Abstract
The proto‐oncogene NOTCH1 is frequently mutated in around 10% of patients with chronic lymphocytic leukemia (CLL). This study analyzed NOTCH1 mutation status of 317 Chinese patients with CLL by Sanger sequencing. The frequencies of NOTCH1 mutation in the PEST (proline (P), glutamic acid (E), serine (S), threonine (T)‐rich protein sequence) domain and the 3′ untranslated regions (UTR) were 8.2% and 0.9%, with the most frequent mutation being c.7541_7542delCT and c.*371A>G, respectively. Clinical and biological associations were determined including NOTCH1 mutations with advanced stage (Binet stage, P = 0.010), unmutated immunoglobulin heavy‐chain variable region (IGHV) gene (P < 0.001) and trisomy 12 (+12) (P = 0.014). NOTCH1‐mutated patients had lower CD20 expression intensity than NOTCH1‐unmutated patients (P = 0.029). In addition, NOTCH1‐mutated patients had shorter overall survival (OS) (P = 0.002) and treatment‐free survival (TFS) (P = 0.002) than NOTCH1‐unmutated patients, especially for patients with NOTCH1 c.7541_7542delCT and/or c.*371A>G mutations. Patients with both mutated NOTCH1 and unmutated IGHV had shorter OS (P < 0.001) and TFS (P < 0.001) than those with unmutated NOTCH1 or mutated IGHV. These data provide a comprehensive view of the clinical relevance and prognostic impact of NOTCH1 mutations on Chinese patients with CLL.
Keywords: Chronic lymphocytic leukemia, cytogenetics, molecular biology, NOTCH1 mutations, prognosis
Introduction
Chronic lymphocytic leukemia (CLL) is a common type of B‐cell chronic lymphoproliferative disorders in adults 1, 2. Some common molecular and cytogenetic abnormities in CLL, such as tumor protein 53 (TP53) mutation, TP53 deletion, immunoglobulin heavy‐chain variable region (IGHV) mutations, and ATM deletion, are associated with clinical features and prognosis of patients with CLL 3, 4, 5, 6, 7, 8.
NOTCH1 mutations are frequent in CLL 9. Encoding domains mutations, mainly in PEST (proline (P), glutamic acid (E), serine (S), threonine (T)‐rich protein sequence) domain 10, 11, 12, are reported to occur in nearly 10% patients with CLL in Western countries 3, 13, 14. The mutations increase with disease progression and drug resistance. In recent years, with the application of next‐generation sequencing technology, a better understanding of NOTCH1 mutations was built 15. Puente et al.16 firstly reported noncoding mutations in 3′ untranslated regions (UTR), including c.*378A>G, c.*371A>G and c.*380A>C, with total mutations rate about 5%. Soon afterward, Larrayoz et al. 17 showed that NOTCH1 noncoding domains mutations occurred in 2.2% of patients with CLL who participated the UK CLL4 trial.
In Western countries, NOTCH1 mutations have a significant association with unmutated IGHV 18. Pozzo et al. 19 found that NOTCH1 mutations suppressed CD20 expression intensity of B‐cell surface by dysregulating histone deacetylase (HDAC)‐mediated epigenetic changes. As a consequence, rituximab seems to have less benefits to patients with NOTCH1 mutations than those without mutation regardless of mutation sites 14, 20. Furthermore, NOTCH1 mutations are associated with trisomy 12 (+12), with nearly 40% mutated patients harboring +12 21, 22, 23. It has been reported that NOTCH1 abnormality represented a poor prognostic factor in CLL on both treatment‐free survival (TFS) and overall survival (OS) 5, 16, 17. The effects of different types of NOTCH1 mutations on prognosis were reported by D'Agaro et al. 24. Although Xia et al. 25 and Wu et al. 26 partly investigated NOTCH1 mutations in Chinese with CLL, so far, there has been no systemic study. It remains unclear that whether NOTCH1 mutations have a similar influence on Chinese patients with CLL as Western countries.
In this retrospective study, we analyzed correlations between NOTCH1 mutations and other clinical and prognostic parameters in 317 Chinese patients with CLL. Besides, survival analysis was performed based on NOTCH1 and other cytogenetic status.
