Abstract
The concept of U’s triangle, which revealed the importance of polyploidization in plant genome evolution, described natural allopolyploidization events in Brassica using three diploids [B. rapa (A genome), B. nigra (B), and B. oleracea (C)] and derived allotetraploids [B. juncea (AB genome), B. napus (AC), and B. carinata (BC)]. However, comprehensive understanding of Brassica genome evolution has not been fully achieved. Here, we performed low-coverage (2–6×) whole-genome sequencing of 28 accessions of Brassica as well as of Raphanus sativus [R genome] to explore the evolution of six Brassica species based on chloroplast genome and ribosomal DNA variations. Our phylogenomic analyses led to two main conclusions. (1) Intra-species-level chloroplast genome variations are low in the three allotetraploids (2~7 SNPs), but rich and variable in each diploid species (7~193 SNPs). (2) Three allotetraploids maintain two 45SnrDNA types derived from both ancestral species with maternal dominance. Furthermore, this study sheds light on the maternal origin of the AC chloroplast genome. Overall, this study clarifies the genetic relationships of U’s triangle species based on a comprehensive genomics approach and provides important genomic resources for correlative and evolutionary studies.
Introduction
Brassicaceae is one of the largest eudicot families; it contains more than 330 genera and 3,800 species. The genomes of species in the tribe Brassiceae share a common whole-genome triplication, which is considered to be a crucial event that drove diversification of the species and intra-species morphotypes1,2. Brassiceae includes several economically important crops that are used for vegetables, oils, and fodders. The basic foundation for the systematic relationship of the six major Brassica species was classically explained as U’s triangle3. U’s triangle proposed that the three tetraploid species B. juncea (AABB genome, 2n = 4x = 36), B. napus (AACC, 2n = 4x = 38), and B. carinata (BBCC, 2n = 4x = 34) are the derived allotetraploids of the diploid species B. rapa (AA, 2n = 2x = 20), B. nigra (BB, 2n = 2x = 16), and B. oleracea (CC, 2n = 2x = 18), respectively, which arose by natural hybridization and chromosome doubling.
Whole-genome sequencing (WGS) analyses of the A, C, AB, and AC genomes has increased our understanding of Brassica genome evolution4–8. It has been suggested that the Brassica genome diverged from Arabidopsis thaliana around 17 million years ago (mya)9, and there is evidence that the B genome first diverged from the Brassica lineage around 9 mya, followed by divergence of the A and C genomes around 4.5 mya10,11. Recent genome sequencing of the two AC and AB genome allotetraploids suggested that they derive from allotetraploidization events that happened approximately 8,000~51,000 years ago6,8.
Cells contain three different genomes (nuclear, mitochondrial, and chloroplast) that follow different evolutionary pathways12. Chloroplast, mitochondrial, and nuclear ribosomal DNA sequences are crucial resources to understand plant genomic diversity due to their highly conserved nature and strong phylogenetic signals. The chloroplast genome is circular, relatively simple, and inherited uniparentally with a highly conserved gene structure and gene order13,14. The chloroplast genome has sufficiently informative nucleotide divergence that it can be utilized to understand genetic diversity, genomic origin, and genetic relationships, as well as for barcode marker development15–19. A few systematic studies have explored the Brassica chloroplast genome; however, these results have yielded a partial and unresolved understanding20–23.
Nuclear ribosomal DNA (nrDNA) sequences are highly homozygous, tandemly repeated transcriptional units that encode important housekeeping functions in nuclear assembly and nuclear function24,25. Two nuclear ribosomal DNA blocks, 5SnrDNA and 45SnrDNA, are generally localized on different chromosomes in plants. The 45SnrDNA units contain a highly conserved multicistronic gene with 18S, 5.8S, and 28S RNA sequences and relatively polymorphic internal transcribed spacer (ITS) regions, which makes 45SnrDNA a preferred target for both phylogenetic and barcoding analyses26–28.
Advances in next-generation sequencing (NGS) technology and bioinformatics algorithms are facilitating the discovery of extensive natural variations in large populations. Most research has focused on identification of intra-species natural variations in the nuclear genome to explore diversity, adaptation, domestication, and evolution, as well as to mine for new alleles29. Our group recently established a method based on ‘genome skimming’ approach called dnaLCW for high-throughput simultaneous de novo assembly of chloroplast and 45SnrDNA transcription unit sequences using low-coverage whole-genome NGS to reveal inter-species and intra-species diversity30–32.
The objective of the current study is to elucidate the genetic diversity and evolution of Brassica species belonging to U’s triangle by performing whole-genome sequencing (WGS). We report the complete sequences of chloroplast genomes and 45SnrDNA transcription units for 28 genotypes. We also investigate genome-wide variation and phylogenomic analysis for chloroplast genomes and 45SnrDNA sequences to revisit the evolution of the six Brassica species in U’s triangle compared with the related species Raphanus sativus.
Results
Characterization of 28 complete chloroplast genomes
The complete chloroplast genomes were obtained for 28 genotypes using the dnaLCW approach (Table 1). Annotation of chloroplast genomes revealed conserved quadripartite structures with coherent gene number and gene order among the 28 genotypes (Fig. 1). The chloroplast genome is highly conserved, with 99–100% sequence similarity within each species, although meaningful variations were observed between species with 98.1–99.5% sequence similarity (Fig. 2; Table S1). Chloroplast genome lengths varied by 607 bp among the 28 genotypes, ranging from 153,037 bp (accession A4) to 153,642 bp (accession B4). Chloroplast genome copy numbers were estimated based on read depth for the haploid genome size, ranging from 453 (accession AB2) to 1,279 (accession BC1) copies per cell (Table 1).
Table 1.
