Abstract
Study question
Can novel meiotic RNA targets of DAZL (deleted in azoospermia-like) be identified in the human foetal ovary?
Summary answer
SYCP1 (synaptonemal complex protein-1), TEX11 (testis expressed 11) and SMC1B (structural maintenance of chromosomes 1B) are novel DAZL targets in the human foetal ovary, thus DAZL may have previously unrecognised roles in the translational regulation of RNAs involved in chromosome cohesion and DNA recombination in the oocyte from the time of initiation of meiosis.
What is known already
The phenotype of Dazl deficiency in mouse is infertility in both sexes and DAZL has also been linked to infertility in humans. Few studies have explored targets of this RNA-binding protein. The majority of these investigations have been carried out in mouse, and have focussed on the male thus the basis for its central function in regulating female fertility is largely unknown.
Study design size, duration
We carried out RNA sequencing after immunoprecipitation of endogenous DAZL from human foetal ovarian tissue (17 weeks of gestation, obtained after elective termination of pregnancy) to identify novel DAZL targets involved in meiosis (n = 3 biological replicates).
Participants/materials, setting, methods
Using quantitative RT-PCR, we examined the expression of selected RNAs identified by our immunoprecipitation across gestation, and visualised the expression of potential target SMC1B in relation to DAZL, with a combination of in situ hybridisation and immunohistochemistry. 3′ untranslated region (3′UTR)-luciferase reporter assays and polysome profile analysis were used to investigate the regulation of three RNA targets by DAZL, representing key functionalities: SYCP1, TEX11 and SMC1B.
Main results and the role of chance
We identified 764 potential RNA targets of DAZL in the human foetal ovary (false discovery rate 0.05 and log-fold change ≥ 2), with functions in synaptonemal complex formation (SYCP1, SYCP3), cohesin formation (SMC1B, RAD21), spindle assembly checkpoint (MAD2L1, TRIP13) and recombination and DNA repair (HORMAD1, TRIP13, TEX11, RAD18, RAD51). We demonstrated that the translation of novel targets SYCP1 (P = 0.004), TEX11 (P = 0.004) and SMC1B (P = 0.002) is stimulated by the presence of DAZL but not by a mutant DAZL with impaired RNA-binding activity.
Large scale data
The raw data are available at GEO using the study ID: GSE81524.
Limitations, reasons for caution
This analysis is based on identification of DAZL targets at the time when meiosis starts in the ovary: it may have other targets at other stages of oocyte development, and in the testis. Representative targets were validated, but detailed analysis was not performed on the majority of putative targets.
Wider implications of the findings
These data indicate roles for DAZL in the regulation of several key functions in human oocytes. Through the translational regulation of novel RNA targets SMC1B and TEX11, DAZL may have a key role in regulating chromosome cohesion and DNA recombination; two processes fundamental in determining oocyte quality and whose establishment in foetal life may support lifelong fertility.
Study funding and competing interest(s)
This study was supported by the UK Medical Research Council (grant no G1100357 to R.A.A. and an intramural MRC programme grant to I.R.A.). The authors declare no competing interests.
Keywords: DAZL, RNA targets, meiosis, female fertility
Introduction
DAZL (deleted in azoospermia-like) and its homologues DAZ (deleted in azoospermia) and BOLL (bol-like RNA-binding protein) comprise a family of RNA-binding proteins that have essential roles in gametogenesis. DAZ gene expression is only found in humans and Old World monkeys; transcription is limited to the male germline (Ruggiu and Cooke, 1999) and DAZ is deleted in 12–15% of azoospermic men (Reijo et al., 1995). In all other vertebrates, DAZ is replaced by an autosomal single-copy gene: DAZ-like (DAZL). Targeted disruption of Dazl in mice results in infertility in both males and females (Ruggiu et al., 1997; Saunders et al., 2003) and in the human DAZL is expressed in germ cells in ovary and testis in both foetal and adult life (Habermann et al., 1998; Dorfman et al., 1999; Anderson et al., 2007). Polymorphisms in the promoter and coding sequence of DAZL have been correlated with total sperm count and sperm motility in infertile men (Teng et al., 2002; Tung et al., 2006a), while altered methylation of the DAZL promoter has been associated with male infertility (Navarro-Costa et al., 2010; Li et al., 2013). In women, however, there has been only one report concerning the relevance of DAZL to fertility, with polymorphisms in DAZL shown to influence the age at premature ovarian insufficiency and menopause (Tung et al., 2006a); however, this has not been reported in other studies (Bartoloni et al., 2004; Zerbetto et al., 2008). These observations suggest that hypomorphic polymorphisms and single nucleotide polymorphisms that quantitatively reduce DAZL expression or function in human germ cells could potentially have consequences for fertility and reproductive lifespan in women.
Deletion of Dazl in mice causes loss of germ cells in the gonads of both sexes, with increased apoptosis, reduced expression of germ-cell markers and aberrant chromatin structure (Ruggiu et al., 1997; Schrans-Stassen et al., 2001; Lin and Page, 2005). Dazl null mice also have a reduction in the number of post-migratory primordial germ cells, and germ cells fail to sexually differentiate and properly erase genomic methylation imprints (Haston et al., 2009; Gill et al., 2011). Of the few cells that are able enter meiosis, none are able to proceed beyond leptotene of Prophase I, as complete synaptonemal complexes fail to form in Dazl−/− mice, thus giving a precise point beyond which meiosis cannot progress without Dazl (Saunders et al., 2003). Furthermore, germ cells fail to induce the expression of meiotic genes in response to retinoic acid (Lin and Page, 2005). Dazl deficiency specifically compromises germ-cell function, as the transplantation of ROSA/LacZ-labelled spermatogonia into the testes of Dazl−/− mice confirms that the somatic tissues are capable of supporting gamete development (Rilianawati et al., 2003).
DAZL acts post-transcriptionally, as demonstrated by its association with actively translating polysomes (Tsui et al., 2000b) and its ability to activate translation of bound mRNAs (Collier et al., 2005). The involvement of DAZL in translational control is key for gametogenesis. During their growth phase, mammalian oocytes are transcriptionally active, however, mRNA synthesis drops to very low or undetectable levels at the onset of meiosis (Clarke, 2012). From this point onwards, gene expression in the oocyte and subsequent protein production is highly dependent upon a well-orchestrated programme of dynamic modulation of mRNA poly(A)-tail length, recruitment to polysomes and translational activation or repression (Seydoux and Braun, 2006; Radford et al., 2008). However, in order to understand the growing spectrum of processes in gametogenesis that this gene may be involved in, a knowledge of DAZL mRNA targets is required. Efforts to uncover in vivo mRNA targets of mammalian DAZL have mainly been focussed on mouse, and the male. Analysis of gene expression in the Dazl−/− mouse foetal ovary has identified mRNAs whose abundance is downstream of Dazl function, but this analysis does not identify mRNAs which are directly regulated by Dazl (Soh et al., 2015). Three direct mRNA targets of Dazl characterised in vivo are Sycp3 (synaptonemal complex protein 3), Mvh (mouse vasa homologue) (formally known as Ddx4 or Vasa) and Tex19.1 (testis expressed 19.1) (Reynolds et al., 2005; Reynolds et al., 2007; Chen et al., 2011), and Tet1 (tet methylcytosine dioxygenase 1) has also been demonstrated to be dependent on Dazl for its translation in cultured mouse embryonic stem cells (Welling et al., 2015). Sycp3-deficient spermatocytes arrest in meiosis due to defects in chromosome synapsis, as do spermatocytes from Mvh and Tex19.1 null males (Tanaka et al., 2000; Ollinger et al., 2008). This is a similar point at which meiotic progression ceases in male mice lacking Dazl (Lin and Page, 2005). This would suggest that a loss or reduced expression of these genes, and possibly also additional transcripts that are dependent on Dazl for their translation, contributes to the infertility phenotype observed in Dazl null male mice. In contrast, whereas Dazl null female mice also arrest meiotic progression in early prophase during foetal development (Saunders et al., 2003), neither Sycp3, Mvh nor Tex19.1 null female mice have such severe defects that oogenesis arrests at this stage (Tanaka et al., 2000; Yuan et al., 2002; Ollinger et al., 2008). Additionally, currently unknown Dazl targets in foetal oocytes may therefore be contributing to the Dazl null phenotype in female mice. In humans, even less is known about DAZL targets and their impact on female fertility (Rosario et al., 2016).