Materials and Methods
Patients
This single‐center retrospective study included 317 patients with CLL diagnosed from November 1991 to December 2016 in our hospital. Diagnosis of CLL was based on the International Workshop on CLL‐National Cancer Institute criteria. This study was approved by the hospital ethics committee, and all patients were provided informed consent according to the Declaration of Helsinki. Among 317 patients, mutation status of PEST domain of 161 patients had been reported by Xia et al. 25.
NOTCH1 mutation detection
Genomic DNA from CLL samples was extracted as previous report 25. Briefly, PCR amplification was performed for NOTCH1 PEST domain forward primer 5′‐CAGATGCAGCAGCAGAACCTG‐3′ and reverse primer 5′‐AAAGGAAGCCGGGGTCTCGT‐3′ and 3′ UTR forward primer 5′‐CCTAACAGGCAGGTGATGCT‐3′ and reverse primer 5′‐ATCTGGCCCCAGGTAGA AAC‐3′. Sanger sequencing was performed in PEST domain and 3′ UTR. Variant allelic fraction threshold to detect mutant alleles by Sanger sequencing was 12% according to a previous study 5.
Cytogenetics and immunophenotyping
Karyotype analysis of CLL cells was performed after CpG‐oligodeoxynucleotide and interleukin‐2 stimulation. Fluorescence in situ hybridization (FISH) was carried out according to the procedures described previously 25. CD38 and ZAP‐70 were detected via flow cytometry, and the cutoff levels for positivity were 30% and 20%, respectively.
Statistical analyses
SPSS 23 was used to analyze data. OS was defined as time from diagnosis to death or last follow‐up, and TFS was calculated as time between diagnosis and first‐line treatment. Survival curves were constructed by Kaplan–Meier method, and log‐rank test was used for statistic associations. Continuous and categorical variables were analyzed by t test and χ 2 test, respectively. P < 0.05 was defined as statistical significance.
Results
Patient characteristics
Totally, 317 patients were enrolled in our study with a male/female ratio of 2.1 (Table 1). Median age was 61 years (range, 20–92), and 53.3% patients were older than 60 years; 163 patients were newly diagnosed, of whom 67 had treatment indication; 97 patients were previously diagnosed elsewhere, of whom 31 patients had treatment indication. Thirty‐three were relapsed, and 24 were refractory. Median time from diagnosis to sampling was 3 months (range, 0–264).
Table 1.
Clinical features of patients
| Characteristics | N |
|---|---|
| Gender (n = 317) | |
| Male | 216 |
| Female | 101 |
| Age (n = 317) | |
| >60 | 169 |
| ≤60 | 148 |
| Rai (n = 302) | |
| 0 | 27 |
| 1–2 | 150 |
| 3–4 | 125 |
| Binet (n = 302) | |
| A | 98 |
| B | 92 |
| C | 112 |
| Disease status (n = 317) | |
| Untreated | 260 |
| Newly diagnosis | |
| With treatment indication | 67 |
| No treatment indication | 96 |
| Diagnosed elsewhere | |
| With treatment indication | 31 |
| No treatment indication | 66 |
| Relapsed | 33 |
| Refractory | 24 |
Median follow‐up was 28 months (range, 3–289), during which 215 patients (67.8%) received treatment including rituximab‐based therapy (72/215, 33.5%), chemotherapy alone (112/215, 52.1%), ibrutinib (4/215, 1.9%), and data not available (27/215, 12.5%). Eventually, 70 patients turned to second‐line treatment because of disease progression, and 25 patients received third‐line therapy.
Sequencing results
Mutation frequencies of NOTCH1 were 8.2% (26/317) and 0.9% (3/317) in the PEST domain and 3′ UTR, respectively (Table S1). Among patients with PEST domain mutations, 21 cases harbored c.7541_7542delCT, a common frameshift deletion. In addition, c.7443delC, c.7210C>T, c.7378G>T, c.7410delC, and c.7222delC were also detected in the study cohort and all 3′UTR mutations were c.*371A>G. The characteristics of these three patients with 3′UTR mutation are shown in Table S2.
Clinical and biological characteristics
There were 27 patients in Rai 0 stage (8.9%), 150 in Rai 1–2 (49.7%), and 125 in Rai 3–4 (41.4%). The number of patients with Binet A, B, and C was 98 (32.4%), 92 (30.5%), and 112 (37.1%), respectively. Totally, 78 patients were CD38 positive (27.1%) and 118 were ZAP‐70 positive (46.1%). In addition, 165 patients harbored mutated IGHV (55.9%).
