Abstract
Mitochondria are double-membrane subcellular organelles with highly conserved metabolic functions including ATP production. Mitochondria shapes change continually through the combined actions of fission and fusion events rendering mitochondrial network very dynamic. Mitochondria are largely implicated in pathologies and mitochondrial dynamics is often disrupted upon muscle degeneration in various models. Currently, the exact roles of mitochondria in the molecular mechanisms that lead to muscle degeneration remain poorly understood. Here we report a role for DRP-1 in regulating apoptosis induced by dystrophin-dependent muscle degeneration. We found that: (i) dystrophin-dependent muscle degeneration was accompanied by a drastic increase in mitochondrial fragmentation that can be rescued by genetic manipulations of mitochondrial dynamics (ii) the loss of function of the fission gene drp-1 or the overexpression of the fusion genes eat-3 and fzo-1 provoked a reduction of muscle degeneration and an improved mobility of dystrophin mutant worms (iii) the functions of DRP-1 in apoptosis and of others apoptosis executors are important for dystrophin-dependent muscle cell death (iv) DRP-1-mediated apoptosis is also likely to induce age-dependent loss of muscle cell. Collectively, our findings point toward a mechanism involving mitochondrial dynamics to respond to trigger(s) of muscle degeneration via apoptosis in Caenorhabditis elegans.
Introduction
Mitochondria are double-membrane organelles that are crucial for many cellular processes such as aerobic ATP generation, calcium homeostasis, lipid biosynthesis or cell death1. Mitochondria are characterized by an important polymorphism of shapes due to permanent fusion and fission events that vary within a considered tissue2. Dynamin family GTPases regulates mitochondrial dynamics. In mammals, Mitofusin-1 and Mitofusin-2 (MFN1 and MFN2)3 allow for mitochondria outer membrane fusion whereas inner membrane fusion implicates Optic atrophy 1 (OPA1)4. Fission machinery of the outer membrane is composed of Dynamin-related protein1 (DRP1) and Dynamin-2 (DYN2)5,6. In Caenorhabiditis elegans (C. elegans), DRP-1 is also required for mitochondrial fission7 whereas EAT-3 (OPA1 homolog in mammals) and FZO-1 (MFN1 homolog in mammals) control inner and outer mitochondrial membrane fusion, respectively8. Mitochondrial fusion is essential for normal mitochondrial activity notably by diluting damaged mitochondrial DNA, lipids or proteins9,10. In mammals, mitochondrial fission is key for mitophagy to eliminate damaged mitochondria11,12. Also, fragmented mitochondria often appear in the early steps of cell death associated with permeabilization of the mitochondrial outer membrane, which induces the release of inter membrane space-stored proapoptotic factors, such as cytochrome c, Apoptosis-Inducing Factor (AIF) or Endonuclease G (EndoG)13–16. In C. elegans, fragmentation of mitochondria during apoptosis has been reported but the exact roles of mitochondria in cell death remain largely unknown17. Whether mitochondrial fusion is directly implicated in apoptosis is still under debate. In mammals, hyperfusion allows for survival under numerous stresses including nutrient deprivation18–21. Furthermore, the fusion proteins MFN1 and MFN2 seem to directly regulate apoptosis suggesting that longer mitochondria are more supportive for cell survival than fragmented mitochondria22,23. Interestingly, molecular links exist between mitochondrial fusion and apoptosis24–26.
Importantly, the main actors of mitochondrial fission and fusion have alternative functions separate from mitochondrial dynamics. It is well established that DRP-1 participates in peroximal division27,28 and in the maintenance of the morphology and the function of the Endoplasmic Reticulum (ER)29,30. In C. elegans, DRP-1 can contribute to apoptotic processes independently from mitochondrial fission31. Furthermore, both DRP-1 and OPA1 allow for mitochondrial cristae remodeling separately from mitochondrial dynamics32–34. The alternative functions of OPA1 also include lipolysis regulation35. The pro-fusion protein MFN2 is an essential component of the contact sites between mitochondria and ER and contributes to mitochondrial Ca2+ uptake and ER morphology maintenance36. Additionally, levels of MFN2 are regulated by ER stress and can affect the ER-Unfolded Protein Response (ER-UPR) and in turn, cell death37,38. Furthermore, axonal mitochondrial transport requires MFN2 but not mitochondrial fusion39.
Strength and function of muscle tissues are highly dependent on the energy produced by mitochondria. Compromised muscle tissues, due to deterioration and loss of muscle cells, and leading to muscle weakness, are hallmarks of muscle degeneration. The primary causes of muscle degeneration are various. For instance, it can be due to numerous genetic mutations causing muscular dystrophies. Muscle degeneration can also be caused by aging processes, and is called sarcopenia. To study muscle degeneration, we used the dys-1(cx18);hlh-1(cc561ts) C. elegans mutant, which exhibits progressive dystrophin-dependent muscle degeneration reflected by muscle weakness and dramatic loss of muscle cells40. DYS-1 is the homolog of dystrophin in mammals and mutations in this gene cause Duchenne Muscular Dystrophy in Human41. HLH-1 is a homolog of Myogenic Differentiation 1 (MyoD), a muscle transcription factor42. Despite the phylogenetic distance between mammals and C. elegans, the nematode presents striated body wall muscle similar to vertebrate skeletal muscle in terms of function and structural components; however, the overall architecture is less complex43,44. Mitochondrial fragmentation increases in muscle cells during dystrophin-dependent muscle degeneration in various animal models including nematode45, zebrafish45, mouse, which exhibits abnormalities of mitochondrial morphology and density as well as an up-regulation of DRP-1 levels46,47, dog48 and in biopsies from Duchenne Muscular Dystrophy patients49. Fragmentation of mitochondria is also observed during muscle aging in nematode50, fly51 and mouse52.
We hypothesized that genetic manipulations of the main actors of mitochondrial dynamics can improve muscle health. We found that decreasing mitochondrial fission or increasing mitochondrial fusion can partially rescue mitochondrial dynamics, reduce muscle degeneration and improve the mobility of dys-1(cx18);hlh-1(cc51ts) mutant worms. Conversely, increased mitochondrial fusion can enhance the muscle degeneration phenotype of dystrophic nematodes. Interestingly, we showed that DRP-1 functions mainly in the apoptosis pathway downstream of the caspase 3 to impact muscle degeneration and that others apoptotic executors are important for muscle health. Finally, apoptosis induced by DRP-1 seems also to be implicated in aging-dependent muscle loss. Collectively, our results point toward a novel mechanism by which, DRP-1-mediated apoptosis provokes muscle degeneration.
Results
Mitochondrial fragmentation increases upon dystrophin-deficiency
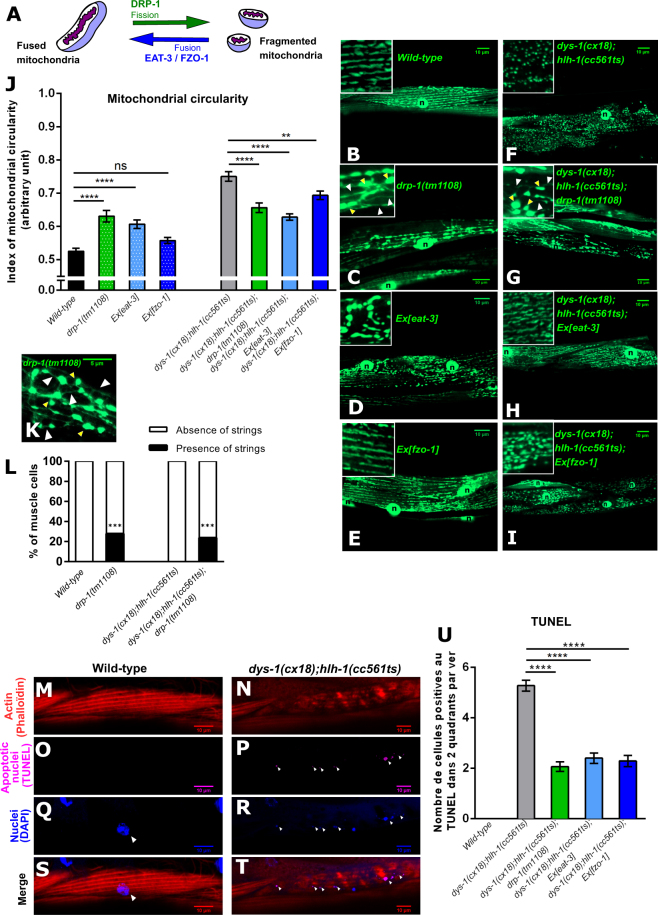
Mitochondrial shapes undergo continual changes through the combined actions of fission and fusion events rendering mitochondrial network very dynamic. In C. elegans, fission is mediated by DRP-17, inner membrane fusion by EAT-3 (OPA1 homolog)53 and outer membrane fusion by FZO-1 (MFN1 homolog)8 (Fig. 1A). As C. elegans is a transparent organism, it is possible to visualize and evaluate mitochondrial network shape of muscle cells in lived worms54. In order to visualize mitochondrial morphologies, we used the ccIs4251 transgene, a genomic integrated GFP construction addressed specifically to the mitochondrial matrix and nuclei of the C. elegans body wall muscle cells (Fig. 1B–I)55. As previously published by Yang et al.54, the index of circularity of individual mitochondrion (a mathematical estimation of circular shapes) was calculated for each of the tested conditions to evaluate mitochondrial dynamics. A value of 1 for the index of mitochondria circularity means that mitochondria are perfectly circular and a value of 0 means that mitochondria are perfectly flat. In wild-type C. elegans muscle cells, tubular and circular mitochondria were in equal proportions (Fig. 1B) as reflected by a mitochondrial circularity index of around 0,5 (Fig. 1J)54. dys-1(cx18);hlh-1(cc561ts) mutant worms, which present progressive muscle degeneration, exhibited mitochondrial dynamics perturbations with more fragmented mitochondria in muscle cells than wild-type worms (Fig. 1B,F) as reflected by a higher index of mitochondria circularity compared to that of wild-type worms (0,75 +/− 0,014 in dys-1(cx18);hlh-1(cc561ts) mutant vs 0,52 +/− 0,009 in wild-type worms) (Fig. 1J). By comparing mitochondrial aspect ratio (major axis/minor axis of each mitochondrion), mitochondrial elongation (1-(minor axis/major axis of each mitochondrion) and mitochondrial size, we also found that dys-1(cx18);hlh-1(cc561ts) mutant worms presented shorter and smaller mitochondria than that of wild-type worms (Fig. S1A–D). Furthermore, dystrophic worms exhibited a less connected mitochondrial network than wild-type animals (Fig. S1D).
Figure 1.
