Abstract
PINK1 mutations that disrupt its kinase activity cause autosomal recessive early onset Parkinson’s disease (PD). Although research in recent years has elucidated a PINK1-Parkin pathway of mitophagy activation that requires PINK1 kinase activity, mitophagy-independent functions of PINK1 and their possible roles in PD pathogenesis have been proposed. Using an unbiased quantitative mass spectrometry approach to analyze the phosphoproteome in primary neurons from wild type and Pink1 knockout mice after mitochondrial depolarization, we uncovered PINK1-regulated phosphorylation sites, which involve coordinated activation of multiple signaling pathways that control cellular response to stress. We further identified the pro-apoptotic protein BAD as a potential mitochondrial substrate of PINK1 both in vitro and in vivo, and found that cells more susceptible to a12poptosis induced by mitochondrial damage can be rescued by phosphorylation mimic BAD. Our results thus suggest that PINK1 kinase activity is important for pro-apoptotic protein function in regulation of cell death.
Introduction
Parkinson’s disease (PD) is a common neurodegenerative disorder with the initial characteristic symptoms including rigidity, bradykinesia, resting tremor, and gait instability. Although the majority of the cases are sporadic, the identification of a number of disease genes through genetic studies of familial cases has shed tremendous light on our understanding of the disease mechanism. Among these genes, PTEN induced putative kinase 1 (PINK1) loss-of-function mutation causes autosomal recessive early-onset Parkinson’s disease [1].
Studies over the past 10 years have established that PINK1 is a mitochondrial localized serine/threonine protein kinase. Early studies showed that PINK1 exerted a protective role against oxidative stress and mitochondrial damage in cells [2, 3]. Recently it was shown that upon mitochondrial damage, PINK1 is stabilized on mitochondrial outer membrane, which promotes the phosphorylation and activation of parkin and ubiquitin. Parkin mediates ubiquitination of mitochondrial outer membrane proteins including TOMM20, mitofusin, miro, and VDAC, forming a recognition site for adaptor proteins and promoting the selective autophagy of damaged mitochondria [4, 5]. This PINK1-parkin pathway of mitophagy plays an important role in organelle quality control and cellular homeostasis. However, whether a malfunction of this system due to PINK1 or parkin mutation is the major cause of selective dopaminergic death or not remains elusive. Therefore, a PINK1 functions in PD pathogenesis beyond mitophagy has been widely proposed [6, 7].
Many studies on the PINK1-parkin mediated mitophagy pathway were conducted in non-neuronal cells; however, several studies on rodent primary neurons or neurons derived from patient iPSCs resulted in controversial conclusions on parkin translocation and mitophagy after mitochondrial depolarization and PINK1 activation [8–10]. It is thus imperative to identify phosphorylation targets downstream of PINK1 in a neuronal context. Quantitative mass spectrometry (MS)-based analysis of PINK1-regulated phosphoproteome has the potential to identify PINK1 targets. Additionally, the analysis can glean a comprehensive insight on cellular pathways that is altered after PINK1 activation. For example, a phosphoproteomic analysis of mitochondrial complex I purified from wild type (WT) and Pink1 knockout (KO) mouse brain identified a critical role of serine-250 phosphorylation in complex I subunit NdufA10 [11]. We have previously analyzed a cell line after transient PINK1 knockdown [12]. In spite of these efforts, it remains elusive how the activation of mitophagy and protection of cell death in neurons are coordinated. Here, we utilized liquid-chromatography fractionation, affinity enrichment of phosphorylated peptides, high-resolution mass spectrometry to quantify the phosphoproteome, and the proteome of primary cortical neurons after mitochondrial depolarization. We find that PINK1 influenced the coordinated activation of multiple signaling pathways that control cellular response to stress, including energy metabolism, response to growth factor stimulation, fine tuning of autophagy, and cell survival. We further identify the pro-apoptotic protein BAD as a potential mitochondrial substrate of PINK1, and suggest a molecular detail on PINK1 regulation of cell death.
Results
Quantitative analysis of PINK1-regulated phosphoproteome upon mitochondrial depolarization
We treated neurons with the protonophore carbonyl cyanide m-chlorophenyl hydrazine (CCCP), which induces persistent depolarization leading to mitochondrial damage [13]. Western blot analysis confirmed the expression of endogenous PINK1 in WT but not KO neurons (Fig. S1b). CCCP treatment resulted in upregulation of endogenous PINK1 in WT neurons (Fig. S1b), consistent with previous studies [14]. We assessed the viability of neurons after CCCP treatment and found that treatment after 2 and 6 h resulted in significantly more death in KO neurons (Fig. S1c). The proteome and phosphoproteome of WT and KO neurons after CCCP treatment were analyzed using high-resolution quantitative MS. We applied a 10-channel tandem mass tag [15] to label trypsin-digested peptides. The labeled peptides were combined and then separated using a basic reversed-phase liquid chromatography, followed by phosphopeptide enrichment using TiO2 (Fig. S1a).
Overall, our phosphoproteome analysis of biological duplicate experiments quantified 12876 and 6422 phosphorylation sites on 3067 and 2413 protein, respectively, from the early (15–45 min) and late (2–6 h) treatment time points (Fig. S1d). The number of singly, doubly, and triply phosphorylated events in both experiments was similar (Fig. S1e); and the percentage of serine, threonine, and tyrosine phosphorylation events is illustrated in Fig. S1f. In addition, we have achieved high mass accuracy measurements in both experiments (Fig. S2a); and the quantification between the biological duplicate experiments was highly correlated (Fig. S2b). Furthermore, gene ontology analysis revealed that among all quantified 3984 phosphoproteins there were 26% organelle proteins and a majority of the rest were categorized as cell part (Fig. S2c). Among organelle proteins nuclear and cytoskeletal proteins occupied 53% and 19%, respectively, whereas mitochondrial and other organelle proteins occupied 4% each (Fig. S2d). These results suggest that our analysis favored soluble proteins but covered less proteins from the membrane structure of organelles. Nevertheless, our analyses have achieved certain depth and quantitative accuracy.
