Abstract
Incidence of glioma is approximately 50% higher in males. Previous analyses have examined exposures related to sex hormones in women as potential protective factors for these tumors, with inconsistent results. Previous glioma genome-wide association studies (GWAS) have not stratified by sex. Potential sex-specific genetic effects were assessed in autosomal SNPs and sex chromosome variants for all glioma, GBM and non-GBM patients using data from four previous glioma GWAS. Datasets were analyzed using sex-stratified logistic regression models and combined using meta-analysis. There were 4,831 male cases, 5,216 male controls, 3,206 female cases and 5,470 female controls. A significant association was detected at rs11979158 (7p11.2) in males only. Association at rs55705857 (8q24.21) was stronger in females than in males. A large region on 3p21.31 was identified with significant association in females only. The identified differences in effect of risk variants do not fully explain the observed incidence difference in glioma by sex.
Introduction
Glioma is the most common type of primary malignant brain tumor in the United States (US), with an average annual age-adjusted incidence rate of 6.0/100,0001. Glioma can be broadly classified into glioblastoma (GBM, 61.9% of gliomas in adults 18+ in the US) and lower-grade glioma (non-GBM glioma, 24.2% of adult gliomas) with tumors such as ependymoma (6.3%), unclassified malignant gliomas (5.1%), and pilocytic astrocytoma (1.9%) making up the majority of other cases1. Many environmental exposures have been investigated as sources of glioma risk, but the only validated risk factors for these tumors are ionizing radiation (which increases risk), and history of allergies or other atopic disease (which decreases risk)2. These tumors are significantly more common in people of European ancestry, in males and in older adults1. The contribution of common low-penetrance SNPs to the heritability of sporadic glioma in persons with no documented family history is estimated to be ~25%3. A recent glioma genome-wide association study (GWAS) meta-analysis validated 12 previously reported risk loci4, and identified 13 new risk loci. These 25 loci in total are estimated to account for ~30% of heritable glioma risk. This suggests that there are both undiscovered environmental risk (which accounts for ~75% of incidence variance) and genetic risk factors (accounting for ~70% of heritable risk)3,4.
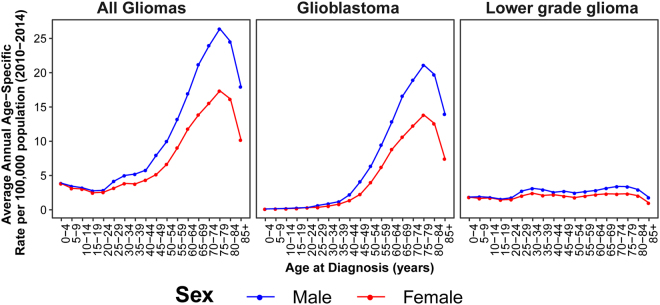
Population-based studies consistently demonstrate that incidence of gliomas varies significantly by sex. Most glioma histologies occur with a 30–50% higher incidence in males, and this male preponderance of glial tumors increases with age in adult glioma (Fig. 1)1. Several studies have attempted to estimate the influence of lifetime estrogen and progestogen exposure on glioma risk in women5,6. Results of these analyses have been mixed, and it is not possible to conclusively determine the impact of hormone exposure on glioma risk. Male predominance in incidence occurs broadly across multiple cancer types and is also evident in cancers that occur in pre-pubertal children and in post-menopausal adults7,8. Together these observations suggest that other mechanisms in addition to acute sex hormone actions must be identified to account for the magnitude of sex difference in glioma incidence.
Figure 1.
Average Annual Incidence of all glioma, glioblastoma and lower grade glioma by sex and age at diagnosis (CBTRUS 2010–2014).
Though sex differences exist in glioma incidence, sex differences have not been interrogated in previous glioma GWAS. Sex-specific analyses have the potential to reveal genetic sources of sexual dimorphism in risk, as well as to increase power for detection of loci where effect size or direction may vary by sex9,10. The aim of this analysis is to investigate potential sex-specific sources of genetic risk for glioma that may contribute to observed sex-specific incidence differences.
Results
Study population
There were 4,831 male cases, 5,216 male controls, 3,206 female cases, and 5,470 female controls (Table 1). A slightly larger proportion of male cases were GBM (58.7% of male cases vs 52.5% of female cases). Controls were slightly older than cases. GBM cases had a higher mean age than non-GBM cases, which was consistent with known incidence patterns of these tumors. Male and female cases within histology groups had similar age at diagnosis. The proportion of non-GBM cases varied by study due to differing recruitment patterns and study objectives (see original publications for details of recruitment patterns and inclusion criteria)4,11–14.
Table 1.
Population characteristics by study and sex.
| Characteristic | Study | Males | Females | ||
|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | ||
| N | Total | 4,831 | 5,216 | 3,206 | 5,470 |
| GICCa | 2,733 | 1,868 | 1,831 | 1,397 | |
| SFAGS-GWASb | 440 | 749 | 237 | 1,611 | |
| MDA-GWASc | 714 | 1,094 | 429 | 1,142 | |
| GliomaScand | 944 | 1,465 | 709 | 1,260 | |
| Mean Age (SD) | Total | 52.5 (14.5)** | 58.2 (15.2)** | 51.8 (14.9)** | 54.7 (14.5)** |
| GICC | 52.5 (14.3) | 56.1 (13.4) | 51.3 (14.6) | 53.4 (14.3) | |
| SFAGS-GWAS | 53.8 (13.0) | 50.6 (14.8) | 53.5 (14.0) | 49.3 (13.2) | |
| MDA-GWAS | 47.1 (13.0) | Modal age group: 60–69e | 47.7 (13.9) | Modal age group: 65–69f | |
| GliomaScan | 56.0 (15.5) | 69.3 (12.7) | 55.1 (15.7) | 64.0 (15.4) | |
| GBM (% of total)g | Total | 2,835 (58.7%)** | — | 1,682 (52.5%)** | — |
| GICC | 1,575 (57.6%) | — | 885 (48.3%) | — | |
| SFAGS-GWAS | 333 (75.7%) | — | 178 (75.1%) | — | |
| MDA-GWAS | 397 (55.6%) | — | 246 (57.3%) | — | |
| GliomaScan | 530 (56.1%) | — | 373 (52.6%) | — | |
| GBM - Mean Age (SD) | Total | 57.3 (12.0) ** | — | 57.8 (12.1) ** | — |
| GICC | 57.7 (11.4) | — | 57.8 (11.6) | — | |
| SFAGS-GWAS | 56.4 (11.5) | — | 56.2 (12.3) | — | |
| MDA-GWAS | 52.0 (11.7) | — | 53.7 (11.3) | — | |
| GliomaScan | 60.4 (13.0) | — | 61.4 (12.5) | — | |
| Non-GBM (% of total)g | Total | 1,716 (35.5%)** | — | 1,320 (41.2%)** | — |
| GICC | 1,036 (37.9%) | — | 862 (47.1%) | — | |
| SFAGS-GWAS | 107 (24.3%) | — | 59 (24.9%) | — | |
| MDA-GWAS | 317 (44.4%) | — | 183 (42.7%) | — | |
| GliomaScan | 256 (27.1%) | — | 216 (30.5%) | — | |
| Non-GBM - Mean Age (SD) | Total | 44.3 (14.4)** | — | 43.9 (14.3)** | — |
| GICC | 44.7 (14.6) | — | 44.6 (14.2) | — | |
| SFAGS-GWAS | 45.7 (14.2) | — | 45.4 (15.8) | — | |
| MDA-GWAS | 41.0 (11.9) | — | 39.6 (12.9) | — | |
| GliomaScan | 46.3 (15.5) | — | 44.4 (15.2) | — | |
aData from Glioma International Case-Control Study (GICC; Melin, et al.4); bData from San Francisco Adult Glioma Study GWAS (SFAGS-GWAS; Wrensch, et al.12); cdata from MD Anderson Cancer Center GWAS (MDA-GWAS; Shete, et al.13); dData from the National Cancer Institute’s GliomaScan (GliomaScan; Rajaraman, et al.14); eData from CGEMS prostate study (Yeager et al.35). Continuous age is not available, age distribution is as follows 50–59: 12.3%, 60–69: 56.7%, 70–79: 30.7%, 80–89: 0.3%; fData from CGEMS breast study (Hunter et al.36). Continuous age is not available, age distribution is as follows: 0–54: 4.3%, 55–59: 15.0%, 60–64: 23.6%, 65–69: 27.5%, 70–74: 19.0%, 75–99: 10.7%; gHistology information not available for all cases and frequencies may not add to 100%.
