Abstract
Coenzyme Q10 (CoQ) or ubiquinone is found in the biological system which is synthesized by the conjugation of benzoquinone ring with isoprenoid chain of variable length. Coenzyme Q10 supplementation energizes the body and increases body energy production in the form of ATP and helps to treat various human diseases such as cardiomyopathy, muscular dystrophy, periodontal disease, etc. Reports of these potential therapeutic advantages of CoQ10 have resulted in its high market demand, which focus the researchers to work on this molecule and develop better bioprocess methods for commercial level production. At the moment, chemical synthesis, semi-synthetic method as well as bio-production utilizing microbes as biofactory are in use for the synthesis of CoQ10. Chemical synthesis involves use of cheap and easily available precursor molecules such as isoprenol, chloromethylquinone, vinylalane, and solanesol. Chemical synthesis methods due to the use of various solvents and chemicals are less feasible, which limits its application. The microbial production of CoQ10 has added advantages of being produced in optically pure form with high yield using inexpensive medium composition. Several bacteria, e.g., Agrobacterium, Paracoccus, Rhodobacterium, and yeast such as Candida, Rhodotorula are the potent ubiquinone producer. Some alternative biosynthetic pathway for designing of CoQ10 production coupled with metabolic engineering might help to increase CoQ10 production. The most common practiced strategy for strain development for commercial CoQ10 production is through natural isolation and chemical mutagenesis. Here, we have reviewed the chemical, semi-synthetic as well as microbial CoQ10 production in detail.
Keywords: Ubiquinone, CoQ10 biosynthesis, Metabolic engineering, Antioxidant, Isoprenoid
Introduction
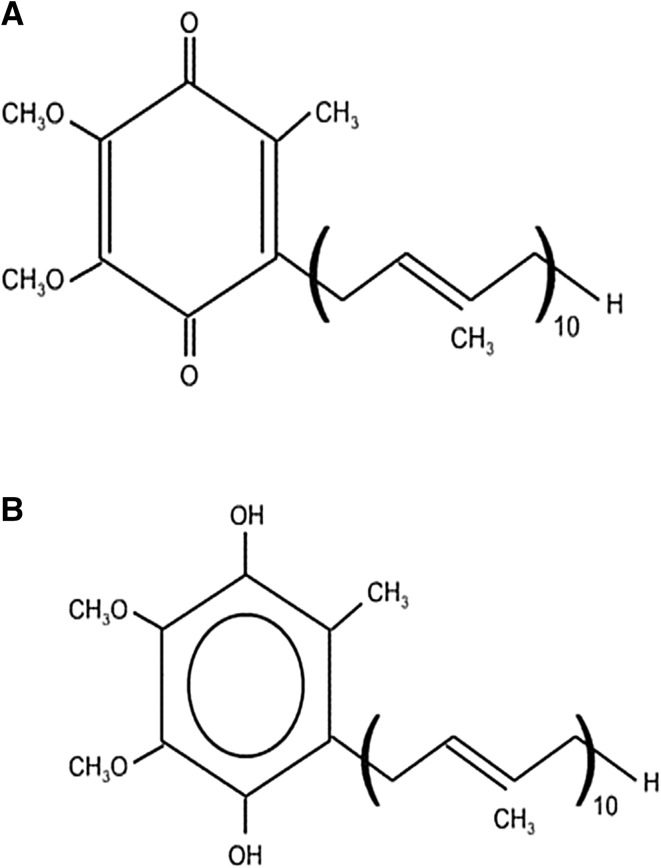
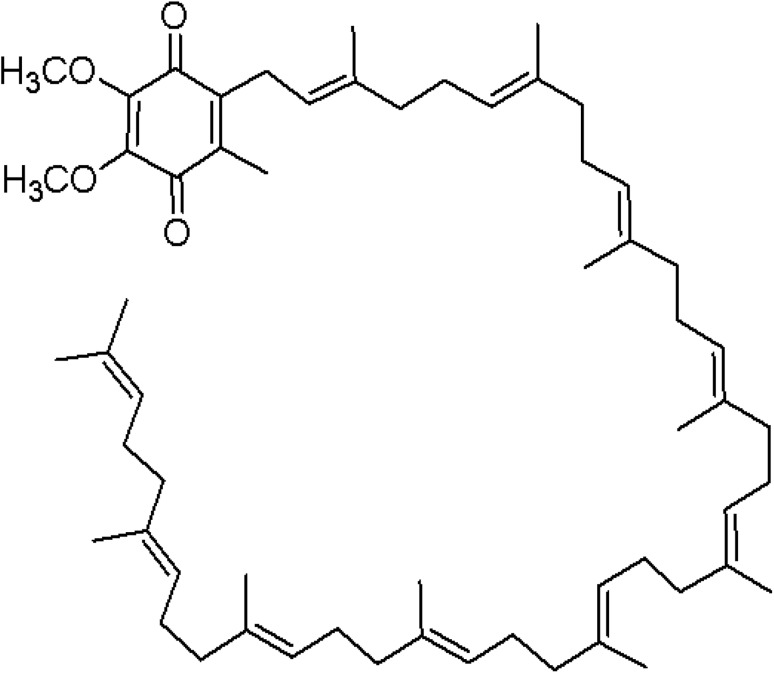
Coenzyme Q (CoQ), also named ubiquinone, ubidecarenone, or coenzyme Q10, is a well-known ubiquitous coenzyme found in the biological system which is synthesized by the conjugation of benzoquinone ring with isoprenoid chain of variable length (Figs. 1a, b, 2). Quinone species like coenzyme Q10 can be artificially synthesized which helps to treat various human diseases like cardiomyopathy, muscular dystrophy, periodontal disease, and in the early stages of congestive heart failure. The increasing frequency of these diseases has led to high demand of coenzyme Q10 and, therefore, continuous requirement of developing better and improved bioprocess methods to meet the requirement of commercial scale production using microbes. Japanese pharmaceutical companies started commercial production of pure CoQ10 during mid-1970. In 1978, Peter Mitchell received the nobel prize for their immense contribution to understand biological energy transfer through the formulation of the chemiosmotic theory, which includes the vital proton motive role of CoQ10 in energy transfer systems (Yasukazu et al. 2006; Klingen et al. 2007; Zhu et al. 2007). In the early 1980s, high-performance liquid chromatography was developed for direct measurement of CoQ10 level in blood and tissue samples. At the same time, the importance of CoQ10 as an antioxidant and free radical scavenger was reported by Lars Ernster of Sweden (Martin et al. 2007).
Fig. 1.
Structure of coenzyme Q10 a Oxidized form (2, 3-dimethoxy-methyl-6-decaprenyl-1, 4-benzoquinone) and b reduced form (2, 3-dimethoxy-methyl-6-decaprenyl-1, 4-benzohydroquinone)
Fig. 2.
3D structure of coenzyme Q10 with “head and tail”
Why coenzyme Q10?
Due to the stress conditions resulting from disease or by busy life style of human, aging factors are continuously increasing and energy production of body system is decreasing. Mitochondria are known as the power houses of cell for energy production. Coenzyme Q10 is very important player in the electron-transport chain of mitochondria. During the stress condition, energy production as well as CoQ10 levels in the body decreases. This can be compensated by giving coenzyme Q10 supplementation orally and health conditions can be improved greatly. Coenzyme Q10 supplementation energizes the body and increases body energy production in the form of ATP.
