Abstract
Background
Germline mutations in the exonuclease domains of the POLE and POLD1 genes are associated with an as yet unquantified increased risk of colorectal cancer (CRC).
Methods
We identified families with POLE or POLD1 variants by searching PubMed for relevant studies prior to October 2016 and by genotyping 669 population-based CRC cases diagnosed <60 years of age from the Australasian Colorectal Cancer Family Registry. We estimated the age-specific cumulative risks (penetrance) using a modified segregation analysis.
Results
We observed 67 CRCs (mean age at diagnosis=50.2 (standard deviation [SD]=13.8) years) among 364 first- and second- degree relatives from 41 POLE families and 6 CRCs (mean age at diagnosis=39.7 (SD=6.83) years) among 69 relatives from 9 POLD1 families. We estimated risks of CRC to age 70 years (95% confidence interval [CI]) for males and females, respectively, to be: 40%(26%–57%) and 32%(20%–47%) for POLE mutation carriers; and 63%(15%–99%) and 52%(11%–99%) for POLD1 mutation carriers.
Conclusion
CRC risks for POLE mutation carriers are sufficiently high warranting consideration of annual colonoscopy screening and management guidelines comparable to Lynch syndrome. Refinement of estimates of CRC risk for POLD1 carriers is needed, however, clinical management recommendations could follow those suggested for POLE carriers.
Keywords: POLE, POLD1, Colorectal cancer, penetrance, polymerase proof reading associated polyposis
INTRODUCTION
Colorectal cancer (CRC) is a leading cause of cancer related mortality worldwide. Identifying people at high risk of developing CRC and optimising their screening can reduce the incidence of, and mortality from, CRC. Approximately 35% of CRC are estimated to be attributable to genetic factors 1, however, known hereditary CRC syndromes caused by high penetrance germline mutations account for only 3–5% of all CRCs 2. In recent years, gene discovery efforts have identified several novel CRC susceptibility genes including rare germline variants within the polymerase proofreading domains of the POLE and POLD1 genes, which are reported to be associated with CRC and polyposis (referred to as polymerase proofreading-associated polyposis)3–5. These novel CRC susceptibility genes are now included on multigene sequencing panels for clinical testing but the age-specific cumulative risks (penetrance) of CRC for people who carry mutations in these genes have not yet been quantified, which is an impediment to optimising personalised clinical management. The aim of this study was to estimate the age-specific cumulative risks of CRC separately for male and female carriers of POLE and POLD1 gene mutations.
METHODS
We searched in PubMed for relevant studies published prior to October 2016 that reported pedigree and cancer data for families with germline POLE or POLD1 variants that were either novel or previously observed at population frequency of ≤0.002 according to the non-Finnish European population in the ExAC database6, given recent evidence that shows low variant allele frequency is an important guide for determining disease causing variants7. Variants identified from the literature were re-annotated, for consistency, via in silico methods using Annovar8 with default settings. We applied a criteria for predicting pathogenicity of missense variants in both genes as recommended by the American College of Medical Genetics and Genomics (ACMG),9 namely using (i) multiple commonly used in silico tools (SIFT, PolyPhen2, MutationTaster, CADD, GERP, REVEL, and M-CAP (Table S1 for references)) and (ii) a high level of consensus between multiple in silico tools for prediction of deleterious effect. For this study, we applied the recommended or default thresholds for prediction of deleteriousness for each of the seven in silico tools (see Table S1 for thresholds). For each variant, the sum of in silico tools that reported the variant to be deleterious was calculated (maximum score of 7). A variant was considered to be likely pathogenic for this study where ≥4 out of 7 in silico tools predicted the variant to have a deleterious effect (Table S1). Families were excluded from the penetrance analysis if they were: (i) families of probands with variants not predicted to be pathogenic by <4 out of the 7 in silico tools used; (ii) discovery families that originally described the POLE c.1270C>G p.Leu424Val and POLD1 c.1433G>A p.Ser478Asn variants3; (iii) families of carriers of de novo mutations; or (iv) uninformative due to missing information on sex or age at cancer diagnosis of probands. For those families included in the analysis, mutation carrier status, sex, cancer or polyp-affected status, age at cancer or polyp/polyposis diagnosis, last known age or death, and country of study of families were extracted, where possible, from identified studies.
