Abstract.
Error estimation in nonlinear medical image registration is a nontrivial problem that is important for validation of registration methods. We propose a supervised method for estimation of registration errors in nonlinear registration of three-dimensional (3-D) images. The method is based on a 3-D convolutional neural network that learns to estimate registration errors from a pair of image patches. By applying the network to patches centered around every voxel, we construct registration error maps. The network is trained using a set of representative images that have been synthetically transformed to construct a set of image pairs with known deformations. The method is evaluated on deformable registrations of inhale–exhale pairs of thoracic CT scans. Using ground truth target registration errors on manually annotated landmarks, we evaluate the method’s ability to estimate local registration errors. Estimation of full domain error maps is evaluated using a gold standard approach. The two evaluation approaches show that we can train the network to robustly estimate registration errors in a predetermined range, with subvoxel accuracy. We achieved a root-mean-square deviation of 0.51 mm from gold standard registration errors and of 0.66 mm from ground truth landmark registration errors.
Keywords: registration error, image registration, supervised learning, convolutional neural networks
1. Introduction
The validation of nonlinear medical image registration is a complex problem. As most registration methods do not provide an indication of accuracy, subsequent processing is required to assess the registration quality. Registration error estimation is necessary to evaluate registration methods and can be used to compare the performances of multiple registration methods for a certain application quantitatively. It is also of importance for error estimation in clinical image-based techniques that (partly) rely on image registration, such as computer-aided diagnosis pipelines, radiation treatment planning, or image-guided interventions. For all these applications, it is valuable to quantify the registration error locally.
Image registration is often validated using overlap measures (e.g., Dice score of tissue overlap), metrics that measure the similarity of the registered images (e.g., normalized correlation coefficient of the intensities), or target registration errors (TREs) measured on corresponding points in the registered images. Overlap and similarity measures fail to measure the registration error directly, and their measurements do not necessarily correlate with the registration error.1 Furthermore, if the tissue segmentations used to calculate the overlap cover a large volume, the local error within that volume cannot be estimated. The preferred way to determine registration accuracy, therefore, is determining TREs on corresponding points in the registered images. These points are commonly relevant anatomical landmarks annotated by experts. For deformable registration problems, these landmarks should cover the entire region of interest to be accurate descriptors of the local registration error. Unfortunately, annotation of dense sets of corresponding landmarks, suitable for validation of deformable registration, is a laborious effort sensitive to inconsistencies by the annotators. A method that can estimate local registration errors is, therefore, very useful in the evaluation of deformable registration algorithms.
Recent work on validation of image registration avoids the need for manual annotations of landmarks, while still aiming to estimate registration errors. An example is the use of registration consistency in sets of triplets of images. Datteri et al.2 used triplet registration consistency to solve a linear system of registration quality metrics to construct error maps for atlas construction of MRI brain data. They show that the registration quality metric they estimate correlates well with the TRE of two landmarks in the brain. Other recent efforts have explored the use of machine learning to estimate the registration error from image features. Muenzing et al. used a machine learning approach to classify the registration error for an arbitrary voxel in thoracic CT images into one of three classes for good, poor, and wrong alignments. This approach uses features from the images, as well as the Jacobian of the deformation field, and reached a classification accuracy of 90%.3 This method was incorporated in a registration boosting algorithm for the combination of multiple registration methods.4 The authors later added to this boosting algorithm the ability to perform regression of the registration error, which they validated also on thoracic CT data. They reported that the estimation error of this regression approach was relatively large and required further development.5 More recently, Sokooti et al.6 proposed a random forest regression-based method for registration error estimation in thoracic CT scans. They compared their method with Muenzing et al. but found that their method suffered from an unbalanced training set, with few samples of poor registration results. A problem with both methods is that they require ground truth registrations of images, which makes it hard to train on new registration problems with different modalities or anatomy.
Registration error estimation is closely related to confidence estimation of registration, where instead of direct estimates of the error, estimates of the uncertainty of the registration result are made. Examples include the use of bootstrap resampling,7 Bayesian methods,8,9 and inspection of a cost space based on image features.10
In this paper, we present a supervised approach that makes estimates of the registration error for the full image domain, giving a full assessment of alignment errors in the registered images. The method does not require any information on the deformation field but estimates the error directly from two registered images. Our approach is based on a sliding window convolutional neural network (CNN). CNNs have been widely used in medical image analysis problems, primarily for segmentation and detection tasks,11 but recently also for image registration,12–15 for steering of the optimization by learning multimodal motion predictors,16 and for learning of multimodal similarity metrics.17,18 Previous work has shown that using machine learning for error estimation in medical images is a viable approach. The fact that CNNs are starting to be used for image registration and have shown promising results suggests that the use of CNNs for error estimation in image registration is feasible.
The registration error estimating CNN is trained without the need for ground truth deformation fields, requiring only a set of representative images. From these images, a training set consisting of synthetically deformed images is constructed. We validate the registration error estimates on deformable image registrations of public data sets of thoracic CTs and compare these to ground truth registration errors on large sets of landmarks. To assess the quality of the algorithm’s error maps for the full image domain, we use gold standard registration error maps.
This paper is an extension of a previous paper, in which we proposed the current method as proof of concept in 2-D image registration for digital subtraction angiography images.19
2. Methods
The method produces registration error estimates for every position in the image, resulting in a registration error map for the full image domain. Let and be two images that are to be registered. is the target image that is held fixed, while the moving image is transformed to align with . Both images are defined on their own -dimensional spatial domain, and , respectively. By registering to , we find an approximation of the transformation . The transformation maps points in the fixed image’s domain to corresponding points in the moving image’s domain, such that the image is aligned to . To evaluate the quality of the estimate , the TRE can be computed, which measures the displacement from the true position of a registered point, i.e.,
| (1) |
The TRE can be computed for a pair of manually annotated corresponding landmarks and , since for these points approaches , subject to the quality of annotation. In this paper, the goal is to fully automatically estimate the TRE for all voxels in the image, which results in a map of registration errors , defined for points in the fixed image’s domain.