Summary of chloroplast and 45SnrDNA assemblies from 28 Brassica and Raphanus genotypes.
| Organism and genome | Genotype IDa | Genome size (Mb) | Total reads (Mb) | Chloroplast genome | 45SnrDNA | ||||
|---|---|---|---|---|---|---|---|---|---|
| Length (bp) | Copy number (x)b | Accession number | Length (bp) | Copy number (x)b | Accession number | ||||
|
B. rapa (A) |
A1 | 529 | 1,557 | 153,483 | 378 | KX681647 | 5,818 | 3,216 | KX709342 |
| A2 | 529 | 1,214 | 153,482 | 305 | KX681648 | 5,818 | 3,770 | KX709343 | |
| A3 | 529 | 1,352 | 153,482 | 363 | KX681649 | 5,818 | 3,872 | KX709344 | |
| A4 | 529 | 1,293 | 153,037 | 496 | KX681650 | 5,818 | 4,183 | KX709345 | |
|
B. nigra (B) |
B1 | 632 | 1,532 | 153,633 | 378 | KT878383 | 5,831 | 1,819 | KX709346 |
| B2 | 632 | 1,632 | 153,641 | 221 | KX681651 | 5,831 | 1,667 | KX709347 | |
| B3 | 632 | 1,489 | 153,623 | 323 | KX681652 | 5,831 | 1,324 | KX709348 | |
| B4 | 632 | 1,631 | 153,642 | 244 | KX681653 | 5,831 | 1,571 | KX709349 | |
|
B. oleracea (C) |
C1 | 630 | 1,489 | 153,364 | 278 | KX681654 | 5,811 | 2,873 | KX709350 |
| C2 | 630 | 1,312 | 153,364 | 510 | KX681655 | 5,848 | 1,384 | KX709351 | |
| C3 | 630 | 1,611 | 153,364 | 285 | KX681656 | 5,818 | 2,768 | KX709352 | |
| C4 | 630 | 2,115 | 153,363 | 347 | KX681657 | 5,819 | 1,957 | KX709353 | |
|
R. sativus (R) |
R1 | 530 | 1,467 | 153,372 | 264 | KX681658 | 5,816 | 3,812 | KX709354 |
| R2 | 530 | 1,487 | 153,444 | 412 | KX681659 | 5,816 | 2,042 | KX709355 | |
| R3 | 530 | 1,440 | 153,376 | 393 | KX681660 | 5,819 | 4,174 | KX709356 | |
| R4 | 530 | 1,470 | 153,370 | 343 | KX681661 | 5,823 | 4,614 | KX709357 | |
|
B. juncea (AB) |
AB1-A | 1,068 | 1,469 | 153,483 | 779 | KX681662 | 5,818 | 2,412 | KX709358 |
| AB1-B | 5,831 | 1,589 | KX709359 | ||||||
| AB2-A | 1,068 | 1,352 | 153,483 | 358 | KX681663 | 5,818 | 1,883 | KX709360 | |
| AB2-B | 5,831 | 690 | KX709361 | ||||||
| AB3-A | 1,068 | 1,528 | 153,490 | 495 | KX681664 | 5,818 | 2,192 | KX709362 | |
| AB3-B | 5,831 | 1,041 | KX709363 | ||||||
| AB4-A | 1,068 | 1,549 | 153,483 | 338 | KX681665 | 5,818 | 3,449 | KX709364 | |
| AB4-B | 5,831 | 1,190 | KX709365 | ||||||
|
B. napus (AC) |
AC1-A | 1,130 | 1,534 | 153,452 | 630 | KX681666 | 5,831 | 1,445 | KX709366 |
| AC1-C | 5,818 | 689 | KX709367 | ||||||
| AC2-A | 1,130 | 1,401 | 153,429 | 890 | KX681667 | 5,831 | 1,169 | KX709368 | |
| AC2-C | 5,819 | 879 | KX709369 | ||||||
| AC3-A | 1,130 | 1,401 | 153,429 | 925 | KX681668 | 5,817 | 1,009 | KX709370 | |
| AC3-C | 5,832 | 865 | KX709371 | ||||||
| AC4-A | 1,130 | 1,579 | 153,453 | 366 | KX681669 | 5,831 | 982 | KX709372 | |
| AC4-C | 5,818 | 741 | KX709373 | ||||||
|
B. carinata (BC) |
BC1-B | 1,284 | 2,156 | 153,636 | 762 | KX681670 | 5,818 | 4,223 | KX709374 |
| BC1-C | 5,818 | 2,409 | KX709375 | ||||||
| BC2-B | 1,284 | 1,457 | 153,636 | 919 | KX681671 | 5,818 | 5,865 | KX709376 | |
| BC2-C | 5,818 | 3,453 | KX709377 | ||||||
| BC3-B | 1,284 | 1,710 | 153,641 | 913 | KX681672 | 5,818 | 2,813 | KX709378 | |
| BC3-C | 5,817 | 1,836 | KX709379 | ||||||
| BC4-B | 1,284 | 1,511 | 153,636 | 540 | KX681673 | 5,818 | 4,791 | KX709380 | |
| BC4-C | 5,818 | 2,551 | KX709381 | ||||||
arDNA from tetraploids was designated as A, B, or C based on the parental genome or sub-genome type. The complete details and list of organisms can be found in Table S6. bCopy numbers of chloroplast and 45SnrDNA were estimated based on average read depth mapping and converted into the corresponding haploid genome size.
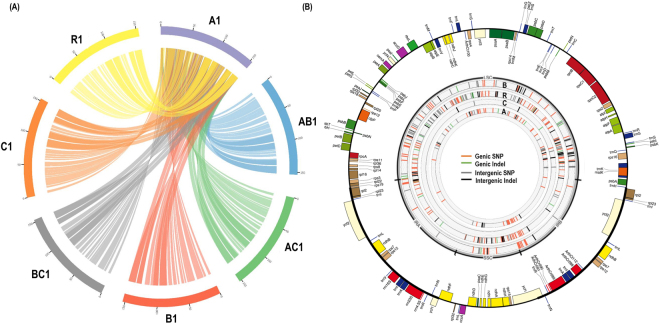
Figure 1.
Chloroplast genome variations and comparative analysis in seven Brassicaceae species. (A) Synteny comparisons of chloroplast genomes in Brassica. Circos-based syntenic comparative map developed for B. rapa (A1) against B. juncea (AB1), B. napus (AC1), B. nigra (B1), B. carinata (BC1), B. oleracea (C1), and Raphanus sativus (R1). Syntenic blocks with minimum length of 1 kb were used for the syntenic analysis. (B) Distribution of intra-species variations in B. nigra (B), R. sativus (R), B. oleracea (C), and B. rapa (R) chloroplast genomes. Outermost chloroplast circular map was developed from the B. rapa chloroplast genome (A1) using OGDRAW. Genes are represented in different colors. Positive and negative gene orientations are shown as outer and inner circles, respectively. Inner circles represent variations in the B, R, C, and A genomes, respectively.