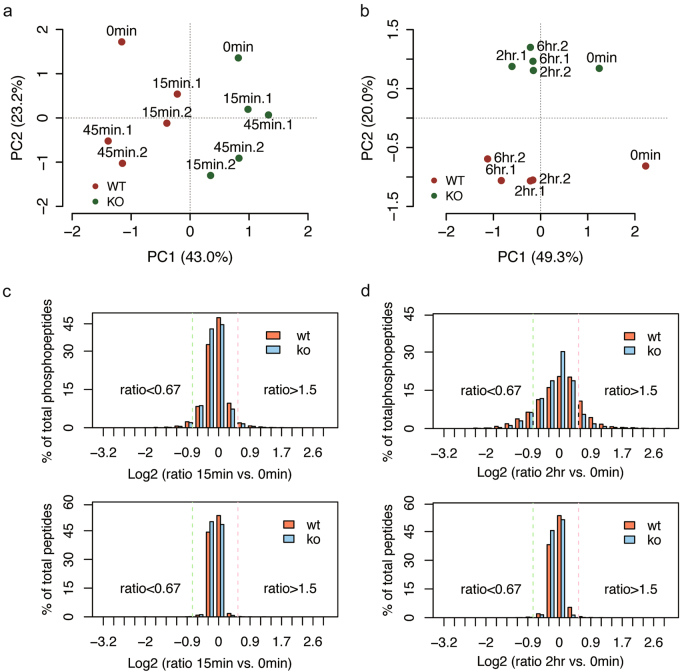
Principal component analysis revealed that the genotype was the major feature that differed across the data at early time points, and the treatment effects were separated in the second component (Fig. 1a). In contrast, at late time points the treatment became the main feature that strongly differed in WT neurons but mild in KO neurons (Fig. 1b). We evaluated changes in phosphorylation in neurons without CCCP treatment (0 min) that might be the result of PINK1 loss-of-function, or the result of adaptation to PINK1 loss. Overall, only 18 serine/threonine residues on 18 proteins showed difference in phosphorylation between KO and WT in both biological replicates (Figs. S3a and b). We then applied the median absolute deviation analysis integrated with a fold change cutoff of 1.5 and a Z score cutoff of 2.5 to identify differential phosphorylation events that happened only in WT neurons. The fold change of treatment against non-treatment was calculated for each time point. At the early time points the percentage of phosphopeptides with fold change more than 1.5 was low, and was not dramatically different between WT and KO neurons (Fig. 1c and Fig. S3c). While at the late time points the percentage became higher with more dramatically increased phosphorylation in WT neurons after treatment (Fig. 1d and Fig. S3d). These results indicate that in response to mitochondrial depolarization PINK1 induced a variety of downstream phosphorylation events, either directly or indirectly.
Fig. 1. Overview of the quantitative phosphoproteome of cortical neurons.
a, b PCA for 12876 and 6422 phosphorylated sites (only those with a localization probability of at least 67% were considered) in WT and Pink1 KO neurons at the early time points a and the late time points b, respectively. WT neurons are represented by red, and KO neurons by green. Around 65~69% of total variance is explained by the first two components for both PCAs. c, d Histograms of phosphoproteome and proteome data distribution at 15 min and 2 h after CCCP treatment. A log2 transformed fold change of 1.5 is marked with a dash line to identify differential phosphorylation events
Clustering analysis of phosphorylation events reveals PINK1 regulated cellular pathways
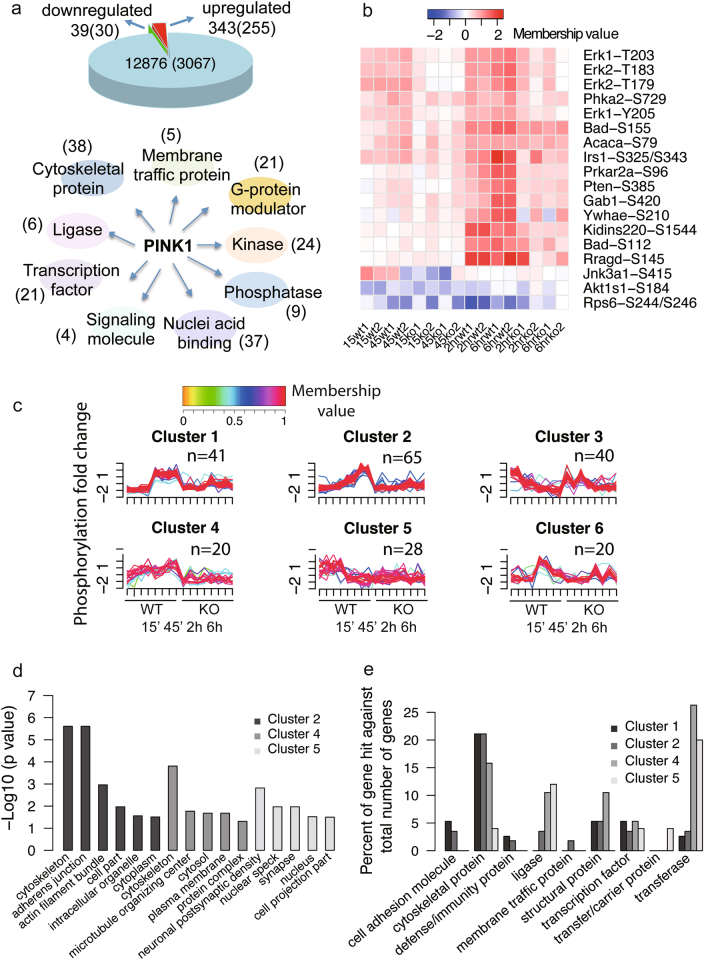
At the early time points, 382 phosphorylation sites from 285 proteins exhibited a differential phosphorylation only in WT neurons, among which only 39 phosphorylation sites from 30 proteins showed reduced phosphorylation (Fig. 2a). At the late time points, 146 phosphorylation sites from 117 proteins exhibited a differential phosphorylation only in WT neurons (Table S1 and S2). Annotation of molecular function indicated that the largest number of proteins fall into the category of cytoskeletal protein, following by nucleic acid binding protein, kinase and transcription factor. Notably, a number of proteins were annotated as G-protein modulators (Fig. 2a). To gain insights at the signaling pathways, we performed a Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis using the online data annotation and integration tool, DAVID [16]. We found that the phosphoproteins were enriched in top 3 pathways including the mTOR pathway (p = 3.16E-05) the neurotrophin signaling pathway (p = 3.16E-05) and the insulin signaling pathway (p = 4.6E-05). The key node proteins of these pathways and the levels of their phosphorylation sites were illustrated in Fig. 2b. The functional diversity suggested that activated PINK1 could mediate phosphorylation of a broad range of proteins.