Coenzyme Q10 is found at the inner mitochondrial membrane, which transfer electron from dehydrogenases bounded to membrane to complex III of ETC and reduce them (Lenaz et al. 2007; Klingenet al. 2007; Zhu et al. 2007). Coenzyme Q10 also act as free radical scavenger which help to protect membrane protein and phospholipids from lipid peroxidation by regeneration of tocopherol (Yasukazu et al. 2006; Martin et al. 2007). Bacteria possess various structurally similar quinones like coenzyme Q (CoQ), e.g., de methyl menaquinone (DMK) and menaquinone (MK). They act as electron carriers during respirational oxidation (Ingledew and Poole 1984; Bader et al. 2000). CoQ is involved in aerobic respiration, whereas DMK and MK are part of anaerobic respiration (Ingledew and Poole 1984). CoQ molecules are classified (CoQ-n) on the basis of side chain length (n). For example, the main CoQ species in humans is CoQ10, whereas, in rodents, Escherichia coli and Saccharomyces cerevisiae, it is CoQ9, CoQ8, and CoQ6, respectively. E. coli and S. cerevisiae are model organisms which are used for deciphering biosynthetic pathway of CoQ (Kwon et al. 2000; Belogrudov et al. 2001; Thai et al. 2001; Gin et al. 2003; Johnson et al. 2005; Tran et al. 2006).
Coenzyme Q10 as miracle molecule/super-vitamin
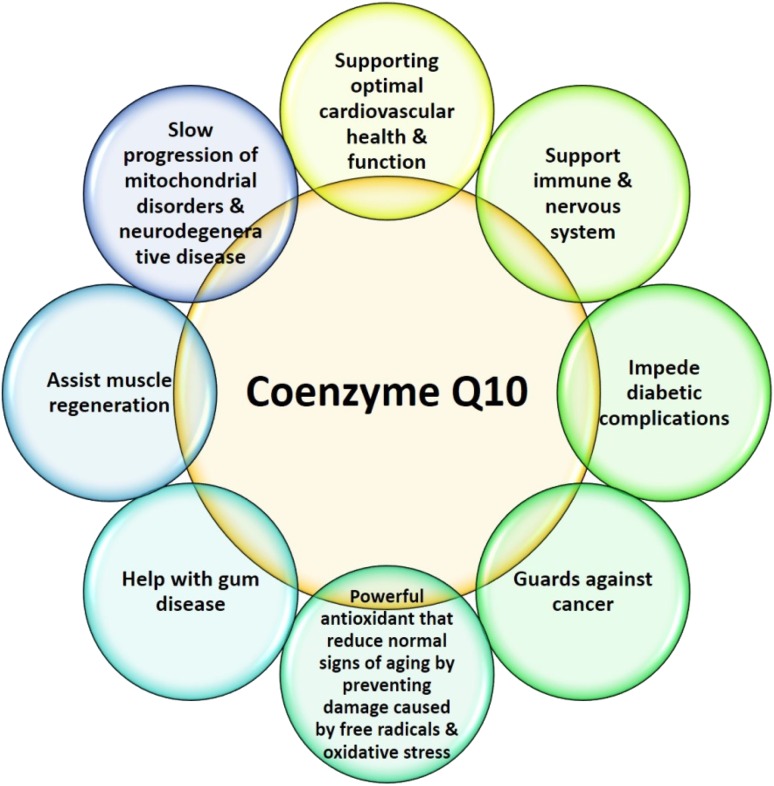
Coenzyme Q is also considered as vitamin Q, because the body cannot synthesize it in sufficient amount for the best of health, and extra amounts of CoQ should be obtained from food. Therefore, CoQ is called as vitamin: a natural, organic substance in food that is required for health and survival. Coenzyme Q10 acts as a supporting medicine (Fig. 3; Table 1). Hernández-Camacho et al. (2018) have summarized the importance of CoQ10 in the treatment of the various human diseases. The oral administration of CoQ10 is reported to be beneficial for the treatment of various human diseases such as cardio-myopathy, Parkinson’s, Alzheimer’s, diabetes, etc.; moreover, they are also found to reduce the risks of statin drugs from myopathy (Hodgson et al. 2002; Dhanasekaran and Ren 2005; Caso et al. 2007). Mutations in CoQ biosynthetic enzymes or associated enzymes lead to low CoQ10 level, which cause encephalomyopathy, Leigh syndrome, and cerebellar ataxia (López et al. 2006; Quinzii et al. 2005, 2006; Mollet et al. 2007). Coenzyme Q10-deficient patients show health improvement upon supplementation of CoQ10, (Quinzii et al. 2006; Artuch et al. 2006).
Fig. 3.
Pictorial representation of CoQ10 as health supplement
Table 1.
CoQ10 application in health care as a supportive medicine and antioxidants
| Disease | References |
|---|---|
| Cardiovascular disorders | Littarru et al. (2011), Flowers et al. (2014) |
| Neurodegenerative diseases and Neuromuscular diseases |
Hodgson et al. (2002) Dhanasekaran and Ren (2005), Asencio et al. (2016) |
| Parkinson’s Disease | Müller et al. (2003), Liu et al. 2011 |
| Age-related decline in immunity | Dhanasekaran and Ren (2005) |
| Mitochondrial cytopathies | Genova and Lenaz (2014), Acosta et al. (2016) Gorman et al. (2016), Rodriguez-Aguilera et al. (2017) |
| Ischemia | Tiano et al. (2007) |
| Ataxias | Quinzii et al. (2005) Artuch et al. (2006) |
| Diabetes | Dhanasekaran and Ren (2005), Fan et al. (2017) |
| Cancer | Dhanasekaran and Ren (2005) Fan et al. (2017) |
| Immune disorders | Langsjoen et al. (1991) |
| Periodontal disease, Periodontal disease Stroke | Hansen (1976) |
| Chronic fatigue syndrome | Campagnolo et al. (2017) |
| Muscular dystrophy | Beal (2004) |
| Alzheimer’s Disease | Beal (2004) |
| Adverse effect of drugs Chemotherapy (especially adriamycin and statins) | Caso et al. (2007) |
Because of its antioxidant properties, CoQ10 is also getting popularity among cosmetic industries (Prakash et al. 2010). Varela-López et al. (2016) has reviewed the current knowledge and evidenced the roles of CoQ cells, its relationship with aging, and possible implications of dietary CoQ in relation to aging, lifespan, or age-related diseases. Reports of these potential therapeutic advantages of CoQ10 have resulted in its high market demand, which focus the researchers to work on this molecule and develop better bioprocess methods for commercial level production. Until now, bacterial mutant selection method has been found to be a predominant approach for improving the production of CoQ10. However, this strategy has almost reached at its threshold level and is insufficient to overcome the demand. Therefore, some alternative approaches such as biosynthetic pathway designing of CoQ10 production coupled with metabolic engineering might help to increase CoQ10 production.
