Abstract
Certain yeasts secrete peptides known as killer toxins or mycocins with a deleterious effect on sensitive yeasts or filamentous fungi, a common phenomenon in environmental species. In a recent work, different Debaryomyces hansenii (Dh) strains isolated from a wide variety of cheeses were identified as producing killer toxins active against Candida albicans and Candida tropicalis. We have analyzed the killer activity of these toxins in C. albicans mutants defective in MAPK signaling pathways and found that the lack of the MAPK Hog1 (but not Cek1 or Mkc1) renders cells hypersensitive to Dh mycocins while mutants lacking other upstream elements of the pathway behave as the wild type strain. Point mutations in the phosphorylation site (T174A-176F) or in the kinase domain (K52R) of HOG1 gene showed that both activities were relevant for the survival of C. albicans to Dh killer toxins. Moreover, Hog1 phosphorylation was also required to sense and adapt to osmotic and oxidative stress while the kinase activity was somehow dispensable. Although the addition of supernatant from the killer toxin- producing D. hansenii 242 strain (Dh-242) induced a slight intracellular increase in Reactive Oxygen Species (ROS), overexpression of cytosolic catalase did not protect C. albicans against this mycocin. This supernatant induced an increase in intracellular glycerol concentration suggesting that this toxin triggers an osmotic stress. We also provide evidence of a correlation between sensitivity to Dh-242 killer toxin and resistance to Congo red, suggesting cell wall specific alterations in sensitive strains.
Keywords: killer toxin, osmotic stress, Debaryomyces hansenii, MAPK, Candida albicans, HOG pathway
Introduction
Killer toxins or mycocins are yeast (glyco)proteins with a toxic effect against other sensitive yeasts. They were first described in Saccharomyces cerevisiae in 1963 and are grouped in three types (K1, K2, and K28) according to the set of sensitive strains and lack of cross-immunity as killer toxin-producing yeasts are resistant to their own toxins (Marquina et al., 2002). More recently, a new killer toxin from wine maker S. cerevisiae strains, named Klus, was reported to kill all other mycocin producing S. cerevisiae strains (Rodríguez-Cousiño et al., 2011). Killer toxins are not exclusive to S. cerevisiae and other yeast genera such as Candida, Cryptococcus, Debaryomyces, Hansenula, Pichia, or Kluyveromyces have been reported to produce them. Killer toxins are protease-sensitive proteins, most of them only stable and active at acidic pH and low temperatures, losing activity above 35°C and a pH above 6. Nevertheless, some killer toxins are stable at a wide range of pH and high temperature like HM-1 from Hansenula makrii which is stable at pH 2–11 and 60°C for 1 h. Many of the genes that encode these killer toxins have been identified and their mechanism of action elucidated (Liu et al., 2015). K1 from S. cerevisiae (Breinig et al., 2002) and PMKT from Pichia membranifaciens (Santos and Marquina, 2004) induce membrane pore formation while K28 from S. cerevisiae and PMKT2 from P. membranifaciens arrest cell cycle at an early S phase (Schmitt et al., 1996; Santos et al., 2013). PMKT2 is also able to induce apoptosis in sensitive yeast (Santos et al., 2013). Kluyveromyces lactis produces a zymocin with tRNase activity (Jablonowski and Schaffrath, 2007) and Hansenula mrakii HM-1 inhibits the beta-1,3-glucan synthase activity thus, perturbing bud formation and conjugation in yeasts (Takasuka et al., 1995).
Mycocin-producing yeasts are ubiquitous in natural habitats, and can be isolated from soil, water or plants as well as in several foods and beverages. A potential application of these killer toxins could be the control of yeast populations during wine or beer fermentation, the inhibition of fungi contaminating food or fungal pathogens in plants (Schmitt and Breinig, 2002; Breinig et al., 2006). Moreover, a potential use in human and animal health has also been proposed as many killer toxins have been reported to have activity against C. albicans (Magliani et al., 1997; Polonelli et al., 2011), the main opportunistic fungal pathogen in humans and the fourth cause of nosocomial systemic infections in many developed countries (Pfaller and Diekema, 2007, 2010). This pathogen forms part of the microbiota in healthy individuals as a harmless commensal, causing diseases only when the immune system or the natural barriers become altered. Many efforts have been made to understand the virulence factors involved in the commensal to pathogen transition in C. albicans (Tang et al., 2016). Among others, the ability to adapt to different niches has been proposed to play a relevant role in C. albicans pathogenicity (Recently reviewed by Mayer et al., 2013; Poulain, 2015; Höfs et al., 2016). The ability to sense and respond to environmental changes allowing adaptation and survival is mediated by conserved signal transduction pathways. MAP Kinase pathways are some of the most studied signaling mechanisms in C. albicans (Alonso-Monge et al., 2006; Román et al., 2007). These MAPK kinase pathways integrate inputs and coordinate responses that allow the survival and adaptation to different environmental conditions. These pathways are integrated by transmembrane proteins, sensors, intermediate proteins and a MAPK module that includes three protein kinases that become sequentially activated by phosphorylation. The HOG pathway has been reported to be activated in response to several stresses and plays a role in cell wall biosynthesis, morphological transitions, tolerance to phagocytes, virulence and adaptation to commensalism (Alonso-Monge et al., 1999; Arana et al., 2007; Prieto et al., 2014). In S. cerevisiae, the phosphorylation of the Hog1 homolog was reported to be triggered by the killer toxin from P. membranifaciens PMKT (Santos et al., 2005). More recently Banjara and co-workers identified 23 Debaryomyces hansenii strains that exerted killer effects over C. albicans and C. tropicalis. These D. hansenii strains were isolated from cheeses of different origins and their toxins displayed different activities depending on the strain, the temperature and pH, thus opening the possibility to control C. albicans infections through a new therapeutic approach (Banjara et al., 2016).
In the present study, the killer activity of seven of the D. hansenii mycocin-producing strains from Banjara's work was tested against signal transduction C. albicans mutants in order to understand their mechanism of action. We show how these toxins have an increased action on specific MAPK mutants (hog1) but not others (cek1 or mkc1) suggesting a role for the HOG MAPK in killer toxin resistance in C. albicans. We also characterize their effect on cellular physiology and the biochemical role of the Hog1 MAP kinase in this process.
Materials and methods
Strains and growth conditions
Candida albicans strains used in the present work are listed in Table 1. D. hansenii strains and their origin are listed in Table 2 (Banjara et al., 2016). D. hansenii strains were routinely incubated at 30°C while C. albicans strains at 37°C. Yeasts were grown in YPD medium (1% yeast extract, 2% peptone, 2% glucose). Usually, overnight cultures were used for sensitivity assays. For this purpose, cells were suspended to an O.D.600nm = 0.8 and 5 μL of 10 fold serial dilutions were plated on YPD solid media supplemented with the different compounds at their indicated concentrations.
Table 1.