DRP-1 induces mitochondrial fragmentation and apoptosis upon dystrophin-dependent muscle degeneration. (A) Schematic representation of the main mitochondrial dynamics actors in C. elegans. Representative confocal images of the mitochondrial network organization visualized with the ccIs4251 transgene in C. elegans body wall muscle cells of: (B) wild-type worms; (C) drp-1(tm1108) mutant worms; (D) worms overexpressing eat-3; (E) worms overexpressing fzo-1; (F) dys-1(cx18);hlh-1(cc561ts) mutant worms; (G) dys-1(cx18);hlh-1(cc561ts);drp-1(tm1108) mutant worms; (H) dys-1(cx18);hlh-1(cc561ts) overexpressing eat-3 mutant worms and (I) dys-1(cx18);hlh-1(cc561ts) overexpressing fzo-1 mutant worms. Scale bar 10 μm (x630). n indicates nuclei. Yellow arrowheads indicate bubble-like shape mitochondria. White arrowheads indicate mitochondria connecting strings. (J) Quantification of mitochondrial circularity in each of the indicated strains (n = 29 worms at least). (K) Representative confocal image of thin mitochondrial filaments in between blebs of mitochondria in the drp-1(tm1108) mutant worms. Scale bar 5 μm (x630). Yellow arrowheads indicate bubble-like shape mitochondria. White arrowheads indicate mitochondria connecting strings. (L) Percentage of muscle cells exhibiting strings in between blebs of mitochondria in each of the indicated strains (n = 54 worms at least). Chi-square test. ***p < 0.001. Representative confocal images: of actin network by phalloïdin staining in (M) wild-type worms and (N) dys-1(cx18);hlh-1(cc561ts) mutant worms; of apoptotic nuclei revealed by TUNEL staining in (O) wild-type worms and (P) dys-1(cx18);hlh-1(cc561ts) mutant worms; of nuclei of muscle cells stained by DAPI in (Q) wild-type worms and (R) dys-1(cx18);hlh-1(cc561ts) mutant worms and of the merge of the phalloidin, TUNEL and DAPI staining in (S) wild-type worms and (T) in dys-1(cx18);hlh-1(cc561ts) mutant worms. Scale bar 10 μm. n indicates nuclei. White arrowheads indicate colocalization of DAPI and TUNEL staining. (x630). (U) Quantification of muscle TUNEL positive cells in each of the indicated strains (n = 55 worms at least). All the experiments were performed on L4 + 3 day-old worms. One-way ANOVA, Tukey’s multiple comparisons test. Data represent the mean obtained by pooling at least three independent assays. Errors bars represent SEM (Standard Error to the Mean) **p < 0.01 ****p < 0.0001 ns. indicates that the mean is not statistically significantly different from the mean obtained in the control condition.
In order to decrease mitochondrial fission, we introduced the drp-1(tm1108) null mutation31 in the dys-1(cx18);hlh-1(cc561ts) mutant worms. Mitochondria of drp-1(tm1108) mutant worms formed bubbles-like shape (Fig. 1C), as previously published7,31,54,56, presumably because fission of the mitochondrial inner membrane still occurs in absence of DRP-17 suggesting that it is possible to separate outer and inner membrane fissions in C. elegans. Accordingly, mitochondria of drp-1(tm1108) mutant worms exhibited increased circularity and size but were shorter compared to that of wild-type worms (Figs 1J and S1A–C)54. In addition, altered DRP-1 expression provoked the apparition of long and thin mitochondrial filaments in between the mitochondrial blebs (Fig. 1C,K) in ~25% of the cells (Fig. 1L)7. Mitochondria of dys-1(cx18);hlh-1(cc561ts);drp-1(tm1108) mutant worms appeared less circular, longer and with the occasional presence of interconnecting mitochondrial strings when compared to those of dys-1(cx18);hlh-1(cc561ts) mutant worms (Figs 1G,J,L and S1A–D), reflecting a rescue of the mitochondrial fragmentation phenotype induced by dystrophin deficiency. Moreover, the index of mitochondrial circularity of dys-1(cx18);hlh-1(cc561ts) mutant worms was significantly decreased in absence of DRP-1 (0,75 +/− 0,014 in dys-1(cx18);hlh-1(cc561ts) mutant vs 0,65 +/− 0,014 in dys-1(cx18);hlh-1(cc561ts);drp-1(tm1108) mutant worms) (Fig. 1J). Similar results were obtained when drp-1 was knock-downed by RNAi (Fig. S1E–I)45. To ensure that the machinery of mitochondrial dynamics was responsible for changes in mitochondrial morphologies upon dystrophin-dependent muscle degeneration, we overexpressed the fusion promoting genes eat-3 or fzo-1. Unexpectedly, in wild-type worms, overexpression of eat-3 increased mitochondrial fragmentation (Fig. 1D) while fzo-1 overexpression had no statistically significant effects on mitochondrial morphology (Fig. 1E). Accordingly, mitochondrial circularity indexes were increased upon eat-3 overexpression and unmodified upon fzo-1 overexpression compared to that of wild-type worms (Fig. 1J). These surprising effects of eat-3 and fzo-1 overexpression have already been observed in C. elegans embryos suggesting that overexpressing fusion machinery in a wild-type background is not sufficient to cause mitochondrial fusion25. Furthermore, EAT-3 may act in both fusion and fission in C. elegans similarly to what was shown in mammals for OPA157. Importantly, both eat-3 and fzo-1 overexpression provoked an apparent increased mitochondrial fusion in dys-1(cx18);hlh-1(cc561ts) mutant worms accompanied by a decreased mitochondrial circularity index when compared to that of dys-1(cx18);hlh-1(cc561ts) mutant worms (0,62 +/− 0,009 in dys-1(cx18);hlh-1(cc561ts);Ex[eat-3] mutant and 0,69 +/− 0,013 in dys-1(cx18);hlh-1(cc561ts);Ex[fzo-1] mutant vs 0,75 +/− 0,014 in dys-1(cx18);hlh-1(cc561ts) mutant worms) (Fig. 1H–J). Overall, absence of the fission protein DRP-1 or increased levels of the fusion proteins EAT-3 and FZO-1 can partially rescue dystrophin-dependent mitochondrial fragmentation in muscles cells. Our data demonstrate that mitochondrial dynamics and its principal key players are impacted by the loss of dystrophin in C. elegans.
DRP-1 induces dystrophin-dependent apoptosis
We next tested whether mitochondrial dynamics could impact muscle cell death in dys-1(cx18);hlh-1(cc561ts) mutant worms. To that extent, we performed TUNEL (Terminal deoxynucleotidyl transferase dUTP nick end labeling) assays along with actin staining on wild-type and dys-1(cx18);hlh-1(cc561ts) mutant worms. TUNEL stains DNA fragments resulting from apoptosis-induced DNA fragmentation58. In wild-type worms, the actin network in muscle cells was nicely and regularly organized in filaments along the diamond-shaped cell with the nuclei, centered along the anterior-posterior axis, showing no staining by TUNEL. By contrast, in dys-1(cx18);hlh-1(cc561ts) mutant worms, muscle cells often presented abnormal phenotypes with actin network perturbations (i.e actin filaments form wavelets and/or aggregates) and with fragments of nuclei displayed over the entire cell and stained by TUNEL (Fig. 1M–T). The number of TUNEL positives muscle cells decreased in dys-1(cx18);hlh-1(cc561ts) mutant worms in absence of the pro-fission protein DRP-1 or when eat-3 and fzo-1 were overexpressed (2.07 +/− 0.19 TUNEL positive cells in dys-1(cx18);hlh-1(cc561ts);drp-1(tm1108) mutant vs 2,4 +/− 0,20 in dys-1(cx18);hlh-1(cc561ts);Ex[eat-3] mutant vs 2,3 +/− 0,21 in dys-1(cx18);hlh-1(cc561ts);Ex[fzo-1] mutant vs 5,28 +/− 0,26 in dys-1(cx18);hlh-1(cc561ts) mutant worms) (Fig. 1U). These data were confirmed using two independent lines overexpressing eat-3 and fzo-1 (Fig. S2). Hence, genetically decreased mitochondrial fission or increased mitochondrial fusion can both reduce apoptosis induced by dystrophin deficiency in muscle.
DRP-1 induces dystrophin-dependent muscle degeneration and locomotion defects
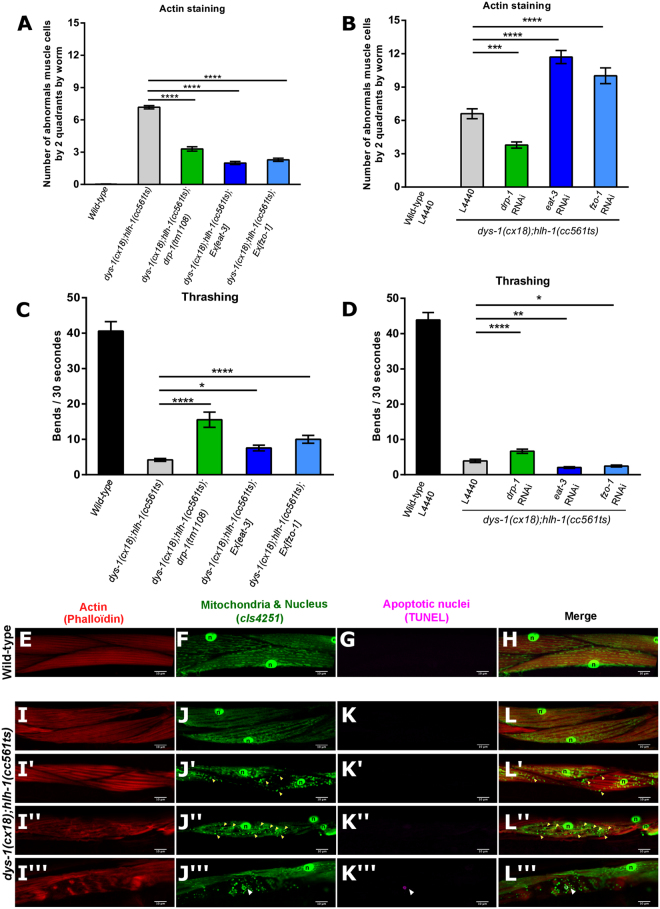
To analyze the impact of mitochondrial dynamics manipulation on muscle health in dys-1(cx18);hlh-1(cc561ts) mutant worms, we further quantified muscle degeneration and estimated muscle activity. In C. elegans, the number of body-wall muscle cells is fixed from one individual to the other (95 muscle cells). By contrast to mammals, C. elegans muscle cells are mononuclear and do not fuse. Moreover, as C. elegans lacks satellite cells and therefore, regenerative capabilities, each muscle cell can be individually followed in vivo throughout its whole life59,60, and muscle degeneration can be quantified by actin staining allowing to visualize and to count abnormal or absent muscle cells. Wild-type and dys-1(cx18) single mutant worms did not present abnormal muscle cells; nor did drp-1(tm1108) mutant worms or eat-3 and fzo-1 overexpressing worms (Figs 2A and S3A)40. hlh-1(cc561ts) mutant worms exhibited a weak number of abnormal muscle cells40. By contrast, dys-1(cx18);hlh-1(cc561ts) mutant worms exhibited a substantial number of abnormal muscle cells (Fig. 2A)40. Interestingly, lack of DRP-1 greatly reduced the number of abnormal muscle cells in dys-1(cx18);hlh-1(cc561ts) mutant worms (7,1 +/− 0,15 abnormal muscle cells by two quadrants in dys-1(cx18);hlh-1(cc561ts) mutant vs 3,3 +/− 0,20 in dys-1(cx18);hlh-1(cc561ts);drp-1(tm1108) mutant worms) (Fig. 2A) similarly to what was observed with drp-1 RNAi (Fig. 2B)45. Both eat-3 and fzo-1 overexpression also reduced the number of abnormal muscle cells of dys-1(cx18);hlh-1(cc561ts) mutant worms (2,0 +/− 0,14 and 2,3 +/− 0,15 abnormal muscle cells by two quadrants in dys-1(cx18);hlh-1(cc561ts);Ex[eat-3] and in dys-1(cx18);hlh-1(cc561ts);Ex[fzo-1] mutant respectively vs 7,1 +/− 0,15 in dys-1(cx18);hlh-1(cc561ts) mutant worms) (Fig. 2A). These data were confirmed using two independent lines overexpressing eat-3 and fzo-1 (Fig. S3A). Conversely, we tested the effects of decreased mitochondrial fusion on muscle cells integrity of dystrophic worms. fzo-1 or eat-3 RNAi treatments had no effect on the actin staining of wild-type animals (data not shown). However, dys-1(cx18);hlh-1(cc561ts) mutant worms fed with either fzo-1 or eat-3 RNAi exhibited enhanced levels of muscle degeneration (number of abnormal muscle cells in dys-1(cx18);hlh-1(cc561ts) mutant worms fed with eat-3 RNAi: 11,7 +/− 0,59 or fzo-1 RNAi: 10,0 +/− 0,70 vs fed with the empty vector L4440: 6,6 +/− 0,44) (Fig. 2B). Collectively, it clearly appears that interfering with the mitochondrial dynamic balance can greatly impact dystrophin-dependent muscle degeneration in C. elegans. Then, we wondered if the protective effects of mitochondrial dynamics on muscle degeneration could lead to positive effects on muscle fitness. To that extent, we quantified worm locomotion with thrashing assays and measured the number of worm bends during 30 seconds swimming sessions. Thrashing of drp-1(tm1108) mutant worms was slightly decreased compared to that of wild-type worms whereas eat-3 and fzo-1 overexpression or eat-3 and fzo-1 RNAi did not affect worm thrashing abilities (Fig. S3B,C). As expected, dys-1(cx18);hlh-1(cc561ts) mutant worms were unable to bend quickly and this poor muscle fitness was partially rescued by lowered DRP-1 levels (Fig. 2C,D). Moreover, eat-3 and fzo-1 overexpression also improved the muscle fitness of dys-1(cx18);hlh-1(cc561ts) mutant worms (Fig. 2C). These data were confirmed using three independent lines overexpressing eat-3 and two independent lines overexpressing fzo-1 (Fig. S3B). By contrast, eat-3 or fzo-1 RNAi could further decrease trashing abilities of dys-1(cx18);hlh-1(cc561ts) mutant worms (Fig. 2D). Together, our data indicate that decreasing mitochondrial fission or increasing mitochondrial fusion can induce a partial functional improvement of muscle cells in dystrophic worms whereas decreasing mitochondrial fusion could worsen muscle degeneration. In conclusion, the main molecular actors of mitochondrial dynamics, namely DRP-1, EAT-3 and FZO-1, appear to be implicated in the establishment of dystrophin-dependent muscle degeneration.