NOTCH1 mutations were more common in patients with Binet C stage (P = 0.010) and unmutated IGHV gene (P < 0.001). In addition, NOTCH1 mutations were related to CD38 ≥ 30% (P = 0.010), with no significant association with ZAP‐70 ≥ 20% (P = 0.293, Table 2).
Table 2.
The relationships between NOTCH1 mutations and clinical features
| Variables | Total (n) | NOTCH1 mutation (n) | NOTCH1 wild type (n) | P value |
|---|---|---|---|---|
| Gender | 317 | 29 | 288 | |
| Male | 216 | 23 | 193 | 0.176 |
| Female | 101 | 6 | 95 | |
| Age | 317 | 29 | 288 | |
| >60 | 169 | 16 | 153 | 0.833 |
| ≤60 | 148 | 13 | 135 | |
| Rai | 302 | 28 | 274 | |
| 0 | 27 | 0 | 27 | 0.086 |
| 1–2 | 150 | 12 | 138 | |
| 3–4 | 125 | 16 | 109 | |
| Binet | 302 | 28 | 274 | |
| A | 98 | 2 | 96 | 0.010 |
| B | 92 | 11 | 81 | |
| C | 112 | 15 | 97 | |
| CD38 | 288 | 27 | 261 | |
| ≥30% | 78 | 13 | 65 | 0.010 |
| <30% | 210 | 14 | 196 | |
| ZAP‐70 | 256 | 23 | 233 | |
| ≥20% | 118 | 13 | 105 | 0.293 |
| <20% | 138 | 10 | 128 | |
| IGHV status | 295 | 29 | 266 | |
| IGHV M | 165 | 6 | 159 | <0.001 |
| IGHV UM | 130 | 23 | 107 |
Cytogenetics
Complex karyotype was defined as ≥3 unrelated chromosome abnormalities in more than one cell on karyotype. Totally, 53 patients harbored complex karyotype (19.7%), of whom 7 had NOTCH1 mutations. The number of patients who harbored del (11) (q22‐23) (11q‐), del (13) (q14) (13q‐), del (17) (p13) (17p‐), and +12 was 39 (15.0%), 39 (13.5%), 79 (38.5%), and 59 (25.4%), respectively. The incidence of 13q‐ was relatively lower than previous study 27, which may due to more Binet C stage patients in our study and the differences of CLL genetic background between Eastern and Western countries patients 26.
The relationship between NOTCH1 mutations and chromosome abnormalities was shown in Table 3. NOTCH1 mutations were enriched in CLL patients with +12 (10 of 59 vs. 11 of 173; P = 0.014). No significant correlation was found between NOTCH1 mutations and 11q‐ (P = 0.210), 17p‐ (P = 1.000), 13q‐ (P = 0.053), or complex karyotype (P = 0.406).
Table 3.
Associations between NOTCH1 mutations and chromosome abnormalities
| Variables | Total (n) | NOTCH1 mutation (n) | NOTCH1 wild type (n) | P value |
|---|---|---|---|---|
| Complex karyotype | 269 | 25 | 244 | |
| Positive | 53 | 7 | 46 | 0.406 |
| Negative | 216 | 18 | 198 | |
| 11q‐ | 260 | 23 | 237 | |
| Positive | 39 | 6 | 33 | 0.210 |
| Negative | 221 | 17 | 204 | |
| 17p‐ | 288 | 26 | 262 | |
| Positive | 39 | 4 | 35 | 1.000 |
| Negative | 249 | 22 | 227 | |
| 13q‐ | 205 | 21 | 184 | |
| Positive | 79 | 4 | 75 | 0.053 |
| Negative | 126 | 17 | 109 | |
| +12 | 232 | 21 | 211 | |
| Positive | 59 | 10 | 49 | 0.014 |
| Negative | 173 | 11 | 162 |
CD20 expression intensity
A total of 13 patients with NOTCH1 mutations had data of CD20 expression intensity, while 138 of NOTCH1‐unmutated patients did. We found that NOTCH1‐mutated patients had lower CD20 expression intensity than unmutated patients (mean fluorescence intensity (MFI): 614.10 ± 430.71 vs. 951.84 ± 994.39, P = 0.029).