C. albicans strains used in this work.
| Microorganism | Strain | Genotype | Nomenclature in manuscript and figures | References |
|---|---|---|---|---|
| C. albicans | SC5314 | wt | Gillum et al., 1984 | |
| C. albicans | CAF2-1 | ura3::imm434/ura3::imm434-URA3 | CAF2 (wt) | Fonzi et al., 1993 |
| C. albicans | CHO4-1 | ura3::imm434/ura3::imm434-URA3 opy2::FRT/opy2::FRT | opy2 | Herrero de Dios, 2013 |
| C. albicans | CK43B-16 | ura3::imm434/ura3::imm434 cek1::hisG/cek1::hisG-URA3-hisG | cek1 | Csank et al., 1998 |
| C. albicans | CSSK21-U-6 | URA3/ura3Δ::imm434 ssk1::hisG/ssk1::hisG-URA3-hisG | ssk1 | Calera et al., 2000 |
| C. albicans | CM1613 | ura3Δ::imm434/ura3Δ::imm434 mkc1::hisG/mkc1::hisG–URA3–hisG | mkc1 | Navarro-García et al., 1995 |
| C. albicans | HI3-21 | ura3Δ::imm434/ura3Δ::imm434 hog1::hisG-URA3-hisG/hog1::hisG | hog1 | Prieto et al., 2014 |
| C. albicans | RM100 | ura3::imm434/ura3::imm434 his1::hisG/his1::hisG-URA3-hisG | RM100 (wt) | Negredo et al., 1997 |
| C. albicans | CNC13 | RM1000 hog1::hisG-URA3-hisG/hog1::hisG | hog1 (RM100) | San José et al., 1996 |
| C. albicans | BRD3 | RM1000 pbs2Δ::cat/pbs2Δ::cat-URA3-cat | pbs2 | Arana et al., 2005 |
| C. albicans | BRD7 | RM1000 hog1::hisG/hog1::hisG pbs2Δ::cat/pbs2Δ::cat-URA3-cat | hog1 pbs2 | Arana et al., 2005 |
| C. albicans | VIC100 | ura3Δ::imm434/ura3Δ::imm434 his1Δ::hisG/his1Δ::hisG sko1Δ::hisG/sko1Δ::hisG-URA3-hisG | sko1 | Alonso-Monge et al., 2010 |
| C. albicans | VIC200 | ura3Δ::imm434/ura3Δ::imm434 his1Δ::hisG/his1Δ::hisG hog1::hisG/hog1::hisG sko1Δ::hisG/sko1Δ::hisG-URA3-hisG | hog1 sko1 | Alonso-Monge et al., 2010 |
| C. albicans | REP3 | ura3::imm434/ura3::imm434 his1::hisG/his1::hisG sho1::hisG/sho1::hisG-URA3-hisG | sho1 | Román et al., 2005 |
| C. albicans | AMB4 | URA3/ura3Δ::imm434 mkk2Δ::FTR/mkk2Δ::FTR | mkk2 | Román et al., 2015 |
| C. albicans | REP18-1 | ura3::imm434/ura3::imm434 his1::hisG/his1::hisG-URA3-hisG msb2::FRT/msb2::FRT | msb2 | Román et al., 2009 |
| C. albicans | COA6 | ura3::imm434/ura3::imm434 ADH1/adh1::tTA pTet-GFP-SAT1 | CAF2-pNIM1R | Prieto et al., 2014 |
| C. albicans | HHH | ura3Δ::imm434/ura3Δ::imm434 hog1::hisG-URA3-hisG/hog1::hisG ADH1/adh1::tTA Ptet–HOG1-SAT1 | hog1-pNIM1R-HOG1 | This work |
| C. albicans | HHA | ura3Δ::imm434/ura3Δ::imm434 hog1::hisG-URA3-hisG/hog1::hisG ADH1/adh1::tTA Ptet–hog1F321L-SAT1 | hog1F321L | This work |
| C. albicans | HKD | ura3Δ::imm434/ura3Δ::imm434 hog1::hisG-URA3-hisG/hog1::hisG ADH1/adh1::tTA Ptet–hog1K52RL-SAT1 | hog1K52R | This work |
| C. albicans | HSP | ura3Δ::imm434/ura3Δ::imm434 hog1::hisG-URA3-hisG/hog1::hisG ADH1/adh1::tTA Ptet–hog1T174A-SAT1 | hog1T174A | This work |
| C. albicans | HDP | ura3Δ::imm434/ura3Δ::imm434 hog1::hisG-URA3-hisG/hog1::hisG ADH1/adh1::tTA Ptet–hog1T174A−Y176F-SAT1 | hog1 T174A−Y176F- | This work |
| C. albicans | PPD7 | ura3Δ::imm434/URA3 ADH1/adh1::tTA Ptet -dTOM2-SAT1 | CAI4-dTOM2 | Prieto et al., 2014 |
| C. albicans | HRFP | ura3Δ::imm434/ura3Δ::imm434 hog1::hisG-URA3-hisG/hog1::hisG ADH1/adh1::tTA Ptet -dTOM2-SAT | hog1-dTOM2 | This work |
| C. albicans | REP41 | ura3Δ::imm434/ura3Δ::imm434 hog1::hisG- /hog1::hisG ADH1/adh1::tTA-URA3 | hog1-pNRU-e | This work |
| C. albicans | REP32 | ura3Δ::imm434/ura3Δ::imm434 hog1::hisG- /hog1::hisG ADH1/adh1::tTA Ptet–CAT1-URA3 | hog1-pNRU-CAT1 | This work |
Table 2.
Killer toxin-producing strains used in this work.
| Microorganism | Origin | |
|---|---|---|
| Dh-65 | Debaryomyces hansenii | Gouda cheese isolate (Netherlands) |
| Dh-72 | D. hansenii | Colby cheese isolate (Wisconsin. US) |
| Dh-220 | D. hansenii | Bel Paese cheese isolate (Italy) |
| Dh-242 | D. hansenii | Parmesan cheese isolate (Parma. Italy) |
| Dh-246 | D. hansenii | Raclette cheese isolate (Wisconsin. US) |
| Dh-262 | D. hansenii | Ricotta cheese isolate (Illinois. US) |
| Dh-274 | D. hansenii | Blue 2 cheese isolate (Wisconsin. US) |
| CBS-767 | Candida boidinii IGC3430 |
Molecular biology procedures and plasmid constructions
The HOG1 gene inserted in a pUC19 vector was mutated using a commercial kit (QuickChange, Stratagene) and specific primers (Table 3). HOG1 mutated versions were amplified using the primers up_HOG_myc (GCCTCGAGATGTCTGCAGATGGAGAATTTACAAGA) and Low_HOG_myc (CTGCGGCCGCTAGCTCCGTTGGCGAATCC), digested with SalI-NotI and integrated in pNIM1R-RFP (Prieto et al., 2014) replacing the RFP gene. The generated plasmids pNIM1R-HOG1#-myc were digested with KpnI-SacII and integrated into the ADH1 locus of C. albicans genome using lithium acetate transformation (Köhler et al., 1997).
Table 3.
Primers used in this work to generate HOG1 mutant versions.
| Mutation | Primer | Sequence |
|---|---|---|
| F321L | F321LHOGU | GAGCCTGTTTGTGAGAGTAAATTGGATTGGAGTTTTAATGACG |
| F321LHOG1 | CGTCATTAAAACTCCAATCCAATTTACTCTCACAAACAGGCTC | |
| K52R | K52R1 | CTGGTCAAAATGTTGCAGTGAGAAAAGTCATGAAACC |
| K52R2 | GGTTTCATGACTTTTCTCACTGCAACATTTTGACCAG | |
| T174A-176F | TGYAGF1 | CTTCAAGATCCACAAATGGCTGGTTTCGTGTCAACCAG |
| TGYAGF2 | CTGGTTGACACGAAACCAGCCATTTGTGGATCTTGAAC |
Fluorescent labeled CAI-4 and hog1 mutant strains were generated by integration of the pNIM1R-dTOM2 plasmid at the ADH1 locus (Prieto et al., 2014). This plasmid carries a Red Fluorescent Protein under the control of the tetracycline-repressible promoter. The strains obtained appear reddish on SD plates (2% glucose, 0.5% ammonium sulfate, 0.17% yeast nitrogen base) allowing quantification (visually) of their relative abundance in competition assays and differentiation from others yeast colonies.
Overexpression of cytosolic catalase in hog1 mutants was achieve by integrating the plasmids pNRU-CAT1 or pNRU-e (empty vector) in the ADH1 region (Román et al., 2016). pNRU-CAT1 plasmid carries the CAT1 gene under the control of the tetracycline repressible promoter and tagged to the myc epitope. The vectors, previously digested with KpnI and SacII, were integrated in the hog1 background generating the hog1-pNRU-CAT1 and hog1-pNRUe corresponding strains (Table 1).
Killer toxin assay
Candida albicans overnight cultures in YPD were refreshed to an optical density of 0.8 (O.D.600nm) in 4 mL of saline solution and spread with a sterile swab on plates of YMB medium (0.3% yeast extract, 0.3% malt extract, 0.5% peptone, 1% glucose) supplemented with 3 % NaCl and buffered with citrate phosphate to pH 4.4 or 4.8. Then, mycocins-producing strains were patched with the help of a sterile swab on the plates and incubated at 30°C (otherwise indicated) for 2 days. Alternatively, mycocins-producing strains were grown for 2 days at 30°C in liquid YPD or YMB pH 4.4; cells were then eliminated through centrifugation plus filtration through a 0.45 μm pore filter (Millipore) and 15 μL of the resulting supernatant was used to soak sterile paper filter discs (6 mm, Filter-Lab). Dried discs were then disposed on YMB 3% NaCl buffered plates previously inoculated with the C. albicans strain to be tested and incubated at 30°C for 2 days. Plates were photographed and the inhibition zone produced by Dh mycocin was measured using the free software ImageJ and expressed in millimeters (mm). Inhibition zone refers the distant between the end of D. hansenii growth (or disk) and the beginning of C. albicans growth.
Protein extracts and immunoblot analysis
Candida albicans overnight cultures were refreshed to an optical density of 0.1 (O.D.600nm) and incubated until cultures reached an optical density of 1. Cultures were then divided in three cultures and either 1 M NaCl or 10 mM hydrogen peroxide was added to the medium, the last culture remained as control. Samples were collected 10 min later and processed for Western-blotting. Protein extracts were obtained as previously indicated (Martin et al., 2000). Even amounts of proteins were loaded onto gels as assessed by protein sample estimates at 280 nm. Blots were probed with anti-phospho-p38 MAP kinase (Thr180/Tyr182) 3D7 monoclonal antibody (Cell Signaling Technology, Inc.), anti-phospho-p44/42 MAP kinase antibody (Thr202/Tyr204) (D13.14.4E) (Cell Signaling Technology, Inc.) and anti-myc Tag antibody, clone 4A6 (Millipore). The detection was performed using the Quantitative Fluorescent Imaging System Odyssey from Li-COR.
Flow cytometry
Flow cytometry was used to quantify intracellular Reactive Oxygen Species (ROS) using dihydrofluorescein diacetate (DHF) and mitochondrial membrane potential using rhodamine 123 (R123). Briefly, overnight cultures from the different C. albicans strains were collected by centrifugation and refreshed either in pre warmed YMB pH 4.4 medium at 30°C or in Dh-242 overnight spent medium. C. albicans cultures were incubated at 30°C for 1 and 2 h. Thirty minutes before this time DHF or R123 was added to the samples to a final concentration of 40 and 20 μM respectively and incubated at 30°C in the dark for the remaining 30 min. Samples were collected, washed twice with PBS and resuspended in PBS at 106 cells per mL. Then, propidium iodide (IP) was added to detect dead cells. Fluorescence intensity was determined by flow cytometry using the FACScan cytometer (Beckton Dickinson) from the Servicio de Citometría (Universidad Complutense de Madrid). The analysis of green (FL1) fluorescence intensity was done on a logarithmic scale and gates were set around debris and intact cells on a FSC vs. SSC dot plot. Positive IP cells (dead cells) were removed from the analysis. Fluorescence histograms corresponding to 10,000 cells were generated using the gated data with data analyses done using Flowing Software 2.5.1.
