships apply to TRM has been somewhat unpredictable.
Figure 1. TRM exhibit a hybrid effector/memory TF-driven differentiation program.

A, Key TFs with known functions in controlling TE vs. MP differentiation are highlighted. B, TFs critical to TRM differentiation are included (inside the circle) or that suppress TRM development relative to circulating memory (outside the circle). Additionally, TFs that are predicted to be required for TRM differentiation based on gene expression [49] (inside the circle) or predicted to suppress TRM differentiation (outside the circle) are included. TFs with validated roles are bolded whereas predicted regulators are not.
TRM differentiation requires a “hybrid” of effector and memory cell transcriptional programs
The observation that early effector populations located in NLTs are predominantly KLRG1lo led to the finding that TRM are preferentially derived from KLRG1lo cells [29,48,49]. Thus, one might expect shared TF programs between TRM precursors and MP cells. However, it was recently demonstrated that early differentiating TRM cells are transcriptionally distinct from MP cells [49]. Instead, and perhaps unexpectedly, it appears that TRM utilize a hybrid TF differentiation program, requiring TFs associated with both memory and effector cell specification (Figure 1) [35,49–51]. For example, T-bet promotes TE/TEM formation [33] but suppresses TRM differentiation [19,26], whereas Blimp1 also promotes TE/TEM formation [44,45] but is required for TRM [52]. Conversely, pro-memory Eomes suppresses TRM formation [19,35], but Nr4a1 which supports formation of MP/TCM cells [53] is required for TRM [54]. Further, other characteristics of TRM seem to fit with this hybrid model. TRM appear to be activated or effector-like by expressing elevated levels of CD69, PD-1, CTLA4, IFNγ, and GzB [49,55], reminiscent of early effector CD8+ T cells or TEM; yet, TRM share some of the “stem-like” properties of TCM in that they are long-lived and not terminally differentiated, giving rise to both TRM and circulating memory cells upon transfer and reinfection into naive recipient mice [55]. Despite these overlapping features, TRM are transcriptionally distinct from TE/TEM and MP/TCM [48,49]. Taken together, TRM appear to “rewire” MP/TCM and TE/TEM TF-driven differentiation programs in order to sustain elevated effector function and long-term survival in NLTs.
It has become evident that TFs regulate multiple aspects of the TRM differentiation pathway, which broadly consists of: 1) NLT trafficking, 2) in situ differentiation and retention, and 3) long-term maintenance within NLTs (Figure 2) [56,57]. Understanding the molecular processes underlying TRM differentiation and maintenance are critical for promoting TRM in a therapeutic setting, and may reveal insights into the seemingly hybrid nature of TRM.
Figure 2. TF regulation of TRM differentiation.
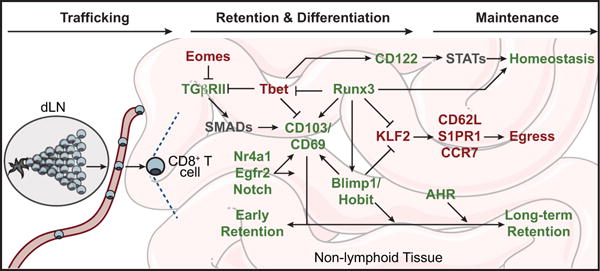
TFs with established roles in promoting TRM are highlighted in green, those repressing TRM are highlighted in red, and putative regulators are gray. dLN; draining lymph node.
Trafficking to NLTs
Within draining lymph nodes (dLNs), naive pathogen-specific CD8+ T cells recognize cognate antigen in the context of MHC class I on antigen-presenting cells, which triggers subsequent CD8+ T cell activation and clonal expansion. Transient suppression of the TF KLF2 results in downregulation of the KLF2-target gene, S1pr1 [58,59]. S1PR1 promotes dLN egress through sphingosine 1 phosphate (S1P) chemotactic gradients, and thus suppressed S1PR1 expression results in prolonged retention in the dLN, allowing for optimal priming of antigen-specific CD8+ T cells [60]. Upon receiving sufficient activation signals, clonally expanded CD8+ T cells are either programmed to traffic to infected sites or express LN homing molecules, such as CD62L and CCR7, to support continued surveillance of secondary lymphoid organs [61,62].
Within dLNs, CD8+ T cell expression of tissue-homing factors facilitates migration to distinct NLTs, which can be regulated by the initial dendritic cell priming [63]. For example, upregulation of CCR9 in the mesenteric LN (gut dLN) [64] sensitizes cells to CCL25 chemotactic gradients secreted in the intestinal epithelium, and α4β7 integrin expression on CD8+ T cells allows binding to the mucosal adressin cell-adhesion molecule (MAdCAM) on intestinal post-capillary venules [62,65]. Alternatively, in skin dLN, chemokine receptors such as CXCR6 and CCR10 [66,67] as well as a variant of the P-selectin ligand, CLA, are associated with CD8+ T cell epidermal homing [62,68]. In peripheral blood, expression of integrin molecules and other cell-cell adhesion molecules facilitate extravasation into infected tissues [6] wherein TRM differentiation ensues.
Retention and differentiation
Upon NLT localization, T cells must adapt to unique microenvironmental cues including diverse cytokine and nutrient milieus as well as low oxygen tension [22,35,69]. Indeed, by day 7 of LCMV infection, effector cells located in non-lymphoid sites are distinct from peripheral blood or splenic-localized effector cells both at the levels of gene-expression and chromatin accessibility [49]. Therefore, entry into NLTs triggers rapid changes in gene-expression programs linked to tissue-residency and adaptation to NLT microenvironments.
Although lymphoid-derived KLRG1lo cells preferentially give rise to TRM [48,49], it is currently unclear which NLT-localized effector cells develop into mature long-lived TRM, exit NLT, or undergo apoptosis. Nevertheless, CD8+ T cells must engage tissue-retention programs and suppress tissue-egress in order to be maintained in NLTs [59]. Parabiosis studies, wherein the vasculature of two mice are conjoined, have demonstrated that CD8+ TRM populations do not equilibrate between NLTs [3,59]. Further, memory CD8+ T cells contained within skin, dorsal root ganglia, or small intestine grafts do not equilibrate with host tissues upon transplant [20,70,71]. Therefore, once CD8+ T cells become lodged in NLTs they do not recirculate, and this is the essence of what defines CD8+ T cells as TRM.