Figure 2.
DRP-1 induces dystrophin-dependent muscle degeneration and locomotion defects. (A) Number of abnormal C. elegans body wall muscle cells by two quadrants quantified by phalloïdin staining in wild-type worms, dys-1(cx18);hlh-1(cc561ts) mutant worms, or dys-1(cx18);hlh-1(cc561ts) mutant worms with drp-1(tm1108) mutation or overexpression of eat-3 or fzo-1 (n = 70 worms at least). (B) Number of abnormal C. elegans body wall muscle cells by two quadrants quantified by phalloïdin staining in dys-1(cx18);hlh-1(cc561ts) mutant worms fed either with the empty vector L4440 or with RNAi constructs targeting drp-1 or eat-3 or fzo-1 (n = 60 worms at least). (C) Quantification of worm trashing in wild-type worms, dys-1(cx18);hlh-1(cc561ts) mutant worms or dys-1(cx18);hlh-1(cc561ts) mutant worms with drp-1(tm1108) mutation or overexpression of eat-3 or fzo-1 (n = 25 worms at least). (D) Quantification of worm trashing in dys-1(cx18);hlh-1(cc561ts) mutant worms fed with the empty vector L4440 or with drp-1 RNAi or eat-3 RNAi or fzo-1 RNAi (n = 100 worms at least). All the experiments were performed on L4 + 3 day-old worms. One-way ANOVA, Tukey’s multiple comparisons test. Data represent the mean obtained by pooling at least three independent assays. Errors bars represent SEM *p < 0.05 **p < 0.01 ***p < 0.001 ****p < 0.0001 ns. indicates that the mean is not statistically significantly different from the mean obtained in the control condition. Representative confocal images of muscles in wild-type worms of: (E) actin network by phalloïdin staining; (F) mitochondrial network and nuclei (n) organization visualized with the ccIs4251 transgene in C. elegans body wall muscle cells; (G) apoptotic nuclei visualized by TUNEL staining and (H) the merge of the phalloidin, mitochondrial and TUNEL staining. (I- L”’) Representative confocal images of body wall muscle cells of dys-1(cx18);hlh-1(cc561ts) mutant worms presenting different degrees of muscle degeneration. (I-I”’) actin network by phalloïdin staining; (J-J”’) mitochondrial network visualized with the ccIs4251 transgene in C. elegans body wall muscle cells; (K-K”’) apoptotic nuclei visualized by TUNEL staining; (L-L”’) merge of the phalloidin, mitochondrial and TUNEL staining. All the experiments were performed in L4 + 3 day-old worms. Yellow arrows indicate circular mitochondria. White arrow indicates apoptotic nuclei. Scale bar 10 μm. n indicates nuclei. (x630).
Alterations of the mitochondrial network are early events of dystrophin-dependent muscle degeneration
We next wondered whether mitochondrial dynamics perturbations or apoptosis appears first in dystrophin-dependent muscle degeneration. We performed actin and TUNEL staining on dys-1(cx18);hlh-1(cc561ts) mutant or wild-type worms carrying the ccIs4251 transgene. In wild-type worms, tubular and circular mitochondria were in equal proportions and organized along the actin network, which is arranged in filaments along the cell (Fig. 2E–H) and we could not detect any TUNEL positive cell (Fig. 2G). In dys-1(cx18);hlh-1(cc561ts) mutant worms, different stages of actin network perturbations were observed, likely reflecting various degrees of muscle degeneration. The actin staining can reveal an apparent totally preserved actin network or the apparition of wavelets or a more or less noticeable aggregation of actin. Muscle cells with mutated dystrophin and unperturbed actin network (Fig. 2I-I’) could be associated with either wild-type-like mitochondria (Fig. 2J) or mitochondria that appeared to be fragmented (Fig. 2J’) and we could not detect any TUNEL staining under these conditions (Fig. 2K-K’). It is noteworthy that weak actin perturbations (Fig. 2I”) were always associated with increased mitochondrial circularity in dys-1(cx18);hlh-1(cc561ts) mutant worms (Fig. 2J”) but without any signs of apoptosis (Fig. 2K”). Finally, strong actin perturbations due to mutated dystrophin (Fig. 2I”’) were concomitant with circular mitochondria (Fig. 2J”’) and TUNEL positive staining (Fig. 2K”’). Together, these observations suggest that mitochondrial dynamics dysfunction is an early event in the molecular mechanisms leading to progressive muscle degeneration. Changes in mitochondrial morphologies are then followed by actin network perturbations and will eventually lead to apoptosis.
Cleavage of DRP-1 by CED-3 is required for dystrophin-dependent muscle degeneration but is dispensable for regulating mitochondrial fission
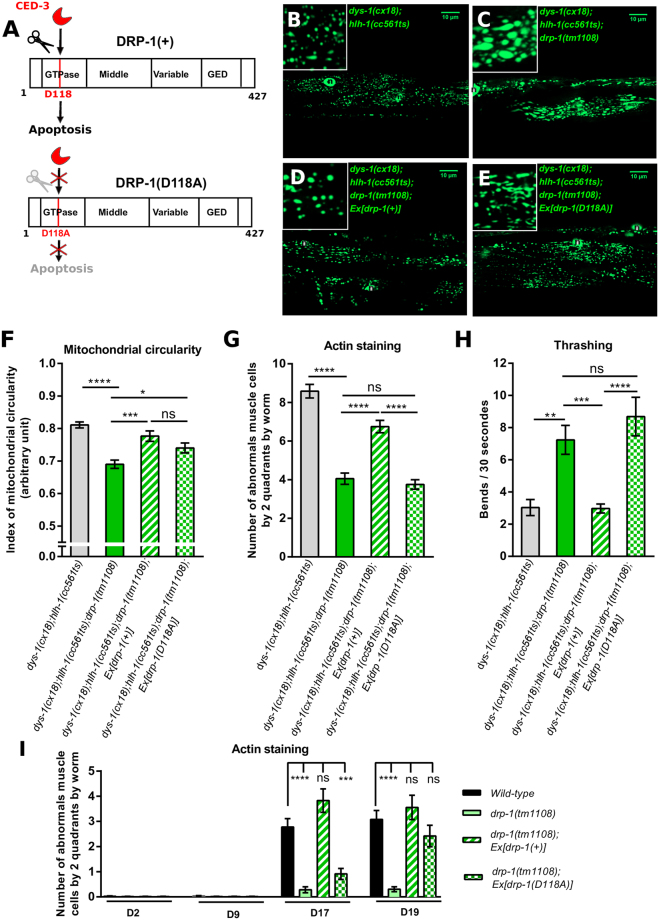
The links between mitochondrial dynamics and cell death have been extensively studied16,31,56,61,62. In C. elegans, the importance and the exact roles of DRP-1 in developmental apoptosis are still controversial31,56. A fraction of DRP-1 can be cleaved in vitro by the caspase CED-3, leading to the formation of DRP-1 complexes containing both, full-length DRP-1 and DRP-1119–712, which are suspected to play a role in apoptosis during worm development. This cleavage by CED-3 may redirect DRP-1 activity towards apoptotic mitochondrial elimination in cell corpses31. Importantly, CED-3 cleavage of DRP-1 is unlikely to be implicated in mitochondrial fission31 suggesting that the roles of DRP-1 in mitochondrial dynamics and cell death might be distinguished. We wondered whether the role of DRP-1 in cell death is required for muscle degeneration in a dystrophin-deficient background. To this end, we used two genetic constructions of DRP-1: (i) a wild-type drp-1 transgene called drp-1(+) and (ii) a transgene of drp-1 with the CED-3 cleavage site mutated called drp-1(D118A) (Fig. 3A). The presence of this mutation prevents the cleavage of DRP-1 by CED-3 rendering DRP-1 presumably unable to act in cell death31. We compared the indexes of mitochondrial circularity of dys-1(cx18);hlh-1(cc561ts) and dys-1(cx18);hlh-1(cc561ts);drp-1(tm1108) mutant worms expressing, or not, drp-1(+) or drp-1(D118A) transgenes. As shown above, dys-1(cx18);hlh-1(cc561ts) mutant worms presented fragmented mitochondria (Figs 1F and 3B) with a high index of mitochondrial circularity that can be lowered in absence of DRP-1 (Figs 1G,J and 3C,F). dys-1(cx18);hlh-1(cc561ts);drp-1(tm1108) mutant worms expressing DRP-1(+) or DRP-1(D118A) showed important mitochondrial fragmentation, loss of the mitochondria bubbles caused by the drp-1(tm1108) mutation7 and an increased index of mitochondrial circularity compared to that of dys-1(cx18);hlh-1(cc561ts);drp-1(tm1108) mutant worms (Fig. 3D–F). These results demonstrate that lack of DRP-1 is responsible for the particular mitochondrial pattern exhibited by dys-1(cx18);hlh-1(cc561ts);drp-1(tm1108) mutant worms and that both drp-1(+) and drp-1(D118A) transgene are able to induce mitochondrial fission. Our data confirmed that cleavage of DRP-1 by CED-3 is at least in part dispensable for its function in regulating mitochondrial fission.
Figure 3.
Cleavage of DRP-1 by CED-3 is required for dystrophin-dependent muscle degeneration but is dispensable for regulating mitochondrial fission. (A) Schematic representation of the wild-type DRP-1 construct (DRP-1(+)) and of the DRP-1 construct with CED-3 cleavage site mutated due to a change of amino acid D118A (DRP-1(D118A)). The wild-type DRP-1 construct DRP-1(+) is supposed to play a role in mitochondrial dynamics and in apoptosis, whereas the DRP-1 construct with D118A mutation DRP-1(D118A) is likely to be dispensable for apoptosis31. Representative confocal images of the mitochondrial network organization visualized with the ccIs4251 transgene in C. elegans body wall muscle cells of: (B) dys-1(cx18);hlh-1(cc561ts) mutant worms; (C) dys-1(cx18);hlh-1(cc561ts);drp-1(tm1108) mutant worms; (D) dys-1(cx18);hlh-1(cc561ts);drp-1(tm1108) mutant worms expressing drp-1(+) and (E) dys-1(cx18);hlh-1(cc561ts);drp-1(tm1108) mutant worms expressing drp-1(D118A). Scale bar 10 μm. (x630). n indicates nuclei. (F) Quantification of mitochondrial circularity in each of the indicated strains (n = 30 worms at least). (G) Number of abnormal C. elegans body wall muscle cells by two quadrants quantified by phalloïdin staining in each of the indicated strains (n = 60 worms). (H) Quantification of worm trashing in each of the indicated strains (n = 45 worms at least). All the experiments were performed on L4 + 3 day-old worms. One-way ANOVA, Tukey’s multiple comparisons test. (I) Number of abnormal C. elegans body wall muscle cells by two quadrants quantified by phalloïdin staining in wild-type worms; drp-1(tm1108) mutant worms; drp-1(tm1108) mutant worms expressing drp-1(+); drp-1(tm1108) mutant worms expressing drp-1(D118A) at day 2, 9, 17 and 19 of adulthood of the nematode (n = 63 worms at least). Student test. Data represent the mean obtained by pooling at least three independent assays. Errors bars represent SEM *p < 0.05 **p < 0.01 ***p < 0.001 ****p < 0,0001 ns. indicates that the mean is not statistically significantly different from the mean obtained in control worms.