Glycerol quantification
To quantify the intracellular glycerol, exponentially growing cells were centrifuged and resuspended in YMB 3% NaCl pH4.4 medium supplemented with 1 M NaCl (positive control) or the medium from Dh-242 strain grown at 30°C during 48 h. Samples were taken before and 1 and 3 h after the challenge. Ten milliliter of the culture was harvested and filtered through a 0.45 μm filter previously weighed. The dry weight was determined by subtracting the weight of the filter from the weight obtained after drying the filtered pellets for 2 days at 45°C until stable weight. For glycerol determination, a 1 mL sample was centrifuged at 10,000 rpm for 5 min; the pellet was resuspended in 1 mL of ultrapure water (Milli Q) and boiled at 99°C for 10 min. Cell debris were removed by centrifugation (10 min 10,000 rpm) and the supernatant was collected. The amount of glycerol was determined using the commercial preparation of Megazyme Glycerol Assay Kit (K-GCEROL following manufacturer instructions. Intracellular glycerol is expressed in μg of glycerol per mg of dry weight.
Results
C. albicans hog1 mutants are sensitive to killer toxins from different D. hansenii strains
Debaryomyces hansenii strains previously reported to have anti-candida activity (Banjara et al., 2016) were tested against the C. albicans SC5314 strain at different pHs and temperatures (Table 4 and Supplementary Figure 1). Although no killer activity was found at 37 or 37°C plus hypoxia, a clear killer effect (as determined by the halo) was observed when different D. hansenii strains were tested at 24 and 30°C, being higher at pH 4.4 than at pH 4.8 (Table 4).
Table 4.
Killer effect of D. hansenii strains against C. albicans strains.
| Dh-65 | Dh-72 | Dh-220 | Dh-242 | Dh-246 | Dh-262 | Dh-274 | CBS-767 | ||
|---|---|---|---|---|---|---|---|---|---|
| SC5314 (wt) | pH 4.4 | 1.79 ± 0.25 | 1.31 ± 0.09 | – | 1.92 ± 0.24 | 1.07 ± 0.19 | 0.97 ± 0.09 | 0.72 ± 0.11 | 0.93 ± 0.20 |
| pH 4.8 | 1.41 ± 0.12 | – | – | 0.77 ± 0.22 | 0.95 ± 0.06 | 0.64 ± 0.15 | – | 0.74 ± 0.13 | |
| mkc1 | pH 4.4 | 1.02 ± 0.2 | – | – | 1.15 ± 0.34 | 1.50 ± 0.70 | 1.18 ± 0.20 | 1.39 ± 0.11 | 1.06 ± 0.32 |
| pH 4.8 | 0.29 ± 0.05 | – | – | – | 0.69 ± 0.08 | 0.84 ± 0.06 | 0.67 ± 0.21 | 0.93 ± 0, 21 | |
| hog1 | pH 4.4 | 4.82 ± 0.15 | 0.88 ± 0.03 | 2.42 ± 021 | 4.70 ± 0.30 | 6.72 ± 0.48 | 6.48 ± 0.06 | 6.23 ± 0.60 | 5.36 ± 0.42 |
| pH 4.8 | 4.32 ± 0.25 | 2,.3 ± 0.34 | 2.49 ± 0.43 | 3.37 ± 0.28 | 3.6 ± 0.30 | 3.37 ± 0.21 | 3.70 ± 0.12 | 3.43 ± 0.04 | |
| cek1 | pH 4.4 | 1.17 ± 0.25 | – | – | 1.12 ± 0.18 | 1.08 ± 0.71 | 1.37 ± 0.67 | 0.86 ± 0.1 | 1.10 ± 0.18 |
| pH 4.8 | – | – | – | 1.27 ± 0.11 | 0.76 ± 0.08 | 0.80 ± 0.10 | 0.73 ± 0.06 | 1.15 ± 0.18 |
Inhibition zone was measured in mm. The assay was performed on YMB 3% NaCl at pH 4.4 or 4.8 plates and incubated at 30°C. Data are the mean of two independent experiments ± Standard deviation. Three measures were performed per experiments.
In order to characterize the mechanism of action and the targets of these Dh killer toxins we examined the behavior of C. albicans MAPKs defective mutants in the presence of the mycocin-producing D. hansenii strains. The mutant lacking the Hog1 MAPK displayed an enhanced susceptibility to most of D. hansenii strains tested while this was not observed for mutants defective in either Cek1 or Mkc1 (Table 4 and Figure 1A). In fact, cek1 and mkc1 mutants were slightly more resistant than the control strain to killer toxins from Dh-65, Dh-72, Dh-220, and –partially- to Dh-242 although these effects were not statistically significant (Table 4 and Figure 1A).
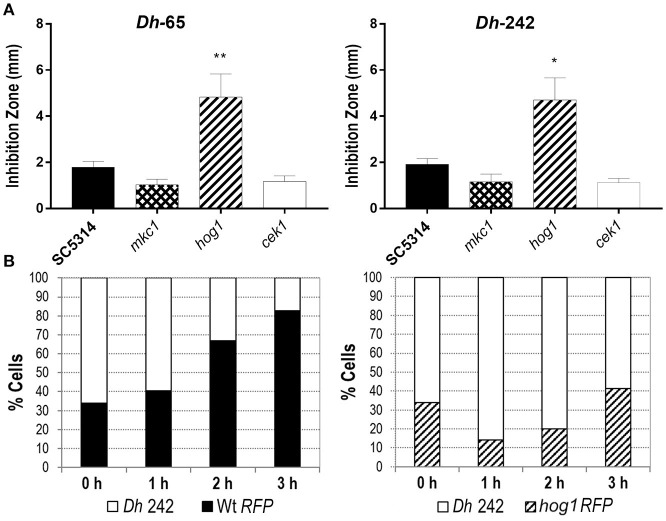
Figure 1.
Effect of D. hansenii (Dh) strains on C. albicans growth. (A) The killer activity of Dh-65 and Dh-242 strains was tested against the indicated C. albicans strains on YMB 3% NaCl pH 4.4 plates at 30°C for 48 h. Graph shows the mean and SD from three independent experiments. t-test analyses were performed to show statistical significant differences. **p < 0.01 and *p < 0.05. (B) C. albicans strains carrying the fluorescent protein dTOM2 (RFP) were mixed with Dh-242 strain to 1:2 proportion in YPD pH 4.4 and incubated at 30°C with shaking. Samples were collected at indicated time points and CFUs were counted and expressed as percentages. RFP label allows differentiating C. albicans (red colonies) from D. hansenii (white colonies).
Dh-242 strain was chosen to analyze its effect on C. albicans liquid cultures since the killing effect of this strain was clear against the strain analyzed. Wild type and hog1 C. albicans strains were tagged with the fluorescent protein dTOM2 in order to discriminate between C. albicans and D. hansenii cells. C. albicans strains tagged with dTOM2 (Prieto et al., 2014) were mixed to a 1:2 proportion with the strain Dh-242 and incubated together at 30°C. Samples were collected after 1, 2 and 3 h, dilutions spread on SD plates and white and red colonies counted and expressed as percentage (Figure 1B). The C. albicans wild type strain was able to grow in the presence of mycocin-producer Dh strain from the first hour, being more than 80 % of the culture after 3 h of incubation. In the case of hog1 cultures, the proportion of hog1 cells decreased to 14% after 1 h and then, resumed growth increasing the percentage of C. albicans cells to the detriment of Dh strain reaching 40% of the total cells in culture. This result indicates that Dh-242 strain has an inhibitory effect on C. albicans hog1 mutant in liquid medium that did not impair the wild type strain. Dh-242 compromised hog1 mutant survival at short time after co-incubation (1–2 h). Thus, the Dh-242 effect is enhanced in the hog1 mutant suggesting that the HOG pathway could be involved in the response to mycocins produced by this strain.
Dh-242 supernatant induces glycerol accumulation but not oxidative stress in C. albicans
In order to check if Dh-242 KT altered cell permeability promoting an osmotic stress that triggers the HOG pathway as it has been suggested for other killer toxins (Santos and Marquina, 2011), we measured the intracellular accumulation of glycerol in response to Dh-242 supernatant. A wild type strain increased the intracellular glycerol in the presence of 1 M NaCl reaching a maximum after 3 h of incubation. I In the presence of Dh-242 supernatant the accumulation was earlier (maximum at 1 h) and transient since it decreased after 3 h of incubation (Figure 2A). The hog1 mutant accumulated intracellular glycerol in the presence of Dh supernatant to a similar level as the wild type strain (Figure 2A). No glycerol accumulation was detected when cells were grown in YMB 3% NaCl pH 4.4 indicating that 3% NaCl (0.5M) is not enough to induce glycerol accumulation in C. albicans and that the observed effect is due to the Dh killer toxin (Supplementary Figure 2). These results suggest that Dh mycocins present in the supernatant of Dh-242 caused an osmotic stress in C. albicans cells triggering a transient accumulation of glycerol which is independent of Hog1.