Prototypical TRM retention factors include the integrin molecule CD103 (encoded by Itgae) and the glycoprotein CD69. CD103 expression, induced by TGFβ [48,72,73], is thought to mediate tissue retention through binding to E-cadherin molecules expressed on epithelial cells [74,75], whereas CD69 antagonizes S1PR1-mediated tissue egress [76,77]; while CD103- or CD69-deficiency can impact TRM formation [48,73], they are not essential for all TRM populations [3,69]. Early upregulation of CD103 may signify a TRM -precursor population as these cells are KLRG1lo, Bcl2hi, and likely have an advantage of enhanced retention/survival [29,48,78]. Conversely, retention also requires downregulation of the KLF2 egress program, including suppressed expression of S1PR1, CCR7, and CD62L [48,59]. Utilizing a KLF2-reporter system, Skon et al. demonstrated that expression of KLF2 is rapidly suppressed upon NLT infiltration, and forced expression of S1PR1 impairs formation of TRM in a variety of NLTs during LCMV infection [59]. Repression of the KLF2-S1PR1 egress program appears to be a common target among multiple TFs supporting TRM differentiation. This is highlighted by the finding that Blimp1 and its homolog Hobit have recently been shown to synergistically regulate TRM differentiation by controlling tissue egress [52]. Mackay et al. demonstrated that dual deletion of Blimp1 and Hobit impaired TRM maintenance in the skin through suppression of genes such as Klf2, S1pr1, and Ccr7. Disruption of Blimp1 and Hobit expression impairs CD8+ TRM formation in multiple NLTs as well as liver NKT cell residency, providing evidence of a shared tissue-residency program among different tissues and cell types [52].
In line with findings that Blimp1/Hobit universally regulate lymphocyte residency, we have recently demonstrated that Runx3 is also required for TRM differentiation in a range of NLTs [49]. Through computational and RNAi screening approaches, Runx3 was identified as a putative regulator of TRM and functionally validated through inducible deletion approaches. Runx3 was critical for promoting expression of key tissue-residency factors such as CD103, CD69, and Blimp1 while suppressing T-bet expression as well as tissue egress molecules including KLF2, S1PR1, and CCR7. Of interest, it has been demonstrated that differentiation of intestinal ILC1 and ILC3 subsets as well as liver NK cells requires Runx3 [79,80], and Runx3 is critical for intestinal localization of CD4+ T cells [81,82]. Further, Runx3 regulates the formation of skin-localized γδ T cells [83]. Taken together, Runx3 also appears to be a central regulator of tissue-residency in diverse immune cell types across multiple NLTs.
Interestingly, T-box transcription factors, T-bet and Eomes, have been shown to suppress TRM formation [19]. Suppression of T-bet and Eomes is critical for optimal TGFβ signaling in differentiating skin TRM, and forced expression of T-bet or Eomes impairs skin TRM formation. Further, it was demonstrated that homozygous or heterozygous deletion of Tbx21 (encoding T-bet) enhanced T cell lodging in the skin [19]. Laidlaw et al. demonstrated that T-bet-deficiency also enhances TRM differentiation in the lung following influenza infection, and forced T-bet expression impaired formation of TRM [26]. The role of T-bet in TRM differentiation is likely multifaceted as it also may directly suppress Itgae (encoding CD103) expression [26].
A number of other TFs are important in the differentiation of TRM. It was demonstrated that loss of AHR in CD8+ T cells impairs the formation of skin TRM [84], and it is likely AHR is important for TRM in multiple NLTs as it is also upregulated in intestinal TRM cells [49,52]. The orphan nuclear receptor TFs Nr4a1, Nr4a2, and Nr4a3 are among the most differentially expressed TFs in intestinal TRM relative to splenic TCM, and are highly upregulated in TRM from other sites [48,49]. In connection, loss of Nr4a1 resulted in impaired TRM formation in the intestine and liver [30]. Further, Nr4a2 and Nr4a3 were also indicated to have important roles in supporting TRM differentiation as RNAi impaired CD8+ T cell accumulation in the intestine relative to splenic memory cells in a loss-of-function screening approach [49]. Further, Egr2 [50] and Notch [85] TFs were required for optimal lung TRM formation following influenza infection. Transcriptional profiling of TRM populations has also yielded insight into additional putative regulators of TRM. Consistent with a hybrid TF program, Zeb2 (pro-TE) and TCF1 (pro-MP) are expressed at lower levels in TRM relative to circulating cells, whereas IRF4 (pro-TE) and Bcl6 (pro-MP) are upregulated in TRM [49]. As novel roles for TFs regulating TRM differentiation are uncovered and their functions elucidated, it will become clear if TRM have rewired their transcriptional profile to support effector and memory TF programs simultaneously, or if their dual nature is indicative of distinct TRM subsets overlooked by bulk transcriptional analyses.
Long-term maintenance
Following pathogen clearance, CD8+ effector cells localized to NLT must maintain expression of retention programs and suppress egress. Although it is unclear at what point TRM precursors become mature TRM, it was shown through principal component analysis of gene expression that the transcriptional signature of long-lived skin TRM was predominantly established by day 25 of vaccinia infection [22]. The homeostatic demands of TRM are also relatively unclear, though access to certain cytokines and changes in cellular metabolism may accompany long-term survival. TRM have been shown to require canonical homeostatic cytokines IL-15 and IL-7 for survival [19,48,86], although IL-15 is not essential for TRM in lymphoid tissues [87]. Despite the suppressive role of T-bet during NLT lodging, some level of expression is necessary for long-term TRM survival through promoting CD122 expression and IL-15 responsiveness [19]. Further, TGFβ-signaling is critical for TRM differentiation in the intestine, skin, and kidney, and thus continual access to TGFβ beyond early TRM differentiation is presumably required for long-term maintenance [19,72,73,88]. Given the importance of TGFβ, IL-7, and IL-15 signaling in promoting and maintaining TRM, it is likely certain downstream SMAD- and STAT-family TFs are required for TRM formation and maintenance; however, SMAD4 was shown to be dispensable for TRM [89].