Fig. 2. Bioinformatics analysis of PINK1-regulated phosphorylation events.
a Pie chart shows the number of changed phosphorylation sites and proteins (parenthesis) (upper panel) at early time points, and protein classification of changed phosphoproteins (lower panel). b Heat map shows the changed phosphoproteins enriched in top 3 pathways in KEGG analysis. c PINK1-regulated phosphorylation sites are grouped into 6 clusters based on temporal changes. The color key indicates membership value with red denoting a high membership and yellow denoting a low membership. The number of phosphorylation sites with membership values greater than 0.5 are shown above each graph. d Gene ontology analysis in the category of biological processes for proteins in clusters 2, 4, and 5. e Phosphoproteins in clusters 1, 2, 4, and 5 were classified with PANTHER
To gain temporal insights into PINK1-dependent phosphorylation events, we applied the c-fuzzy algorithm [17] to cluster the phosphorylation data across all time points. The changed phosphorylation sites were grouped into six clusters (Fig. 2c and Table S3). Each cluster exhibited distinct pattern of phosphorylation change, with clusters 1, 2, 4, and 5 showing late response, gradual increase, subtle increase, and early response in phosphorylation, respectively. Except for clusters 3 and 6, all changes in phosphorylation in these clusters showed PINK1 dependency. We then applied the PANTHER classification system [18] to analyze the biological functions of the PINK1-dependent phosphoproteins. Proteins in clusters 1, 2, and 4 were enriched in cytoskeletal proteins, while ligases and transferases occupied high percentage in clusters 4 and 5 (Figs. 2d, e). Analysis in the category of cellular component revealed that faster response proteins in cluster 5 were enriched in synapse and nucleus.
Analysis of PINK1 regulated phosphorylation site motif indicates potentially activated kinases and signal pathways
In order to obtain sequence features surrounding the phosphorylation sites, and to infer kinases responsible for their phosphorylation, we extracted sequence motifs using motif-x [19] from the 382 and 146 PINK1 regulated phosphorylation sites. A motif shared by both time points was the proline-directed motif, while there was a distinct basophilic motif containing an arginine at the -2 position of the central serine residue at early time points. An acidophilic motif containing a glutamine at the +1 position of the central serine residue stood out at late time points (Figs. 3a, b).
Fig. 3. Analysis of PINK1-regulated phosphorylation site motif.
a, b Sequence motifs from the 382 and 146 PINK1-regulated phosphorylation sites after mitochondrial depolarization at the early and the late time points are shown respectively. NetworKIN predicted kinases for these motif-enriched sites are visualized in a network format, where c, d represent kinases predicted for sP and R..s. sites, respectively, at the early time points, e, f represent kinases predicted for sP and sE. sites respectively, at the late time points. g Differential phosphorylation sites in major signaling pathways after CCCP treatment in a PINK1-regulated manner are shown in a pathway format
To further understand the kinase-substrate relationship from the PINK1-regulated phosphorylation events, we applied NetworKIN [20] that predicts kinases based on the motif and experimentally documented kinase-substrate interactions. The PINK1-regulated phosphosites and their predicted kinases were summarized in Supplementary Table 4. The kinase-substrate pairs were further classified based on motifs (Figs. 3c–f). Of note, insulin activated signaling nodes including AKT1, IRS1, MAPKs, all were activated after mitochondrial depolarization to a greater extent in WT neurons than KO neurons. We observed increased phosphorylation of EIF4G1 (Table S1), an essential component of translation initiation complex, which is consistent with mTOR activation by growth factors to promote protein synthesis. Although controversy exists regarding the causal relationship between mutation of EIF4G1 and onset of PD [21], a recent study identified genetic interaction and convergence among PD related proteins VPS35, EIF4G1 and α-synuclein [22]. Our data suggested a likely biochemical interaction between PINK1 and EIF4G1. We also observed elevated phosphorylation of BNIP3L after mitochondrial depolarization in a PINK1-dependent manner (Table S1). Interestingly, BNIP3 and BNIP3L have been linked to the PINK1-Parkin mediated mitophagy pathway [23, 24]. Thus, these proteins may form an intricate interaction loop to control mitophagy.
We then constructed major signaling pathways covering PINK1-regulated phosphorylation sites after CCCP treatment (Fig. 3g) with differential phosphorylation sites marked brownish yellow. These pathways play important roles regulating multiple cellular functions, including growth, survival, and autophagy.
Identification and validation of BAD as a potential PINK1 target
We tested whether PINK1-regulated proteins could be phosphorylated by PINK1. We selected IRS1, TSC2, ULK1, YWHAZ (14-3-3), BAD, and DAPK1, proteins that are involved in growth factor signaling, mTOR, autophagy and cell death. We co-expressed tagged IRS1, TSC2 and ULK1 with either wild type or kinase-dead PINK1 in cells, and used phos-Tag gel to detect phosphorylation. We did not observe band shift for these proteins (Figs. S4a-d). We constructed GST or His fusion protein of BAD, YWHAZ, and a truncated form of DAPK1. These proteins play a direct or indirect role in cell death [25–27]. In vitro kinase assay using an enzymatically active, insect-derived TcPINK1 [28] followed by phos-Tag gel showed distinct band shift in BAD and DAPK1, but not in YWHAZ (Figs. S4e and f).