Figure 2.
Comparative analysis based on complete chloroplast genomes identify similar and variable regions among the 28 Brassica and Raphanus genotypes.
The chloroplast genomes showed different levels of intra-species polymorphism (Tables 2, S2, S3). The chloroplast genomes from C genome species had very low intra-species diversity with seven SNPs and four InDels, whereas other chloroplast genomes had relatively high intra-species diversity with 88 SNPs and 16 InDels in the A species genomes, 99 and 24 in the B species genomes, and 193 and 112 in the R genome, respectively (Tables S2, S3). Polymorphism was richer in genic regions than in intergenic regions (Table 3). Abundant polymorphisms were detected on the inter-species level (Tables 2, S4). The highest number of inter-species variations was 2,502 SNPs and 294 InDels between the B and C chloroplast genomes, and the lowest was 257 SNPs and 65 InDels between the A and C chloroplast genomes (Table S4). The three tetraploids showed fewer variations in the chloroplast genome compared to the diploid species.
Table 2.
Summary of inter-species and intra-species variations based on chloroplast genomes.
| SNP/Indel | A | B | C | R |
|---|---|---|---|---|
| A | 88/16a | 280b | 65 | 167 |
| B | 2,402b | 99/24 | 294 | 245 |
| C | 257 | 2,502 | 7/4 | 183 |
| R | 1,203 | 2,259 | 1,293 | 193/112 |
a18/16 denotes the number of SNP/Indel variations in the A genome. bB genome has 2,402 and 280 SNP and InDel variations, respectively, compared with the A genome.
Table 3.
Summary and distribution of intraspecies SNP and Indel variations based on chloroplast genomes.
| Genome | SNP | Indel | ||||
|---|---|---|---|---|---|---|
| Genic | Intergenic | Total | Genic | Intergenic | Total | |
| A | 58 | 30 | 88 | 8 | 8 | 16 |
| B | 68 | 31 | 99 | 13 | 11 | 24 |
| C | 2 | 5 | 7 | 3 | 1 | 4 |
| R | 137 | 56 | 193 | 57 | 55 | 112 |
Characterization of 45SnrDNA sequences
The complete 45SnrDNA sequences of the four diploid species ranged from 5,816 to 5,831 bp (Table 1). Only one representative 45S was identified for each of the 16 diploid accessions of the A, B, C, and R genomes. By contrast, two different 45S sequences were identified for each of the tetraploid accessions. Therefore, 24 different 45SnrDNA sequences were identified for all 12 genotypes of the three allotetraploids (AB, AC, and BC genomes) (Table 1). Comparative analysis of the 40 types of 45SnrDNA sequences revealed 39 bp length variations in the 5,818 bp sequence (Figs 3, 4). Compared with the chloroplast genome, 45SnrDNA sequences were less diverse, with 22 SNPs and one InDel among 40 types of 45SnrDNA sequences from 28 genotypes. These variations were distributed among genic and intergenic regions.
Figure 3.
Summary of nucleotide variations based on 45SnrDNA sequences from 28 genotypes.
Figure 4.

Structure and similarity analyses of 45SnrDNA sequences from 28 Brassica and Raphanus genotypes. (A) Complete structure and gene annotation of 45SrDNA sequences from the A1 genome. (B) Red and black arrowheads indicate the SNP and InDel variations, respectively. (C) Coverage of 45SnrDNA-based read mapping. Red lines indicate the proportion of G + C in the 4S 45SnrDNA. (D) Comparative analysis of similar and variable regions using mVISTA. Red arrowheads indicate inter-species variations.
Our analysis identified two types of 45SnrDNA (both parental) in three allotetraploids (AB, AC, and BC genomes). Each 45SnrDNA type in the three allotetraploids showed 100% sequence similarity with their corresponding parental diploid genome. For example, the A and B types of 45SnrDNA in the AB genome were 100% identical with those in the A and B genomes, respectively (Figs 3, 4). Read depth approach was used to estimate the copies of each 45SnrDNA type. Copy numbers differed among the allotetraploid sub-genomes, with 3,000–6,500 copies in the AB genome, 6,000–11,000 copies in the AC genome, and 2,200–2,900 copies in the BC genome. Copy numbers of each 45SnrDNA type in sub-genomes displayed a biased proportion up to 1.5-3-fold, with a higher proportion always occurring in the maternal ancestors of the AB, AC, and BC genomes (Table S5).
Validation and utilization of species-specific variations
Although chloroplast and 45SnrDNA sequences are highly conserved, our comparative analyses revealed a considerable number of variations (Fig. 5). There were more SNPs in chloroplast sequences than in 45SnrDNA sequences, with an average of 15 SNPs identified for every 1 kb of chloroplast genome, but only 3 for every 1 kb of 45SnrDNA. We tested the utility of this information on the diversity in chloroplast and 45SnrDNA sequences for identification and authentication of species or cultivars. We began with comprehensive analysis of the SNP and InDel variations in the chloroplast genome and 45SnrDNA to facilitate the development of barcode markers that enable the discrimination of each species. A total of 2,796 chloroplast variations were identified in 28 genotypes, and many of them were potential candidates for species-specific marker development (Tables S2, S3). We performed PCR analysis to validate the sequence polymorphism against several diversity-containing regions, and identified two InDel variations based on the chloroplast genome that could differentiate each diploid genome (A, B, C, and R genomes) (Figure S1). By contrast, only 23 variations (including 22 SNPs and one InDel) were identified based on 40 different 45SnrDNA sequences from 28 genotypes (Fig. 3). The 18S and ITS regions had relatively rich diversity and provide potential targets to differentiate the A, B, C, and R genomes by PCR analysis (Figure S2).
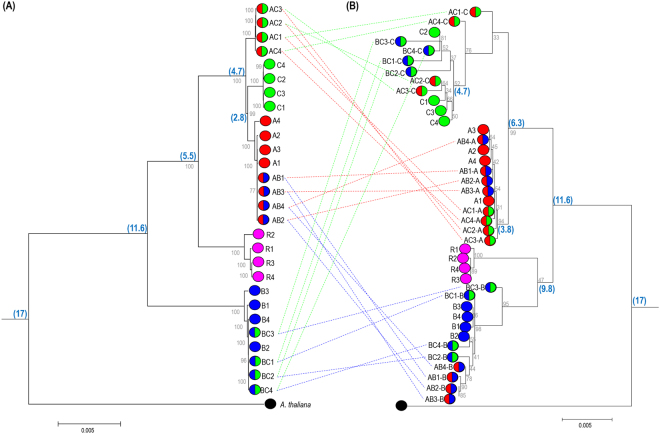
Figure 5.