CoQ10 production methods
Chemical synthetic and semi-synthetic methods for CoQ10 production
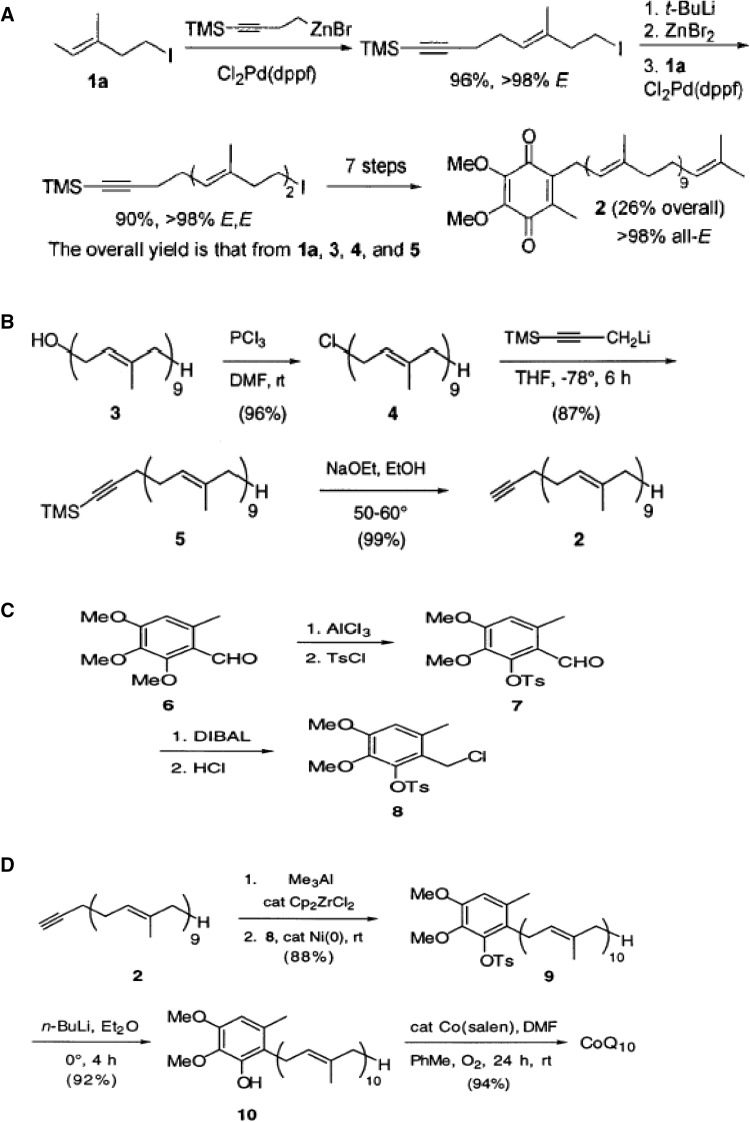
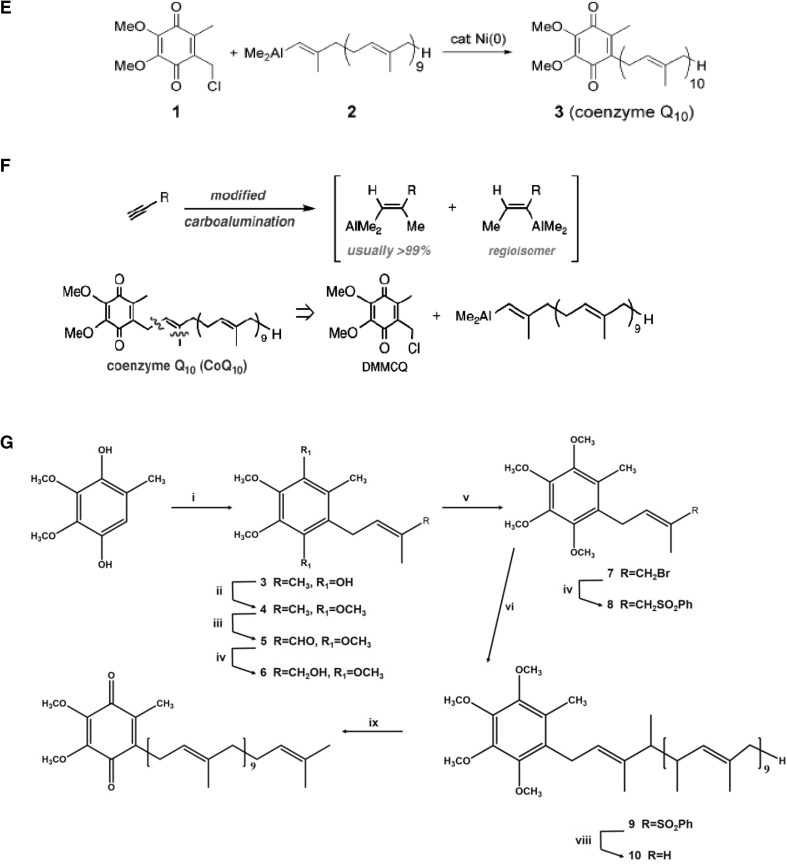
In naturally occurring CoQ10, the tail part consists of isoprene units and all of them exist in E-conformation. Therefore, all the CoQ10 synthesized by chemical method must also be stereoselectively in E conformation. Negishi et al. (2002) reported a novel, highly selective, and general methodology for the synthesis of 1,5-diene-containing oligoisoprenoids of all possible geometrical combinations exemplified by an iterative and convergent synthesis of coenzyme Q10. In this chemical synthesis method, terpenoids containing 1,5-diene units of E and/or Z geometry involved Pd-catalyzed homoallyl- and homopropargyl-alkenyl coupling and Zr-catalyzed carboalumination of alkynes (Negishi et al. 2002). They developed an efficient and selective convergent synthesis of coenzyme Q10 (2) from E-1,4-diiodo-2-methyl-1-butenes (1a), 4-iodo-1-trimethylsilyl-1-butyne (3), (E)- 1-iodo-2,6-dimethyl-1,5-heptadiene (4), and 2,3-dimethoxy- 4-chloromethyl-5-methyl-1,4-benzoquinone (5) in nine steps with 26% overall yield (Negishi et al. 2002), where Pd catalyzes homoallyl- and homopropargyl-alkenyl coupling reaction (Fig. 4a).
Fig. 4.
a Chemical synthesis of coenzyme Q10 (Negishi et al. 2002); b preparation of Alkynes (Lipshutz et al. 2002); c generation of coupling partner (Lipshutz et al. 2002); d cross coupling and final conversion of CoQ10 (Lipshutz et al. 2002); e single-step coenzyme Q10 synthesis (Lipshutz et al. 2005); f new improved synthesis process by (Lipshutz et al. 2007); and g synthesis of CoQ10 (Ravada et al. 2009)
Lipshutz et al. (2002) reported the easy chemical synthesis method (Fig. 4 b, c d) for CoQ10 production from 2- alkoxy-3, 4-dimethoxy-6-methylbenzyl chloride, 1-TMS propyne, and solanesol. They utilized solanesol (3) and nonaprenol derived from tobacco leaves as the source of isoprenoid side chain (2) in CoQ semi-synthetic method (Fig. 4b) (Lipshutz et al. 2002). The head part of the CoQ was synthesized from a readily available inexpensive crystalline benzaldehyde (6) (Lipshutz et al. 2002). After tosylation (98%), the resulting crystalline aldehyde (7) (Fig. 4c) was smoothly converted to the coupling partner, i.e., derived benzylic chloride (8) (Lipshutz et al. 2002). Therefore, by preparing “2” in three steps (Lipshutz et al. 2002) and “8” available commercially, the coupling was carried out using nickel as catalyst with overall 64% yield without any impurities of cis-isomer and coenzyme Q9 (Lipshutz et al. 2002) (Fig. 4d).
Lipshutz et al. (2005) reported an improved synthesis of miracle nutrient CoQ10. They discovered a new route to the key coupling partner (Fig. 4e), chloromethylated CoQ0 (1), which allows for the direct formation of CoQ10 (3) via a nickel-catalyzed cross coupling with the side chain in the form of an in situ-derived vinyl alane (2).
Lipshutz et al. (2007) further addressed the limitation of chemical synthesis methodology as it requires the − 70 °C temperature, and moreover, the regioisomeric product produces hurdle in the separation of isomeric form of CoQ10. Lipshutz et al. (2007) proposed the new “generation” advanced methodology for the terminal alkynes carboalumination (Fig. 4e). They found that using simple, inexpensive additives which alter the Al–Zr complex formed between Me3Al and Cp2ZrCl2 (Fig. 4f) result in the effective reagent mix which provides the complete control of regiochemistry on 1-alkynes carboalumination (Lipshutz et al. 2007). They stated that regioisomers from subsequent coupling are avoided, which otherwise will be difficult to separate (Lipshutz et al. 2007).
Ravada et al. (2009) proposed CoQ10 production starting from a relatively inexpensive isoprenol as precursor. They carried out the prenylation of 2, 3-dimethoxy-5-methylhydroquinone (2) (Fig. 4g) using isoprenol in the presence of a lewis acid, which upon subsequent selective oxidation of trans-methyl group of isoprenyl side chain and subsequent allylic bromination yielded a bromide precursor (Ravade et al. 2009). The ptoluenesulfination of the bromide followed by coupling with solanesyl bromide and de-ptoluenesulfination resulted in the production of dimethyl derivative of the CoQ10-quinol (Fig. 4g). Finally, CAN oxidation of dimethyl quinol followed by purification produced CoQ10 with 13% overall yield (Fig. 4g) (Ravade et al. 2009). Luo et al. 2017 have summarized the latest details in CoQ10 synthesis. They have reviewed the detailed discussion of recent advances in the semi-synthesis of CoQ10.