We then focused on BAD, a pro-apoptotic protein whose in vivo phosphorylation has been demonstrated to raise the mitochondrial threshold for apoptosis [29]. Phosphorylation on serine 112, 136, and 155 of BAD can be catalyzed by a variety of kinases including Raf-1, PKA, Akt, PAK or RSK [30, 31]; and their phosphorylation leads to relocalization from the mitochondria to the cytosol and subsequent sequestration by 14-3-3 [32, 33]. We constructed His-tagged wild type and S112A mutant BAD and performed in vitro kinase assay. In the presence of PINK1, wild type but not the mutant BAD was phosphorylated, and the phosphorylation was largely reduced by calf intestinal phosphatase (CIP) (Fig. 4a). Ubiquitin, a known PINK1 substrate, was phosphorylated at serine 65 and the phosphorylation was also greatly reduced by CIP (Fig. 4b). The S112 phosphorylation on BAD and S65 phosphorylation on ubiquitin were observed at a time-dependent fashion, clearly observable starting at 10 min and gradually increased until 60 min (Figs. 4c, d). We then used a targeted mass spectrometry to observe temporal pattern of BAD phosphorylation on multiple sites, and found that both S112 and S136 were gradually phosphorylated across a one-hour time period (Figs. 4e, f). Thus, we have demonstrated that two serine residues on BAD can be directly phosphorylated by PINK1 in vitro.
Fig. 4. PINK1 phosphorylates BAD in vitro.
a In vitro kinase assay using recombinant BAD as the substrate and TcPINK1 as the kinase. Western blot analysis was performed to determine BAD phosphorylation at S112. CIP calf intestinal phosphatase. b In vitro kinase assay using known PINK1 substrate ubiquitin as a positive control. c, d Time-course analysis of TcPINK1-catalyzed substrate phosphorylation. e, f In vitro kinase assay and parallel reaction monitoring (PRM) measurement of the time course of BAD phosphorylation at S112 and S136
We explored the in vivo phosphorylation states of the serine residues on BAD. Quantitative mass spectrometry showed an elevated phosphorylation on S112 at 2 and 6 h after mitochondrial depolarization in WT neurons, as well as an elevated phosphorylation on S155 at 45 min (Fig. 5a). These results were further validated by western blot using phosphorylation site-specific antibodies. Mitochondrial depolarization induced by 20 μM CCCP at 45 min and 2 h robustly increased phosphorylation on S112 and S136, and slightly on S155, as demonstrated in Figs. 5b, c. Applying CCCP at a lower dose of 2 μM that caused mild mitochondrial depolarization also increased BAD phosphorylation on S112 and S136, and potentially S155 although no statistical significance was reached (Figs. S5a and b). The effect on BAD phosphorylation appears to be specific to CCCP treatment since mitochondrial toxin MPP+ failed to affect BAD phosphorylation (Fig. S5c). We co-expressed BAD with either WT or KD PINK1, and then induced mitochondrial depolarization. Wild type but not kinase dead mutant PINK1 robustly increased BAD phosphorylation on S112, and faintly on S155, although CCCP treatment itself had little effect on these phosphorylation sites (Figs. 5d, e). Nevertheless, our data demonstrated that kinase active PINK1 can mediate phosphorylation of endogenous BAD in primary neurons.
Fig. 5. PINK1-dependent BAD phosphorylation induced by CCCP in mouse neurons and in human cells.
a Two phosphopeptides of BAD (Ser112 and Ser155) were quantified with mass spectrometry in mouse primary cortical neurons after CCCP (20 μM) treatment. b, c Western blot analysis of primary cortical neurons from WT and Pink1 KO mice for phospho-BAD (S112), (S136), (S155) and BAD after treatment with CCCP (20 μM). Data represents three independent experiments. d, e PINK1 maintains the phosphorylation of BAD. BAD was co-expressed with wild type or kinase dead PINK1 in HEK293T cells and then treated with CCCP (20 μM) for indicated time. Error bars represent the S.E. of the mean of 3 independent experiments. Student’s t-test: * p < 0.05, ** p < 0.01, *** p < 0.001
PINK1 mediated BAD phosphorylation reduces BAD localization on mitochondria
Because BAD exerts a pro-apoptotic function through increased localization on mitochondria [34], we examined whether BAD localization on mitochondria can be regulated by PINK1. Triple immunofluorescence labeling and confocal microscopy showed that PINK1 and a mitochondrial outer membrane protein TOMM20 co-localized well in the presence and absence of mitochondrial depolarization. BAD showed a defused staining pattern with little localization on mitochondria after co-expression with wild type PINK1 as well as co-expression with KD PINK1 under basal culture conditions. In contrast, in the presence of kinase dead PINK1 after CCCP treatment, BAD dramatically increased mitochondrial localization (Figs. 6a, b). Similar results were obtained when expressing GFP-tagged BAD in WT or KO mouse embryonic fibroblasts (MEFs). Under basal conditions there is little BAD co-localization with TOMM20. After CCCP treatment there were less than 20% WT cells with BAD on mitochondria, while nearly 80% of KO cells showed BAD localization on mitochondria (Fig. S6a and b). Co-immunoprecipitation of overexpressed BAD and PINK1 also demonstrated that the two proteins interact with each other after mitochondrial depolarization (Fig. 6c). Comparing wild type, phosphorylation null (3SA) and phosphorylation mimic (3 SD) mutant BAD demonstrated that the phosphorylation null BAD had nearly 50% of cells showing strong mitochondrial localization, the wild type BAD had small percentage (<20%) of cells showing mitochondrial localization, and the phosphorylation mimic BAD had almost no cell showing mitochondrial localization (Figs. 6d, e). Based on these results, we propose a model whereby PINK1 promotes the phosphorylation of BAD and subsequently expels BAD from the mitochondria.
Fig. 6. PINK1 co-localizes with BAD in mitochondria and dynamically regulates BAD localization in mitochondria.