We next tested the effects of expressing drp-1(+) or drp-1(D118A) transgenes on muscle degeneration and fitness of dys-1(cx18);hlh-1(cc561ts);drp-1(tm1108) mutant worms. As shown above, dys-1(cx18);hlh-1(cc561ts) mutant worms exhibited an important number of abnormal muscle cells and weak thrashing abilities (Figs 2A,C and 3G,H) and both phenotypes were attenuated by the absence of DRP-1 (Figs 2A,C and 3G,H). The introduction of drp-1(+) transgene into dys-1(cx18);hlh-1(cc561ts);drp-1(tm1108) mutant worms provoked an increased number of abnormal muscle cells and a drastic thrashing decrease (6,75 +/− 0,32 abnormal muscle cells and 2,9+/−0,28 bends/30 seconds in dys-1(cx18);hlh-1(cc561ts);drp-1(tm1108);Ex[drp-1(+)] mutant vs 8,59 +/− 0,35 abnormal muscle cells and 7,4 +/− 0,90 bends/30 seconds in dys-1(cx18);hlh-1(cc561ts);drp-1(tm1108) mutant worms) (Fig. 3G,H). This demonstrates that DRP-1 takes active part in both the muscle cells integrity and movement defects phenotypes of dys-1(cx18);hlh-1(cc561ts) mutant worms. We hypothesized that if drp-1(D118A) construct is unable to increase hallmarks of muscle degeneration in the dys-1(cx18);hlh-1(cc561ts);drp-1(tm1108) mutant worms that would indicate that the role of DRP-1 in cell death is required for dystrophin-dependent muscle degeneration. We found that introducing drp-1(D118A) construct into dys-1(cx18);hlh-1(cc561ts);drp-1(tm1108) mutant worms affected neither the number of abnormal muscle cells nor thrashing (Fig. 3G,H). Similar results were obtained with at least two independent lines expressing DRP-1(D118A) (Fig. S4A,B). The mutated form of DRP-1 is unable to counteract the positive effects of the absence of DRP-1 on the muscle degeneration of dys-1(cx18);hlh-1(cc561ts) mutant worms. This strongly suggests that caspase cleavage of DRP-1 is required for dystrophin-dependent muscle degeneration in C. elegans.
Cleavage of DRP-1 by CED-3 is required for age-dependent muscle degeneration
Physiological processes such as aging can also induce a type of muscle degeneration called sarcopenia. Sarcopenia, which results in the gradual loss of skeletal muscle mass in the elderly, is a major factor determining the decline in overall health and loss of autonomy of the aging population. In wild-type C. elegans, we revealed the spontaneous apparition of more than two abnormal muscle cells by muscle quadrant at days 17 and 19 of adulthood (Fig. 3I) accompanied by a gradual loss of locomotion as the animal ages (Fig. S5). The drp-1(tm1108) mutant worms showed impaired mobility early in adulthood life, which could be explained by energy production impairment63,64. Importantly, in absence of DRP-1, the apparition of abnormal muscle cells, over aging, was a very rare event (Fig. 3I). Yet, locomotion defects due to age cannot be improved by the absence of DRP-1. Our data demonstrate that DRP-1 is involved in both dystrophin-dependent and age-dependent muscle degeneration but is unlikely to be the key factor for locomotion defects that are observed over aging. To understand if the apoptotic functions of DRP-1 are important for age-dependent muscle degeneration as it is for dystrophin-dependent muscle degeneration, we analyzed drp-1(tm1108) mutant worms expressing either the wild-type drp-1 transgene (drp-1(+)) or the drp-1 transgene with the CED-3 cleavage site mutated (drp-1(D118A)) supposed to be inefficient in apoptosis (Fig. 3A). At day 17 and 19 of adulthood, drp-1(tm1108) mutant worms with drp-1(+) transgene exhibited apparition of abnormal muscle cells to the same extent as wild-type worms, demonstrating a role for DRP-1 in age-related muscle degeneration. At day 17 of adulthood, drp-1(tm1108) mutant worms expressing the drp-1(D118A) transgene presented very few abnormal muscle cells (Fig. 3I). By contrast, at day 19 of adulthood, drp-1(tm1108) mutant worms expressing the drp-1(D118A) transgene exhibited nearly the same level of muscle degeneration than wild-type worms (Fig. 3I). Collectively, our data suggest a role for DRP-1 through apoptotic-like processes in both dystrophin-dependent and age-related muscle degeneration.
The canonical apoptosis pathway mediates dystrophin-dependent muscle degeneration
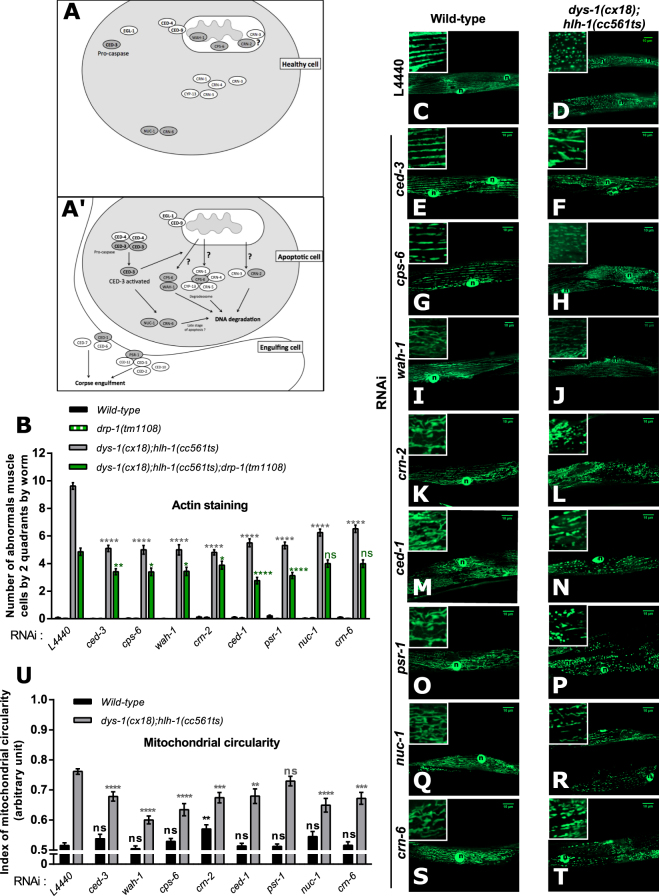
Having established that the functions of DRP-1 in apoptosis play a key role in dystrophin-dependent muscle degeneration, we asked whether other executors of apoptosis could be important. For this, we performed RNAi experiments against several executors of cell death and tested the effects of each RNAi constructs on mitochondrial morphology and the number of abnormal cells in dys-1(cx18);hlh-1(cc561ts) mutant worms. We thus down-regulated CED-3, required for the execution of apoptosis via its activation by CED-4 (APAF-1 homolog in mammals) and for DRP-1 to act in apoptosis (Fig. 4A-A’)65,66. We have also tested the effects of RNAi-mediated down-regulation of five actors implicated in the three sub-pathways involved in DNA degradation. First, WAH-1 (AIF homolog in mammals) and CPS-6 (EndoG homolog in mammals) which, upon cell death signal, are released from mitochondria and can interact together to translocate to the nucleus where they participate to DNA degradation67. CPS-6 can also interact with CRN-1, CRN-4, CRN-5 and CYP-13 to form the degradeosome68. Second, CRN-2 likely to encode an homolog of the TatD nuclease (TATDN1 homolog in mammals) and predicted to be mitochondrial68. CRN-2 acts in the same pathway as CRN-368. Third, NUC-1 and CRN-6 (two DNase II homologs in mammals) that seem involved in the degradation of DNA debris from apoptotic cells in late stage of apoptosis68,69. Concerning corpse engulfment, important for the phagocytic process70, we have tested the effects of knocking-down CED-1, which is involved in cell-corpse recognition with CED-6 and CED-771 and PSR-1, which participates in the migration of engulfing cell72,73.
Figure 4.
Genetic manipulation of apoptosis impacts muscle degeneration and mitochondrial dynamics of dys-1(cx18);hlh-1(cc561ts) mutant worms. Simplified schematic of the sub-cellular localization, in healthy (A) and apoptotic (A’) cells, of the main apoptotic factors involved in the inducing and execution phases of apoptosis in C. elegans. Gene names in dark gray indicate the genes, which knock-downs by RNAi, were tested on different phenotypes of dys-1(cx18);hlh-1(cc561ts) mutant and wild-type worms. (B) Number of abnormal C. elegans body wall muscle cells by two quadrants quantified by phalloïdin staining in wild-type worms or drp-1(tm1108) mutant worms or dys-1(cx18);hlh-1(cc561ts) mutant worms or dys-1(cx18);hlh-1(cc561ts);drp-1(tm1108) mutant worms fed either with the empty vector L4440 or with the indicated RNAi against executors of cell death (n = 90 worms at least). Representative confocal images of the mitochondrial network organization visualized with the ccIs4251 transgene in C. elegans body wall muscle cells of: (C) wild-type worms or (D) dys-1(cx18);hlh-1(cc561ts) mutant worms fed with empty vector L4440; of (E) wild-type worms or (F) dys-1(cx18);hlh-1(cc561ts) mutant worms fed with ced-3RNAi; of (G) wild-type worms or (H) dys-1(cx18);hlh-1(cc561ts) mutant worms fed with cps-6 RNAi; of (I) wild-type worms or (J) dys-1(cx18);hlh-1(cc561ts) mutant worms fed with wah-1 RNAi; of (K) wild-type worms or (L) dys-1(cx18);hlh-1(cc561ts) mutant worms fed with crn-2 RNAi; of (M) wild-type worms or (N) dys-1(cx18);hlh-1(cc561ts) mutant worms fed with ced-1 RNAi; of (O) wild-type worms or (P) dys-1(cx18);hlh-1(cc561ts) mutant worms fed with psr-1 RNAi; of (Q) wild-type worms or (R) dys-1(cx18);hlh-1(cc561ts) mutant worms fed with nuc-1 RNAi and of (S) wild-type worms or (T) dys-1(cx18);hlh-1(cc561ts) mutant worms fed with crn-6 RNAi. Scale bar 10 μm. (x630). n indicates nuclei. (U) Quantification of mitochondrial circularity in each of the indicated conditions (n = 27 worms at least). Note that quantitative PCR data showed that nuc-1 and crn-6 mRNA expression were reduced, in all strains, by ~50% and 70% upon nuc-1 and crn-6 RNAi treatments, respectively (Fig. S6). All the experiments were performed on L4 + 3 day-old worms. One-way ANOVA, Dunnett’s multiple comparisons test. Data represent the mean obtained by pooling at least three independent assays. Errors bars represent SEM **p < 0.01 ***p < 0.001 ****p < 0.0001 ns. indicates that the mean is not statistically significantly different from the mean obtained in control conditions.