Biosynthesis
CoQ10-producing microorganisms
Paracoccus denitrificans, Agrobacterium tumefaciens, and Rhodobacter sphaeroides are the naturally high producers of CoQ10 (Tokdar and Khora 2017). Notably, these strains produce only one class of quinine, i.e., CoQ in contrast to E. coli, which produces various classes of quinines, i.e., CoQ, MK and DMK (Yoshida et al. 1998). These strains produce CoQ10 in the range of 30–130 mg/L (Yoshida et al. 1998). A minimum of 500 mg/L yield is expected from a commercially viable strain which implies that these natural strains have failed to meet the industrial requirement for production of CoQ10. Optimization of growth-conditions and metabolic regulation has been examined in these natural producer strains so as to improve their CoQ biosynthesis efficiency. Tokdar et al. (2014a) developed an induced mutants from Paracoccus denitrificans ATCC 19,367 showing the antibiotic resistant markers using protoplast fusion technology. They developed an induced mutants PF-P1 which showed 1.73-fold (1.51 mg/g) enhancement in the specific CoQ10 content than wild-type strain (Todker et al. 2014a). They further developed a process technology for the production of CoQ10 at 2L bioreactor level and achieved 2.44 mg/g specific CoQ10 yield (Tokdar and Khora 2017). A superior mutant strain P-87 was also generated from Paracoccus denitrificans ATCC 19,367 (Tokdar et al. 2015) through the iterative rounds of mutagenesis using gamma rays and NTG, followed by selection on various inhibitors like CoQ10 structural analogues and antibiotics which showed 1.25-fold improvement (Tokdar et al. 2014b) in specific CoQ10 content higher than the wild-type strain at shake flask level (1.63 mg/g specific CoQ10).
The most important factors which regulate the fermentation process during CoQ10 biosynthesis in these bacterial strains are oxidation–reduction potential (ORP). By limiting the supply of O2, they are able to maintain low ORP condition which in turn results in the increment of CoQ10 production in A. tumefaciens and R. sphaeroides strains (Yoshida et al. 1998; Choi et al. 2005; Ha et al. 2007). This outcome might result due to the reduction of ORP which results in shifting of ratio of oxidized CoQ and reduced CoQH2 towards reduced CoQH2. This shifting results in imbalance of respiratory system and/or scavenge reactive electrons can cause harm to their own membranes; therefore, to compensate this effect, cells trigger themselves to produce more of CoQ10 (Choi et al. 2005). Ha et al. (2008) improved the CoQ production in A. tumefaciens KCCM 10,413 by 22.8% to give 562.3 mg/L CoQ10 production using corn steep powder as a nitrogen source and lactate as a supplement. Improved CoQ10 production arising by lactate supplementation was ascribed to the high cell concentration and specific CoQ10 content derived from the acceleration of TCA cycle and energy production (Ha et al. 2008). Balakumaran and Meenakshisundaram (2015) significantly enhanced CoQ10 production from 10 mgL (in control) to 39.2 mg/L in batch mode with RSM-optimized precursor concentration. Using fed-batch mode, PHB and soybean oil-feeding strategy they obtained enhanced CoQ10 production (78.2 mg/L) (Balakumaran and Meenakshisundaram 2015). Lee et al. (2017) have recently discussed various strategies currently adopted to overproduce CoQ10 and have discussed some of the potential novel areas for future research. Current development in the production of CoQ10 is going on continuously from plant, recombinant Escherichia coli, and metabolically engineered Escherichia coli sources (Pahari et al. 2016). Heterologous expression of various rate limiting key regulatory enzymes in E. coli has resulted in the upgraded CoQ10 bio-production (Choi et al. 2009; Huang et al. 2011).
Biosynthesis of coenzyme Q10 in prokaryotes and eukaryotes
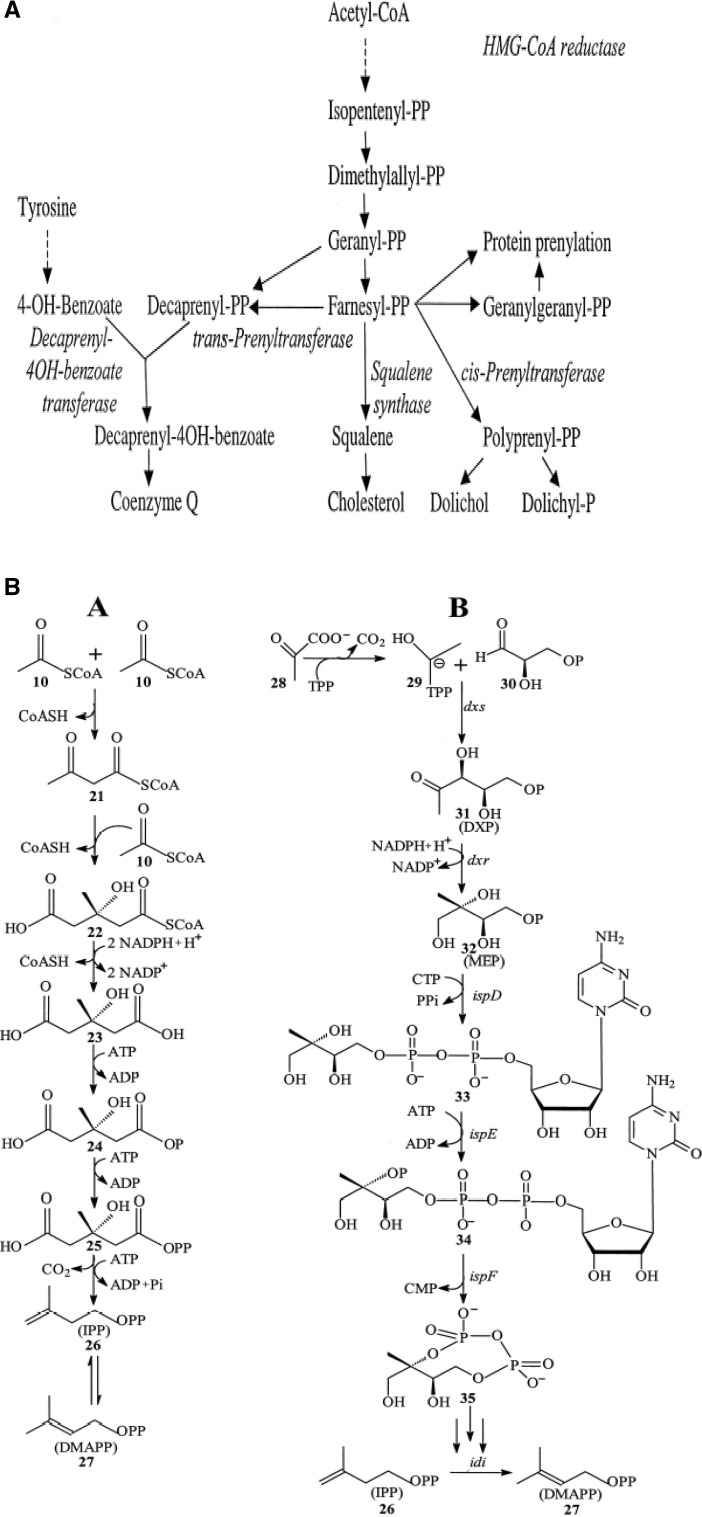
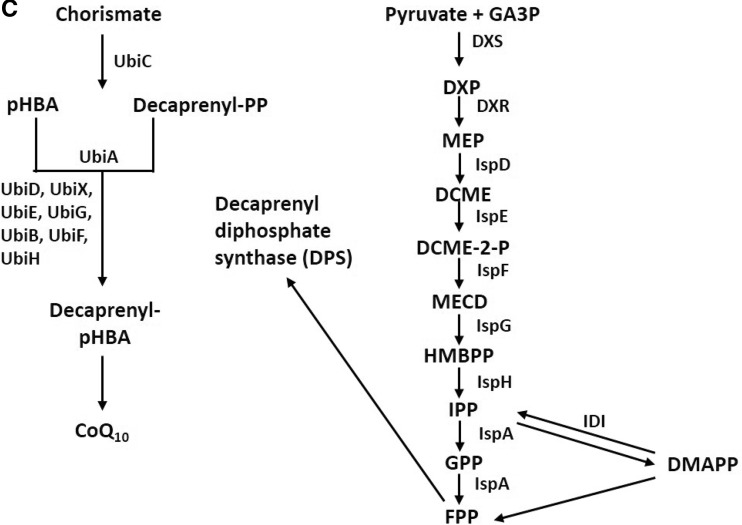
The biosynthesis of CoQ10 has been extensively studied in prokaryotic and eukaryotic organisms. CoQ metabolic synthesis is coupled with two discrete biosynthestic pathways. The first step results in the formation of aromatic quinone head group and in a separate pathway production of isoprene tail occurs. In the final step, the the attachment of various length of condensed isoprene units to the aromatic head group results in the formation of CoQ. In eukaryotes, CoQ10 is embedded in plasma membrane and plays a crucial role in various cellular metabolic pathways including mitochondrial respiratory chain. Acetyl co-A undergoes series of enzymatic reaction resulting in the production of the isoprenoid side chain of CoQ10, whereas benzoquinone is derived from amino acid tyrosine (Fig. 5a). CoQ10 biosynthetic pathway has common steps with that of cholesterol, isopentenyl pyrophosphate and its isomer dimethyl allyl pyrophosphate. However, in case of CoQ10 synthesis, it is linked alternately to form polyprenyl chain, which is also called isoprene (Fig. 5b). The biosynthesis of isoprenoids occurs via two distinct pathways: the mevalonate pathways and non-mevalonate pathways (deoxy-xylulose 5-phosphate) known