a, b Confocal microscopy images of HeLa cells transfected with PINK1 constructs together with BAD constructs. CCCP (20 μM) treatment lasted for 6 h. Scale bar, 10 μm. Error bars indicate the S.E. of the mean of 3 independent experiments with counting of 100 cells per treatment. c PINK1 interaction with BAD on depolarized mitochondria was detected by co-immunoprecipitation (co-IPs) of overexpressed PINK1 and BAD in HEK293T cells. d, e Wild type, phosphorylation null (3SA) or phosphorylation mimic (3 SD) mutant BAD was transfected into HeLa cells and then treated with DMSO or CCCP (20 μM) for 6 h. Error bars indicate the S.E. of the mean of 3 independent experiments with counting of 100 cells per treatment. Scale bar, 10 μm. Student’s t-test: * p < 0.05, ** p < 0.01
PINK1 regulates neuronal cell death through BAD
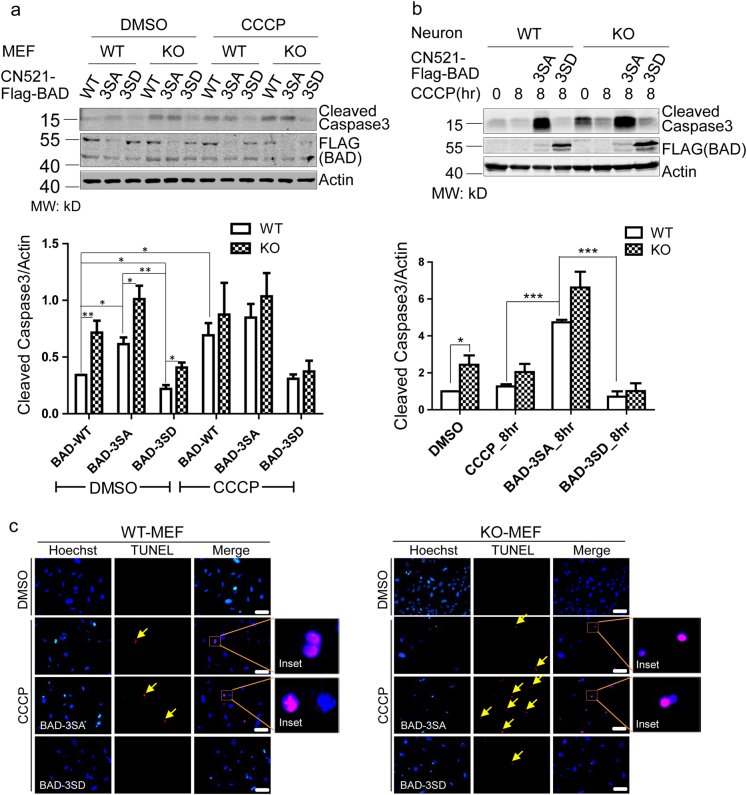
What will be the cellular consequences of BAD phosphorylation after mitochondrial damage? CCCP treatment induced more caspase-3 cleavage in KO than WT cells (Fig. S7a), consistent with established function of PINK1 to protect cells from apoptotic damage. The results were further supported by cell viability assay showing that KO cells were significantly less viable 48 h after CCCP treatment (Fig. S7b). After virus-mediated expression of wild type, 3SA, or 3 SD mutant BAD into WT or KO MEFs, we found that BAD-3SA expression resulted in more cleaved caspase-3 even in WT MEFs, regardless of CCCP treatment. On the other hand, BAD-3SD expression dramatically reduced caspase-3 cleavage in KO MEFs, both under basal conditions and after CCCP treatment (Fig. 7a). Similar results were obtained in primary cortical neurons from WT and KO mice (Fig. 7b). Using the TUNEL assay to detect apoptosis [35], we found that prolonged CCCP treatment resulted in increased TUNEL signal to a greater extent in KO MEFs than in WT MEFs, suggesting that more cells had gone through apoptosis. Consistent with biochemical results, BAD-3SA expression further increased TUNEL signal in both MEFs but much more dramatic in KO MEFs. In contrast, BAD-3SD expression effectively eliminated TUNEL positive cells, even in KO MEFs (Fig. 7c and Fig. S7c).
Fig. 7. PINK1-dependent BAD phosphorylation promotes survival of MEFs and neurons.
a Western blot analysis of caspase-3 cleavage after CCCP treatment for 24 h in cultured MEFs. WT and Pink1 KO cells, as well as these cells over-expressed with wild type, phosphorylation null and phosphorylation mimic mutant BAD were compared. Bar graph in the lower panel shows the expression levels of cleaved caspase-3 in each condition. b The same analysis as in a except that primary neurons were used. c TUNEL staining comparing WT and Pink1 KO MEF cells with or without CCCP treatment for 24 h, as well as comparing MEF cells overexpressed with wild type, phosphorylation null and phosphorylation mimic mutant BAD. Arrows indicate positive TUNEL signal. Scale bar, 50 μm.Data represent mean ± S.E.M. Student’s t-test: *p < 0.05, ** p< 0.01, *** p < 0.001
Discussion
To our knowledge, this study is the first to describe global phosphorylation changes in cultured primary neurons after mitochondrial damage. By comparing WT and Pink1 KO neurons, we identified PINK1-dependent activation of MAP kinase signaling, insulin signaling, and mTOR signaling as the major activated pathways induced by CCCP. Through biochemical validation and functional studies, we found the pro-apoptotic protein BAD to be a direct PINK1 substrate in vitro. In vivo experiments further demonstrated that S112 and S136 on BAD were phosphorylated after CCCP treatment in cells in a PINK1 dependent manner. Finally, we found that phosphorylation mimic BAD rescued apoptotic cell death induced by CCCP. Thus, our study provides a new layer of molecular mechanism on PINK1 regulation of cell death.
A previous study compared the proteome and phosphoproteome of Pink1 deficient mouse brain with that of wild-type and found that signaling networks, energy metabolism, and proteostasis were altered in PINK1 deficient brain [36]. Similarly, our phosphoproteomic analysis of cortical neurons captured robust phosphorylation changes in multiple signaling pathways after mitochondrial depolarization (Fig. 3g). On the other hand, because our phosphoproteome dataset had low coverage of organelle proteins, we did not identify phosphorylation of well-known PINK1 targets including parkin, MIRO, or NCLX [37–39]. We did identify S57 phosphorylation on ubiquitin but it did not show any difference between WT and KO neurons, nor did it show response to CCCP treatment. However, from our proteome dataset we identified MIRO and parkin-independent mitophagy proteins, FKBP8 and PLSCR3 [40–42], without identifying phosphorylation on these proteins. Thus, our strategy focused on the setting of mitochondrial membrane potential disruption, which might be in favor of identifying non-mitochondrial targets. Regardless, our phosphoproteomic analysis provides a broad view of cell signaling pathways that are influenced by PINK1 kinase.