We searched for POLE or POLD1 mutation carrier families by genotyping 669 population-based probands diagnosed with CRC before 60 years of age from the Australasian Colorectal Cancer Family Registry (ACCFR)10,11 (Table S2) for 17 rare germline variants within the exonuclease domains of the POLE and POLD1 genes (Table S3 and Supplementary Materials and Methods). These 17 variants were selected based on multiple sources namely: 1) rare germline variants in the exonuclease domains of POLE and POLD1 reported in the ExAC database (≤0.002 allele frequency), 2) variants identified from our in-house whole genome and whole exome sequencing studies of 100 multiple-case CRC-affected families from the clinic-based recruitment arm of the Australasian Colorectal Cancer Family Registry (ACCFR) that were either rare or novel variants according to ExAC database (≤0.002 allele frequency), 3) were reported in the discovery paper by Palles et al3, and 4) were predicted to be deleterious by ≥4 out of the 7 in silico tools.
Using data from both published studies and the ACCFR, we estimated the hazard ratio (HR) and corresponding 95% confidence interval (CI) of CRC for mutation carriers compared with the general population (based on age, sex- and country-specific incidences) and the age-specific cumulative risks (penetrance), using a modified segregation analysis that incorporated data of all family members whether genotyped or not, and whether affected or not. We properly adjusted for ascertainment of families in which each pedigree’s data was conditioned on the proband’s genotype, cancer status and age at diagnosis (for population-based families) or on the proband’s genotype, and the cancer statuses and ages at diagnoses of all family members (for clinic-based families) to produce unbiased estimates (see details in Supplementary Materials and Methods). All statistical tests were two-sided, and P-values less than 0.05 were considered statistically significant.
RESULTS
From the literature, we identified 15 studies reporting 37 rare variants (MAF ≤0.002) within the exonuclease domains of POLE and POLD1. Of these 37 variants, 32 were predicted to be pathogenic by in silico criteria (Table S1). Of the 89 families with POLE (n=70) or POLD1 (n=19) mutations, 42 were excluded (7 families with variants not predicted to be pathogenic, 3 families from Palles et al3 study which initially reported the association of POLE and POLD1 with CRC, 3 families of de novo mutation carriers, 29 families of probands without age or sex information). From the ACCFR, two unrelated carriers of the POLE c.861T>A p.Asp287Glu variant and one carrier of the POLE c.1336C>T p.Arg446Trp variant were identified (Pedigrees shown in Figure S1).
We included 47 families (38 POLE and 9 POLD1) from published studies, and 3 families with POLE mutations from the ACCFR (Figure S1), in the penetrance analysis. Of these, 28 (21 POLE and 7 POLD1) families were ascertained because they had a family history of cancer while 22 (20 POLE and 2 POLD1) were ascertained via population-based cancer registries regardless of family history. We observed 67 CRCs with a mean age at diagnosis of 50.2 (standard deviation [SD]=13.8) years among 364 first- and second- degree relatives (53% female) from 41 families with POLE mutations, and 6 CRCs with a mean age at diagnosis of 39.7 (SD=6.83) years among 69 first- and second- degree relatives (45% female) from 9 families with POLD1 mutations (Table 1).
Table 1.
Numbers and mean ages at diagnosis of cancers in the first- and second-degree male and female relatives of probands from POLE and POLD1 mutation families.
