Abstract
Background
Conjunctival melanoma is a potentially deadly eye tumour. Despite effective local therapies, tumour recurrence and metastasis remain frequent. The genetics of conjunctival melanomas remain incompletely understood.
Methods
A large cohort of 63 conjunctival melanomas was screened for gene mutations known to be important in other melanoma subtypes by targeted next-generation sequencing. Mutation status was correlated with patient prognosis.
Results
Frequent mutations in genes activating the MAP kinase pathway were identified. NF1 mutations were most frequent (n = 21, 33%). Recurrent activating mutations were also identified in BRAF (n = 16, 25%) and RAS genes (n = 12, 19%; 11 NRAS and 1 KRAS).
Conclusions
Similar to cutaneous melanomas, conjunctival melanomas can be grouped genetically into four groups: BRAF-mutated, RAS-mutated, NF1-mutated and triple wild-type melanomas. This genetic classification may be useful for assessment of therapeutic options for patients with metastatic conjunctival melanoma
Subject terms: Cancer genetics, Eye cancer
Introduction
Conjunctival melanoma accounts for 5–10% of all ocular melanomas with a 10-year local recurrence rate of 38–69% and disease-related mortality of 13–38%.1–6 A better understanding of the genetics of conjunctival melanoma may help identify improved therapeutic options for patients with advanced disease.
In recent years, major melanoma subtypes have been genetically characterised. Cutaneous melanomas frequently harbour activating mutations in BRAF (~50%)7 or NRAS (~20%), as well as mutations in NF1.8–11 BRAF, NRAS and NF1 mutations lead to activation of the mitogen-activated protein (MAP) kinase pathway.9, 12, 13 Based on these findings, a genetic classification of cutaneous melanomas has been proposed distinguishing four genetic groups: BRAF-mutated, RAS-mutated, NF1-mutated or triple wild type.11
Uveal melanomas exhibit a different mutation profile, and harbour mutations in GNAQ,14 GNA11,15 CYSLTR2,16 PLCB4,17 EIF1AX,18 SF3B119 and BAP1,20 which are rarely found in other melanomas.15, 21–23
Conjunctival melanomas have not been characterised genetically as well as other melanoma subtypes. BRAFV600E and NRAS mutations are present in 14–50%24–27 and 18%,28 respectively, of conjunctival melanomas. TERT promoter mutations were identified in 32–41% of conjunctival melanomas.29, 30 One study reported a KIT mutation in 1/14 (7%) tumours.31 Copy number analysis identified alterations reminiscent of cutaneous and mucosal melanomas, including CDKN2A and PTEN losses.28 These data suggest that conjunctival melanomas are genetically similar to cutaneous melanomas, but aside from BRAF, NRAS and TERT promoter mutations, recurrent mutations in other genes have not been identified.
There are two main therapeutic avenues for metastatic melanoma. Firstly, targeted small inhibitors dampening pathologically activated cell-intrinsic signalling mechanisms, with the most effective to date being a combination of BRAF and MEK inhibitors in BRAF-mutated melanoma.32 Secondly, immunotherapies applying anti-CTLA-4 and anti-PD-1 antibodies have shown impressive response rates in cutaneous and mucosal melanoma.33–35 Both approaches may be clinically useful in advanced conjunctival melanoma.36
Our study aimed to further elucidate genetic events in conjunctival melanoma by analysing a large tumour cohort with a targeted next-generation sequencing assay covering genes that are recurrently mutated in cutaneous and uveal melanoma.
Materials and methods
Sample selection and histopathology
Sixty-seven conjunctival melanoma samples were obtained from the tissue archives of the Departments of Ophthalmology, Dermatology and Pathology of the University Hospital Essen, and the Department of Ophthalmology, University Hospital Tübingen, Germany. The study was approved by the local ethics committee of the University of Duisburg-Essen.
DNA isolation
Formalin-fixed, paraffin-embedded tumour tissues were sectioned, deparaffinised and manually microdissected as previously described.37 Genomic DNA was isolated using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany).