The microenvironment of NLTs is distinct from that of lymphoid tissues or blood, including differences in nutrient availability and oxygen tension [90]. It was recently demonstrated that mature TRM acquire exogenous free fatty acids through expression of FABP4 and FABP5, which are subsequently oxidized to fuel energetic demands required for TRM survival in the lung and skin [22]. Further, mTOR activity is essential for optimal TRM generation as rapamycin treatment blunts their formation [91]. However, it is unclear what other metabolic adaptations are required for TRM survival in diverse NLT microenvironments or which TF programs sustain altered expression and activity of key metabolic enzymes.
One difficulty with identifying regulators of TRM maintenance is that many genes required for long-term TRM homeostasis are also linked to NLT retention and early differentiation. Therefore, disruption of key TFs often impairs differentiation, which impedes the assignment of functional roles in maintenance and longevity. Utilizing a tamoxifen-inducible deletion system, it was recently demonstrated that Runx3 is also important for TRM maintenance in addition to controlling early differentiation [49]. Hobit and AHR also appear to be critical for later maintenance phases of TRM development [52,84]. A deeper understanding of TRM heterogeneity may provide insight into the dynamic requirements for certain TFs, helping to answer questions such as: 1) Do TRM continuously mature over time, changing the kinetic requirements of certain TF? or 2) Are there long-lived and short-lived subsets of TRM that have distinct TF requirements?
Targeting transcriptional programming of TRM to combat disease
Given the sentinel role of TRM in host protection, developing approaches to induce TRM is an attractive avenue for enhancing vaccine efficacy [62]. Further, it has become evident that promoting or repressing tissue-residency of CD8+ T cells may be relevant in non-infectious settings ranging from cancer to inflammatory diseases and autoimmunity [62]. The tumor microenvironment holds parallels to NLTs (relative to lymphoid organs) including lower oxygen tension, nutrient availability, and a distinct cytokine milieu (e.g. TGFβ) [90,92]. In connection, it has been demonstrated that tumor infiltrating lymphocytes (TILs) share certain features of TRM [93,94], and that a TRM-like gene expression program in TILs is linked to better prognoses in lung cancer patients [95]. TRM express elevated levels of effector molecules such as GzB, IFNγ, and FasL relative to circulating cells [49,55], which is another characteristic of TRM that would benefit the anti-tumor function of TILs [95]. In connection, we have demonstrated that Runx3 is critical for CD8+ T cell accumulation in tumors and is key in programming expression of genes that support tissue-residency and cytotoxic function of TILs [49]. Therefore, targeting TFs that orchestrate multiple aspects of the TRM program could enhance the efficacy of adoptive cell therapies against malignancies.
In other instances, the potent inflammatory and cytotoxic capacity of CD8+ T cells in NLTs can be harmful [62]. In autoimmune or inflammatory diseases such as diabetes, psoriasis, or inflammatory bowel disease, CD8+ T cells residing in the pancreas [96], skin [22,61], or intestine [92] likely acquire aspects of tissue-residency reprogramming, albeit they likely are not true memory cells (as the term TRM implies) given the presence of persistent self-antigen. Indeed, conditions that impair TRM formation such as CD103-deficiency [97] or rapamycin treatment [91] have also been shown to alleviate pathology in models of intestinal inflammation/autoimmunity. Further, TRM have been shown to mediate rapid contact hypersensitivity in the skin [61]. Notably, other related conditions include metabolic diseases in which CD8+ T cells localize to inflamed white adipose tissue [98] or atherosclerotic lesions [99] and further potentiate inflammation. Taken together, manipulating TRM TF programs to block engagement of tissue-residency features could be beneficial in these contexts.
Conclusions
TRM have emerged as key mediators of long-lived immunity. Given the expansive role of TFs in regulating expression of diverse molecules critical to CD8+ T cell residency in NLTs, defining novel functions for TFs in controlling TRM differentiation will likely enhance our ability to treat complex T cell mediated-diseases and yield important information regarding the ontogeny, function, and hybrid nature of TRM. Further, parsing out overlapping requirements of CD8+ T cell tissue-residency in healthy (following acute infection) and diseased (e.g. tumors or autoimmunity) non-lymphoid settings may be conceptually useful in studying TRM development and informing approaches to improve adoptive cell therapies or suppress inflammatory/auto-reactive CD8+ T cell populations.
Highlights.