Phylogenetic relationships of the genus Brassica inferred from complete chloroplast (A) and 45SnrDNA (B) sequences of 28 Brassica and Raphanus genotypes. Tree was developed using MEGA7 with 1,000 bootstrap replications. The bootstrap values for clades are shown in corresponding branches of the tree. Taxon markers with single and double circles represent diploid and tetraploid genomes, respectively. The circled legend for 45SnrDNA and chloroplast corresponds to each species. Dotted line connects the corresponding allotetradiploid genomes of chloroplast and 45SnrDNA. Species divergence times were inferred from Bayesian analysis, and are shown at the side of the node in million years (my). A. thaliana was used as an outgroup.
Phylogenomic exploration of U’s triangle
Separate phylogenetic analyses based on chloroplast and 45SnrDNA sequences identified conserved genetic relationships and displayed essentially identical topologies among the Brassica species in U’s triangle. The high bootstrap values on the nodes support the reliability of the phylogenies produced based on both chloroplast and 45SnrDNA sequences (Fig. 5).
The chloroplast phylogenetic tree displayed five different clades, with clear discrimination between the four diploid genomes but an ambiguous clade in the AC genome (Figs 5A; S3). The four AC genomes were clustered with each other, but did not group with the parental A or C genomes. The other two allotetraploids (AB and BC) were clustered with one of their parental genomes; AB clustered with the A genome, and BC clustered with the B genome indicating that the A and B genomes were the maternal ancestors for the AB and BC genomes, respectively. However, the AC genome followed neither the A nor C genome as a source of maternal origin and showed an enigmatic relationship with its diploid parental genomes. Furthermore, we did not observe any reciprocal hybridization patterns in any of the three tetraploids.
The 45SnrDNA phylogenetic tree displayed clear classifications (Figs 5B, S4). The four diploid species diverged into four distinct clades, and each of the tetraploid genomes contained two independent 45SnrDNAs according to their corresponding ancestral maternal/paternal genomes. The AB genome harbored both A-type and B-type 45SnrDNA (AB-A and AB-B, respectively) with AB-A from the maternal genome and AB-B from the paternal. The BC and AC genomes also harbored two original subgenomic 45SnrDNA types; the B and C type in the BC genome (BC-B and BC-C, respectively), and the A and B type in the AB genome (AB-A and AB-B, respectively). Overall, the 45SnrDNA phylogenetic analysis of three alloteraploids revealed the expected parental association with the three diploids.
A phylogenetic tree generated by BEAST analysis showed similar topology as that generated by MEGA. The molecular dating based on chloroplast and 45SnrDNA sequences generated essentially similar divergence times (Figures S3, S4). Tree topologies with inferred speciation dates clearly identified three major divergence periods in both analyses (chloroplast and 45SnrDNA) of Brassica. The trees indicated that divergence and speciation in the B genome occurred 11 mya, followed by R genome divergence at 9 mya, and speciation of A and C genomes about 4.5 mya. The allotetraploids appear to have arisen from their diploid ancestors from 0.001 to 0.03 mya (Figure S5).
Discussion
Inter-species nucleotide diversity of chloroplast and 45SnrDNA
Chloroplast genomes are highly stable, have low mutation rates, and produce highly reliable phylogenetic trees that help elucidate plant evolutionary history15,18,30,33. Nuclear ribosomal DNAs can remain highly homozygous, whereas nuclear genomes are subject to cross-hybridization and cross-over during meiosis32. We produced a comprehensive diversity map for Brassica based on 28 complete chloroplast genomes and 40 types of complete 45SnrDNA sequences of the major species listed in the classical U’s triangle. The chloroplast and 45SnrDNA sequences exhibited highly conserved gene structures and gene orders at the inter-species and intra-species levels. However, considerable numbers of nucleotide variations were observed in both chloroplast and 45SnrDNA, which represent genus- and species-specific variations that can be developed for barcode markers and molecular breeding analysis. The B genome was highly diverged from the other genomes, suggesting prolonged, independent evolution. The estimated speciation time of the B genome is consistent with this result (Table 2). Phylogenetic analyses based on chloroplast genomes and 45SnrDNA sequences showed general agreement, with the B genome as a sister group to the A and C genomes and the derived allotetraploids following their corresponding progenitor genomes6,34. The chloroplast and 45SnrDNA sequences indicate that the R genome was closer to the A and C genomes then the B genome.
Chloroplast genomes of seven species have different intra-species nucleotide diversity levels
Four genotypes for each species all showed different levels of divergent intra-species chloroplast genome polymorphism, although there were fewer intra-species polymorphisms than inter-species polymorphisms. The three allotetraploids rarely showed intra-species diversity (as expected) because those species were generated by allotetraploidization less than 0.05 mya6,8. Although four genotypes cannot represent the full diversity of each species, our results indicate that the C genome is less diverse than the maternal ancestor genome, the A and B genomes are moderately diverse, and the R genome is more diverse than the maternal ancestor genome. These results are consistent with a recent report of very low diversity in the C genome and relatively rich diversity in the A and B chloroplast genomes23,35. Similarly, rich variations were identified in the mitochondrial genome of R. sativus. Our previous work showed that dynamic mitochondrial genome rearrangements caused cytoplasmic male sterility and large variations among radish lines36 compared with the relatively conserved mitochondrial genome structures in Brassica30,37.
Two types of 45SnrDNA are derived from two diploid ancestors in allotetraploids
Some polyploid plants maintain both parental ribosomal DNA genomes (5SnrDNA and 45SnrDNA) after allopolyploidization38–41. However, many allopolyploids express nucleolar dominance (ND), in which rRNA from one parent is transcriptionally silenced or recessively expressed42,43. ND is anticipated to have a significant role in chromatin modification and genome evolution44,45. Homogenization into one of two rDNA types also occurs via concerted evolution, mediated by rearrangements such as repeat loss, replacement, and recombination46–49.