First, we performed actin staining and counted the number of abnormal muscles cells in dys-1(cx18);hlh-1(cc561ts) mutant worms fed with bacteria expressing either the empty vector L4440 or RNAi constructs against ced-3 or each of the above described cell death executors tested. ced-3 RNAi treatment dramatically decreased the number of abnormal muscle cells compared to that observed in dys-1(cx18);hlh-1(cc561ts) mutant worms fed with the empty vector L4440 (5,1 +/− 0,22 abnormal muscle cells for dys-1(cx18);hlh-1(cc561ts) fed with ced-3 RNAi vs 9,63 +/− 0,24 abnormal muscle cells for dys-1(cx18);hlh-1(cc561ts) fed with L4440) (Fig. 4B) confirming that CED-3 plays an important role in dystrophin-dependent muscle degeneration. Interestingly, degenerin-dependent muscle degeneration in C. elegans also depends on CED-374. All the RNAi against executors of cell death tested provoked a decrease in the number of abnormal muscle in dys-1(cx18);hlh-1(cc561ts) mutant worms (dys-1(cx18);hlh-1(cc561ts) mutant worms fed with: cps-6 RNAi: 5,01 +/− 0,32 abnormal muscle cells; wah-1 RNAi: 5,00 +/− 0,37 abnormal muscle cells; crn-2 RNAi: 4,81+/0,19 abnormal muscle cells; nuc-1 RNAi: 6,25 +/− 0,25 abnormal muscle cells; crn-6 RNAi: 6,53 +/− 0,26 abnormal muscle cells; ced-1 RNAi: 5,50 +/− 0,29 abnormal muscle cells; psr-1 RNAi: 5,31 +/− 0,24 abnormal muscle cells vs dys-1(cx18);hlh-1(cc561ts) mutant worms fed with the empty vector L4440: 9,63 +/− 0,24 abnormal muscle cells) (Fig. 4B). Our data indicates that the canonical apoptotic pathway, which has been extensively described in developmental apoptosis75, also participates in cell death upon dystrophin-muscle degeneration.
Then, we wondered whether DRP-1 was required for CED-3 and others cell death executors to impact dystrophin-dependent muscle degeneration. For that, we tested the effects of the different RNAi presented above on muscle degeneration of dys-1(cx18);hlh-1(cc561ts);drp-1(tm1108) mutant worms. The effects of ced-3 RNAi and of the drp-1(tm1108) mutation on muscle cells of dys-1(cx18);hlh-1(cc561ts) were cumulative indicating that CED-3 probably has an additional role in dystrophin-dependent muscle degeneration, which is DRP-1-independent. Moreover, knock-down of ced-1 and psr-1, involved in corpse engulfment, could also further decrease the muscle degeneration of dys-1(cx18);hlh-1(cc561ts);drp-1(tm1108) mutant worms (dys-1(cx18);hlh-1(cc561ts) mutant worms fed with: ced-1 RNAi: 2,77 +/− 0,24 abnormal muscle cells; psr-1 RNAi: 3,13 +/− 0,22 abnormal muscle cells vs dys-1(cx18);hlh-1(cc561ts) mutant worms fed with the empty vector L4440: 4,86 +/− 0,26 abnormal muscle cells) (Fig. 4B). Similarly, knock-down of three genes that are involved in DNA degradation (cps-6, wah-1 and crn-2) could further decrease muscle degeneration of dys-1(cx18);hlh-1(cc561ts);drp-1(tm1108) mutant worms fed with the empty vector L4440 (dys-1(cx18);hlh-1(cc561ts) mutant worms fed with: cps-6 RNAi: 2,33 +/− 0,29 abnormal muscle cells; wah-1 RNAi: 3,68 +/− 0,33 abnormal muscle cells; crn-2 RNAi: 3,88 +/− 0,29 abnormal muscle cells vs dys-1(cx18);hlh-1(cc561ts) mutant worms fed with L4440: 4,86 +/− 0,26 abnormal muscle cells) (Fig. 4B). By contrast, NUC-1 and CRN-6, final actors of DNA degradation during the apoptotic process, appeared to act in the same pathway and downstream of DRP-1 as their inactivation had no significant effects on muscle degeneration of dys-1(cx18);hlh-1(cc561ts);drp-1(tm1108) mutant worms although nuc-1 and crn-6 RNAi decreased muscle degeneration in dys-1(cx18);hlh-1(cc561ts) mutant worms (Fig. 4B). None of the tested RNAi treatments had an effect on the muscle degeneration of wild-type or drp-1(tm1108) mutant worms (Fig. 4B). Based on these data, we propose two hypothesis: (i) DRP-1 may act in parallel of the DNA degradation pathway in dystrophin-dependent muscle cell death, (ii) DRP-1 may act upstream of the DNA degradation pathways.
We then tested the effect of cell death executors on mitochondrial dynamics. None of the RNAi tested (except for crn-2 RNAi) had an effect on mitochondrial morphologies in wild-type worms carrying the ccIs4251 transgene (Fig. 4C–U). In a dys-1(cx18);hlh-1(cc561ts) mutant background, all the tested RNAi, with exception of psr-1 RNAi (Fig. 4O,P), diminished mitochondrial fragmentation and circularity indexes when compared to mutant worms fed with empty vector (index of mitochondrial circularity in dys-1(cx18);hlh-1(cc561ts) mutant worms fed with: wah-1 RNAi: 0,60 +/− 0,013; cps-6 RNAi: 0,63 +/− 0,020; crn-2 RNAi: 0,67 +/− 0,017; nuc-1 RNAi: 0,64 +/− 0,022; crn-6 RNAi: 0,67 +/− 0,019; ced-1 RNAi: 0,68 +/− 0,024 vs dys-1(cx18);hlh-1(cc561ts) mutant worms fed with the empty vector L4440: 0,76 +/− 0,009) (Fig. 4C–U). Thus, decreasing the expression of apoptotic factors can partially rescue mitochondrial circularity of dys-1(cx18);hlh-1(cc561ts) mutant worms.
Discussion
Apoptotic muscle fibers have been frequently associated with Duchenne Muscular Dystrophy76–79 and sarcopenia80–83 in various species including Human. However, the exact molecular mechanisms leading to muscle apoptosis upon dystrophin deficiency or aging remain unclear. Here, we revealed a DRP-1-dependent apoptotic mechanism that participates in dystrophin-dependent and age-dependent muscle degeneration. DRP-1 is the principal and universal actor of mitochondrial fission. In mammals, mitochondrial fragmentation induced by DRP-1 can trigger apoptosis84. In C. elegans, a previous study suggested a marginal role for DRP-1 in apoptosis during development using the mutation drp-1(tm1108)31, whereas another study showed that the dominantly interfering mutation drp-1(K40A) caused mitochondria to form large blebs and induced excessive apoptosis in embryos56. Our study reveals that DRP-1 can play a key role in apoptosis that takes place during cellular stresses provoked either by the loss of dystrophin function or by aging. Our data also demonstrate that EAT-3 and FZO-1 overexpression in dystrophin deficient worms can reduce both mitochondrial fragmentation and muscle degeneration. A recent study showed that in mice, muscle-specific deletion of OPA1 (EAT-3 homolog in C. elegans) induces a precocious aging phenotype with muscle loss and weakness85 and a reduced muscle mass has been also reported in mice ablated for both Mfn1 (FZO-1 homolog in C. elegans) and Mfn285. These observations are nicely corroborating our findings and the fact that levels of mitochondrial fusion greatly impact muscle degeneration. Our data also suggest that manipulating mitochondrial fusion is sufficient to decrease apoptosis in dystrophic worms. In C. elegans, the direct links between mitochondrial dynamics and developmental apoptosis are highly controversial25,26; our study suggests that, upon cellular stress such as dystrophin deficiency, the major actors of mitochondrial dynamics in worms, namely DRP-1, EAT-3 and FZO-1, can directly interplay with the apoptotic pathways.
We also showed that DRP-1 cleavage by the pro-caspase CED-3 is required for dystrophin-dependent and age-dependent muscle degeneration. Hence, our study demonstrates that DRP-1 sits downstream of CED-3 and strongly suggests that DRP-1 acts mostly via apoptosis rather than through mitochondrial fission to induce muscle cell death. In mammals, DRP-1-induced outer membrane fragmentation allows for release of mitochondrial pro-apoptotic factors such as cytochrome c, EndoG (CPS-6 in worms) or AIF (WAH-1 in worms)13–15,86. In C. elegans muscle cells, cytochrome c could be released from mitochondria to the cytosol during muscle degeneration45,74 but its role in activating APAF-1 (CED-4 homolog in worms) to form the apoptosome is unlikely to be conserved87,88. Strong evidence suggests that both WAH-1 (AIF homolog in mammals) and CSP-6 (EndoG homolog in mammals) can be released from the mitochondria to the nucleus to degrade DNA upon apoptotic stimuli in C. elegans67,89. Our data emphasize a function for WAH-1/CSP-6 in dystrophin-dependent muscle degeneration potentially downstream of DRP-1. One possibility is that DRP-1, once activated by CED-3 induces mitochondrial outer membrane permeabilization that will, in turn, allows for WAH-1/CSP-6 release from the mitochondria and translocation into the nucleus. However, we cannot exclude that DRP-1 acts in a parallel pathway of WAH-1/CSP-6. Our data strongly suggest that DRP-1 does not participate in corpse engulfment during apoptosis and is likely to act upstream of NUC-1 and CRN-6. Collectively, our data converge toward a mechanism that underlies DRP-1-dependent mitochondrial pathways activated downstream of CED-3 to impact dystrophin-dependent muscle degeneration. The cleavage of DRP-1 by caspase CED-3 seems to be restricted to C. elegans31 suggesting that it could be an ancestral role of CED-3 that was lost over evolution. However, we also found that diminution of CED-3 expression can decrease muscle degeneration of a dystrophin C. elegans mutant in a DRP-1-independent manner. Our data are consistent with the fact that maintenance of C. elegans muscle integrity during aging depends on CED-390. Interestingly, skeletal muscles of aged rats exhibit a high level of activated caspase-391 and muscles of Duchenne Muscular Dystrophy patients present an increase of both caspase-3 expression92 and activity93. Collectively, the role of the caspase-3 in muscle degeneration appears to be likely conserved among species.
Having established that DRP-1 acts mostly in apoptosis to impact muscle degeneration upon dystrophin deficiency, it would be interesting to investigate whether others molecular mechanisms than cleavage by caspase-3 can regulate DRP-1 activity94. Control of intracellular concentration of Ca2+ ([Ca2+]i) homeostasis is critical for the maintenance of muscle contraction and relaxation. Elevated [Ca2+]i increases production of reactive oxygen species (ROS)95, activates calpain96, impairs autophagy97, increases mitochondrial Ca2+ accumulation96 and induces cell death98,99. Intriguingly, a drastic increase in the [Ca2+]i in myofibers and myotubes deficient in dystrophin was observed in several studies100,101. In mdx mice, abnormal elevation of [Ca2+]i is due to SERCA (sarco(endo)plasmic reticulum Ca2+ ATPase) inhibition suggesting an importance of the ER-dependent regulation of Ca2+ in Duchenne Muscular Dystrophy102,103. Moreover, muscle degeneration of the dys-1(cx18);hlh-1(cc561ts) mutants worms is likely to be a calcium-dependent104,105. Calcium is also a well-known activator of DRP-1. For instance, ER-calcium release induces uptake of calcium by mitochondria and DRP-1 activation allowing for calcium-dependent mitochondrial fission106. Calcineurin, a calcium-dependent phosphatase, dephosphorylates DRP1 and promotes mitochondrial fragmentation and cell vulnerability to apoptosis107,108. In neurons of C. elegans, phosphorylation of DRP-1 by Ca/calmodulin-dependent kinase II (CaMKII) inhibits DRP-1 activity109. One possibility is that the loss of dystrophin function could induce changes in intracellular calcium that would activate DRP-1 and cell death.
In conclusion, our findings point to mitochondrial dynamics as an early signaling hub that controls cell death in dystrophin-dependent muscle degeneration. A better understanding of the role of DRP-1 in muscle degeneration could allow in the long term to identify new targets for treatments of muscular dystrophies or to improve muscle degeneration in the elderly.