Here we identify novel RNA targets of DAZL in the human ovary, using foetal tissue to allow analysis during the onset and early stages of Meiosis I. Our data confirm the meiotic role of DAZL in the human foetal ovary, and also reveal novel potential functions for DAZL through translational regulation of RNA targets involved in establishment of chromosome cohesion and DNA recombination during early meiosis in the foetal oocyte, key pathways implicated in the determination of lifelong oocyte quality.
Materials and Methods
Ethics statement
Ethical approval for this study was obtained from Lothian Research Ethics Committee (study code LREC 08/S1101/1), and women gave informed written consent. Experiments involving mice were approved by the University of Edinburgh Animal Research Ethics Committee and performed according to the UK Animal (Scientific Procedures) Act 1986.
Collection of human foetal ovaries
Human fetuses (8–20 weeks gestational age (wga)) were obtained after elective termination of pregnancy, and all fetuses used in this study were morphologically normal. Gestational age was determined by ultrasound scan, and confirmed (for second trimester fetuses) by direct measurement of foot length. The sex of first trimester foetal gonads was determined by PCR for the SRY gene (Childs and Anderson, 2012). Extra-ovarian tissue was removed from dissected ovaries, which were then either snap frozen on dry ice and stored at −80°C for subsequent RNA extraction, or fixed in Bouins or 4% neutral buffered formalin for 2–3 h before processing into paraffin blocks for immunohistochemical analysis. Thirty-two foetal ovarian samples were obtained and used in this study.
RNA immunoprecipitation
RNA immunoprecipitation was carried out using the Magna RIP™ Kit (Millipore, Livingstone, UK) according to manufacturer's instructions. Immunoprecipitations were performed in lysates made from 2 homogenised 17-week human foetal ovaries, from different fetuses. Separate lysates were made for 3 biological replicates, thus 12 ovaries from a total of 6 fetuses were used for these experiments. RNA–protein complexes were immunoprecipitated using an antibody against human DAZL (dilution 1:100) (#8042, Cell Signalling Technology, Leiden, The Netherlands) (see Supplementary Fig. S1 for antibody validation) or a control matched-IgG. Following RNA purification, the Ovation® RNA-Seq system V2 (NuGEN, The Netherlands) was utilised to generate sufficient quantities of oligo-T primed cDNA (average size 350 bp), which was then subjected to paired-end RNA sequencing using the Illumina HiSeq platform, performed by Edinburgh Genomics, University of Edinburgh, UK. Paired-end sequencing data were processed using TrimGalore v0.4.1 (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) to remove the CTTTGTGTTTGA Ovation RNA-Seq adapter using—stringency 5 and—length 30 command line parameters, then mapped to the hg19 assembly of the human genome and Ensembl transcriptome using Tophat v2.1.0 (http://ccb.jhu.edu/software/tophat) with command line parameters-g 1—mate-inner-dist 260—mate-std-dev 110—no-coverage-search—b2-sensitive. Mapped reads that were primary alignments and overlapped Ensembl transcript co-ordinates were counted using Bedtools intersect v2.25.0 (https://github.com/arq5x/bedtools) in-split mode. Paired reads that mapped within the same transcript were counted as a single read. Read counts were imported into R v3.3.2 (https://www.r-project.org/) and biomaRt v2.30.0 (https://bioconductor.org/packages/release/bioc/html/biomaRt.html) used to map Ensembl transcripts to Ensembl genes using the Homo sapiens GRCh38.p7 data set. Genes with multiple transcripts were collapsed into the transcript containing the most read counts, the total number of reads mapped to Ensembl genes counted. Low abundance genes containing fewer than one count per million mapped reads in at least three samples were discarded. Differential analysis was performed using DESeq2 v1.14.0 (https://bioconductor.org/packages/release/bioc/html/DESeq2.html) using the biological replicate as a blocking factor in the design model to identify differences between control and DAZL immunoprecipitations. False discovery rate (FDR)-adjusted P-values of ≤0.05 were considered significant, and genes significantly enriched more than 4-fold in DAZL relative to control immunoprecipitations were considered DAZL targets. This was filtered further to include only Ensembl genes with an HGNC gene symbol in the bioMart database. Relationships between significantly differentially expressed RNAs were further explored using GeneSetDB (Araki et al., 2012) using all genes represented in the control and DAZL immunoprecipitations as the universe.
Site-directed mutagenesis
The R115G mutant DAZL overexpression construct was generated by PCR of the R115G coding sequence from pMS2-hDAZL (Collier et al., 2005) harbouring the R115G mutation (a gift from Prof. Nicola Gray), which was introduced with the QuikChange Lightening Site-Directed Mutagenesis kit (Agilent Technologies, Edinburgh, UK). The PCR product was cloned into pCMV6-entry (referred to as vector only control) (OriGene Technologies, Rockville, MD, USA) using SgfI and XhoI. The mutant SMC1B 3′ untranslated region (3′UTR)-luciferase construct and the mutant TEX11 3′UTR-luciferase construct were created by mutating G in the mouse Dazl-binding site to A using the QuikChange Lightening Site-Directed Mutagenesis kit (Agilent Technologies, Edinburgh, UK). A second mutant SMC1B 3′UTR-luciferase construct was also created using site-directed mutagenesis to delete the entire mouse Dazl-binding site. All constructs were confirmed to be correct using an ABI 3730XL capillary Sanger sequencing instrument.
Luciferase assays
HEK293T cells (human embryonic kidney cells) were cultured in DMEM + GlutaMAX™ supplemented with 10% foetal bovine serum and maintained at 37°C in 5% CO2. For the luciferase assays, cells were seeded at a density of 20 000 cells per well of a 96-well plate. Cells were transfected with 1ng of the 3′UTR-luciferase construct (wildtype or mutant) or a 3′UTR-luciferase empty vector control (Genecopoeia, Rockville, MD, USA), plus 50 ng of either a wildtype DAZL overexpression construct, a R115G mutant DAZL overexpression construct or a vector only control (OriGene Technologies, Rockville, MD, USA), using TransIT-LT1 transfection reagent (Mirus Bio, Cambridge, UK). Luciferase expression was detected 48 h post transfection using the Luc-Pair Luciferase Assay Kit (Genecopoeia, Rockville, MD, USA) according to manufacturer's instructions.