**Differs between included studies at the p < 0.05 level.
Previously discovered glioma risk regions
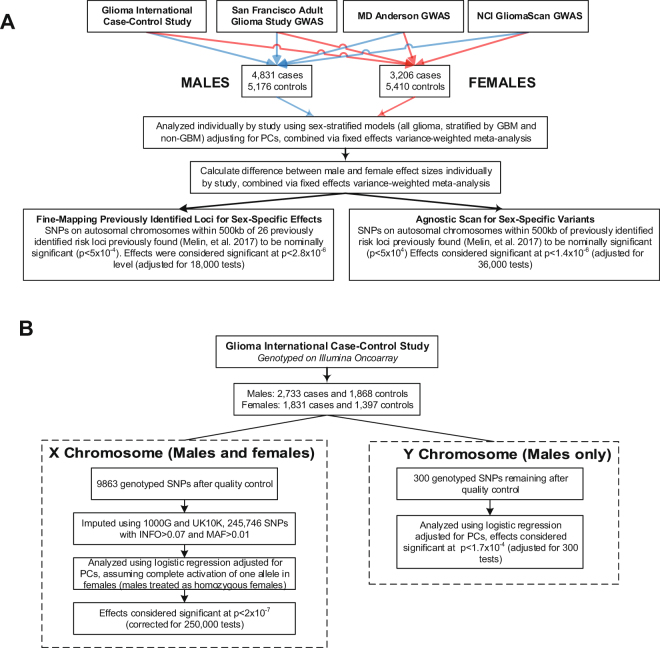
There were 5,934 SNPs within 500 kb of 26 previously discovered glioma risk loci with IMPUTE2 information score (INFO) > 0.7 and MAF > 0.01 that were previously found to have at least a nominal (p < 5 × 10−4) association with glioma4, and results were considered significant at p < 2.8 × 10−6 level (adjusted for 6,000 tests in each of three histologies [18,000 tests], see Fig. 2A for schematic of study design). Among the 25 previously validated glioma risk loci, nine loci contained 10 SNPs with pM < 2.8 × 10−6 and/or pF < 2.8 × 10−6 in any histology: 1p31.3 (RAVER2), 5p15.33 (TERT), 7p11.2 (EGFR, two independent loci), 8q24.21 (intergenic region near MYC), 9p21.3 (CDKN2B-AS1), 11q23.3 (PHLDB1), 16p13.3 (RHBDF1), 17p13.1 (TP53), and 20q13.33 (RTEL1) (Table 2). ORM and ORF were similar in the majority of these loci.
Figure 2.
Study Schematic for analyses of (A) autosomal SNPs and (B) SNPs on sex chromosomes.
Table 2.
Previously identified glioma risk loci and histology-specific odds ratios (OR) and 95% confidence intervals (95% CI) stratified by sex.
| SNP (Locus) | Risk Allele | Histology | Males | Females | PD | ||
|---|---|---|---|---|---|---|---|
| PM | ORM (95% CI) | PF | ORF (95% CI) | ||||
| rs12752552 (1p31.3) | T/C | All glioma | 1.40 × 10−6 | 1.25 (1.14–1.37) | 3.22 × 10−4 | 1.21 (1.09–1.34) | 0.7280 |
| GBM | 3.27 × 10−6 | 1.28 (1.15–1.42) | 8.41 × 10−4 | 1.24 (1.09–1.41) | 0.7535 | ||
| Non-GBM | 0.0235 | 1.15 (1.02–1.30) | 0.0036 | 1.23 (1.07–1.42) | 0.4252 | ||
| rs9841110 (3p21.31) | C /G | All glioma | 0.5885 | 1.02 (0.96–1.08) | 5.55 × 10–8 | 1.22 (1.14–1.32) | 1.77 × 10–4 |
| GBM | 0.3429 | 1.04 (0.96–1.11) | 1.44 × 10−7 | 1.27 (1.16–1.38) | 6.04 × 10−4 | ||
| Non-GBM | 0.4816 | 0.97 (0.89–1.06) | 0.0160 | 1.13 (1.02–1.24) | 0.0186 | ||
| rs10069690 (5p15.33) | C/T | All glioma | 7.58 × 10−31 | 1.49 (1.39–1.60) | 4.88 × 10−20 | 1.45 (1.34–1.57) | 0.5688 |
| GBM | 3.38 × 10−35 | 1.64 (1.52–1.78) | 6.29 × 10–22 | 1.60 (1.45–1.76) | 0.7049 | ||
| Non-GBM | 1.20 × 10−6 | 1.27 (1.15–1.40) | 1.67 × 10−6 | 1.31 (1.17–1.46) | 0.7036 | ||
| rs75061358 (7p11.2) | T/G | All glioma | 6.93 × 10−12 | 1.43 (1.29–1.59) | 1.71 × 10−9 | 1.46 (1.29–1.66) | 0.8114 |
| GBM | 2.66 × 10−16 | 1.65 (1.46–1.86) | 1.16 × 10−11 | 1.68 (1.45–1.96) | 0.8211 | ||
| Non-GBM | 0.0079 | 1.23 (1.06–1.43) | 0.0129 | 1.25 (1.05–1.49) | 0.9246 | ||
| rs11979158 (7p11.2) | A/G | All glioma | 4.87 × 10−12 | 1.33 (1.23–1.44) | 0.0187 | 1.12 (1.02–1.22) | 0.0055 |
| GBM | 1.26 × 10−12 | 1.40 (1.28–1.54) | 1.33 × 10−4 | 1.24 (1.11–1.39) | 0.1184 | ||
| Non-GBM | 2.74 × 10−5 | 1.27 (1.13–1.41) | 0.9014 | 0.99 (0.88–1.12) | 0.0034 | ||
| rs55705857 (8q24.21) | A/G | All glioma | 1.09 × 10−14 | 1.56 (1.40–1.75) | 1.22 × 10−39 | 2.45 (2.14–2.80) | 3.46 × 10−7 |
| GBM | 0.0344 | 1.17 (1.01–1.34) | 4.16 × 10−7 | 1.61 (1.34–1.94) | 0.0066 | ||
| Non-GBM | 8.13 × 10−36 | 2.66 (2.28–3.10) | 1.85 × 10−65 | 4.71 (3.94–5.63) | 8.44 × 10−7 | ||
| rs634537 (9p21.3) | T/G | All glioma | 2.37 × 10−21 | 1.33 (1.25–1.41) | 6.38 × 10−14 | 1.30 (1.21–1.39) | 0.6496 |
| GBM | 1.00 × 10−20 | 1.38 (1.29–1.48) | 1.92 × 10−16 | 1.41 (1.30–1.53) | 0.6544 | ||
| Non-GBM | 2.63 × 10−8 | 1.26 (1.16–1.37) | 4.88 × 10−4 | 1.18 (1.08–1.30) | 0.3131 | ||
| rs12803321 (11q23.3) | G/C | All glioma | 3.96 × 10−4 | 1.12 (1.05–1.19) | 8.49 × 10−6 | 1.18 (1.10–1.26) | 0.2680 |
| GBM | 0.4497 | 0.97 (0.91–1.04) | 0.6463 | 1.02 (0.94–1.11) | 0.3667 | ||
| Non-GBM | 1.82 × 10−14 | 1.41 (1.29–1.53) | 8.88 × 10−13 | 1.43 (1.30–1.57) | 0.7207 | ||
| rs3751667 (16p13.3) | C/T | All glioma | 2.98 × 10−6 | 1.18 (1.10–1.26) | 0.0297 | 1.09 (1.01–1.19) | 0.1779 |
| GBM | 2.22 × 10−4 | 1.16 (1.07–1.26) | 0.1130 | 1.08 (0.98–1.19) | 0.2729 | ||
| Non-GBM | 2.62 × 10−6 | 1.26 (1.14–1.38) | 0.0060 | 1.17 (1.05–1.31) | 0.3241 | ||
| rs78378222 (17p13.1) | T/G | All glioma | 3.36 × 10−17 | 2.41 (1.97–2.96) | 1.75 × 10−12 | 2.43 (1.90–3.12) | 0.8483 |
| GBM | 1.27 × 10−14 | 2.65 (2.07–3.40) | 2.28 × 10−9 | 2.67 (1.93–3.68) | 0.8731 | ||
| Non-GBM | 1.10 × 10−10 | 2.79 (2.04–3.80) | 4.40 × 10−8 | 2.70 (1.89–3.85) | 0.9385 | ||
| rs2297440 (20q13.33) | T/C | All glioma | 4.09 × 10−21 | 1.42 (1.32–1.52) | 1.34 × 10−13 | 1.37 (1.26–1.49) | 0.5299 |
| GBM | 1.22 × 10−19 | 1.47 (1.35–1.59) | 1.15 × 10−16 | 1.53 (1.39–1.70) | 0.5159 | ||
| Non-GBM | 2.92 × 10−7 | 1.29 (1.17–1.43) | 0.0040 | 1.18 (1.05–1.32) | 0.1916 | ||
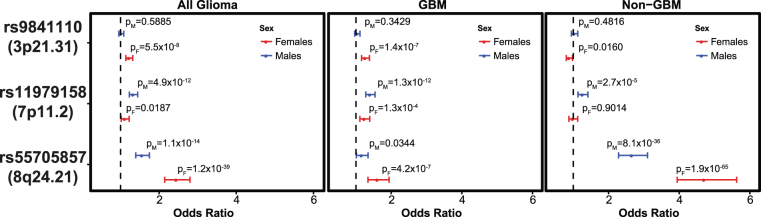
For one of two independent loci at 7p11.2 (rs11979158), there was a significant association only in males for all glioma (ORM = 1.33 [95% CI = 1.23–1.44], pM = 4.87 × 10−12) and GBM (ORM = 1.40 [95% CI = 1.28–1.54], pM = 1.26 × 10−12) but the sex differences did not meet the significance threshold (overall pD = 0.0055, and GBM pD = 0.1184) (Fig. 3, Table 2).