The method we propose estimates the TRE for any voxel from a pair of small image patches centered around the voxel. Each pair consists of a patch from the fixed image and a patch from the registered moving image . In the current implementation, both patches have a size of voxels. We train a CNN to estimate displacement magnitude from this pair of image patches. This displacement magnitude is estimated in voxel distances. To enable error estimates in millimeters and to account for different voxel sizes, we first resample the images to (Sec. 2.1). The network is trained on pairs of synthetically deformed images (Sec. 2.2). We describe the network’s architecture and the training process in Secs. 2.3 and 2.4.
2.1. Image Resampling
The registration error is expressed as a physical distance, measured in millimeters. Because the pairs of image patches do not provide the convolutional network with any means to estimate the voxel size in millimeters, we resample the images such that they have voxels. The images used in the training set are resampled in the same way, such that the error is estimated from cubic regions in the image. In all cases, resampling is performed using fourth-order B-spline interpolation.
2.2. Training Set Construction
The training set consists of pairs of synthetically deformed pulmonary CT images that simulate a fixed image and a registered image . The training set is constructed using the assumption that the registration errors for lung registration in pulmonary CTs are relatively small, i.e., in the order of a few millimeters, which is motivated by the range of registration errors in state-of-the-art lung registration algorithms.20 It is furthermore assumed that registration errors have a high spatial frequency, i.e., the value of the error can fluctuate a lot from position to position.
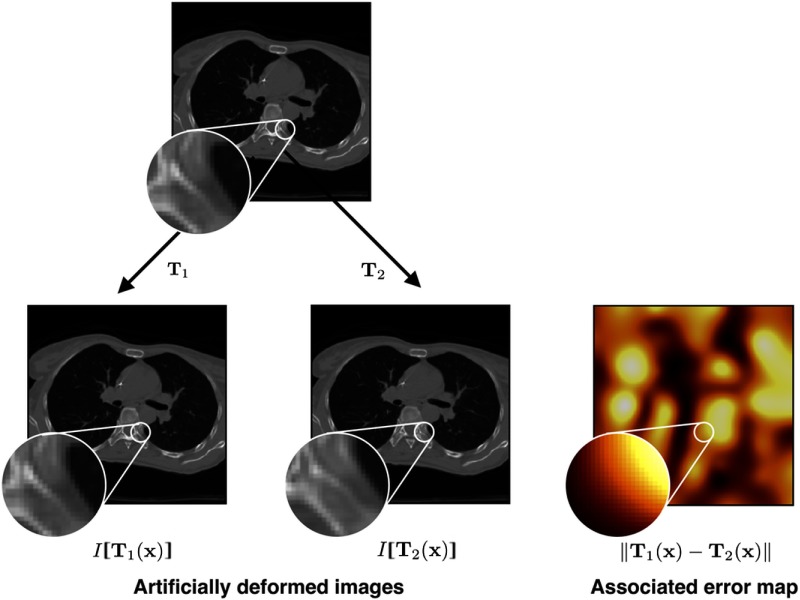
We simulate registration errors as differences between two transformations, denoted as and . Both transformations are applied to the same image , resulting in images and . These two images simulate two registered images with error map (Fig. 1).
Fig. 1.
From one image in the training set, two deformed versions are constructed using random TPS transformations.
The synthesized deformations and are thin plate spline (TPS) transformations, defined by displacements on a grid of equidistant control points. The control point displacement vectors are sampled from three uniform distributions in the [0, 2] mm range. The resultant deformation from , therefore, has a range of [0, 4] mm. The TPS that is fitted through the displacements of the control points is defined as
| (2) |
where is an affine matrix, is a translation vector, is the kernel function , and are the spline coefficients. The parameters , , and are computed from the displacements , using the implementation in Ref. 21. To increase the number of training pairs, this process for creating image pairs and associated error maps is repeated 10 times for every image. Using multiple transformations, we obtain a natural augmentation of the data. Each pair of images is divided into patches. The corresponding true registration error for a pair of patches around voxel can be found in the simulated error map . In addition to the augmentation provided by the spatial transformations of the images, we also perform augmentation of the image intensities in the patches. We scale the patches with a random value from the normal distribution and add a random offset from the normal distribution .
2.3. Network Architecture
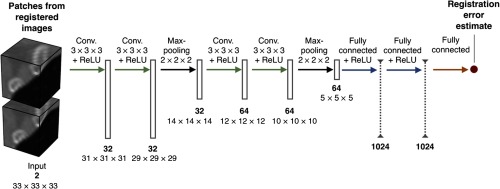
The error is estimated by a three-dimensional (3-D) CNN (Fig. 2). The network has four 3-D convolutional layers that only differ in the number of feature maps they output: 32, 32, 64, and 64 feature maps, respectively. These layers learn kernels and perform convolution without zero padding. After every two convolutional layers, the feature maps are downsampled by max-pooling layers with stride 2. The second pooling layer is followed by three fully connected layers with 1024 units and a fully connected layer with a single scalar output. All convolutional layers and the first two fully connected layers are followed by a rectified linear unit (ReLU) activation function.
Fig. 2.
The CNN architecture is comprised of two sequences of two convolutional layers and a pooling layer and followed by three fully connected layers that compute the error estimate.
