Abstract
Antimicrobial resistance (AMR) in bacterial pathogens threatens global health, though the spread of AMR bacteria and AMR genes between humans, animals, and the environment is still largely unknown. Here, we investigated the role of wild birds in the epidemiology of AMR Escherichia coli. Using next-generation sequencing, we characterized cephalosporin-resistant E. coli cultured from sympatric gulls and bald eagles inhabiting a landfill habitat in Alaska to identify genetic determinants conferring AMR, explore potential transmission pathways of AMR bacteria and genes at this site, and investigate how their genetic diversity compares to isolates reported in other taxa. We found genetically diverse E. coli isolates with sequence types previously associated with human infections and resistance genes of clinical importance, including blaCTX-M and blaCMY. Identical resistance profiles were observed in genetically unrelated E. coli isolates from both gulls and bald eagles. Conversely, isolates with indistinguishable core-genomes were found to have different resistance profiles. Our findings support complex epidemiological interactions including bacterial strain sharing between gulls and bald eagles and horizontal gene transfer among E. coli harboured by birds. Results suggest that landfills may serve as a source for AMR acquisition and/or maintenance, including bacterial sequence types and AMR genes relevant to human health.
Introduction
Antimicrobial resistance (AMR) in bacterial pathogens is a growing threat to human and animal health, with an increasing number of infections no longer responding to once-standard treatments1,2. Cephalosporin-resistant bacteria, including those producing extended spectrum β-lactamases (ESBLs), are particularly concerning to public health, due to their resistance to commonly prescribed beta-lactam antibiotics, their common co-resistance to other antimicrobial agents, and their increasing global prevalence3,4. While exposure to AMR pathogens presents obvious human and animal health risks due to the potential of contracting difficult-to-treat bacterial afflictions, non-pathogenic bacteria harbouring resistance determinants may also represent a more cryptic threat to public health given that genes conferring AMR can be transferred to bacterial pathogens via horizontal gene transfer5. Furthermore, bacteria and genes conferring AMR have the potential to spread and proliferate through humans, animals, and the environment6,7, prompting the need for a coordinated One Health approach to understand direct and indirect pathways of dissemination and to inform risk management8,9.
Most antibiotic compounds are naturally occurring in the environment10 and AMR in soil-dwelling bacteria predates antibiotic use by humans11. However, there is a large body of evidence that widespread use of antibiotics by humans and their application in livestock production has increased the prevalence of AMR in bacterial communities of humans, animals, and the environment12–17. Significant data gaps regarding the acquisition, distribution, and proliferation of AMR determinants have hampered efforts at quantifying the human health risk posed by AMR bacterial pathogens and AMR determinants from environmental sources5. Thus, understanding risk to human health requires active surveillance for AMR bacteria in diverse environments to elucidate transmission pathways and to provide information on the extent of horizontal gene transfer among environmental sources and human microbial communities.
Free-ranging wildlife represents one environmental source by which AMR bacteria may emerge and/or be maintained. While there is substantial support for the premise that anthropogenic inputs into the local environment18–21, or a relative lack thereof16,22,23, influence the prevalence of AMR bacteria among wildlife inhabiting an area, the role of free-ranging animals in maintaining and dispersing such bacteria is less clear24. Wild birds have been a common focus for investigations to understand the occurrence of AMR bacteria in the environment because many species are relatively abundant, use anthropogenically influenced habitats, and disperse over relatively long distances25. Thus, birds may be informative taxa for understanding the ecology of AMR bacteria and for gaining insight into proliferation and dispersal. Gulls (family Laridae) in particular appear to represent useful model species for such research because of their tendency to forage in response to human activities26, their apparent propensity to be colonized with AMR Escherichia coli at relatively high rates27–30, and circumstantial evidence indicating that some species may transport AMR bacteria through migratory movements23,28,31. Additionally, wild raptors may provide an alternative indicator of AMR in the environment based on their predatory nature, and thus their potential to acquire AMR bacteria harboured by diverse prey32–34.
The use of high resolution molecular approaches, such as next-generation whole genome sequencing (WGS), can help resolve pathways by which AMR bacteria are maintained and dispersed in different environments. Evaluation of the significance of genetically similar AMR genes between hosts and locations35,36 can inform risk assessments regarding AMR threats to human and animal population health5,37. However, attributing the source and transmission routes of AMR bacteria and AMR determinants that are clinically relevant to human and animal health is currently confounded by the paucity of molecular epidemiological data on AMR determinants outside of the clinic. Therefore, in this study, we genomically characterized cephalosporin-resistant E. coli in sympatric gulls and bald eagles inhabiting a landfill habitat to gain insight into the genetic determinants conferring AMR in these environmental sources, to explore potential transmission pathways and evolutionary mechanisms contributing to AMR at this site, and to investigate how bacterial diversity compares to isolates previously reported in other taxa and clinical settings. Results provide insight into how AMR E. coli and AMR determinants are maintained and shared among sympatric birds inhabiting an anthropogenically influenced habitat and suggest plausible sources of exposure.
Results
Phenotypic characterization of cephalosporin-resistant E. coli isolates
A total of 27 cephalosporin-resistant E. coli isolates were recovered from CHROMagar C3GR plates, 13 and 14 of which originated from faecal samples collected from bald eagles and gulls, respectively. Phenotypic resistance was tested against 18 antibiotics, with all 27 isolates resistant to Ampicillin and Cefadroxil. Most isolates were resistant to between four and seven antibiotics (range = 4–13; median = 6), but one isolate (A1_007_Gull) was resistant to 13 antibiotics (Supplementary Table S1). According to the definition of multidrug resistance proposed by Schwarz et al.38 (i.e. bacteria exhibiting resistance to at least one agent in at least three antimicrobial classes), 11 isolates displayed phenotypic multidrug resistance.
Whole genome sequencing and de novo assembly
The conservatively estimated average depth of coverage for 27 cephalosporin-resistant E. coli isolates originating from gull and bald eagle faecal samples on which WGS was performed was 29× and there was an average of 149 contigs >500-base pairs after genome assembly using SPAdes (detailed assembly metrics are provided in Supplementary Table S2). Sequencing coverage was low (an average of 7.5-fold coverage) for one bald eagle faecal isolate (A1_020_BaldEagle), which was therefore excluded from further analyses. Thus, genomic sequencing data from a total of 26 E. coli isolates originating from 14 gull and 12 bald eagle faecal samples was further analysed.
E. coli sequence types and core genome phylogeny
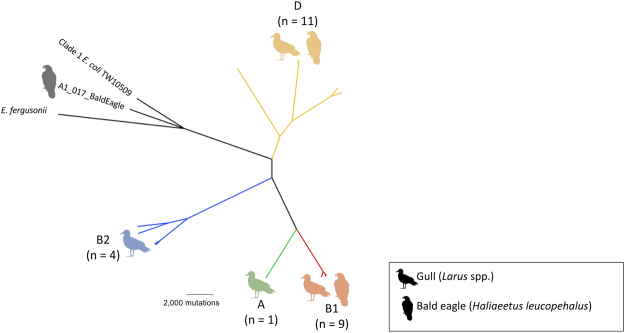
The maximum likelihood phylogeny based on the core genome (i.e. orthologous regions present in all genomes), and estimated using ClonalFrameML to account for mutation and recombination events, revealed a diverse population of E. coli isolates (Fig. 1). All four major E. coli phylotypes were identified through in silico phylotyping. Most isolates belonged to phylotype D, which accounted for 42% (n = 11) of isolates, including E. coli recovered from samples collected from both bald eagles (n = 4) and gulls (n = 7). Phylotypes B1, B2, and A accounted for 35% (n = 9), 15% (n = 4), and 4% (n = 1) of isolates, respectively. Phylotypes A and B2 were represented only by isolates from gulls, whereas those assigned to phylotype B1 included both isolates from bald eagles and gulls. One additional isolate, A1_017_BaldEagle, was most closely related to the reference strain E. coli TW10509, which represents a divergent, “cryptic” lineage termed Clade 139.