In addition, we analyzed the relationship between +12 and CD20 expression intensity. The number of patients who had data of CD20 expression intensity with and without +12 was 29 and 93, respectively. We found that +12 patients had higher CD20 expression intensity than patients without +12 (MFI: 796.90 ± 903.92 vs. 1294.70 ± 1167.18, P = 0.018).
Furthermore, we studied the synergetic effect of the status of NOTCH1 mutations and +12 on CD20 expression. We stratified our patients into four groups: (1) NOTCH1‐mutated and +12 (n = 4) (NOTCH1 M & +12); (2) NOTCH1‐unmutated and +12 (n = 25) (NOTCH1 UM & +12); (3) NOTCH1‐mutated and +12 negative (n = 6) (NOTCH1 M & +12 neg); and (4) NOTCH1‐unmutated and +12 negative (n = 87) (NOTCH1 UM & +12 neg). NOTCH1‐unmutated and +12 patients had higher CD20 expression than those with NOTCH1‐mutated and +12 negative (MFI: 1401.36 ± 1219.30 vs. 524.61 ± 265.72, P = 0.049) or NOTCH1‐unmutated and +12 negative patients (MFI: 1401.36 ± 1219.30 vs. 815.68 ± 929.76, P = 0.009) (Tables 4 and 5).
Table 4.
CD20 expression intensity of different groups stratified according to the status of NOTCH1 mutations and +12
| Groups (n) | MFI (mean ± SD) |
|---|---|
| NOTCH1 M & +12 (4) | 628.10 ± 365.98 |
| NOTCH1 UM & +12 (25) | 1401.36 ± 1219.30 |
| NOTCH1 M & +12 neg (6) | 524.61 ± 265.72 |
| NOTCH1 UM & +12 neg (87) | 815.68 ± 929.76 |
MFI, mean fluorescence intensity; SD, standard deviation; M, mutated; UM, unmutated; neg, negative.
Table 5.
Hierarchical stratification of CD20 expression intensity according to the status of NOTCH1 mutations and +12
| Pairwise comparisons | NOTCH1 M & +12 | NOTCH1 UM & +12 | NOTCH1 M & +12 neg |
|---|---|---|---|
| NOTCH1 UM & +12 | P = 0.141 | – | – |
| NOTCH1 M & +12 neg | P = 0.869 | P = 0.049 | – |
| NOTCH1 UM & +12 neg | P = 0.706 | P = 0.009 | P = 0.478 |
M, mutated; UM, unmutated; neg, negative.
Survival
Patients with NOTCH1 mutation had worse TFS (Fig. 1A, median: 3 vs. 18 months; P = 0.002) and OS (Fig. 1D, median: 52 vs. 216 months; P = 0.002) than those with wild‐type NOTCH1. Taking the status of NOTCH1 mutations and +12 into consideration at the same time, we further stratified our patients into four groups as previously mentioned: (1) NOTCH1 M & +12 (n = 10); (2) NOTCH1 UM & +12 (n = 49); (3) NOTCH1 M & +12 neg (n = 11); and (4) NOTCH1 UM & +12 neg (n = 162). There was no TFS (P = 0.235) and OS (P = 0.375) difference among four cohorts (Fig. 1B and E).
Figure 1.
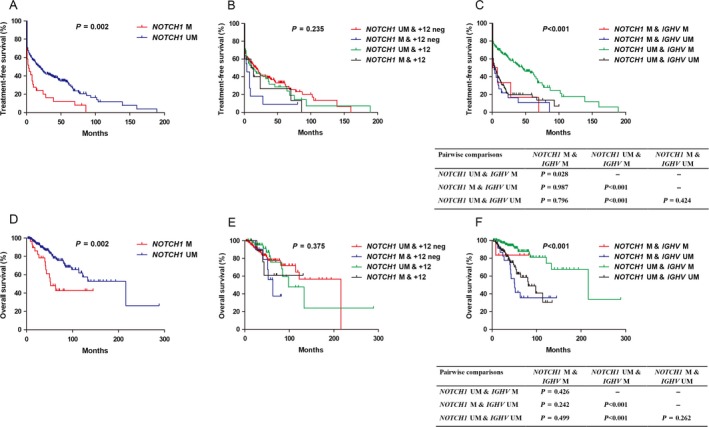
A and D: Kaplan–Meier curves of TFS (A) and OS (D) for NOTCH1 mutation status. B and E: Hierarchical stratification of TFS (B) and OS (E) according to +12 and NOTCH1 mutation status. C and F: Hierarchical stratification of TFS (C) and OS (F) according to IGHV and NOTCH1 mutation status. Abbreviations: UM: unmutated; M: mutated; neg: negative.