2.4. Training
The network is trained by minimizing the norm of the difference between the network’s estimate of the registration error and the actual registration error for a pair of patches centered around voxel
| (3) |
The loss is minimized using the stochastic gradient descent optimizer. We used mini-batches of 32 pairs of patches and a learning rate that decreases every 10,000 iterations with a factor 10, starting at 0.001. To further prevent overfitting, the first two fully connected layers were trained with Dropout22 with a dropout probability of . Additionally, batch normalization is applied to all convolutional and fully connected layers.23
3. Experiments
To test our approach, the method has been applied to estimate registration errors in inhale-to-exhale registration of thoracic CTs. The network was trained on similar images and synthetic deformations as outlined in Sec. 2.2. The aim was to estimate registration errors up to 4 mm in affine registrations and B-spline registrations with varying grid spacing. To validate the estimated errors, we used two approaches. In the first, we used a gold standard approach that generates registration errors that are compared to the network’s estimate to validate the method’s ability to estimate error maps for the full image domain. In the second approach, we compared the results to ground truth TREs on manually annotated landmarks.
3.1. Materials
The network was trained and the full method was evaluated on publicly available thoracic CT scans. We used data from four data sets: the DIRLAB 4DCT set, the DIRLAB COPDgene set, the POPI-model data set, and the CREATIS data set. The name of each image in the rest of this paper is derived from the file name in the downloaded material. All images come with a set of expert-annotated corresponding points for landmarks inside the lungs.
3.1.1. Data set 1: DIRLAB 4DCT data
The DIRLAB 4DCT data set contains data from four-dimensional (4-D) CT scan sequences of five patients free of pulmonary disease24 and five patients treated for thoracic malignancies.25 For each patient, the 4-D CT consists of five 3-D CT images covering the respiratory cycle from end inhalation to end exhalation. The 3-D CTs were cropped and subsampled to voxels in-plane, with voxel dimensions ranging from to .24,25 The images consist of between 94 and 136 slices with a 2.5-mm slice thickness. For each 4-D CT sequence, the extreme inhale and exhale phases were made available from the DIRLAB website. For each pair of inhale and exhale scans, a set of 300 corresponding landmarks for both images was provided. The landmarks were annotated by a single observer using a semiautomatic tool. Data on interobserver reproducibility have been made available in the form of average errors per image, which range from to .24,25
3.1.2. Data set 2: DIRLAB COPDgene study data
The COPDgene set is an extension of the DIRLAB 4DCT data set and contains pairs of inspiratory and expiratory breath-hold CT scans of 10 subjects reconstructed to a in-plane resolution and a 2.5-mm slice thickness, with in-plane voxel dimensions between and .26 For every pair of inspiratory and expiratory images, 300 corresponding landmark pairs are provided, which have been annotated in a similar way as for the 4DCT data. Average reproducibility errors of landmark annotations per image range from to .
3.1.3. Data set 3: POPI model
The POPI-model data set is a public 4-D CT scan consisting of 10 frames covering the full respiratory cycle, with 41 annotated corresponding points in every frame. The data were published together with deformation fields that model the breathing motion of the thorax. The 3-D images are made up of 90 to 110 slices with a 3-mm slice thickness, a in-plane voxel size, and each slice having a resolution.27 The landmarks were annotated by multiple experts using a procedure developed for a previous study, in which interobserver variability was on average .28
3.1.4. Data set 4: CREATIS data set
The POPI-model data set was later extended with six more 4-D CTs, each with a voxel size of , and 100 anatomical landmarks for the first and sixth frame, which correspond to end of expiration and end of inspiration, respectively. The landmarks were annotated by two observers with a mean interobserver error of .29 These six additional sets were published as the CREATIS data set.
3.2. Training and Test Sets
The ten odd-numbered images from the DIRLAB 4DCT and the COPDgene data sets were used to train the network. As described in Sec. 2.2, random TPS transformations are applied 10 times for every image. From these transformed images, 115,860 patches were selected that make up the training set. Although the displacements on the TPS grid were sampled from uniform distributions, the errors in this set are not uniformly distributed due to the TPS interpolation. To balance the training set, a uniform distribution was sampled from the original training set. The network was trained and tested on a system with a 3.50-GHz hexacore Intel Core i7-5930K CPU, 64 GB of memory, and an Nvidia Titan X graphics card with 12 GB of GPU memory. The neural network was implemented in Lasagne30 and Theano31 and used the Nvidia CUDA and CUDNN toolboxes. We trained the network for 200,000 iterations. The even-numbered image pairs from the DIRLAB 4DCT and COPDgene data sets, as well as the POPI and CREATIS data sets, were registered and used for evaluation of the trained network.
3.3. Registration
Both the gold standard approach and the ground truth approach require realistic registrations of the inhale–exhale pairs. Pairs of corresponding images were registered using an algorithm based on a registration algorithm that ranked third in the EMPIRE thoracic CT registration challenge.20,32 The registration algorithm performs registration in three stages. The first stage performs affine registration to get a coarse alignment. The second stage uses a coarse B-spline transformation, and the third stage uses a fine B-spline transformation. In each stage, the normalized cross-correlation similarity metric is optimized using adaptive stochastic gradient descent. A multiresolution strategy is used with four resolutions in each stage. For stages 2 and 3, the grid spacing of the B-splines was decreased in each step as well. The last stage uses a lung mask to improve registration inside the lungs. Lung masks for the images were made by segmenting all voxels with Hounsfield units below , which results in a segmentation of lung tissue and the exterior of the patient. After setting the largest morphological component, i.e., the exterior of the patient, to zero, we are left with a rough segmentation of the lungs. For further details on the registration algorithm, see Ref. 32. The intermediate as well as the final stage’s results are used for validation.
3.4. Gold Standard Evaluation Approach
In this case, we simulate a registration error map as the -norm of the difference of two transformations and , i.e., . Hence, the network will make an estimate for from and . We used the net transformation of the full registration sequences (affine, coarse B-spline, and fine B-spline) as and the net transformation after the first two stages (affine and coarse B-spline) as . White Gaussian noise was added to both images to simulate noise difference between the two acquisitions. This noise simulates a signal-to-noise ratio (SNR) of 10, where the SNR is defined as the ratio between the standard deviations of the voxels inside the lungs and the noise: . The network was run on the full images using the shift-and-stitch approach.33 The estimated errors were compared to .