Figure 1.
Unrooted clonal core genome phylogeny of 26 cephalosporin-resistant E. coli isolates originating from gulls and bald eagles in Alaska, E. fergusonii (NC_011740.1) and a Clade 1 E. coli sequence (TW10509; NZ_GL872204.1). Phylotypes A (green), B1 (red), B2 (blue) and D (yellow) are indicated at the tips with an image of a gull, bald eagle, or both, according to the species from which the isolates were obtained. The number of cephalosporin-resistant E. coli isolates belonging to each phylotype is indicated in parentheses. Silhouette images of gulls (credit: Rebecca Groom, https://creativecommons.org/licenses/by/3.0/legalcode) and bald eagles were sourced from PhyloPic (www.phylopic.org).
When reference strains and the divergent A1_017_BaldEagle isolate were excluded, a total of 176,043 SNPs were detected in the 4,734,259-base pair core genome alignment of 25 cephalosporin-resistant E. coli recovered from gulls and bald eagles. The ClonalFrameML log-likelihood ratio indicated there was evidence of recombination, with an estimated mean rate of recombination to mutation (R/θ) of 0.66. Accounting for recombination, an estimated clonal phylogeny of the 25 core genomes revealed extensive genetic diversity between small clusters of genetically closely related isolates (i.e. >99.95% nucleotide identity; Fig. 2a).
Figure 2.
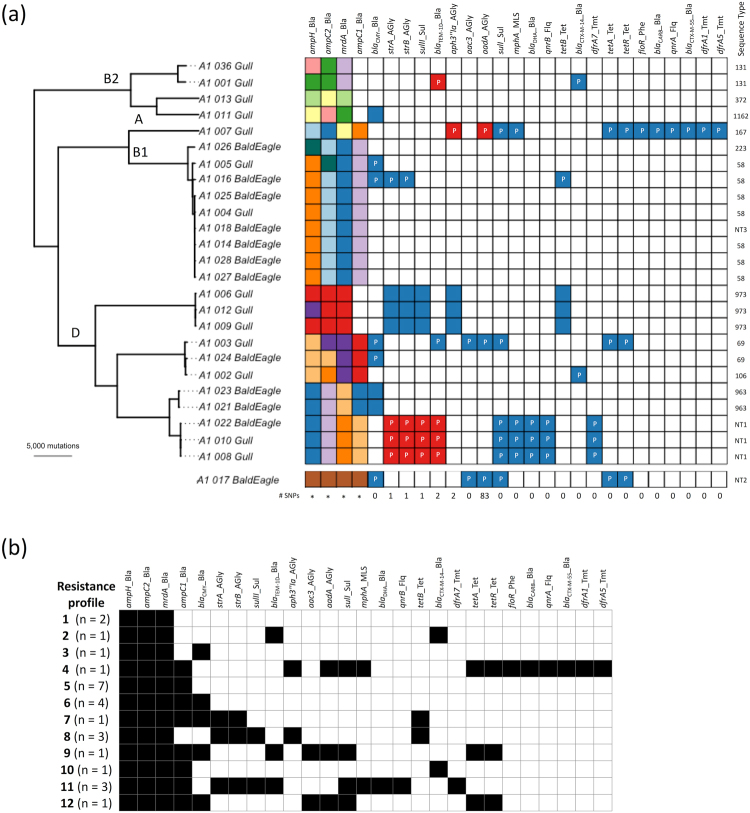
(a) Midpoint rooted clonal core genome phylogeny of 25 cephalosporin-resistant E. coli isolates originating from gulls and bald eagles in Alaska. Phylotypes (A, B1, B2, D) are indicated on branches. Presence of each of 27 identified AMR genes is shown as a matrix, with gene names indicated at the top of the matrix underscored by the antibiotics class (AGly: aminoglycosides, Bla: beta-lactamases, Flq: fluoroquinolones, MLS: macrolide-lincosamide-streptogramin, Phe: phenicols, Sul: sulfonamides, Tet: tetracyclines and Tmt: trimethoprim). Coloured squares indicate presence and white indicates that the gene was not found. Each colour indicates a different allele for each gene. The number of SNPs differentiating AMR genes with two alleles is shown below the matrix, while the phylogenetic diversity of genes with more than two alleles (indicated by *) can be seen in Fig. 3. AMR genes found on plasmids identified by PlasmidSPAdes are indicated with the letter “P”. The final column of the matrix indicates in silico identified E. coli sequence types. The divergent A1_017_BaldEagle isolate is included at the bottom of the matrix, as it was omitted from the midpoint rooted clonal core genome phylogeny. (b) Unique resistance profiles identified through in silico AMR gene detection in cephalosporin-resistant E. coli isolates. A presence/absence matrix of 27 identified AMR genes is shown, with black shading indicating presence, and no shading indicating that the gene was not found. The number of isolates found with each resistance profile is indicated in parentheses.
In silico MLST analysis identified 13 sequence types (Table 1). Sequence type 58 was the most common type, representing 27% (n = 7) of all isolates, followed by ST973 (n = 3), NT1 (n = 3), ST69 (n = 2), ST131 (n = 2), and ST963 (n = 2). The remaining sequence types, ST106, ST167, ST223, ST372, ST1162, NT2, and NT3, were represented by a single isolate each. Three new sequence types (NT) were identified, one of which was represented by three isolates. Based on the core genome, the NT3 isolate was closely related to ST58 isolates and differed from the ST58 MLST profile by a single SNP in purA. We compared the two ST131 isolates from gulls in our study to previously sequenced ST131 isolates that represented the three reported ST131 clades35. A1_036_Gull was identified as clade B and A1_001_Gull was identified as clade A, based on a separate core genome analysis using Parsnp (Supplementary Fig. S1).
Table 1.