The TRM transcriptional differentiation program employs drivers of both effector and memory T cell subsets
Early differentiating TRM are transcriptionally distinct from circulating memory-precursor cells
Blimp1, Hobit, and Runx3 are central regulators of tissue-residency across multiple immune cell types and non-lymphoid sites
Promoting or suppressing CD8+ T cell residency in non-lymphoid tissues may be a therapeutic strategy to treat cancers or prevent inflammatory diseases
Acknowledgments
This review was supported by the US National Institutes of Health U19AI109976 to AWG and the UCSD Molecular Biology Cancer Fellowship to JJM. We thank the Goldrath lab for their insightful input, especially Clara Toma, Laura Shaw, and Quyhn Nguyen.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wherry EJ, Teichgräber V, Becker TC, Masopust D, Kaech SM, Antia R, von Andrian UH, Ahmed R. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat Immunol. 2003;4:225–234. doi: 10.1038/ni889. [DOI] [PubMed] [Google Scholar]
- 2.Masopust D, Vezys V, Marzo AL, Lefrançois L. Preferential localization of effector memory cells in nonlymphoid tissue. Science. 2001;291:2413–2417. doi: 10.1126/science.1058867. [DOI] [PubMed] [Google Scholar]
- ••3.Steinert EM, Schenkel JM, Fraser KA, Beura LK, Manlove LS, Igyártó BZ, Southern PJ, Masopust D. Quantifying Memory CD8 T Cells Reveals Regionalization of Immunosurveillance. Cell. 2015;161:737–749. doi: 10.1016/j.cell.2015.03.031. Steinert et al. demonstrate that current approaches for isolation of T cells from non-lymphoid tissues fail to recover most cells and can bias results. Further, the authors provide compelling data that our current understanding of the migration and localization patterns of memory CD8 T cell subsets is incomplete. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sallusto F, Lenig D, Förster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 5.Carbone FR, Mackay LK, Heath WR, Gebhardt T. Distinct resident and recirculating memory T cell subsets in non-lymphoid tissues. Curr Opin Immunol. 2013;25:329–333. doi: 10.1016/j.coi.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Mueller SN, Gebhardt T, Carbone FR, Heath WR. Memory T cell subsets, migration patterns, and tissue residence. Annu Rev Immunol. 2013;31:137–161. doi: 10.1146/annurev-immunol-032712-095954. [DOI] [PubMed] [Google Scholar]
- 7.Gebhardt T, Whitney PG, Zaid A, Mackay LK, Brooks AG, Heath WR, Carbone FR, Mueller SN. Different patterns of peripheral migration by memory CD4+ and CD8+ T cells. Nature. 2011;477:216–219. doi: 10.1038/nature10339. [DOI] [PubMed] [Google Scholar]
- 8.Klonowski KD, Williams KJ, Marzo AL, Blair DA, Lingenheld EG, Lefrançois L. Dynamics of blood-borne CD8 memory T cell migration in vivo. Immunity. 2004;20:551–562. doi: 10.1016/s1074-7613(04)00103-7. [DOI] [PubMed] [Google Scholar]
- 9.Jiang X, Clark RA, Liu L, Wagers AJ, Fuhlbrigge RC, Kupper TS. Skin infection generates non-migratory memory CD8+ TRM cells providing global skin immunity. Nature. 2012;483:227–231. doi: 10.1038/nature10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ariotti S, Hogenbirk MA, Dijkgraaf FE, Visser LL, Hoekstra ME, Song J-Y, Jacobs H, Haanen JB, Schumacher TN. Skin-resident memory CD8+ T cells trigger a state of tissue-wide pathogen alert. Science. 2014;346:101–105. doi: 10.1126/science.1254803. [DOI] [PubMed] [Google Scholar]
- 11.Schenkel JM, Fraser KA, Masopust D. Cutting edge: resident memory CD8 T cells occupy frontline niches in secondary lymphoid organs. J Immunol. 2014;192:2961–2964. doi: 10.4049/jimmunol.1400003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••12.Kumar BV, Ma W, Miron M, Granot T, Guyer RS, Carpenter DJ, Senda T, Sun X, Ho SH, Lerner H, et al. Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep. 2017;20:2921–2934. doi: 10.1016/j.celrep.2017.08.078. This study and others by Farber and colleagues demonstrate that human non-lymphoid tissues have abundant populations of CD69+CD103+ Trm cells, and human Trm cells are transcriptionally and phenotypically similar to mouse Trm cells in many contexts. Further, it was also demonstarted that human lymph nodes and spleen have an abundant population of CD8+CD69+ cells further indicating that Trm populations may also exist in lymphoid sites. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beura LK, Wijeyesinghe S, Thompson EA, Macchietto MG, Rosato PC, Pierson MJ, Schenkel JM, Mitchell JS, Vezys V, Fife BT, et al. T Cells in Nonlymphoid Tissues Give Rise to Lymph-Node-Resident Memory T Cells. Immunity. 2018;48:327–338.e325. doi: 10.1016/j.immuni.2018.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enamorado M, Iborra S, Priego E, Cueto FJ, Quintana JA, Martínez-Cano S, Mejías-Pérez E, Esteban M, Melero I, Hidalgo A, et al. Enhanced anti-tumour immunity requires the interplay between resident and circulating memory CD8+ T cells. Nat Commun. 2017;8:16073. doi: 10.1038/ncomms16073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schenkel JM, Fraser KA, Beura LK, Pauken KE, Vezys V, Masopust D. Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science. 2014;346:98–101. doi: 10.1126/science.1254536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schenkel JM, Fraser KA, Vezys V, Masopust D. Sensing and alarm function of resident memory CD8+ T cells. Nat Immunol. 2013;14:509–513. doi: 10.1038/ni.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beura LK, Mitchell JS, Thompson EA, Schenkel JM, Mohammed J, Wijeyesinghe S, Fonseca R, Burbach BJ, Hickman HD, Vezys V, et al. Intravital mucosal imaging of CD8+ resident memory T cells shows tissue-autonomous recall responses that amplify secondary memory. Nat Immunol. 2018;19:173–182. doi: 10.1038/s41590-017-0029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park SL, Zaid A, Hor JL, Christo SN, Prier JE, Davies B, Alexandre YO, Gregory JL, Russell TA, Gebhardt T, et al. Local proliferation maintains a stable pool of tissue-resident memory T cells after antiviral recall responses. Nat Immunol. 2018;19:183–191. doi: 10.1038/s41590-017-0027-5. [DOI] [PubMed] [Google Scholar]