3.5. Ground Truth Evaluation Approach
The ground truth validation approach uses the corresponding landmarks provided with the data sets. These landmarks are used to compute target registrations error as Euclidean distance between points in the fixed image and the registered moving image. For every landmark and the corresponding landmark , we compute the . These values are compared to the error estimate at position .
To show the performance of our algorithm on different kinds of transformations, we let the algorithm estimate these registration errors for the final results of all three stages, i.e., for the affine registration, the sequence of the affine and coarse B-spline registration, and the full sequence of three stages.
4. Results
4.1. Validation Set: Registration Results
All images from the three public data sets were registered using the method detailed in Sec. 3.3. The TREs of all registrations were computed using the landmark sets supplied with the images. Average TREs are shown in Table 2. On average, the TREs for images in the COPDgene set were larger than those in the DIRLAB 4DCT and CREATIS sets. This corresponds to the much larger deformations prior to registration for the COPDgene images compared with the DIRLAB 4DCT set (displacements of for COPDgene versus for DIRLAB 4DCT and CREATIS).
Table 2.
Performance statistics for error estimation at landmarks compared to ground truth TREs for the final stage of registration (i.e., complete sequences of affine, coarse B-spline, and fine B-spline). RMSD and normalized RMSD are given for landmarks with ground truth TRE below 4 mm. Landmark properties are given for all landmarks in the set, and averages are followed by standard deviations in parentheses.
| Error estimation results | Landmark properties | |||||
|---|---|---|---|---|---|---|
| Data set | Image pair | RMSD (mm) | Normalized RMSD (%) | Number of landmarks with TRE | Registration TRE (mm) | Annotation error (mm)a |
| DIRLAB | case02 | 0.55 | 21 | 300 | 0.94 (0.52) | 0.70 (0.99) |
| case04 | 0.76 | 21 | 298 | 1.42 (1.00) | 1.13 (1.27) | |
| case06 | 0.72 | 19 | 296 | 1.17 (0.88) | 0.97 (1.38) | |
| case08 | 0.58 | 21 | 296 | 1.12 (0.89) | 1.03 (2.19) | |
| case10 | 0.58 | 21 | 298 | 1.02 (0.72) | 0.86 (1.45) | |
| COPDgene | copd02 | 0.84 | 23 | 256 | 2.67 (3.82) | 1.06 (1.51) |
| copd04 | 0.79 | 21 | 266 | 2.21 (3.45) | 0.71 (0.96) | |
| copd06 | 0.58 | 16 | 220 | 4.08 (6.04) | 1.06 (2.38) | |
| copd08 | 0.66 | 18 | 279 | 1.55 (2.49) | 0.96 (3.07) | |
| copd10 | 0.75 | 19 | 260 | 2.23 (3.10) | 0.87 (1.65) | |
| POPI | popi | 0.65 | 18 | 39 | 1.38 (1.24) | 2.30 (1.30) |
| CREATIS | creatis01 | 0.57 | 21 | 99 | 1.06 (0.58) | 0.50 (0.90) |
| creatis02 | 0.56 | 17 | 97 | 1.15 (0.87) | 0.50 (0.90) | |
| creatis03 | 0.40 | 19 | 99 | 0.77 (0.42) | 0.50 (0.90) | |
| creatis04 | 0.46 | 17 | 98 | 0.74 (0.87) | 0.50 (0.90) | |
| creatis05 | 0.47 | 27 | 104 | 0.95 (0.80) | 0.50 (0.90) | |
| creatis06 | 0.58 | 22 | 112 | 0.89 (0.50) | 0.50 (0.90) | |
4.2. Gold Standard Evaluation Approach
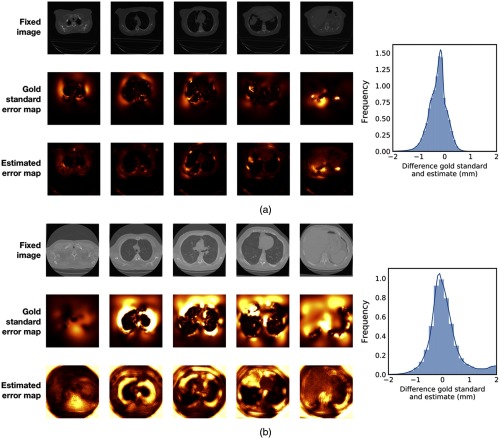
Qualitative comparisons between gold standard error maps and estimated error maps by the convolutional network for two examples are shown in Fig. 3. These examples show that the network estimates error maps that resemble the gold standard error maps, which is reflected in the histograms of the differences between the gold standard and estimated errors. The histograms in Fig. 3 show that the distribution of these differences is close to symmetric. No clear over- or underestimation of the error was found. The error maps for the examples in Fig. 3 show that while the estimates mimic the gold standard error map, the actual gold standard has a higher spatial frequency. More abrupt fluctuations in the error map are missed by the network.
Fig. 3.
Comparisons of a gold standard error map and error map estimate for six axial slices of (a) DIRLAB image case08 and (b) COPDgene image copd06 for which the error maps have smallest and largest RMSD values (see Table 1). The registrations’ fixed images are shown as a reference.
The root-mean-square deviation (RMSD) between the gold standard and estimated error maps over all voxels inside the lung masks that were used for registration (Sec. 3.3) in the test image pairs was 0.51 mm. The normalized RMSD, computed by dividing the RMSD by the range of all actual registration error maps combined, has a value of 6.85%. Table 1 also shows the normalized RMSD values per image, where the RMSD was normalized by dividing by the range of the image’s error map. Computation of the gold standard error maps took min on average, using a shift-and-stitch technique.
Table 1.