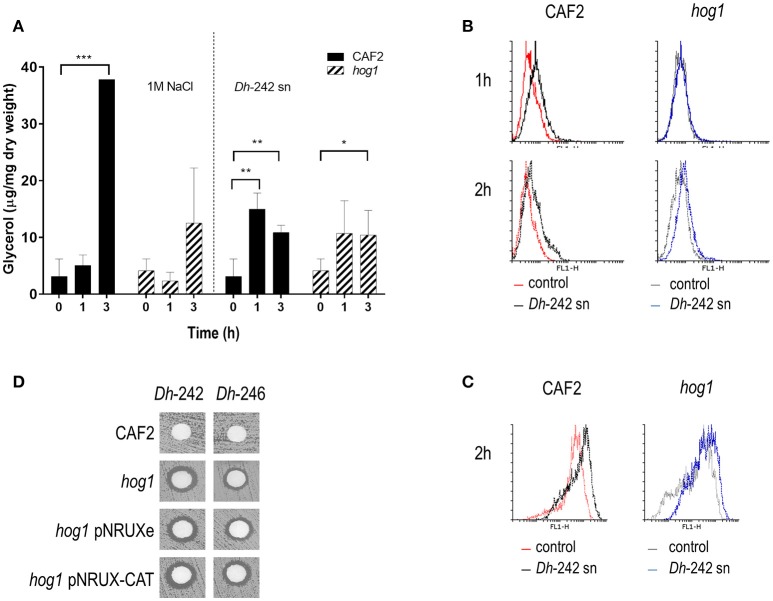
Figure 2.
Effect of Dh-242 supernatant on intracellular glycerol accumulation and oxidative stress. (A) C. albicans wild type and hog1 mutant cells were resuspended in either a supernatant from Dh-242 strain grown for 48 h in YMB 3% NaCl pH 4.4 or fresh medium pre-warmed at 30°C supplemented with 1M NaCl (1.5 M final concentration). Samples were collected at 0, 1, and 3 h and intracellular glycerol was quantified. Graph shows the mean and SD from 3 independent experiments. t-test analyses were performed to show statistical significant differences ***p < 0.001, **p < 0.01, and *p < 0.05 (B) CAF2 and hog1 mutants strains were incubated in the presence of supernatant from Dh-242 strain (+) or YMB pH 4.4 (−) liquid medium at 30°C. Intracellular oxidative stress was quantified by flow cytometry using Rhodamine 123. A histogram from a representative experiment is shown. (C) Mitochondrial membrane potential was quantified using DHF after 2 h of incubation in supernatant from Dh-242 strain (+) or YMB pH 4.4 (−) liquid medium. A histogram from a representative experiment is shown. (D) A killer toxin assay was performed using supernatant from Dh-242 and Dh-246 strains grown in YMB 3% NaCl pH4.4 liquid medium for 48 h. The hog1 mutant strain carrying the empty vector (pNRUXe) or overexpressing the Catalase (pNRUX-CAT) were tested.
Since the HOG pathway is also needed to respond to oxidative stress, the intracellular ROS amount was measured using flow cytometry analysis in response to a supernatant from the Dh-242 strain. Wild type and hog1 strains showed a similar percentage of cells stained with IP (dead, IP+) within the first 2 h. In the presence of Dh supernatant, cultures of the CAF2 wild type strain increased intracellular ROS at 1 and 2 h while in the case of the hog1 mutant was less evident even after 2 h of incubation (Figure 2B). Since intracellular ROS accumulation requires mitochondrial activity the mitochondrial membrane potential was also quantified by flow cytometry. The mitochondrial membrane potential in the presence of the Dh-242 supernatant increased equally in both strains after 2 h in the presence of the Dh-242 supernatant when measured with DHF (dihydrofluorescein) (Figure 2C).
Since the differences in intracellular ROS and mitochondrial membrane potential were not substantial, we checked if a hog1 mutant overexpressing catalase (CAT1) could overcome the effect of Dh-KT (Figure 2D). C. albicans cells overexpressing CAT1 are more resistant to hydrogen peroxide, phagocytes, and decrease intracellular ROS generated by antifungal such as amphotericin B (Román et al., 2016). The hog1 mutant strain was transformed with the empty vector or with a vector carrying the CAT1 gene under the control of tetracycline promoter in its repressible version. The overexpression of CAT1 did not revert the sensibility to Dh-KT displayed by the hog1 mutant.
Altogether, we can conclude that the increased sensitivity of the hog1 mutant to Dh mycocins is not due to the impairment of the response to osmotic or oxidative stress mediated by HOG1, pointing to other type of defects.
The MAPK Hog1 but not the HOG pathway is essential for killer toxin activity
As certain MAPK mutants displayed altered susceptibilities to killer toxins, mutants in other elements of the HOG, cell wall integrity (MKC1) and Cek1-mediated pathways were analyzed under similar conditions (Table 5 and Supplementary Figure 3). Remarkably, the deletion of the response regulator of two-component system (Ssk1), the MAPKK (Pbs2), or the transcription factor (Sko1) which participate in the HOG pathway displayed variable susceptibility to Dh mycocins compared to the parental wild type strain (RM100) (Table 5). Interestingly, none of the mutants showed sensitivities to Dh-mycocins to the same extend that the hog1 mutant in the same background (CNC13 strain). The CNC13 strain displayed an enhanced sensitivity compared to the parental strain RM100, suggesting that the sensitivity to Dh killer toxins does not depend on the genetic background but, rather, it is specific to HOG1 deletion. As expected, the integration of the HOG1 gene into the hog1 mutant restored wild type phenotype. When pbs2 hog1 and sko1 hog1 double mutants were tested, the susceptibility to mycocins depended on the Dh strain analyzed (Table 5). pbs2 hog1 and sko1 hog1 double mutants were more susceptible than the hog1 mutant to Dh-220 and Dh-274 mycocins. The inhibition zone in the presence of the control strain CSB-767 was reduced in pbs2 hog1 and sko1 hog1 double mutants compared to hog1 strain. These data suggest that Hog1 is crucial for the tolerance to D. hansenii killer toxins, while, others elements of the pathways such as Ssk1, Pbs2 and Sko1 play a relatively minor role in killer toxin tolerance. Mutants in upstream elements of the Cek1 pathway such as the transmembrane proteins Msb2, Opy2 or the MAPKK of the Mkc1 pathway, Mkk2 were more resistant to certain Dh killer toxins than the parental strain (Table 5) suggesting that these mutants lack (or have in lesser proportion) a putative receptor(s) required to bind or activate Dh killer toxins.
Table 5.
Killer effect of D. hansenii strains against C. albicans strains.
| Dh-65 | Dh-72 | Dh-220 | Dh-242 | Dh-246 | Dh-262 | Dh-274 | CBS-767 | |
|---|---|---|---|---|---|---|---|---|
| RM100 | 0.68 ± 0.23 | – | 0.75 ± 0.06 | 0.83 ± 0.21 | 0.95 ± 0.14 | 1.01±.16 | 0.73 ± 0.09 | 0.95 ± 0.1 |
| ssk1 | 0.65 ± 0.14 | – | – | 0.87 ± 0.17 | 1.32 ± 0.09 | 0.67 ± 0.08 | 1.24 ± 0.19 | 0.67 ± 0.07 |
| pbs2 | 0.67 ± 0.07 | – | – | 1.84 ± 0.10 | 1.86 ± 0.05 | 1.83 ± 0.27 | 2.99 ± 0.23 | 0.51 ± 0, 26 |
| hog1 (RM100) | 6.78 ± 0.01 | 2.71 ± 0.19 | 2.97 ± 0.49 | 4.34 ± 0.28 | 6.37 ± 0.16 | 6.36 ± 0.38 | 5.12 ± 0.59 | 6.00 ± 0.16 |
| hog1reint (CNC16-13) | 1.54 ± 0.32 | – | – | 1.77 ± 0.17 | 1.09 ± 0.13 | 1.02 ± 0.17 | 0.93 ± 0.16 | 0.63 ± 0.07 |
| hog1 pbs2 | 4.26 ± 0.14 | 3.60 ± 0.26 | 6.25 ± 0.26 | 4.67 ± 0.03 | 6.20 ± 0.46 | 6.87 ± 0.38 | 6.51 ± 0.45 | 5.31 ± 0.25 |
| sko1 | 0.86 ± 0.33 | – | 0.57 ± 0.38 | 1.45 ± 0.59 | 1.49 ± 0.69 | 1.63 ± 0.69 | 1.26 ± 0.61 | 0.83 ± 0.185 |
| hog1 sko1 | 2.02 ± 0.24 | 3.31 ± 0.62 | 8.1 ± 0.23 | 1.82 ± 0.09 | 5.93 ± 0.39 | 6.98 ± 0.35 | 5.59 ± 0.32 | 1.59 ± 0.24 |
| msb2 | – | – | – | 0.37 ± 0.01 | 0.57 ± 0.11 | 0.5 ± 0.14 | 0.42 ± 0.03 | 0.44 ± 0.07 |
| opy2 | – | – | – | 1.63 ± 0.40 | 1.19 ± 0.11 | 1.856 ± 0.12 | 0.82 ± 0.05 | – |
| mkk2 | – | – | – | 0.60 ± 0.06 | 0.50 ± 0.19 | 0.58 ± 0.19 | – | – |
Inhibition zone was measured in mm. The assay was performed on YMB 3% NaCl at pH 4.4 plates and incubated at 30°C. Data are the mean of two independent experiments ± Standard deviation. Three measures were performed per experiments.