Targeted sequencing
A custom amplicon-based sequencing panel covering 29 genes known to be mutated in melanoma was used (genes listed in Supplemental Table 1), as previously described.37 Mean coverage of 2094 reads, with a minimum coverage of 30 reads in >80% of the target loci, was achieved. Four samples were excluded from analysis due to low coverage.
Sequence analysis
CLC Cancer Research Workbench from QIAGEN® was used for sequence analysis, as previously reported.37 Mutations were considered if coverage of the mutation site was ≥30 reads, ≥10 reads reported the mutated variant and the frequency of mutated reads was ≥10%.
Associations of mutation status with clinical and pathological parameters
Associations of mutation status with available clinico-pathological parameters (listed in Table 1) were explored. Analyses were performed with IBM SPSS Statistics software (version 20.0; International Business Machines Corp., Armonk NY, USA). A p-value of <0.05 was considered statistically significant.
Table 1.
Correlation between mutation status and clinical features in conjunctival melanomas
| Total | BRAF WT | BRAF V600E | P-value (p < 0.05) | RAS WT | RAS MUT | P-value (p < 0.05) | NF1 WT | NF1 MUT | P-value (p < 0.05) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % | (n) | % | (n) | % | (n) | % | (n) | % | (n) | % | (n) | % | (n) | |||||
| Total | 63 | 74.6 | 47 | 25.4 | 16 | 81.0 | 51 | 19.0 | 12 | 66.7 | 42 | 33.3 | 21 | |||||
| Sex | Female | 50.8 | 32 | 38.1 | 24 | 12.7 | 8 | 0.94 | 44.4 | 28 | 6.3 | 4 | 0.18 | 36.5 | 23 | 14.3 | 9 | 0.37 |
| Male | 49.2 | 31 | 36.5 | 23 | 12.7 | 8 | 36.5 | 23 | 12.7 | 8 | 30.2 | 19 | 19 | 12 | ||||
| Eye | Right | 55.6 | 35 | 39.7 | 25 | 15.9 | 10 | 0.63 | 42.9 | 27 | 12.7 | 8 | 0.59 | 34.9 | 22 | 20.6 | 13 | 0.52 |
| Left | 41.3 | 26 | 31.7 | 20 | 9.5 | 6 | 34.9 | 22 | 6.3 | 4 | 28.6 | 18 | 12.7 | 8 | ||||
| N/A | 3.2 | 2 | 3.2 | 2 | 0 | 0 | 3.2 | 2 | 0 | 0 | 3.2 | 2 | 0 | 0 | ||||
| TNM | 1 | 55.6 | 35 | 41.3 | 26 | 14.3 | 9 | 0.77 | 44.4 | 28 | 11.1 | 7 | 0.44 | 38.1 | 24 | 17.5 | 11 | 0.56 |
| 2 | 23.8 | 15 | 17.5 | 11 | 6.3 | 43 | 22.2 | 14 | 1.6 | 1 | 14.3 | 9 | 9.5 | 6 | ||||
| 3 | 15.9 | 10 | 11.1 | 7 | 4.8 | 0 | 11.1 | 7 | 4.8 | 3 | 9.5 | 6 | 6.3 | 4 | ||||
| N/A | 4.8 | 3 | 4.8 | 3 | 0 | 3.2 | 2 | 1.6 | 1 | 4.8 | 3 | 0 | 0 | |||||
| Tumour origin | PAM | 52.4 | 33 | 41.3 | 26 | 11.1 | 7 | 0.1 | 44.4 | 28 | 7.9 | 5 | 0.35 | 37.5 | 24 | 14.3 | 9 | 0.43 |
| Naevus | 17.5 | 11 | 7.9 | 5 | 9.5 | 6 | 15.9 | 10 | 1.6 | 1 | 11.1 | 7 | 6.3 | 4 | ||||