The numbers of patients with NOTCH1‐mutated and IGHV‐mutated (NOTCH1 M & IGHV M), NOTCH1‐unmutated and IGHV‐mutated (NOTCH1 UM & IGHV M), NOTCH1‐mutated and IGHV‐unmutated (NOTCH1 M & IGHV UM), and NOTCH1‐unmutated and IGHV‐unmutated (NOTCH1 UM & IGHV UM) were 6, 159, 23, and 107, respectively. Patients with mutated NOTCH1 and unmutated IGHV had worse TFS (Fig. 1C, median: 3 vs. 45 months, P < 0.001) and OS (Fig. 1F, median: 51 vs. 216 months, P < 0.001) than those with unmutated NOTCH1 and mutated IGHV. In addition, patients with mutated NOTCH1 and mutated IGHV (median: 1 vs. 45 months, P = 0.028) or unmutated NOTCH1 and unmutated IGHV (median: 4 vs. 45 months, P < 0.001) showed shorter TFS than unmutated NOTCH1 and mutated IGHV. Patients with unmutated NOTCH1 and unmutated IGHV had worse OS than those with unmutated NOTCH1 and mutated IGHV (median: 81 vs. 216 months, P < 0.001).
It was unknown whether different mutation types had different impacts on prognosis. We allocated patients into four groups: (1) NOTCH1‐unmutated (n = 288); (2) c.7541_7542delCT (n = 21); (3) c.*371A>G (n = 3); and (4) other NOTCH1 mutation types (n = 5). NOTCH1‐unmutated patients had longer TFS than those with c.7541_7542delCT (median: 18 vs. 3 months, P = 0.004) or other mutation types (median: 18 vs. 1 months, P = 0.048). Moreover, patients with unmutated NOTCH1 had longer OS than those with c.7541_7542delCT (median: 216 vs. 52 months, P = 0.003) or c.*371A>G (median: 216 vs. 18 months, P = 0.009) (Fig. 2).
Figure 2.
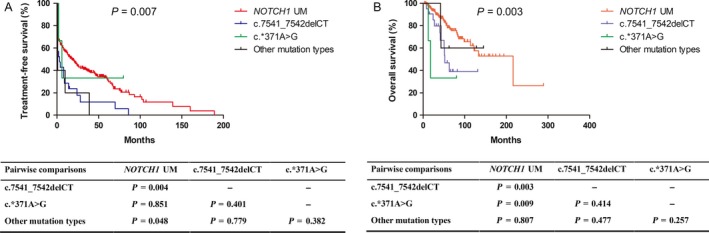
Kaplan–Meier curves of TFS (A) and OS (B) for NOTCH1 mutation types. Other NOTCH1 mutation types include c.7443delC, c.7210C>T, c.7378G>T, c.7410delC, and c.7222delC. Abbreviations: UM: unmutated.
Discussion
In the present research, we analyzed clinical characteristics and outcome of 317 Chinese CLL patients with different NOTCH1 mutational status. The mutation frequencies of PEST domain and noncoding domains were 8.2% and 0.9%, respectively, which were lower than those reported in Western countries 28. Moreover, only c.*371A>G was detected in noncoding domains, with no c.*378A>G or c.*380A>C. Several reasons might account for these different results: first, the differences of CLL genetic background between Eastern and Western countries patients. Second, the frequencies of 3′ UTR mutations were relatively low, with 2.0% for c.*378A>G and 0.8% for c.*380A>C 5. Third, the low sensitivity of Sanger sequencing may contribute. Previous studies have demonstrated that some NOTCH1 mutations, especially those in noncoding domains, had too low variant frequency to be detected by Sanger sequencing, but could be found by next‐generation sequencing 5. Consequently, mutation frequencies reported by our study might be lower than the true level, and to better study NOTCH1 mutations, these limitations should be taken into consideration.