Sucrose gradient analysis
HEK293T cells were transfected as described above. At 48 h post transfection, cells were treated with 150 µg/ml of cycloheximide before lysis in a polysome lysis buffer (20 mM Hepes pH 7.6, 2 mM MgCl2, 150 mM KCl, 0.5% Nonidet P40, 2 mM dithiothreitol (DTT), 100 U/ml RNasin (Promega, South Hampton, UK)), 150 µg/ml cycloheximide, 1× protease inhibitor (Sigma Aldrich, Dorset, UK) and 1× Halt phosphatase inhibitor (Thermo Fisher Scientific, Paisley, UK). The KCl concentration of the lysates was adjusted before being loaded onto a 10-ml linear sucrose gradient (10–50%) containing 20 mM Hepes pH7.6, 2 mM MgCl2, 250 mM KCl, 0.5% Nonidet p40, 2.5 mM DTT and 0.5 µg/ml heparin. Gradients were centrifuged for 120 min at 4°C in a Beckman SW41 Rotor at 181 781g. Following centrifugation, 10 fractions of 1 ml were collected from the gradient with a KD Scientific peristaltic pump (Holliston, MA, USA) and a Foxy Jr® fraction collector (Teledyne Isco, Lincoln, NE, USA). Absorption traces were recorded with a UA6 Absorbance Detector with an A254 filter (Teledyne Isco, Lincoln, NE, USA). Fractions 1–2 and 6–10 were pooled, respectively, for luciferase RNA analysis.
RNA extraction, cDNA synthesis and quantitative RT-PCR
RNA from human foetal ovaries was extracted using the RNeasy Micro Kit (Qiagen, Crawley, UK) according to manufacturer's instructions. Cytoplasmic RNA from luciferase-transfected HEK293T cells was extracted with the Cytoplasmic and Nuclear RNA Purification Kit (Norgen-Biotek, Ontario, Canada) according to manufacturer's instructions. RNA was precipitated from sucrose gradients using phenol–chloroform (Life Technologies, Paisley, UK), 0.3 M sodium acetate and isopropanol. RNA was reverse transcribed to cDNA using concentrated random primers and Superscript III reverse transcriptase (Life Technologies) according to manufacturer's instructions, and the cDNA synthesis reaction was diluted appropriately before proceeding. Primers for quantitative RT-PCR (RT-qPCR) were designed to amplify all transcript variants and are exon-spanning. Primer pair efficiencies were calculated with the LinReg PCR applet (Ramakers et al., 2003). Each reaction was performed in a final volume of 10 µl, with 1× Brilliant III SYBR Green qPCR Master Mix (Agilent, Santa Clara, CA, USA), 20 pmol of each primer and 2 µl of diluted cDNA. Each cDNA sample was analysed in triplicate. For expression analyses in human foetal ovary, target genes were normalised to the geometric mean expression of B2M (Beta-2 microglobulin) and RPL32 (ribosomal protein L32). For Luciferase expression, the Renilla sequence which was located on the same construct was used for normalisation. Data analysis for relative quantification of gene expression and calculation of SDs was performed as outlined (Livak and Schmittgen, 2001; Vandesompele et al., 2002).
RNA in situ hybridisation and immunohistochemistry
Multiplex detection of SMC1B and DAZL protein was performed by Aquila Histoplex, University of Edinburgh. RNA in situ hybridisation was carried out using RNAscope® designed probes and reagents (Advanced Cell Diagnostics, Hayward, CA, USA). Briefly, slides were dewaxed and tissue permeabilised with hydrogen peroxide, heat retrieval and proteinase K. RNA probes (SMC1B or DapB (RNAscope® universal negative control)) were hybridised at 40°C for 2 h, followed by six amplification steps to amplify the signal. For detection, tissue was stained with FITC tyramide at 1:100 for 10 min. Immunohistochemistry was carried out using a mouse DAZL antibody (Abd Serotec, Oxford, UK) at 1:200, a peroxidase secondary antibody at 1:200 and Cy3 tyramide signal amplification at 1:50; counterstained with DAPI.
Results
RNA immunoprecipitation and sequencing identifies novel RNA targets of DAZL
To discover novel and human DAZL targets in the human foetal ovary, we carried out RNA-Seq after immunoprecipitation with foetal ovaries at 17 weeks gestation, when many oocytes are in the early stages of meiosis. Differential gene expression analysis identified a total of 764 RNAs that were significantly enriched by DAZL immunoprecipitation relative to the IgG control (FDR ≤ 0.05 and log-fold change ≥ 2) (see Supplementary Table S1). Among these were validated RNA targets of murine Dazl, SYCP3 and TEX14, indicating technical success.
We performed a gene set enrichment analysis using GeneSetDB (Araki et al., 2012) to investigate whether any disease/phenotype or gene ontology was overrepresented in our list of DAZL-bound RNAs (FDR < 0.05)(Table I and Supplementary Table S2). The majority of the disease/phenotypes that were enriched were related to germ cells, meiosis, gamete production and infertility, but only in males, which likely reflects the lack of female/ovary-related high-throughput data available for enrichment analyses. The role of DAZL in gametogenesis was also evident in the gene ontologies, as the ontology meiosis was significantly enriched (GO:0007126). Within this ontology, the DAZL-enriched RNAs could be divided into different functionalities in meiosis such as synaptonemal complex formation (SYCP1, SYCP3), cohesin formation (SMC1B, RAD21), spindle assembly checkpoint (MAD2L1, TRIP13), and recombination and DNA repair (HORMAD1, TRIP13, TEX11, RAD18, RAD51). The enrichment analysis also highlighted the role DAZL targets may have in the mitotic cell cycle (GO:0000278) and the G1/S transition (GO:0000082).
Table I.
Gene set enrichment analysis.
| Class | Set name | FDR |
|---|---|---|
| Disease/phenotype | Arrest of male meiosis | 7.80E-06 |
| Abnormal male meiosis | 1.70E-03 | |
| Azoospermia | 2.50E-03 | |
| Decreased testis weight | 4.80E-03 | |
| Abnormal spermatocyte morphology | 5.80E-03 | |
| Male infertility | 5.90E-03 | |
| Small testis | 6.80E-03 | |
| Male germ-cell apoptosis | 7.30E-03 | |
| Testicular atrophy | 8.10E-03 | |
| Abnormal apoptosis | 1.70E-02 | |
| Abnormal synaptonemal complex | 3.90E-02 | |
| Gene ontology | Mitotic cell cycle (GO:0000278) | 1.40E-04 |
| G1/S transition of mitotic cell cycle (GO:0000082) | 2.50E-03 | |
| DNA repair (GO:0006281) | 8.10E-03 | |
| Protein K63-linked ubiquitination (GO:0070534) | 1.70E-02 | |
| GTP catabolic process (GO:0006184) | 1.70E-02 | |
| Protein K48-linked ubiquitination (GO:0070936) | 2.60E-02 | |
| Meiosis (GO:0007126) | 2.60E-02 | |
| G1 phase of mitotic cell cycle (GO:0000080) | 4.70E-02 | |
| Ubiquitin-dependent protein catabolic process (GO:0006511) | 4.80E-02 |
False discovery rate (FDR) is <0.05. For a list of RNAs associated with different Subclasses, refer to Supplementary Table S2.
