Abstract
Although early stage ovarian cancer is in most cases a curable disease, some patients relapse even with appropriate adjuvant treatment. Therefore, the identification of patient and tumor characteristics to better stratify risk and guide rational drug development is desirable. Using transcriptomic functional annotation followed by protein–protein interacting (PPI) network analyses, we identified functions that were upregulated and associated with detrimental outcome in patients with early stage ovarian cancer. Some of the identified functions included cell cycle, cell division, signal transduction/protein modification, cellular response to extracellular stimuli or transcription regulation, among others. Genes within these functions included AURKA, AURKB, CDK1, BIRC5, or CHEK1 among others. Of note, the histone‐lysine N‐methyltransferase (EZH2) and the ubiquitin‐conjugating enzyme E2C (UBE2C) genes were found to be upregulated and amplified in 10% and 6% of tumors, respectively. Of note, EZH2 and UBE2C were identified as principal interacting proteins of druggable networks. In conclusion, we describe a set of genes overexpressed in ovarian cancer with potential for therapeutic intervention including EZH2 and UBE2C.
Keywords: Clinical outcome, druggable proteins, EZH2, Ovarian cancer, protein–protein interaction, UBE2C
Introduction
Disseminated ovarian cancer is an incurable disease 1. However, if diagnosed in its early stage, resection and adjuvant chemotherapy can reduce the probability of the tumor to relapse and spread 2. Unfortunately, some patients with early stage ovarian cancer (mainly stage 1 and 2) are still at high risk of relapse, even after being treated with adequate surgical and adjuvant chemotherapy 2. In this context, the identification of patients who have high risk of recurrence is desirable as it can influence adjuvant treatment and guide future drug development.
Similar to other cancers, in ovarian cancer, different molecular mechanisms are responsible for cancer initiation and progression. Uncontrolled proliferation, migration, evasion from immunological regulation, or the capacity to generate new vessels are, among others, oncogenic hallmarks of ovarian cancer 3. Of note, agents that mitigate these functions, such as antimitotic chemotherapies, DNA damaging agents or anti‐angiogenic compounds, have reached the clinical practice 3, 4. Among agents that target classical deregulated functions such as cell division or proliferation, novel vulnerabilities with potential for therapeutic capacity are under evaluation, including protein modifications or epigenetic events. New drugs targeting the proteasome, ubiquitination, or bromodomains are currently under evaluation in several solid tumors 5.
In this context, it will be desirable to identify biological functions that are characteristically deregulated in ovarian cancer at a transcriptomic and proteomic level. Genomic signatures and protein–protein interacting networks could be used to select patients with higher risk of relapse in the long term. Furthermore, molecular elements involved in these biological functions could be potentially druggable, opening the door to evaluate new compounds against these alterations in the clinical setting. With this approach in mind, we have described genes and gene signatures associated with mitosis that predicted poor outcome specifically in patients with early stage ovarian tumors 6. However, we envision that an analysis based of functional genomics and protein–protein interactions could provide more robust prediction outcome in ovarian cancers, and a more general overview of the biological characteristics of this disease.
In this project using an in silico approach using public transcriptomic data, we identified deregulated functions in early stage ovarian cancer that were associated with worse outcome. Expression of some of these signatures identified patients at a higher risk. A protein–protein interaction analysis revealed hubs of proteins with oncogenic implications that could be inhibited pharmacologically. Of note, a relevant finding was the identification of the histone‐lysine N‐methyltransferase EZH2, and the ubiquitin‐conjugating enzyme E2C as key upregulated interacting proteins. In addition, these proteins were amplified in 10% and 6% of the ovarian tumors. The data presented opens the door to the further assessment of these signatures in clinical studies, and for the evaluation of novel therapies against the mentioned proteins or pathways.
Material and Methods
Transcriptomic and gene expression analyses
To identify differences at a transcriptomic level, we used a public dataset (GEO DataSet accession number: GSE14407) of mRNA levels from twelve isolated ovarian epithelial cell lines and twelve isolated serous ovarian cancer epithelial (CEPI) cells. Affymetrix CEL files were downloaded and analyzed with Affymetrix Transcriptome Analysis Console 3.0. Differential gene expression profile for both groups was performed using a minimum fourfold change. Oncomine™ Platform was used to confirm the GEO DataSet findings (https://www.oncomine.org/resource/login.html).
Evaluation of clinical outcome
The publicly available Kaplan–Meier (KM) Plotter Online Tool (http://kmplot.com/analysis/) was used to evaluate the relationship between gene expression levels and patient's clinical outcome in early stage ovarian cancer (stage I and II). Only genes significantly associated with detrimental outcome (Hazard Ratio ≥1 and P‐value ≤0.05) were used for subsequent analysis (n = 131). This tool was also used to determine progression‐free survival (PFS) and overall survival (OS) in functional combined analyses. All the analyses were performed independently by two authors (SMC and MLR) and reviewed by a third author (EMGM) (Accession date January 8th 2018). No discrepancies were observed.
Protein–protein interactions maps and functional evaluation
Using the String Online Tool (http://www.string-db.org), we constructed the interactome. The PPI map was based on the list of genes associated with poor PFS. Proteins showing less than two interactions were not considered. Subsequently, we performed a functional screening using Ensembl (http://www.ensembl.org), and Gene Ontology (GO) by biological function.
Selection of potential drug candidates
We used information from Selleckchem (http://www.selleckchem.com) and Genecards (http://www.genecards.org) to select potentially druggable genes. Then, as described above, we used the STRING tool to build the druggable ovarian cancer interactome. Based on interacting groups, we divided the PPI map in three functional clusters: cell cycle (n = 19), DNA damage (n = 4), and angiogenesis (n = 3). PPI hubs proteins were determined as those with a higher number of interactions than the average (Edges ≥17.2).
Identification of molecular alterations
We used data contained at cBioportal (http://www.cbioportal.org; TCGA Ovarian Serous Cystadenocarcinoma, n = 603) to identify potential copy number alterations (amplification or deletion), and the presence of mutations in the identified genes.
Results
Selection of deregulated genes and functional analyses
To identify deregulated functions in ovarian cancer cells, we used public transcriptomic data (GSE14407), to compare isolated serous ovarian cancer epithelial (CEPI) cells with isolated ovarian epithelial cell lines. Using a minimum fold change of four, we identified 2925 genes of which 131 were associated with poor clinical outcome (Fig. 1A and Table 1). The upregulation of the genes was confirmed using data from human samples contained at Oncomine (Table 1). Protein–protein interaction network showed 130 nodes and a cluster coefficient of 0.62 (Fig. S1).
Figure 1.