Genomic characteristics of 26 cephalosporin-resistant E. coli isolates originating from gull and bald eagle faecal samples in Alaska.
| Isolate ID | Phylotype | Sequence Type | K-Pax2 cluster | # unique CDS |
|---|---|---|---|---|
| A1_001_Gull | B2 | 131 | 8 | 57 |
| A1_002_Gull | D | 106 | 2 | 34 |
| A1_003_Gull | D | 69 | 2 | 143 |
| A1_004_Gull | B1 | 58 | 1 | 0 |
| A1_005_Gull | B1 | 58 | 5 | 68 |
| A1_006_Gull | D | 973 | 3 | 2 |
| A1_007_Gull | A | 167 | 9 | 102 |
| A1_008_Gull | D | NT1† | 4 | 1 |
| A1_009_Gull | D | 973 | 3 | 60 |
| A1_010_Gull | D | NT1† | 4 | 2 |
| A1_011_Gull | B2 | 1162 | 6 | 170 |
| A1_012_Gull | D | 973 | 3 | 103 |
| A1_013_Gull | B2 | 372 | 6 | 158 |
| A1_014_BaldEagle | B1 | 58 | 1 | 1 |
| A1_016_BaldEagle | B1 | 58 | 5 | 36 |
| A1_017_BaldEagle | A | NT2† | 10 | 179 |
| A1_018_BaldEagle | B1 | NT3† | 1 | 11 |
| A1_021_BaldEagle | D | 963 | 7 | 100 |
| A1_022_BaldEagle | D | NT1† | 4 | 65 |
| A1_023_BaldEagle | D | 963 | 7 | 99 |
| A1_024_BaldEagle | D | 69 | 2 | 9 |
| A1_025_BaldEagle | B1 | 58 | 1 | 0 |
| A1_026_BaldEagle | B1 | 223 | 5 | 0 |
| A1_027_BaldEagle | B1 | 58 | 1 | 0 |
| A1_028_BaldEagle | B1 | 58 | 1 | 0 |
| A1_036_Gull | B2 | 131 | 8 | 0 |
†NT = new sequence type.
Accessory genome characteristics
AMR gene detection using SRST2 identified a total of 27 different AMR genes, three of which (ampH, ampC2, mrdA [encoding Penicillin binding protein 2]) were found in all isolates. These three genes, as well as ampC1 that was identified in 19 isolates, are chromosomal E. coli genes with point mutations associated with AMR. Genes conferring resistance to beta-lactams predominated (37% of genes), followed by those conferring resistance to aminoglycosides (19% of genes). One blaCTX-M-55 and two blaCTX-M-14 positive isolates were found and eight isolates harboured the pAmpC gene blaCMY-2. Twelve unique presence/absence resistance profiles were identified (Fig. 2b). Identical profiles were observed in up to seven isolates from both gulls and bald eagles and in highly genetically divergent isolates based on the core genome phylogeny (Fig. 2a). Conversely, some isolates (e.g. A1_003_Gull vs A1_024_BaldEagle or A1_002_Gull) with highly similar core genomes had very different resistance profiles. A total of 61 individual, non-intrinsic AMR genes were detected in putative plasmids assembled using PlasmidSPAdes, whereas 18 acquired AMR genes were not detected in plasmids using this method (Fig. 2a).
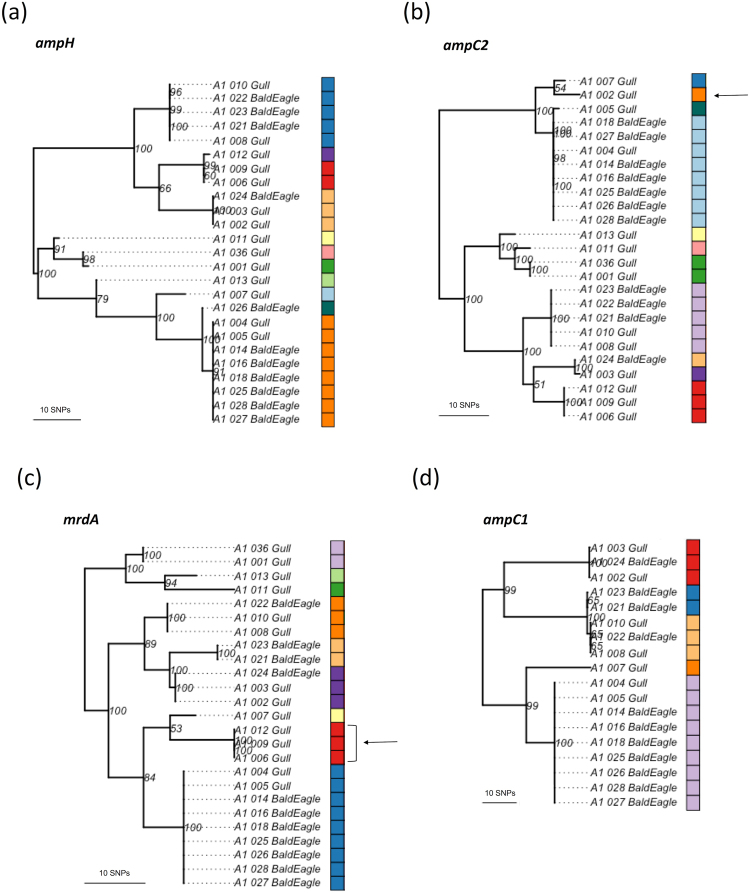
We compared individual AMR gene sequences between isolates, identifying more than one genotype in 10 genes and more than two genotypes in four genes (Fig. 2a). Individual maximum likelihood phylogenies were estimated for the four intrinsic chromosomal genes with more than two genotypes (Fig. 3). In most cases, individual gene phylogenies clustered isolates similarly as compared to the core genome phylogeny; however, this did not occur on two occasions. The A1_002_Gull phylotype D ampC2 nucleotide sequence was most closely related to a phylotype A isolate, differing by only seven SNPs. Additionally, three phylotype D isolates (A1_006_Gull, A1_009_Gull, and A1_012_Gull) were more closely related to the phylogroup A and phylogroup B1 isolates than to other phylogroup D isolates based on mrdA.
Figure 3.
Midpoint rooted maximum likelihood phylogenies of cephalosporin-resistant E. coli isolates originating from gulls and bald eagles in Alaska based on individual chromosomally-encoded AMR genes – (a) ampH, (b) ampC2, (c) mrdA, and (d) ampC1. Each colour indicates a different allele for each gene sequence. Arrows indicate incongruence with the core genome phylogeny. Branch support values are indicated and based on 100 bootstrap pseudoreplicates.
Biocide and heavy metal resistance genes were identified in silico in isolates collected from gulls and bald eagles by querying against a database of 704 experimentally confirmed resistance genes, resulting in the identification of a total of 96 different biocide/heavy metal resistance genes in our collection of 26 cephalosporin-resistant E. coli isolates. A range of 22–60 biocide and heavy metal resistance genes were found per isolate (median = 34) and the presence of individual genes ranged from detection in 1–26 isolates (median = 6.5) (Supplementary Fig. S2). Ten of the 96 biocide/heavy metal resistance genes detected among gull and bald eagle isolates are reported to be plasmid-borne, with the remainder being chromosomally encoded. The 96 biocide/heavy metal resistance genes detected were predicted to confer resistance to 64 different compounds, with resistance to hydrogen peroxide and zinc being the most common among our isolates. To investigate the possibility of plasmid co-localization of AMR genes and biocide/heavy metal resistance genes, the isolate with the highest number of resistance genes (A1_007_Gull) was investigated in detail by visualizing detected genes on the 81 PlasmidSPAdes-assembled contigs. All 12 mobile AMR gene sequences were located within an estimated five nodes (i.e. assembled contigs) of a biocide/heavy metal resistance gene, qacEdelta1, which was located on the same node as the AMR gene SulI (Supplementary Fig. S3). Additionally, we identified the class 1 integron-integrase gene intI1 in three assemblies (A1_003_Gull, A1_007_Gull, and A1_017_BaldEagle) using in silico PCR amplification.