Figure 3.
Sex-specific odds ratios overall and by histology grouping, 95% CI and p values for selected previous GWAS hits and 3p21.31 (rs9841110) for all glioma, GBM, and non-GBM.
The previously identified SNP at 8q24.21 (rs55705857) was the most significant SNP in both males and females. Odds ratio for rs55705857 in all glioma was significantly higher in females (ORF = 2.45 [95% CI = 2.14–2.80], pF = 1.22 × 10−39) as compared to males (ORM = 1.56 [95% CI = 1.40–1.75], pM = 1.09 × 10−14) with pD = 3.46 × 10−7. In non-GBM only, ORF (ORF = 4.71 [95% CI = 3.94–5.63], pF = 1.85 × 10−65) was also elevated as compared to ORM (ORM = 2.66 [95% CI = 2.28–3.10)], pM = 8.13 × 10−36) with pD = 8.44 × 10−7 (Fig. 3, Table 2). This association was further explored in a case-only analysis, where there was a significant difference between males and females overall (p = 0.0012), and in non-GBM (p = 0.0084) (Supplemental Table 1).
Previous studies have found a strong association between rs55705857 and oligodendroglial tumors (particularly tumors with isocitrate dehydrogenase 1/2 (IDH1/2) mutation and loss of the short arm of chromosome 1 [1p] and the long arm of chromosome 19 [19q]), so this association was further explored in the non-GBM (lower grade glioma [LGG]) histology groups (Table 3). For World Health Organization (WHO) grade II-grade III astrocytoma, effect was stronger in females (ORF = 4.64 [95% CI = 3.53–6.09], pF = 2.15 × 10−28) as compared to males (ORM = 2.87 [95% CI = 2.31–3.56], pM = 1.19 × 10−21) with pD = 0.0065. For WHO grade II–III oligodendrogliomas effect was stronger than observed in WHO grade II–III astrocytomas, and effect size was stronger in females (ORF = 12.15 [95% CI = 8.96–16.48], pF = 3.68 × 10−58) as compared to males (ORM = 5.47 [95% CI = 4.16–7.19], pM = 5.37 × 10−34) with pD = 6.60 × 10−5. Oligoastrocytic tumors were not included in sub-analyses due to recent research that suggests that these tumors are not an entity that is molecularly distinct from oligodendrogliomas or astrocytomas15.
Table 3.
Sex-specific odds ratios (OR), 95% confidence intervals (95% CI), and p values from meta-analysis for rs11979158, rs55705857 and rs9841110 by specific non-GBM histologies.
| RSID (Locus) | Histology | Males | Females | PD | ||||
|---|---|---|---|---|---|---|---|---|
| PM | ORM (95% CI) | Phet | PF | ORF (95% CI) | Phet | |||
| rs9841110 (3p21.31) | Astrocytoma (Non-GBM) (WHO grade II-III) | 0.5304 | 1.04 (0.92–1.17) | 0.751 | 0.0407 | 1.15 (1.01–1.32) | 0.549 | 0.2409 |
| Oligodendroglioma (WHO grade II-III) | 0.4190 | 0.94 (0.81–1.09) | 0.694 | 0.0973 | 1.14 (0.98–1.34) | 0.360 | 0.0649 | |
| rs11979158 (7p11.2) | Astrocytoma (Non-GBM) (WHO grade II-III) | 0.0023 | 0.79 (0.68–0.92) | 0.056 | 0.9363 | 0.99 (0.83–1.18) | 0.418 | 0.0500 |
| Oligodendroglioma (WHO grade II-III) | 0.0221 | 0.81 (0.68–0.97) | 0.865 | 0.6561 | 1.05 (0.86–1.28) | 0.262 | 0.0471 | |
| rs55705857 (8q24.21) | Astrocytoma (Non-GBM) (WHO grade II-III) | 1.19 × 10−21 | 2.87 (2.31–3.56) | 0.073 | 2.15 × 10−28 | 4.64 (3.53–6.09) | 0.237 | 0.0065 |
| Oligodendroglioma (WHO grade II-III) | 5.37 × 10−34 | 5.47 (4.16–7.19) | 0.103 | 3.68 × 10−58 | 12.15 (8.96–16.48) | 0.027 | 6.60 × 10−5 | |
Genome-wide scan of nominally significant regions
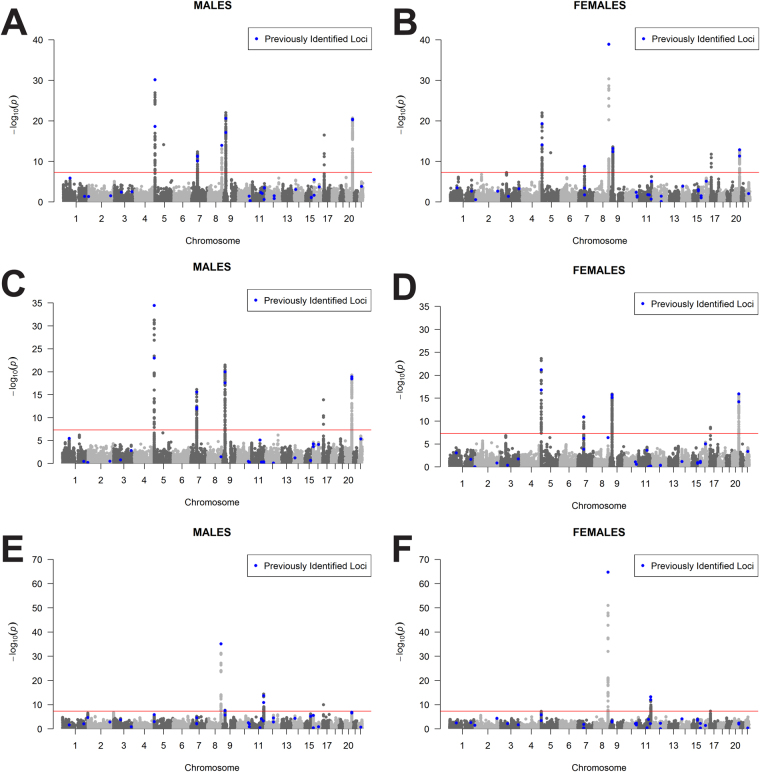
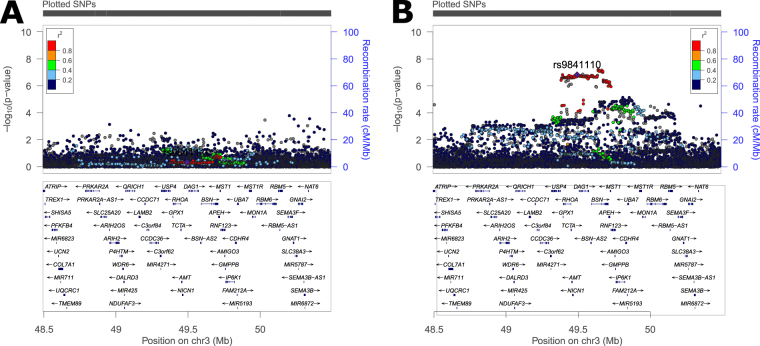
In a previous eight study meta-analysis, ~12,000 SNPs (INFO > 0.7, MAF > 0.01) were identified as having a nominally significant (p < 5 × 10−4) association with all glioma, GBM, or non-GBM4. A sex-stratified genome-wide scan was conducted within this set of SNPs and results were considered significant at pD < 1.4 × 10−6 (adjusted for 12,000 tests in each of three histologies [36,000 tests], see Fig. 2A for schematic of study design). Similar genome-wide peaks were observed between males and females (Fig. 4). One large region within 3p21.31 (49400kb–49600kb, ~200 kb) was identified as being significantly associated with glioma and GBM in females only (Fig. 5, Supplemental Fig. 1). There were 243 SNPs with nominally significant associations within this region in the previous eight-study meta-analysis (p < 5 × 10−4), and 32 of these had nominally significant sex associations (pF < 5 × 10−6 or pM < 5 × 10−6) in all glioma or GBM. The strongest association in females within this region was at rs9841110, in both all glioma (ORF = 1.22 [95% CI = 1.14–1.32], pF = 5.55 × 10−8) with pD = 1.77 × 10−4) and GBM only (ORF = 1.27 [95% CI = 1.16–1.38], pF = 3.86 × 10−7) with pD = 6.04 × 10−4), while there were no significant associations detected in males (Fig. 3). No SNPs in this region were significantly associated with non-GBM. In a case-only analysis a marginally significant difference was detected between males and females overall (p = 0.0520) and in GBM (p = 0.0428) (Supplemental Table 1).