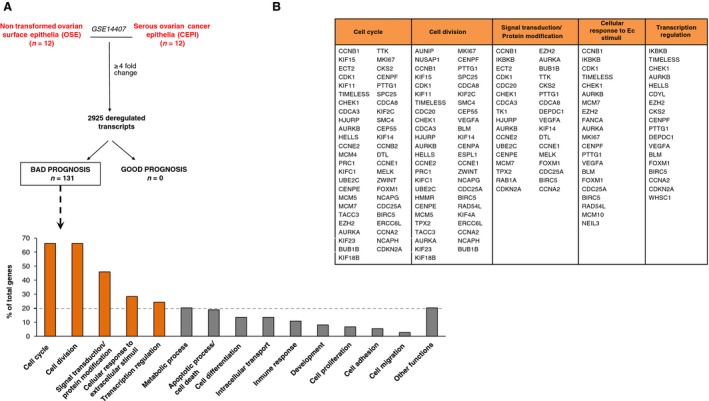
Transcriptomic analyses comparing isolated serous ovarian cancer epithelial (CEPI) cells with isolated ovarian epithelial cells. (A) Identification of deregulated genes (fold change ≥4) which are associated with bad prognosis in CEPI. (B) Functional enrichment analyses identify cell cycle, cell division, signal transduction/protein modification, cellular response to extracellular stimuli and transcription regulation, as the most altered functions in CEPI.
Table 1.
List of deregulated genes associated with bad prognosis
| Probe ID | Transcript ID | Gene symbol | AFFYMETRIX | ONCOMINE | KMPLOTTER | |||
|---|---|---|---|---|---|---|---|---|
| PFS | ||||||||
| Fold change | P‐Value ANOVA | Fold change | P‐Value | HR | P‐Value | |||
| 211767_at | g13543688 | GINS4 | 4.01 | 0.002866 | 2.404 | 1.11E‐05 | 2.87 (1.54–5.35) | 0.0005 |
| 228729_at | Hs.23960.0 | CCNB1 | 4.03 | 8.24E‐07 | 6.152 | 5.33E‐06 | 2.85 (1.3–6.22) | 0.0062 |
| 1569241_a_at | Hs2.149839.1 | ZNF93 | 4.06 | 0.004685 | 2.296 | 6.40E‐06 | 2.41 (1.14–5.08) | 0.0176 |
| 205869_at | g4506144 | PRSS1 | 4.06 | 0.000394 | 2.008 | 2.12E‐07 | 1.91 (1.06–3.45) | 0.0296 |
| 213100_at | Hs.13350.0 | UNC5B | 4.07 | 0.000451 | 1.351 | 0.006 | 1.82 (1.01–3.3) | 0.0432 |
| 216615_s_at | Hs.2142.1 | HTR3A | 4.11 | 0.001305 | 3.505 | 4.76E‐16 | 2.23 (1.19–4.15) | 0.0098 |
| 40020_at | 4858618_RC | CELSR3 | 4.13 | 9.48E‐08 | 1.502 | 7.89E‐07 | 3.86 (1.98–7.54) | 0 |
| 219306_at | g9910265 | KIF15 | 4.17 | 0.000147 | 3.696 | 2.29E‐08 | 2.88 (1.54–5.38) | 0.0005 |
| 209342_s_at | g4185274 | IKBKB | 4.17 | 0.007424 | 1.31 | 2.12E‐04 | 2.44 (1.31–4.56) | 0.0038 |
| 213759_at | Hs.111554.1 | ARL4C | 4.21 | 0.008012 | 3.624 | 8.81E‐06 | 1.89 (1.06–3.39) | 0.0289 |
| 206134_at | g7657318 | ADAMDEC1 | 4.21 | 0.003324 | 1.87 | 0.012 | 1.88 (1.04–3.39) | 0.0337 |
| 219787_s_at | g8922431 | ECT2 | 4.23 | 0.000033 | 10.209 | 2.81E‐08 | 2.62 (1.42–4.83) | 0.0014 |
| 210559_s_at | g3126638 | CDK1 | 4.23 | 0.000803 | 7.317 | 4.85E‐07 | 1.8 (1.01–3.23) | 0.0447 |
| 204444_at | g13699823 | KIF11 | 4.25 | 0.000272 | 5.467 | 7.52E‐07 | 2.73 (1.48–5.03) | 0.0008 |
| 209053_s_at | Hs.110457.3 | WHSC1 | 4.25 | 0.000004 | 3.805 | 3.67E‐10 | 3.03 (1.57–5.84) | 0.0005 |
| 209198_s_at | g13279139 | SYT11 | 4.27 | 0.000005 | 2.974 | 1.43E‐07 | 3.4 (1.81–6.39) | 0.0001 |
| 207156_at | g10800131 | HIST1H2AG | 4.3 | 0.006192 | 1.554 | 5.81E‐06 | 1.96 (1.1–3.5) | 0.0201 |
| 205544_s_at | g4503026 | CR2 | 4.34 | 0.027892 | 1.428 | 6.37E‐06 | 3.37 (1.74–6.53) | 0.0001 |
| 203046_s_at | g4507506 | TIMELESS | 4.36 | 4.84E‐07 | 3.434 | 2.30E‐09 | 1.86 (1.03–3.37) | 0.0365 |
| 202870_s_at | g4557436 | CDC20 | 4.41 | 0.000004 | 11.259 | 2.44E‐06 | 3.87 (2.01–7.46) | 0 |
| 202860_at | g7662151 | DENND4B | 4.43 | 0.000019 | 1.563 | 3.19E‐04 | 3.3 (1.71–6.37) | 0.0002 |
| 214933_at | Hs.96253.2 | CACNA1A | 4.45 | 0.000001 | 2.563 | 1.47E‐05 | 2.6 (1.39–4.85) | 0.0018 |
| 210587_at | g13477368 | INHBE | 4.47 | 0.042977 | 1.32 | 7.38E‐05 | 2.22 (1.2–4.09) | 0.0089 |
| 214005_at | Hs.77719.1 | GGCX | 4.5 | 2.02E‐08 | 1.837 | 2.04E‐04 | 2.45 (1.35–4.46) | 0.0025 |
| 205660_at | g11321576 | OASL | 4.53 | 0.006553 | 2.449 | 9.95E‐05 | 2.06 (1.13–3.78) | 0.0165 |
| 219454_at | g13124887 | EGFL6 | 4.54 | 0.000439 | 2.582 | 5.00E‐03 | 2.34 (1.27–4.31) | 0.0049 |
| 212816_s_at | Hs.84152.2 | CBS | 4.55 | 0.000561 | 2.658 | 1.00E‐03 | 2.06 (1.14–3.74) | 0.0146 |
| 205394_at | g4502802 | CHEK1 | 4.6 | 0.000004 | 4.147 | 2.43E‐07 | 2.04 (1.13–3.66) | 0.015 |
| 221436_s_at | g13876383 | CDCA3 | 4.65 | 0.001047 | 4.847 | 1.30E‐09 | 2.09 (1.15–3.77) | 0.0128 |
| 207109_at | g7657408 | POU2F3 | 4.66 | 0.024312 | 1.741 | 4.43E‐04 | 2.81 (1.53–5.18) | 0.0005 |
| 202219_at | g5032096 | SLC6A8 | 4.68 | 0.000195 | 1.94 | 1.76E‐07 | 2.17 (1.18–4) | 0.0108 |