Hog1 phosphorylation and kinase activity are crucial for killer activity resistance
Since upstream and downstream elements of the HOG pathway played a limited role in sensitivity to Dh-242 supernatant, we checked if the regulation module or the kinase activity of Hog1p could be important for this effect by obtaining different mutants and testing their killer toxin susceptibility. The lysine in position 52 was replaced by an arginine to generate a mutant defective in its kinase activity (Hog1K52R). Similarly, the phosphorylation site, threonine 174 and tyrosine 176, were replaced by alanine and phenylalanine, respectively, to generate a mutant unable to be phosphorylated in its regulation domain (Hog1T174A Y176F). In addition, a single substitution of one of those phosphorylation sites was also obtained (Hog1T174A). Finally, a phosphomimetic Hog1 version was generated replacing the phenylalanine 321 by a leucine (Hog1F321L). Both kinase dead (Hog1K52R) and non-phosphorylatable (Hog1T174A Y176F) strains were found to be susceptible to killer toxins in a similar magnitude to the hog1 mutant (Figure 3A). Moreover, phosphomimetic Hog1 version (Hog1F321L) displayed an intermediate susceptibility between the hog1 mutant and wild type strain, suggesting that dephosphorylation of Hog1 must be relevant to Hog1 function (Figure 3A). Also, HOG1 overexpression reverted the sensitivity to mycocins to wild type levels, while the Hog1T174A strain behaved similarly to the kinase dead mutant (Figure 3C). Similar results were obtained in the presence of other mycocin-producing D. hansenii strains (Data not shown).
Figure 3.
Effect of D. hansenii killer strains on different hog1 mutants. (A) Killer toxin assay of different D. hansenii strains (Dh) against different C. albicans strains. Equal suspensions of C. albicans strains were spread on YMB plates supplemented with 3% NaCl pH 4.4 with a sterile swab and, D. hansenii strains were immediately spotted with a swab; the plates were thenincubated at 30°C (B) Killer toxin assay of supernatants of different D. hansenii strains against the indicated C. albicans strains. Dh strains were grown in YMB 3% NaCl pH 4.4 liquid medium for 48 h, centrifuged and 15 μl of the filtered supernatant were inoculated in a paper filters. Paper filters impregnated with the supernatant were deposited on YMB 3% NaCl pH 4.4 plates previously inoculated with the indicated C. albicans strains using a sterile swab. (C) Killer activity of the Dh-246 strain against different C. albicans strains was measured (in mm) from at least three independent experiments. t-test analyses were performed to show statistical significant differences. ****p < 0.001, ***p < 0.001, and **p < 0.05.
In parallel, we tested the killer activity exerted by supernatants from Dh mycocin-producing strains to confirm that Dh mycocin were secreted to the culture medium (Figure 3B). Although no killer effect was detected against wild type C. albicans, hog1, and Hog1T174A Y176F strains displayed a clear inhibition zone (Figure 3B). The kinase dead strain (Hog1K52R) showed a weak inhibition halo, suggesting that Hog1 phosphorylation is more important than kinase activity to tolerate Dh mycocins.
Mutations in HOG1 induce an altered congo red sensitivity
Mycocins can bind different components of the fungal cell wall as first step to get the final target: β-1,3, β-1,6-D-glucan, chitin or mannans have been reported as primary receptors (Liu et al., 2015). Since alterations in cell wall biogenesis have been described in hog1 mutants (Alonso-Monge et al., 1999), we wondered if there was any correlation between sensitivity to Dh killer toxins and others cell wall-related phenotypes. Susceptibility to caspofungin and Congo red was tested on plates at 30 and 37°C. hog1 mutants did not display caspofungin sensitivity compared to wild type at any temperature and condition analyzed (Supplementary Figure 4). Nevertheless, hog1 and Hog1K52R strains are resistant to Congo red at both temperatures (30 and 37°C) (Figure 4A). The non-phosphorylatable Hog1T174A Y176F strain displayed Congo red resistance at 37°C and sensitivity at 30°C (the temperature used to perform killer toxin activity). Therefore, the lack of Hog1 renders cells sensitive to Dh killer toxins and resistant to Congo red and this phenotype depends on its kinase activity.
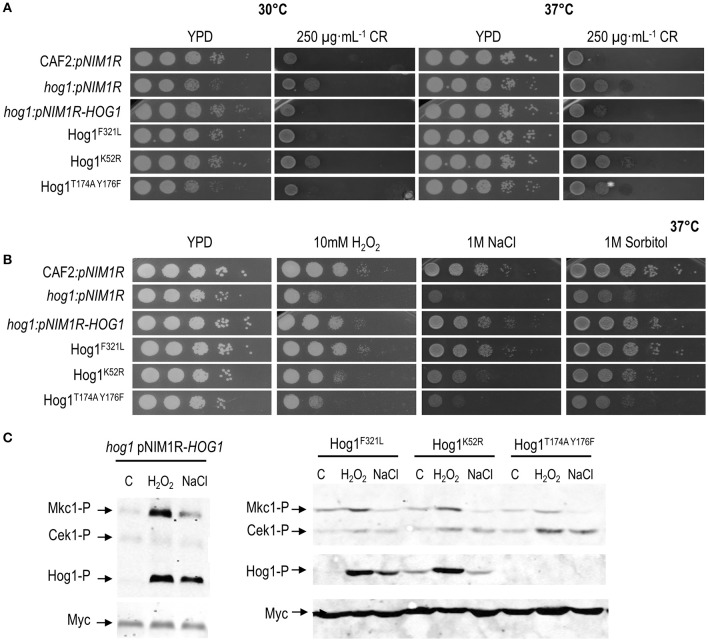
Figure 4.
Role of different domains of Hog1 in tolerance and response to stress. (A) C. albicans strains carrying different HOG1 gene versions were spotted on YPD plates supplemented or not with Congo red and incubated for 24 h at 30 or 37°C. (B) Tenfold cell suspensions of the indicated strains were spotted on YPD solid medium supplemented with the indicated compounds and incubated at 37°C for 24 h. (C) Exponentially growing cells were exposed to 10 mM H2O2 or 1M NaCl during 10 min and processed for immunoblot detection. Mkc1-P, Cek1-P, and Hog1-P indicate the phosphorylated forms of the MAPKs. Load control was detected with Anti-Myc antibody.
The susceptibility to cell wall disturbing compounds was also analyzed for others signaling mutants (mkc1, mkk2, cek1, and opy2) resistant to Dh-mycocins. In agreement with the correlation between resistance to killer toxin and Congo red sensitivity, these mutants are more sensitive to Congo red than the wild type strain (Herrero de Dios et al., 2013; Román et al., 2015) (Supplementary Figure 5).
Since the kinase activity of Hog1 seems to be important for Congo red resistance, we tested the role played by specific Hog1 domains in the canonical Hog1 functions, signaling and adaptation to osmotic and oxidative stress. Sensitivity assays were performed by spotting cell suspensions on YPD plates supplemented with H2O2, NaCl or sorbitol, and incubated at 37°C. As shown in Figure 4B, the non-phosphorylatable Hog1T174A Y176F strain displayed an increased susceptibility to oxidative and osmotic stress similar to the hog1 deletion mutant. Oddly, the kinase-dead strain expressing Hog1K52R displayed an intermediate phenotype between wild type and the hog1 deletion mutant. Mutants carrying the phosphomimetic Hog1 version or overexpressing HOG1 behaved as the wild type strain and did not increase the tolerance to osmotic and oxidative stress.
The MAPK phosphorylation pattern was also analyzed in response to standard stimuli that trigger the HOG pathway: hydrogen peroxide and NaCl (Figure 4C). In response to oxidative stress, Hog1 was phosphorylated in phosphomimetic and the kinase-dead Hog1 strains. In response to osmotic stress, Hog1 phosphorylation was clear in Hog1F321L strain although the kinase dead (Hog1K52R) strain showed a lower phosphorylation level in response to NaCl. As expected, no Hog1 phosphorylation was detected in non-phosphorylatable Hog1174A Y176F strain. Moreover, Mkc1 was phosphorylated in response to hydrogen peroxide in both the phosphomimetic and the kinase-dead Hog1 strains. The non-phosphorylatable Hog1 strain displayed higher Cek1 phosphorylated level and hardly activated Mkc1 in response to H2O2. This MAPK phosphorylation pattern matches with hog1 deletion mutant (Arana et al., 2005; Alonso-Monge et al., 2009). These data indicate that Hog1 phosphorylation is required to trigger osmotic and oxidative stress responses that enable C. albicans to grow in the presence of these stresses. The HOG1 overexpressing strain (hog1 pNIM1R-HOG1 strain) behaved as the wild type strain (Figure 4B left panel).
Discussion
Candida albicans infections represent a major health problem worldwide. In spite of the introduction of extended-spectrum triazole and echinocandin antifungal agents in the last decades, the frequency of candidemia and others invasive candidiasis, as well as the associated mortality (reaching 49% of patients) have not decreased. Current research efforts focus on finding faster and more sensitive diagnostic methods to avoid unnecessary treatments, fighting against antifungal resistance and reducing economic costs (Pfaller and Castanheira, 2016). Searching for novel antimycotics or new natural compounds with anti-Candida activity is an interesting alternative approach. For example, several phytocompounds have been proved to exert anti-Candida activity, some of them with synergistic effect in combination with fluconazole (Lu et al., 2017). In addition, different killer toxins have been discovered to have activity against C. albicans (Liu et al., 2015).
We focused here on some D. hansenii strains isolated from cheese displaying activity against Candida (Banjara et al., 2016), although the molecular basis for this activity have not been reported. Our results indicate that these killer toxins present an optimum inhibitory effect at pH 4.4, 30°C and in NaCl-supplemented medium, similarly to what occurs with most of the killer toxins reported (with few exceptions) (Liu et al., 2015).