Figure 4.
Manhattan plot of -log(p) values for all glioma in (A) males and (B) females, for GBM in (C) males and (D) females, and for non-GBM in (E) males and (F) females.
Figure 5.
Plot of region on chromosome 3 identified as having a sex-specific association with GBM for (A) males and (B) females.
Agnostic scan of sex chromosome loci
SNPs on the sex chromosomes were analyzed in GICC only. There were 245,746 SNPs with INFO > 0.7 and MAF > 0.01 on the X chromosome after quality control and imputation, and results were considered significant at p < 2 × 10−7 (corrected for 250,000 tests, see Fig. 2B for a schematic of study design). No SNPs met this significance threshold. After quality control procedures were complete, there were 300 SNPs remaining on the Y chromosome. There was no imputation performed of the Y chromosome data and only the 300 genotyped SNPs were evaluated. No significant signals were detected on the Y chromosome.
Combined analysis of germline variants and somatic characterization
Due to the lack of molecular classification data included in the GICC, MDA-GWAS, SFAGS-GWAS and GliomaScan datasets, glioma data obtained from TCGA datasets (GBM and LGG) were used to explore the potential confounding due to molecular subtype variation with histologies. There were 758 individuals from the TCGA dataset available for analysis with available germline genotyping, molecular characterization, sex, and age data (Supplemental Table 2). Overall, slightly more females (53.2%) as compared to males (47.2%) had IDH1/2 mutant glioma, but this difference was not statistically significant (p = 0.1104) (Supplemental Fig. 2). When tumors were stratified by histological type, approximately equal proportions of males and females had IDH1/2 mutations present in their tumors (GBM: 6.0% in males, and 5.2% in females; LGG: 17.9% in males, and 17.7% in females). There were also no significant differences by sex in IDH/TERT/1p19q subtype (Supplemental Fig. 3, overall p = 0.2859), or pan-glioma methylation subgroup (Supplemental Fig. 4, overall p = 0.4153).
SNPs found to be nominally significant (p < 5 × 10−4) in a previous 8 study meta-analysis, with imputation quality (r2) ≥ 0.7 were identified within the TCGA germline genotype data and D’ and r2 values in CEU were used to select proxy SNPs (Supplemental Table 3)16. A case-only analysis was conducted using sex as a binary phenotype for proxy SNPs in the TCGA dataset. In the overall meta-analysis, there was a nominally significant signal in the case-only meta-analysis for the proxy SNP in 3p21.31 in glioblastoma (Table 4). There was no significant association in the TCGA set, but RAF was elevated in females as compared to males in the GBM set, as well as in all IDH1/2 wild type gliomas. MAF in LGG and IDH1/2 mutant glioma was similar among males and females. There was a nominally significant signal in the case-only meta-analysis for the proxy SNP at 7p11.2, but no significant association in the TCGA, but RAF was elevated in males as compared to females in the GBM set, as well as in all IDH1/2 wild type gliomas. There was no significant signal detected in the overall case-only meta-analysis for the proxy SNP at 8q24.21, or within the TCGA set. Among both LGG and IDH1/2 mutant, RAF was elevated in females as opposed to males.
Table 4.
Risk allele frequencies (RAF) Case-only odds ratios, 95% confidence intervals (95% CI), and p values for marker SNPs from four study meta-analysis and the Cancer Genome Atlas genotyping data.
| Marker SNP | Histology | Four-study Meta-Analysis | The Cancer Genome Atlas | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Males | Females | Case-only analysis (males:females) | INFO | Males | Females | Case-only analysis (males:females) | ||||
| RAFcases | RAFcases | P | OR (95% CI) | RAFcases | RAFcases | p | OR (95% CI) | |||
| rs9814873 (3p21.31) | All glioma | 0.692 | 0.707 | 0.0577 | 1.07 (1.00–1.15) | 1.00 | 0.701 | 0.716 | 0.5321 | 0.93 (0.75–1.16) |
| GBM | 0.694 | 0.716 | 0.0371 | 1.11 (1.01–1.22) | 1.00 | 0.697 | 0.742 | 0.2003 | 0.80 (0.58–1.12) | |
| LGG (non-GBM) | 0.686 | 0.691 | 0.6446 | 1.03 (0.92–1.15) | 1.00 | 0.705 | 0.697 | 0.8039 | 1.04 (0.77–1.40) | |
| IDH1/2 wild type | — | — | — | — | 1.00 | 0.704 | 0.731 | 0.4343 | 0.88 (0.64–1.21) | |
| IDH1/2 mutant | — | — | — | — | 1.00 | 0.705 | 0.692 | 0.7023 | 1.06 (0.77–1.47) | |
| rs7785013 (7p11.2) | All glioma | 0.864 | 0.847 | 0.0058 | 0.88 (0.80–0.96) | 0.99 | 0.855 | 0.850 | 0.7813 | 1.04 (0.78–1.39) |
| GBM | 0.872 | 0.861 | 0.2141 | 0.92 (0.81–1.05) | 0.99 | 0.854 | 0.840 | 0.6073 | 1.12 (0.73–1.72) | |
| LGG (non-GBM) | 0.855 | 0.832 | 0.0109 | 0.83 (0.72–0.96) | 0.99 | 0.856 | 0.857 | 0.9585 | 0.99 (0.67–1.47) | |
| IDH1/2 wild type | — | — | — | — | 0.99 | 0.864 | 0.837 | 0.3132 | 1.24 (0.82–1.87) | |
| IDH1/2 mutant | — | — | — | — | 0.99 | 0.846 | 0.875 | 0.2447 | 0.77 (0.50–1.19) | |
| rs4636162 (8q24.21) | All glioma | 0.358 | 0.365 | 0.5161 | 1.02 (0.96–1.09) | 0.93 | 0.392 | 0.424 | 0.2113 | 0.88 (0.71–1.08) |
| GBM | 0.343 | 0.338 | 0.6001 | 0.98 (0.89–.07) | 0.93 | 0.374 | 0.404 | 0.4456 | 0.89 (0.66–1.20) | |
| LGG | 0.383 | 0.401 | 0.1594 | 1.08 (0.97–1.20) | 0.94 | 0.410 | 0.438 | 0.3891 | 0.88 (0.67–1.17) | |
| IDH1/2 wild type | — | — | — | — | 0.92 | 0.371 | 0.373 | 0.9480 | 0.99 (0.73–1.34) | |
| IDH1/2 mutant | — | — | — | — | 0.94 | 0.419 | 0.460 | 0.2613 | 0.84 (0.63–1.14) | |
Sex-stratified genotypic risk scores
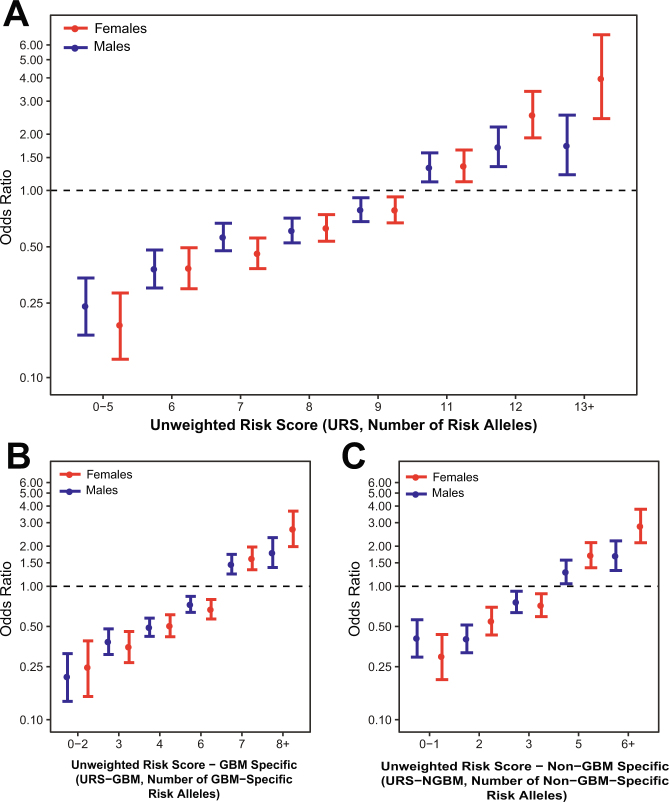
In order to estimate the cumulative effects of significant variants by sex, unweighted risk scores (URS) were calculated by summing all risk alleles for each individual using the 10 SNPs (rs12752552, rs9841110, rs10069690, rs11979158, rs55705857, rs634537, rs12803321, rs3751667, rs78378222, and rs2297440) found to be significantly associated with glioma in this analysis. GBM (URS-GBM) and non-GBM (URS-NGBM) specific URS were calculated only using sets of 6 SNPs in this set that were significantly associated with these histologies (URS-GBM: rs9841110, rs10069690, rs11979158, rs634537, rs78378222, and rs2297440, and URS-NGBM: rs10069690, rs55705857, rs634537, rs12803321, rs78378222, and rs2297440). See Methods for additional information on score calculation. Median URS, URS-GBM, and URS-NGBM were significantly different (p < 0.0001) between cases and controls in both males and females in all histology groups (Supplemental Fig. 5). There was no significant difference in median risk scores between male and female cases for any histology group. Glioma risk increased with increasing number of alleles in both males and females for the 10 SNPs included in the overall URS, as well as the 6 SNPs in the URS-GBM and 6 SNPs in URS-NGBM (Fig. 6, Supplemental Table 4). Risk was higher in females (OR = 3.97 [95% CI = 2.42–6.80]) as compared to males (OR = 1.74 [95% CI = 1.21–2.53]) in all glioma for individuals for with 13–16 alleles, though the difference between these estimates were not statistically significant. Risk was also higher among females (OR = 2.69 [95% CI = 1.98–3.66]) as compared to males (OR = 1.79 [95% CI = 1.38–2.32]) in GBM for individuals with 8–11 risk alleles, as well as in non-GBM for individuals with 6–11 risk alleles (females: OR = 2.83 [95% CI = 2.12–3.78], males: OR = 1.70 [95% CI = 1.31–2.19]), though the difference between these estimates were not statistically significant. The estimates may underestimate actual risk due to varying effect sizes and alleles frequencies between risk variants.