| 217025_s_at | Hs.89434.1 | DBN1 | 4.69 | 0.007175 | 2.141 | 1.84E‐06 | 2.11 (1.13–3.93) | 0.0163 |
| 202338_at | g4507518 | TK1 | 4.73 | 0.00005 | 4.968 | 1.55E‐08 | 2.07 (1.13–3.77) | 0.0156 |
| 222251_s_at | Hs.28906.1 | GMEB2 | 4.81 | 0.004936 | 1.339 | 3.27E‐04 | 5.5 (2.57–11.76) | 0 |
| 210697_at | g4454677 | ZNF257 | 4.82 | 0.001284 | 1.673 | 1.66E‐05 | 2.06 (1.15–3.7) | 0.0135 |
| 214339_s_at | Hs.86575.2 | MAP4K1 | 4.87 | 0.000074 | 1.866 | 1.13E‐05 | 2.07 (1.13–3.79) | 0.0154 |
| 203022_at | g5454009 | RNASEH2A | 4.94 | 0.000147 | 2.785 | 1.11E‐06 | 2.06 (1.13–3.76) | 0.016 |
| 206280_at | g4826670 | CDH18 | 4.96 | 0.006096 | 1.396 | 0.004 | 2.05 (1.14–3.7) | 0.0143 |
| 211343_s_at | g180828 | COL13A1 | 5 | 0.000822 | 1.318 | 3.17E‐04 | 2.05 (1.13–3.75) | 0.0165 |
| 206513_at | g4757733 | AIM2 | 5.02 | 0.007612 | 1.547 | 6.09E‐04 | 2.73 (1.46–5.11) | 0.0011 |
| 204994_at | g11342663 | MX2 | 5.03 | 0.001647 | 3.916 | 3.32E‐04 | 2.2 (1.19–4.06) | 0.0098 |
| 205163_at | g7019426 | MYLPF | 5.04 | 0.000789 | 1.397 | 4.94E‐06 | 1.97 (1.1–3.54) | 0.0201 |
| 218726_at | g8922180 | HJURP | 5.07 | 0.010918 | 5.547 | 2.20E‐09 | 1.95 (1.08–3.5) | 0.023 |
| 239219_at | Hs.221197.0 | AURKB | 5.1 | 0.001028 | 2.818 | 2.34E‐05 | 2.22 (1.04–4.76) | 0.0353 |
| 202575_at | g6382069 | CRABP2 | 5.27 | 0.000004 | 3.216 | 9.08E‐05 | 2.08 (1.16–3.74) | 0.0124 |
| 35160_at | 4870487_RC | LDB1 | 5.29 | 0.00032 | 1.5 | 1.00E‐03 | 2.35 (1.27–4.32) | 0.0048 |
| 212556_at | Hs.239784.0 | SCRIB | 5.31 | 5.24E‐07 | 2.578 | 8.65E‐07 | 2 (1.1–3.65) | 0.021 |
| 203439_s_at | g12653744 | STC2 | 5.31 | 0.001003 | 2.509 | 1.53E‐06 | 1.85 (1.03–3.35) | 0.0379 |
| 234040_at | Hs.287543.0 | HELLS | 5.35 | 0.004925 | 2.352 | 3.86E‐05 | 2.39 (1.11–5.11) | 0.0209 |
| 221125_s_at | g7657250 | KCNMB3 | 5.47 | 0.000016 | 1.637 | 2.06E‐06 | 2.03 (1.11–3.71) | 0.0183 |
| 205569_at | g7657660 | LAMP3 | 5.48 | 0.040774 | 3.979 | 3.15E‐04 | 3.56 (1.85–6.85) | 0.0001 |
| 213520_at | Hs.31442.0 | RECQL4 | 5.48 | 0.00012 | 1.358 | 0.002 | 2.61 (1.4–4.87) | 0.0018 |
| 205034_at | g4757931 | CCNE2 | 5.49 | 0.00002 | 1.344 | 2.00E‐03 | 2.29 (1.27–4.11) | 0.0046 |
| 222037_at | Hs.154443.1 | MCM4 | 5.52 | 0.013214 | 4.726 | 8.00E‐08 | 2.9 (1.55–5.41) | 0.0005 |
| 218494_s_at | g13236503 | SLC2A4RG | 5.64 | 0.000878 | 2.118 | 5.86E‐06 | 2.48 (1.33–4.62) | 0.0031 |
| 212235_at | Hs.301685.0 | PLXND1 | 5.64 | 0.000117 | 1.496 | 4.67E‐04 | 2.05 (1.13–3.71) | 0.0152 |
| 218296_x_at | g8922469 | MSTO1 | 5.72 | 0.000026 | 1.381 | 0.018 | 1.83 (1–3.33) | 0.0458 |
| 218009_s_at | g4506038 | PRC1 | 5.74 | 9.19E‐07 | 7.214 | 5.32E‐08 | 2.85 (1.53–5.32) | 0.0006 |
| 209680_s_at | g12653842 | KIFC1 | 5.78 | 0.000129 | 3.845 | 3.64E‐08 | 2.33 (1.28–4.24) | 0.0046 |
| 202954_at | g5902145 | UBE2C | 5.81 | 2.13E‐08 | 10.184 | 2.24E‐07 | 3.03 (1.62–5.66) | 0.0003 |
| 205240_at | g9558734 | GPSM2 | 6.01 | 0.000545 | 3.965 | 2.97E‐08 | 1.81 (1.01–3.25) | 0.0435 |
| 209262_s_at | g12803666 | NR2F6 | 6.05 | 2.41E‐07 | 1.61 | 2.18E‐05 | 2.26 (1.23–4.17) | 0.0071 |
| 203632_s_at | g7706450 | GPRC5B | 6.12 | 0.000022 | 1.672 | 0.004 | 2.22 (1.21–4.04) | 0.0078 |
| 207165_at | g7108350 | HMMR | 6.14 | 0.000011 | 3.819 | 1.48E‐10 | 2.35 (1.3–4.25) | 0.0037 |
| 205046_at | g4502780 | CENPE | 6.16 | 0.00057 | 2.711 | 1.59E‐07 | 2.55 (1.36–4.75) | 0.0024 |
| 208394_x_at | g13259505 | ESM1 | 6.2 | 0.000021 | 1.496 | 0.009 | 2.05 (1.13–3.71) | 0.0152 |
| 216237_s_at | Hs.77171.1 | MCM5 | 6.22 | 0.001843 | 1.795 | 7.33E‐05 | 2.33 (1.25–4.34) | 0.0063 |
| 205449_at | g9558738 | SAC3D1 | 6.31 | 0.000029 | 1.891 | 3.53E‐05 | 2.19 (1.2–4) | 0.0086 |
| 203099_s_at | g4558755 | CDYL | 6.33 | 0.000004 | 1.889 | 4.26E‐05 | 2.1 (1.15–3.83) | 0.013 |
| 210983_s_at | g12751125 | MCM7 | 6.48 | 0.008102 | 3.523 | 2.31E‐07 | 2.23 (1.21–4.1) | 0.0084 |
| 210052_s_at | g6073830 | TPX2 | 6.5 | 2.90E‐08 | 13.887 | 1.65E‐07 | 2.55 (1.38–4.69) | 0.0019 |