There are few reports of the complete 45SnrDNA sequence in plant genomes. Here, we obtained the complete 45S rDNA transcription sequences for 28 accessions. We found only one highly homologous 45SnrDNA sequence in each accession of four diploids, but detected two types of 45SnrDNAs derived from the parental diploid ancestors in all three allotetraploids. Copy number analysis revealed that 45SnrDNA sequence bias toward the maternal genome occurred in the order of A > B > C genomes (Table S5), suggesting that there was genome-specific expansion of 45SnrDNA, which might be caused by sub-genome dominance6. However, further studies are required to address the consequences of rDNA copy number variation in allotetraploid Brassica (Table 1).
Origin of the chloroplast genome in the AC genome accessions
Phylogenetic analyses conducted with only one or a few loci can misrepresent the derived phylogenic history, and complete information on genetic diversity is required for accurate analysis50. Unlike 45SnrDNA, chloroplast-based phylogenetic analysis indicates that the AC genome chloroplasts did not follow either of the parental nuclear genomes (A or C genome). Studies have been performed to clarify the genetic relationships of the major diploid and tetraploid Brassica species, but the origin of the chloroplast in the AC genome species is still unclear15,20,21,51. Initially, maternal parent of the AC genome was thought to be derived from the C genome due to their similarities in their chloroplast DNA restriction digest patterns; however, analysis with a wider range of accessions suggested that A genome was the maternal source20. Moreover, analysis with both chloroplast and nuclear markers suggested that the AC genome arose from several independent hybridization events including artificial introgression of A-genome51. A survey of the rpo locus revealed that >90% of 488 AC accessions displayed different genotypes than the parental accessions (A and C), but they were classified as an independent group with different origin20. Comparison with the recent findings of the Brassica chloroplast genome shows overall agreement, such as grouping based on species and the maternal and paternal origin of the allotetraploids23. Though there were two different A genome sources for the AC and AB genomes, we did not observe any divergence based on 45SnrDNA, suggesting that 45SnrDNA has been conserved in the Brassica genome8. Furthermore, chloroplast genomes from nine and seven different A and C genome morphotypes, respectively, formed a single cluster to confirm that the chloroplast genome and 45SnrDNA are stable even upon divergence of different sub-species and morphotypes23. In addition, sub-genome parallel selection played a crucial role in evolution of different morphotypes52.
Furthermore, A recent chloroplast genome survey of more diverse A genotypes revealed two different types of chloroplast genomes. The rapa-type1 chloroplast genome is generally found in all B. rapa, whereas the rapa-type2 is unique for some Italian Broccoletto genotypes of B. rapa23. Phylogenetic analysis indicated that the rapa-type2 clustered with the chloroplast of the AC genome, which explains why the Italian Broccoletto genotype is the donor for the most abundant AC chloroplast genome. The rapa-type2 chloroplast genome diverged 4.7 mya, which coincides with the currently known A and C genome divergence around 5.4 to 2.7 mya (Fig. 5). By contrast, analysis of the AC genome indicated that allotetraploidization occurred 7,500 years ago6. Both of these results indicate that the rapa-type2 chloroplast genome was maintained in the Italian Broccoletto genotype by geographical isolation or maternal dominance since 4.7 mya, and the Italian Broccoletto genome was utilized as the matenal parent to generate the AC genome 7,500 years ago. However, there are still questions about the evolution of the maternal genomes for the A and AC genomes. It is still not known how the rapa-type2 chloroplast genome became associated with the common maternal parent for most AC genomes, although the Italian Broccoletto genotype is not widespread in the A genome.
Conclusion
This study analyzed the genetic relationships and diversity among Brassica species using chloroplast genome and 45SnrDNA sequences. Phylogenetic analysis revealed that the B genome diverged first in the Brassica clade, followed by R, A, and C, and with three allotetraploids forming during last 0.1 to 0.01 mya. We cataloged the complete variants in chloroplast and 45SrDNA sequences, which will serve as excellent resources for the development of barcode markers and species identification. Comparative genome analyses of species-specific variations would facilitate the study of genome evolution and morphological divergence of Brassica. The combined results of this study reveal comprehensive genetic relationships of U’s triangle species and provide insights into genome evolution in Brassica. The results of this study will be extensively applicable for species identification and evolutionary studies.
Materials and Methods
Plant materials and DNA sequencing
Seeds of four genotypes representing each A, B, C, R, AB, AC, and BC genome were obtained from the RDA Genebank Center, Suwon, South Korea. All plants were grown at 22 °C (day)/18 °C (night) with a 16 h light/8 h dark photoperiod at the RDA experimental farm, Suwon, South Korea, during the spring of 2014. High-quality total genomic DNA was isolated from young leaves using a modified CTAB method53. Whole-genome shotgun libraries were generated using the TrueSeq DNA PCR-Free Library Preparation kit (Illumina) according to the manufacturer’s instructions. Briefly, 5 ng of high-quality DNA from each accession was fragmented via sonication. Then, the fragments were end-repaired and A-tailed. Adapters were ligated, including the barcoding and multiplex identifier adapters, and the fragments were amplified with 10 PCR cycles. Finally, a paired-end (PE) library with inserts of 400−500 bp was generated. The library was sequenced with the MiSeq System (Illumina) at LabGenomics (www.labgenomics.co.kr, South Korea). Multiplex adapters were used to separate the 28 genotypes from the bulked raw reads, and the sequence reads were trimmed for adaptors and low quality and utilized for further analysis. All trimmed high quality sequences (NN3658-NN3685) for the 28 accessions were deposited into the National Agricultural Biotechnology Information Center (http://nabic.rda.go.kr) public database54 (Table S6).
Assembly and annotation of chloroplast genome and 45SrDNA sequences
Complete chloroplast genome and 45SnrDNA sequences were simultaneously assembled for all 28 Brassica and Raphanus genotypes using the dnaLCW method32. The dnaLCW method is a fast and comparatively easy method that does not require a PCR based gap filling to assemble the chloroplast and rDNA sequences. With slight modification, dnaLCW also allows the characterization of the major repeats in the Brassica genome55. Briefly, high-quality Illumina paired-end reads were denovo assembled using the CLC genome assembler (ver. 4.06 beta, CLC Inc., Aarhus, Denmark) with autonomously controlled overlap size (200–500 bp). After gap closing, the resulting contigs were homology searched against the Arabidopsis thaliana chloroplast reference genome (GenBank accession: NC_000932) using mummer. Contigs related to the chloroplast genome were ordered according to the reference genome. Gaps and other errors such as false SNPs, copies of tandem repeats and homopolymer errors were corrected according to the dnaLCW approach32. Likewise, Arabidopsis thaliana 45SrDNA sequence (GenBank accession: X52322.1) was used as a reference to assemble the 45SnrDNA sequences of 28 genotypes. Due to the number of variations in intergenic spacer sequences (up to six types in B. oleracea), only the unique 45SnrDNA transcription units were assembled. We also identified both parental types of 45SnrDNA in an allotetraploid genome [i.e., B. napus (AC) genome] containing parental or sub-genomes of B. rapa (A) and B. oleracea (C), which were represented as AC-A and AC-C, respectively.