- ••19.Mackay LK, Wynne-Jones E, Freestone D, Pellicci DG, Mielke LA, Newman DM, Braun A, Masson F, Kallies A, Belz GT, et al. T-box Transcription Factors Combine with the Cytokines TGF-β and IL-15 to Control Tissue-Resident Memory T Cell Fate. Immunity. 2015;43:1101–1111. doi: 10.1016/j.immuni.2015.11.008. This study demonstrated that T-box transcription factors, T-bet and Eomes, suppress TRM differentiation in the skin. Further, T-bet-deficiency was shown to enhance CD8+ T cell lodging in the skin. However, Tbet was also important for the maintenance of TRM through promoting CD122 expression and IL-15 responsiveness. [DOI] [PubMed] [Google Scholar]
- 20.Gebhardt T, Wakim LM, Eidsmo L, Reading PC, Heath WR, Carbone FR. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat Immunol. 2009;10:524–530. doi: 10.1038/ni.1718. [DOI] [PubMed] [Google Scholar]
- 21.Shin H, Iwasaki A. A vaccine strategy that protects against genital herpes by establishing local memory T cells. Nature. 2012;491:463–467. doi: 10.1038/nature11522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••22.Pan Y, Tian T, Park CO, Lofftus SY, Mei S, Liu X, Luo C, O’Malley JT, Gehad A, Teague JE, et al. Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature. 2017;543:252–256. doi: 10.1038/nature21379. Pan et al. demonstrate that human and mouse skin TRM express elevated levels of fatty acid binding proteins, FABP4 and FABP5, relative to circulating memory cells. Their data show that uptake and metabolism of free fatty acids is essential for the long-term survival of TRM. This study is the first to provide evidence of the unique metabolic adaptations required for TRM maintenance in non-lymphoid tissues. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu L, Zhong Q, Tian T, Dubin K, Athale SK, Kupper TS. Epidermal injury and infection during poxvirus immunization is crucial for the generation of highly protective T cell-mediated immunity. Nat Med. 2010;16:224–227. doi: 10.1038/nm.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khan TN, Mooster JL, Kilgore AM, Osborn JF, Nolz JC. Local antigen in nonlymphoid tissue promotes resident memory CD8+ T cell formation during viral infection. J Exp Med. 2016;213:951–966. doi: 10.1084/jem.20151855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steinbach K, Vincenti I, Kreutzfeldt M, Page N, Muschaweckh A, Wagner I, Drexler I, Pinschewer D, Korn T, Merkler D. Brain-resident memory T cells represent an autonomous cytotoxic barrier to viral infection. J Exp Med. 2016;213:1571–1587. doi: 10.1084/jem.20151916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laidlaw BJ, Zhang N, Marshall HD, Staron MM, Guan T, Hu Y, Cauley LS, Craft J, Kaech SM. CD4+ T cell help guides formation of CD103+ lung-resident memory CD8+ T cells during influenza viral infection. Immunity. 2014;41:633–645. doi: 10.1016/j.immuni.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zens KD, Chen JK, Farber DL. Vaccine-generated lung tissue–resident memory T cells provide heterosubtypic protection to influenza infection. JCI Insight. 2016;1:e85832. doi: 10.1172/jci.insight.85832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slütter B, Van Braeckel-Budimir N, Abboud G, Varga SM, Salek-Ardakani S, Harty JT. Dynamics of influenza-induced lung-resident memory T cells underlie waning heterosubtypic immunity. Sci Immunol. 2017;2:eaag2031. doi: 10.1126/sciimmunol.aag2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheridan BS, Pham QM, Lee YT, Cauley LS, Puddington L, Lefrançois L. Oral infection drives a distinct population of intestinal resident memory CD8+ T cells with enhanced protective function. Immunity. 2014;40:747–757. doi: 10.1016/j.immuni.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernandez-Ruiz D, Ng WY, Holz LE, Ma JZ, Zaid A, Wong YC, Lau LS, Mollard V, Cozijnsen A, Collins N, et al. Liver-resident memory CD8+ T cells form a front-line defense against malaria liver-stage infection. Immunity. 2016;45:889–902. doi: 10.1016/j.immuni.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 31.Nizard M, Roussel H, Diniz MO, Karaki S, Tran T, Voron T, Dransart E, Sandoval F, Riquet M, Rance B, et al. Induction of resident memory T cells enhances the efficacy of cancer vaccine. Nat Comm. 2017;8:15221. doi: 10.1038/ncomms15221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malik BT, Byrne KT, Vella JL, Zhang P, Shabaneh TB, Steinberg SM, Molodtsov AK, Bowers JS, Angeles CV, Paulos CM, et al. Resident memory T cells in the skin mediate durable immunity to melanoma. Sci Immunol. 2017;2:eaam6346. doi: 10.1126/sciimmunol.aam6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, Gapin L, Kaech SM. Inflammation directs memory precursor and short-lived effector CD8+ T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27:281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarkar S, Kalia V, Haining WN, Konieczny BT, Subramaniam S, Ahmed R. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J Exp Med. 2008;205:625–640. doi: 10.1084/jem.20071641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang JT, Wherry EJ, Goldrath AW. Molecular regulation of effector and memory T cell differentiation. Nat Immunol. 2014;15:1104–1115. doi: 10.1038/ni.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olson JA, McDonald-Hyman C, Jameson SC, Hamilton SE. Effector-like CD8+ T cells in the memory population mediate potent protective immunity. Immunity. 2013;38:1250–1260. doi: 10.1016/j.immuni.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji Y, Pos Z, Rao M, Klebanoff CA, Yu Z, Sukumar M, Reger RN, Palmer DC, Borman ZA, Muranski P, et al. Repression of the DNA-binding inhibitor Id3 by Blimp-1 limits the formation of memory CD8+ T cells. Nat Immunol. 2011;12:1230–1237. doi: 10.1038/ni.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang CY, Best JA, Knell J, Yang E, Sheridan AD, Jesionek AK, Li HS, Rivera RR, Lind KC, D’Cruz LM, et al. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat Immunol. 2011;12:1221–1229. doi: 10.1038/ni.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou X, Yu S, Zhao DM, Harty JT, Badovinac VP, Xue HH. Differentiation and persistence of memory CD8+ T cells depend on T cell factor 1. Immunity. 2010;33:229–240. doi: 10.1016/j.immuni.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, Banica M, DiCioccio CB, Gross DA, Mao CA, et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 41.Banerjee A, Gordon SM, Intlekofer AM, Paley MA, Mooney EC, Lindsten T, Wherry EJ, Reiner SL. Cutting edge: The transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J Immunol. 2010;185:4988–4992. doi: 10.4049/jimmunol.1002042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cui W, Liu Y, Weinstein JS, Craft J, Kaech SM. An interleukin-21-interleukin-10-STAT3 pathway is critical for functional maturation of memory CD8+ T cells. Immunity. 2011;35:792–805. doi: 10.1016/j.immuni.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cannarile MA, Lind NA, Rivera R, Sheridan AD, Camfield KA, Wu BB, Cheung KP, Ding Z, Goldrath AW. Transcriptional regulator Id2 mediates CD8+ T cell immunity. Nat Immunol. 2006;7:1317–1325. doi: 10.1038/ni1403. [DOI] [PubMed] [Google Scholar]
- 44.Kallies A, Xin A, Belz GT, Nutt SL. Blimp-1 transcription factor is required for the differentiation of effector CD8+ T cells and memory responses. Immunity. 2009;31:283–295. doi: 10.1016/j.immuni.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 45.Rutishauser RL, Martins GA, Kalachikov S, Chandele A, Parish IA, Meffre E, Jacob J, Calame K, Kaech SM. Transcriptional repressor Blimp-1 promotes CD8+ T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 2009;31:296–308. doi: 10.1016/j.immuni.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Omilusik KD, Best JA, Yu B, Goossens S, Weidemann A, Nguyen JV, Seuntjens E, Stryjewska A, Zweier C, Roychoudhuri R, et al. Transcriptional repressor ZEB2 promotes terminal differentiation of CD8+ effector and memory T cell populations during infection. J Exp Med. 2015;212:2027–2039. doi: 10.1084/jem.20150194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dominguez CX, Amezquita RA, Guan T, Marshall HD, Joshi NS, Kleinstein SH, Kaech SM. The transcription factors ZEB2 and T-bet cooperate to program cytotoxic T cell terminal differentiation in response to LCMV viral infection. J Exp Med. 2015;212:2041–2056. doi: 10.1084/jem.20150186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mackay LK, Rahimpour A, Ma JZ, Collins N, Stock AT, Hafon M-L, Vega-Ramos J, Lauzurica P, Mueller SN, Stefanovic T, et al. The developmental pathway for CD103+ CD8+ tissue-resident memory T cells of skin. Nat Immunol. 2013;14:1294–1301. doi: 10.1038/ni.2744. [DOI] [PubMed] [Google Scholar]
- ••49.Milner JJ, Toma C, Yu B, Zhang K, Omilusik K, Phan AT, Wang D, Getzler AJ, Nguyen T, Crotty S, et al. Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumors. Nature. 2017;552:253–257. doi: 10.1038/nature24993. In this study, it was demonstrated that early differentiating TRM are transcriptionally distinct from circulating memory precursor cells, and Runx3 was identified as a central regulator of TRM differentiation in mutliple non-lymphoid sites. It was also shown that tumor infiltrating lymphocytes (TILs) share certain tissue-residency qualities with TRM, and Runx3 was required for TIL accumulation. Overexpression of Runx3 resulted in enhanced TIL accumulation in tumors and enhanced protection in an adoptive therapy model. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Du N, Kwon H, Li P, West EE, Oh J, Liao W, Yu Z, Ren M, Leonard WJ. EGR2 is critical for peripheral naïve T-cell differentiation and the T-cell response to influenza. Proc Natl Acad Sci U S A. 2014;111:16484–16489. doi: 10.1073/pnas.1417215111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mathieu M, Duval F, Daudelin J-F, Labrecque N. The Notch signaling pathway controls short-lived effector CD8+ T cell differentiation but is dispensable for memory generation. J Immunol. 2015;194:5654–5662. doi: 10.4049/jimmunol.1402837. [DOI] [PubMed] [Google Scholar]
- ••52.Mackay LK, Minnich M, Kragten NA, Liao Y, Nota B, Seillet C, Zaid A, Man K, Preston S, Freestone D, et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science. 2016;352:459–463. doi: 10.1126/science.aad2035. This is the first study to demonstrate that distinct immune cell types localized in non-lymphoid tissues share a universal tissue-residency program. Further, it was demonstrated that Blimp1 and its homolog Hobit are central regulators of tissue-residency features in multiple cell types and tissue sites. [DOI] [PubMed] [Google Scholar]
- 53.Nowyhed HN, Huynh TR, Thomas GD, Blatchley A, Hedrick CC. Cutting edge: the orphan nuclear receptor Nr4a1 regulates CD8+ T cell expansion and effector function through direct repression of Irf4. J Immunol. 2015;195:3515–3519. doi: 10.4049/jimmunol.1403027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boddupalli CS, Nair S, Gray SM, Nowyhed HN, Verma R, Gibson JA, Abraham C, Narayan D, Vasquez J, Hedrick CC, et al. ABC transporters and NR4A1 identify a quiescent subset of tissue-resident memory T cells. J Clin Invest. 2016;126:3905–3916. doi: 10.1172/JCI85329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Masopust D, Vezys V, Wherry EJ, Barber DL, Ahmed R. Cutting edge: gut microenvironment promotes differentiation of a unique memory CD8 T cell population. J Immunol. 2006;176:2079–2083. doi: 10.4049/jimmunol.176.4.2079. [DOI] [PubMed] [Google Scholar]
- 56.Mackay LK, Kallies A. Transcriptional Regulation of Tissue-Resident Lymphocytes. Trends Immunol. 2016;38:94–103. doi: 10.1016/j.it.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 57.Mueller SN, Mackay LK. Tissue-resident memory T cells: local specialists in immune defence. Nat Rev Immunol. 2016;16:79–89. doi: 10.1038/nri.2015.3. [DOI] [PubMed] [Google Scholar]