Figure 6.
Odds ratios and 95% confidence intervals for unweighted risk (URS) score in (A) all glioma, (B) GBM-specific URS (URS-G) in GBM, and (C) and non-GBM-specific URS (URS-NGBM) for in non-GBM.
Discussion
This is the first analysis of inherited risk variants in sporadic glioma focused specifically on sex differences, and the first agnostic unbiased scan for glioma risk variants on the X and Y sex chromosomes. One SNP at the 7p11.2 locus (rs11979158) showed significant association in males only, in both all glioma and GBM (Table 2, see Supplemental Table 5 and Supplemental Fig. 6 for study-specific estimates). This variant is within one of two previously identified independent glioma risk loci located near epidermal growth factor receptor (EGFR) and is most strongly associated with risk for GBM4,17. Though EGFR is implicated in many cancer types and is a target for many anti-cancer therapies, this risk locus has not been previously associated with any other cancer type. While estrogen has been demonstrated to interact with EGFR as well as other growth factors, previous epidemiological studies have not consistently found an association between proxy markers for endogenous estrogen exposure and decreased glioma risk18. Cell intrinsic, hormone independent sex differences in EGF effects have been observed in a murine model of gliomagenesis, where EGF treatment was transforming for male but not female astrocytes that had been rendered null for neurofibromin and p53 function19. While this SNP was not genotyped on the germline genotyping array used for TCGA, a SNP in strong LD with rs11979158 (rs7785013, D’ = 1, r2 = 1 in CEU16) was evaluated using a case-only approach. The association was not statistically significant in any histology group, but a similar trend to that observed in the overall meta-analysis in sex-specific RAF was observed in both the overall GBM and the IDH1/2 wild type groups.
The association at 8q24.21 (rs55705857) is the strongest that has been identified by glioma GWAS to date4, with an odds ratio of 1.99 (95% CI = 1.85–2.13, p = 9.53 × 10−79) in glioma overall, and an odds ratio of 3.39 (95% CI = 3.09–3.71, p = 7.28 × 10−149) in non-GBM (see Supplemental Table 5 and Supplemental Fig. 7 for study-specific estimates). The identified SNP, rs55705857, is located in an intergenic region near coiled-coil domain containing 26 (CCDC26, a long non-coding RNA). This analysis found a stronger association in females than males in all glioma and non-GBM, where female odds ratio estimates are ~2 × those of males (Table 2). ORs were higher in women than men in all studies included in the analysis (see Supplemental Tables 5 and 6 for study-specific estimates and MAF). A sensitivity analysis was conducted to assess the effect of study heterogeneity on this estimate in non-GBM using only the GICC, MDA-GWAS, and GliomaScan datasets. The exclusion of SFAGS-GWAS did not substantially change the results (Main analysis pD = 1.20 × 10−6 and sensitivity pD = 1.49 × 10−5). A histology-specific analysis found a similar sex differences in ORs for rs55705957 for both non-GBM astrocytoma, and oligodendroglioma (Table 3, see Supplemental Table 7 for study-specific estimates). This variant is strongly associated with IDH1/2 mutant and 1p/19q codeleted glioma tumors, but data on these molecular markers was not available for the four GWAS datasets used20,21. The TCGA GBM and LGG datasets22–24 were used to assess potential sex differences in frequency of IDH1/2 mutation within histologies. Approximately the same proportion of males as females with histologically confirmed GBM had IDH1/2 mutations (5.2% vs 6.0%, respectively, Supplemental Fig. 2). While this SNP was not genotyped on the germline genotyping array used for TCGA, a SNP in weak LD with rs55705857 (rs4636162, D’ = 1; r2 = 0.104, in CEU16) was evaluated using a case-only approach. There was no significant association in the overall meta-analysis for this SNP, and the association in the analysis of TCGA cases was not statistically significant in any group.
A large region in 3p21.31 was identified that was associated with all glioma and GBM in females only (Table 2, see Supplemental Table 5 and Supplemental Fig. 8 for study-specific estimates). The strongest association in this region was rs9841110, an intronic variant located upstream of dystroglycan 1 (DAG1) within an enhancer region. While this SNP was not genotyped on the germline genotyping array used for TCGA, a SNP in strong LD with rs9841110 (rs9814873, D’ = 1, r2 = 1 in CEU16) was able to be evaluated using a case-only approach. The association was not statistically significant in any group, but a similar trend in RAF was observed in the overall GBM and IDH1/2 wild type groups. Though this region has not previously been associated with glioma, previous GWAS have detected associations at 3p21.31 for a large variety of traits, including several autoimmune diseases as well as increased age at menarche25–28. If increased lifetime estrogen exposure decreases glioma risk, as some have hypothesized, it is reasonable that variants which increase age at menarche (potentially decreasing total lifetime estrogen exposure) may increase glioma risk in females. Due to the complexity of measuring lifetime estrogen exposure (which is affected by age at menarche, age at menopause, parity, breast feeding patterns, and estrogen replacement therapy post-menopause) it is difficult to determine the ‘true’ effect that this exposure might have on glioma risk.
As compared to a model containing age at diagnosis and sex alone, the three SNPs (rs55705857, rs9841110 and rs11979158) identified as having sex-specific effects explain an additional 1.4% of trait variance within the GICC set. The variance explained by these SNPs varies by histology (0.6% in GBM, and 3.3% in Non-GBM). The variance explained by the addition of these three SNPs was higher in females for all glioma (1.3% in males and 2.2% in females), and non-GBM glioma (2.3% in males and 5.3% in females), and slightly higher in males for GBM (0.9% in males and 0.7% in females). Unweighted risk scores (URS) were generated to compare the cumulative effects of glioma risk variants by sex by summing all risk alleles using the 10 SNPs found to be significantly associated with glioma in this analysis. GBM (URS-GBM) and non-GBM (URS-NGBM) specific URS were calculated using sets of six SNPs in this set that were associated with significantly associated with each histology. Individuals with lower numbers of risk alleles had significantly lower odds of glioma, and those with higher numbers of alleles had increased odds of glioma, with statistically significant trends in each histology group. Males and females with low risk scores had similar odds of glioma, while females had increased odds in the upper strata of scores as compared to males. Development of risk scores that weight alleles by effect size, and use sex-specific estimates for variants for which effect size varies by sex (such as 7p11.2 and 8q24.21), may lead to better predictive values.