| Male | Female | |||
|---|---|---|---|---|
|
|
||||
| Site of cancer | N | Mean age (SD) | N | Mean age (SD) |
| POLE | (n=172) | (n=192) | ||
|
| ||||
| Colon/rectum | 36 | 48.5 (14.9) | 31 | 52.1 (12.5) |
| Cutaneous malignant melanoma | 5 | 46.8 (29.9)* | 11 | 54.4 (18.7) |
| Duodenum | 1 | 45 | 0 | — |
| Renal | 1 | 86 | 0 | — |
| Ureter | 1 | 46 | 0 | — |
| Oesophagus | 1 | 85 | 1 | 77 |
| Pancreas | 1 | 46 | 1 | 78 |
| Brain | 1 | 69 | 2 | 68* |
| Bone | 1 | UK | 0 | — |
| Lung | 2 | 61.0 (5.66) | 1 | 92 |
| Liver | 0 | — | 1 | UK |
| Leukaemia | 2 | 74.0 (12.7) | 0 | — |
| Astrocytoma | 3 | 50.7 (30.9) | 0 | — |
| Ewing’s sarcoma | 0 | — | 1 | 14 |
| Intra-abdominal | 0 | — | 1 | UK |
| Head and neck | 1 | 52 | 3 | 74.0 (2.65) |
| Breast | 0 | — | 4 | 64.5 (21.4) |
| Endometrium | NR | NR | 3 | 53.0 (9.85) |
| Ovary | NR | NR | 4 | 43.8 (6.29) |
| Prostate | 5 | 74.0 (14.2)* | NA | NA |
|
| ||||
| POLD1 | (n=38) | (n=31) | ||
|
| ||||
| Colon/Rectum | 4 | 42.3 (7.04) | 2 | 34.5 (2.12) |
| Stomach | 1 | 72 | 1 | 85 |
| Renal | 0 | — | 1 | 74 |
| Urinary bladder | 0 | — | 1 | UK |
| Hodgkin lymphoma | 0 | — | 1 | 30 |
| Head and neck | 1 | 56 | 0 | — |
| Breast | 0 | — | 3 | 58.3 (10.7) |
| Endometrium | NR | NR | 3 | 55.0 (2.65) |
| Ovary | NR | NR | 1 | 31 |
N, total number of affected relatives; SD, standard deviation; UK, unknown age at diagnosis; NR, not relevant; —, not applicable
unknown age at diagnosis for one person
We estimated cumulative risks of CRC to age 70 years for males and females, respectively, to be: 28% (95% CI, 10%–42%) and 21% (95% CI, 7%–33%) for POLE mutation carriers; and 90% (95% CI, 33%–99%) and 82% (95% CI, 26%–99%) for POLD1 mutation carriers (Figure 1). The CRC HR was estimated to be 12.2 (95% CI, 7.35–20.2) and 87.2 (95% CI, 15.3–495) for POLE and POLD1 mutation carriers, respectively (Table 2). The HRs decreased with age, being 38.7 (95% CI, 17.5–85.4) for <50 years compared with 8.21 (95% CI, 4.24–15.9) for ≥50 years for POLE mutation carriers (P =0.003), and 201 (95% CI, 62.0–651) for <50 years compared with 3.34 (95% CI, 0.22–50.1) for ≥50 years for POLD1 mutation carriers (P =0.007; Table 2). There was no evidence for a difference in HRs by sex (all P >0.1).
Figure 1.
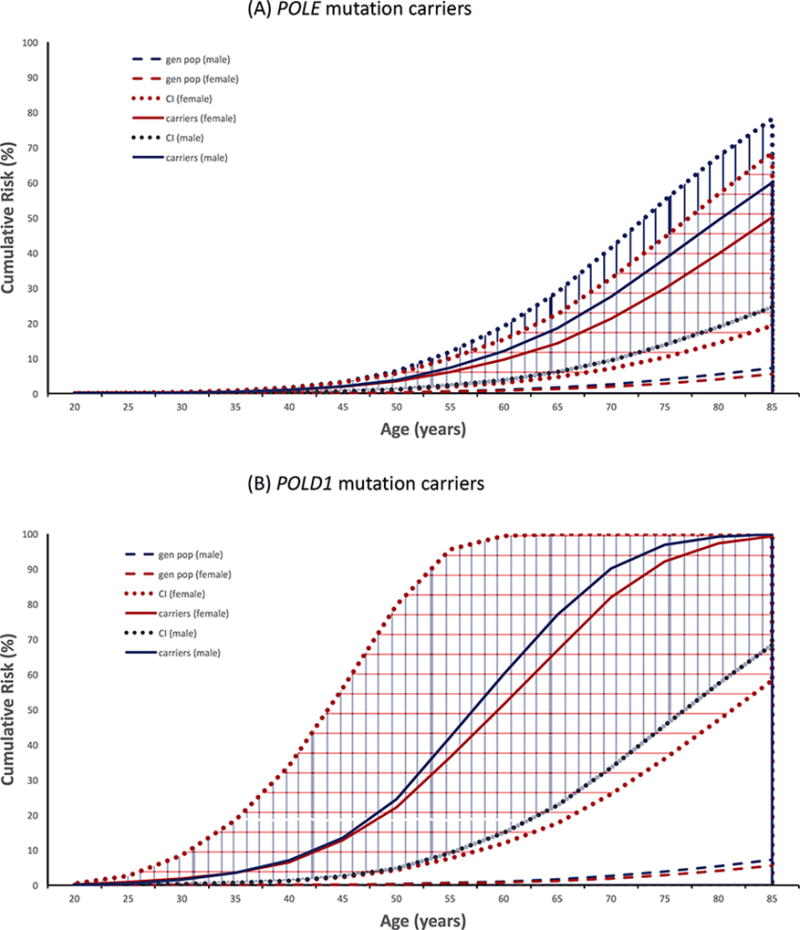
Cumulative risks (unbroken lines) and corresponding 95% confidence intervals (dotted lines) of colorectal cancer for (A) POLE mutation carriers, (B) POLD1 mutation carriers, and for the general population* (dashed lines). Blue and red represent males and females, respectively. *based on SEER (9 Registries): White (2003–2007) incidence data. SEER, Surveillance, Epidemiology and End Results database.