LS-BSR identified 3,487 core, 7,332 accessory, and 1,400 unique (i.e. present in a single genome) CDS among all 26 genomes (Table 1). Isolate A1_017_BaldEagle had the largest number of unique CDS (n = 179). K-Pax2 partitioned isolates into ten distinct clusters based on the presence/absence of 1,844 accessory CDS that were determined to be significantly discriminatory (Table 1). The number of isolates per cluster ranged from one to six and isolates were distributed among clusters similarly to the topology inferred from the core genome phylogeny.
Discussion
We genomically characterized 26 cephalosporin resistant E. coli isolates from gulls and bald eagles sampled at a landfill in southcentral Alaska. The clonal core genome phylogeny of the major E. coli phylotypes identified in our isolates, as well as the divergent Clade 1 bald eagle isolate, was consistent with the general branching pattern of phylogenetic trees previously reconstructed for E. coli isolated from humans and animals40,41. However, we found a predominance of phylotype D E. coli isolates among our sample collection, followed by B1 isolates, whereas previous studies reported that B1 strains predominated in domestic and wild animals40, and specifically in birds42. This may be attributed to our culture methodology that selected for cephalosporin-resistant E. coli. Previous observations found that phylotypes A and D were more permissive to develop resistance to third-generation cephalosporins40, which is further supported in our study where phylotype A and D isolates generally harboured a higher number of AMR genes compared to phylotypes B1 and B2. The core genome of phylotype B1 isolates in our study showed the lowest degree of genetic diversity, as has been previously reported42.
We recovered closely related cephalosporin-resistant E. coli isolates from faecal samples of bald eagles and gulls inhabiting the same landfill, including two groups of nearly genetically indistinguishable E. coli isolates. The finding that five bald eagles and one gull harboured E. coli isolates with over 99.95% core genome nucleotide identity (sequence types 58 and NT3) and one bald eagle and two gulls carried similarly genetically closely related E. coli isolates (sequence type NT1) suggests acquisition via two possible scenarios: 1) common point source (via similar foraging behaviour by gulls and eagles at the landfill or pirating behaviour) or 2) through an inter-species transmission pathway (via faecal-oral route or predation of gulls by eagles). Further efforts are needed to understand the precise mechanisms by which cephalosporin-resistant E. coli may be shared among these sympatric taxa, and the potential implications this has for the spatial dissemination of AMR enteric bacteria by wild birds.
All 10 MLST sequence types found in our study that could be assigned to an existing type have previously been isolated from humans - nine of which have caused human disease (http://enterobase.warwick.ac.uk). These 10 MLST sequence types found in gulls and bald eagles included globally widespread, clinically important E. coli sequence types, including the often-multi-drug resistant and pathogenic ST131 and ST69 clones43–45. ESBL-producing ST131 E. coli isolates have also been isolated from several wild and domestic animal species, including gulls in Barrow (now Utqiaġvik), Alaska30 and Winnipeg, Canada46, the latter of which carried, among others, the two blaCTX-M genotypes found in our study (blaCTX-M-14 and blaCTX-M-55). The two ST131 isolates from our study belonged to ST131 clade A and clade B, with the clade A isolate harbouring acquired ESBL genes blaCTX-M-14 and blaTEM. This supports previous findings that blaCTX-M-14 is associated with clades A and C1 and that blaCTX-M genes are rarely found in clade B isolates47. CTX-M enzymes hydrolyse a wide variety of newer generation β-lactam antibiotics48 and have been increasing in incidence globally. CTX-M-14 and CTX-M-15 genotypes are particularly widespread, and the prevalence of CTX-M-55 is increasing in China49. The isolate in our study with the highest number of resistant genes, A1_007_Gull, belonged to ST167. In clinical human derived isolates, ST167 has been found to harbour resistance genes to last-line of defense antibiotics, such as carbapenems and colistin50. However, we did not find evidence for resistance to these two antibiotics in the ST167 gull isolate recovered in our sample collection. Additional WGS of E. coli isolates from humans and birds, combined with MLST results, would be useful for providing further inference as to the utility of commonly employed genetic approaches (e.g., MLST) for elucidating potential epidemiological connections across the human-animal interface.
The cephalosporin-resistant E. coli sequence types and AMR gene alleles found in gulls and bald eagles in our study have previously been reported to be widespread, making it difficult to identify specific sources for these isolates and associated resistance genes. However, cephalosporin-resistant E. coli isolates were previously recovered from gull faeces at the Soldotna landfill in 2014, but were not isolated from spatially-proximate samples collected at the mouth of the Kenai River or more spatially-distant samples collected from Middleton Island21. Thus, it is plausible that the Soldotna landfill either serves as a point source for cephalosporin-resistant E. coli or at least provides selection pressures to maintain AMR E. coli in the environment. This inference is substantiated by previous research that found increased AMR bacteria in gulls foraging in anthropogenic environments51.
Cephalosporin-resistant E. coli isolates genomically characterized in our study harboured genes conferring resistance to 64 different biocide and heavy metal compounds, many of which are reported to be plasmid-borne. The presence of heavy metals and biocides in contaminated environments, such as landfills52, can facilitate horizontal gene transfer53,54 and co-select for AMR genes, since genes conferring resistance to heavy metals and antibiotics are often physically linked55,56. We found evidence of this in the one isolate we investigated in detail, with qacEdelta1, a biocide/heavy metal resistance gene, co-located with SulI, an AMR gene, on a plasmid. The association of these two genes has been observed previously57,58, and although correlative and anecdotal in the current study, the finding of co-occurrence of AMR determinants and genes conferring resistance to biocides and heavy metals among cephalosporin-resistant E. coli isolated from gulls and bald eagles is consistent with the premise that the Soldotna landfill may serve as a source for acquisition and maintenance of AMR bacteria by wild birds.
Three (11.5%) isolates, two from gulls and one from a bald eagle, harboured the class 1 integron-integrase gene, intI1, a target that is often linked to genes conferring AMR and that has previously been used as a proxy for anthropogenic pollution59. Four resistance genes (three with identical nucleotide sequences), all predicted to be plasmid-borne, were found only in these three isolates, suggesting horizontal gene transfer as a likely mechanism of dissemination of intI1 among bacteria harboured by birds. Additional research regarding the prevalence of intI1 in bacteria harboured by wild birds in environments differently impacted by anthropogenic inputs would be helpful for interpretation of the significance of these findings.
Phylogenetic clustering of isolates in each of the chromosomal AMR gene phylogenies generally closely matched the core genome phylogeny; however, the finding that phylotype D isolates clustered with phylotype A isolates in two of the four AMR gene sequence phylogenies is indicative of homologous recombination. Our ClonalFrameML results provides additional evidence for recombination. Contrary to our findings, previous analysis of inter-phylotype recombination found phylotype D strains more likely to recombine with phylotype B2 strains, whereas phylotype A strains were more likely to recombine with types B1 and E41. Nevertheless, most resistance determinants found in the isolates investigated in this study were predicted to be plasmid-mediated and therefore, we infer that horizontal gene transfer was likely more influential than homologous recombination in the distribution of resistance genes among isolates in the current study.