| De novo | 22.2 | 14 | 19 | 12 | 3.2 | 2 | 15.9 | 10 | 4.8 | 4 | 11.1 | 7 | 11.1 | 7 | ||||
| N/A | 7.9 | 5 | 6.3 | 4 | 1.6 | 1 | 4.8 | 3 | 3.2 | 2 | 6.3 | 4 | 1.6 | 1 | ||||
| Relapses | No | 46 | 29 | 36.5 | 23 | 9.5 | 6 | 0.26 | 33.3 | 21 | 12.7 | 8 | 0.22 | 27 | 17 | 19 | 12 | 0.46 |
| Yes | 47.6 | 30 | 31.7 | 20 | 15.9 | 10 | 42.9 | 27 | 4.8 | 3 | 34.9 | 22 | 12.7 | 8 | ||||
| N/A | 6.3 | 4 | 6.3 | 4 | 0 | 0 | 4.8 | 3 | 1.6 | 1 | 4.8 | 3 | 1.6 | 1 | ||||
| Metastasis | No | 68.3 | 43 | 49.2 | 31 | 19.0 | 12 | 0.32 | 54.0 | 34 | 14.3 | 9 | 0.34 | 42.9 | 27 | 25.4 | 16 | 0.55 |
| Yes | 22.2 | 14 | 15.9 | 10 | 6.3 | 4 | 20.6 | 13 | 1.6 | 1 | 17.5 | 11 | 4.8 | 3 | ||||
| N/A | 9.5 | 6 | 9.5 | 6 | 0 | 0 | 6.3 | 4 | 3.2 | 2 | 6.3 | 4 | 3.2 | 2 | ||||
| Exenteration | No | 76.2 | 48 | 57.1 | 36 | 19.0 | 12 | 0.36 | 60.3 | 38 | 15.9 | 10 | 0.64 | 47.6 | 30 | 28.6 | 18 | 0.44 |
| Yes | 17.5 | 11 | 11.1 | 7 | 6.3 | 4 | 15.9 | 10 | 1.6 | 1 | 14.3 | 9 | 3.2 | 2 | ||||
| N/A | 6.3 | 4 | 6.3 | 4 | 0 | 0 | 4.8 | 3 | 1.6 | 1 | 4.8 | 3 | 1.6 | 1 | ||||
| Age at diagnose |
Median 67.4 years , Range 40.1–88.8 years |
|||||||||||||||||
Clinical and pathological stage is according to TNM 7th edition AJCC 2010 for conjunctival melanoma
N/A not assessable, PAM primary acquired melanosis
Results
Tumours and patients
Conjunctival melanomas occurred equally in male and female with a median age of 67 years (range 40–89 years). Of the samples for which information was available, 52% (33/63) originated from primary acquired melanosis (PAM), 18% (11/63) from naevi and 22% (14/63) arose de novo. Clinical stage at initial presentation was stage 1, stage 2 and stage 3 in 56% (35/63), 24% (15/63) and 16% (10/63) of patients, respectively (American Joint Committee on Cancer staging system for conjunctival melanoma, 7th edition, 2010). Adjuvant treatment was received by 87% (55/63) of patients (21 ruthenium, 17 proton, 6 percutaneous radiotherapy, 7 cryotherapy and 3 mitomycin C). Tumours recurred in 47% (30/63) and metastasised in 22% (14/63) of cases. Additional information is listed in Table 1.
Identified mutations
Activating BRAFV600E (c.1799A>T) mutations were detected in 16/63 (25%) tumours. Additionally, 4 BRAF mutations with unknown functional consequences were identified (Supplemental Table 2).
Activating RAS mutations (11 NRAS and 1 KRAS mutation) were identified in 12/63 (19%) tumours (Table 2). We also detected 4 NRAS, 3 KRAS and 5 HRAS mutations with unknown functional consequences (Supplemental Table 2).
Table 2.