We found that the absence of the Hog1 MAPKK promoted sensitivity to mycocins from several D. hansenii strains. Unexpectedly, this was only detected in the hog1 mutant and not in others mutants of the pathway such as pbs2, ssk1 or sko1. Hog1 phosphorylation depends directly on the MAPKK Pbs2 (Arana et al., 2005) and consequently, hog1 and pbs2 mutants share sensitivity to those stresses that trigger Hog1 phosphorylation such as osmotic and oxidative stress. This suggests that the susceptibility to Dh killer toxins seems to be more complex and may involve different triggering stimuli. What differentiates hog1 from pbs2 mutants? One possibility is the cell wall architecture: hog1 and pbs2 mutants share several phenotypes but not sensitivity to cell wall disturbing compounds. The lack of Hog1 renders the mutant more resistant to calcofluor white and Congo red than the wild type, pbs2 mutant (Arana et al., 2005) and the sko1 mutant (Alonso-Monge et al., 2010). Killer toxins require two receptors to exert their effect: the primary receptor is usually located on the cell wall and the secondary receptor on the plasma membrane or cytosol. The first step for killing activity involves the binding of the mycocin to a yet unknown cell wall component. This component can be any of the constituents that integrate the fungal cell wall such as β-1,3 or β-1,6-D-glucan, chitin or mannans (Liu et al., 2015). Our data indicate that there is no relationship between caspofungin and Dh killer toxin sensitivity but there is with Congo red resistance. Since no difference could be found in caspofungin susceptibility between Hog1 mutants and wild type strains, the compensatory mechanism of cell wall construction that increase chitin accumulation should not be responsible for the different sensitivity to Dh killer toxin (Lee et al., 2012). C. albicans hog1 mutants have been shown to be more sensitive to zymolyase (Eisman et al., 2006), a β-1,3-glucanase preparation with a residual protease activity that is essential to allow the access of the β-1,3-glucanase to its target (Zlotnik et al., 1984; García et al., 2009). hog1 mutants have an increased β-1,3-glucan exposure (D. Prieto unpublished data), suggesting an altered cell wall that could in turn be responsible for increased access of zymolyase (and maybe also mycocins) to its target, probably β-1,3-glucan.
After reaching the cytoplasmic membrane certain killer toxins, such as the K1 toxin produced by S. cerevisiae, exert its lethal activity by ion channel formation and therefore, perturbing the permeability of the membrane (Flegelová et al., 2003). Similarly, the killer toxin from P. membranifaciens PMKT has been suggested to bind to a receptor in the cell wall such as β-D-(1,6)-glucan that would somehow allow crossing the cell wall and binding to a second receptor in the plasma membrane (Santos et al., 2005; Santos and Marquina, 2011). PMKT causes the loss of ions and metabolites which is sensed by yeast cells as a hyperosmotic stress therefore triggering Hog1p phosphorylation (Santos and Marquina, 2011). Interestingly, the supernatant from Dh-242 strain also induces a transient intracellular glycerol increase in C. albicans, suggesting that Dh-242 mycocin may trigger an osmotic stress response which would be Hog1-independent. In agreement with this, no phosphorylation of Hog1 was detected when C. albicans was exposed to Dh supernatants (data not shown). This could be caused by the reduced mycocin concentrations in the supernatant, not enough to trigger the response. However, it can be also possible that Dh mycocins do not trigger Hog1 phosphorylation in C. albicans. Glycerol accumulation could be due to the closure of channels which would in turn prevent glycerol efflux through a mechanism independent of Hog1 as occurs in S. cerevisiae (Saxena and Sitaraman, 2016). Although these results lead us to conclude that the participation of Hog1 in the response to the osmotic stress generated by Dh-242 mycocin is not crucial, the assays performed on solid medium indicate that Hog1 phosphorylation is needed. The continued presence of Dh cells and therefore, Dh mycocins, may generate a sustained osmotic stress that requires Hog1 phosphorylation.
The relevance of the phosphorylation domain for Hog1 activity was reported previously (Cheetham et al., 2011; Chang et al., 2016) but these previous works did not address the role of the kinase domain as shown here. Interestingly, the Hog1 kinase dead mutant behaves closer to the wild type strain under osmotic and oxidative stress but closer to the hog1 deletion mutant on Congo red supplemented plates. Thus, cell wall related phenotypes require Hog1 kinase activity although Hog1 phosphorylation is not so relevant. This suggests that non-phosphorylated (or basal Hog1 phosphorylation) is enough to control cell wall biogenesis, maybe controlling others MAPK phosphorylation (in the absence of Hog1, Cek1 is constitutively phosphorylated) or its own pathway as reported in S. cerevisiae (Sharifian et al., 2015). Survival of C. albicans cells when exposed to Dh-killer toxins requires both phosphorylation and kinase activities indicating that both the role of Hog1 in cell wall biogenesis and signaling are compulsory to face Dh-mycocins.
The phosphorylation of the kinase dead Hog1 mutant may, somehow, be enough to trigger a downstream response and bypass the requirement for kinase activity. This observation is in agreement with results reported previously that showed that the nuclear localization of Hog1 is dispensable for many of the functions exerted by this MAPK (Day et al., 2017) such as growth in the presence of osmotic or oxidative stressors, virulence or gene transcription. Hog1 localization is, however, important to maintain signal fidelity, avoiding others MAPKs being phosphorylated that could change cell wall architecture. How Hog1 can trigger a downstream cascade without being translocated to the nucleus or without kinase activity remains unknown. The phosphomimetic version of Hog1 (Hog1F321L) did not increase the resistance to different stresses nor triggered Hog1 constitutive phosphorylation but became phosphorylated when exposed to osmotic or oxidative stress. A similar behavior was described by Cheetham and co-workers analyzing a phosphomimetic version of the MAPKK Pbs2 (Pbs2DD) (Cheetham et al., 2011); a Pbs2DD allele did not phosphorylate Hog1 constitutively, exhibited a partial sensitivity to stress and triggered Hog1 phosphorylation upon different stresses to significantly lower levels compared to wild type and this phosphorylation was independent of Ssk2 (Cheetham et al., 2011). Hyperactivation of Hog1 leads to a lethal phenotype in S. cerevisiae (Wurgler-Murphy et al., 1997; Yaakov et al., 2003), but not in C. albicans, suggesting that C. albicans may have evolved to prevent Hog1 constitutive activation and evidencing substantial differences in signal transduction between both yeast.
Since relationships among microorganisms in natural environments are complex and involve synergistic and antagonistic interactions, the production of killer toxins could provide evolutionary advantages for mycocin-producing strains like D. hansenii, a safe microorganism present in cheeses and fermented sausages as part of their production processes. D. hansenii strains have been isolated from the human gastrointestinal tract following ingestion of dairy products (Cosentino et al., 2001; Vasdinyei and Deák, 2003; Borelli et al., 2006; Desnos-Ollivier et al., 2008; Hallen-Adams et al., 2015). The interaction of D. hansenii and C. albicans could take place in the human gastrointestinal tract through feeding, although killer activity in the gastrointestinal tract has not been yet proved. The MAPK Hog1 seems to mediate the tolerance to killer toxins first avoiding the binding of killer toxins to cell surface and then, triggering a response to counterbalance their effects. All these data reinforce the relevance of signaling pathways in intra-species relationships and cell survival as well as the search for killer toxins that could be active at physiological temperatures in the human body.
Author contributions
AM-M, IC, JG-A, FN-G performed the experiments. FN-G, ER, DP JP, RA-M analyzed the data, results discussion and supervising the manuscript. JP did funding acquisition. RA-M did experimental design, supervising, and writing manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr. Hallen-Adams for sharing D. hansenii strains used in this study. Work in our laboratory is supported by Grants PCIN-2014-052 (Infect-ERA) and BIO2015-64777-P from Ministerio de Economía y Competitividad (MINECO, Spain).
Glossary
Abbreviations
- MAPK
Mitogen Activated Protein Kinase.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2018.00135/full#supplementary-material
Dh Kiler Toxin activity against C. albicans strains: The killer activity was analyzed on YMB plates supplemented with 3% NaCl pH 4.4 against SC5314 and the hog1 mutant strain under different environmental conditions.
Quantification of intracellular glycerol from CAF2 and hog1 mutant exposed to YMB plates plus 3% NaCl pH 4.4 (as control), supplemented with 1 M NaCl or Dh-242 supernatant at 0, 1 and 3 h. Fold increase is shown compared to glycerol level at time 0 for each strain.
Schematic graph of C. albicans MAPK signal transduction pathways analyzed in the present work. Transmembrane proteins and sensors are shown as rectangles, intermediate molecules are depicted as circles and MAP Kinase modules are ovals. The transcription factor Sko1 is shown as a white rectangle. Numbers indicate the inhibition zone (in mm) displayed by defective mutants in the specific elements in the presence of Dh-242 strain in YMB NaCl pH 4.4 at 30°C.
Susceptibility of different strains to caspofungin. The indicated strains were spotted on YPD supplemented or not with caspofungin (Casp) and incubated at 30 or 37°C for 24 h before scanning.
The indicated strains were spotted on YPD supplemented or not with Congo red and incubated at 30 or 37°C for 24 h before scanning.