We classified 77% of individual AMR genes detected in our study as plasmid-borne, however, this may not reflect the true number of plasmid-encoded genes due to challenges in plasmid sequence assembly60. Complete plasmid sequence reconstruction from short-read sequencing data is problematic, as resistance genes are often flanked by repetitive elements making them difficult to assemble61,62. Interestingly, no plasmid-borne AMR genes were detected in phylotype D isolates A1_006_Gull, A1_012_Gull, and A1_009_Gull, despite all three isolates having identical resistance profiles, characterized by five acquired AMR genes. It is possible that PlasmidSPAdes failed to recognize putative plasmid sequences in these isolates, and others where plasmid-encoded genes were expected (e.g. blaCMY-positive isolates), or these genes may have been integrated into the E. coli chromosome via integrons. These findings exemplify the need for improved bioinformatics tools to investigate accessory genomes of bacteria in order to understand mechanisms by which AMR determinants may be acquired and dispersed.
Several of the AMR genes identified in our study had 100% nucleotide identity to genes found in human clinical isolates, including two (tetA and sulI) of the seven AMR genes found in soil bacteria that matched human clinical isolates reported by Forsberg et al.63. While this could be interpreted as evidence of recent transmission at the soil-human-animal interface, this could also indicate that some gene alleles are particularly widespread and/or under purifying selection pressures. Such detailed molecular characterization of AMR genes can, conceptually, help resolve the epidemiology of resistance and identify the relative importance of different sources and transmission pathways. However, bacteria harbouring identical AMR genes, or even the genes themselves, may still be unrelated on an epidemiologically-relevant timescale. Thus, increased molecular surveillance of AMR genes in multiple hosts and environments to assess the relative prevalence of AMR gene alleles in different hosts/environments is critical if we are to gain rigorous inference regarding the dissemination of AMR genes and transmission pathways.
In summary, we found evidence that cephalosporin-resistant E. coli isolates were abundant among gulls and bald eagles at a landfill in southcentral Alaska. Identified sequence types of E. coli isolates included those previously associated with human infections, as well as presence of AMR determinants of clinical importance. Furthermore, genomic analyses provided evidence that gulls and bald eagles acquired bacteria via common point sources and/or through transmission among individuals, with horizontal gene transfer likely playing some role in the evolution of E. coli resistance maintained by the taxa sampled. Without environmental sampling at the landfill and other locations frequented by gulls and bald eagles, it cannot be conclusively determined whether the landfill is a source of AMR genes, whether this habitat provides selection pressure for AMR E. coli to persist, or whether the landfill is simply a foraging site for previously colonized birds. However, our results are consistent with previous studies that support the premise that anthropogenically influenced habitats play a role in the maintenance of AMR determinants in the environment. Future work to clarify point sources of AMR E. coli in wild birds and associated transmission pathways should incorporate sampling for AMR determinants in the physical environment and through space and time to better assess the transfer of AMR genes among bacteria associated with soil, water, and wildlife.
Materials and Methods
Sample collection and culture
Faecal material was collected from 20 bald eagles (Haliaeetus leucopehalus) and 56 gulls, which included glaucous-winged gulls (Larus glaucescens), American herring gulls (Larus argentatus), and hybrids, at the Soldotna landfill in southcentral Alaska (60.448°N, 151.118°W) from 7–9 June 2016. Samples were collected by inserting a sterile swab into recently deposited faecal material (all samples from bald eagles and 50 of those from gulls) or directly into the cloaca of live-captured gulls (n = 6) caught using noose carpets (authority granted under Alaska Department of Fish and Game permit #16–109, U.S. Fish and Wildlife permit #MB789758-5, U.S. Geological Survey Alaska Science Center Animal Care and Use Committee approval #2016-6). All methods were performed in accordance with the relevant guidelines and regulations. Swabs were subsequently placed into a vial with chilled Luria broth (BD, Sparks, USA), and kept cool on ice packs, for approximately 4–48 hours until frozen at −80 °C.
For E. coli culture, samples were thawed and inoculated in 2 ml brain heart infusion broth (BHI; Becton Dickinson, USA), supplemented with vancomycin (16 mg/L; ICN Biomedicals Inc., USA) to select for gram negative bacteria, using a sterile swab. Following incubation for 18–24 hours at 36 °C for enrichment, 10 μl of broth was streaked onto CHROMagar C3GR plates (CHROMagar, France), a medium that supports growth of bacteria with reduced susceptibility to third generation cephalosporins. E. coli CCUG 17620 and K. pneumoninae CCUG 45421 were included as negative and positive controls, respectively. All plates were incubated in aerobic conditions for 18–24 hours at 36 °C. Putative E. coli isolates (one per plate) were analysed by matrix-assisted laser desorption ionization time of flight mass spectrometry (Bruker Corporation, Germany), as described previously64 and antimicrobial susceptibility testing was subsequently performed on all isolates confirmed to be E. coli.
Phenotypic antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed on E. coli isolates according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) disc diffusion method using the following antibiotic discs, selected to represent commonly used agents for E. coli infections in human and veterinary medicine: Nalidixic acid (30 µg), Nitrofurantoin (100 µg), Piperazillin-tazobactam (36 µg), Tetracycline (30 µg), Trimethoprim (5 µg), Trimethoprim-sulfamethoxazole (25 µg), Meropenem (10 µg), Ciprofloxacin (5 µg), Ampicillin (10 µg), Cefadroxil (30 µg), Chloramphenicol (30 µg), Gentamicin (10 µg) and Mecillinam (10 µg) (Thermo Fisher Scientific Oxoid Ltd, Hants, UK). The inhibition zone diameters were interpreted according to EUCAST breakpoints65, or, for antibiotics with no defined clinical breakpoints (i.e. Nalidixic acid, Tetracycline and Chloramphenicol), the inhibition zone diameters were interpreted by breakpoints defined by the Normalized Resistance Interpretation method66.
Phenotypic characterization to identify ESBL-producing isolates was performed using the following five antibiotic discs: Ceftazidime (10 µg), Cefotaxime (5 µg), Cefepime (30 µg), Cefoxitin (30 µg) and Amoxicillin/clavulanic acid (30/1 µg) (Thermo Fisher Scientific Oxoid Ltd, Hants, UK). Inhibition zone diameters were used to determine the phenotypes, according to EUCAST guidelines67.
Library preparation and whole genome sequencing
DNA was extracted from all ESBL-producing E. coli isolates using the MagnaPure compact nucleic acid isolation kit (Roche, Mannheim, Germany). Multiplexed DNA libraries were prepared using the NexteraXT library preparation kit (Illumina, San Diego, USA) according to manufacturer’s instructions. Paired-end WGS was performed using the MiSeq platform (Illumina, San Diego, USA) using either 250 or 300 base pair read lengths. Average read depth was determined by reference mapping to the E. coli K12 genome (NC_000913.3).
De novo assembly
ConDeTri was used to remove reads with low quality scores, trim high quality reads and remove duplicate reads68. High-quality, trimmed, unique reads were assembled with SPAdes69 using default parameters and k-values of 21, 33, 55, 77, 99, and 127. A second assembly excluding the k-value 127 was also performed to improve assemblies with shorter read lengths. Assembly quality was evaluated with QUAST70 and the assembly with the fewest number of contigs and highest N50 was used in downstream analyses. Additionally, PlasmidSPAdes was implemented with default settings to detect and assemble putative plasmid sequences from trimmed reads71. See File S1 for specific commands and settings used for each program.