MAP kinase pathway activating mutations in conjunctival melanoma
| Gene | Mutation type | Tumours harbouring mutation | ||
|---|---|---|---|---|
| N | % | |||
| BRAF | All mutations | 16 | 25 | |
| V600E | 16 | 25 | ||
| RAS | All mutations | 12 | 19 | |
| NRAS | Q61R | 5 | 8 | |
| Q61K | 2 | 3 | ||
| Q61H | 1 | 2 | ||
| Q61L | 1 | 2 | ||
| G13D | 1 | 2 | ||
| G12N, G12C | 1 | 2 | ||
| KRAS | G12A | 1 | 2 | |
| NF1 | All mutations | 21 | 33 | |
| T60del | 1 | 2 | ||
| R262C | 1 | 2 | ||
| C42Y, G2397R, S2587L | 1 | 2 | ||
| S2751N, L552P, G2392E | 1 | 2 | ||
| D176E | 2 | 3 | ||
| L847P, P866S, V1762I | 1 | 2 | ||
| C1899Y | 1 | 2 | ||
| M1180I, S52F; T60I | 1 | 2 | ||
| A2715V; A2208T | 1 | 2 | ||
| G2397R, R2517fs | 1 | 2 | ||
| I1824fs | 1 | 2 | ||
| L1892a | 1 | 2 | ||
| N1451L | 1 | 2 | ||
| Q1815a | 1 | 2 | ||
| Q756fs | 1 | 2 | ||
| R1362a | 1 | 2 | ||
| R440a, Q2239a; S1497F; V1393A | 1 | 2 | ||
| S168L | 1 | 2 | ||
| S1786a, L1102a; Q1815fs | 1 | 2 | ||
| Y1678fs | 1 | 2 | ||
| Wild type | 14 | 22 | ||
| Total | 63 |
MAP mitogen-activated protein, fs frameshift mutations
aNonsense mutations
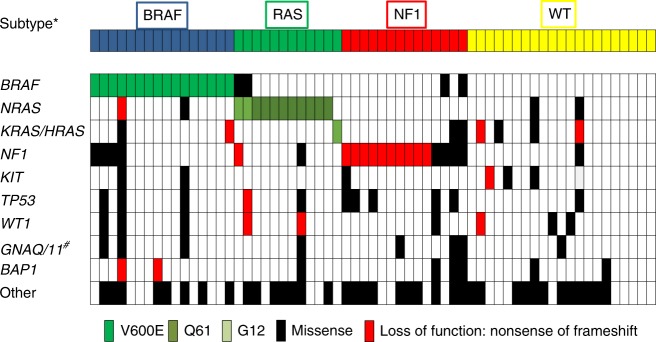
NF1 mutations were identified in 21/63 (33%) tumours. Clearly inactivating NF1 mutations were observed in 10 tumours. NF1 mutations co-occurred with BRAF and RAS gene mutations in some tumours, but also frequently occurred alone. All identified mutations are shown in Fig. 1 and listed in Table 2, and shown in Supplemental Figs. 1 and 2.
Fig. 1.
Mutations in conjunctival melanoma. Distribution of mutations identified by amplicon panel next-generation sequencing. Green: mutations known or assumed to be activating; Red: nonsense or frameshift loss-of-function mutations; Black: missense mutation with unknown functional consequences. Mutations listed as “Other” include mutations detected in CDK4, FLT4, PIK3CA, PIK3R1, FBXW7, MITF, MAP2K1, MAP2K2, ARID1A, ARID2, SF3B1, CTNNB1, PTEN, CDKN2A, SMARCA4A, EZH2, IDH1 and the protein-coding area of TERT (the promoter region of TERT was not covered by the amplicon-based sequencing panel used in this study). *Subtype according to TCGA genomic classification of cutaneous melanoma. #None of the GNAQ or GNA11 mutations identified were the known activating Q209 or R183 mutations recurrently identified in uveal melanomas (details in Supplemental Table 2)
Additionally, mutations in various genes frequently mutated in cutaneous melanoma were detected. The majority of these mutations were of unknown functional consequences (Supplemental Table 2). While a few GNAQ and GNA11 mutations were identified (Fig. 1, Supplemental Table 2), they presumably represent functionally non-relevant bystander mutations, as none of the identified mutations were the activating R183 or Q209 mutations known to occur in uveal melanomas.14, 15, 21
Statistical analysis
There were no statistically significant associations between clinico-pathological parameters with BRAF, RAS and NF1 mutation status (Table 1).
Discussion
To our knowledge, the present study represents the most detailed analysis of gene mutations in conjunctival melanoma to date.