The chloroplast genomes of the 28 genotypes were annotated for protein-coding genes, transfer RNA (tRNA), and ribosomal RNA (rRNA) using DOGMA (https://dogma.ccbb.utexas.edu/)56. The accuracy of the start and stop codons and intron–exon boundaries were manually annotated based on previously annotated information from the close relative A. thaliana. The complete structure of tRNA genes was validated using tRNAscan-SE v1.2.157. The systematic circular view of the chloroplast genome was created using OGDRAW and in-house customized perl script58. Comparative syntenic maps were generated using circos following the BlastZ annotation. A chloroplast-based browser was developed for systematic analysis of the chloroplast genomes of the 28 Brassica and Raphanus genotypes, which can be accessed at www.phyzen.co.kr/cpbrowser. The chloroplast browser also contains sequence and gene annotation information for all 28 genotypes. Similarly, 45SnrDNA genes (18S, 5.8S, and 26S) were annotated based on Blast analyses and reported reference units. The mvista tool was used to visualize comparative syntenic relationships with other genotypes59. Complete chloroplast genomes and 40 complete 45SnrDNA sequences from 28 genotypes were deposited in GenBank (Table 1).
Structural variations and PCR analysis of chloroplast and 45SnrDNA
Extensive manual curation of chloroplast and 45SnrDNA revealed different kinds of non-redundant sequence variations (SV) such as SNPs, InDels, and copy number variations. Inter-species and intra-species structural variations were analyzed for chloroplast and 45SnrDNA sequences from 28 Brassica and Raphanus genotypes (Tables S2–S5). Putative SNPs and InDels were manually analyzed using the file aligned with MEGA7. Tandem repeats were identified using the Tandem repeats finder (TRF) tool. To detect highly reliable variations, all predicted variations were manually curated for both chloroplast and 45SnrDNA. Some of the randomly selected and highly informative variations were validated by PCR analysis.
To validate the polymorphic regions of chloroplast and 45SnrDNA sequences, specific primers were developed for high-quality structural variations such as SNPs and InDels (Table S7). DNA templates from 28 genotypes were used for target analysis. Each PCR reaction contained 10 ng template DNA, 10 pM primers, 0.5 µM dNTPs, 2 units of Taq polymerase (TAKARA, Japan), and the final volume brought to 20 µl with sterile distilled water. The PCR reactions were 10 min at 95 °C; followed by 36 cycles of 30S at 94 °C, 30S at 55−62 °C, and 30S at 72 °C; with a final extension at 72 °C for 5 min. Amplified fragments were checked with 2% agarose gel electrophoresis to estimate the product size.
Phylogenetic analysis and divergence estimation based on chloroplast genomes and 45SnrDNA sequences
Complete chloroplast genomes and 45SnrDNA sequences were independently explored for phylogenetic and divergence analysis. Chloroplast sequences of 28 Brassica and Raphanus genotypes were aligned with a previously reported Brassica chloroplast sequence using MAFFT (http://mafft.cbrc.jp/alignment/software/). Phylogenetic trees were constructed in MEGA7 using the neighbor-joining iterative model with 1,000 bootstrap replications60. Phylogenetic analysis was performed for 40 types of 45SnrDNA sequences based on 28 genotypes. A. thaliana chloroplast and 45SnrDNA sequences were used as an outgroup for the phylogenetic analysis. The reference chloroplast sequence with its annotation of A. thaliana, B. rapa, B. oleracea, B. nigra, B. juncea, B. carinata, and Raphanus sativus was obtained from GenBank.
Chloroplast and 45SnrDNA sequences from 28 genotypes were subjected to tree topology analysis and divergence time estimation using Bayesian methods implemented in BEAST (http://beast.bio.ed.ac.uk/)61. The BEAST program assumes auto-correlation, and is widely used to estimate the uncertainty of divergence dates and branch lengths, to estimate divergence using known speciation dates, and to accommodate the branching rate. The GTR + I + G substitution model was used to construct the tree topology and divergence time. We used an uncorrelated lognormal relaxed clock model to perform 10,000,000 generations of Markov chain Monte Carlo (MCMC) analysis with sampling every 1,000 generations. A Yule tree prior were used to generate the random starting tree. Tracer v. 1.6 was used to obtain the BEAST run after discarding 10% of the generations as burn-in. The remaining BEAST runs were used for the posterior possibilities. The divergence time was estimated using Tree annotator. A. thaliana was constrained as the outgroup, and the age of divergence between A. thaliana and Brassica lineages was constrained by a normal distribution with a mean of 17 million years (my) and standard deviation of 2 my9.
Availability of data and materials
All data generated or analysed during this study were obtained from the accession numbers provide at Tables 1 and S6.
Electronic supplementary material
Acknowledgements
This study was conducted with support from the Research Program for Agricultural Science and Technology Development (Project no. PJ010112) of the NAAS, and from the Cooperative Research Program for Agriculture Science & Technology Development (SSAC, Grant no. PJ011650), RDA, Republic of Korea. This work was also partially supported by a Golden Seed Project (Center for Horticultural Seed Development, No. 213003-04-4-SB430), Ministry of Agriculture, Food and Rural Affairs (MAFRA), Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ01100801), Rural Development Administration, Republic of Korea. The funding body has no role in the design of the study, analysis, and writing the manuscript.