Core genome analysis
An alignment of orthologous sequences conserved in all genomes was generated from de novo assemblies using Parsnp72, with the –c option invoked to force inclusion of all genomes. Reference genomes E. fergusonii (NC_011740.1) and E. coli TW10509 (NZ_GL872204.1) were included to provide phylogenetic context for divergent strains. The resulting core genome alignment and SNP tree was used as input into ClonalFrameML73, with default settings, to detect recombination and reconstruct the phylogeny based on an evolutionary model that accounts for both mutation and recombination events. The previously described methodology was re-performed excluding reference and divergent genomes to improve phylogenetic resolution of the 25 most closely related isolates. In silico phylotyping, based on the Clermont et al. typing scheme74, was performed using a previously described methodology75. The program SRST2 (v0.2.0)76 was used to identify MLST types by matching reads to the Escherichia coli #1 database77 downloaded from pubmlst.org.
Accessory genome analysis
AMR genes were detected from trimmed, unique high quality reads using SRST276 and matched to the ARGannot resistance gene database78. This manually curated AMR gene database includes sequences found in both the ResFinder database79 and the Comprehensive Antibiotic Resistance Database (CARD)80, and includes both acquired resistance genes and point mutations in chromosomal target genes. AMR gene sequences from each isolate were extracted using the “report_new_consensus” function in SRST2, which outputs all reads mapped to the gene sequence. Gene alignments were visualized in Geneious (v10.1.3)81 and insertions/deletions (indels) were confirmed or rejected by de novo assembly of mapped reads. Sequences differing by one or more confirmed SNP or indel were considered a unique allele. Maximum likelihood phylogenetic trees of AMR genes with more than two alleles were estimated in the statistical programming language R (v3.3.3)82 using the package phangorn (v2.2.0)83. Separate nucleotide substitution model tests84 were performed on each gene alignment and the model with the lowest Akaike information criterion value was used to estimate phylogenies with 100 bootstrap pseudoreplicates. AMR gene presence was mapped onto the core genome phylogeny using the -phydataplot function in the R package ape (v4.1)85, with different colours representing different alleles. SRST2-detected AMR genes were queried and visualized against putative plasmid contigs that were assembled with PlasmidSPAdes from select isolates using Bandage86. Biocide and metal resistance genes were detected from assembled contigs using BacMetScan87, using the experimentally confirmed resistance genes dataset (v1.1), and visualized on putative plasmid contigs using Bandage, as described above. Predicted resistance to particular compounds and gene location (chromosomal or plasmid) was obtained from the BacMet database. In silico PCR amplification of the class 1 integron-integrase gene intI1 was performed using the program seqpoet (v0.3.4) using previously described primers (IntiIf: TTCGAATGTCGTAACCGC and IntiIr: CGAGGCATAGACTGTAC)88 and the SPAdes-assembled contigs.
Accessory genomes were further explored using the program LS-BSR89 to determine the relative level of relatedness among isolates of each detected coding sequence (CDS). Core sequences were excluded using the filter_BSR_variome script. Accessory CDS with BLAST score ratio (BSR) values ≥0.70 were considered present and a binary present/absent matrix was created as input for the Bayesian clustering tool K-Pax290 to partition isolates based on their accessory genome content. Default prior settings were used with an initial partition of 26 units (one for each isolate). All data that support the findings of this publication can be found in Ahlstrom et al.91.
Data Availability
Raw WGS reads have been deposited in the Sequence Read Archive; accession number: SRP126755.
Electronic supplementary material
Acknowledgements
We appreciate field and laboratory support provided by John Reed, Andrew Reeves, Matthew Smith, Lee Tibbitts, and Anna Petersson. We thank Rolf Kaas for providing in silico phylotyping scripts. We appreciate reviews provided by John Pearce, Taya Forde, and two anonymous reviewers. This research used resources of the Core Science Analytics and Synthesis Advanced Research Computing program at the U.S. Geological Survey. This project was funded, in part, by the U.S. Geological Survey through the Contaminants Program of the Environmental Health Mission Area. None of the authors have any financial interests or conflict of interest with this article. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Author Contributions
C.A., J.B., and A.R. drafted the manuscript and C.A., J.B., B.O., and A.R. made lead contributions in the conception, design, field work, analysis, and interpretation of the work. H.W. and J.H. made contributions to laboratory work, analysis, and interpretation of the work. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-25474-w.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Christina A. Ahlstrom, Email: cahlstrom@usgs.gov
Andrew M. Ramey, Email: aramey@usgs.gov
References
- 1.WHO. Antimicrobial resistance: global report on surveillance 2014. http://www.who.int/drugresistance/documents/surveillancereport/en/ (2014).
- 2.GHSA. Global Health Security Agenda: GHSA Antimicrobial Resistance Action Package (GHSA Action Package Prevent-1). https://www.ghsagenda.org/packages/p1-antimicrobial-resistance (2015).
- 3.McDanel J, et al. Incidence of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli and Klebsiella infections in the United States: A systematic literature review. Infect. Control Hosp. Epidemiol. 2017;38:1209–1215. doi: 10.1017/ice.2017.156. [DOI] [PubMed] [Google Scholar]
- 4.Dhillon RHP, Clark J. ESBLs: A clear and present danger? Crit. Care Res. Pract. 2012;2012:625170. doi: 10.1155/2012/625170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashbolt NJ, et al. Human Health Risk Assessment (HHRA) for environmental development and transfer of antibiotic resistance. Environ. Health Perspect. 2013;121:993–1001. doi: 10.1289/ehp.1206316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Brien TF. Emergence, spread, and environmental effect of antimicrobial resistance: how use of an antimicrobial anywhere can increase resistance to any antimicrobial anywhere else. Clin. Infect. Dis. 2002;34:S78–S84. doi: 10.1086/340244. [DOI] [PubMed] [Google Scholar]
- 7.VonWintersdorff CJH, et al. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 2016;7:173. doi: 10.3389/fmicb.2016.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woolhouse, M. & Ward, M. Sources of antimicrobial resistance. Science. 341 (2013). [DOI] [PubMed]
- 9.Hiltunen, T., Virta, M. & Laine, A. L. Antibiotic resistance in the wild: an eco-evolutionary perspective. Philos. Trans. R. Soc. London B Biol. Sci. 372 (2016). [DOI] [PMC free article] [PubMed]
- 10.Allen HK, et al. Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010;8:251–259. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- 11.D’Costa VM, et al. Antibiotic resistance is ancient. Nature. 2011;477:457–461. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 12.Finley RL, et al. The scourge of antibiotic resistance: the important role of the environment. Clin. Infect. Dis. 2013;57:704–710. doi: 10.1093/cid/cit355. [DOI] [PubMed] [Google Scholar]
- 13.Knapp CW, Dolfing J, Ehlert PAI, Graham DW. Evidence of increasing antibiotic resistance gene abundances in archived soils since 1940. Environ. Sci. Technol. 2010;44:580–587. doi: 10.1021/es901221x. [DOI] [PubMed] [Google Scholar]