Enrichment of specific disease/phenotypes and gene ontologies was performed using GeneSetDB (Araki et al., 2012).
Given the well-established role of DAZL in meiosis (Saunders et al., 2003), we decided to focus our investigation by identifying other RNA targets of DAZL with meiotic roles in the human foetal ovary. SYCP1, TEX11 and SMC1B, which represent RNAs from different meiotic functions, were selected for further investigation (Supplementary Fig. S2). We validated these novel RNA targets in vitro using RT-qPCR on a separate immunoprecipitation experiment to confirm that DAZL can bind these RNAs, and calculated the enrichment ratios for each target (Supplementary Fig. S3). Furthermore, we assessed the abundance of reference genes RPL32 and GAPDH, and the granulosa cell marker FOXL2, showing these were not enriched after DAZL immunoprecipitation, thus highlighting the specificity of our RNA immunoprecipitation experiment.
DAZL and novel target RNA expression across gestation
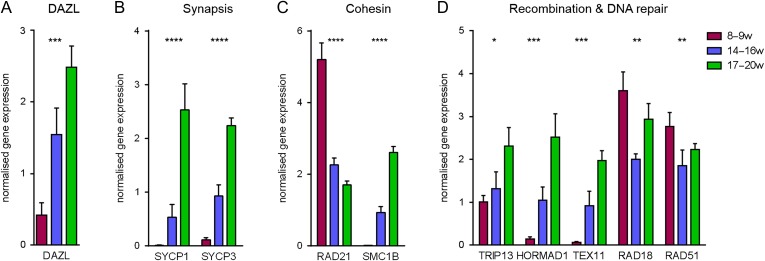
Expression of a selected number of novel meiotic RNA targets in the human foetal ovary identified by DAZL immunoprecipitation was analysed across gestation, which was divided into three periods, including germ cells in mitosis before the onset of meiosis (8–9 wga), at the commencement of meiosis (14–16 wga) and later in meiosis including the onset of meiotic arrest and primordial follicle formation (17–20 wga). DAZL transcript levels dramatically increased at 14–16 wga, and continued to rise at 17–20 wga (Fig. 1A). This pattern of expression across gestation was also observed in RNAs that have a role in synaptonemal complex formation (Fig. 1B), as well as with the meiotic cohesin SMC1B. The expression of the mitotic cohesin RAD21, however, was highest at 8–9 wga and fell significantly during gestation (Fig. 1C). Those RNAs with functions in recombination and DNA repair showed two different patterns of expression (Fig. 1D). TRIP13, HORMAD1 and TEX11 mimicked the expression trend observed for the other meiotic RNAs, while RAD18 and RAD51 transcript levels dropped between 8–9 and 14–16 wga, but then increased again at 17–20 wga.
Figure 1.
Selected RNA expression in the human foetal ovary across gestation. RNAs have been grouped into different functions based on published literature (A–D); however, some RNAs have overlapping functions. Target gene expression has been normalised to the geometric mean expression of RPL32 (ribosomal protein L32) and B2M (beta-2 microglobulin). Mean ± SEM, n = 5. *P < 0.02, **P < 0.002, ***P < 0.0002, ****P < 0.0001 by ANOVA.
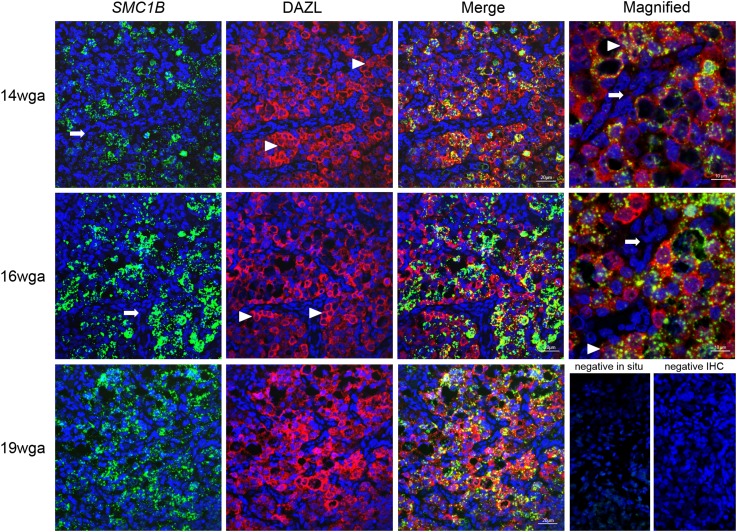
As chromosome cohesion is an example of a role not previously associated with DAZL, we visualised expression of the meiotic cohesin subunit SMC1B across gestation in relation to DAZL using RNA in situ hybridisation for SMC1B with immunohistochemistry for DAZL (Fig. 2). At all gestational ages examined, SMC1B RNA was exclusively present in germ cells, and predominantly co-localised with DAZL protein, although there are areas of germ cells expressing neither, and a few DAZL positive cells that lacked SMC1B were also identified. At 14 wga, SMC1B staining was punctate, with expression being mainly located in the germ-cell cytoplasm; only a few germ cells showed some nuclear staining. This changed at 16 wga, as SMC1B transcripts were detected in markedly more germ cells. There was no significant difference in SMC1B expression between 16 wga and 19 wga, however in the 19 wga tissue, SMC1B staining appeared more punctate again.
Figure 2.
Localisation of SMC1B mRNA with DAZL in human foetal ovary, by RNA in situ hybridisation in combination with immunohistochemistry.
Columns show SMC1B (structural maintenance of chromosomes 1B) or DAZL (deleted in azoospermia-like), and merged images as indicated, with the right hand column showing higher power images at the indicated gestations and negative controls. Arrows identify somatic cell streams. Arrowheads identify germ-cell nests. Merge image; scale bar is 20 µm (the scale is the same for columns 1 and 2). Magnified image; scale bar is 10 µm.