References
- Alonso-Monge R., Carvaihlo S., Nombela C., Rial E., Pla J. (2009). The Hog1 MAP kinase controls respiratory metabolism in the fungal pathogen Candida albicans. Microbiology 155, 413–423. 10.1099/mic.0.023309-0 [DOI] [PubMed] [Google Scholar]
- Alonso-Monge R., Navarro-García F., Molero G., Díez-Orejas R., Gustin M., Pla J., et al. (1999). Role of the mitogen-activated protein kinase Hog1p in morphogenesis and virulence of Candida albicans. J. Bacteriol. 181, 3058–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Monge R., Román E., Arana D. M., Prieto D., Urrialde V., Nombela C., et al. (2010). The Sko1 protein represses the yeast-to-hypha transition and regulates the oxidative stress response in Candida albicans. Fungal Genet. Biol. 47, 587–601. 10.1016/j.fgb.2010.03.009 [DOI] [PubMed] [Google Scholar]
- Alonso-Monge R. A., Román E., Nombela C., Pla J. (2006). The MAP kinase signal transduction network in Candida albicans. Microbiology 152, 905–912. 10.1099/mic.0.28616-0 [DOI] [PubMed] [Google Scholar]
- Arana D. M., Alonso-Monge R., Du C., Calderone R., Pla J. (2007). Differential susceptibility of mitogen-activated protein kinase pathway mutants to oxidative-mediated killing by phagocytes in the fungal pathogen Candida albicans. Cell. Microbiol. 9, 1647–1659. 10.1111/j.1462-5822.2007.00898.x [DOI] [PubMed] [Google Scholar]
- Arana D. M., Nombela C., Alonso-Monge R., Pla J. (2005). The Pbs2 MAP kinase kinase is essential for the oxidative-stress response in the fungal pathogen Candida albicans. Microbiology 151, 1033–1049. 10.1099/mic.0.27723-0 [DOI] [PubMed] [Google Scholar]
- Banjara N., Nickerson K. W., Suhr M. J., Hallen-Adams H. E. (2016). Killer toxin from several food-derived Debaryomyces hansenii strains effective against pathogenic Candida yeasts. Int. J. Food Microbiol. 222, 23–29. 10.1016/j.ijfoodmicro.2016.01.016 [DOI] [PubMed] [Google Scholar]
- Borelli B. M., Ferreira E. G., Lacerda I. C. A., Franco G. R., Rosa C. A. (2006). Yeast populations associated with the artisanal cheese produced in the region of Serra da Canastra, Brazil. World J. Microb. Biot. 22, 1115–1119. 10.1007/s11274-006-9151-3 [DOI] [Google Scholar]
- Breinig F., Sendzik T., Eisfeld K., Schmitt M. J. (2006). Dissecting toxin immunity in virus-infected killer yeast uncovers an intrinsic strategy of self-protection. Proc. Natl. Acad. Soc. U.S.A. 103, 3810–3815. 10.1073/pnas.0510070103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breinig F., Tipper D. J., Schmitt M. J. (2002). Kre1p, the plasma membrane receptor for the yeast K1 viral toxin. Cell 108, 395–405. 10.1016/S0092-8674(02)00634-7 [DOI] [PubMed] [Google Scholar]
- Calera J. A., Herman D., Calderone R. (2000). Identification of YPD1, a gene of Candida albicans which encodes a two-component phosphohistidine intermediate protein. Yeast 16, 1053–1059. [DOI] [PubMed] [Google Scholar]
- Chang W. H., Liang S. H., Deng F. S., Lin C. H. (2016). The conserved dual phosphorylation sites of the Candida albicans Hog1 protein are crucial for white-opaque switching, mating, and pheromone-stimulated cell adhesion. Med. Mycol. 54, 628–640. 10.1093/mmy/myw015 [DOI] [PubMed] [Google Scholar]
- Cheetham J., MacCallum D. M., Doris K. S., da Silva Dantas A., Scorfield S., Odds F., et al. (2011). MAPKKK-independent regulation of the Hog1 stress-activated protein kinase in Candida albicans. J. Biol. Chem. 286, 42002–42016. 10.1074/jbc.M111.265231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosentino S., Fadda M. E., Deplano M., Mulargia A. F., Palmas F. (2001). Yeasts associated with Sardinian ewe's dairy products. Int. J. Food Microbiol. 69, 53–58. 10.1016/S0168-1605(01)00572-4 [DOI] [PubMed] [Google Scholar]
- Csank C., Schröppel K., Leberer E., Harcus D., Mohamed O., Meloche S., et al. (1998). Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect. Immun. 66, 2713–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day A. M., Herrero-de-Dios C. M., MacCallum D. M., Brown A. J. P., Quinn J. (2017). Stress-induced nuclear accumulation is dispensable for Hog1-dependent gene expression and virulence in a fungal pathogen. Sci. Rep. 7, 14340. 10.1038/s41598-017-14756-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnos-Ollivier M., Ragon M., Robert V., Raoux D., Gantier J. C., Dromer F. (2008). Debaryomyces hansenii (Candida famata), a rare human fungal pathogen often misidentified as Pichia guilliermondii (Candida guilliermondii). J. Clin. Microbiol. 46, 3237–3242. 10.1128/JCM.01451-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisman B., Alonso-Monge R., Román E., Arana D., Nombela C., Pla J. (2006). The Cek1 and Hog1 mitogen-activated protein kinases play complementary roles in cell wall biogenesis and chlamydospore formation in the fungal pathogen Candida albicans. Eukaryot Cell 5, 347–358. 10.1128/EC.5.2.347-358.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flegelová H., Chaloupka R., Novotná D., Malác J., Gásková D., Sigler K., et al. (2003). Changes in plasma membrane fluidity lower the sensitivity of S. cerevisiae to killer toxin K1. Folia Microbiol. 48, 761–766. 10.1007/BF02931510 [DOI] [PubMed] [Google Scholar]
- Fonzi W. A., Saporito-Irwin S. M., Chen J. Y., Sypherd P. (1993). Genetic basis for dimorphism and pathogenicity in Candida albicans, in Dimorphic Fungi in Biology and Medicine, eds vanden Bossche H., Odds F., Kerridge D. (New York, NY: Plenum Press; ), 37–50. [Google Scholar]
- García R., Rodríguez-Peña J. M., Bermejo C., Nombela C., Arroyo J. (2009). The high osmotic response and cell wall integrity pathways cooperate to regulate transcriptional responses to zymolyase-induced cell wall stress in Saccharomyces cerevisiae. J. Biol. Chem. 284, 10901–10911. 10.1074/jbc.M808693200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillum A. M., Tsay E. Y., Kirsch D. R. (1984). Isolation of the Candida albicans gene for orotidine-5'- phosphate decarboxylase by complementation of S. cerevisiae ura3, and, E. coli pyrF, mutations. Mol. Gen. Genet. 198, 179–182. 10.1007/BF00328721 [DOI] [PubMed] [Google Scholar]
- Hallen-Adams H. E., Kachman S. D., Kim J., Legge R. M., Martinez I. (2015). Fungi inhabiting the healthy human gastrointestinal tract: a diverse and dynamic community. Fungal. Ecol. 15, 9–17. 10.1016/j.funeco.2015.01.006 [DOI] [Google Scholar]
- Herrero de Dios C. (2013). Opy2, un nuevo elemento implicado en la señalización vía quinasas de tipo MAP en el hongo patógeno Candida albicans. Departmento de Microbiología II, Universidad Complutense de Madrid, Facultad de Farmacia. [Google Scholar]
- Herrero de Dios C., Román E., Diez C., Alonso-Monge R., Pla J. (2013). The transmembrane protein Opy2 mediates activation of the Cek1 MAP kinase in Candida albicans. Fungal Genet. Biol. 50, 21–32. 10.1016/j.fgb.2012.11.001 [DOI] [PubMed] [Google Scholar]
- Höfs S., Mogavero S., Hube B. (2016). Interaction of Candida albicans with host cells: virulence factors, host defense, escape strategies, and the microbiota. J. Microbiol. 54, 149–169. 10.1007/s12275-016-5514-0 [DOI] [PubMed] [Google Scholar]
- Jablonowski D., Schaffrath R. (2007). Zymocin, a composite chitinase and tRNase killer toxin from yeast. Biochem. Soc. Trans. 35, 1533–1537. 10.1042/BST0351533 [DOI] [PubMed] [Google Scholar]
- Köhler G. A., White T. C., Agabian N. (1997). Overexpression of a cloned IMP dehydrogenase gene of Candida albicans confers resistance to the specific inhibitor mycophenolic acid. J. Bacteriol. 179, 2331–2338. 10.1128/jb.179.7.2331-2338.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. K., Maccallum D. M., Jacobsen M. D., Walker L. A., Odds F. C., Gow N. A., et al. (2012). Elevated cell wall chitin in Candida albicans confers echinocandin resistance in vivo. Antimicrob. Agents Chemother. 56, 208–217. 10.1128/AAC.00683-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G. L., Chi Z., Wang G. Y., Wang Z. P., Li Y., Chi Z. M. (2015). Yeast killer toxins, molecular mechanisms of their action and their applications. Crit. Rev. Biotechnol. 35, 222–234. 10.3109/07388551.2013.833582 [DOI] [PubMed] [Google Scholar]
- Lu M., Li T., Wan J., Li X., Yuan L., Sun S. (2017). Antifungal effects of phytocompounds on Candida species alone and in combination with fluconazole. Int. J. Antimicrob. Agents 49, 125–136. 10.1016/j.ijantimicag.2016.10.021 [DOI] [PubMed] [Google Scholar]
- Magliani W., Conti S., de Bernardis F., Gerloni M., Bertolotti D., Mozzoni P., et al. (1997). Therapeutic potential of antiidiotypic single chain antibodies with yeast killer toxin activity. Nat. Biotechnol. 15, 155–158. 10.1038/nbt0297-155 [DOI] [PubMed] [Google Scholar]
- Marquina D., Santos A., Peinado J. M. (2002). Biology of killer yeasts. Int. Microbiol. 5, 65–71. 10.1007/s10123-002-0066-z [DOI] [PubMed] [Google Scholar]