- 14.Martinez JL. Antibiotics and antibiotic resistance genes in natural environments. Science. 2008;321:365–367. doi: 10.1126/science.1159483. [DOI] [PubMed] [Google Scholar]
- 15.Woolhouse M, Ward M, van Bunnik B, Farrar J. Antimicrobial resistance in humans, livestock and the wider environment. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2015;370:20140083. doi: 10.1098/rstb.2014.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thaller MC, et al. Tracking acquired antibiotic resistance in commensal bacteria of Galápagos land iguanas: no man, no resistance. PLoS One. 2010;5:e8989. doi: 10.1371/journal.pone.0008989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noyes NR, et al. Characterization of the resistome in manure, soil and wastewater from dairy and beef production systems. Sci. Rep. 2016;6:24645. doi: 10.1038/srep24645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rolland RM, Hausfater G, Marshall B, Levy SB. Antibiotic-resistant bacteria in wild primates: increased prevalence in baboons feeding on human refuse. Appl. Environ. Microbiol. 1985;49:791–794. doi: 10.1128/aem.49.4.791-794.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allen SE, et al. Antimicrobial resistance in generic Escherichia coli isolates from wild small mammals living in swine farm, residential, landfill, and natural environments in Southern Ontario, Canada. Appl. Environ. Microbiol. 2011;77:882–888. doi: 10.1128/AEM.01111-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gómez P, et al. Detection of MRSA ST3061-t843- mecC and ST398-t011- mecA in white stork nestlings exposed to human residues. J. Antimicrob. Chemother. 2016;71:53–57. doi: 10.1093/jac/dkv314. [DOI] [PubMed] [Google Scholar]
- 21.Atterby C, et al. Increased prevalence of antibiotic-resistant E. coli in gulls sampled in Southcentral Alaska is associated with urban environments. Infect Ecol Epidemiol. 2016;6:32334. doi: 10.3402/iee.v6.32334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Österblad M, Norrdahl K, Korpimäki E, Huovinen P. Antibiotic resistance: How wild are wild mammals? Nature. 2001;409:37–38. doi: 10.1038/35051173. [DOI] [PubMed] [Google Scholar]
- 23.Ramey, A. M. et al. Antibiotic-resistant Escherichia coli in migratory birds inhabiting remote Alaska. Ecohealth10.1007/s10393-017-1302-5 (2017). [DOI] [PubMed]
- 24.Arnold, K. E., Williams, N. J. & Bennett, M. ‘Disperse abroad in the land’: the role of wildlife in the dissemination of antimicrobial resistance. Biol. Lett. 12 (2016). [DOI] [PMC free article] [PubMed]
- 25.Guenther S, Ewers C, Wieler LH. Extended-spectrum beta-lactamases producing E. coli in wildlife, yet another form of environmental pollution? Front. Microbiol. 2011;2:246. doi: 10.3389/fmicb.2011.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weiser EL, Powell AN. Does garbage in the diet improve reproductive output of Glaucous Gulls? Condor. 2010;112:530–538. doi: 10.1525/cond.2010.100020. [DOI] [Google Scholar]
- 27.Dolejská M, Cizek A, Literak I. High prevalence of antimicrobial-resistant genes and integrons in Escherichia coli isolates from Black-headed Gulls in the Czech Republic. J. Appl. Microbiol. 2007;103:11–19. doi: 10.1111/j.1365-2672.2006.03241.x. [DOI] [PubMed] [Google Scholar]
- 28.Bonnedahl J, et al. Dissemination of Escherichia coli with CTX-M Type ESBL between humans and Yellow-Legged Gulls in the South of France. PLoS One. 2009;4:e5958. doi: 10.1371/journal.pone.0005958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dolejská M, Bierošová B, Kohoutová L, Literák I, Čížek A. Antibiotic-resistant Salmonella and Escherichia coli isolates with integrons and extended-spectrum beta-lactamases in surface water and sympatric black-headed gulls. J. Appl. Microbiol. 2009;106:1941–1950. doi: 10.1111/j.1365-2672.2009.04155.x. [DOI] [PubMed] [Google Scholar]
- 30.Bonnedahl J, et al. Extended-spectrum β-lactamases in Escherichia coli and Klebsiella pneumoniae in gulls, Alaska, USA. Emerg. Infect. Dis. 2014;20:897–899. doi: 10.3201/eid2005.130325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alm EW, et al. Potential for gulls to transport bacteria from human waste sites to beaches. Sci. Total Environ. 2018;615:123–130. doi: 10.1016/j.scitotenv.2017.09.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guenther S, et al. Comparable high rates of extended-spectrum-beta-lactamase-producing Escherichia coli in birds of prey from Germany and Mongolia. PLoS One. 2012;7:e53039. doi: 10.1371/journal.pone.0053039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molina-Lopez RA, et al. Wild raptors as carriers of antimicrobial-resistant Salmonella and Campylobacter strains. Vet. Rec. 2011;168:565. doi: 10.1136/vr.c7123. [DOI] [PubMed] [Google Scholar]
- 34.Pinto L, et al. Genetic detection of extended-spectrum beta-lactamase-containing Escherichia coli isolates from birds of prey from Serra da Estrela Natural Reserve in Portugal. Appl. Environ. Microbiol. 2010;76:4118–4120. doi: 10.1128/AEM.02761-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McNally A, et al. Combined analysis of variation in core, accessory and regulatory genome regions provides a super-resolution view into the evolution of bacterial populations. Plos Genet. 2016;12:e1006280. doi: 10.1371/journal.pgen.1006280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kahn LH. Antimicrobial resistance: A One Health perspective. Trans. R. Soc. Trop. Med. Hyg. 2017;111:255–260. doi: 10.1093/trstmh/trx050. [DOI] [PubMed] [Google Scholar]
- 37.Singer RS, Ward MP, Maldonado G. Can landscape ecology untangle the complexity of antibiotic resistance? Nat. Rev. Microbiol. 2006;4:943–952. doi: 10.1038/nrmicro1553. [DOI] [PubMed] [Google Scholar]
- 38.Schwarz S, et al. Editorial: Assessing the antimicrobial susceptibility of bacteria obtained from animals. J. Antimicrob. Chemother. 2010;65:601–604. doi: 10.1093/jac/dkq037. [DOI] [PubMed] [Google Scholar]
- 39.Luo C, et al. Genome sequencing of environmental Escherichia coli expands understanding of the ecology and speciation of the model bacterial species. Proc. Natl. Acad. Sci. USA. 2011;108:7200–7205. doi: 10.1073/pnas.1015622108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tenaillon O, Skurnik D, Picard B, Denamur E. The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 2010;8:207–217. doi: 10.1038/nrmicro2298. [DOI] [PubMed] [Google Scholar]
- 41.Chaudhuri RR, Henderson IR. The evolution of the Escherichia coli phylogeny. Infect. Genet. Evol. 2012;12:214–226. doi: 10.1016/j.meegid.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 42.White AP, et al. Intergenic sequence comparison of Escherichia coli isolates reveals lifestyle adaptations but not host specificity. Appl. Environ. Microbiol. 2011;77:7620–7632. doi: 10.1128/AEM.05909-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nicolas-Chanoine MH, et al. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 2007;61:273–281. doi: 10.1093/jac/dkm464. [DOI] [PubMed] [Google Scholar]
- 44.Blanco J, et al. National survey of Escherichia coli causing extraintestinal infections reveals the spread of drug-resistant clonal groups O25b:H4-B2-ST131, O15:H1-D-ST393 and CGA-D-ST69 with high virulence gene content in Spain. J. Antimicrob. Chemother. 2011;66:2011–2021. doi: 10.1093/jac/dkr235. [DOI] [PubMed] [Google Scholar]
- 45.Doumith M, et al. Rapid identification of major Escherichia coli sequence types causing urinary tract and bloodstream infections. J. Clin. Microbiol. 2015;53:160–166. doi: 10.1128/JCM.02562-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bonnedahl J, et al. Comparison of extended-spectrum β-lactamase (ESBL) CTX-M genotypes in Franklin Gulls from Canada and Chile. PLoS One. 2015;10:e0141315. doi: 10.1371/journal.pone.0141315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stoesser N, et al. Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. MBio. 2016;7:e02162. doi: 10.1128/mBio.02162-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jacoby GA, Munoz-Price LS. The New β-Lactamases. N Engl J Med. 2005;352:380–391. doi: 10.1056/NEJMra041359. [DOI] [PubMed] [Google Scholar]