Stimulation of translation by DAZL via 3′UTR sequences
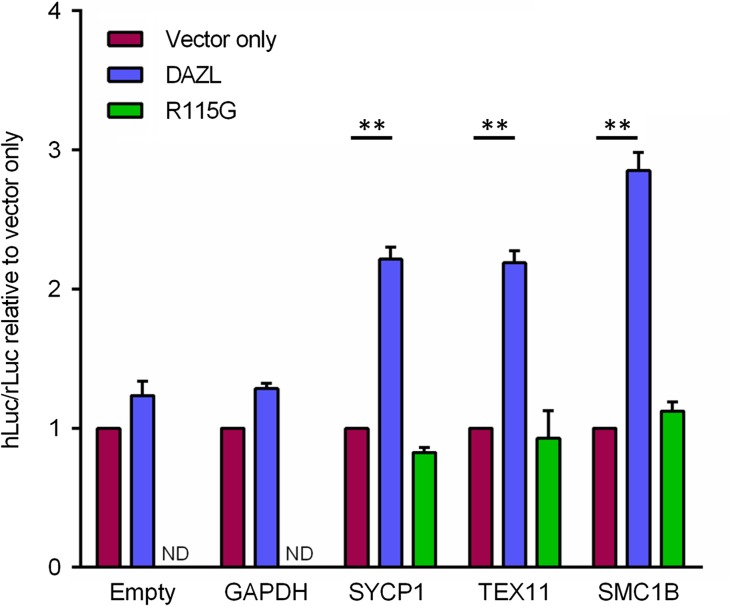
Previous investigations into the function of Dazl have shown this RNA-binding protein to either stimulate (Reynolds et al., 2005; Reynolds et al., 2007) or, more rarely, to repress (Chen et al., 2014) the translation of specific transcripts. To ascertain what translational effect DAZL had on the selected targets SYCP1, TEX11 and SMC1B, representing members of different functional groups, we used a 3′UTR-luciferase reporter assay in HEK293T cells. All three novel RNA targets showed significantly increased luciferase activity in the presence of DAZL, compared to the vector only control (Fig. 3). No stimulation of translation was observed with a GAPDH 3′UTR or in the absence of a 3′UTR, indicating this effect is specific to the novel target 3′UTRs. The DAZL R115G mutant, identified in a woman with spontaneous premature ovarian failure (Tung et al., 2006b), has a significantly impaired ability to bind RNA, as the mutation is located within the DAZL RNA recognition motif (RRM) (Jenkins et al., 2011). Expression of the R115G mutant in HEK293T cells resulted in no increase in luciferase activity compared with the vector only control for all the novel RNA targets (Fig. 3). This indicates that the translation of SYCP1, TEX11 and SMC1B is dependent on the RNA-binding ability of DAZL.
Figure 3.
3′UTR-luciferase reporter assay. Increased luciferase activity is observed for target RNAs, but not an empty 3′ untranslated region (3′UTR) or GAPDH 3′UTR, in the presence of DAZL (blue bar) relative to an empty vector control (magenta bar). No stimulation is observed when the R115G DAZL mutant is overexpressed (green bar). Firefly luciferase signals were normalised to Renilla luciferase signals. Mean ± SEM, n = 4, **P < 0.01 (Student's t-test).
As the human DAZL-binding site has not been identified, we mutated instances of the consensus mouse Dazl-binding site, i.e. U(2–10)G/CU(2–10) (Venables et al., 2001) in these novel target 3′UTR's to elucidate whether DAZL is acting via this site in humans. To do this, we mutated the G base to an A in SMC1B and TEX11, which has previously been demonstrated to significantly weaken DAZL binding (Venables et al., 2001). However, upon using these mutant 3′UTR constructs in a luciferase reporter assay, we were unable to see a significant difference in the stimulation of translation in comparison to the wildtype 3′UTR (Supplementary Fig. S4). In addition, we repeated this assay using a mutant SMC1B 3′UTR-luciferase construct with deleted mouse Dazl-binding sites, and were also unable to observe any significant differences (Supplementary Fig. S4).
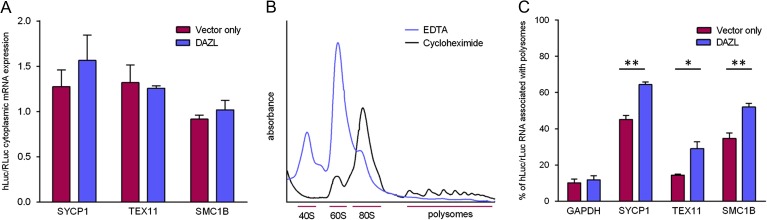
As mRNA translation and mRNA stability are often linked, and DAZL has been reported to mediate the stabilisation of HuB mRNA in germ cells of zebrafish embryos (Wiszniak et al., 2011), we examined the steady-state levels of luciferase mRNA in the cytoplasm to determine whether the change in luciferase activity in the presence of DAZL (Fig. 3) was a consequence of altered luciferase mRNA stability. For each of the target mRNAs that showed a dependence on DAZL for increased luciferase activity, there appeared to be no significant change in luciferase mRNA levels, as assessed by RT-qPCR, when compared to the control (Fig. 4A), indicating a specific effect on translation.
Figure 4.
(A) DAZL stimulates the translation of SYCP1, TEX11 and SMC1B. Quantitative RT-PCR of 3′UTR-luciferase expression shows that RNA stability is not affected by the presence of DAZL. Mean ± SEM, n = 4. (B) Polysome profiles. With the use of cycloheximide, polysomes are ‘frozen’ on RNA and can be seen as small bumps in the trace. Polysomes are no longer present with EDTA. (C) Percentage of 3′UTR-luciferase RNA associated with translating polysomes. In the presence of DAZL, the 3′UTRs of SYCP1 (synaptonemal complex protein-1), TEX11 (testis expressed 11) and SMC1B were significantly more ribosome bound than ribosome free, implicating DAZL in the translation of these targets. Mean ± SEM, n = 4–6. *P < 0.05, **P < 0.01 (Student's t-test).
We complemented the mRNA stability analysis with polysome profiling to study the translational state (i.e. ribosome loading) of novel target RNAs in the presence and absence of DAZL. We used the 3′UTR-luciferase reporter system with added cycloheximide to ‘freeze’ actively translating ribosomes on mRNAs (Fig. 4B). After ultracentrifugation to separate the cellular extracts into efficiently translated (ribosome bound) and untranslated (ribosome free) RNAs, we used RT-qPCR to calculate the proportion of luciferase RNA being translated. The proportion of SYCP1, TEX11 and SMC1B luciferase RNA that was ribosome bound was significantly higher in the presence of DAZL (Fig. 4C) while the control GAPDH 3′UTR mRNA was unaffected, suggesting this effect is specific to the novel target 3′UTRs and confirms the ability of DAZL to stimulate the translation of these RNAs.
Discussion
We adopted a global approach combining the immunoprecipitation of endogenous DAZL with RNA-sequencing technology to identify novel transcripts bound by DAZL in the human foetal ovary, and identified 764 potential RNA targets of DAZL. Among these targets were RNAs involved in synaptonemal complex formation, including SYCP1. More importantly, these data identified novel potential roles for DAZL function during early meiosis in the human oocyte, specifically in chromosome cohesion establishment, through regulation of targets including SMC1B, and in DNA recombination, via targets including TEX11. These are fundamental processes in determining oocyte quality and whose establishment in foetal life may support lifelong fertility. We report changes in the levels of a range of novel DAZL mRNA targets across gestation in the human foetal ovary, over the period including germ-cell mitosis, entry into meiosis and subsequent meiotic arrest and follicle formation. Furthermore, using translation assays we showed that DAZL stimulates translation of its novel targets SYCP1, TEX11 and SMC1B (representing different meiotic processes) via their 3′UTRs. This effect was not seen using a DAZL mutant with impaired RNA-binding activity, indicating that translation of these mRNAs is indeed regulated by DAZL.