Table 2.
Hazards Ratio (95% confidence interval) of colorectal cancer for carriers of a germline mutation in POLE or POLD1
|
|
||||
|---|---|---|---|---|
| POLE | POLD1 |
POLE p.Leu424Val |
||
|
| ||||
| Overall HR (95%CI) | 12.2 (7.35–20.2) | 87.2 (15.3–495) | 131 (71.3–242) | |
| Sex | ||||
| Male | 12.3 (5.35–28.1) | 723 (102–5140) | 75.0 (37.9–149) | |
| Female | 12.1 (5.64–16.1) | 75.0 (19.2–292) | 269 (111–651) | |
| P-difference | 0.9 | 0.1 | 0.03 | |
|
| ||||
| Age | ||||
| <50 years | 38.7 (17.5–85.4) | 201 (62.0–651) | 238 (95.6–593) | |
| ≥50 years | 8.21 (4.24–15.9) | 3.34 (0.22–50.1) | 98.0 (44.1–218) | |
| P-difference | 0.003 | 0.007 | 0.2 | |
The POLE c.1270C>G, p.Leu424Val mutation has been reported in 19 families. For these specific mutation carriers, the estimated cumulative risks of CRC to age 70 years were 97% (95% CI, 85%–99%) for males and 92% (95% CI, 75%–99%) for females. The corresponding HR was 131 (95% CI, 71.3–242) when both sexes were combined, and 75.0 (95% CI, 37.9–149) for males and 269 (95% CI, 111–650) for females. Results for penetrance and HR estimates were not materially different between analyses with and without imputing missing ages.
DISCUSSION
To our knowledge, this is the first report of both relative and cumulative risks of CRC for people who carry germline mutations within the exonuclease domains of the POLE or POLD1 genes. Our current analysis included all of the reported rare germline variants within the exonuclease domains of POLE and POLD1 predicted to be pathogenic by multiple commonly used in silico tools, however, it is possible that not all variants result in the same level of risk as evidenced by the recurring POLE c.1270C>G, p.Leu424Val mutation.
CRC has been the predominant cancer identified in POLE and POLD1 mutation carriers to date (perhaps due to the way the families had been selected for genetic testing), however a broader extracolonic spectrum of cancers is being revealed as additional carrier families are identified. In addition to CRC, cancers of the endometrium, ovaries, pancreas, brain and small intestine have been reported for carriers, similar to what has been observed for DNA mismatch repair (MMR) gene mutation carriers (Lynch syndrome).12 The presence of 10 to <100 adenomas and/or the presence of duodenal polyps could be a distinguishing feature of POLE or POLD1 mutation carriers from MMR gene mutation carriers5. Interestingly, carriers of the POLE c.1270C>G, p.Leu424Val mutation showed no predilection for site and a mucinous adenocarcinoma histological type for a subset of tumors, suggesting variability in phenotype even for the same mutation, implicating potential environmental or genetic modifiers.
Additional phenotypic variability was observed with regards to tumor DNA mismatch repair status where the majority of CRCs from POLE and POLD1 carriers have been MMR-proficient or microsatellite stable, however, a small subset of CRCs in POLE mutation carriers have shown tumor MMR-deficiency, without evidence of a germline MMR gene mutation 13,14. Therefore, germline POLE exonuclease domain variants may account for a proportion of people with tumor MMR-deficient phenotype not explained by germline MMR gene mutations or acquired MLH1 promoter hypermethylation (suspected Lynch syndrome)15. Furthermore, somatic POLE and POLD1 mutations have been reported in both colorectal and endometrial cancers 16,17, supporting the hypothesis that loss of polymerase proofreading and the resultant hypermutation tumor phenotype can underlie inactivation of the MMR genes through somatic mutations.