| 225846_at | Hs.24743.1 | ESRP1 | 6.53 | 0.000005 | 2.135 | 2.68E‐04 | 2.3 (1.04–5.06) | 0.0335 |
| 218308_at | g5454101 | TACC3 | 6.54 | 0.000462 | 4.047 | 9.61E‐06 | 4.1 (2.04–8.24) | 0 |
| 239570_at | Hs.144137.0 | RAB1A | 6.76 | 0.000581 | 1.31 | 3.07E‐04 | 2.48 (1.1–5.6) | 0.0242 |
| 203358_s_at | g4758323 | EZH2 | 6.84 | 0.000002 | 6.584 | 1.44E‐06 | 3.63 (1.93–6.8) | 0 |
| 203806_s_at | g4503654 | FANCA | 6.87 | 0.00001 | 1.793 | 7.55E‐05 | 2.69 (1.42–5.08) | 0.0016 |
| 219502_at | g8922721 | NEIL3 | 6.91 | 0.000004 | 1.519 | 1.08E‐05 | 2.36 (1.3–4.28) | 0.0035 |
| 208079_s_at | g4507278 | AURKA | 7 | 3.64E‐08 | 6.504 | 6.53E‐08 | 2.95 (1.6–5.45) | 0.0003 |
| 204709_s_at | g13699831 | KIF23 | 7 | 0.000061 | 4.68 | 2.17E‐06 | 2.7 (1.48–4.95) | 0.0008 |
| 203755_at | g5729749 | BUB1B | 7.09 | 6.83E‐10 | 8.04 | 2.56E‐07 | 2.86 (1.55–5.29) | 0.0004 |
| 222039_at | Hs.274448.1 | KIF18B | 7.2 | 6.26E‐09 | 2.135 | 4.91E‐06 | 2.4 (1.31–4.37) | 0.0032 |
| 204822_at | g4507718 | TTK | 7.21 | 7.51E‐07 | 15.153 | 2.06E‐09 | 2.52 (1.38–4.61) | 0.0019 |
| 212023_s_at | Hs.80976.1 | MKI67 | 7.25 | 1.04E‐07 | 4.023 | 5.17E‐10 | 1.94 (1.07–3.51) | 0.0256 |
| 204170_s_at | g4502858 | CKS2 | 7.39 | 6.82E‐08 | 5.956 | 3.85E‐05 | 2.06 (1.14–3.73) | 0.0147 |
| 207183_at | g5453665 | GPR19 | 7.43 | 0.000063 | 2.901 | 8.95E‐09 | 3.07 (1.64–5.72) | 0.0002 |
| 207828_s_at | g4885132 | CENPF | 7.52 | 0.000002 | 3.811 | 1.75E‐06 | 2.64 (1.43–4.87) | 0.0013 |
| 206157_at | g4506332 | PTX3 | 7.56 | 0.000006 | 2.79 | 0.004 | 2.93 (1.6–5.37) | 0.0003 |
| 218039_at | g7705950 | NUSAP1 | 7.63 | 5.89E‐09 | 9.731 | 7.45E‐07 | 2.08 (1.16–3.75) | 0.0123 |
| 203554_x_at | g11038651 | PTTG1 | 7.68 | 0.000002 | 5.99 | 1.80E‐05 | 3.34 (1.76–6.34) | 0.0001 |
| 209891_at | g9963834 | SPC25 | 7.73 | 0.000027 | 2.928 | 9.73E‐24 | 2.45 (1.34–4.47) | 0.0026 |
| 221520_s_at | g12804484 | CDCA8 | 7.78 | 0.000076 | 3.705 | 5.44E‐07 | 2.63 (1.41–4.91) | 0.0016 |
| 218755_at | g5032012 | KIF20A | 7.9 | 1.87E‐08 | 9.021 | 9.21E‐08 | 2.56 (1.37–4.78) | 0.0021 |
| 201761_at | g13699869 | MTHFD2 | 7.92 | 0.000004 | 3.82 | 1.20E‐04 | 2.68 (1.45–4.94) | 0.001 |
| 204649_at | g4885624 | TROAP | 7.95 | 0.000014 | 3.096 | 5.11E‐08 | 2.83 (1.49–5.35) | 0.0008 |
| 209408_at | g1695881 | KIF2C | 8.01 | 2.31E‐08 | 2.834 | 6.75E‐11 | 2.43 (1.33–4.44) | 0.003 |
| 201663_s_at | g4885112 | SMC4 | 8.12 | 0.008518 | 7.44 | 9.32E‐09 | 2.55 (1.38–4.71) | 0.0019 |
| 218542_at | g8922501 | CEP55 | 8.28 | 0.000158 | 8.075 | 1.50E‐08 | 1.89 (1.05–3.4) | 0.0304 |
| 222958_s_at | Hs.133260.0 | DEPDC1 | 8.47 | 0.000182 | 3.833 | 2.12E‐07 | 2.61 (1.19–5.7) | 0.0127 |
| 222008_at | Hs.154850.0 | COL9A1 | 8.48 | 0.000545 | 1.946 | 2.30E‐16 | 1.93 (1.08–3.47) | 0.0249 |
| 210512_s_at | g3719220 | VEGFA | 8.51 | 6.97E‐09 | 2.741 | 1.17E‐07 | 3.37 (1.75–6.48) | 0.0001 |
| 205733_at | g4557364 | BLM | 8.53 | 0.000002 | 2.88 | 3.59E‐06 | 1.99 (1.1–3.59) | 0.0205 |
| 236641_at | Hs.116649.0 | KIF14 | 8.88 | 0.000311 | 3.139 | 5.27E‐06 | 2.28 (1.01–5.13) | 0.0414 |
| 204962_s_at | g4585861 | CENPA | 8.9 | 0.000014 | 11.775 | 2.63E‐09 | 2.48 (1.35–4.57) | 0.0026 |
| 202705_at | g10938017 | CCNB2 | 9.15 | 1.13E‐07 | 10.154 | 1.59E‐06 | 1.87 (1.04–3.37) | 0.0329 |
| 218585_s_at | g7705575 | DTL | 9.2 | 2.49E‐10 | 6.089 | 1.58E‐07 | 1.89 (1.06–3.38) | 0.0289 |
| 38158_at | 4852842_RC | ESPL1 | 9.2 | 8.96E‐09 | 4.341 | 6.11E‐07 | 3.19 (1.69–6.04) | 0.0002 |
| 213523_at | Hs.9700.0 | CCNE1 | 9.36 | 2.03E‐07 | 7.062 | 8.28E‐09 | 1.84 (1.02–3.33) | 0.0407 |
| 222946_s_at | g12652906 | AUNIP | 9.41 | 0.00175 | 2.956 | 7.47E‐08 | 2.11 (0.99–4.52) | 0.0485 |
| 213075_at | Hs.94795.0 | OLFML2A | 9.44 | 0.000481 | 1.423 | 3.00E‐03 | 1.86 (1.04–3.34) | 0.0348 |
| 204825_at | g7661973 | MELK | 9.59 | 0.000007 | 10.6 | 2.98E‐07 | 1.95 (1.08–3.5) | 0.0233 |
| 212563_at | Hs.30736.0 | BOP1 | 9.61 | 0.000165 | 1.669 | 3.35E‐06 | 2.03 (1.11–3.69) | 0.0186 |