as DXP pathway (Fig. 5b). Mevalonate pathways are associated with archaebacteria and eukaryotes, whereas DXP pathway is found in eubacteria (Croteau et al. 2000). In prokaryotes, the benzoquinone is synthesized by p-hydroxybenzoate (PHB) via shikimate pathway a key pathway in the biosynthesis of aromatic amino acid. Chorismate is converted to PHB and is used for prenylation and ring modification by a reaction catalyzed by chorismate pyruvate lyase encoded by ubiC in E. coli (Fig. 5c). The terminal step of CoQ10 biosynthesis is different in prokaryotes as compared to that of eukaryotes. In prokaryotes’ ring modifications by side chain, attachment is followed by decarboxylation step prior to hydroxylation and methylation step, whereas in case of eukaryotes, hydroxylation and methylation are followed by decarboxylation step.
Fig. 5.
a Biosynthesis of coenzyme Q10 in eukaryote (Dallner and Sindelar 2000); b biosynthetic pathways for isopentenyl diphosphate and dimethylallyl diphosphate synthesis (Meganathan 2001) A: Mevalonate pathway. 10, acetyl-CoA; 21, acetoacetyl-CoA; 22, HMG-CoA; 23, mevalonate; 24, mevalonate 5-phosphate; 25, mevalonate 5-diphosphate; 26, isopentenyl diphosphate; 27, dimethylallyl diphosphate. B: 2-C-Methyl-d-erythritol 4-phosphate (MEP/DOXP) pathway. 28, pyruvate; 29, hydroxyethyl-TPP anion; 30, D-glyceraldehyde-3-phosphate; 31,1-deoxy-d-xylulose 5-phosphate (DXP); 32, 2-C-methyl-d-erythritol 4-phosphate (MEP); 33, 4-diphosphocytidyl-2-C-methylerythritol; 34, 4-diphosphocytidyl-2-C-methyl-d-erythritol 2-phosphate; 35, 2-C-methyl-d-erythritol 2,4-cyclodiphosphate; 26, isopentenyl diphosphate; 27, dimethylallyl diphosphate; c biosynthesis of coenzyme Q10 in prokaryotes (Jeya et al. 2010). Decaprenyl diphosphate synthesized by DPS combines with pHBA and undergoes a series of reactions for the production of CoQ10. 4-Diphosphocytidyl-2-C-methyl-d-erythritol (DCME) 2-phosphate, 2-Cmethyl-d-erythritol 2,4-cyclodiphosphate (MECD)
CoQ is present in all subcellular compartments, and therefore, its transport is required within the cell. Acetyl-CoA is the initial substrate for the product which is FPP. Most of the enzymes of CoQ biosynthesis are present in the cytosol except the HMG-CoA reductase. HMG-Co A reductase is localized in microsomes and peroxisomes is the key regulatory enzyme of the mevalonate pathway which preferentially affects the CoQ synthesis (Turunen et al. 2004). Statin is the main inhibitor of HMG-Co-A reductase (Istvan 2003). The initial steps of mevalonate pathway result in formation of farnesyl-PP (FPP) from acetyl-CoA. FPP is the last common substrate which can lead to formation of several different end products including CoQ10 (Turunen et al. 2004). In addition to HMG-CoA reductase, various branch point enzymes after FPP synthase like squalene synthase, Cis phenyl transferase, and trans-prenyl transferase for cholesterol, dolichol, and CoQ10, respectively, are the main regulatory enzymes for lipid biosynthesis (Turunen et al. 2002).
Technoeconomic feasibility
CoQ production by chemical and microbial methods is well practiced. CoQ production by chemical synthesis predominately forms “cis” isomer in the side chain (pharmacologically less active form) and also produces coenzyme Q9 having nine isoprene unites in the side chain as a bi-products which needs extra purification step for its elimination. On the other hand, CoQ production by natural bacterial fermentation process produces only “trans” form of the coenzyme Q10 (pharmacologically active form), with trace of coenzyme Q9. The chemical synthesis method produces CoQ10 in low yield, whereas microbes produce high levels of CoQ10 naturally.
Due to the growing demand by pharmaceutical industry, the economical production of CoQ10 using biological processes is becoming more important. To increase the CoQ10 yield, there have been frequent attempts to engineer key enzymes within the CoQ10 pathway. For CoQ10 production, potential future engineering approaches have been used. By chemical mutagenesis, improvements in UQ-10 production were achieved. Attempts to increase the production level and specific CoQ10 content in recombinant hosts by metabolic engineering have to be made. There are various rate limiting steps in the CoQ10 biosynthesis such as Ubi A, DPS, and pHBA, and by targeting these steps, higher CoQ yield could be achieved (Cluis et al. 2011).
Inhibiors and stimulators of CoQ10 metabolic biosynthetic pathway
Antimycin
Antimycin is the most potent inhibitors of the ubiquinone reduction site (Qi site) of complex III. This inhibitor consists of salicylic acid and dilactane moiety. Salicylic acid moiety is recognized by the enzyme. The dilactane ring moiety plays a supporting role in inhibition binding to the Qi site by increasing the hydrophobicity of the molecules (Miyoshi 2005).
Stigmatellin
Stigmatellin is one of the most potent inhibitors of the ubiquinone oxidation site (Qo site) of complex III (Miyoshi 2005).
Paraquat
Paraquat (1, 1 dimethyl-4, 4 bipyridium di chloride) is an oxidative stressor by the process of lipid per-oxidation (Fukushima et al. 2002).