The correlation between NOTCH1 mutations and chromosome abnormalities was assessed in the current study. NOTCH1 mutations were more common in advanced stage patients and were associated with CD38 positivity and unmutated IGHV 22. Complex karyotype, 11q‐, 13q‐, and 17p‐ were not associated with NOTCH1 mutations. However, patients with NOTCH1 mutations usually carried +12 21.
The relationships between NOTCH1 mutations or +12 and CD20 expression intensity were also performed in our study. Patients with NOTCH1 mutations had lower CD20 expression intensity, and +12 was associated with higher expression intensity of CD20. These findings had clinical value to guide the usage of rituximab for patients who had those two abnormities.
NOTCH1 mutation represented an unfavorable prognosis factor, which reflected on both TFS and OS 29. By subgroup analysis, no statistic difference for TFS or OS was found among four groups stratified by NOTCH1 and +12 status. Both NOTCH1 mutations and unmutated IGHV gene had unfavorable effects on survival, but their synergistic effects remained unknown. In our research, we studied survivals of different mutation status of NOTCH1 and IGHV. We found that patients with both mutated NOTCH1 and unmutated IGHV had shorter OS and TFS than those with unmutated NOTCH1 or mutated IGHV. Frameshift deletion c.7541_7542delCT represented unfavorable outcomes for TFS and OS, and c.*371A>G had a negative effect on OS. As a result, CLL patients with c.7541_7542delCT and c.*371A>G mutation deserve more attention in the clinic practice.
In summary, we analyzed clinical and molecular features of NOTCH1 mutations in 317 Chinese patients with CLL. The frequencies of mutations in the PEST domain and 3′ UTR of NOTCH1 were lower than those reported by Western countries. NOTCH1 mutations were more common in patients with advanced stage, unmutated IGHV gene, and +12. Moreover, NOTCH1 mutation represented an unfavorable prognostic factor, especially for c.7541_7542delCT and c.*371A>G. Further researches are needed to better understand the molecular mechanisms of NOTCH1 mutations and study the appropriate strategies to treat Chinese patients with CLL who harbor NOTCH1 mutations.
Conflict of Interest
The authors declare no conflict of interest.
Supporting information
Table S1. NOTCH1 mutations identified by Sanger sequencing in 317 Chinese CLL cases.
Table S2. Clinical characteristics of patients with NOTCH1 3′UTR mutation.
Acknowledgments
This study was supported by National Natural Science Foundation of China (81370657, 81470328, 81600130, 81600162, 81770166, 8170010636 and 81720108002), Project of National Key Clinical Specialty, the National Science & Technology Pillar Program (2014BAI09B12), Jiangsu Province's Medical Elite Programme (ZDRCA2016022), Jiangsu Provincial Special Program of Medical Science (BL2014086 and BE2017751), National Science and Technology Major Project (2017ZX09304032), Excellent Youth Foundation Project of Jiangsu Province (BK20160099), Natural Science Foundation of Jiangsu Province (BK20141028, BK20171079), and “Liu Da Ren Cai Gao Feng” of Jiangsu Province (2015‐WSN‐050).
Cancer Medicine 2018; 7(5):1689–1696
Contributor Information
Wei Xu, Email: xuwei10000@hotmail.com.
Jianyong Li, Email: lijianyonglm@medmail.com.cn.