In order to understand the role of DAZL in fertility, several groups have carried out investigations to identify in vivo mRNA targets of DAZL. These inquiries have mainly relied on microarray analysis with or without immunoprecipitation of recombinant/endogenous Dazl from wildtype and Dazl−/− male mice (Jiao et al., 2002; Reynolds et al., 2005). The limited information on the function of Dazl in the female indicates that it is important in later stages of oocyte maturation through zygote formation, where Dazl-dependent translation is necessary for spindle assembly, the Metaphase I–II transition and early embryo development (Chen et al., 2011). More recently, expression profiling of the Dazl−/− mouse foetal ovary has identified 104 genes expressed specifically in pre-meiotic to pachytene germ cells and the use of single-cell, single-transcript measurements has demonstrated that Dazl was required for the initial induction of nearly all identified meiotic prophase genes (Soh et al., 2015), but whether the mRNA transcripts of these genes were bound and/or translated by Dazl was not investigated. Both these studies were conducted in mouse, and no studies have addressed DAZL and its mRNA targets in human female meiosis.
In this investigation, the limited availability of fresh human foetal ovaries and the need to pool snap-frozen foetal ovaries to create a lysate for immunoprecipitation precluded the use of CLIP-Seq (UV cross-linking prior to immunoprecipitation and sequencing), currently the gold standard for large-scale identification of protein–RNA interactions (Murigneux et al., 2013). While the potential for post-lysis association of RNAs is a concern, Supplementary Fig. S2 shows no enrichment of the reference RNAs GAPDH and RPL32, or the granulosa cell marker FOXL2 with DAZL. Furthermore, our use of RNA in situ hybridisation with immunohistochemistry confirms that SMC1B is co-localised with DAZL in germ cells. However, we cannot exclude the possibility of post-lysis interactions between germ-cell expressed mRNAs containing a DAZL-binding site and DAZL protein. In addition, DAZL may bind different variations of its binding site with different affinities, and mRNAs containing low affinity sites may be missed in the absence of cross-linking.
Interestingly, only a few of the validated RNAs that have been previously identified as direct Dazl targets in mouse also appear in our human data set; namely Sycp3 and Tex14 but not Tex19, Mvh or Tet1. However, the majority of the previous work was done using testis, and the differences we observe may reflect species differences (mouse versus human), but also possibly reflect the different requirements of female and male germ cells. Indeed, there is some evidence of a sex-dependent function of DAZL: the Dazl−/− phenotype can be partially rescued by a human DAZL transgene in mouse testis, but not ovary (Vogel et al., 2002). Additionally, several DAZL allelic variants have been correlated with different phenotypic effects in men and women (Tung et al., 2006b). In their study, Soh et al. (2015) identified 104 genes whose expression at the mRNA level during prophase is Dazl-dependent (Soh et al., 2015). Intriguingly, a small number of these transcripts, including SYCP1, TEX11 and SMC1B, were also identified in our study. This indicates that full expression of a subset of genes may be dependent on DAZL at two distinct stages: early on for initial induction and accumulation of mRNA via a poorly understood DAZL activity (Lin et al., 2008; Soh et al., 2015) and at a later stage for efficient activation of translation (Fig. 3), as previously demonstrated for mouse Dazl targets (Reynolds et al., 2005; Reynolds et al., 2007; Chen et al., 2011).
The human DAZL-binding element has yet to be elucidated, therefore we carried out analysis of the effect of mutating the mouse Dazl-binding site in SMC1B and TEX11, but found that the specific mutations we introduced, based on work by Venables et al. (2001), did not affect the ability of DAZL to stimulate translation. However, a few RNAs identified by our DAZL immunoprecipitation did not contain the murine Dazl-binding site, one of these being SYCP3, a major component of the lateral elements of the synaptonemal complex in meiotic germ cells (Schramm et al., 2011). This is intriguing given the evidence implicating it as a direct Dazl target in mouse (Reynolds et al., 2007). In primordial germ cells derived from human embryonic stem cells, overexpression of DAZL resulted in the upregulation of SYCP3 and formation of synaptonemal complexes, consistent with SYCP3 being a DAZL target in human (Kee et al., 2009). Potentially, translational regulation of SYCP3 mRNA in humans occurs via a non-consensus DAZL-binding site located in the 3′UTR, or regulation may occur indirectly through the association of DAZL with a partner protein that is also an mRNA-specific binding protein. A known example is DAZL associated protein-1 (Tsui et al., 2000a; Smith et al., 2011). This may offer a possible explanation for the absence of a significant effect of our Dazl-binding site mutagenesis. While a detailed identification and analysis of the molecular interaction between DAZL and the novel targets identified here is outwith the scope of this study, both these lines of evidence indicate the need for such studies in the human.
These data confirm the involvement of DAZL in synaptonemal complex formation in humans through the translation of SYCP1, and evidence suggests this may have important implications for lifelong female fertility. SYCP1 is thought to function as a molecular framework, which enables other proteins to attach and complete synaptonemal complex assembly and progression of recombination (Schramm et al., 2011). Indeed, Sycp1 null mice of both sexes are infertile due to germ-cell loss through apoptosis despite normal axial element formation and chromosome alignment (de Vries et al., 2005). Importantly, these data also suggest novel roles for DAZL (which may be indirect) during early meiosis in the foetal oocyte, namely in establishment of chromosome cohesion through the regulation of SMC1B, and in DNA recombination via TEX11. There are no previous data implicating DAZL in the translation of RNAs with these functions, which are increasingly recognised to be key pathways that underpin lifelong oocyte quality. Recent data have highlighted the cohesin complex as a major contributor to age-related aneuploidy. Mice that are deficient in Smc1β show loss of cohesion and aneuploidy in their oocytes (Gilliland and Hawley, 2005; Hodges et al., 2005), while oocytes of older women have decreased SMC1β (Tsutsumi et al., 2014) show an increased inter-kinetochore distance (Duncan et al., 2012) and increasingly show premature predivision of sister chromatids (Handyside, 2012; Ottolini et al., 2015). Intriguingly, the cohesion complex is also implicated in DNA repair (Watrin and Peters, 2006). Similarly, mutations in TEX11 have been reported in infertile men with non-obstructive azoospermia (Yang et al., 2015). Meiotic arrest in these patients resembled the phenotype of Tex11−/− male mice (Yang et al., 2008), which have spermatocytes with asynapsed chromosomes and reduced cross-over formation, leading to apoptosis at the pachytene stage of Prophase I. Thus, these data suggest the potential involvement of DAZL in the subsequent risk of age-dependent aneuploidy through translational regulation of SMC1B in early meiosis, and as well as through its regulation of several other factors of established importance in synapsis and recombination.
Conclusion
The RNA-binding protein DAZL is well established as an important regulator of germ-cell development in both male and female although most data derive from studies of mouse testis. Here we have identified 764 potential RNA targets of DAZL in the human foetal ovary, and confirmed in translational assays that three of these, SYCP1, TEX11 and SMC1B are DAZL-dependent targets. Through regulation of these RNAs, we have uncovered novel potential functions for DAZL in the foetal oocyte, in establishment of chromosome cohesion and in DNA recombination, in addition to roles in synaptonemal complex formation. Consequently, we suggest DAZL has a key role in regulating fundamental processes that underpin oocyte quality, which may in turn influence a woman's fertility and reproductive lifespan.