Para-hydroxy benzoic acid
Ubiquinone biosynthesis pathways consist of two steps, first results in benzaquinone nucleus formation and second step results in formation of isoprenoid side chain. Tyrosine-derived PHB is act as precursor for benzaquinone nucleus. The isoprene pathway probably up to the formation of isopentenyl pyrophosphate is shared by ubiquinone and sterol (Herebian et al. 2017).
Lovastatin
Lovastatin inhibits the synthesis of cholesterol and other non sterol end products such as CoQ10. Lovastatin treatment diminishes ubiquinone and alpha tocopherol content (Rousseau et al. 1998).
Conclusion
Strategy of mutagenesis and/or selection on inhibitor has proven to be most result oriented until date if compared with other strategies used for enhancing CoQ10 production and strain selection. However, the scope for further improvement with this strategy is less, because the probability of positive mutations enabling growth using the selection media for higher coenzyme Q10 yields is very low. Moreover, there is need for higher throughput techniques for screening and detection of increased CoQ10 producing strains using random mutagenesis approach. On the other hand, metabolic engineering in contrast to random mutagenesis provides handy control for genetic modification in specific biochemical pathways. The initial attempts of engineered CoQ10 metabolic pathways for higher yield in E. coli provided rigid evidence for potential rational design for improving production yield of CoQ10 in biological systems. Despite these promising results of metabolic engineering, still, current yields obtained are not so efficient so as to fulfill the need of commercial scale production, and moreover, there are still needs of careful assessment of bottlenecks limitation in physiological and metabolic pathways of CoQ10 biosynthesis. The main riddle in this case is that whether the yield is low due to limited flux through these pathways or there are still some additional or unidentified physiological factors left to detect which limits CoQ10 production in E. coli and other mutants. These results signify that P. denitrificans along with other high yielding strains have some additional factors in their biochemical or physiological pathways that allow them for higher production and accumulation of CoQ10.
For this, most probable targeted sites could be the composition and the efficiency of electron-transport chain and/or the absence of other quinines which limits the flux of precursor, such as MK in case of P. denitrificans (Anraku 1988). On careful analysis of higher yield producing strains, we can pick up the bottleneck points and then implement them in engineering of strain improvement for development of robust industrial strains to meet industrial scale need (Cluis et al. 2011).
Acknowledgements
The authors thank the Department of Biotechnology (DBT), Government of India, for providing financial support. The authors are thankful to Ms. Karuna Yadav for assisting in the preparation of this review article.
Compliance with ethical standards
Conflict of interest
The author declares no competing financial interest.
Contributor Information
Shraddha Shukla, Email: shraddha@iitg.ernet.in.
Kashyap Kumar Dubey, Email: kashyapdubey@gmail.com.
References
- Acosta MJ, Vazquez Fonseca L, Desbats MA, Cerqua C, Zordan R, Trevisson E, et al. Coenzyme Q biosynthesis in health and disease. Biochim Biophys Acta. 2016;1857(18):1079–1085. doi: 10.1016/j.bbabio.2016.03.036. [DOI] [PubMed] [Google Scholar]
- Anraku Y. Bacterial electron transport chains. Annu Rev Biochem. 1988;57:101–132. doi: 10.1146/annurev.bi.57.070188.000533. [DOI] [PubMed] [Google Scholar]
- Artuch R, et al. Cerebellar ataxia with coenzyme Q10 deficiency: diagnosis and follow-up after coenzyme Q10 supplementation. J Neurol Sci. 2006;246:153–158. doi: 10.1016/j.jns.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Asencio C, Rodriguez-Hernandez MA, Briones P, Montoya J, Cortes A, Emperador S, et al. Severe encephalopathy associated to pyruvate dehydrogenase mutations and unbalanced coenzyme Q10 content. Eur J Hum Genet. 2016;24:367–372. doi: 10.1038/ejhg.2015.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader MW, Xie T, Yu CA, James CA. Bardwell disulfide bonds are generated by quinone reduction. J BiolChem. 2000;275:26082–26088. doi: 10.1074/jbc.M003850200. [DOI] [PubMed] [Google Scholar]
- Balakumaran PA, Meenakshisundaram S. Modeling of process parameters for enhanced production of Coenzyme Q10 from Rhodotorula glutinis. Prep Biochem Biotechnol. 2015;45:398–410. doi: 10.1080/10826068.2014.923447. [DOI] [PubMed] [Google Scholar]
- Beal MF. Mitochondrial dysfunction and oxidative damage in Alzheimer’s and Parkinson’s diseases and coenzyme Q10 as a potential treatment. J Bioenerg Biomembr. 2004;36:381–386. doi: 10.1023/B:JOBB.0000041772.74810.92. [DOI] [PubMed] [Google Scholar]
- Belogrudov GI, Lee PT, Jonassen T, Hsu AY, Gin P, Clarke CF. Yeast COQ4 encodes a mitochondrial protein required for coenzyme Q synthesis. Arch Biochem Biophys. 2001;392:48–58. doi: 10.1006/abbi.2001.2448. [DOI] [PubMed] [Google Scholar]
- Campagnolo N, Johnston S, Collatz A, Staines D, Marshall-Gradisnik S. Dietary and nutrition interventions for the therapeutic treatment of chronic fatigue syndrome/myalgic encephalomyelitis: a systematic review. J Hum Nutr Diet. 2017;30:247–259. doi: 10.1111/jhn.12435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caso G, Kelly P, McNurlan MA, Lawson WE. Effect of coenzyme Q10 on myopathic symptoms in patients treated with statins. Am J Cardiol. 2007;99:1409–1412. doi: 10.1016/j.amjcard.2006.12.063. [DOI] [PubMed] [Google Scholar]
- Choi GS, Kim YS, Seo JH, Ryu YW. Restricted electron flux increases coenzyme Q10 production in Agrobacterium tumefaciens ATCC4452. Process Biochem. 2005;40:3225–3229. doi: 10.1016/j.procbio.2005.03.038. [DOI] [Google Scholar]
- Choi J, Ryu Y, Park Y, Seo J. Synergistic effects of chromosomal ispB deletion and dxs overexpression on coenzyme Q10 production in recombinant Escherichia coli expressing Agrobacterium tumefaciens dps gene. J Biotechnol. 2009;144:64–69. doi: 10.1016/j.jbiotec.2009.04.010. [DOI] [PubMed] [Google Scholar]
- Cluis CP, Ekins A, Narcross L, Jiang H, Gold ND, Burja AM, Martin VJ. Identification of bottlenecks in Escherichia coli engineered for the production of CoQ10. Metab Eng. 2011;13:733–744. doi: 10.1016/j.ymben.2011.09.009. [DOI] [PubMed] [Google Scholar]
- Croteau R, Lange MB, Rujan T, Martin W. Isoprenoid biosynthesis:the evolution of two ancient and distinct pathways across genomes.PronatlacadSci. USA. 2000;97:13172–13177. doi: 10.1073/pnas.240454797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallner G, Sindelar PJ. Regulation of ubiquinone metabolism. Free Radic Biol Med. 2000;29(3–4):285–294. doi: 10.1016/S0891-5849(00)00307-5. [DOI] [PubMed] [Google Scholar]