| 204026_s_at | g6857828 | ZWINT | 9.92 | 1.65E‐09 | 7.001 | 1.71E‐05 | 2.05 (1.14–3.7) | 0.015 |
| 202580_x_at | g11386144 | FOXM1 | 10.06 | 0.000022 | 5.982 | 8.64E‐09 | 3.03 (1.6–5.73) | 0.0003 |
| 205694_at | g4507756 | TYRP1 | 10.15 | 0.000772 | 1.624 | 2.09E‐34 | 1.79 (1–3.23) | 0.0486 |
| 204584_at | Hs.1757.0 | L1CAM | 10.27 | 0.000008 | 3.985 | 7.02E‐15 | 3.02 (1.63–5.58) | 0.0002 |
| 218662_s_at | g11641252 | NCAPG | 10.35 | 4.18E‐08 | 3.207 | 2.13E‐10 | 3.28 (1.76–6.14) | 0.0001 |
| 204695_at | Hs.1634.0 | CDC25A | 10.39 | 0.000002 | 2.633 | 2.49E‐05 | 2.39 (1.31–4.34) | 0.0033 |
| 212807_s_at | Hs.281706.1 | SORT1 | 10.54 | 4.63E‐07 | 1.977 | 9.60E‐06 | 2.05 (1.12–3.75) | 0.0172 |
| 202094_at | Hs.1578.0 | BIRC5 | 10.82 | 0.000018 | 4.83 | 2.20E‐10 | 2.85 (1.53–5.31) | 0.0006 |
| 204558_at | g4506396 | RAD54L | 11.24 | 0.000856 | 2.09 | 6.38E‐06 | 2.85 (1.53–5.32) | 0.0006 |
| 218355_at | g7305204 | KIF4A | 11.53 | 5.42E‐08 | 2.359 | 1.00E‐03 | 2.82 (1.51–5.27) | 0.0007 |
| 219650_at | g8923111 | ERCC6L | 12.29 | 6.30E‐07 | 2.245 | 8.13E‐06 | 2.24 (1.24–4.04) | 0.0063 |
| 204437_s_at | g9257206 | FOLR1 | 12.72 | 0.000017 | 1.696 | 2.00E‐03 | 2.05 (1.13–3.7) | 0.0155 |
| 203418_at | g4502612 | CCNA2 | 13.14 | 0.00196 | 4.795 | 2.28E‐07 | 1.82 (1.02–3.26) | 0.0403 |
| 205242_at | g5453576 | CXCL13 | 16.29 | 0.0001 | 2.091 | 2.94E‐08 | 1.83 (1.02–3.28) | 0.0411 |
| 205572_at | g4557314 | ANGPT2 | 19.81 | 0.000006 | 1.312 | 0.029 | 2.02 (1.12–3.63) | 0.0164 |
| 212949_at | Hs.1192.0 | NCAPH | 19.94 | 7.17E‐07 | 2.497 | 1.51E‐06 | 3.06 (1.62–5.78) | 0.0003 |
| 206772_at | g4826953 | PTH2R | 21.94 | 0.000027 | 5.579 | 8.58E‐10 | 3.14 (1.66–5.97) | 0.0002 |
| 222962_s_at | g11527601 | MCM10 | 29.44 | 6.72E‐09 | 2.718 | 1.28E‐07 | 2.33 (1.06–5.1) | 0.0296 |
| 207039_at | g4502748 | CDKN2A | 45.1 | 0.000022 | 6.481 | 5.60E‐14 | 2.01 (1.11–3.63) | 0.0186 |
| 206373_at | g4507970 | ZIC1 | 99.77 | 2.13E‐08 | 3.712 | 8.24E‐07 | 2.19 (1.22–3.93) | 0.0073 |
Functional gene signatures associated with poor outcome
Functional annotation of the identified genes demonstrated several altered functions (Fig. 1A and B). By selecting those more represented (with a more than 20% of total genes expression), we identified cell cycle, cell division, signal transduction/protein modification, cellular response to extracellular (EC) stimuli, and transcription regulation. Table S1 provides detailed information of all functions and genes included within each function.
Using the KM Plotter online tool, we explored the association with clinical outcome of genes within each function. We did so to observe the role of each group with clinical prognosis. Genes within the cell cycle and cell division were associated with detrimental PFS and OS (PFS: HR = 4.07 (95% CI 1.66–9.98), P = 0.00086 and OS: HR = 3.33 (95% CI 0.94–11.81), P = 0.048 for cell cycle and PFS: HR = 3.58 (95% CI 1.46–8.78), P = 0.0029 and OS HR = 3.52 (95% CI 0.99–12.46), P = 0.038, for cell division) (Fig. 2). Results in the same range were observed for signal transduction/protein modification (PFS HR = 3.73 (95% CI 1.52–9.14), P = 0.002 and OS HR = 3.33 (95% CI 0.94–11.81), P = 0.048) and for transcription regulation PFS data (PFS: HR = 3.69 (95% CI 1.51–9.03), P = 0.0022). Interestingly, a poorer outcome for OS was found for this latter group (OS: HR = 12.55 (95% CI 1.65–95.48), P = 0.0017) (Fig. 3). Finally, the group of genes within the cellular response to EC stimuli function showed the worse outcome for both PFS and OS (PFS: HR = 6.37 (95% CI 2.22–18.28), P = 7.7e‐05 and OS: HR = 13.25 (95% CI 1.74–100.79), (Fig. 3).
Figure 2.
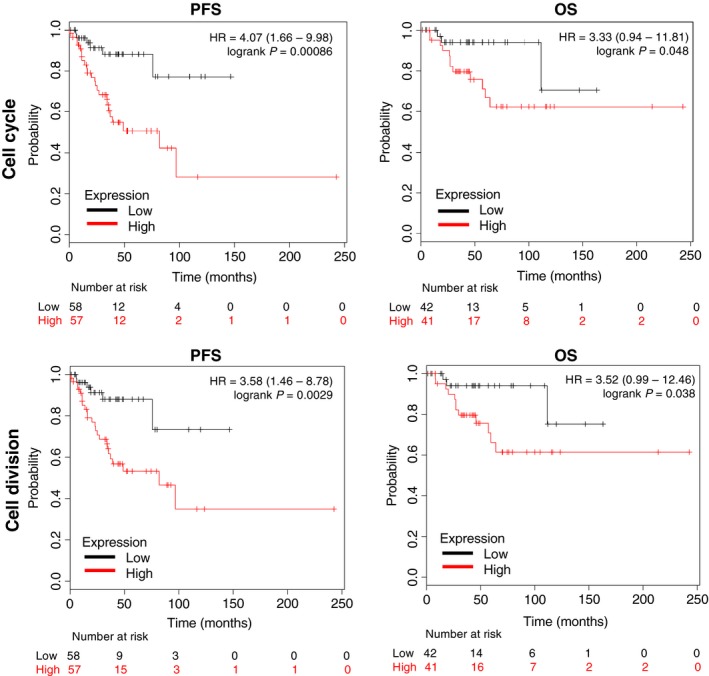
Association with progression‐free survival (PFS) and overall survival (OS) in stage I and II ovarian cancer of gene sets included in the cell cycle and cell division function.