- 49.Bevan ER, Jones AM, Hawkey PM. Global epidemiology of CTX-M β-lactamases: temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 2017;72:2145–2155. doi: 10.1093/jac/dkx146. [DOI] [PubMed] [Google Scholar]
- 50.Yao X, Doi Y, Zeng L, Lv L, Liu JH. Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect. Dis. 2016;16:288–289. doi: 10.1016/S1473-3099(16)00057-8. [DOI] [PubMed] [Google Scholar]
- 51.Vittecoq M, et al. VIM-1 carbapenemase-producing Escherichia coli in gulls from southern France. Ecol. Evol. 2017;7:1224–1232. doi: 10.1002/ece3.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reinhart D. A Review of recent studies on the sources of hazardous compounds emitted from solid waste landfills: A U.S. experience. Waste Manag. Res. 1993;11:257–268. doi: 10.1177/0734242X9301100307. [DOI] [Google Scholar]
- 53.Beaber JW, Hochhut B, Waldor MK. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature. 2004;427:72–74. doi: 10.1038/nature02241. [DOI] [PubMed] [Google Scholar]
- 54.Guerin E, et al. The SOS response controls integron recombination. Science. 2009;324:1034–1034. doi: 10.1126/science.1172914. [DOI] [PubMed] [Google Scholar]
- 55.Knapp CW, et al. Antibiotic resistance gene abundances correlate with metal and geochemical conditions in archived Scottish soils. PLoS One. 2011;6:e27300. doi: 10.1371/journal.pone.0027300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baker-Austin C, Wright MS, Stepanauskas R, McArthur JV. Co-selection of antibiotic and metal resistance. Trends Microbiol. 2006;14:176–182. doi: 10.1016/j.tim.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 57.Pal C, Bengtsson-Palme J, Kristiansson E, Larsson DGJ. Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genomics. 2015;16:964. doi: 10.1186/s12864-015-2153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wyrsch ER, et al. Genomic microbial epidemiology is needed to comprehend the global problem of antibiotic resistance and to improve pathogen diagnosis. Front. Microbiol. 2016;7:843. doi: 10.3389/fmicb.2016.00843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gillings MR, et al. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 2015;9:1269–1279. doi: 10.1038/ismej.2014.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arredondo-Alonso, S., Willems, R. J., van Schaik, W. & Schürch, A. C. On the (im)possibility of reconstructing plasmids from whole-genome short-read sequencing data. Microb. Genomics3 (2017). [DOI] [PMC free article] [PubMed]
- 61.Treangen TJ, Salzberg SL. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat. Rev. Genet. 2011;13:36–46. doi: 10.1038/nrg3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Orlek A, et al. Plasmid classification in an era of whole-genome sequencing: Application in studies of antibiotic resistance epidemiology. Front. Microbiol. 2017;8:182. doi: 10.3389/fmicb.2017.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Forsberg KJ, et al. The shared antibiotic resistome of soil bacteria and human pathogens. Science. 2012;337:1107–1111. doi: 10.1126/science.1220761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seng P, et al. Ongoing revolution in bacteriology: Routine identification of bacteria by matrix‐assisted laser desorption ionization time‐of‐flight mass spectrometry. Clin. Infect. Dis. 2009;49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 65.EUCAST. Breakpoint tables for interpretation of MICs and zone diameters. Version 6. 0; http://www.eucast.org (2016).
- 66.Kronvall G, Smith P. Normalized resistance interpretation, the NRI method. APMIS. 2016;124:1023–1030. doi: 10.1111/apm.12624. [DOI] [PubMed] [Google Scholar]
- 67.EUCAST. EUCAST guidelines for detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance. Version 1. 0. http://www.amcli.it/wp-content/uploads/2015/10/EUCAST_detection_resistance_mechanisms_V1.pdf. (2013).
- 68.Smeds L, Künstner A, Wysoker A, Fennell T, Ruan J. ConDeTri - A content dependent read trimmer for Illumina data. PLoS One. 2011;6:e26314. doi: 10.1371/journal.pone.0026314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bankevich A, et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Antipov D, et al. PlasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics. 2016;32:btw493. doi: 10.1093/bioinformatics/btw493. [DOI] [PubMed] [Google Scholar]
- 72.Treangen TJ, et al. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Didelot X, Wilson DJ. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput. Biol. 2015;11:e1004041. doi: 10.1371/journal.pcbi.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 2000;66:4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kaas RS, Friis C, Ussery DW, Aarestrup FM. Estimating variation within the genes and inferring the phylogeny of 186 sequenced diverse Escherichia coli genomes. BMC Genomics. 2012;13:577. doi: 10.1186/1471-2164-13-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Inouye M, et al. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014;6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wirth T, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 2006;60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gupta SK, et al. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014;58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zankari E, et al. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jia B, et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2016;45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kearse M, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.RCoreTeam. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. (2017).
- 83.Schliep KP. phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27:592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Posada D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 85.Paradis E, Claude J, Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 86.Wick RR, Schultz MB, Zobel J, Holt KE. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 2015;31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pal C, Bengtsson-Palme J, Rensing C, Kristiansson E, Larsson DGJ. BacMet: antibacterial biocide and metal resistance genes database. Nucleic Acids Res. 2014;42:D737–D743. doi: 10.1093/nar/gkt1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Orman BE, et al. Evolution of multiresistance in nontyphoid Salmonella serovars from 1984 to 1998 in Argentina. Antimicrob. Agents Chemother. 2002;46:3963–3970. doi: 10.1128/AAC.46.12.3963-3970.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sahl JW, Caporaso JG, Rasko DA, Keim P. The large-scale blast score ratio (LS-BSR) pipeline: a method to rapidly compare genetic content between bacterial genomes. PeerJ. 2014;2:e332. doi: 10.7717/peerj.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pessia A, Grad Y, Cobey S, Puranen JS, Corander J. K-Pax2: Bayesian identification of cluster-defining amino acid positions in large sequence datasets. Microb. genomics. 2015;1:e000025. doi: 10.1099/mgen.0.000025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ahlstrom, C. A. & Ramey A. M. Sampling and Resistance and Genomic Typing of Cephalosporin-resistant E. coli in Gulls (Larus spp.) and Bald Eagles (Haliaeetus leucocephalus) in Southcentral Alaska, 2016: U.S. Geological Survey data release, 10.5066/F70V8C2Q (2018).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw WGS reads have been deposited in the Sequence Read Archive; accession number: SRP126755.