Activating BRAF mutations were detected in 25% of samples, lying within the range of previous studies reporting 14–50%.24–27, 38 This variation may be due to sample bias or technical differences. In view of the recent development of effective BRAF and MEK inhibitors, the presence of BRAF V600 mutations in conjunctival melanomas is of considerable therapeutic relevance.39
In addition to known activating NRAS mutations in 18% (11/63) of tumours, we identified an activating KRAS G12A mutation. Being the first report on these mutations in conjunctival melanoma, this finding is reminiscent of cutaneous melanoma, in which KRAS mutations are rare but occur in a mutually exclusive fashion with NRAS mutations.11 In the proposed TCGA (The Cancer Genome Atlas) genomic classification of cutaneous melanoma, mutations in all three RAS genes are grouped together as RAS-mutated melanomas.
Our study is the first to identify NF1 as a frequently mutated oncogene (33%) in conjunctival melanoma. NF1 has recently been recognised as the third most commonly mutated gene (after BRAF and RAS) in cutaneous melanoma, activating the MAP kinase pathway.11 In our conjunctival melanoma cohort, NF1 mutations were also present in samples harbouring activating RAS or BRAF mutations (Fig. 1). This is similar to the situation in cutaneous melanoma where the co-occurrence of NF1 with BRAF, RAS and other mutations is well recognised.11, 12, 40
NF1 mutations are particularly frequent in melanoma subtypes rarely harbouring BRAF and NRAS mutations,8, 12, 41 including melanomas associated with high sun exposure.8, 12 Ultraviolet exposure is a known pathogenic factor in conjunctival melanoma and could explain the high number of NF1 mutations detected. NF1 mutations have been associated with high tumour mutational load and affected patients have been reported to benefit from anti-PD-1 therapy in cutaneous melanoma.42 This suggests that NF1 mutation status has potential as a biomarker for immunotherapy in conjunctival melanoma.
In summary, our study identifies a range of mutations in conjunctival melanoma. The distribution of activating mutations, with RAS gene mutations occurring not only in NRAS but also KRAS, and NF1 mutations being frequent in tumours lacking BRAF or RAS mutations, suggests that the proposed genetic classification of cutaneous melanomas into BRAF-mutated, RAS-mutated, NF1-mutated or triple-wild-type tumours is also applicable to conjunctival melanoma.
Electronic supplementary material
Acknowledgements
We would like to thank Nadine Stadler and Nicola Bielefeld for their excellent technical support. The research was supported by a grant from Dr. Werner-Jackstädt-Stiftung (www.jackstaedt-stiftung.de) and Hiege-Stiftung gegen Hautkrebs. This work was also funded in part through the NIH/NCI Cancer Center Support Grant P30 CA008748 and received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No 641458. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author contributions
Literature search: S.L.S., K.G.G., B.S., H.W., R.M.; study design: K.G.G., K.P.S., D.S.; data collection: S.L.S., I.C., D.S., R.M., I.M., H.R., S.T., S.L., E.H., C.F., K.G.G., H.W., A.P., D.S.; data analysis: S.L.S., K.G.G., R.M., T.S., H.W., A.S., A.P.; data interpretation: S.L.S., K.G.G., A.P., A.S., H.W.; manuscript writing: all authors.
Competing interests
Dirk Schadendorf is on the advisory board or has received honararia from Roche, Genetech, Novartis, Amgen, GSK, Boehringer Ingelheim and Merck. The other authors declare no competing interests.
Footnotes
These authors contributed equally: H. Westekemper, K. G. Griewank.
These authors contributed equally: K. G. Griewank, S. L. Scholz.
Contributor Information
S. L. Scholz, Email: simone.scholz@uk-essen.de
K. G. Griewank, Email: klaus.griewank@uk-essen.de
Electronic supplementary material
Supplementary information is available for this paper at 10.1038/s41416-018-0046-5.