- 58.Bai A, Hu H, Yeung M, Chen J. Krüppel-like factor 2 controls T cell trafficking by activating L-selectin (CD62L) and sphingosine-1-phosphate receptor 1 transcription. J Immunol. 2007;178:7632–7639. doi: 10.4049/jimmunol.178.12.7632. [DOI] [PubMed] [Google Scholar]
- 59.Skon CN, Lee JY, Anderson KG, Masopust D, Hogquist KA, Jameson SC. Transcriptional downregulation of S1pr1 is required for the establishment of resident memory CD8+ T cells. Nat Immunol. 2013;14:1285–1293. doi: 10.1038/ni.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schenkel JM, Masopust D. Tissue-resident memory T cells. Immunity. 2014;41:886–897. doi: 10.1016/j.immuni.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gaide O, Emerson RO, Jiang X, Gulati N, Nizza S, Desmarais C, Robins H, Krueger JG, Clark RA, Kupper TS. Common clonal origin of central and resident memory T cells following skin immunization. Nat Med. 2015;21:647–653. doi: 10.1038/nm.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •62.Park CO, Kupper TS. The emerging role of resident memory T cells in protective immunity and inflammatory disease. Nat Med. 2015;21:688–697. doi: 10.1038/nm.3883. This is a comprehensive and insightful review that highlights key features of TRM ontogeny and function. Further, the authors provide novel insight into the potential roles of TRM in other non-infectious diseases such as cancer and autoimmunity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Iwata M, Hirakiyama A, Eshima Y, Kagechika H, Kato C, Song S-Y. Retinoic acid imprints gut-homing specificity on T cells. Immunity. 2004;21:527–538. doi: 10.1016/j.immuni.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 64.Zabel BA, Agace WW, Campbell JJ, Heath HM, Parent D, Roberts AI, Ebert EC, Kassam N, Qin S, Zovko M, et al. Human G protein–coupled receptor GPR-9-6/CC chemokine receptor 9 is selectively expressed on intestinal homing T lymphocytes, mucosal lymphocytes, and thymocytes and is required for thymus-expressed chemokine–mediated chemotaxis. J Exp Med. 1999;190:1241–1256. doi: 10.1084/jem.190.9.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Briskin M, Winsor-Hines D, Shyjan A, Cochran N, Bloom S, Wilson J, McEvoy LM, Butcher EC, Kassam N, Mackay CR. Human mucosal addressin cell adhesion molecule-1 is preferentially expressed in intestinal tract and associated lymphoid tissue. Am J Pathol. 1997;151:97–110. [PMC free article] [PubMed] [Google Scholar]
- 66.Tubo NJ, McLachlan JB, Campbell JJ. Chemokine receptor requirements for epidermal T-cell trafficking. Am J Pathol. 2011;178:2496–2503. doi: 10.1016/j.ajpath.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zaid A, Hor JL, Christo SN, Groom JR, Heath WR, Mackay LK, Mueller SN. Chemokine Receptor–Dependent Control of Skin Tissue–Resident Memory T Cell Formation. J Immunol. 2017;199:2451–2459. doi: 10.4049/jimmunol.1700571. [DOI] [PubMed] [Google Scholar]
- 68.Fuhlbrigge RC, Kieffer JD, Armerding D, Kupper TS. Cutaneous lymphocyte antigen is a specialized form of PSGL-1 expressed on skin-homing T cells. Nature. 1997;389:978–981. doi: 10.1038/40166. [DOI] [PubMed] [Google Scholar]
- 69.Bergsbaken T, Bevan MJ. Proinflammatory microenvironments within the intestine regulate the differentiation of tissue-resident CD8+ T cells responding to infection. Nat Immunol. 2015;16:406–414. doi: 10.1038/ni.3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Masopust D, Choo D, Vezys V, Wherry EJ, Duraiswamy J, Akondy R, Wang J, Casey KA, Barber DL, Kawamura KS, et al. Dynamic T cell migration program provides resident memory within intestinal epithelium. J Exp Med. 2010;207:553–564. doi: 10.1084/jem.20090858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wakim LM, Waithman J, van Rooijen N, Heath WR, Carbone FR. Dendritic cell-induced memory T cell activation in nonlymphoid tissues. Science. 2008;319:198–202. doi: 10.1126/science.1151869. [DOI] [PubMed] [Google Scholar]
- 72.Zhang N, Bevan MJ. Transforming growth factor-β signaling controls the formation and maintenance of gut-resident memory T cells by regulating migration and retention. Immunity. 2013;39:687–696. doi: 10.1016/j.immuni.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Casey KA, Fraser KA, Schenkel JM, Moran A, Abt MC, Beura LK, Lucas PJ, Artis D, Wherry EJ, Hogquist K, et al. Antigen-independent differentiation and maintenance of effector-like resident memory T cells in tissues. J Immunol. 2012;188:4866–4875. doi: 10.4049/jimmunol.1200402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hadley GA, Higgins JM. Integrin αEβ7: molecular features and functional significance in the immune system. Adv Exp Med Bio. 2014;19:97–110. doi: 10.1007/978-94-017-9153-3_7. [DOI] [PubMed] [Google Scholar]
- 75.Schön MP, Arya A, Murphy EA, Adams CM, Strauch UG, Agace WW, Marsal J, Donohue JP, Her H, Beier DR, et al. Mucosal T lymphocyte numbers are selectively reduced in integrin αE (CD103)-deficient mice. J Immunol. 1999;162:6641–6649. [PubMed] [Google Scholar]
- 76.Mackay LK, Braun A, Macleod BL, Collins N, Tebartz C, Bedoui S, Carbone FR, Gebhardt T. Cutting edge: CD69 interference with sphingosine-1-phosphate receptor function regulates peripheral T cell retention. J Immunol. 2015;194:2059–2063. doi: 10.4049/jimmunol.1402256. [DOI] [PubMed] [Google Scholar]
- 77.Shiow LR, Rosen DB, Brdičková N, Xu Y, An J, Lanier LL, Cyster JG, Matloubian M. CD69 acts downstream of interferon-α/β to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature. 2006;440:540–544. doi: 10.1038/nature04606. [DOI] [PubMed] [Google Scholar]