While often not included in GWAS, sex-stratified analyses can reveal genetic sources of sexual dimorphism in risk9,10. Sex-stratified analyses not only contribute to understanding of sources of sex difference in incidence, but may also suggest mechanisms and pathways disease development that vary by sex. Sex variation in genetic susceptibility to disease is likely not due to sex differences in actual DNA sequence, but is thought to be the result of sex-specific regulatory functions29–31. In addition to genetic sources of difference, there are likely several additional factors acting in combination which contribute to sex differences in glioma incidence. Sex differences in disease can also be linked to in-utero development, during which time gene expression and risk phenotypes are patterned through the action of X chromosome alleles that escape inactivation and genes on the non-pseudo-autosomal component of the Y chromosome, as well as the epigenetic effects of in utero testosterone32. A previous analysis estimating heritability of brain and CNS tumors by sex using twins attempted to estimate sex-specific relative risks, but these analyses were limited by a small sample size33. Further investigation of the inheritance patterns of familial glioma by sex may also provide additional information about sex differences in this disease.
There are several limitations to this analysis. Individuals included in these datasets were recruited during different time periods from numerous institutions, with no central review of pathology. Molecular tumor markers were unavailable for all datasets, and as a result classifications are based on the treating pathologist using the prevailing histologic criteria at time of diagnosis. The variant at 8q24.21 has been shown to have significant association with particular molecular subtypes, and without molecular data it was not possible to determine whether the observed result is an artifact of varying molecular features by sex. Oligodendroglioma as a histology is highly enriched for IDH1/2 and 1p/19q co-deleted tumors (117/174, or ~67% within TCGA) and it is therefore likely that the analysis using only tumors classified as oligodendroglioma captured most of this molecular subtype. Males and females within histology groups have different frequencies of IDH1/2 mutation22, which may have confounded the estimates for 8q24.21. The TCGA dataset was used to explore sex differences in allele frequency within molecular groups, but none of the identified SNPs were able to be directly validated within this set; however SNPs in strong LD were evaluated except for in 8q24.21. The 8q24.21 region is not well characterized on the array used for the TCGA genotyping, and as a result this region imputed poorly. No proxy SNP in strong LD with rs55705857 was able to be identified. Similar trends in RAF to those observed in the overall meta-analysis were seen in the TCGA set, though these differences were not statistically significant. Further interrogation in datasets with molecular classification where direct genotyping of these regions is warranted in order to confirm the sex-specific associations observed in this analysis.
Conclusions
Sex and other demographic differences in cancer susceptibility can provide important clues to etiology, and these differences can be leveraged for discovery in genetic association studies. This analysis identified potential sex-specific effects in 2 previous identified glioma risk loci (7p11.2, and 8q24.21), and 1 newly identified autosomal locus (3p21.31). Odds ratios for the highest strata of an unweighted risk score calculated by summing total risk alleles was higher in females as compared to males in all three histology groups. These significant differences in effect size may be a result of differing biological function of these variants by sex due to biological sex differences, or interaction between these variants and unidentified risk factors that vary in prevalence or effect by sex.
Materials and Methods
Study cohorts
This study was approved locally by the institutional review board (IRB) at University Hospitals Cleveland Medical Center and by each participating study site’s IRB. Written informed consent was obtained from all participants. All research was performed in accordance with relevant guidelines and regulation. In this study, data was combined from four prior glioma GWAS: Glioma International Case-Control Study (GICC), San Francisco Adult Glioma Study GWAS (SFAGS-GWAS), MD Anderson Glioma GWAS (MDA-GWAS), and National Cancer Institute’s GliomaScan (Fig. 4A)4,11–14. The SFAGS-GWAS includes controls from the Illumina iControls dataset, and MDA-GWAS includes controls from Cancer Genetic Markers of Susceptibility (CGEMS) breast and prostate studies34–36. Details of data collection and classification are available in previous publications4,11–14.
Genotyping and imputation of GWAS datasets
GICC cases and controls were genotyped on the Illumina Oncoarray37. The array included 37,000 beadchips customized to include previously-identified glioma-specific candidate single nucleotide polymorphisms (SNPs). SFAGS-GWAS cases and some controls were genotyped on Illumina’s HumanCNV370-Duo BeadChip, and the remaining controls were genotyped on the Illumina HumanHap300 and HumanHap550. MDA-GWAS cases were genotyped on the Illumina HumanHap610 and controls using the Illumina HumanHap550 (CGEMS breast34,36) or HumanHap300 (CGEMS prostate35). GliomaScan cases were genotyped on the Illumina 660 W, while controls were selected from cohort studies and were genotyped on Illumina 370D, 550 K, 610Q, or 660 W (See Rajaraman et al. for specific details of genotyping)14. Details of DNA collection and processing are available in previous publications4,12–14. Individuals with a call rate (CR) <99% were excluded, as well as all individuals who were of non-European ancestry (<80% estimated European ancestry using the FastPop38 procedure developed by the GAMEON consortium). For all apparent first-degree relative pairs were removed (identified using estimated identity by descent [IBD] ≥ .5), for example, the control was removed from a case-control pair; otherwise, the individual with the lower call rate was excluded. SNPs with a call rate < 95% were excluded as were those with a minor allele frequency (MAF) <0.01, or displaying significant deviation from Hardy-Weinberg equilibrium (HWE) (p < 1 × 10−5). Additional details of quality control procedures have been previously described in Melin et al.4. All datasets were imputed separately using SHAPEIT v2.837 and IMPUTE v2.3.2 using a merged reference panel consisting of data from phase three of the 1,000 genomes project and the UK10K39–44.
TCGA cases were genotyped on the Affymetrix Genomewide 6.0 array using DNA extracted from whole blood (see previous manuscript for details of DNA processing23,24), and underwent standard GWAS QC, and duplicate and related individuals within datasets have been excluded4. Ancestry outliers were identified in TCGA using principal components analysis in plink 1.945. Resulting files were imputed using Eagle 2 and Minimac3 as implemented on the Michigan imputation server (https://imputationserver.sph.umich.edu) using the Haplotype Reference Consortium Version r1.1 2016 as a reference panel46–48. Somatic characterization of TCGA cases was obtained from the final dataset used for the TCGA pan-glioma analysis22, and classification schemes were adopted from Eckel-Passow, et al.49 and Ceccarelli, et al.22.
Sex-stratified scan of the autosomal chromosomes
The data were analyzed using sex-stratified logistic regression models in SNPTEST for all SNPs on autosomal chromosomes within 500 kb of previously identified risk loci, and/or those found to be nominally significant (p < 5 × 10−4) in a previous meta-analysis (Fig. 2A)4,50. Sex-specific betas (βM and βF), standard errors (SEM and SEF), and p-values (pM and pF) were generated using sex-stratified logistic regression models that were adjusted for number of principal components found to significant differed between cases and controls within each study in a previous meta-analysis4,50. Genomic inflation factors were calculated After excluding SNPs with MAF < 0.05, INFO score < 0.7, and that significantly violated Hardy-Weinberg equilibrium in controls (p < 5 × 10−8), genomic inflation factors (Males: GICC: λadjusted = 1.04, SFAGS-GWAS: λadjusted = 1.01 MDA-GWAS: λadjusted = 1.02; Gliomascan: λadjusted = 1.01. Females: GICC: λadjusted = 1.03; SFAGS-GWAS: λadjusted = 1.02; MDA-GWAS: λadjusted = 1.04; Gliomascan: λadjusted = 1.01).
Estimation of sex difference and test of statistical significance
βD and SED were estimated using the sex-specific betas and standard errors separately for each dataset, as follows:
| 1 |
| 2 |
The difference between the groups was then tested using a z test51,52. Sex-stratified results and differences estimates from the four studies were separately combined via inverse-variance weighted fixed effects meta-analysis in META53. See Fig. 2A for schematic of autosomal analysis methods. Case only-analyses were performed for SNPs found to be significant in agnostic analyses using sex as outcome for all glioma, GBM, and non-GBM by study and betas and standard errors were combined via inverse-variance weighted fixed effects meta-analysis in META53.
Sex chromosome analysis
X and Y chromosome data were available from GICC set only. Males and females were imputed separately for the X chromosome using the previously described merged reference panel. X chromosomes were analyzed using logistic regression model in SNPTEST module ‘newml’ assuming complete inactivation of one allele in females, and males are treated as homozygous females (Fig. 2B). For prioritized SNPs in the combined model, sex-specific effect estimates were generated using stratified logistic regression models. Y chromosome data were analyzed using logistic regression in SNPTEST (Fig. 2B)54. Figures were generated using LocusZoom and R 3.3.2 using GenABEL, qqman, and ggplot55–59.