A few PINK1 substrates that mediate cell death have been reported. TRAP1 is the first PINK1 substrate identified, and phosphorylation of TRAP1 by PINK1 promotes cell survival in a cytochrome-c-dependent manner [3]. Later, a PD susceptibility locus-encoded mitochondrial serine protease HtrA2 is found to be phosphorylated in a PINK1-dependent manner, which contributes to cell’s increased resistance to mitochondrial stress [43]. A recent study provides evidence showing that PINK1 interacts with and phosphorylates Bcl-xl, a member of the Bcl-2 family of apoptotic checkpoint proteins [44]. In our study, we failed to identify TRAP1, HtrA2, or Bcl-xl, presumably due to their low expression levels. However, we identified BAD as a new PINK1 substrate through an unbiased phosphoproteomic screen. Our study highlights the importance of BAD phosphorylation by PINK1 in promoting cell survival.
Previous studies have demonstrated that upon survival factor stimulation, endogenous BAD phosphorylation at S112, S136, and S155 residues is able to resist apoptotic stimuli [45], making these three sites attractive targets for modulating cell survival. BAD is a BH3 only protein with the BH3 domain encompassing the S112 site. Phosphorylation of S112 together with S136 and S155 regulates BAD’s ability to dimerize with Bcl-xl to form apoptotic complex in mitochondrial outer membrane, allowing cytochrome-c release and activation of caspase-3 [46]. Our observation that activated PINK1 mediates the phosphorylation of mitochondrion-localized BAD after mitochondrial damage, potentially preventing the formation of apoptotic complex and promoting cell survival, highlights the pleiotropic effect of key proteins regulating the cell decision on survival or death.
Despite the extensively use of CCCP to reveal the PINK1/Parkin-mediated mitophagy pathway, it has several limitations. First, the complete loss of membrane potential triggered by CCCP would not likely be encountered under physiological conditions. Second, CCCP causes a global damage to the mitochondrial network and bypass the ROS signaling [47]. Last, CCCP has protonophoric activity on other membranes such as lysosome and inhibits autophagy at both the initiation and degradation stages [48]. Therefore, further phosphoproteomic studies applying more physiologically relevant challenges such as ROS induction or mitochondrial complex inhibition, on human-derived dopaminergic neurons in the context of PINK1 mutation would shed further insights on the pathogenesis of PD. On the other hand, although phosphorylation-mediated BAD inactivation promoting cell survival is a general principle applying to many cell types [45], recent studies have directly linked defects in mitophagy caused by mutations in VPS35, another familial PD-linked gene involved in endosomal trafficking, to dopaminergic neuronal death [49]. Fine-tuning the balance between mitophagy and mitochondrial-mediated apoptotic cell death might hold the key for neuronal survival, and Bcl-2 family of apoptotic regulators emerged as a crossroad of autophagy initiation and apoptosis. Given our finding that activated PINK1 phosphorylates mitochondrion-localized BAD to prevent apoptotic cell death, our study sheds new light on molecular details of neural degeneration caused by PINK1 loss of function.
Materials and methods
Animals
All experiments involving animal treatment and care were performed following the Institutional Animal Care and Use Committee protocols from the East China Normal University (ECNU). Pink1 KO mice were kindly provided by Dr. ZhuoHua Zhang (State key laboratory of Medical Genetics, XiangYa School of Medicine, CHN).
In vitro kinase assay
Recombinant T. castaneum 6xHis-MBP-PINK1 was purchased from R&D Systems (#AP-180). Kinase assays were performed immediately after His-BAD purification, with 1 uM PINK1 and 2 μg of recombinant substrate protein and 2 mM ATP in kinase assay buffer (50 mM Tris-HCl, 10 mM MgCl2, 10 mM DTT). Reactions were incubated at 37 °C for 1 h. For dephosphorylation reaction, the calf intestinal phosphatase (CIP, New England Biolabs, M0290S) were added. The reaction system was incubated in 90 °C for 5 min to inactivate PINK1.
Immunofluorescence
For immunofluorescence staining of exogenous Flag-BAD, HeLa cells were washed with 1xPBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4 and 2 mM KH2PO4) prior to fixation in 4% paraformaldehyde at room temperature for 60 min, incubated with 1% Triton X-100 on ice for 10 min, blocked with 5% BSA in 37 °C incubator for 60 min and incubated with mouse anti-Flag antibody overnight. The coverslips were washed three times with PBST, followed by incubation with a secondary antibody (Life Technology, Alexa Fluor 405-blue, 546-red, 488-green). Images were acquired with a confocal microscopy (Leica TCS SP5).
Apoptosis assays
WT and Pink1 KO MEF cells were seeded in a six-well dish, and then overexpressed with BAD constructs, treated with 20 μM CCCP or DMSO for indicated times.
The TUNEL assay (Roche Applied Science 12156792910, Indianapolis, IN) was performed according to manufacturer’s instructions.
Statistics
Data are presented as standard error of the mean (SEM) of multiple replicates. All data presented are representative of at least three independent experiments. P-values were generated by two-tailed t-test assuming equal variance using Prism Graphpad Software (San Diego, CA). The value 0.05 (*), 0.01 (**) and 0.001 (***) was assumed as the level of significance for the statistic tests.
Other details including reagents, cell culture, transfection, gel electrophoresis, phosphoproteomics workflow, LC-MS/MS data acquisition, MS raw data processing, bioinformatics analysis can be found in Supplementary Information.
Data availability
The mass spectrometry data has been deposited to ProteomeXchange with the accession number PXD006304.
Electronic supplementary material
Acknowledgements
The authors would like to acknowledge financial support from the Shanghai Pujiang Talent Project (14PJ1402900), and ECNU National “985” Project grant.
Author's contributions
LL, ZZ, and LX conceived the project; HW, QZ performed the experiments. BT, HW, and LL analyzed data; LL, BT, and HW wrote the paper.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
Edited by N. Chandel
Electronic supplementary material
The online version of this article (10.1038/s41418-017-0027-x) contains supplementary material, which is available to authorized users.
Contributor Information
Bin Tang, Email: thtang25@163.com.
Lujian Liao, Email: ljliao@bio.ecnu.edu.cn.