Gold standard statistics. For every image in the test set, the RMSD and normalized RMSD between the gold standard error map and estimated error map for all voxels inside the lung mask are reported.
| Data set | Image pair | RMSD (mm) | Normalized RMSD (%) |
|---|---|---|---|
| DIRLAB | case02 | 0.44 | 9.8 |
| case04 | 0.58 | 12.4 | |
| case06 | 0.43 | 6.6 | |
| case08 | 0.32 | 5.4 | |
| case10 | 0.38 | 3.3 | |
| COPDgene | copd02 | 0.29 | 6.8 |
| copd04 | 0.30 | 4.8 | |
| copd06 | 0.35 | 8.0 | |
| copd08 | 0.37 | 5.0 | |
| copd10 | 0.25 | 7.9 | |
| POPI | popi | 0.25 | 6.2 |
| CREATIS | creatis01 | 0.96 | 7.7 |
| creatis02 | 0.73 | 6.7 | |
| creatis03 | 0.90 | 6.4 | |
| creatis04 | 0.69 | 8.1 | |
| creatis05 | 0.89 | 6.5 | |
| creatis06 | 0.52 | 4.9 | |
| Average | 0.51 | 6.85 | |
4.3. Ground Truth Evaluation Approach
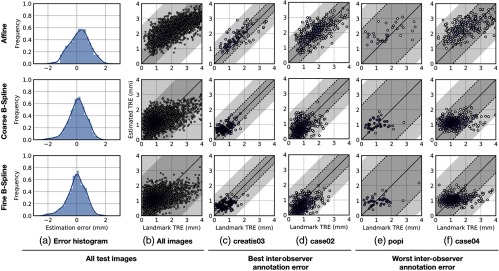
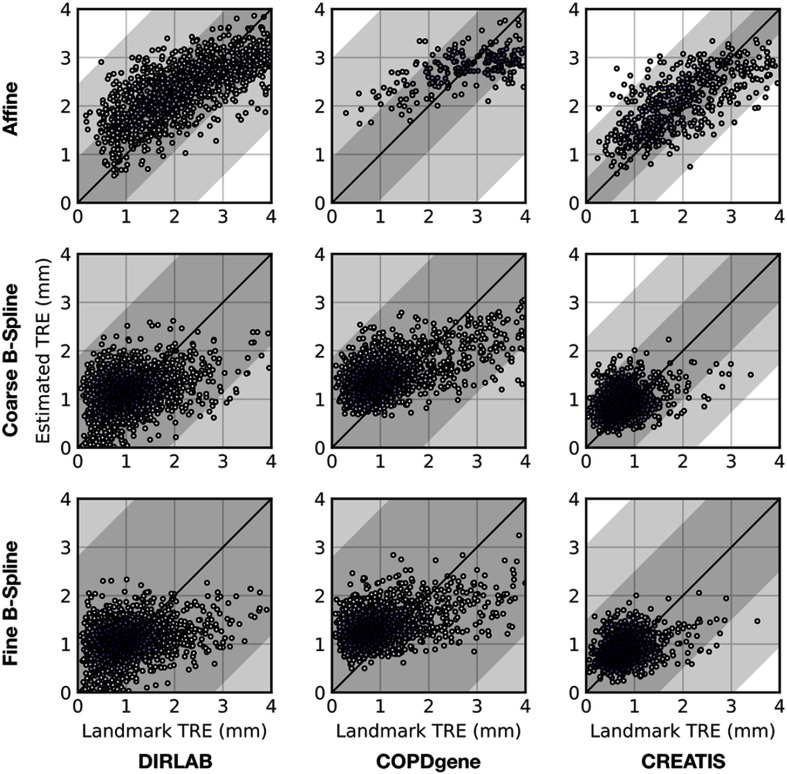
We compare the network’s registration error estimates evaluated at the landmarks to TREs for the landmarks. In all cases, we evaluate the performance for landmarks inside the lung masks used in Sec. 3.3. Figure 4 shows correlation plots and a histogram of differences between estimated and ground truth registration errors. These plots are shown for all three stages of registration and contain the errors for landmarks with a TRE below 4 mm, i.e., the range of errors in the training set. Performance statistics for error estimation of the full registration algorithm (three stages) are shown in Table 2. RMSD values between the TRE computed on the landmarks and the corresponding estimate in the error map show that estimation error in terms of RMSD and normalized RMSD is similar for all three stages of registration (Table 3). The same can be concluded qualitatively from the correlation plots in Fig. 4 and the correlation plots per data set in Fig. 5. Note that the RMSD and normalized RMSD values in Tables 2 and 3 are computed only on the landmarks that have a TRE below 4 mm, as this corresponds to the range present in the training set. To show the effect of intensity augmentation, we computed the bias of the estimated errors for the network trained with and without the intensity augmentations described in Sec. 2.2 in Table 4. The bias is computed by subtracting the ground truth errors from the estimated errors and averaging the results. When using the intensity augmentations, the results are much less biased toward larger errors.
Fig. 4.
(a) Histogram of all differences between registration error estimates and ground truth. (b) Estimated TRE versus true TRE for every landmark in the test set. Darker colors indicate higher density of points. (c), (d), (e), and (f) Show similar correlation plots for specific registration pairs with good and poor interobserver landmark annotation errors. In these plots, the black dotted lines show the average annotation error on either side of the identity, with the gray areas indicating the average annotation error plus the standard deviation.
Table 3.
Performance statistics for error estimation at all landmarks in the test set compared to ground truth TREs per registration stage. Results for landmarks with TREs below 4 mm.
| Registration stage | Number of landmarks | RMSD (mm) | Normalized RMSD |
|---|---|---|---|
| Affine | 1863 | 0.70 | 0.18 |
| Coarse B-spline | 3344 | 0.65 | 0.17 |
| Fine B-spline | 3417 | 0.66 | 0.17 |
Fig. 5.
Correlation plots of estimated registration errors against TREs for all landmarks per data set.
Table 4.