- Dhanasekaran M, Ren J. The emerging role of coenzyme Q-10 in aging, neurodegeneration, cardiovascular disease, cancer and diabetes mellitus. Curro Neurovasc Res. 2005;2:447–459. doi: 10.2174/156720205774962656. [DOI] [PubMed] [Google Scholar]
- Fan L, Feng Y, Chen GC, Qin LQ, Fu CL, Chen LH. Effects of coenzyme Q10 supplementation on inflammatory markers: a systematic review and meta-analysis of randomized controlled trials. Pharmacol Res. 2017;119:128–136. doi: 10.1016/j.phrs.2017.01.032. [DOI] [PubMed] [Google Scholar]
- Flowers N, Hartley L, Todkill D, Stranges S, Rees K. Co-enzyme Q10 supplementation for the primary prevention of cardiovascular disease. Cochrane Database Syst Rev. 2014;12:CD010405. doi: 10.1002/14651858.CD010405.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima T, Tanaka K, Lim H, Mariyama M (2002) Mechanism of cytotoxicity of Paraquat. Environ Health Prev Med (7): 89–94 [DOI] [PMC free article] [PubMed]
- Genova ML, Lenaz G. Functional role of mitochondrial respiratory supercomplexes. Biochim Biophys Acta. 2014;1837:427–443. doi: 10.1016/j.bbabio.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Gin P, Hsu AY, Rothman SC, Jonassen T, Lee PT, Tzagoloff A, Clarke CF. The Saccharomyces cerevisiae COQ6 gene encodes a mitochondrial flavin-dependent monooxygenase required for coenzyme Q biosynthesis. J Biol Chem. 2003;278:25308–25316. doi: 10.1074/jbc.M303234200. [DOI] [PubMed] [Google Scholar]
- Gorman GS, Chinnery PF, Dimauro S, Hirano M, Koga Y, Mcfarland R, et al. Mitochondrial diseases. Nat Rev Dis Primers. 2016;2:16080. doi: 10.1038/nrdp.2016.80. [DOI] [PubMed] [Google Scholar]
- Ha SJ, Kim SY, Seo JH, Oh DK, Lee JK. Optimization of culture conditions and scale-up to pilot and plant scales for coenzyme Q10 production by Agrobacterium tumefaciens. Appl Microbiol Biotechnol. 2007;74:974–980. doi: 10.1007/s00253-006-0744-4. [DOI] [PubMed] [Google Scholar]
- Ha SJ, Kim SY, Seo JH, Sim W, Moon HJ, Lee JK. Lactate increases coenzyme Q10 production by Agrobacterium tumefacians. World J MicrobiolBiotechnol. 2008;24:887–890. doi: 10.1007/s11274-007-9547-8. [DOI] [Google Scholar]
- Hansen IL. Bioenergetics in clinical medicine. Gingival leucocytic deficiencies of coenzyme Q10 in patients with periodontal disease. Res Commun Chempatholpharmacol. 1976;14(4):729–738. [PubMed] [Google Scholar]
- Herebian D, Seibt A, Smits SHJ, Rodenburg RJ, Mayatepek E, Distelmaier F. 4-Hydroxybenzoic acid restores CoQ10 biosynthesis in human COQ2 deficiency. Ann Clin Transl Neurol. 2017;4(12):902–908. doi: 10.1002/acn3.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Camacho JD, Bernier M, Guillermo López-Lluch G, Plácido NP (2018) Coenzyme Q10 supplementation in aging and disease. Front Physiol. 10.3389/fphys.2018.00044 [DOI] [PMC free article] [PubMed]
- Hodgson JM, Watts GF, Playford DA, Burke V, Croft KD. Coenzyme Q10 improves blood pressure and, glycaemic control: a controlled trial in subjects with Type 2 diabetes. Ear J ClinNutl. 2002;56:1137–1142. doi: 10.1038/sj.ejcn.1601464. [DOI] [PubMed] [Google Scholar]
- Huang M, Wang Y, Liu J, Mao Z. Multiple strategies for metabolic engineering of Escherichia coli for efficient production of coenzyme Q10. Chin J Chem Eng. 2011;19:316–326. doi: 10.1016/S1004-9541(11)60171-7. [DOI] [Google Scholar]
- Ingledew WJ, Poole RK. The respiratory chains of Escherichia coli. Microbial Rev. 1984;48:222–271. doi: 10.1128/mr.48.3.222-271.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Istvan E. Statin inhibition of HMG-CoA reductase: a 3-dimensional view. Atheroscler Suppl. 2003;4(1):3–8. doi: 10.1016/S1567-5688(03)00003-5. [DOI] [PubMed] [Google Scholar]
- Jeya M, Moon HJ, Lee JL, Kim IW, Lee JK. Current state of coenzyme Q(10) production and its applications. Appl Microbiol Biotechnol. 2010;85(6):1653–1663. doi: 10.1007/s00253-009-2380-2. [DOI] [PubMed] [Google Scholar]
- Johnson A, Gin P, Marbois BN, Hsieh EJ, Wu M, Barros MH, Clarke CF, Tzagoloff A. COQ9, a new gene required for the biosynthesis of coenzyme Q in Saccharomyces cerevisiae. J BiolChem. 2005;280:31397–31404. doi: 10.1074/jbc.M503277200. [DOI] [PubMed] [Google Scholar]
- Klingen AR, Palsdottir H, Hunte C, Ullmann GM (2007) Redox-linked protonation state changes in Cytochrome bc1 identified by Poisson–Boltzmann electrostatics calculations. Biochim Biophys Acta 1767: 204–221 [DOI] [PubMed]
- Kwon O, Kotsakis A, Meganathan R. Ubiquinone (coenzyme Q) biosynthesis. inEscherichia coli: identification of the ubiFgene. FEMS Microbiol Lett. 2000;186:157–161. doi: 10.1111/j.1574-6968.2000.tb09097.x. [DOI] [PubMed] [Google Scholar]
- Langsjoen PH, Langsjoen PH, Folkers K, Richardson P. Treatment of patients with human immunodeficiency virus infection with coenzyme Q10. In: Folkers K, Littarru GP, Yamagami T, editors. Biomedical and Clinical Aspects of Coenzyme Q. New York: Elsevier Science Publishers; 1991. pp. 409–415. [Google Scholar]
- Lee SQE, Tan TS, Kawamukai M, Chen ES (2017) Cellular factories for coenzyme Q10 production. Microb Cell Fact (2017): 16–39 [DOI] [PMC free article] [PubMed]
- Lenaz G, Fato R, Formiggini G, Genova ML. The role of Coenzyme Q in mitochondrial electron transport. Mitochondrion. 2007;7:S8-S33. doi: 10.1016/j.mito.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Lipshutz BH, Mollard P, Pfeiffer S, Chrisman W. A short, highly efficient synthesis of Coenzyme Q10. J Am ChemSoc. 2002;124(48):14282–14283. doi: 10.1021/ja021015v. [DOI] [PubMed] [Google Scholar]
- Lipshutz BH, Lower A, Berl V, Schein K, Wetterich F. An improved synthesis of the “miracle nutrient” coenzyme Q10. Org Lett. 2005;7(19):4095–4097. doi: 10.1021/ol051329y. [DOI] [PubMed] [Google Scholar]
- Lipshutz BH, Butler T, Lower A, Servesko J. Enhancing regiocontrol in carboaluminations of terminal alkynes.application to the one-pot synthesis of coenzyme Q10. Org Lett. 2007;9(19):3737–3740. doi: 10.1021/ol701469e. [DOI] [PubMed] [Google Scholar]
- Littarru GP, Tiano L, Belardinelli R, Watts GF. Coenzyme Q10, endothelial function, and cardiovascular disease. BioFactors. 2011;37:366–373. doi: 10.1002/biof.154. [DOI] [PubMed] [Google Scholar]
- Liu J, Wang L, Zhan SY, Xia Y. Coenzyme Q10 for Parkinson’s disease. Cochrane Database Syst Rev. 2011;12:CD008150. doi: 10.1002/14651858.CD008150.pub2. [DOI] [PubMed] [Google Scholar]
- López LC, Schuelke M, Quinzii CM, Kanki T, Rodenburg RJT, Naini A, DiMauro S, Hirano M. Leigh syndrome with nephropathy and CoQ10 deficiency due to decaprenyl diphosphate synthase subunit 2 (PDSS2) mutations. Am J Hum Genet. 2006;79:1125–1129. doi: 10.1086/510023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Yang X, Hu J, Ruan X, Mu F, Fu Y. The Synthesis of Coenzyme Q10. Curr Org Chem. 2017;21(6):489–450. doi: 10.2174/1385272820666160811123714. [DOI] [Google Scholar]