Analysis of TCGA germline and somatic data
Only newly diagnosed cases from TCGA GBM and LGG with no neo-adjuvant treatment or prior cancer were used. Demographic characteristics, molecular classification and somatic alterations data was obtained from Ceccarelli, et al.22. Chi-square tests were used to compare the frequency of somatic alterations between age groups. SNPs found to be nominally significant (p < 5 × 10−4) in a previous 8 study meta-analysis4, with imputation quality ≥ 0.7 were identified within the TCGA genotype data and D’ and r2 values in CEU were used to select proxy SNPs16. Using these SNPs, a case-only analysis using sex as a binary phenotype was conducted using logistic regression in SNPTEST assuming an additive model to estimate beta, standard error, and p values50. Results were considered significant at p < 0.003 (Bonferroni correction for 15 tests, for the three assessed loci in each of five histology groups).
Calculation of unweighted genetic risk scores
In order to estimate the cumulative effects of significant variants by sex, histology-specific unweighted risk scores were calculated using the SNPs found to be significantly associated with each outcome. Data from all four studies was merged, and any imputed genotypes with genotype probability >0.8 were converted to hard calls. An overall unweighted risk score (URS) was generated using the sum of risk alleles at rs12752552, rs9841110, rs10069690, rs11979158, rs55705857, rs634537, rs12803321, rs3751667, rs78378222, and rs2297440. As risk alleles are known to have histology specific associations4, histologic specific scores were generated for GBM and non-GBM using only the SNPs found to have a significant association with each histology. GBM-specific URS (URS-G) was calculated by summing the number of risk alleles at rs9841110, rs10069690, rs11979158, rs634537, rs78378222, and rs2297440. Non-GBM-specific (URS-N) specific URS was calculated by summing the number of risk alleles at rs10069690, rs55705857, rs634537, rs12803321, rs78378222, and rs2297440. Unweighted risk scores (URS) were calculated by summing all risk alleles for each individual. Differences in median scores between groups using were tested using Wilcoxon rank sum tests. Scores were compared against the median score for each set (URS: ten alleles, URS-GBM: six alleles, URS-NGBM: four alleles). Odds ratios and 95% confidence intervals for each level of the score using sex-stratified logistic regression adjusted for age at diagnosis (for controls where only an age range was available, the mean value of the range was used), where each score was compared to the median score within the entire population as described in Shete et al.13.
Calculation of trait variance explained by SNPs with sex-specific effects
In order to determine whether the identified SNPs with sex-specific effects more accurate estimate odds of glioma than sex alone, logistic regression models were used to estimate odds of all glioma, GBM, and non-GBM glioma based on sex using the GICC data only. Proportion of variance in odds of glioma explained by sex-specific SNPs was calculated using R2 estimated using the log likelihood of the null model (sex, age at diagnosis, and the first two principal components only) and the full model (including identified SNPs, rs9841110, rs11979158, rs55705857)60, calculated as follows:
| 3 |
Proportion of variance explained was also calculated separately by sex for each histology (null model adjusted for age at diagnosis, and the first two principal components only).
Electronic supplementary material
Supplemental Tables 1-7, Supplemental Figures 1-8
Acknowledgements
The results of this study were previously presented at the 2016 Annual Meeting of the Society for Neuro-Oncology, and the 2017 Annual Meeting of the American Association for Cancer Research. A pre-publication version of this manuscript was previously made available on bioRxiv (10.1101/229112). QTO is supported by a Research Training Grant from the Cancer Prevention and Research Institute of Texas (CPRIT; RP160097T). The GICC was supported by grants from the National Institutes of Health, Bethesda, Maryland (R01CA139020, R01CA52689, P50097257, P30CA125123, P30CA008748, C. Thompson PI). Additional support was provided by the McNair Medical Institute and the Population Sciences Biorepository at Baylor College of Medicine. In Sweden work was additionally supported by Acta Oncologica through the Royal Swedish Academy of Science (BM salary) and The Swedish Research council and Swedish Cancer foundation. We are grateful to the National clinical brain tumor group, all clinicians and research nurses throughout Sweden who identified all cases. The UCSF Adult Glioma Study was supported by the National Institutes of Health (grant numbers R01CA52689, P50CA097257, R01CA126831, and R01CA139020), the Loglio Collective, the National Brain Tumor Foundation, the Stanley D. Lewis and Virginia S. Lewis Endowed Chair in Brain Tumor Research, the Robert Magnin Newman Endowed Chair in Neuro-oncology, and by donations from families and friends of John Berardi, Helen Glaser, Elvera Olsen, Raymond E. Cooper, and William Martinusen. This project also was supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through UCSF-CTSI Grant Number UL1 RR024131. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. The collection of cancer incidence data used in this study was supported by the California Department of Public Health as part of the statewide cancer reporting program mandated by California Health and Safety Code Section 103885; the National Cancer Institute’s Surveillance, Epidemiology and End Results Program under contract HHSN261201000140C awarded to the Cancer Prevention Institute of California, contract HHSN261201000035C awarded to the University of Southern California, and contract HHSN261201000034C awarded to the Public Health Institute; and the Centers for Disease Control and Prevention’s National Program of Cancer Registries, under agreement # U58DP003862-01 awarded to the California Department of Public Health. The ideas and opinions expressed herein are those of the author(s) and endorsement by the State of California Department of Public Health, the National Cancer Institute, and the Centers for Disease Control and Prevention or their Contractors and Subcontractors is not intended nor should be inferred. Other significant contributors for the UCSF Adult Glioma Study include: M Berger, P Bracci, S Chang, J Clarke, A Molinaro, A Perry, M Pezmecki, M Prados, I Smirnov, T Tihan, K Walsh, J Wiemels, S Zheng. UK10K data generation and access was organized by the UK10K consortium and funded by the Wellcome Trust. We are grateful to all the patients and individuals for their participation and we would also like to thank the clinicians and other hospital staff, cancer registries and study staff in respective centers who contributed to the blood sample and data collection.
Author Contributions
QTO and JSB-S conceptualized and designed the analyses. QTO wrote the main manuscript text and prepared all tables and figures. QTO, BK, YC, GA, and TR prepared the analytic files. MRW, JEE-P, JKW, LSM, HMH, CIA, JLB, EBC, DI, CJ, DHL, RKL, RTM, SHO, SS, JMS, SS, UA, PR, SJC, MS, ZW, MY, LEBF, SK, DA, KV, VLS, RH, DSM, MF, AA, GGG, RM, RM-C, LLM, MS, AM, TC, GH, AZ-J, JMG, HDS, MPP, EW, UP, JB, RSH, RBJ, BM, MLB, and JSB-S provided data. JSB-S supervised all analyses. BK, MRW, JEE-P, EC, SHO, JBR, JDL, MEB, RSH, RBJ, BM, MLB, and JSB-S critically revised the manuscript for intellectual content. All authors reviewed the manuscript.
Competing Interests
This study was approved locally by the institutional review board (IRB) at University Hospitals Cleveland Medical Center and by each participating study site’s IRB. Written informed consent was obtained from all participants. There are no conflicts of interest to report. The GliomaScan (phs000652.v1.p1), CGEMS (phs000812.v1.p1), San Francisco Adult Glioma Study (phs001497.v1.p1), and GICC (phs001319.v1.p1) data used for these analyses are available through dbGap. Individual genotypes and phenotypes from the MD Anderson Glioma GWAS are being submitted to dbGAP, and final identifiers to these data will be provided in the final manuscript. The results here are in part based upon data generated by the TCGA Research Network: (http://cancergenome.nih.gov/).
Footnotes
A comprehensive list of consortium members appears at the end of the paper
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-24580-z.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jill. S. Barnholtz-Sloan, Email: jsb42@case.edu
GliomaScan consortium:
Laura E. Beane Freeman, Stella Koutros, Demetrius Albanes, Kala Visvanathan, Victoria L. Stevens, Roger Henriksson, Dominique S. Michaud, Maria Feychting, Anders Ahlbom, Graham G. Giles, Roger Milne, Roberta McKean-Cowdin, Loic Le Marchand, Meir Stampfer, Avima M. Ruder, Tania Carreon, Göran Hallmans, Anne Zeleniuch-Jacquotte, J. Michael Gaziano, Howard D. Sesso, Mark P. Purdue, Emily White, Ulrike Peters, and Julie Buring
References
- 1.Ostrom QT, et al. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2009–2013. Neuro-oncology. 2016;18:v1–v75. doi: 10.1093/neuonc/now207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ostrom QT, et al. The epidemiology of glioma in adults: a “state of the science” review. Neuro-oncology. 2014;16:896–913. doi: 10.1093/neuonc/nou087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kinnersley B, et al. Quantifying the heritability of glioma using genome-wide complex trait analysis. Scientific reports. 2015;5:17267. doi: 10.1038/srep17267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Melin BS, et al. Genome-wide association study of glioma subtypes identifies specific differences in genetic susceptibility to glioblastoma and non-glioblastoma tumors. Nature genetics. 2017;49:789–794. doi: 10.1038/ng.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson VS, Kirichek O, Beral V, Green J. Menopausal hormone therapy and central nervous system tumor risk: large UK prospective study and meta-analysis. International journal of cancer. 2015;136:2369–2377. doi: 10.1002/ijc.29274. [DOI] [PubMed] [Google Scholar]
- 6.Zong H, et al. Reproductive factors in relation to risk of brain tumors in women: an updated meta-analysis of 27 independent studies. Tumor biology. 2014;35:11579–11586. doi: 10.1007/s13277-014-2448-1. [DOI] [PubMed] [Google Scholar]
- 7.Howlader N, N. A. et al (eds). SEER Cancer Statistics Review, 1975-2014, based on November 2016 SEER data submission. (National Cancer Institute, Bethesda, MD, 2017).