- 78.Wakim LM, Woodward-Davis A, Bevan MJ. Memory T cells persisting within the brain after local infection show functional adaptations to their tissue of residence. Proc Natl Acad Sci U S A. 2010;107:17872–17879. doi: 10.1073/pnas.1010201107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ebihara T, Song C, Ryu SH, Plougastel-Douglas B, Yang L, Levanon D, Groner Y, Bern MD, Stappenbeck TS, Colonna M, et al. Runx3 specifies lineage commitment of innate lymphoid cells. Nat Immunol. 2015;16:1124–1133. doi: 10.1038/ni.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rapp M, Lau CM, Adams NM, Weizman OE, O’Sullivan TE, Geary CD, Sun JC. Core-binding factor β and Runx transcription factors promote adaptive natural killer cell responses. Sci Immunol. 2017;2 doi: 10.1126/sciimmunol.aan3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reis BS, Rogoz A, Costa-Pinto FA, Taniuchi I, Mucida D. Mutual expression of the transcription factors Runx3 and ThPOK regulates intestinal CD4+ T cell immunity. Nat Immunol. 2013;14:271–280. doi: 10.1038/ni.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Reis BS, Hoytema van Konijenburg DP, Grivennikov SI, Mucida D. Transcription factor T-bet regulates intraepithelial lymphocyte functional maturation. Immunity. 2014;41:244–256. doi: 10.1016/j.immuni.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Woolf E, Brenner O, Goldenberg D, Levanon D, Groner Y. Runx3 regulates dendritic epidermal T cell development. Dev Biol. 2007;303:703–714. doi: 10.1016/j.ydbio.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 84.Zaid A, Mackay LK, Rahimpour A, Braun A, Veldhoen M, Carbone FR, Manton JH, Heath WR, Mueller SN. Persistence of skin-resident memory T cells within an epidermal niche. Proc Natl Acad Sci U S A. 2014;111:5307–5312. doi: 10.1073/pnas.1322292111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •85.Hombrink P, Helbig C, Backer RA, Piet B, Oja AE, Stark R, Brasser G, Jongejan A, Jonkers RE, Nota B, et al. Programs for the persistence, vigilance and control of human CD8+ lung-resident memory T cells. Nat Immunol. 2016;17:1467–1478. doi: 10.1038/ni.3589. This study demonstrated genetic deletion or pharmacological inhibition of notch activity impairs TRM differentiation in the lungs of influenza-infected mice. [DOI] [PubMed] [Google Scholar]
- 86.Adachi T, Kobayashi T, Sugihara E, Yamada T, Ikuta K, Pittaluga S, Saya H, Amagai M, Nagao K. Hair follicle-derived IL-7 and IL-15 mediate skin-resident memory T cell homeostasis and lymphoma. Nat Med. 2015;21:1272–1279. doi: 10.1038/nm.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schenkel JM, Fraser KA, Casey KA, Beura LK, Pauken KE, Vezys V, Masopust D. IL-15–Independent Maintenance of Tissue-Resident and Boosted Effector Memory CD8 T Cells. J Immunol. 2016;196:3920–3926. doi: 10.4049/jimmunol.1502337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ma C, Mishra S, Demel EL, Liu Y, Zhang N. TGF-β Controls the Formation of Kidney-Resident T Cells via Promoting Effector T Cell Extravasation. J Immunol. 2017;198:749–756. doi: 10.4049/jimmunol.1601500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hu Y, Lee YT, Kaech SM, Garvy B, Cauley LS. Smad4 promotes differentiation of effector and circulating memory CD8 T cells but is dispensable for tissue-resident memory CD8 T cells. J Immunol. 2015;194:2407–2414. doi: 10.4049/jimmunol.1402369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pearce EL, Poffenberger MC, Chang C-H, Jones RG. Fueling immunity: insights into metabolism and lymphocyte function. Science. 2013;342:1242454. doi: 10.1126/science.1242454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sowell RT, Rogozinska M, Nelson CE, Vezys V, Marzo AL. Cutting edge: generation of effector cells that localize to mucosal tissues and form resident memory CD8 T cells is controlled by mTOR. J Immunol. 2014;193:2067–2071. doi: 10.4049/jimmunol.1400074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Anderson KG, Stromnes IM, Greenberg PD. Obstacles posed by the tumor microenvironment to t cell activity: a case for synergistic therapies. Cancer Cell. 2017;31:311–325. doi: 10.1016/j.ccell.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Webb JR, Milne K, Watson P, Deleeuw RJ, Nelson BH. Tumor-infiltrating lymphocytes expressing the tissue resident memory marker CD103 are associated with increased survival in high-grade serous ovarian cancer. Clin Cancer Res. 2014;20:434–444. doi: 10.1158/1078-0432.CCR-13-1877. [DOI] [PubMed] [Google Scholar]
- 94.Djenidi F, Adam J, Goubar A, Durgeau A, Meurice G, de Montpréville V, Validire P, Besse B, Mami-Chouaib F. CD8+ CD103+ Tumor–Infiltrating Lymphocytes Are Tumor-Specific Tissue-Resident Memory T Cells and a Prognostic Factor for Survival in Lung Cancer Patients. J Immunol. 2015;194:3475–3486. doi: 10.4049/jimmunol.1402711. [DOI] [PubMed] [Google Scholar]
- ••95.Ganesan AP, Clarke J, Wood O, Garrido-Martin EM, Chee SJ, Mellows T, Samaniego-Castruita D, Singh D, Seumois G, Alzetani A, et al. Tissue-resident memory features are linked to the magnitude of cytotoxic T cell responses in human lung cancer. Nat Immunol. 2017;18:940–950. doi: 10.1038/ni.3775. Ganesan et al. demonstrate that human tumor infiltrating lymphocytes resemble certain qualities of TRM. Further, they demonstrate that TIL exhibiting a TRM-like gene program may confer better anti-tumor protection as this correlated with positive prognoses in lung cancer patients. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Feng Y, Wang D, Yuan R, Parker CM, Farber DL, Hadley GA. CD103 expression is required for destruction of pancreatic islet allografts by CD8+ T cells. J Exp Med. 2002;196:877–886. doi: 10.1084/jem.20020178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Annacker O, Coombes JL, Malmstrom V, Uhlig HH, Bourne T, Johansson-Lindbom B, Agace WW, Parker CM, Powrie F. Essential role for CD103 in the T cell–mediated regulation of experimental colitis. J Exp Med. 2005;202:1051–1061. doi: 10.1084/jem.20040662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Reilly SM, Saltiel AR. Adapting to obesity with adipose tissue inflammation. Nat Rev Endocrinol. 2017;13:633–643. doi: 10.1038/nrendo.2017.90. [DOI] [PubMed] [Google Scholar]
- 99.Gisterå A, Hansson GK. The immunology of atherosclerosis. Nat Rev Nephrol. 2017;13:368–380. doi: 10.1038/nrneph.2017.51. [DOI] [PubMed] [Google Scholar]