- Martin SF, Burón I, Espinosa JC, Castilla J, Villalba JM, Torres JM. Coenzyme Q and proteinllipid oxidation in a BSE-infected transgenic mouse model. Free Radic Bioi Med. 2007;42:1723–1729. doi: 10.1016/j.freeradbiomed.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Meganathan R. Ubiquinone biosynthesis in microorganisms. FEMS Microbiol. 2001;203(2):131–139. doi: 10.1111/j.1574-6968.2001.tb10831.x. [DOI] [PubMed] [Google Scholar]
- Miyoshi H. Inhibitor of mitochondrial respiratory enzymes. J pesticidal sci. 2005;30(2):120–121. doi: 10.1584/jpestics.30.120. [DOI] [Google Scholar]
- Mollet J, Giurgea I, Schlemmer D, Dallner G, Chretien D, Delahodde A, Bacq D, de Lonlay P, Munnich A, Rötig A. Prenyldiphosphate synthase. subunit IIPDSSJI and OH-benzoate polyprenyltransferase I (COQ2) mutations in ubiquinone deficiency and oxidative phosphorylation disorders. J Clin Invest Mar. 2007;117(3):765–772. doi: 10.1172/JCI29089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller T, Büttner T, Gholipour AF, Kuhn W. Coenzyme Q10 supplementation provides mild symptomatic benefit in patients with Parkinson’s disease. Neurosci Lett. 2003;341(3):201–204. doi: 10.1016/S0304-3940(03)00185-X. [DOI] [PubMed] [Google Scholar]
- Negishi E, Liou S-Y, Xu C, Huo S. A novel, highly selective, and general methodology for the synthesis of 1,5-diene-containing oligoisoprenoids of all possible geometrical combinations exemplified by an iterative and convergent synthesis of Coenzyme Q10. Organic Lett. 2002;4(2):261–264. doi: 10.1021/ol010263d. [DOI] [PubMed] [Google Scholar]
- Pahari SK, Ghosh S, Halder S, Jana M. Role of Coenzyme Q10 in human life. RJ PT. 2016;9(6):635–640. [Google Scholar]
- Prakash S, Sunitha J, Hans M. Role of coenzyme Q10 as an antioxidant and bioenergizer in periodontal diseases. Indian J Pharmacol. 2010;42(6):334–337. doi: 10.4103/0253-7613.71884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinzii CM, Kattah AG, Naini A, Akman HO, Mootha VK, DiMauro S, Hirano M. Coenzyme Q deficiency and cerebellar ataxia associated with an aprataxin mutation. Neurology. 2005;64:539–541. doi: 10.1212/01.WNL.0000150588.75281.58. [DOI] [PubMed] [Google Scholar]
- Quinzii C, Naini A, Salviati L, Trevisson E, Navas P, DiMauro S, Hirano M. A mutation in para-hydroxybenzoatepolyprenyl transferase (COQ2) causes primary Coenzyme Q10 deficiency. Am J Hum Genet. 2006;78:345–349. doi: 10.1086/500092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravada SR, Emani LR, Garaga MR, Meka B, Golakoti T. Synthesis of Coenzyme Q10. Am J Infect Dis. 2009;5(2):83–89. doi: 10.3844/ajidsp.2009.83.89. [DOI] [Google Scholar]
- Rodriguez-Aguilera JC, Cortes AB, Fernandez-Ayala DJ, Navas P. Biochemical assessment of coenzyme Q10 deficiency. J Clin Med. 2017;6:E27. doi: 10.3390/jcm6030027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau G, Desrosiers C, Varin F. A comparison of the effects of lovastatin and pravastatin on ubiquinone tissue levels in rats. Curr Ther Res. 1998;59(9):666–679. doi: 10.1016/S0011-393X(98)85064-9. [DOI] [Google Scholar]
- Thai QD, Adam YH, Jonassen T, Lee PT, Catherine FC. A defect in coenzyme Q biosynthesis is responsible for the respiratory deficiency in Saccharomyces cerevisiae abe1 mutants. J BiolChem. 2001;276:18161–18168. doi: 10.1074/jbc.M100952200. [DOI] [PubMed] [Google Scholar]
- Tiano L, Belardinelli R, Carnevali P, Principi F, Seddaiu G, Littarru GP. Effect of coenzyme Q10 administration on endothelial function and extra cellular super oxide dismutase in patients with ischaemic heart disease: a double-blind, randomized controlled study. Eur Heart. 2007;28(18):2249–2255. doi: 10.1093/eurheartj/ehm267. [DOI] [PubMed] [Google Scholar]
- Tokdar P, Khora SS. Optimization of fermentation process condition for the production of CoQ10 using Paracoccus denitrificans ATCC 19367 fusant strain PF-P1. Int J Eng Res Technol. 2017;6(7):135–143. [Google Scholar]
- Tokdar P, Vanka R, Ranadive P, George S, Khora SS, Deshmukh SK. Protoplast fusion technology for improved production of coenzyme Q10 using Paracoccus denitrificans ATCC 19367 mutant strains. Biochem Tech. 2014;5(2):685–692. [Google Scholar]
- Tokdar P, Ranadive P, Kshirsagar R, Khora SS, Deshmukh SK. Influence of substrate feeding and process parameters on production of coenzyme Q10 Using Paracoccus denitrificans ATCC 19367 mutant strain P-87. Adv Biosci Biotechnol. 2014;5:966–977. doi: 10.4236/abb.2014.512110. [DOI] [Google Scholar]
- Tokdar P, Sanakal A, Ranadive P, Khora SS, George S, Deshmukh SK. Molecular, physiological and phenotypic characterization of Paracoccus denitrificans ATCC 19367 mutant strain P-87 producing improved coenzyme Q10. Indian J Microbiol. 2015;55(2):184–193. doi: 10.1007/s12088-014-0506-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran UC, Marbois B, Gin P, Gulmezian M, Jonassen T, Clarke CF. Complementation of Saccharomyces cerevisiae coq7 mutants by mitochondrial targeting of the Escherichia. coliUbiF polypeptide: two functions of yeast Coq7 polypeptide in coenzyme Q biosynthesis. J Biol Chem. 2006;281:16401–16409. doi: 10.1074/jbc.M513267200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turunen M, Swiezewska E, Chojnacki T, Sindelar P, Dallner G. Regulatory aspects of coenzyme Q metabolism. Free Radic Res. 2002;36:437–443. doi: 10.1080/10715760290021298. [DOI] [PubMed] [Google Scholar]
- Turunen M, Olsson J, Dallner G. Metabolism and function of coenzyme Q. Biochim Biophys Acta. 2004;660(2):171–199. doi: 10.1016/j.bbamem.2003.11.012. [DOI] [PubMed] [Google Scholar]
- Varela-López A, Giampieri F, Battino M, Quiles JL. Coenzyme Q and its role in the dietary therapy against Aging. Molecules. 2016;21(3):373. doi: 10.3390/molecules21030373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukazu Y, Mieko H, Yoko H, Etsuo N. evaluation of the dietary effects of coenzyme Q in vivo by the oxidative stress marker, hydroxyoctadecadienoic acid and its stereoisomer ratio. Biochim Biophys Acta. 2006;1760:1558–1568. doi: 10.1016/j.bbagen.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Kotani Y, Ochiai K, Araki K. Production of ubiquione-10 using bacterium. J Gen ApplMicrobiol. 1998;44:19–26. doi: 10.2323/jgam.44.19. [DOI] [PubMed] [Google Scholar]
- Zhu J, Egawa T, Yeh S-R, Yu L, Yu C-A. Simultaneous reduction of iron-sulfur protein and Cytochrome beL) during ubiquinol oxidation in Cytochrome bc1 complex. Proc Nail AcadSci USA. 2007;104:4864–4869. doi: 10.1073/pnas.0607812104. [DOI] [PMC free article] [PubMed] [Google Scholar]