This study has several limitations including the lack of detailed information on how cancer histories of family members in published studies were verified. A large proportion of families were excluded from the analysis due to missing information on age and sex of probands, thereby reducing the precision of our risk estimates. We used in silico predictions to assign pathogenicity for variant inclusion in the penetrance analysis, however, it has been shown that in silico tools and their algorithms for missense variant effect prediction are only 65–80% accurate when examining known disease causing missense variants18. The POLE and POLD1 variants included in the analysis were predicted to be deleterious by multiple in silico variant effect prediction tools (predicted deleterious by at least four out of the seven tools used in this study), as recommended by the American College of Medical Genetics and Genomics,9 and further selected based on a variant allele frequency filter of ≤0.002, adding confidence that our CRC risk estimates were based on only those variants likely to be pathogenic. We genotyped 17 POLE and POLD1 rare, likely pathogenic variants to identify additional carriers, however, we cannot exclude the possibility that other POLE and POLD1 variants outside of those genotyped may exist within the ACCFR CRC-affected individuals. Apart from CRC, we were unable to estimate risks of other cancers due to insufficient numbers of cancer diagnoses. Finally, our estimates might not necessarily be applicable to non-Caucasians.
In summary, the increased CRC risks for all carriers of a POLE pathogenic or likely pathogenic exonuclease domain variant, particularly for the recurrent POLE c.1270C>G, p.Leu424Val mutation, warrant consideration of annual colonoscopy screening and clinical management guidelines comparable to those currently recommended for people with Lynch syndrome or Familial Adenomatous Polyposis. As yet the risk of metachronous CRC is not known, but is likely to be similarly increased, raising consideration of subtotal colectomy rather than segmental resection for POLE mutation carriers. Functional studies to support variant classification may help to further refine the CRC risk estimates as will additional carrier families. For POLD1 mutation carriers, refinement of penetrance estimates for CRC is needed, however, clinical management recommendations could follow those suggested for POLE mutation carriers.
Supplementary Material
Acknowledgments
The authors thank Susan Preston and Neil O’Callaghan from the Colorectal Oncogenomics Group and staff from the Genetic Epidemiology Laboratory for their contributions to this project. The authors thank all study participants of the Australasian Colorectal Cancer Family Registry and staff for their contributions to this project, in particular Judi Maskiell, Maggie Angelakos and Allyson Templeton.
Financial support: This work was supported by grant UM1 CA167551 from the National Cancer Institute and through cooperative agreements with Australasian Colorectal Cancer Family Registry (U01 CA074778 and U01/U24 CA097735) and was conducted under Colon-CFR approval C-AU-0312-01. Aung Ko Win is an Australian National Health and Medical Council (NHMRC) Early Career Fellow. John L. Hopper is a NHMRC Senior Principal Research Fellow and Distinguished Visiting Professor at Seoul National University, Korea. Melissa C. Southey and Mark A. Jenkins are NHMRC Senior Research Fellows. Christophe Rosty is the Jass Pathology Fellow. Daniel D. Buchanan is a University of Melbourne Research at Melbourne Accelerator Program (R@MAP) Senior Research Fellow and NHMRC R.D. Wright Career Development Fellow.
Footnotes
Author contributions:
DDB - study concept and design; acquisition of data; analysis and interpretation of data; drafting of the manuscript; critical revision of the manuscript for important intellectual content; study supervision
JS - acquisition of data; analysis and interpretation of data; drafting of the manuscript; critical revision of the manuscript for important intellectual content; technical support
MC - acquisition of data; analysis and interpretation of data; drafting of the manuscript; critical revision of the manuscript for important intellectual content; technical support
CR - study concept and design; acquisition of data; analysis and interpretation of data; drafting of the manuscript; critical revision of the manuscript for important intellectual content; technical support; study supervision
KM - analysis and interpretation of data; drafting of the manuscript; critical revision of the manuscript for important intellectual content; technical support
BJP - analysis and interpretation of data; drafting of the manuscript; critical revision of the manuscript for important intellectual content; technical support
MAJ - study concept and design; acquisition of data; analysis and interpretation of data; drafting of the manuscript; critical revision of the manuscript for important intellectual content; statistical analysis; obtained funding; technical, or material support; study supervision
JLH - study concept and design; acquisition of data; analysis and interpretation of data; drafting of the manuscript; critical revision of the manuscript for important intellectual content; statistical analysis; obtained funding; technical, or material support; study supervision
MCS - study concept and design; analysis and interpretation of data; drafting of the manuscript; critical revision of the manuscript for important intellectual content; technical, or material support; study supervision
FAM - analysis and interpretation of data; drafting of the manuscript; critical revision of the manuscript for important intellectual content; study supervision
IMW - analysis and interpretation of data; drafting of the manuscript; critical revision of the manuscript for important intellectual content; study supervision
AKW - study concept and design; acquisition of data; analysis and interpretation of data; drafting of the manuscript; critical revision of the manuscript for important intellectual content; statistical analysis
Conflict of Interest Statement: The authors declare they hold no conflict of interest with respect to this work.