Supplementary Material
Acknowledgements
We are grateful to Anne Saunderson and the staff of the Bruntsfield Suite, Royal Infirmary of Edinburgh for recruitment.
Authors’ roles
R.R. and R.A.A. designed experiments. R.R. carried out experiments and drafted the manuscript. All authors contributed to data interpretation and editing the manuscript, and its final approval.
Funding
Medical Research Council (G1100357 to R.A.A. and an intramural programme grant to I.R.A.).
Conflict of interest
The authors’ declare no conflict of interest.
References
- Anderson RA, Fulton N, Cowan G, Coutts S, Saunders PT. Conserved and divergent patterns of expression of DAZL, VASA and OCT4 in the germ cells of the human fetal ovary and testis. BMC Dev Biol 2007;7:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki H, Knapp C, Tsai P, Print C. GeneSetDB: a comprehensive meta-database, statistical and visualisation framework for gene set analysis. FEBS Open Bio 2012;2:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoloni L, Cazzadore C, Ferlin A, Garolla A, Foresta C. Lack of the T54A polymorphism of the DAZL gene in infertile Italian patients. Mol Hum Reprod 2004;10:613–615. [DOI] [PubMed] [Google Scholar]
- Chen HH, Welling M, Bloch DB, Munoz J, Mientjes E, Chen X, Tramp C, Wu J, Yabuuchi A, Chou YF et al. . DAZL limits pluripotency, differentiation, and apoptosis in developing primordial germ cells. Stem Cell Rep 2014;3:892–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Melton C, Suh N, Oh JS, Horner K, Xie F, Sette C, Blelloch R, Conti M. Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Genes Dev 2011;25:755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs AJ, Anderson RA. Experimental approaches to the study of human primordial germ cells. Methods Mol Biol 2012;825:199–210. [DOI] [PubMed] [Google Scholar]
- Clarke HJ. Post-transcriptional control of gene expression during mouse oogenesis. Results Probl Cell Differ 2012;55:1–21. [DOI] [PubMed] [Google Scholar]
- Collier B, Gorgoni B, Loveridge C, Cooke HJ, Gray NK. The DAZL family proteins are PABP-binding proteins that regulate translation in germ cells. EMBO J 2005;24:2656–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries FA, de Boer E, van den Bosch M, Baarends WM, Ooms M, Yuan L, Liu JG, van Zeeland AA, Heyting C, Pastink A. Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev 2005;19:1376–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman DM, Genest DR, Reijo Pera RA. Human DAZL1 encodes a candidate fertility factor in women that localizes to the prenatal and postnatal germ cells. Hum Reprod 1999;14:2531–2536. [DOI] [PubMed] [Google Scholar]
- Duncan FE, Hornick JE, Lampson MA, Schultz RM, Shea LD, Woodruff TK. Chromosome cohesion decreases in human eggs with advanced maternal age. Aging Cell 2012;11:1121–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill ME, Hu YC, Lin Y, Page DC. Licensing of gametogenesis, dependent on RNA binding protein DAZL, as a gateway to sexual differentiation of fetal germ cells. Proc Natl Acad Sci USA 2011;108:7443–7448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland WD, Hawley RS. Cohesin and the maternal age effect. Cell 2005;123:371–373. [DOI] [PubMed] [Google Scholar]
- Habermann B, Mi HF, Edelmann A, Bohring C, Backert IT, Kiesewetter F, Aumuller G, Vogt PH. DAZ (Deleted in AZoospermia) genes encode proteins located in human late spermatids and in sperm tails. Hum Reprod 1998;13:363–369. [DOI] [PubMed] [Google Scholar]
- Handyside AH. Molecular origin of female meiotic aneuploidies. Biochim Biophys Acta 2012;1822:1913–1920. [DOI] [PubMed] [Google Scholar]
- Haston KM, Tung JY, Reijo Pera RA. Dazl functions in maintenance of pluripotency and genetic and epigenetic programs of differentiation in mouse primordial germ cells in vivo and in vitro. PLoS One 2009;4:e5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges CA, Revenkova E, Jessberger R, Hassold TJ, Hunt PA. SMC1beta-deficient female mice provide evidence that cohesins are a missing link in age-related nondisjunction. Nat Genet 2005;37:1351–1355. [DOI] [PubMed] [Google Scholar]
- Jenkins HT, Malkova B, Edwards TA. Kinked beta-strands mediate high-affinity recognition of mRNA targets by the germ-cell regulator DAZL. Proc Natl Acad Sci U S A 2011;108:18266–18271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X, Trifillis P, Kiledjian M. Identification of target messenger RNA substrates for the murine deleted in azoospermia-like RNA-binding protein. Biol Reprod 2002;66:475–485. [DOI] [PubMed] [Google Scholar]
- Kee K, Angeles VT, Flores M, Nguyen HN, Reijo Pera RA. Human DAZL, DAZ and BOULE genes modulate primordial germ-cell and haploid gamete formation. Nature 2009;462:222–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Li JB, Xiao XF, Ma YF, Wang J, Liang XX, Zhao HX, Jiang F, Yao YQ, Wang XH. Altered DNA methylation patterns of the H19 differentially methylated region and the DAZL gene promoter are associated with defective human sperm. PLoS ONE 2013;8:e71215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Gill ME, Koubova J, Page DC. Germ cell-intrinsic and -extrinsic factors govern meiotic initiation in mouse embryos. Science 2008;322:1685–1687. [DOI] [PubMed] [Google Scholar]
- Lin Y, Page DC. Dazl deficiency leads to embryonic arrest of germ cell development in XY C57BL/6 mice. Dev Biol 2005;288:309–316. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods (San Diego, Calif) 2001;25:402–408. [DOI] [PubMed] [Google Scholar]
- Murigneux V, Saulière J, Roest Crollius H, Le Hir H. Transcriptome-wide identification of RNA binding sites by CLIP-seq. Methods 2013;63:32–40. [DOI] [PubMed] [Google Scholar]
- Navarro-Costa P, Nogueira P, Carvalho M, Leal F, Cordeiro I, Calhaz-Jorge C, Goncalves J, Plancha CE. Incorrect DNA methylation of the DAZL promoter CpG island associates with defective human sperm. Hum Reprod 2010;25:2647–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollinger R, Childs AJ, Burgess HM, Speed RM, Lundegaard PR, Reynolds N, Gray NK, Cooke HJ, Adams IR. Deletion of the pluripotency-associated Tex19.1 gene causes activation of endogenous retroviruses and defective spermatogenesis in mice. PLoS Genet 2008;4:e1000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottolini CS, Newnham LJ, Capalbo A, Natesan SA, Joshi HA, Cimadomo D, Griffin DK, Sage K, Summers MC, Thornhill AR et al. . Genome-wide maps of recombination and chromosome segregation in human oocytes and embryos show selection for maternal recombination rates. Nat Genet 2015;47:727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford HE, Meijer HA, de Moor CH. Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochim Biophys Acta 2008;1779:217–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C, Ruijter J, Deprez R, Moorman A. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 2003;339:62–66. [DOI] [PubMed] [Google Scholar]