Figure 3.
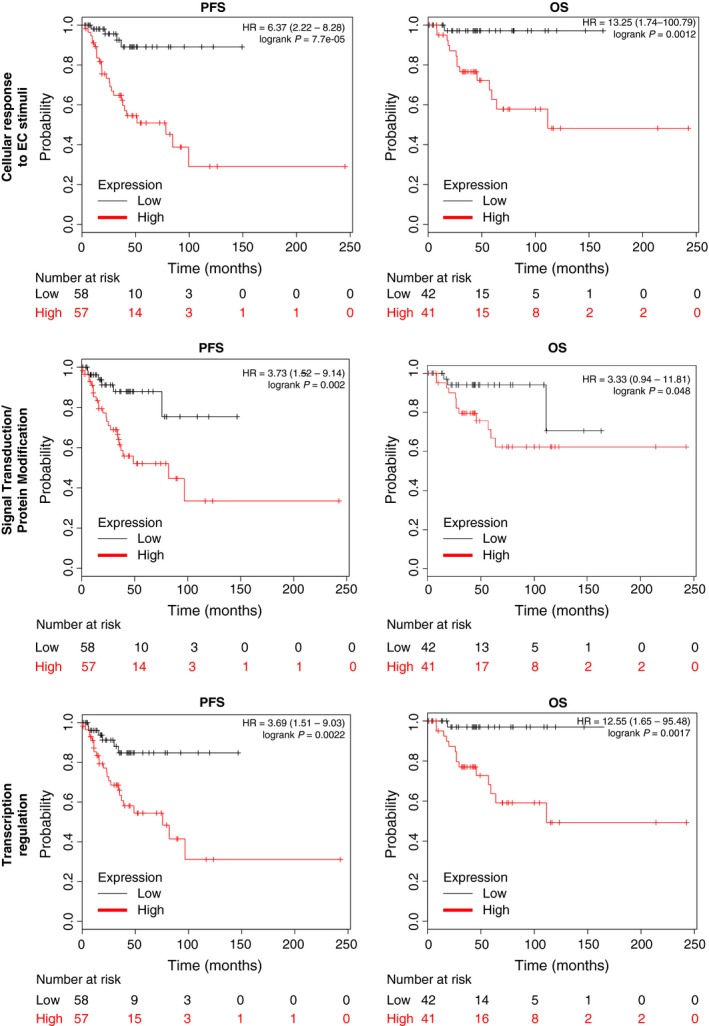
Association with progression‐free survival (PFS) and overall survival (OS) in stage I and II ovarian cancer of gene sets included in the cellular response to extracellular (EC) stimuli, signal transduction/protein modification and transcription regulation function.
Druggable opportunities within the identified functions
The description of functional signatures has the advantage of identifying relevant molecular alterations that have a potential oncogenic role in this disease, and therefore are susceptible to be inhibited. To get insights into potential therapies for those patients harboring these signatures, we used the drug gene interaction database available in Genecards and confirmed by other sources as described in Material and Methods. We therefore selected 26 genes that could potentially be inhibited pharmacologically (Table S2). We next used the proteins coded by these genes to build a protein–protein interaction network. We found 223 interactions (edges) linking 26 proteins (nodes). As expected, the clustering coefficient in this druggable network was high (0.85), confirming that most of the proteins act as a functional unit. We identified three different functional clusters with special affinity: Cell cycle (n = 19 genes), DNA damage (n = 4 genes), and angiogenesis (n = 3 genes) (Fig. 4A). Of note, DNA damage was included as part of the cellular response to EC stimuli in our initial functional annotation, and angiogenesis was one of the functions identified in the functional annotation studies, although was less represented (Table S1). These results suggest an important role of this in the druggable PPI. Next, based on the number of interactions, we selected the hub proteins of the interactome, defined as those with a higher number of interactions than the average (Edges ≥17.2; n = 18) (Fig. 4B).
Figure 4.
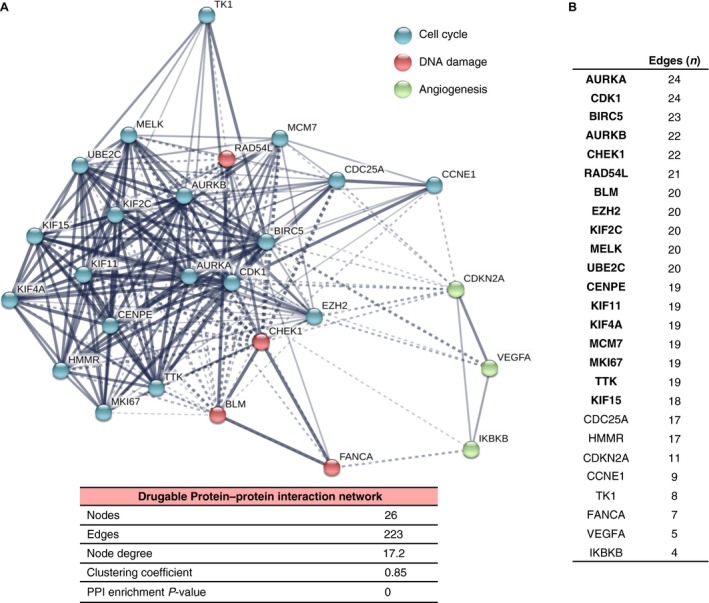
Protein–protein interaction (PPI) map of the 26 potential druggable targets. (A) Potentially druggable targets were used to construct a PPI network using the online tool STRING. Blue nodes represent proteins involved in cell cycle. Red and green nodes represent proteins associated with DNA damage and angiogenesis, respectively. The nodes indicate proteins coded by the identified druggable targets and edges indicate the number of interactions. The number of average interactions per node is represented by the node degree. The clustering coefficient indicates the average node density of the map. (B) List of hub proteins according the number of interactions (edges) in the druggable PPI network.