References
- 1.Valente EM, Abou-Sleiman PM, Caputo V, Muqit MMK, Harvey K, Gispert S, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 2.Abeliovich A. Parkinson’s disease: Pro-survival effects of PINK1. Nature. 2007;448:759–60. doi: 10.1038/448759a. [DOI] [PubMed] [Google Scholar]
- 3.Pridgeon JW, Olzmann JA, Chin LS, Li L. PINK1 Protects against Oxidative Stress by Phosphorylating Mitochondrial Chaperone TRAP1. PLoS Biol. 2007;5:e172. doi: 10.1371/journal.pbio.0050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–14. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron. 2015;85:257–73. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arena G, Valente EM. PINK1 in the limelight: multiple functions of an eclectic protein in human health and disease. J Pathol. 2017;241:251–63. doi: 10.1002/path.4815. [DOI] [PubMed] [Google Scholar]
- 7.Voigt A, Berlemann LA, Winklhofer KF. The mitochondrial kinase PINK1: functions beyond mitophagy. J Neurochem. 2016;139:232–9. doi: 10.1111/jnc.13655. [DOI] [PubMed] [Google Scholar]
- 8.Van Laar VS, Arnold B, Cassady SJ, Chu CT, Burton EA, Berman SB. Bioenergetics of neurons inhibit the translocation response of Parkin following rapid mitochondrial depolarization. Hum Mol Genet. 2011;20:927–40. doi: 10.1093/hmg/ddq531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai Q, Zakaria HM, Simone A, Sheng ZH. Spatial Parkin Translocation and Degradation of Damaged Mitochondria Via Mitophagy in Live Cortical Neurons. Current Biol. 2012;22:545–52. doi: 10.1016/j.cub.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rakovic A, Shurkewitsch K, Seibler P, Grünewald A, Zanon A, Hagenah J, et al. Phosphatase and Tensin Homolog (PTEN)-induced Putative Kinase 1 (PINK1)-dependent Ubiquitination of Endogenous Parkin Attenuates Mitophagy: STUDY IN HUMAN PRIMARY FIBROBLASTS AND INDUCED PLURIPOTENT STEM CELL-DERIVED NEURONS. J Biol Chem. 2013;288:2223–37. doi: 10.1074/jbc.M112.391680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morais VA, Haddad D, Craessaerts K, De Bock PJ, Swerts J, Vilain S, et al. PINK1 loss-of-function mutations affect mitochondrial complex I activity via NdufA10 ubiquinone uncoupling. Science. 2014;344:203–7. doi: 10.1126/science.1249161. [DOI] [PubMed] [Google Scholar]
- 12.Qin X, Zheng C, Yates JR, 3rd, Liao L. Quantitative phosphoproteomic profiling of PINK1-deficient cells identifies phosphorylation changes in nuclear proteins. Mol Biosyst. 2014;10:1719–29. doi: 10.1039/C3MB70565J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moon Y, Lee KH, Park JH, Geum D, Kim K. Mitochondrial membrane depolarization and the selective death of dopaminergic neurons by rotenone: protective effect of coenzyme Q10. J Neurochem. 2005;93:1199–208. doi: 10.1111/j.1471-4159.2005.03112.x. [DOI] [PubMed] [Google Scholar]
- 14.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dayon L, Hainard A, Licker V, Turck N, Kuhn K, Hochstrasser DF, Burkhard PR, Sanchez JC. Relative quantification of proteins in human cerebrospinal fluids by MS/MS using 6-plex isobaric tags. Anal Chem. 2008;80:2921–31. doi: 10.1021/ac702422x. [DOI] [PubMed] [Google Scholar]
- 16.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protocols. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 17.Futschik LKaM. Mfuzz: A software package for soft clustering of microarray data. Bioinformation. 2007;2:3. doi: 10.6026/97320630002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41:D377–D386. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol. 2005;23:1391–8. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- 20.Horn H, Schoof EM, Kim J, Robin X, Miller ML, Diella F, et al. KinomeXplorer: an integrated platform for kinome biology studies. Nat Methods. 2014;11:603–4. doi: 10.1038/nmeth.2968. [DOI] [PubMed] [Google Scholar]
- 21.Deng H, Wu Y, Jankovic J. The EIF4G1 gene and Parkinson’s disease. Acta Neurol Scand. 2015;132:73–78. doi: 10.1111/ane.12397. [DOI] [PubMed] [Google Scholar]
- 22.Dhungel N, Eleuteri S, Li Lb, Kramer NJ, Chartron J, Spencer B, et al. Parkinson’s disease genes VPS35 and EIF4G1 interact genetically and converge on α–synuclein. Neuron. 2015;85:76–87. doi: 10.1016/j.neuron.2014.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao F, Chen D, Si J, Hu Q, Qin Z, Fang M, et al. The mitochondrial protein BNIP3L is the substrate of PARK2 and mediates mitophagy in PINK1/PARK2 pathway. Hum Mol Genet. 2015;24:2528–38. doi: 10.1093/hmg/ddv017. [DOI] [PubMed] [Google Scholar]
- 24.Zhang T, Xue L, Li L, Tang C, Wan Z, Wang R, et al. BNIP3 Protein Suppresses PINK1 Kinase Proteolytic Cleavage to Promote Mitophagy. J Biol Chem. 2016;291:21616–29. doi: 10.1074/jbc.M116.733410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gozuacik D, Bialik S, Raveh T, Mitou G, Shohat G, Sabanay H, et al. DAP-kinase is a mediator of endoplasmic reticulum stress-induced caspase activation and autophagic cell death. Cell Death Differ. 2008;15:1875–86. doi: 10.1038/cdd.2008.121. [DOI] [PubMed] [Google Scholar]
- 26.Vaux DL, Korsmeyer SJ. Cell Death in Development. Cell. 1999;96:245–54. doi: 10.1016/S0092-8674(00)80564-4. [DOI] [PubMed] [Google Scholar]
- 27.Zha J, Harada H, Yang E, Jockel J, Korsmeyer SJ. Serine Phosphorylation of Death Agonist BAD in Response to Survival Factor Results in Binding to 14-3-3 Not BCL-XL. Cell. 1996;87:619–28. doi: 10.1016/S0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