Publisher's Disclaimer: Disclaimer: The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Cancer Family Registries, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government or the Cancer Family Registry. Authors had full responsibility for the design of the study, the collection of the data, the analysis and interpretation of the data, the decision to submit the manuscript for publication, and the writing of the manuscript.
Supplementary information is available at the Genetics in Medicine website
References
- 1.Lichtenstein P, Holm NV, Verkasalo PK, et al. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343(2):78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 2.Jasperson KW, Tuohy TM, Neklason DW, Burt RW. Hereditary and familial colon cancer. Gastroenterology. 2010;138(6):2044–2058. doi: 10.1053/j.gastro.2010.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palles C, Cazier JB, Howarth KM, et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet. 2013;45(2):136–144. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spier I, Holzapfel S, Altmuller J, et al. Frequency and phenotypic spectrum of germline mutations in POLE and seven other polymerase genes in 266 patients with colorectal adenomas and carcinomas. Int J Cancer. 2015;137(2):320–331. doi: 10.1002/ijc.29396. [DOI] [PubMed] [Google Scholar]
- 5.Bellido F, Pineda M, Aiza G, et al. POLE and POLD1 mutations in 529 kindred with familial colorectal cancer and/or polyposis: review of reported cases and recommendations for genetic testing and surveillance. Genet Med. 2016;18(4):325–332. doi: 10.1038/gim.2015.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobayashi Y, Yang S, Nykamp K, Garcia J, Lincoln SE, Topper SE. Pathogenic variant burden in the ExAC database: an empirical approach to evaluating population data for clinical variant interpretation. Genome Med. 2017;9(1):13. doi: 10.1186/s13073-017-0403-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newcomb PA, Baron J, Cotterchio M, et al. Colon Cancer Family Registry: an international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol Biomarkers Prev. 2007;16(11):2331–2343. doi: 10.1158/1055-9965.EPI-07-0648. [DOI] [PubMed] [Google Scholar]
- 11.Buchanan DD, Clendenning M, Rosty C, et al. Tumor testing to identify lynch syndrome in two Australian colorectal cancer cohorts. J Gastroenterol Hepatol. 2017;32(2):427–438. doi: 10.1111/jgh.13468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96(4):261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elsayed FA, Kets CM, Ruano D, et al. Germline variants in POLE are associated with early onset mismatch repair deficient colorectal cancer. Eur J Hum Genet. 2015;23(8):1080–1084. doi: 10.1038/ejhg.2014.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jansen AM, van Wezel T, van den Akker BE, et al. Combined mismatch repair and POLE/POLD1 defects explain unresolved suspected Lynch syndrome cancers. Eur J Hum Genet. 2016;24(7):1089–1092. doi: 10.1038/ejhg.2015.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buchanan DD, Rosty C, Clendenning M, Spurdle AB, Win AK. Clinical problems of colorectal cancer and endometrial cancer cases with unknown cause of tumor mismatch repair deficiency (suspected Lynch syndrome) The application of clinical genetics. 2014;7:183–193. doi: 10.2147/TACG.S48625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshida R, Miyashita K, Inoue M, et al. Concurrent genetic alterations in DNA polymerase proofreading and mismatch repair in human colorectal cancer. Eur J Hum Genet. 2011;19(3):320–325. doi: 10.1038/ejhg.2010.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim TM, Laird PW, Park PJ. The landscape of microsatellite instability in colorectal and endometrial cancer genomes. Cell. 2013;155(4):858–868. doi: 10.1016/j.cell.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thusberg J, Olatubosun A, Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum Mutat. 2011;32(4):358–368. doi: 10.1002/humu.21445. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