Materials and Methods
Strains
Most of the strains used were obtained from the Caenorhabditis Genetic Center with the exception of the drp-1(tm1108) strain provided by the National Bioresource Project for the Nematode to Sylvia Lee’s lab (Cornell University-Ithaca, NY-USA). N2 was used as the wild-type (WT) strain. The dys-1(cx18);hlh-1(cc561ts)40, dys-1(cx18);hlh-1(cc561ts);drp-1(tm1108) and the dys-1(cx18);hlh-1(cc561ts);ccIs4251 strains were constructed using standard genetic methods. The drp-1(tm1108) strain, which has no detectable DRP-1 protein expression, is likely to represent a strong loss-of-function or null mutant31. The integrated ccIs4251 transgene contains Pmyo-3::Ngfp-lacZ (nuclear localization of GFP) and Pmyo-3::Mtgfp (mitochondrial localization of GFP)55.
All strains were maintained at 15 °C on standard NGM (Nematode Growth Medium) agar plates seeded with Escherichia coli strain OP50 or with HT115 bacteria for RNAi experiments.
Generation of drp-1, eat-3, fzo-1 constructs and transgenic lines
All constructs were verified by sequencing and restriction fragment length. Sequences of the primers used for PCR are available upon request.
drp-1(+) transgene under control of endogenous drp-1 promoter
A first 4.3 Kbp genomic fragment containing the 950 bp sequence upstream and the 2.5 Kbp coding region, and the 1.1 Kbp sequence downstream of the drp-1 coding region was amplified from genomic DNA by PCR. A second 3.6 Kbp genomic fragment containing the 650 bp sequence upstream, the 2.5 Kbp coding region, and the 490 bp sequence downstream of the drp-1 coding region was amplified from the first 4.3 Kbp fragment. Gibson Assembly® Master Mix and KpnI enzyme was used to introduce this second fragment in pBSC vector.
drp-1(D118A) under control of endogenous drp-1 promoter
A first 4.3 Kbp genomic fragment containing the 950 bp sequence upstream, the 2.5 Kbp coding region, and the 1.1 Kbp sequence downstream of the drp-1 coding region was amplified from genomic DNA by PCR. Two fragments of 1050 pb and 2640 pb with overlapping end in drp-1 containing a mutation (A > G) on the CED-3 cleavage site of DRP-1 were amplified by PCR from the first 4.3 Kbp fragment. A 3.6 Kpb fragment containing the 650 bp sequence upstream, the 2.5 Kbp coding region with a mutation on the CED-3 cleavage site of DRP-1, and the 490 bp sequence downstream of the drp-1 coding region was amplified from the two fragments described above by Fusion-PCR. Gibson Assembly® Master Mix and KpnI enzyme was used to introduce this second fragment in pBSC vector.
eat-3(+) transgene under control of endogenous eat-3 promoter
First, two genomic fragments of 2.9 Kbp and 3.3 Kpb containing the 701 bp sequence upstream, the 4.7 Kbp coding region, and the 474 bp sequence downstream of the eat-3 coding region were amplified from genomic DNA by PCR. Secondly, two fragments of 2.3 Kpb and 2.8 Kpb with overlapping end in eat-3 were amplified by PCR from the first two fragments described above. A 5.1 Kpb fragment containing the 182 bp sequence upstream, the 4.7 Kbp coding region, and the 216 bp sequence downstream of the eat-3 coding region was amplified from the two fragments described above by Fusion-PCR.
fzo-1(+) transgene under control of endogenous fzo-1 promoter
A first 3.8 Kbp genomic fragment containing the 163 bp sequence upstream, the 2.8 Kbp coding region, and the 762 bp sequence downstream of the fzo-1 coding region was amplified from genomic DNA by PCR. A second 3.6 Kbp genomic fragment containing the 113 bp sequence upstream, the 2.8 Kbp coding region, and the 702 bp sequence downstream of the fzo-1 coding region was amplified from the first fragment described above.
For generation of transgenic animals: 10 ng/μl of transgene (drp-1(+) or drp-1(D118A) or eat-3(+) or fzo-1(+)); 2,5 ng/μl of the co-injection marker pCFJ90 (Pmyo-2::MCherry::unc-54utr) and 116,5 ng/μl Bluescript plasmid filler DNA were microinjected into the gonads of adult wild-type, dys-1(cx18);hlh-1(cc561ts);drp-1(tm1108) or dys-1(cx18);hlh-1(cc561ts) mutant worms using standard methods110. F1 progeny were selected on the basis of MCherry fluorescence. Individual F2 animals were isolated to establish independent lines.
RNA interference
RNAi bacteria clones from the commercial C. elegans RNAi collection (Ahringher laboratory-Gene Service Inc) were cultured overnight in LB medium supplemented with 100 mg/ml ampicillin and 12.5 mg/ml tetracycline (LBAT) at 37 C. The day after, 0.01 volume of this pre-culture was added in 1 volume of LBAT and incubated at 37 C. At 0.6–0.8 DO, bacteria were concentrated 5 times and then 300 μL of the bacteria culture was seeded on NGM plates containing ampicillin (100 ug/mL) and tetracycline (12,5 μg/mL). Plates were allowed to dry at room temperature for at least 3 days. Then, plates were stored at 4 C during 2 months utmost. The day before the experiment, 4 mM IPTG was added to induce bacteria overnight at room temperature. Gravid worms were allowed to lay eggs during 6 hours on NGM plates seeded with RNAi bacteria carrying specific dsRNA-expressing bacteria and progeny grew on RNAi plates until the day of experiment. Unless indicated differently, experiments were performed at the age of 3 days after the L4 larval stage (L4 + 3 days). As RNAi efficiency control, dpy-13 RNAi was performed in parallel and resulted in nearly 100% dumpy. As RNAi-negative control, plates seeded with HT115 carrying the empty vector RNAi L4440 were used.
Phalloidin staining
Phalloidin staining was carried out as described previously111. Synchronized adult (day 2, 9, 17 and 19 of adulthood) animals were fixed in 1 ml of PBS supplemented with 20 μl of 37% formaldehyde, extracted with acetone at −20 °C, and incubated in 3U of phalloidinAlexa Fluor® 488 (Molecular Probes) for 2 h. Imager Z1 microscope was used for microscopic observations. Only the two most visible quadrants of each animal were counted for the quantification of muscle degeneration.
For aging experiments, 1.3 μg/mL 5-FU (5-Fluorouracil, Sigma) was added when the worms reached the L4 stage.
Thrashing assay
L4 worms were transferred to a plate during 3 days. In the case of transgenic lines, only the MCherry-fluorescent L4 worms (expressing the co-injection marker Pmyo-2::mCherry::unc-54utr) were transferred. Synchronized adult animals (day 2, 9, 17 and 21 of adulthood), were moved onto a fresh 2% agarose 12 wells-plates containing M9 bufferpre-equilibrated at a temperature of 15 °C. Thrashing frequency was measured during the swimming sessions in the liquid. Videos of animal movements were acquired using a Macro Zoom microscope. The videos were recorded for 30 seconds at 30 frames per second. The thrashing frequency of the worms, measured in body bends per 30 seconds, was quantified using the open-source wrMTrck plugin for Fiji software112 (http://www.phage.dk/plugins/wrmtrck.html). One body bend was defined as a change indirection of bending at the midbody. For aging experiments, 1.3 μg/mL 5-FU (5-Fluorouracil, Sigma) was added when the worms reached the L4 stage.
Mitochondrial morphology analysis
Gravid adults with integrated ccIs4251 transgene were allowed to lay eggs on NGM plates for 6 hours before being removed from the plate. L4 + 3 day-old progeny worms were immobilized in 3.3 mM levamisole on a 2% agarose pad for acquisition. Confocal images of the vulva area of lived paralyzed worms were taken on a Zeiss LSM 510 Meta or Zeiss LSM 800 using a 63X oil objective. The percentage of muscle cells exhibiting interconnecting filaments in between mitochondria blebs was calculated after observation, with the unaided human eye of the experimenter, of each muscle cells with an increase of brightness of the confocal images. To quantify mitochondrial circularity, Fiji software and a homemade macro were used to process and to analyze captured images. To fix threshold, the inverse Fourier transformation (Inverse FFT) was used in order to minimize the noise and to sharpen the contrast between mitochondria and the background. Nucleus of each image was removed by hand (with the unaided human eye). « Analyze particles » command was used to obtain values for circularity (calculated by 4πArea/perimeter²) for each mitochondrion in an image, ignoring mitochondria <5 square-pixels or on the edge of the image. Mitochondrial circularity at 0 refers to a straight line whereas mitochondrial circularity at 1 refers to a perfect circle.
In order to determine the accuracy and consistency of the circularity measurements obtained with Fiji, we have compared the circularity indexes of 96 confocal images measured by visual counting and the circularity indexes measured by the macro processed. No statistical differences were found between the two methods.
TUNEL assay
Synchronized L4 + 3 day-old worms were fixed in 1 ml of PBS supplemented with 20 μl of 37% formaldehyde, extracted with acetone at −20 °C, and incubated in 50 μL of TUNEL solution (In Situ Cell Death Detection Kit, Fluorescein Roche) for 1.5 hours at 37 C. After incubation, worms were washed 3 times in PBST for 10 min. Then, phalloïdin staining with phalloïdin Alexa Fluor® 633 (Molecular Probes) was performed like previously described above. Worms were then mounted in Vectorshield mounting medium with DAPI (Vector Laboratories) and visualized using either a Zeiss Axiophot microscope or Zeiss LSM 710 confocal microscope. Only the two most visible quadrants of each animal were counted for the quantification of TUNEL positives muscle cells.
Statistical analyses
One-way ANOVA was used to test statistical differences between independent groups within the same experiment. Statistical significance was tested with Tukey and Dunnett post tests. Two-tailed unpaired Student’s t-test was used to examine direct differences between two independent groups.
No data sets were generated or analyzed during the current study.
Electronic supplementary material
Acknowledgements
We thank the Caenorhabditis Genetics Center (CGC-University of Minnesota) and the Lee laboratory (Cornell University) for providing nematode strains used in this study. We are very grateful to the members of the Delattre and Palladino laboratories (ENS-Lyon), Bessereau and Boulin laboratories (INMG-Lyon) for their logistical and scientific assistance. We thank all the members of the Mollereau laboratory (ENS-Lyon) and Pr. Carole Kretz (INMG-Lyon) for helpful scientific discussions during the course of this work and the CTMµ (Lyon 1 University), the PLATIM and Letitia Arias (ENS-Lyon) for their help with microscopy experiments. This work was supported by grants from the FP7 Marie Curie (IRG 256479) and from the Fondation pour la Recherche Médicale (FRM-DPM20121125561) to LW and grant from the Association Française Contre les Myopathies (AFM - contract 16109) to KG. CS is supported by a PhD Fellowship from the Fondation Groupama pour la Santé - Espoir 2014.
Author Contributions
L.W. and C.S. wrote the main manuscript text. L.W. designed the project. C.S. contributed to experiments and prepared all figures. S.B., E.M. and M.C.P. helped for experiments. L.W., C.S., K.G. and B.M. participated in the interpretation of the results. All authors have read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-25727-8.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kathrin Gieseler, Email: kathrin.gieseler@univ-lyon1.fr.
Ludivine Walter, Email: ludivine.walter@univ-lyon1.fr.