References
- 1.Harooni H, Schoenfield LR, Singh AD. Current appraisal of conjunctival melanocytic tumors: classification and treatment. Future Oncol. 2011;7:435–446. doi: 10.2217/fon.11.12. [DOI] [PubMed] [Google Scholar]
- 2.Shields CL, et al. Conjunctival melanoma: risk factors for recurrence, exenteration, metastasis, and death in 150 consecutive patients. Arch. Ophthalmol. 2000;118:1497–1507. doi: 10.1001/archopht.118.11.1497. [DOI] [PubMed] [Google Scholar]
- 3.Paridaens AD, McCartney AC, Minassian DC, Hungerford JL. Orbital exenteration in 95 cases of primary conjunctival malignant melanoma. Br. J. Ophthalmol. 1994;78:520–528. doi: 10.1136/bjo.78.7.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Werschnik C, Lommatzsch PK. Long-term follow-up of patients with conjunctival melanoma. Am. J. Clin. Oncol. 2002;25:248–255. doi: 10.1097/00000421-200206000-00009. [DOI] [PubMed] [Google Scholar]
- 5.Missotten GS, Keijser S, De Keizer RJ, De Wolff-Rouendaal D. Conjunctival melanoma in the Netherlands: a nationwide study. Invest. Ophthalmol. Vis. Sci. 2005;46:75–82. doi: 10.1167/iovs.04-0344. [DOI] [PubMed] [Google Scholar]
- 6.Shildkrot Y, Wilson MW. Conjunctival melanoma: pitfalls and dilemmas in management. Curr. Opin. Ophthalmol. 2010;21:380–386. doi: 10.1097/ICU.0b013e32833b7aab. [DOI] [PubMed] [Google Scholar]
- 7.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 8.Wiesner T, et al. NF1 mutations are common in desmoplastic melanoma. Am. J. Surg. Pathol. 2015;39:1357–1362. doi: 10.1097/PAS.0000000000000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shain AH, et al. Exome sequencing of desmoplastic melanoma identifies recurrent NFKBIE promoter mutations and diverse activating mutations in the MAPK pathway. Nat. Genet. 2015;47:1194–1199. doi: 10.1038/ng.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Viskochil D, et al. Deletions and a translocation interrupt a cloned gene at the neurofibromatosis type 1 locus. Cell. 1990;62:187–192. doi: 10.1016/0092-8674(90)90252-A. [DOI] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Network. Genomic classification of cutaneous melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krauthammer M, et al. Exome sequencing identifies recurrent mutations in NF1 and RASopathy genes in sun-exposed melanomas. Nat. Genet. 2015;47:996–1002. doi: 10.1038/ng.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griewank KG, et al. Genetic alterations and personalized medicine in melanoma: progress and future prospects. J. Natl. Cancer Inst. 2014;106:djt435. doi: 10.1093/jnci/djt435. [DOI] [PubMed] [Google Scholar]
- 14.Van Raamsdonk CD, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Raamsdonk CD, et al. Mutations in GNA11 in uveal melanoma. N. Engl. J. Med. 2010;363:2191–2199. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore AR, et al. Recurrent activating mutations of G-protein-coupled receptor CYSLTR2 in uveal melanoma. Nat. Genet. 2016;48:675–680. doi: 10.1038/ng.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johansson P, et al. Deep sequencing of uveal melanoma identifies a recurrent mutation in PLCB4. Oncotarget. 2016;7:4624–4631. doi: 10.18632/oncotarget.6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin M, et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat. Genet. 2013;45:933–936. doi: 10.1038/ng.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harbour JW, et al. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat. Genet. 2013;45:133–135. doi: 10.1038/ng.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harbour JW, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robertson AG, et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell. 2017;32:204–220. doi: 10.1016/j.ccell.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griewank KG, et al. SF3B1 and BAP1 mutations in blue nevus-like melanoma. Mod. Pathol. 2017;30:928–939. doi: 10.1038/modpathol.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van de Nes JAP, et al. Activating CYSLTR2 and PLCB4 mutations in primary leptomeningeal melanocytic tumors. J. Invest. Dermatol. 2017;137:2033–2035. doi: 10.1016/j.jid.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spendlove HE, et al. BRAF mutations are detectable in conjunctival but not uveal melanomas. Melanoma Res. 2004;14:449–452. doi: 10.1097/00008390-200412000-00003. [DOI] [PubMed] [Google Scholar]