Average bias in error estimates for training with and without intensity augmentation.
| Phase | Bias with intensity augmentation (mm) | Bias without intensity augmentation (mm) |
|---|---|---|
| Affine | 0.16 (0.68) | 0.32 (0.70) |
| Coarse B-Spline | 0.14 (0.64) | 0.61 (0.88) |
| Fine B-Spline | 0.11 (0.65) | 0.59 (0.93) |
5. Discussion
In this paper, we propose a supervised algorithm for the estimation of image registration errors. We propose this method as an alternative for measuring TREs on corresponding landmarks that require expert annotations that are hard to come by and are sensitive to inconsistencies by the annotators. The proposed algorithm is based on a 3-D CNN that learns to estimate errors from a set of pairs of training images. The network estimates the registration error as a physical distance. Validation on the gold standard error maps shows a good agreement between the estimates and gold standard, with RMSD of 0.51 mm, showing subvoxel accuracy for registration error estimation. Using the gold standard error maps, we consistently show that CNNs can compute deformation norms and are, therefore, suitable for estimation of registration errors. The gold standard validation method uses realistic deformation models that mimic true registration by taking the difference of two known deformation fields. This imitates a real registration error map, which is also the norm of the difference between the transformation estimated by the registration method and the true, but unknown transformation between the images. In one aspect, the gold standard validation data are not realistic, because both images are transformations from one and the same image, namely the same inspiration image. To compensate for that, we have added white Gaussian noise to both input images.
Validation on ground truth landmarks shows that the network can learn errors to within a small deviation from the TRE calculated on the landmarks. On average, the RMSD between the estimates and ground truth TRE is 0.66 mm for the full registration sequence. From this, we conclude that the estimates are close to the true error, with a subvoxel accuracy. The comparison to TREs is subject to uncertainty in the landmark placement as well. The interobserver localization error in the landmark annotation is on the order of 0.5 to 1 mm and exceeding 2 mm for the POPI data set. The average error made by the algorithm falls within this range, which is supported by the error bars in Fig. 4. Given the better performance on the gold standard validation set, we suspect the larger errors made when comparing to landmark TREs are in part caused by errors in the landmark annotations. It can further be explained by possible differences in the gold standard and ground truth data sets. The deformations in the gold standard validation set may be smoother compared to those in the ground truth set, which makes them resemble the training set more. In addition, the gold standard deformations have been applied to the same image, which makes the estimation of the remaining deformation easier compared to the ground truth validation problem. To make the network more generic, we applied intensity augmentations in the training set. Without these augmentations, the error estimates are overestimated on average, as shown by the increased bias in the estimates, as shown in Table 4.
The network’s architecture was optimized empirically. We experimented with deeper and more shallow networks, as well as a varying number of units per layer. The network’s input patch size has been set to . We found that smaller patch sizes reduce the amount of context and, consequently, reduce the performance of the network. However, larger patch sizes only include more voxels at a larger distance from the center voxels. This means that for very regular transformations, e.g., affine transformations, these voxels may add information, but they will contribute far less to the error estimation for error maps with a lot of local variations. In our experience, larger patch sizes actually decreased the accuracy of the estimations in the current setting. One further point that can determine the optimal patch size is the range of registration errors the network has to estimate, as the patch size should be large enough to leave sufficient contextual information relative to the maximal size of the errors. After initial experiments, we found the current patch size was large enough to estimate errors up to 4 mm, which is adequate for the majority of the registration errors in the thoracic CT registrations. These assumptions may not hold for other registration problems or other anatomical structures, such as interpatient brain MRI registration, where larger errors can occur that will likely require larger patch sizes. To train a network for a larger range of errors that at the same time are spatially fluctuating, it may be necessary to switch to architectures with multiple input patches at different sizes35 or incorporating information at multiple scales.36
The TPS deformations used in the training set and the deformation determined by the registration algorithm (B-spline deformations) lead to different deformation fields. The choice for TPSs was motivated by the fact that an error estimation algorithm should be able to model deformations not captured by the registration algorithm. The TPSs in the training set and the B-splines used in registration are different in nature. From Fig. 3, it seems that the network has created smoother error maps compared to the gold standard error maps. This could be explained by the relative smoothness of the training set deformations. The current implementation has been tested on Elastix’ registrations, and the applicability of the methodology to other registration methodologies and frameworks deserves further attention. Possible difficulties could occur when the registration suffers from shape collapse, where structures disappear or appear because of extreme but wrong transformations caused by structures that have insufficient overlap in the images.37 This will result in areas that have a high registration error while looking very similar. Further experiments are required to investigate this potential problem, which often occurs in brain MRI registration.38
The images used in training and testing have all been resampled to voxel sizes of . This necessity stems from the fact that the network has no information on the scale of image features. However, resampling the images before estimating the error map has a clear advantage: it means that images that initially have vastly different voxel sizes can be used as input to the network, both for training and application. Since the anatomy in the images will be roughly the same size in all scans, the network can more easily train to recognize transformations of relevant anatomical features. A disadvantage is that the images can get quite large (up to for image pair creatis06), which means that using a shift-and-stitch technique to compute the error map on full images requires more memory than available in current GPUs, or that the network needs to be run on multiple overlapping patches, which is not time efficient.
The current method is not the first to use supervised learning to estimate registration errors. The method by Muenzing et al.3 performs supervised classification of the registration error into good, poor, and wrong alignment classes, rather than estimating continuous values for the error. In contrast, our method estimates the registration error directly using a regression approach and only uses the registered images. The two methods are not easily compared since the method by Muenzing et al. is optimized to classify errors and minimizes the misclassifications between the poor and wrong alignment classes, and the good and poor alignment classes. Our convolutional network has not been optimized to distinguish three alignment classes and a proper comparison is, therefore, not possible. The registration error estimation method by Sokooti et al.6 does perform regression using random forests. Both methods require registered images for training, using annotated landmarks at distinctive points. As discussed in Sec. 1, these sets are hard to come by and do not exist for every registration problem. In contrast, our method does not require a true ground truth for training but uses known random transformation to make training data, requiring no manual annotation.