Author Contributions
C.K., Y.S., S.P. and T.J.Y. planned and designed the research. Y.S., C.K., S.S., J.L., N.E.W., M.J., S.C.L., S.J. Y.Y., H.K., J.C., K.R., and B.C. contributed materials and chloroplast genome assembly and comparative analysis. Y.S. and C.K. conducted genome sequencing. S.P. and T.J.Y. analyzed data and interpreted the results. S.P., C.K., Y.S. and T.J.Y. wrote the manuscript and I.P. and S.S. edited the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Chang-Kug Kim, Young-Joo Seol and Sampath Perumal contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-25585-4.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cheng F, Wu J, Wang X. Genome triplication drove the diversification of Brassica plants. Horticulture Research. 2014;1:14024. doi: 10.1038/hortres.2014.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng F, et al. Genome sequencing supports a multi-vertex model for Brassiceae species. Current opinion in plant biology. 2017;36:79–87. doi: 10.1016/j.pbi.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 3.U, N. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jap J Bot7, 389-452 (1935).
- 4.Wang X, et al. The genome of the mesopolyploid crop species Brassica rapa. Nature genetics. 2011;43:1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- 5.Liu S, et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nature communications. 2014;5:3930. doi: 10.1038/ncomms4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chalhoub B, et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 2014;345:950–953. doi: 10.1126/science.1253435. [DOI] [PubMed] [Google Scholar]
- 7.Parkin IA, et al. Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome biology. 2014;15:R77. doi: 10.1186/gb-2014-15-6-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J, et al. The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nature genetics. 2016;48:1225–1232. doi: 10.1038/ng.3657. [DOI] [PubMed] [Google Scholar]
- 9.Yang T-J, et al. Sequence-level analysis of the diploidization process in the triplicated FLOWERING LOCUS C region of Brassica rapa. The Plant cell. 2006;18:1339–1347. doi: 10.1105/tpc.105.040535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koenig D, Weigel D. Beyond the thale: comparative genomics and genetics of Arabidopsis relatives. Nature reviews. Genetics. 2015;16:285–298. doi: 10.1038/nrg3883. [DOI] [PubMed] [Google Scholar]
- 11.Mun JH, et al. Genome-wide comparative analysis of the Brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication. Genome biology. 2009;10:R111. doi: 10.1186/gb-2009-10-10-r111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palmer, J. D. In Cell organelles 99–133 (Springer, 1992).
- 13.Birky CW. Uniparental inheritance of mitochondrial and chloroplast genes: mechanisms and evolution. Proceedings of the National Academy of Sciences. 1995;92:11331–11338. doi: 10.1073/pnas.92.25.11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reboud X, Zeyl C. Organelle inheritance in plants. Heredity. 1994;72:132–140. doi: 10.1038/hdy.1994.19. [DOI] [Google Scholar]
- 15.Palmer JD, Shields C, Cohen D, Orton T. Chloroplast DNA evolution and the origin of amphidiploid Brassica species. Theoretical and Applied Genetics. 1983;65:181–189. doi: 10.1007/BF00308062. [DOI] [PubMed] [Google Scholar]
- 16.Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proceedings of the National Academy of Sciences. 2010;107:4623–4628. doi: 10.1073/pnas.0907801107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, et al. Plant DNA barcoding: from gene to genome. Biological Reviews. 2015;90:157–166. doi: 10.1111/brv.12104. [DOI] [PubMed] [Google Scholar]
- 18.Nikiforova SV, Cavalieri D, Velasco R, Goremykin V. Phylogenetic analysis of 47 chloroplast genomes clarifies the contribution of wild species to the domesticated apple maternal line. Molecular biology and evolution. 2013;30:1751–1760. doi: 10.1093/molbev/mst092. [DOI] [PubMed] [Google Scholar]
- 19.Zhang, Y. et al. The complete chloroplast genome sequences of five Epimedium species: lights into phylogenetic and taxonomic analyses. Frontiers in plant science7 (2016). [DOI] [PMC free article] [PubMed]
- 20.Qiao J, et al. High‐throughput multiplex cpDNA resequencing clarifies the genetic diversity and genetic relationships among Brassica napus, Brassica rapa and Brassica oleracea. Plant biotechnology journal. 2016;14:409–418. doi: 10.1111/pbi.12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma S, et al. Two plastid DNA lineages—Rapa/Oleracea and Nigra—within the tribe Brassiceae can be best explained by reciprocal crosses at hexaploidy: evidence from divergence times of the plastid genomes and R-block genes of the A and B genomes of Brassica juncea. PloS one. 2014;9:e93260. doi: 10.1371/journal.pone.0093260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franzke A, Lysak MA, Al-Shehbaz IA, Koch MA, Mummenhoff K. Cabbage family affairs: the evolutionary history of Brassicaceae. Trends in plant science. 2011;16:108–116. doi: 10.1016/j.tplants.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 23.Li P, et al. A Phylogenetic Analysis of Chloroplast Genomes Elucidates the Relationships of the Six Economically Important Brassica Species Comprising the Triangle of U. Frontiers in plant science. 2017;8:111. doi: 10.3389/fpls.2017.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koo DH, et al. Rapid divergence of repetitive DNAs in Brassica relatives. Genomics. 2011;97:173–185. doi: 10.1016/j.ygeno.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 25.Lim KB, et al. Characterization of rDNAs and tandem repeats in the heterochromatin of Brassica rapa. Molecules and cells. 2005;19:436–444. [PubMed] [Google Scholar]
- 26.Waminal, N. E. et al. In The Brassica rapa Genome 83–96 (Springer, 2015).