- 25.Lake SL, et al. Multiplex ligation-dependent probe amplification of conjunctival melanoma reveals common BRAF V600E gene mutation and gene copy number changes. Invest. Ophthalmol. Vis. Sci. 2011;52:5598–5604. doi: 10.1167/iovs.10-6934. [DOI] [PubMed] [Google Scholar]
- 26.Goldenberg-Cohen N, et al. T1799A BRAF mutations in conjunctival melanocytic lesions. Invest. Ophthalmol. Vis. Sci. 2005;46:3027–3030. doi: 10.1167/iovs.04-1449. [DOI] [PubMed] [Google Scholar]
- 27.Gear H, Williams H, Kemp EG, Roberts F. BRAF mutations in conjunctival melanoma. Invest. Ophthalmol. Vis. Sci. 2004;45:2484–2488. doi: 10.1167/iovs.04-0093. [DOI] [PubMed] [Google Scholar]
- 28.Griewank K, et al. Conjunctival melanomas harbor BRAF and NRAS mutations and copy number changes similar to cutaneous and mucosal melanomas. Clin. Cancer Res. 2013;19:3143–3152. doi: 10.1158/1078-0432.CCR-13-0163. [DOI] [PubMed] [Google Scholar]
- 29.Griewank KG, et al. TERT promoter mutations in ocular melanoma distinguish between conjunctival and uveal tumours. Br. J. Cancer. 2013;109:497–501. doi: 10.1038/bjc.2013.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koopmans AE, et al. Prevalence and implications of TERT promoter mutation in uveal and conjunctival melanoma and in benign and premalignant conjunctival melanocytic lesions. Invest. Ophthalmol. Vis. Sci. 2014;55:6024–6030. doi: 10.1167/iovs.14-14901. [DOI] [PubMed] [Google Scholar]
- 31.Beadling C, et al. KIT gene mutations and copy number in melanoma subtypes. Clin. Cancer Res. 2008;14:6821–6828. doi: 10.1158/1078-0432.CCR-08-0575. [DOI] [PubMed] [Google Scholar]
- 32.Robert C, et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N. Engl. J. Med. 2015;372:30–39. doi: 10.1056/NEJMoa1412690. [DOI] [PubMed] [Google Scholar]
- 33.Robert C, et al. Pembrolizumab versus Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2015;372:2521–2532. doi: 10.1056/NEJMoa1503093. [DOI] [PubMed] [Google Scholar]
- 34.Larkin J, et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N. Engl. J. Med. 2015;373:23–34. doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schadendorf D, et al. Pooled analysis of long-term survival data from phase II and phase III trials of ipilimumab in unresectable or metastatic melanoma. J. Clin. Oncol. 2015;33:1889–1894. doi: 10.1200/JCO.2014.56.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Griewank KG, et al. Conjunctival melanomas harbor BRAF and NRAS mutations--response. Clin. Cancer Res. 2013;19:6331–6332. doi: 10.1158/1078-0432.CCR-13-2368. [DOI] [PubMed] [Google Scholar]
- 37.van de Nes J, et al. Targeted next generation sequencing reveals unique mutation profile of primary melanocytic tumors of the central nervous system. J. Neuro oncol. 2016;127:435–444. doi: 10.1007/s11060-015-2052-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larsen AC, et al. BRAF mutations in conjunctival melanoma: investigation of incidence, clinicopathological features, prognosis and paired premalignant lesions. Acta Ophthalmol. 2016;94:463–470. doi: 10.1111/aos.13007. [DOI] [PubMed] [Google Scholar]
- 39.Cao J, et al. Targeting of the MAPK and AKT pathways in conjunctival melanoma shows potential synergy. Oncotarget. 2017;8:58021–58036. doi: 10.18632/oncotarget.10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nissan MH, et al. Loss of NF1 in cutaneous melanoma is associated with RAS activation and MEK dependence. Cancer Res. 2014;74:2340–2350. doi: 10.1158/0008-5472.CAN-13-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cosgarea I, et al. Targeted next generation sequencing of mucosal melanomas identifies frequent NF1 and RAS mutations. Oncotarget. 2017;8:40683–40692. doi: 10.18632/oncotarget.16542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson DB, et al. Targeted next generation sequencing identifies markers of response to PD-1 blockade. Cancer Immunol. Res. 2016;4:959–967. doi: 10.1158/2326-6066.CIR-16-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.