References
- 1.McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Curr. Biol. CB. 2006;16:R551–560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 2.Wai T, Ler T. Mitochondrial Dynamics and Metabolic Regulation. Trends Endocrinol. Metab. TEM. 2016;27:105–117. doi: 10.1016/j.tem.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Chen H, et al. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delettre C, et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat. Genet. 2000;26:207–210. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- 5.Mears JA, et al. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat. Struct. Mol. Biol. 2011;18:20–26. doi: 10.1038/nsmb.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JE, Westrate LM, Wu H, Page C, Voeltz GK. Multiple dynamin family members collaborate to drive mitochondrial division. Nature. 2016;540:139–143. doi: 10.1038/nature20555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Labrousse AM, Zappaterra MD, Rube DA, van der Bliek AM. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell. 1999;4:815–826. doi: 10.1016/s1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- 8.van der Bliek, A. M., Shen, Q. & Kawajiri, S. Mechanisms of mitochondrial fission and fusion. Cold Spring Harb. Perspect. Biol. 5 (2013). [DOI] [PMC free article] [PubMed]
- 9.Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J. Biol. Chem. 2005;280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- 10.Chen H, et al. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141:280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamacher-Brady A, Brady NR. Mitophagy programs: mechanisms and physiological implications of mitochondrial targeting by autophagy. Cell. Mol. Life Sci. 2016;73:775–795. doi: 10.1007/s00018-015-2087-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X, Kim CN, Yang J, Jemmerson R, Wang X. Induction of Apoptotic Program in Cell-Free Extracts: Requirement for dATP and Cytochrome c. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- 14.Susin SA, et al. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397:441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 15.Li LY, Luo X, Wang X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature. 2001;412:95–99. doi: 10.1038/35083620. [DOI] [PubMed] [Google Scholar]
- 16.Martinou J-C, Youle RJ. Mitochondria in Apoptosis: Bcl-2 Family Members and Mitochondrial Dynamics. Dev. Cell. 2011;21:92–101. doi: 10.1016/j.devcel.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conradt B, Wu Y-C, Xue D. Programmed Cell Death During Caenorhabditis elegans Development. Genetics. 2016;203:1533–1562. doi: 10.1534/genetics.115.186247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tondera D, et al. SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J. 2009;28:1589–1600. doi: 10.1038/emboj.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rambold AS, Kostelecky B, Elia N, Lippincott-Schwartz J. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc. Natl. Acad. Sci. USA. 2011;108:10190–10195. doi: 10.1073/pnas.1107402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rossignol R, et al. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res. 2004;64:985–993. doi: 10.1158/0008-5472.can-03-1101. [DOI] [PubMed] [Google Scholar]
- 21.Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 2011;13:589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pyakurel A, Savoia C, Hess D, Scorrano L. Extracellular regulated kinase phosphorylates mitofusin 1 to control mitochondrial morphology and apoptosis. Mol. Cell. 2015;58:244–254. doi: 10.1016/j.molcel.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jahani-Asl A, et al. Mitofusin 2 Protects Cerebellar Granule Neurons against Injury-induced Cell Death. J. Biol. Chem. 2007;282:23788–23798. doi: 10.1074/jbc.M703812200. [DOI] [PubMed] [Google Scholar]
- 24.Karbowski M, Norris KL, Cleland MM, Jeong S-Y, Youle RJ. Role of Bax and Bak in mitochondrial morphogenesis. Nature. 2006;443:658–662. doi: 10.1038/nature05111. [DOI] [PubMed] [Google Scholar]
- 25.Rolland SG, Lu Y, David CN, Conradt B. The BCL-2-like protein CED-9 of C. elegans promotes FZO-1/Mfn1,2- and EAT-3/Opa1-dependent mitochondrial fusion. J. Cell Biol. 2009;186:525–540. doi: 10.1083/jcb.200905070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breckenridge DG, Kang B-H, Xue D. Bcl-2 proteins EGL-1 and CED-9 do not regulate mitochondrial fission or fusion in Caenorhabditis elegans. Curr. Biol. CB. 2009;19:768–773. doi: 10.1016/j.cub.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koch A, et al. Dynamin-like protein 1 is involved in peroxisomal fission. J. Biol. Chem. 2003;278:8597–8605. doi: 10.1074/jbc.M211761200. [DOI] [PubMed] [Google Scholar]
- 28.Ishihara N, et al. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat. Cell Biol. 2009;11:958–966. doi: 10.1038/ncb1907. [DOI] [PubMed] [Google Scholar]
- 29.Pitts KR, Yoon Y, Krueger EW, McNiven MA. The Dynamin-like Protein DLP1 Is Essential for Normal Distribution and Morphology of the Endoplasmic Reticulum and Mitochondria in Mammalian Cells. Mol. Biol. Cell. 1999;10:4403–4417. doi: 10.1091/mbc.10.12.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wikstrom JD, et al. AMPK Regulates ER Morphology and Function in Stressed Pancreatic β-Cells via Phosphorylation of DRP1. Mol. Endocrinol. 2013;27:1706–1723. doi: 10.1210/me.2013-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Breckenridge DG, et al. Caenorhabditis elegans drp-1 and fis-2 regulate distinct cell death execution pathways downstream of ced-3 and independent of ced-9. Mol. Cell. 2008;31:586–597. doi: 10.1016/j.molcel.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Germain M, Mathai JP, McBride HM, Shore GC. Endoplasmic reticulum BIK initiates DRP1-regulated remodelling of mitochondrial cristae during apoptosis. EMBO J. 2005;24:1546–1556. doi: 10.1038/sj.emboj.7600592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frezza C, et al. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 2006;126:177–189. doi: 10.1016/j.cell.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 34.Landes T, et al. The BH3‐only Bnip3 binds to the dynamin Opa1 to promote mitochondrial fragmentation and apoptosis by distinct mechanisms. EMBO Rep. 2010;11:459–465. doi: 10.1038/embor.2010.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pidoux G, et al. Optic atrophy 1 is an A-kinase anchoring protein on lipid droplets that mediates adrenergic control of lipolysis. EMBO J. 2011;30:4371–4386. doi: 10.1038/emboj.2011.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 37.Ngoh GA, Papanicolaou KN, Walsh K. Loss of Mitofusin 2 Promotes Endoplasmic Reticulum Stress. J. Biol. Chem. 2012;287:20321–20332. doi: 10.1074/jbc.M112.359174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Debattisti V, Pendin D, Ziviani E, Daga A, Scorrano L. Reduction of endoplasmic reticulum stress attenuates the defects caused by Drosophila mitofusin depletion. J Cell Biol. 2014;204:303–312. doi: 10.1083/jcb.201306121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Misko A, Jiang S, Wegorzewska I, Milbrandt J, Baloh RH. Mitofusin 2 is necessary for transport of axonal mitochondria and interacts with the Miro/Milton complex. J. Neurosci. Off. J. Soc. Neurosci. 2010;30:4232–4240. doi: 10.1523/JNEUROSCI.6248-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gieseler K, Grisoni K, Ségalat L. Genetic suppression of phenotypes arising from mutations in dystrophin-related genes in Caenorhabditis elegans. Curr. Biol. 2000;10:1092–1097. doi: 10.1016/s0960-9822(00)00691-6. [DOI] [PubMed] [Google Scholar]
- 41.Bessou C, Giugia JB, Franks CJ, Holden-Dye L, Ségalat L. Mutations in the Caenorhabditis elegans dystrophin-like gene dys-1 lead to hyperactivity and suggest a link with cholinergic transmission. Neurogenetics. 1998;2:61–72. doi: 10.1007/s100480050053. [DOI] [PubMed] [Google Scholar]
- 42.Chen L, Krause M, Sepanski M, Fire A. The Caenorhabditis elegans MYOD homologue HLH-1 is essential for proper muscle function and complete morphogenesis. Dev. Camb. Engl. 1994;120:1631–1641. doi: 10.1242/dev.120.6.1631. [DOI] [PubMed] [Google Scholar]
- 43.Lecroisey C, Ségalat L, Gieseler K. The C. elegans dense body: anchoring and signaling structure of the muscle. J. Muscle Res. Cell Motil. 2007;28:79–87. doi: 10.1007/s10974-007-9104-y. [DOI] [PubMed] [Google Scholar]
- 44.Gieseler, K. Development, structure, and maintenance of C. elegans body wall muscle. WormBook 1–59, 10.1895/wormbook.1.81.2 (2017) [DOI] [PMC free article] [PubMed]
- 45.Giacomotto J, et al. Chemical genetics unveils a key role of mitochondrial dynamics, cytochrome c release and IP3R activity in muscular dystrophy. Hum. Mol. Genet. 2013;22:4562–4578. doi: 10.1093/hmg/ddt302. [DOI] [PubMed] [Google Scholar]
- 46.Percival JM, Siegel MP, Knowels G, Marcinek DJ. Defects in mitochondrial localization and ATP synthesis in the mdx mouse model of Duchenne muscular dystrophy are not alleviated by PDE5 inhibition. Hum. Mol. Genet. 2013;22:153–167. doi: 10.1093/hmg/dds415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pant M, et al. Metabolic Dysfunction and Altered Mitochondrial Dynamics in the Utrophin-Dystrophin Deficient Mouse Model of Duchenne Muscular Dystrophy. PLOS ONE. 2015;10:e0123875. doi: 10.1371/journal.pone.0123875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valentine BA, Cooper BJ, Cummings JF, de Lahunta A. Canine X-linked muscular dystrophy: morphologic lesions. J. Neurol. Sci. 1990;97:1–23. doi: 10.1016/0022-510x(90)90095-5. [DOI] [PubMed] [Google Scholar]
- 49.Scholte HR, Busch HF. Early changes of muscle mitochondria in Duchenne dystrophy. Partition and activity of mitochondrial enzymes in fractionated muscle of unaffected boys and adults and patients. J. Neurol. Sci. 1980;45:217–234. doi: 10.1016/0022-510x(80)90167-7. [DOI] [PubMed] [Google Scholar]
- 50.Regmi SG, Rolland SG, Conradt B. Age-dependent changes in mitochondrial morphology and volume are not predictors of lifespan. Aging. 2014;6:118–130. doi: 10.18632/aging.100639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Demontis F, Piccirillo R, Goldberg AL, Perrimon N. Mechanisms of skeletal muscle aging: insights from Drosophila and mammalian models. Dis. Model. Mech. 2013;6:1339–1352. doi: 10.1242/dmm.012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pietrangelo L, et al. Age-dependent uncoupling of mitochondria from Ca2+ release units in skeletal muscle. Oncotarget. 2015;6:35358–35371. doi: 10.18632/oncotarget.6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kanazawa Takayuki, et al. The C. elegans Opa1 Homologue EAT-3 Is Essential for Resistance to Free Radicals. PLoS Genetics. 2008;4:e1000022. doi: 10.1371/journal.pgen.1000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang CC, Chen D, Lee SS, Walter L. The dynamin-related protein DRP-1 and the insulin signaling pathway cooperate to modulate C. elegans longevity. Aging Cell. 2011;10:724–728. doi: 10.1111/j.1474-9726.2011.00711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 56.Jagasia R, Grote P, Westermann B, Conradt B. DRP-1-mediated mitochondrial fragmentation during EGL-1-induced cell death in C. elegans. Nature. 2005;433:754–760. doi: 10.1038/nature03316. [DOI] [PubMed] [Google Scholar]
- 57.Anand R, et al. The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J. Cell Biol. 2014;204:919–929. doi: 10.1083/jcb.201308006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grasl-Kraupp B, et al. In situ detection of fragmented DNA (tunel assay) fails to discriminate among apoptosis, necrosis, and autolytic cell death: A cautionary note. Hepatology. 1995;21:1465–1468. doi: 10.1002/hep.1840210534. [DOI] [PubMed] [Google Scholar]