References
- 1. Fernandez‐Martinez, J. L. , deAndres‐Galiana E. J., and Sonis S. T.. 2017. Genomic data integration in chronic lymphocytic leukemia. J. Gene Med. 19. [DOI] [PubMed] [Google Scholar]
- 2. Kim, J. A. , Hwang B., Park S. N., Huh S., Im K., Choi S., et al. 2016. Genomic profile of chronic lymphocytic leukemia in Korea identified by targeted sequencing. PLoS ONE 11:e0167641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stilgenbauer, S. , Schnaiter A., Paschka P., Zenz T., Rossi M., Dohner K., et al. 2014. Gene mutations and treatment outcome in chronic lymphocytic leukemia: results from the CLL8 trial. Blood 123:3247–3254. [DOI] [PubMed] [Google Scholar]
- 4. Amaya‐Chanaga, C. I. , and Rassenti L. Z.. 2016. Biomarkers in chronic lymphocytic leukemia: Clinical applications and prognostic markers. Best Pract. Res. Clin. Haematol. 29:79–89. [DOI] [PubMed] [Google Scholar]
- 5. Nadeu, F. , Delgado J., Royo C., Baumann T., Stankovic T., Pinyol M., et al. 2016. Clinical impact of clonal and subclonal TP53, SF3B1, BIRC3, NOTCH1, and ATM mutations in chronic lymphocytic leukemia. Blood 127:2122–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Quijano, S. , Lopez A., Rasillo A., Sayagues J. M., Barrena S., Sanchez M. L., et al. 2008. Impact of trisomy 12, del(13q), del(17p), and del(11q) on the immunophenotype, DNA ploidy status, and proliferative rate of leukemic B‐cells in chronic lymphocytic leukemia. Cytometry B Clin. Cytom. 74:139–149. [DOI] [PubMed] [Google Scholar]
- 7. Rasi, S. , Khiabanian H., Ciardullo C., Terzi‐di‐Bergamo L., Monti S., Spina V., et al. 2016. Clinical impact of small subclones harboring NOTCH1, SF3B1 or BIRC3 mutations in chronic lymphocytic leukemia. Haematologica 101:e135–e138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pozzo, F. , Dal Bo M., Peragine N., Bomben R., Zucchetto A., Rossi F., et al. 2013. Detection of TP53 dysfunction in chronic lymphocytic leukemia by an in vitro functional assay based on TP53 activation by the non‐genotoxic drug Nutlin‐3: a proposal for clinical application. J. Hematol. Oncol. 6:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pozzo, F. , Bittolo T., Vendramini E., Bomben R., Bulian P., Rossi F. M., et al. 2017. NOTCH1‐mutated chronic lymphocytic leukemia cells are characterized by a MYC‐related overexpression of nucleophosmin 1 and ribosome‐associated components. Leukemia 31:2407–2415. [DOI] [PubMed] [Google Scholar]
- 10. Campregher, P. V. , Petroni R. C., Muto N. H., Sitnik R., de Carvalho F. P., Bacal N. S., et al. 2016. A novel assay for the identification of NOTCH1 PEST domain mutations in chronic lymphocytic leukemia. Biomed. Res. Int. 2016:4247908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arruga, F. , Gizdic B., Bologna C., Cignetto S., Buonincontri R., Serra S., et al. 2017. Mutations in NOTCH1 PEST domain orchestrate CCL19‐driven homing of chronic lymphocytic leukemia cells by modulating the tumor suppressor gene DUSP22. Leukemia 31:1882–1893. [DOI] [PubMed] [Google Scholar]
- 12. Xu, J. J. , Yao F. R., Jiang M., Zhang Y. T., and Guo F.. 2017. High‐resolution melting analysis for rapid and sensitive NOTCH1 screening in chronic lymphocytic leukemia. Int. J. Mol. Med. 39:415–422. [DOI] [PubMed] [Google Scholar]
- 13. Fabbri, G. , Holmes A. B., Viganotti M., Scuoppo C., Belver L., Herranz D., et al. 2017. Common nonmutational NOTCH1 activation in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U S A. 114:E2911–E2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bo, M. D. , Del Principe M. I., Pozzo F., Ragusa D., Bulian P., Rossi D., et al. 2014. NOTCH1 mutations identify a chronic lymphocytic leukemia patient subset with worse prognosis in the setting of a rituximab‐based induction and consolidation treatment. Ann. Hematol. 93:1765–1774. [DOI] [PubMed] [Google Scholar]