- Reijo R, Lee TY, Salo P, Alagappan R, Brown LG, Rosenberg M, Rozen S, Jaffe T, Straus D, Hovatta O et al. . Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein gene. Nat Genet 1995;10:383–393. [DOI] [PubMed] [Google Scholar]
- Reynolds N, Collier B, Bingham V, Gray NK, Cooke HJ. Translation of the synaptonemal complex component Sycp3 is enhanced in vivo by the germ cell specific regulator Dazl. RNA 2007;13:974–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds N, Collier B, Maratou K, Bingham V, Speed RM, Taggart M, Semple CA, Gray NK, Cooke HJ. Dazl binds in vivo to specific transcripts and can regulate the pre-meiotic translation of Mvh in germ cells. Hum Mol Genet 2005;14:3899–3909. [DOI] [PubMed] [Google Scholar]
- Rilianawati, Speed R, Taggart M, Cooke HJ. Spermatogenesis in testes of Dazl null mice after transplantation of wild-type germ cells. Reproduction 2003;126:599–604. [DOI] [PubMed] [Google Scholar]
- Rosario R, Adams IR, Anderson RA. Is there a role for DAZL in human female fertility. Mol Hum Reprod 2016;22:377–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiu M, Cooke H. Y bind RNA for spermatogenesis. Int J Androl 1999;22:19–27. [DOI] [PubMed] [Google Scholar]
- Ruggiu M, Speed R, Taggart M, McKay SJ, Kilanowski F, Saunders P, Dorin J, Cooke HJ. The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature 1997;389:73–77. [DOI] [PubMed] [Google Scholar]
- Saunders PT, Turner JM, Ruggiu M, Taggart M, Burgoyne PS, Elliott D, Cooke HJ. Absence of mDazl produces a final block on germ cell development at meiosis. Reproduction 2003;126:589–597. [DOI] [PubMed] [Google Scholar]
- Schramm S, Fraune J, Naumann R, Hernandez-Hernandez A, Hoog C, Cooke HJ, Alsheimer M, Benavente R. A novel mouse synaptonemal complex protein is essential for loading of central element proteins, recombination, and fertility. PLoS Genet 2011;7:e1002088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrans-Stassen BH, Saunders PT, Cooke HJ, de Rooij DG. Nature of the spermatogenic arrest in Dazl-/- mice. Biol Reprod 2001;65:771–776. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Braun RE. Pathway to totipotency: lessons from germ cells. Cell 2006;127:891–904. [DOI] [PubMed] [Google Scholar]
- Smith RW, Anderson RC, Smith JW, Brook M, Richardson WA, Gray NK. DAZAP1, an RNA-binding protein required for development and spermatogenesis, can regulate mRNA translation. RNA 2011;17:1282–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh YQ, Junker JP, Gill ME, Mueller JL, van Oudenaarden A, Page DC. A gene regulatory program for meiotic prophase in the fetal ovary. PLoS Genet 2015;11:e1005531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka SS, Toyooka Y, Akasu R, Katoh-Fukui Y, Nakahara Y, Suzuki R, Yokoyama M, Noce T. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev 2000;14:841–853. [PMC free article] [PubMed] [Google Scholar]
- Teng YN, Lin YM, Lin YH, Tsao SY, Hsu CC, Lin SJ, Tsai WC, Kuo PL. Association of a single-nucleotide polymorphism of the deleted-in-azoospermia-like gene with susceptibility to spermatogenic failure. J Clin Endocrinol Metab 2002;87:5258–5264. [DOI] [PubMed] [Google Scholar]
- Tsui S, Dai T, Roettger S, Schempp W, Salido EC, Yen PH. Identification of two novel proteins that interact with germ-cell-specific RNA-binding proteins DAZ and DAZL1. Genomics 2000. a;65:266–273. [DOI] [PubMed] [Google Scholar]
- Tsui S, Dai T, Warren ST, Salido EC, Yen PH. Association of the mouse infertility factor DAZL1 with actively translating polyribosomes. Biol Reprod 2000. b;62:1655–1660. [DOI] [PubMed] [Google Scholar]
- Tsutsumi M, Fujiwara R, Nishizawa H, Ito M, Kogo H, Inagaki H, Ohye T, Kato T, Fujii T, Kurahashi H. Age-related decrease of meiotic cohesins in human oocytes. PLoS ONE 2014;9:e96710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung JY, Rosen MP, Nelson LM, Turek PJ, Witte JS, Cramer DW, Cedars MI, Pera RA. Variants in Deleted in AZoospermia-Like (DAZL) are correlated with reproductive parameters in men and women. Hum Genet 2006. a;118:730–740. [DOI] [PubMed] [Google Scholar]
- Tung JY, Rosen MP, Nelson LM, Turek PJ, Witte JS, Cramer DW, Cedars MI, Reijo-Pera RA. Novel missense mutations of the Deleted-in-AZoospermia-Like (DAZL) gene in infertile women and men. Reprod Biol Endocrinol 2006. b;4:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002;3:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables JP, Ruggiu M, Cooke HJ. The RNA-binding specificity of the mouse Dazl protein. Nucleic Acids Res 2001;29:2479–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel T, Speed RM, Ross A, Cooke HJ. Partial rescue of the Dazl knockout mouse by the human DAZL gene. Mol Hum Reprod 2002;8:797–804. [DOI] [PubMed] [Google Scholar]
- Watrin E, Peters JM. Cohesin and DNA damage repair. Exp Cell Res 2006;312:2687–2693. [DOI] [PubMed] [Google Scholar]
- Welling M, Chen HH, Munoz J, Musheev MU, Kester L, Junker JP, Mischerikow N, Arbab M, Kuijk E, Silberstein L et al. . DAZL regulates Tet1 translation in murine embryonic stem cells. EMBO Rep 2015;16:791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiszniak SE, Dredge BK, Jensen KB. HuB (elavl2) mRNA is restricted to the germ cells by post-transcriptional mechanisms including stabilisation of the message by DAZL. PLoS ONE 2011;6:e20773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Gell K, van der Heijden GW, Eckardt S, Leu NA, Page DC, Benavente R, Her C, Hoog C, McLaughlin KJ et al. . Meiotic failure in male mice lacking an X-linked factor. Genes Dev 2008;22:682–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Silber S, Leu NA, Oates RD, Marszalek JD, Skaletsky H, Brown LG, Rozen S, Page DC, Wang PJ. TEX11 is mutated in infertile men with azoospermia and regulates genome-wide recombination rates in mouse. EMBO Mol Med 2015;7:1198–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Liu JG, Hoja MR, Wilbertz J, Nordqvist K, Hoog C. Female germ cell aneuploidy and embryo death in mice lacking the meiosis-specific protein SCP3. Science 2002;296:1115–1118. [DOI] [PubMed] [Google Scholar]
- Zerbetto I, Gromoll J, Luisi S, Reis FM, Nieschlag E, Simoni M, Petraglia F. Follicle-stimulating hormone receptor and DAZL gene polymorphisms do not affect the age of menopause. Fertil Steril 2008;90:2264–2268. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.