A useful extension to the current algorithm would be the development of a similar method for multimodal registration. In the multimodal case, creating a training set would require simulating other modalities from a given image. Training multimodal registration errors using the current framework would require pairs of perfectly registered multimodal images that would then be deformed with known transformations. One way to solve this would be to let the training and testing of the network rely on modality-independent feature maps of the images instead of the images themselves.39
6. Conclusion
An automatic supervised method for estimation of nonlinear registration errors has been presented. We have shown that CNNs are a viable approach for estimating continuous registration errors for the full image domain. We consider this a generic approach for mono-modal data that only require a small number of images to be applied to different anatomies and modalities. Error estimation methods, such as the one presented in this paper, are important to further development and evaluation of registration algorithms, to combine multiple registration methods, and can be of use in clinical image-guided procedures that rely on nonlinear image registration.
Biographies
Koen A. J. Eppenhof is a PhD candidate at Eindhoven University of Technology. He received his BSc and MSc degrees in biomedical engineering from the same university in 2012 and 2015, respectively. His research interests include medical image registration, deep learning, and medical image analysis.
Josien P. W. Pluim is a professor of medical image analysis at Eindhoven University of Technology and holds a part-time appointment at the University Medical Center (UMC) Utrecht. She received her MSc degree in computer science from the University of Groningen in 1996 and her PhD from UMC Utrecht in 2001. She was the chair of SPIE Medical Imaging Image Processing from 2006 to 2009, is an associate editor of five international journals (including The Journal of Medical Imaging), and a member of the MICCAI board of directors.
Disclosures
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors. The authors declare that there are no conflicts of interests regarding this paper.
References
- 1.Rohlfing T., “Image similarity and tissue overlaps as surrogates for image registration accuracy: widely used but unreliable,” IEEE Trans. Med. Imaging 31(2), 153–163 (2012).https://doi.org/10.1109/TMI.2011.2163944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Datteri R. D., et al. , “Validation of a nonrigid registration error detection algorithm using clinical MRI brain data,” IEEE Trans. Med. Imaging 34(1), 86–96 (2015).https://doi.org/10.1109/TMI.2014.2344911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muenzing S. E. A., et al. , “Supervised quality assessment of medical image registration: application to intra-patient CT lung registration,” Med. Image Anal. 16(8), 1521–1531 (2012).https://doi.org/10.1016/j.media.2012.06.010 [DOI] [PubMed] [Google Scholar]
- 4.Muenzing S. E. A., et al. , “DIRBoost—an algorithm for boosting deformable image registration: application to lung CT intra-subject registration,” Med. Image Anal. 18(3), 449–459 (2014).https://doi.org/10.1016/j.media.2013.12.006 [DOI] [PubMed] [Google Scholar]
- 5.Muenzing S. E. A., “Learning-based approaches to deformable image registration,” PhD Thesis, Utrecht University (2014). [Google Scholar]
- 6.Sokooti H., et al. , “Accuracy estimation for medical image registration using regression forests,” Lect. Notes Comput. Sci. 9902, 107–115 (2016).https://doi.org/10.1007/978-3-319-46726-9 [Google Scholar]
- 7.Kybic J., “Bootstrap resampling for image registration uncertainty estimation without ground truth,” IEEE Trans. Image Process. 19(1), 64–73 (2010).https://doi.org/10.1109/TIP.2009.2030955 [DOI] [PubMed] [Google Scholar]
- 8.Risholm P., et al. , “Bayesian characterization of uncertainty in intra-subject non-rigid registration,” Med. Image Anal. 17(5), 538–555 (2013).https://doi.org/10.1016/j.media.2013.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janoos F., Risholm P., Wells W. M., III, “Bayesian characterization of uncertainty in multi-modal image registration,” Lect. Notes Comput. Sci. 7359, 50–59 (2012).https://doi.org/10.1007/978-3-642-31340-0 [Google Scholar]
- 10.Saygili G., Staring M., Hendriks E. A., “Confidence estimation for medical image registration based on stereo confidences,” IEEE Trans. Med. Imaging 35(2), 539–549 (2016).https://doi.org/10.1109/TMI.2015.2481609 [DOI] [PubMed] [Google Scholar]
- 11.Greenspan H., van Ginneken B., Summers R. M., “Guest editorial deep learning in medical imaging: overview and future promise of an exciting new technique,” IEEE Trans. Med. Imaging 35(5), 1153–1159 (2016).https://doi.org/10.1109/TMI.2016.2553401 [Google Scholar]
- 12.Miao S., Wang Z. J., Liao R., “A CNN regression approach for real-time 2D/3D registration,” IEEE Trans. Med. Imaging 35(5), 1352–1363 (2016).https://doi.org/10.1109/TMI.2016.2521800 [DOI] [PubMed] [Google Scholar]
- 13.Sokooti H., et al. , “Nonrigid image registration using multi-scale 3D convolutional neural networks,” Lect. Notes Comput. Sci. 10433, 232–239 (2017).https://doi.org/10.1007/978-3-319-66182-7 [Google Scholar]
- 14.Krebs J., et al. , “Robust non-rigid registration through agent-based action learning,” Lect. Notes Comput. Sci. 10433, 344–352 (2017).https://doi.org/10.1007/978-3-319-66182-7 [Google Scholar]