- 8.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA: a cancer journal for clinicians. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 9.Liu LY, Schaub MA, Sirota M, Butte AJ. Sex differences in disease risk from reported genome-wide association study findings. Human genetics. 2012;131:353–364. doi: 10.1007/s00439-011-1081-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorak MT, Karpuzoglu E. Gender differences in cancer susceptibility: an inadequately addressed issue. Frontiers in genetics. 2012;3:268. doi: 10.3389/fgene.2012.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amirian ES, et al. The Glioma International Case-Control Study: A Report From the Genetic Epidemiology of Glioma International Consortium. American journal of epidemiology. 2016;183:85–91. doi: 10.1093/aje/kwv235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wrensch M, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nature genetics. 2009;41:905–908. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shete S, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nature genetics. 2009;41:899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajaraman P, et al. Genome-wide association study of glioma and meta-analysis. Human Genetics. 2012;131:1877–1888. doi: 10.1007/s00439-012-1212-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sahm F, et al. Farewell to oligoastrocytoma: in situ molecular genetics favor classification as either oligodendroglioma or astrocytoma. Acta neuropathologica. 2014;128:551–559. doi: 10.1007/s00401-014-1326-7. [DOI] [PubMed] [Google Scholar]
- 16.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31:3555–3557. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanson M, et al. Chromosome 7p11.2 (EGFR) variation influences glioma risk. Human molecular genetics. 2011;20:2897–2904. doi: 10.1093/hmg/ddr192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Filardo EJ. Epidermal growth factor receptor (EGFR) transactivation by estrogen via the G-protein-coupled receptor, GPR30: a novel signaling pathway with potential significance for breast cancer. The Journal of steroid biochemistry and molecular biology. 2002;80:231–238. doi: 10.1016/S0960-0760(01)00190-X. [DOI] [PubMed] [Google Scholar]
- 19.Sun T, et al. Sexually dimorphic RB inactivation underlies mesenchymal glioblastoma prevalence in males. J. Clin. Invest. 2014;124:4123–4133. doi: 10.1172/JCI71048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enciso-Mora V, et al. Deciphering the 8q24.21 association for glioma. Human molecular genetics. 2013;22:2293–2302. doi: 10.1093/hmg/ddt063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jenkins RB, et al. A low-frequency variant at 8q24.21 is strongly associated with risk of oligodendroglial tumors and astrocytomas with IDH1 or IDH2 mutation. Nature genetics. 2012;44:1122–1125. doi: 10.1038/ng.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ceccarelli M, et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell. 2016;164:550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brennan CW, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The Cancer Genome Atlas Research Network et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. The New England journal of medicine372, 2481–2498, 10.1056/NEJMoa1402121 (2015). [DOI] [PMC free article] [PubMed]
- 25.Elks CE, et al. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nature genetics. 2010;42:1077–1085. doi: 10.1038/ng.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perry JR, et al. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature. 2014;514:92–97. doi: 10.1038/nature13545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pokrajac-Bulian A, Toncic M, Anic P. Assessing the factor structure of the Body Uneasiness Test (BUT) in an overweight and obese Croatian non-clinical sample. Eating and weight disorders: EWD. 2015;20:215–222. doi: 10.1007/s40519-014-0166-8. [DOI] [PubMed] [Google Scholar]
- 28.Raelson JV, et al. Genome-wide association study for Crohn’s disease in the Quebec Founder Population identifies multiple validated disease loci. Proceedings of the national academy of sciences of the United States of America. 2007;104:14747–14752. doi: 10.1073/pnas.0706645104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reinius B, et al. An evolutionarily conserved sexual signature in the primate brain. PLoS genetics. 2008;4:e1000100. doi: 10.1371/journal.pgen.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rinn JL, Snyder M. Sexual dimorphism in mammalian gene expression. Trends in genetics: TIG. 2005;21:298–305. doi: 10.1016/j.tig.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 31.Ellegren H, Parsch J. The evolution of sex-biased genes and sex-biased gene expression. Nature reviews. Genetics. 2007;8:689–698. doi: 10.1038/nrg2167. [DOI] [PubMed] [Google Scholar]
- 32.Sun T, Plutynski A, Ward S, Rubin JB. An integrative view on sex differences in brain tumors. Cellular and molecular life sciences. 2015;72:3323–3342. doi: 10.1007/s00018-015-1930-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lichtenstein P, et al. Environmental and heritable factors in the causation of cancer–analyses of cohorts of twins from Sweden, Denmark, and Finland. The New England journal of medicine. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 34.Ahmed S, et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nature genetics. 2009;41:585–590. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeager M, et al. Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nature genetics. 2009;41:1055–1057. doi: 10.1038/ng.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hunter DJ, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nature genetics. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amos, C. I. et al. The OncoArray Consortium: a Network for Understanding the Genetic Architecture of Common Cancers. Cancer epidemiology, biomarkers & prevention, 10.1158/1055-9965.EPI-16-0106 (2016). [DOI] [PMC free article] [PubMed]
- 38.Li Y, et al. FastPop: a rapid principal component derived method to infer intercontinental ancestry using genetic data. BMC Bioinformatics. 2016;17:122. doi: 10.1186/s12859-016-0965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang J, et al. Improved imputation of low-frequency and rare variants using the UK10K haplotype reference panel. Nature communications. 2015;6:8111. doi: 10.1038/ncomms9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nature methods. 2012;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 41.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS genetics. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Genomes Project C, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Golding J, Pembrey M, Jones R, Team AS. ALSPAC–the Avon Longitudinal Study of Parents and Children. I. Study methodology. Paediatric and perinatal epidemiology. 2001;15:74–87. doi: 10.1046/j.1365-3016.2001.00325.x. [DOI] [PubMed] [Google Scholar]
- 44.Moayyeri A, Hammond CJ, Hart DJ, Spector TD. The UK Adult Twin Registry (TwinsUK Resource) Twin research and human genetics: the official journal of the International Society for Twin Studies. 2013;16:144–149. doi: 10.1017/thg.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Purcell, S. & Chang, C. PLINK 1.9, https://www.cog-genomics.org/plink2.
- 46.Das S, et al. Next-generation genotype imputation service and methods. Nature genetics. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loh PR, et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nature genetics. 2016;48:1443–1448. doi: 10.1038/ng.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCarthy S, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nature genetics. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eckel-Passow JE, et al. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. The New England journal of medicine. 2015;372:2499–2508. doi: 10.1056/NEJMoa1407279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nature genetics. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 51.Paternoster R, Brame R, Mazerolle P, Piquero A. Using The Correct Statisticaltest For The Equality of Regression Coefficients. Criminology. 1998;36:859–866. doi: 10.1111/j.1745-9125.1998.tb01268.x. [DOI] [Google Scholar]
- 52.Mittelstrass K, et al. Discovery of Sexual Dimorphisms in Metabolic and Genetic Biomarkers. PLoS Genet. 2011;7:e1002215. doi: 10.1371/journal.pgen.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu JZ, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nature genetics. 2010;42:436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chow JC, Yen Z, Ziesche SM, Brown CJ. Silencing of the mammalian X chromosome. Annual review of genomics and human genetics. 2005;6:69–92. doi: 10.1146/annurev.genom.6.080604.162350. [DOI] [PubMed] [Google Scholar]
- 55.R Core Team. R: A language and environment for statistical computing, http://www.R-project.org/ (2017).
- 56.Wickham, H. ggplot2: elegant graphics for data analysis, http://had.co.nz/ggplot2/book (2009).
- 57.Karssen LC, van Duijn CM, Aulchenko YS. The GenABEL Project for statistical genomics. F1000Research. 2016;5:914. doi: 10.12688/f1000research.8733.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. biorXiv, 10.1101/005165 (2014).
- 59.Pruim RJ, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mittlbock, M. & Schemper, M. Explained variation for logistic regression. Statistics in medicine15, 1987–1997, 10.1002/(SICI)1097-0258 (19961015)15:19 1987::AID-SIM318 3.0.CO;2-9 (1996). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Tables 1-7, Supplemental Figures 1-8