Some of the genes identified here have been described previously in ovarian cancer as deregulated, including AURKA, AURKB, CDK1, BIRC5, and CHEK1 among others 6. Of note, the histone‐lysine N‐methyltransferase EZH2 is a novel epigenetic target not previously described, and the ubiquitin‐conjugating enzyme E2C (UBE2C), which belongs to the ubiquitin ligase family of enzymes is also a potentially druggable protein with limited evaluation in ovarian cancer. Interestingly, these two genes strongly associate with worse prognosis for OS (Table S3)
Molecular alterations in the identified signatures
To complete our study, we used the cancer genomics database (cBioportal 7) to obtain information about copy number alterations or mutations of the identified druggable genes. Most of genes that code for the identified druggable hubs were found to be amplified in ovarian cancer (Table 2). Of note, the new potential targets EZH2 and UBE2C were amplified in around 10% and 6% of ovarian cancers, respectively. Deletions and mutations were present at a very low frequency. Amplifications of other genes such as RAD54L, AURKA, KIF2C, or BIRC5 were also observed.
Table 2.
Molecular alterations of the identified hub proteins
| 311 Ovarian serous cystadenocarcinoma samples | |||
|---|---|---|---|
| Gene Name | Amplification | Deletion | Mutation |
| EZH2 | 10.30% | 0.30% | – |
| RAD54L | 9.00% | – | 0.60% |
| AURKA | 8.70% | – | – |
| KIF2C | 6.40% | – | 0.30% |
| BIRC5 | 6.10% | 0.60% | – |
| UBE2C | 5.80% | – | – |
| BLM | 5.50% | 0.30% | 1.30% |
| CHEK1 | 3.90% | 0.60% | – |
| MKI67 | 3.50% | 1.00% | 1.30% |
| MCM7 | 3.20% | – | – |
| KIF4A | 1.90% | 0.30% | 0.60% |
| CDK1 | 1.90% | – | 0.60% |
| TTK | 1.60% | 0.30% | 0.60% |
| MELK | 1.30% | – | 0.60% |
| KIF15 | 1.00% | 0.30% | 0.30% |
| CENPE | 0.60% | 1.30% | 0.60% |
| AURKB | 0.60% | 0.60% | – |
| KIF11 | 0.60% | 0.30% | – |
Discussion
In the present article, we describe functional gene signatures and PPI networks associated with adverse outcome in early stage ovarian cancer. These signatures and interacting protein networks provide information about druggable opportunities that could be validated preclinically.
As ovarian cancer is an incurable disease, the identification of oncogenic functions and protein interacting networks associated with detrimental outcome is expected to improve the therapeutic landscape of this disease. In early stage ovarian cancer, the identification of patients with worse outcome is even more relevant as it may help in the selection of patients for additional adjuvant therapy, and even guide the evaluation of novel therapies.
In our study, we have identified five functions linked with detrimental PFS and OS in early stage ovarian cancer. Within cell cycle and cell division, we found genes such as AURKA, AURKB, CDK1, BIRC5, and CHEK1 that are associated with control of mitosis and cell cycle regulation 8. Of note, some of these genes have been reported previously to be linked with detrimental outcome 6. Inhibitors against proteins coded by these genes, such as AURKA and B or CHEK1, are currently in clinical development, so our findings provide support for the specific development of those agents in ovarian cancer.
An interesting finding was the identification of protein modifications and transcription regulation as upregulated functions. Protein modification and degradation is a vulnerability of tumor cells as has been demonstrated by the clinical activity of proteasome inhibitors in some hematological malignancies 9, 10. Ubiquitination is a necessary pathway to target proteins for degradation 11. The ubiquitin‐conjugating enzyme E2C is required for the destruction of mitotic cyclins and for cell cycle progression 12. UBE2C has been found to be overexpressed in esophageal squamous cell carcinoma playing a role in cancer progression 13, 14, as well as, in other tumor types such as nonsmall cell lung cancer 15. However, there are no published data regarding the role of this protein in ovarian cancer. As this family of proteins can be inhibited pharmacologically 11, the study of such agents in ovarian cancer is warranted.
Other relevant findings include the identification of EZH2 as upregulated and involved in the PPI network. EZH2 has been associated with epithelial to mesenchymal transition in ovarian cancers 16. Of note, EZH2 inhibitors seem to be particularly active in malignant rhabdoid tumors, which are deficient in the Switch/Sucrose NonFermentable (SWI/SNF) chromatin remodeling complexes INI1 (SMARCB1). Of interest, a subgroup of ovarian tumors has a similar phenotype and has shown responses to inhibitors of this complex 17. In our study, we observe that EZH2 is a relevant component of the PPI network therefore confirming a potentially druggable vulnerability. Of note, drugs such as tazemetostat, a potent and selective EZH2 inhibitor is currently in phase II testing 18. Other molecular alteration includes RAD54L that is amplified in 9% of patients. The protein associated by this gene is involved in the homologous recombination repair of DNA double‐strand breaks 19. Finally, genes such as KIF2C or AURKA are involved in mitotic formation and chromosome segregation 20.
Our analysis highlights several druggable functions in early stage ovarian cancer for which new agents are currently in preclinical or clinical evaluation. However, we should acknowledge that our study has some limitations. This is an in silico analysis that need confirmatory studies using human samples. In addition, functional assessment has the limitation for the redundancy of functions, as many genes can be classified in many different annotations. Finally, there are limitations for the existed software that help identifying druggable opportunities mainly for redundancy.
In conclusion, we have identified biological functions and PPI networks that are prognostic in early stage ovarian cancer and may guide future drug development (Fig. 5). Some of the identified genes such as EZH2 or UBE2C have not been described previously in ovarian cancer but are amplified, linked with detrimental prognosis and potentially druggable, and warrant preclinical and clinical assessment.
Figure 5.
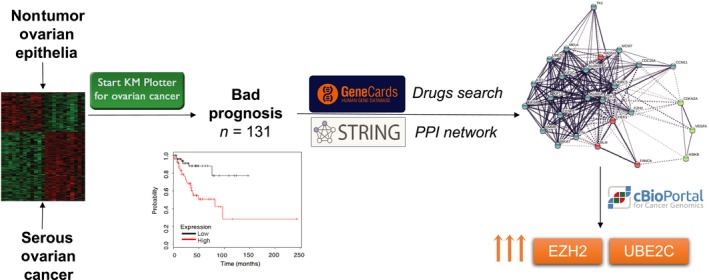
Study graphical abstract.
Conflict of Interest
None declared.
Supporting information
Table S1. Functional classification of the deregulated genes.
Table S2. List of potentially druggable genes.
Table S3. Association with progression free survival (PFS) and overall survival (OS) of the identified hub proteins.
Figure S1. Protein‐protein interaction network of the 130 deregulated genes associated with detrimental prognosis.
Acknowledgments
This work has been funded by Instituto de Salud Carlos III (PI16/01121), Diputación de Albacete and CRIS Cancer Foundation (to AO) and the framework agreement between University of Castilla‐La Mancha and Albacete Provincial Council (UCLM‐Excma. Diputación de Albacete) in support to research activity (to EMGM). We would like to also thanks to the cancer associations AMUMA and ACEPAIN for supporting part of this work. EMGM is funded by the implementation research program of the UCLM (UCLM resolution date: 31/07/2014), with a contract for accessing the Spanish System of Science, Technology and Innovation‐Secti (cofunded by the European Commission/FSE funds).