- 15.Yang X., et al. , “Quicksilver: fast predictive image registration a deep learning approach,” NeuroImage 158, 378–396 (2017).https://doi.org/10.1016/j.neuroimage.2017.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutiérrez-Becker B., et al. , “Learning optimization updates for multimodal registration,” Lect. Notes Comput. Sci. 9902, 19–27 (2016).https://doi.org/10.1007/978-3-319-46726-9 [Google Scholar]
- 17.Simonovsky M., et al. , “A deep metric for multimodal registration,” Lect. Notes Comput. Sci. 9902, 10–18 (2016).https://doi.org/10.1007/978-3-319-46726-9 [Google Scholar]
- 18.Wu G., et al. , “Scalable high-performance image registration framework by unsupervised deep feature representations learning,” IEEE Trans. Biomed. Eng. 63(7), 1505–1516 (2016).https://doi.org/10.1109/TBME.2015.2496253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eppenhof K. A. J., Pluim J. P. W., “Supervised local error estimation for nonlinear image registration using convolutional neural networks,” Proc. SPIE 10133, 101331U (2017).https://doi.org/10.1117/12.2253859 [Google Scholar]
- 20.Murphy K., et al. , “Evaluation of registration methods on thoracic CT: the EMPIRE10 challenge,” IEEE Trans. Med. Imaging 30(11), 1901–1920 (2011).https://doi.org/10.1109/TMI.2011.2158349 [DOI] [PubMed] [Google Scholar]
- 21.Klein S., et al. , “Elastix: a toolbox for intensity-based medical image registration,” IEEE Trans. Med. Imaging 29(1), 196–205 (2010).https://doi.org/10.1109/TMI.2009.2035616 [DOI] [PubMed] [Google Scholar]
- 22.Srivastava N., et al. , “Dropout: a simple way to prevent neural networks from overfitting,” J. Mach. Learn. Res. 15(1), 1929–1958 (2014). [Google Scholar]
- 23.Ioffe S., Szegedy C., “Batch normalization: accelerating deep network training by reducing internal covariate shift,” in Proc. Int. Conf. on Machine Learning (ICML), pp. 448–456 (2015). [Google Scholar]
- 24.Castillo R., et al. , “A framework for evaluation of deformable image registration spatial accuracy using large landmark point sets,” Phys. Med. Biol. 54(7), 1849–1870 (2009).https://doi.org/10.1088/0031-9155/54/7/001 [DOI] [PubMed] [Google Scholar]
- 25.Castillo E., et al. , “Four-dimensional deformable image registration using trajectory modeling,” Phys. Med. Biol. 55(1), 305–327 (2009).https://doi.org/10.1088/0031-9155/55/1/018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castillo R., et al. , “A reference dataset for deformable image registration spatial accuracy evaluation using the COPDgene study archive,” Phys. Med. Biol. 58(9), 2861–2877 (2013).https://doi.org/10.1088/0031-9155/58/9/2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vandemeulebroucke J., Sarrut D., Clarysse P., “The POPI model, a point-validated pixel-based breathing thorax model,” in Proc. Int. Conf. on the Use of Computers in Radiation Therapy (ICCR) (2007). [Google Scholar]
- 28.Sarrut D., et al. , “Simulation of four-dimensional CT images from deformable registration between inhale and exhale breath-hold CT scans,” Med. Phys. 33(3), 605–617 (2006).https://doi.org/10.1118/1.2161409 [DOI] [PubMed] [Google Scholar]
- 29.Vandemeulebroucke J., et al. , “Spatiotemporal motion estimation for respiratory-correlated imaging of the lungs,” Med. Phys. 38(1), 166–178 (2011).https://doi.org/10.1118/1.3523619 [DOI] [PubMed] [Google Scholar]
- 30.Dieleman S., et al. , “Lasagne: first release” (2015).https://doi.org/10.5281/zenodo.27878
- 31.Bergstra J., “Theano: a CPU and GPU Math Expression Compiler,” in Proc. of the Python for Scientific Computing Conference (SciPy) (2010). [Google Scholar]
- 32.Staring M., et al. , “Pulmonary image registration with elastix using a standard intensity-based algorithm,” in Proc. Medical Image Analysis For The Clinic—A Grand Challenge, Workshop Held in Conjunction with MICCAI (2010). [Google Scholar]
- 33.Sermanet P., et al. , “Overfeat: integrated recognition, localization and detection using convolutional networks,” in Proc. Int. Conf. on Learning Representations (ICLR) (2014). [Google Scholar]
- 34.Vandemeulebroucke J., et al. , “Automated segmentation of a motion mask to preserve sliding motion in deformable registration of thoracic CT,” Med. Phys. 39(2), 1006–1015 (2012).https://doi.org/10.1118/1.3679009 [DOI] [PubMed] [Google Scholar]
- 35.Moeskops P., et al. , “Automatic segmentation of MR brain images with a convolutional neural network,” IEEE Trans. Med. Imaging 35(5), 1252–1261 (2016).https://doi.org/10.1109/TMI.2016.2548501 [DOI] [PubMed] [Google Scholar]
- 36.Farabet C., et al. , “Learning hierarchical features for scene labeling,” IEEE Trans. Pattern Anal. Mach. Intell. 35(8), 1915–1929 (2013).https://doi.org/10.1109/TPAMI.2012.231 [DOI] [PubMed] [Google Scholar]
- 37.Durumeric O. C., Oguz I., Christensen G. E., “The shape collapse problem in image registration,” in Proc. Mathematical Foundations of Computational Anatomy (MFCA), Workshop Held in Conjunction with MICCAI, pp. 95–106 (2013). [Google Scholar]
- 38.Shao W., et al. , “Population shape collapse in large deformation registration of MR brain images,” in Proc. IEEE Conf. on Computer Vision and Pattern Recognition Workshops (CVPRW), pp. 549–557 (2016).https://doi.org/10.1109/CVPRW.2016.75 [Google Scholar]
- 39.Heinrich M. P., et al. , “MIND: modality independent neighbourhood descriptor for multi-modal deformable registration,” Med. Image Anal. 16(7), 1423–1435 (2012).https://doi.org/10.1016/j.media.2012.05.008 [DOI] [PubMed] [Google Scholar]