- 27.Hasterok R, Jenkins G, Langdon T, Jones RN, Maluszynska J. Ribosomal DNA is an effective marker of Brassica chromosomes. Theoretical and Applied Genetics. 2001;103:486–490. doi: 10.1007/s001220100653. [DOI] [Google Scholar]
- 28.Warwick SI, Mummenhoff K, Sauder CA, Koch MA, Al-Shehbaz IA. Closing the gaps: phylogenetic relationships in the Brassicaceae based on DNA sequence data of nuclear ribosomal ITS region. Plant Systematics and Evolution. 2010;285:209–232. doi: 10.1007/s00606-010-0271-8. [DOI] [Google Scholar]
- 29.Atwell S, et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465:627–631. doi: 10.1038/nature08800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang J, et al. Comparative mitochondrial genome analysis reveals the evolutionary rearrangement mechanism inBrassica. Plant biology. 2015;18:527–536. doi: 10.1111/plb.12414. [DOI] [PubMed] [Google Scholar]
- 31.Kim K, et al. Comprehensive Survey of Genetic Diversity in Chloroplast Genomes and 45S nrDNAs within italic Panax ginseng /italic Species. PloS one. 2015;10:e0117159. doi: 10.1371/journal.pone.0117159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim K, et al. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Scientific reports. 2015;5:15655. doi: 10.1038/srep15655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mandáková T, Lysak MA. Chromosomal phylogeny and karyotype evolution in x = 7 crucifer species (Brassicaceae) The Plant cell. 2008;20:2559–2570. doi: 10.1105/tpc.108.062166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitsui Y, et al. The radish genome and comprehensive gene expression profile of tuberous root formation and development. Scientific reports. 2015;5:10835. doi: 10.1038/srep10835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seol, Y.-J. et al. The complete chloroplast genome of two Brassica species, Brassica nigra and B. oleracea. Mitochondrial DNA, 1-2, 10.3109/19401736.2015.1115493 (2015). [DOI] [PubMed]
- 36.Park JY, et al. Complete mitochondrial genome sequence and identification of a candidate gene responsible for cytoplasmic male sterility in radish (Raphanus sativus L.) containing DCGMS cytoplasm. TAG. Theoretical and applied genetics. Theoretische und angewandte. Genetik. 2013;126:1763–1774. doi: 10.1007/s00122-013-2090-0. [DOI] [PubMed] [Google Scholar]
- 37.Chang S, et al. Mitochondrial genome sequencing helps show the evolutionary mechanism of mitochondrial genome formation in Brassica. BMC genomics. 2011;12:497. doi: 10.1186/1471-2164-12-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bennett RI, Smith AG. Use of a genomic clone for ribosomal RNA from Brassica oleracea in RFLP analysis of Brassica species. Plant molecular biology. 1991;16:685–688. doi: 10.1007/BF00023432. [DOI] [PubMed] [Google Scholar]
- 39.Sang T, Crawford DJ, Stuessy TF. Documentation of reticulate evolution in peonies (Paeonia) using internal transcribed spacer sequences of nuclear ribosomal DNA: implications for biogeography and concerted evolution. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:6813–6817. doi: 10.1073/pnas.92.15.6813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Kane Jr, S. L., Schaal, B. A. & Al-Shehbaz, I. A. The origins of Arabidopsis suecica (Brassicaceae) as indicated by nuclear rDNA sequences. Systematic Botany, 559–566 (1996).
- 41.Poczai P, Hyvonen J. Nuclear ribosomal spacer regions in plant phylogenetics: problems and prospects. Molecular biology reports. 2010;37:1897–1912. doi: 10.1007/s11033-009-9630-3. [DOI] [PubMed] [Google Scholar]
- 42.Idziak D, Hasterok R. Cytogenetic evidence of nucleolar dominance in allotetraploid species of Brachypodium. Genome. 2008;51:387–391. doi: 10.1139/G08-017. [DOI] [PubMed] [Google Scholar]
- 43.Tucker S, Vitins A, Pikaard CS. Nucleolar dominance and ribosomal RNA gene silencing. Current opinion in cell biology. 2010;22:351–356. doi: 10.1016/j.ceb.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Earley K, et al. Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes & development. 2006;20:1283–1293. doi: 10.1101/gad.1417706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Preuss SB, et al. Multimegabase silencing in nucleolar dominance involves siRNA-directed DNA methylation and specific methylcytosine-binding proteins. Mol Cell. 2008;32:673–684. doi: 10.1016/j.molcel.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fuertes Aguilar J, Rosselló J, Nieto Feliner G. Nuclear ribosomal DNA (nrDNA) concerted evolution in natural and artificial hybrids of Armeria (Plumbaginaceae) Molecular ecology. 1999;8:1341–1346. doi: 10.1046/j.1365-294X.1999.00690.x. [DOI] [PubMed] [Google Scholar]
- 47.Bao Y, Wendel JF, Ge S. Multiple patterns of rDNA evolution following polyploidy in Oryza. Molecular phylogenetics and evolution. 2010;55:136–142. doi: 10.1016/j.ympev.2009.10.023. [DOI] [PubMed] [Google Scholar]
- 48.Kotseruba V, et al. The evolution of the hexaploid grass Zingeriakochii (Mez) Tzvel. (2n = 12) was accompanied by complex hybridization and uniparental loss of ribosomal DNA. Molecular phylogenetics and evolution. 2010;56:146–155. doi: 10.1016/j.ympev.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 49.Dobesova E, et al. Silenced rRNA genes are activated and substitute for partially eliminated active homeologs in the recently formed allotetraploid, Tragopogon mirus (Asteraceae) Heredity (Edinb) 2015;114:356–365. doi: 10.1038/hdy.2014.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bailey CD, et al. Toward a global phylogeny of the Brassicaceae. Molecular biology and evolution. 2006;23:2142–2160. doi: 10.1093/molbev/msl087. [DOI] [PubMed] [Google Scholar]
- 51.Allender CJ, King GJ. Origins of the amphiploid species Brassica napus L. investigated by chloroplast and nuclear molecular markers. BMC plant biology. 2010;10:54. doi: 10.1186/1471-2229-10-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng F, et al. Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and Brassica oleracea. Nature genetics. 2016;48:1218–1224. doi: 10.1038/ng.3634. [DOI] [PubMed] [Google Scholar]
- 53.Allen G, Flores-Vergara M, Krasynanski S, Kumar S, Thompson W. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nature protocols. 2006;1:2320–2325. doi: 10.1038/nprot.2006.384. [DOI] [PubMed] [Google Scholar]
- 54.Seol Y-J, Lee T-H, Park D-S, Kim C-K. NABIC: A New Access Portal to Search, Visualize, and Share Agricultural Genomics Data. Evolutionary bioinformatics online. 2016;12:51. doi: 10.4137/EBO.S34493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perumal S, et al. Elucidating the major hidden genomic components of the A, C, and AC genomes and their influence on Brassica evolution. Scientific reports. 2017;7:17986. doi: 10.1038/s41598-017-18048-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- 57.Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic acids research. 2005;33:W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Current genetics. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- 59.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic acids research. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular biology and evolution, 10.1093/molbev/msw054 (2016). [DOI] [PMC free article] [PubMed]
- 61.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular biology and evolution. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study were obtained from the accession numbers provide at Tables 1 and S6.