- 28.Woodroof HI, Pogson JH, Begley M, Cantley LC, Deak M, Campbell DG, et al. Discovery of catalytically active orthologues of the Parkinson’s disease kinase PINK1: analysis of substrate specificity and impact of mutations. Open Biol. 2011;1:110012. doi: 10.1098/rsob.110012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zha J, Harada H, Yang E, Jockel J, Korsmeyer SJ. Serine Phosphorylation of Death Agonist BAD in Response to Survival Factor Results in Binding to 14-3-3 Not BCL-XL. Cell. 1996;87:619–28. doi: 10.1016/S0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
- 30.Hirai I, Wang HG. Survival-factor-induced phosphorylation of Bad results in its dissociation from Bcl-x(L) but not Bcl-2. Biochem J. 2001;359(Pt 2):345–52. doi: 10.1042/bj3590345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Desai S, Pillai P, Win-Piazza H, Acevedo-Duncan M. PKC-iota promotes glioblastoma cell survival by phosphorylating and inhibiting BAD through a phosphatidylinositol 3-kinase pathway. Biochim Biophys Acta. 2011;1813:1190–7. doi: 10.1016/j.bbamcr.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 32.Tan Y, Demeter MR, Ruan H, Comb MJ. BAD Ser-155 phosphorylation regulates BAD/Bcl-XL interaction and cell survival. J Biol Chem. 2000;275:25865–9. doi: 10.1074/jbc.M004199200. [DOI] [PubMed] [Google Scholar]
- 33.Datta SR, Katsov A, Hu L, Petros A, Fesik SW, Yaffe MB, et al. 14-3-3 Proteins and Survival Kinases Cooperate to Inactivate BAD by BH3 Domain Phosphorylation. Mol Cell. 2000;6:41–51. doi: 10.1016/S1097-2765(05)00012-2. [DOI] [PubMed] [Google Scholar]
- 34.Datta SR, Brunet B, Greenberg ME. Cellular survival: a play in three Akts. Genes Dev. 1999;13:2905–27. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- 35.Darzynkiewicz Z, Galkowski D, Zhao H. Analysis of apoptosis by cytometry using TUNEL assay. Methods. 2008;44:250–4. doi: 10.1016/j.ymeth.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Triplett JC, Zhang Z, Sultana R, Cai J, Klein JB, Büeler H, Butterfield DA. Quantitative expression proteomics and phosphoproteomics profile of brain from PINK1 knockout mice: insights into mechanisms of familial Parkinson’s disease. J Neurochem. 2015;133:750–65. doi: 10.1111/jnc.13039. [DOI] [PubMed] [Google Scholar]
- 37.Kondapalli C, Kazlauskaite A, Zhang N, Woodroof HI, Campbell DG, Gourlay R, et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2012;2:120080. doi: 10.1098/rsob.120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shlevkov E, Kramer T, Schapansky J, LaVoie MJ, Schwarz TL. Miro phosphorylation sites regulate Parkin recruitment and mitochondrial motility. Proc Natl Acad Sci U S A. 2016;113:E6097–E6106. doi: 10.1073/pnas.1612283113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kostic M, Ludtmann MHR, Bading H, Hershfinkel M, Steer E, Chu CT, et al. PKA Phosphorylation of NCLX Reverses Mitochondrial Calcium Overload and Depolarization, Promoting Survival of PINK1-Deficient Dopaminergic Neurons. Cell Rep. 2015;13:376–86. doi: 10.1016/j.celrep.2015.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhujabal Z, Birgisdottir AringB, Sjøttem E, Brenne HB, Øvervatn A, Habisov S, et al. FKBP8 recruits LC3A to mediate Parkin-independent mitophagy. EMBO Rep. 2017;18:947–61. doi: 10.15252/embr.201643147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chu CT, Ji J, Dagda RK, Jiang JF, Tyurina YY, Kapralov AA, et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat Cell Biol. 2013;15:1197–205. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strappazzon F, Nazio F, Corrado M, Cianfanelli V, Romagnoli A, Fimia GM, et al. AMBRA1 is able to induce mitophagy via LC3 binding, regardless of PARKIN and p62/SQSTM1. Cell Death Differ. 2015;22:419–32. doi: 10.1038/cdd.2014.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plun-Favreau H, Klupsch K, Moisoi N, Gandhi S, Kjaer S, Frith D, et al. The mitochondrial protease HtrA2 is regulated by Parkinson’s disease-associated kinase PINK1. Nat Cell Biol. 2007;9:1243–52. doi: 10.1038/ncb1644. [DOI] [PubMed] [Google Scholar]
- 44.Chao DT, Korsmeyer SJ. BCL-2 FAMILY: Regulators of Cell Death. Annu Rev Immunol. 1998;16:395–419. doi: 10.1146/annurev.immunol.16.1.395. [DOI] [PubMed] [Google Scholar]
- 45.Datta SR, Ranger AM, Lin MZ, Sturgill JF, Ma YC, Cowan CW, et al. Survival Factor-Mediated BAD Phosphorylation Raises the Mitochondrial Threshold for Apoptosis. Dev Cell. 2002;3:631–43. doi: 10.1016/S1534-5807(02)00326-X. [DOI] [PubMed] [Google Scholar]
- 46.Wang IHaHG. Survival-factor-induced phosphorylation of Bad results in its dissociation from Bcl-xL but not Bcl-2. Biochem J. 2001;359:345–52. doi: 10.1042/bj3590387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Nartiss Y, Steipe B, McQuibban GA, Kim PK. ROS-induced mitochondrial depolarization initiates PARK2/PARKIN-dependent mitochondrial degradation by autophagy. Autophagy. 2012;8:1462–76. doi: 10.4161/auto.21211. [DOI] [PubMed] [Google Scholar]
- 48.Padman BS, Bach M, Lucarelli G, Prescott M, Ramm G. The protonophore CCCP interferes with lysosomal degradation of autophagic cargo in yeast and mammalian cells. Autophagy. 2013;9:1862–75. doi: 10.4161/auto.26557. [DOI] [PubMed] [Google Scholar]
- 49.Tang FL, Liu W, Hu JX, Erion Joanna R, Ye J, Mei L, et al. VPS35 Deficiency or Mutation Causes Dopaminergic Neuronal Loss by Impairing Mitochondrial Fusion and Function. Cell Rep. 2015;12:1631–43. doi: 10.1016/j.celrep.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry data has been deposited to ProteomeXchange with the accession number PXD006304.