Cancer Medicine 2018; 7(5):1896–1907
Contributor Information
Eva María Galán‐Moya, Email: EvaMaria.Galan@uclm.es.
Alberto Ocaña, Email: albertoo@sescam.jccm.es.
References
- 1. Wilson, M. K. , Pujade‐Lauraine E., Aoki D., Mirza M. R., Lorusso D., Oza A. M., Bois A., Vergote I., Reuss A., Bacon M., and Friedlander M.. 2017. Fifth Ovarian cancer consensus conference of the gynecologic cancer intergroup: recurrent disease. Ann. Oncol. 28:727–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Karam, A. , Ledermann J. A., Kim J. W., Sehouli J., Lu K., Gourley C., Katsumata N., Burger R. A., Nam B. H., Bacon M., and Ng C.. 2017. Fifth Ovarian cancer consensus conference of the gynecologic cancer intergroup: first‐line interventions. Ann. Oncol. 28:711–717. [DOI] [PubMed] [Google Scholar]
- 3. George, A. , Kaye S., and Banerjee S.. 2017. Delivering widespread BRCA testing and PARP inhibition to patients with ovarian cancer. Nat. Rev. Clin. Oncol. 14:284–296. [DOI] [PubMed] [Google Scholar]
- 4. Jayson, G. C. , Kerbel R., Ellis L. M., and Harris A. L.. 2016. Antiangiogenic therapy in oncology: current status and future directions. Lancet 388:518–529. [DOI] [PubMed] [Google Scholar]
- 5. Ocana, A. , and Pandiella A.. 2017. Targeting oncogenic vulnerabilities in triple negative breast cancer: biological bases and ongoing clinical studies. Oncotarget 8:22218–22234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ocaña, A. , Pérez‐Peña J., Alcaraz‐Sanabria A., Sánchez‐Corrales V., Nieto‐Jiménez C., Templeton A. J., Seruga B., Pandiella A., and Amir E.. 2016. In silico analyses identify gene‐sets, associated with clinical outcome in ovarian cancer: role of mitotic kinases. Oncotarget 7:22865–22872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cerami, E. , Gao J., Dogrusoz U., Gross B. E., Sumer S. O., Aksoy B. A., Jacobsen A., Byrne C. J., Heuer M. L., Larsson E., and Antipin Y.. 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dominguez‐Brauer, C. , Thu K. L., Mason J. M., Blaser H., Bray M. R., and Mak T. W.. 2015. Targeting mitosis in cancer: emerging strategies. Mol. Cell 60:524–536. [DOI] [PubMed] [Google Scholar]
- 9. San‐Miguel, J. F. , Hungria V. T., Yoon S. S., Beksac M., Dimopoulos M. A., Elghandour A., Jedrzejczak W. W., Günther A., Nakorn T. N., Siritanaratkul N., and Corradini P.. 2014. Panobinostat plus bortezomib and dexamethasone versus placebo plus bortezomib and dexamethasone in patients with relapsed or relapsed and refractory multiple myeloma: a multicentre, randomised, double‐blind phase 3 trial. Lancet Oncol. 15:1195–1206. [DOI] [PubMed] [Google Scholar]
- 10. San Miguel, J. F. , Schlag R., Khuageva N. K., Dimopoulos M. A., Shpilberg O., Kropff M., Spicka I., Petrucci M. T., Palumbo A., Samoilova O. S., and Dmoszynska A.. 2008. Bortezomib plus melphalan and prednisone for initial treatment of multiple myeloma. N. Engl. J. Med. 359:906–917. [DOI] [PubMed] [Google Scholar]
- 11. Gallo, L. H. , Ko J., and Donoghue D. J.. 2017. The importance of regulatory ubiquitination in cancer and metastasis. Cell Cycle 16:634–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bajaj, S. , Alam S. K., Roy K. S., Datta A., Nath S., and Roychoudhury S.. 2016. E2 Ubiquitin‐conjugating Enzyme, UBE2C Gene, Is Reciprocally Regulated by Wild‐type and Gain‐of‐Function Mutant p53. J. Biol. Chem. 291:14231–14247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Palumbo, A. Jr , Da Costa N. M., De Martino M., Sepe R., Pellecchia S., de Sousa V. P. L., Neto P. N., Kruel C. D., Bergman A., Nasciutti L. E., and Fusco A.. 2016. UBE2C is overexpressed in ESCC tissues and its abrogation attenuates the malignant phenotype of ESCC cell lines. Oncotarget 7:65876–65887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lin, J. , Raoof D. A., Wang Z., Lin M. Y., Thomas D. G., Greenson J. K., Giordano T. J., Orringer M. B., Chang A. C., Beer D. G., and Lin L.. 2006. Expression and effect of inhibition of the ubiquitin‐conjugating enzyme E2C on esophageal adenocarcinoma. Neoplasia 8:1062–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang, Z. , Liu P., Wang J., Gong T., Zhang F., Ma J., and Han N.. 2015. Ubiquitin‐conjugating enzyme E2C regulates apoptosis‐dependent tumor progression of non‐small cell lung cancer via ERK pathway. Med. Oncol. 32:149. [DOI] [PubMed] [Google Scholar]
- 16. Cardenas, H. , Zhao J., Vieth E., Nephew K. P., and Matei D.. 2016. EZH2 inhibition promotes epithelial‐to‐mesenchymal transition in ovarian cancer cells. Oncotarget 7:84453–84467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chan‐Penebre, E. , Armstrong K., Drew A., Grassian A. R., Feldman I., Knutson S. K., Kuplast‐Barr K., Roche M., Campbell J., Ho P., and Copeland R. A.. 2017. Selective killing of SMARCA2‐ and SMARCA4‐deficient small cell carcinoma of the ovary, hypercalcemic type cells by inhibition of EZH2: in vitro and in vivo preclinical models. Mol. Cancer Ther. 16:850–860. [DOI] [PubMed] [Google Scholar]
- 18. Christofides, A. , Karantanos T., Bardhan K., and Boussiotis V. A.. 2016. Epigenetic regulation of cancer biology and anti‐tumor immunity by EZH2. Oncotarget 7:85624–85640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mazina, O. M. , Rossi M. J., Thomaa N. H., and Mazin A. V.. 2007. Interactions of human rad54 protein with branched DNA molecules. J. Biol. Chem. 282:21068–21080. [DOI] [PubMed] [Google Scholar]
- 20. Ritter, A. , Sanhaji M., Friemel A., Roth S., Rolle U., Louwen F., and Yuan J.. 2015. Functional analysis of phosphorylation of the mitotic centromere‐associated kinesin by Aurora B kinase in human tumor cells. Cell Cycle 14:3755–3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Functional classification of the deregulated genes.
Table S2. List of potentially druggable genes.
Table S3. Association with progression free survival (PFS) and overall survival (OS) of the identified hub proteins.
Figure S1. Protein‐protein interaction network of the 130 deregulated genes associated with detrimental prognosis.