- 59.Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 60.Brouilly N, et al. Ultra-structural time-course study in the C. elegans model for Duchenne muscular dystrophy highlights a crucial role for sarcomere-anchoring structures and sarcolemma integrity in the earliest steps of the muscle degeneration process. Hum. Mol. Genet. 2015;24:6428–6445. doi: 10.1093/hmg/ddv353. [DOI] [PubMed] [Google Scholar]
- 61.Suen D-F, Norris KL, Youle RJ. Mitochondrial dynamics and apoptosis. Genes Dev. 2008;22:1577–1590. doi: 10.1101/gad.1658508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karbowski M, Youle RJ. Dynamics of mitochondrial morphology in healthy cells and during apoptosis. Cell Death Differ. 2003;10:870–880. doi: 10.1038/sj.cdd.4401260. [DOI] [PubMed] [Google Scholar]
- 63.Luz AL, et al. Mitochondrial Morphology and Fundamental Parameters of the Mitochondrial Respiratory Chain Are Altered in Caenorhabditis elegans Strains Deficient in Mitochondrial Dynamics and Homeostasis Processes. PloS One. 2015;10:e0130940. doi: 10.1371/journal.pone.0130940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bess, A. S. et al. Effects of mutations in mitochondrial dynamics-related genes on the mitochondrial response to ultraviolet C radiation in developing Caenorhabditis elegans. Worm2 (2013). [DOI] [PMC free article] [PubMed]
- 65.Seshagiri S, Miller LK. Caenorhabditis elegans CED-4 stimulates CED-3 processing and CED-3-induced apoptosis. Curr. Biol. CB. 1997;7:455–460. doi: 10.1016/s0960-9822(06)00216-8. [DOI] [PubMed] [Google Scholar]
- 66.Ellis HM, Horvitz HR. Genetic control of programmed cell death in the nematode C. elegans. Cell. 1986;44:817–829. doi: 10.1016/0092-8674(86)90004-8. [DOI] [PubMed] [Google Scholar]
- 67.Wang X, Yang C, Chai J, Shi Y, Xue D. Mechanisms of AIF-mediated apoptotic DNA degradation in Caenorhabditis elegans. Science. 2002;298:1587–1592. doi: 10.1126/science.1076194. [DOI] [PubMed] [Google Scholar]
- 68.Parrish JZ, Xue D. Functional genomic analysis of apoptotic DNA degradation in C. elegans. Mol. Cell. 2003;11:987–996. doi: 10.1016/s1097-2765(03)00095-9. [DOI] [PubMed] [Google Scholar]
- 69.Wu Y-C, Stanfield GM, Horvitz HR. NUC-1, a Caenorhabditis elegans DNase II homolog, functions in an intermediate step of DNA degradation during apoptosis. Genes Dev. 2000;14:536–548. [PMC free article] [PubMed] [Google Scholar]
- 70.Fadok VA, Bratton DL, Henson PM. Phagocyte receptors for apoptotic cells: recognition, uptake, and consequences. J. Clin. Invest. 2001;108:957–962. doi: 10.1172/JCI14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Su HP, et al. Interaction of CED-6/GULP, an adapter protein involved in engulfment of apoptotic cells with CED-1 and CD91/low density lipoprotein receptor-related protein (LRP) J. Biol. Chem. 2002;277:11772–11779. doi: 10.1074/jbc.M109336200. [DOI] [PubMed] [Google Scholar]
- 72.Zhou Z, Hartwieg E, Horvitz HR. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell. 2001;104:43–56. doi: 10.1016/s0092-8674(01)00190-8. [DOI] [PubMed] [Google Scholar]
- 73.Ellis RE, Jacobson DM, Horvitz HR. Genes required for the engulfment of cell corpses during programmed cell death in Caenorhabditis elegans. Genetics. 1991;129:79–94. doi: 10.1093/genetics/129.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gaffney CJ, et al. Degenerin channel activation causes caspase-mediated protein degradation and mitochondrial dysfunction in adult C. elegans muscle. J. Cachexia Sarcopenia Muscle. 2016;7:181–192. doi: 10.1002/jcsm.12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Conradt, B. Programmed cell death. WormBook10.1895/wormbook.1.32.1 (2005). [DOI] [PMC free article] [PubMed]
- 76.Sandri M, et al. Apoptosis, DNA damage and ubiquitin expression in normal and mdx muscle fibers after exercise. FEBS Lett. 1995;373:291–295. doi: 10.1016/0014-5793(95)00908-r. [DOI] [PubMed] [Google Scholar]
- 77.Tidball JG, Albrecht DE, Lokensgard BE, Spencer MJ. Apoptosis precedes necrosis of dystrophin-deficient muscle. J. Cell Sci. 1995;108:2197–2204. doi: 10.1242/jcs.108.6.2197. [DOI] [PubMed] [Google Scholar]
- 78.Sandri M, Minetti C, Pedemonte M, Carraro U. Apoptotic myonuclei in human Duchenne muscular dystrophy. Lab. Investig. J. Tech. Methods Pathol. 1998;78:1005–1016. [PubMed] [Google Scholar]
- 79.Tews DS. Characterization of initiator and effector caspase expressions in dystrophinopathies. Neuropathol. Off. J. Jpn. Soc. Neuropathol. 2006;26:24–31. doi: 10.1111/j.1440-1789.2006.00650.x. [DOI] [PubMed] [Google Scholar]
- 80.Park SY, et al. Differential expression of apoptosis-related factors induces the age-related apoptosis of the gracilis muscle in humans. Int. J. Mol. Med. 2014;33:1110. doi: 10.3892/ijmm.2014.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dirks A, Leeuwenburgh C. Apoptosis in skeletal muscle with aging. Am. J. Physiol. - Regul. Integr. Comp. Physiol. 2002;282:R519–R527. doi: 10.1152/ajpregu.00458.2001. [DOI] [PubMed] [Google Scholar]
- 82.Alway SE, Degens H, Krishnamurthy G, Smith CA. Potential role for Id myogenic repressors in apoptosis and attenuation of hypertrophy in muscles of aged rats. Am. J. Physiol. Cell Physiol. 2002;283:C66–76. doi: 10.1152/ajpcell.00598.2001. [DOI] [PubMed] [Google Scholar]
- 83.Strasser H, et al. Age dependent apoptosis and loss of rhabdosphincter cells. J. Urol. 2000;164:1781–1785. [PubMed] [Google Scholar]
- 84.Frank S, et al. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev. Cell. 2001;1:515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- 85.Tezze C, et al. Age-Associated Loss of OPA1 in Muscle Impacts Muscle Mass, Metabolic Homeostasis, Systemic Inflammation, and Epithelial Senescence. Cell Metab. 2017;25:1374–1389.e6. doi: 10.1016/j.cmet.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bhola PD, Letai A. Mitochondria-Judges and Executioners of Cell Death Sentences. Mol. Cell. 2016;61:695–704. doi: 10.1016/j.molcel.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yan N, et al. Structure of the CED-4–CED-9 complex provides insights into programmed cell death in Caenorhabditis elegans. Nature. 2005;437:831–837. doi: 10.1038/nature04002. [DOI] [PubMed] [Google Scholar]
- 88.Zou H, Henzel WJ, Liu X, Lutschg A, Wang X. Apaf-1, a Human Protein Homologous to C. elegans CED-4, Participates in Cytochrome c–Dependent Activation of Caspase-3. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 89.Parrish J, et al. Mitochondrial endonuclease G is important for apoptosis in C. elegans. Nature. 2001;412:90–94. doi: 10.1038/35083608. [DOI] [PubMed] [Google Scholar]
- 90.Fisher K, et al. Maintenance of muscle myosin levels in adult C. elegans requires both the double bromodomain protein BET-1 and sumoylation. Biol. Open. 2013;2:1354–1363. doi: 10.1242/bio.20136007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Song W, Kwak H-B, Lawler JM. Exercise Training Attenuates Age-Induced Changes in Apoptotic Signaling in Rat Skeletal Muscle. Antioxid. Redox Signal. 2006;8:517–528. doi: 10.1089/ars.2006.8.517. [DOI] [PubMed] [Google Scholar]
- 92.Sandri M, et al. Caspase 3 expression correlates with skeletal muscle apoptosis in Duchenne and facioscapulo human muscular dystrophy. A potential target for pharmacological treatment? J. Neuropathol. Exp. Neurol. 2001;60:302–312. doi: 10.1093/jnen/60.3.302. [DOI] [PubMed] [Google Scholar]
- 93.Alexander MS, et al. Regulation of DMD pathology by an ankyrin-encoded miRNA. Skelet. Muscle. 2011;1:27. doi: 10.1186/2044-5040-1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Otera, H. & Mihara, K. Mitochondrial Dynamics: Functional Link with Apoptosis. International Journal of Cell Biology, 10.1155/2012/821676 (2012). [DOI] [PMC free article] [PubMed]
- 95.Whitehead NP, Yeung EW, Froehner SC, Allen DG. Skeletal muscle NADPH oxidase is increased and triggers stretch-induced damage in the mdx mouse. PloS One. 2010;5:e15354. doi: 10.1371/journal.pone.0015354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Goonasekera SA, et al. Mitigation of muscular dystrophy in mice by SERCA overexpression in skeletal muscle. J. Clin. Invest. 2011;121:1044–1052. doi: 10.1172/JCI43844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pal R, et al. Src-dependent impairment of autophagy by oxidative stress in a mouse model of Duchenne muscular dystrophy. Nat. Commun. 2014;5:4425. doi: 10.1038/ncomms5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Orrenius S, Gogvadze V, Zhivotovsky B. Calcium and mitochondria in the regulation of cell death. Biochem. Biophys. Res. Commun. 2015;460:72–81. doi: 10.1016/j.bbrc.2015.01.137. [DOI] [PubMed] [Google Scholar]
- 99.Zhivotovsky B, Orrenius S. Calcium and cell death mechanisms: a perspective from the cell death community. Cell Calcium. 2011;50:211–221. doi: 10.1016/j.ceca.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 100.Robert V, et al. Alteration in calcium handling at the subcellular level in mdx myotubes. J. Biol. Chem. 2001;276:4647–4651. doi: 10.1074/jbc.M006337200. [DOI] [PubMed] [Google Scholar]
- 101.Duncan CJ. Role of intracellular calcium in promoting muscle damage: a strategy for controlling the dystrophic condition. Experientia. 1978;34:1531–1535. doi: 10.1007/BF02034655. [DOI] [PubMed] [Google Scholar]
- 102.Mázala DAG, et al. SERCA1 overexpression minimizes skeletal muscle damage in dystrophic mouse models. Am. J. Physiol. Cell Physiol. 2015;308:C699–709. doi: 10.1152/ajpcell.00341.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schneider JS, et al. Increased sarcolipin expression and decreased sarco(endo)plasmic reticulum Ca2+ uptake in skeletal muscles of mouse models of Duchenne muscular dystrophy. J. Muscle Res. Cell Motil. 2013;34:349–356. doi: 10.1007/s10974-013-9350-0. [DOI] [PubMed] [Google Scholar]
- 104.Mariol MC, Ségalat L. Muscular degeneration in the absence of dystrophin is a calcium-dependent process. Curr. Biol. CB. 2001;11:1691–1694. doi: 10.1016/s0960-9822(01)00528-0. [DOI] [PubMed] [Google Scholar]
- 105.Joyce PI, Satija R, Chen M, Kuwabara PE. The Atypical Calpains: Evolutionary Analyses and Roles in Caenorhabditis elegans Cellular Degeneration. PLOS Genet. 2012;8:e1002602. doi: 10.1371/journal.pgen.1002602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Breckenridge DG, Stojanovic M, Marcellus RC, Shore GC. Caspase cleavage product of BAP31 induces mitochondrial fission through endoplasmic reticulum calcium signals, enhancing cytochrome c release to the cytosol. J. Cell Biol. 2003;160:1115–1127. doi: 10.1083/jcb.200212059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cribbs JT, Strack S. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep. 2007;8:939–944. doi: 10.1038/sj.embor.7401062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cereghetti GM, et al. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc. Natl. Acad. Sci. USA. 2008;105:15803–15808. doi: 10.1073/pnas.0808249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jiang H-C, et al. Neural activity and CaMKII protect mitochondria from fragmentation in aging Caenorhabditis elegans neurons. Proc. Natl. Acad. Sci. USA. 2015;112:8768–8773. doi: 10.1073/pnas.1501831112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Berkowitz, L. A., Knight, A. L., Caldwell, G. A. & Caldwell, K. A. Generation of Stable Transgenic C. elegans Using Microinjection. JoVE J. Vis. Exp. e833–e833, 10.3791/833 (2008). [DOI] [PMC free article] [PubMed]
- 111.Reiner DJ, Weinshenker D, Thomas JH. Analysis of Dominant Mutations Affecting Muscle Excitation in Caenorhabditis Elegans. Genetics. 1995;141:961–976. doi: 10.1093/genetics/141.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.