- Martin H., Rodriguez-Pachon J. M., Ruiz C., Nombela C., Molina M. (2000). Regulatory mechanisms for modulation of signaling through the cell integrity Slt2-mediated pathway in Saccharomyces cerevisiae. J. Biol. Chem. 275, 1511–1519. [DOI] [PubMed] [Google Scholar]
- Mayer F. L., Wilson D., Hube B. (2013). Candida albicans pathogenicity mechanisms. Virulence 4, 119–128. 10.4161/viru.22913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-García F., Sánchez M., Pla J., Nombela C. (1995). Functional characterization of the MKC1 gene of Candida albicans, which encodes a mitogen-activated protein kinase homolog related to cell integrity. Mol. Cell. Biol. 15, 2197–2206. 10.1128/MCB.15.4.2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negredo A., Monteoliva L., Gil C., Pla J., Nombela C. (1997). Cloning, analysis and one-step disruption of the ARG5,6, gene of Candida albicans. Microbiology 143, 297–302. 10.1099/00221287-143-2-297 [DOI] [PubMed] [Google Scholar]
- Pfaller M. A., Castanheira M. (2016). Nosocomial candidiasis: antifungal stewardship and the importance of rapid diagnosis. Med. Mycol. 54, 1–22. 10.1093/mmy/myv076 [DOI] [PubMed] [Google Scholar]
- Pfaller M. A., Diekema D. J. (2007). Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20, 133–163. 10.1128/CMR.00029-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller M. A., Diekema D. J. (2010). Epidemiology of invasive mycoses in North America. Crit. Rev. Microbiol. 36, 1–53. 10.3109/10408410903241444 [DOI] [PubMed] [Google Scholar]
- Polonelli L., Magliani W., Ciociola T., Giovati L., Conti S. (2011). From Pichia anomala killer toxin through killer antibodies to killer peptides for a comprehensive anti-infective strategy. Antonie Van Leeuwenhoek 99, 35–41. 10.1007/s10482-010-9496-3 [DOI] [PubMed] [Google Scholar]
- Poulain D. (2015). Candida albicans, plasticity and pathogenesis. Crit. Rev. Microbiol. 41, 208–217. 10.3109/1040841X.2013.813904 [DOI] [PubMed] [Google Scholar]
- Prieto D., Román E., Correia I., Pla J. (2014). The HOG pathway is critical for the colonization of the mouse gastrointestinal tract by Candida albicans. PLoS ONE 9:e87128. 10.1371/journal.pone.0087128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Cousiño N., Maqueda M., Ambrona J., Zamora E., Esteban R., Ramirez M. (2011). A new wine Saccharomyces cerevisiae killer toxin (Klus), encoded by a double-stranded rna virus, with broad antifungal activity is evolutionarily related to a chromosomal host gene. Appl. Environ. Microbiol. 77, 1822–1832. 10.1128/AEM.02501-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Román E., Alonso-Monge R., Miranda A., Pla J. (2015). The Mkk2 MAPKK regulates cell wall biogenesis in cooperation with the Cek1-pathway in Candida albicans. PLoS One 10:e0133476. 10.1371/journal.pone.0133476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Román E., Arana D. M., Nombela C., Alonso-Monge R., Pla J. (2007). MAP kinase pathways as regulators of fungal virulence. Trends Microbiol. 15, 181–190. 10.1016/j.tim.2007.02.001 [DOI] [PubMed] [Google Scholar]
- Román E., Cottier F., Ernst J. F., Pla J. (2009). Msb2 signaling mucin controls activation of Cek1 mitogen-activated protein kinase in Candida albicans. Eukaryotic Cell 8, 1235–1249. 10.1128/EC.00081-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Román E., Nombela C., Pla J. (2005). The Sho1 adaptor protein links oxidative stress to morphogenesis and cell wall biosynthesis in the fungal pathogen Candida albicans. Mol. Cell. Biol. 25, 10611–10627. 10.1128/MCB.25.23.10611-10627.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Román E., Prieto D., Martin R., Correia I., Mesa Arango A. C., Alonso-Monge R., et al. (2016). Role of catalase overproduction in drug resistance and virulence in Candida albicans. Future Microbiol. 11, 1211–1215. 10.2217/fmb-2016-0067 [DOI] [PubMed] [Google Scholar]
- San José C., Alonso-Monge R., Pérez-Díaz R. M., Pla J., Nombela C. (1996). The mitogen-activated protein kinase homolog HOG1 gene controls glycerol accumulation in the pathogenic fungus Candida albicans. J. Bacteriol. 178, 5850–5852. 10.1128/jb.178.19.5850-5852.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos A. M., Del Mar Alvarez Mauro M. S., Abrusci C., Marquina D. (2005). The transcriptional response of Saccharomyces cerevisiae to Pichia membranifaciens killer toxin. J. Biol. Chem. 280, 41881–41892. 10.1074/jbc.M507014200 [DOI] [PubMed] [Google Scholar]
- Santos A., Alonso A., Belda I., Marquina D. (2013). Cell cycle arrest and apoptosis, two alternative mechanisms for PMKT2 killer activity. Fungal. Genet. Biol. 50, 44–54. 10.1016/j.fgb.2012.10.006 [DOI] [PubMed] [Google Scholar]
- Santos A., Marquina D. (2004). Killer toxin of Pichia membranifaciens and its possible use as a biocontrol agent against grey mould disease of grapevine. Microbiology 150, 2527–2534. 10.1099/mic.0.27071-0 [DOI] [PubMed] [Google Scholar]
- Santos A., Marquina D. (2011). The transcriptional response of Saccharomyces cerevisiae to proapoptotic concentrations of Pichia membranifaciens killer toxin. Fungal Genet. Biol. 48, 979–989. 10.1016/j.fgb.2011.07.002 [DOI] [PubMed] [Google Scholar]
- Saxena A., Sitaraman R. (2016). Osmoregulation in Saccharomyces cerevisiae via mechanisms other than the high-osmolarity glycerol pathway. Microbiology 162, 1511–1526. 10.1099/mic.0.000360 [DOI] [PubMed] [Google Scholar]
- Schmitt M. J., Breinig F. (2002). The viral killer system in yeast: from molecular biology to application. FEMS Microbiol. Rev. 26, 257–276. 10.1111/j.1574-6976.2002.tb00614.x [DOI] [PubMed] [Google Scholar]
- Schmitt M. J., Klavehn P., Wang J., Schönig I., Tipper D. J. (1996). Cell cycle studies on the mode of action of yeast K28 killer toxin. Microbiology 142 (Pt 9), 2655–2662. 10.1099/00221287-142-9-2655 [DOI] [PubMed] [Google Scholar]
- Sharifian H., Lampert F., Stojanovski K., Regot S., Vaga S., Buser R., et al. (2015). Parallel feedback loops control the basal activity of the HOG MAPK signaling cascade. Integr. Biol. 7, 412–422. 10.1039/C4IB00299G [DOI] [PubMed] [Google Scholar]
- Takasuka T., Komiyama T., Furuichi Y., Watanabe T. (1995). Cell wall synthesis specific cytocidal effect of Hansenula mrakii toxin-1 on Saccharomyces cerevisiae. Cell. Mol. Biol. Res. 41, 575–581. [PubMed] [Google Scholar]
- Tang S. X., Moyes D. L., Richardson J. P., Blagojevic M., Naglik J. R. (2016). Epithelial discrimination of commensal and pathogenic Candida albicans. Oral Dis. 22(Suppl. 1), 114–119. 10.1111/odi.12395 [DOI] [PubMed] [Google Scholar]
- Vasdinyei R., Deák T. (2003). Characterization of yeast isolates originating from Hungarian dairy products using traditional and molecular identification techniques. Int. J. Food Microbiol. 86, 123–130. 10.1016/S0168-1605(03)00251-4 [DOI] [PubMed] [Google Scholar]
- Wurgler-Murphy S. M., Maeda T., Witten E. A., Saito H. (1997). Regulation of the, Saccharomyces cerevisiae HOG1, mitogen-activated protein kinase by the, PTP2, and, PTP3, protein tyrosine phosphatases. Mol. Cell. Biol. 17, 1289–1297. 10.1128/MCB.17.3.1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaakov G., Bell M., Hohmann S., Engelberg D. (2003). Combination of two activating mutations in one HOG1 gene forms hyperactive enzymes that induce growth arrest. Mol. Cell. Biol. 23, 4826–4840. 10.1128/MCB.23.14.4826-4840.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotnik H., Fernandez M. P., Bowers B., Cabib E. (1984). Saccharomyces cerevisiae mannoproteins form an external cell wall layer that determines wall porosity. J. Bacteriol. 159, 1018–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dh Kiler Toxin activity against C. albicans strains: The killer activity was analyzed on YMB plates supplemented with 3% NaCl pH 4.4 against SC5314 and the hog1 mutant strain under different environmental conditions.
Quantification of intracellular glycerol from CAF2 and hog1 mutant exposed to YMB plates plus 3% NaCl pH 4.4 (as control), supplemented with 1 M NaCl or Dh-242 supernatant at 0, 1 and 3 h. Fold increase is shown compared to glycerol level at time 0 for each strain.
Schematic graph of C. albicans MAPK signal transduction pathways analyzed in the present work. Transmembrane proteins and sensors are shown as rectangles, intermediate molecules are depicted as circles and MAP Kinase modules are ovals. The transcription factor Sko1 is shown as a white rectangle. Numbers indicate the inhibition zone (in mm) displayed by defective mutants in the specific elements in the presence of Dh-242 strain in YMB NaCl pH 4.4 at 30°C.
Susceptibility of different strains to caspofungin. The indicated strains were spotted on YPD supplemented or not with caspofungin (Casp) and incubated at 30 or 37°C for 24 h before scanning.
The indicated strains were spotted on YPD supplemented or not with Congo red and incubated at 30 or 37°C for 24 h before scanning.