- 15. Quijada‐Alamo, M. , Hernandez‐Sanchez M., Robledo C., Hernandez‐Sanchez J. M., Benito R., Montano A., et al. 2017. Next‐generation sequencing and FISH studies reveal the appearance of gene mutations and chromosomal abnormalities in hematopoietic progenitors in chronic lymphocytic leukemia. J. Hematol. Oncol. 10:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Puente, X. S. , Bea S., Valdes‐Mas R., Villamor N., Gutierrez‐Abril J., Martin‐Subero J. I., et al. 2015. Non‐coding recurrent mutations in chronic lymphocytic leukaemia. Nature 526:519–524. [DOI] [PubMed] [Google Scholar]
- 17. Larrayoz, M. , Rose‐Zerilli M. J., Kadalayil L., Parker H., Blakemore S., Forster J., et al. 2017. Non‐coding NOTCH1 mutations in chronic lymphocytic leukemia; their clinical impact in the UK CLL4 trial. Leukemia 31:510–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kristensen, L. , Kristensen T., Abildgaard N., Royo C., Frederiksen M., Mourits‐Andersen T., et al. 2016. LPL gene expression is associated with poor prognosis in CLL and closely related to NOTCH1 mutations. Eur. J. Haematol. 97:175–182. [DOI] [PubMed] [Google Scholar]
- 19. Pozzo, F. , Bittolo T., Arruga F., Bulian P., Macor P., Tissino E., et al. 2016. NOTCH1 mutations associate with low CD20 level in chronic lymphocytic leukemia: evidence for a NOTCH1 mutation‐driven epigenetic dysregulation. Leukemia 30:182–189. [DOI] [PubMed] [Google Scholar]
- 20. Bittolo, T. , Pozzo F., Bomben R., D'Agaro T., Bravin V., Bulian P., et al. 2017. Mutations in the 3’ untranslated region of NOTCH1 are associated with low CD20 expression levels chronic lymphocytic leukemia. Haematologica 102:e305–e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Del Giudice, I. , Rossi D., Chiaretti S., Marinelli M., Tavolaro S., Gabrielli S., et al. 2012. NOTCH1 mutations in +12 chronic lymphocytic leukemia (CLL) confer an unfavorable prognosis, induce a distinctive transcriptional profiling and refine the intermediate prognosis of +12 CLL. Haematologica 97:437–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Riches, J. C. , O'Donovan C. J., Kingdon S. J., McClanahan F., Clear A. J., Neuberg D. S., et al. 2014. Trisomy 12 chronic lymphocytic leukemia cells exhibit upregulation of integrin signaling that is modulated by NOTCH1 mutations. Blood 123:4101–4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lopez, C. , Delgado J., Costa D., Conde L., Ghita G., Villamor N., et al. 2012. Different distribution of NOTCH1 mutations in chronic lymphocytic leukemia with isolated trisomy 12 or associated with other chromosomal alterations. Genes Chromosom. Cancer 51:881–889. [DOI] [PubMed] [Google Scholar]
- 24. D'Agaro, T. , Bittolo T., Bravin V., Dal Bo M., Pozzo F., Bulian P., et al. 2017. NOTCH1 mutational status in chronic lymphocytic leukaemia: clinical relevance of subclonal mutations and mutation types. Br. J. Haematol. [DOI] [PubMed] [Google Scholar]
- 25. Xia, Y. , Fan L., Wang L., Gale R. P., Wang M., Tian T., et al. 2015. Frequencies of SF3B1, NOTCH1, MYD88, BIRC3 and IGHV mutations and TP53 disruptions in Chinese with chronic lymphocytic leukemia: disparities with Europeans. Oncotarget 6:5426–5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu, S. J. , Lin C. T., Agathangelidis A., Lin L. I., Kuo Y. Y., Tien H. F., et al. 2017. Distinct molecular genetics of chronic lymphocytic leukemia in Taiwan: clinical and pathogenetic implications. Haematologica 102:1085–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dohner, H. , Stilgenbauer S., Benner A., Leupolt E., Krober A., Bullinger L., et al. 2000. Genomic aberrations and survival in chronic lymphocytic leukemia. N. Engl. J. Med. 343:1910–1916. [DOI] [PubMed] [Google Scholar]
- 28. Rossi, D. , Rasi S., Fabbri G., Spina V., Fangazio M., Forconi F., et al. 2012. Mutations of NOTCH1 are an independent predictor of survival in chronic lymphocytic leukemia. Blood 119:521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Willander, K. , Dutta R. K., Ungerback J., Gunnarsson R., Juliusson G., Fredrikson M., et al. 2013. NOTCH1 mutations influence survival in chronic lymphocytic leukemia patients. BMC Cancer 13:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. NOTCH1 mutations identified by Sanger sequencing in 317 Chinese CLL cases.
Table S2. Clinical characteristics of patients with NOTCH1 3′UTR mutation.


