Abstract
The aim of this study is to determine the contribution of 2 single nucleotide polymorphisms (SNPs) in thrombospondin 2 (THBS2) gene to the development of intervertebral disc degeneration (IDD) in a Chinese Han population.
We studied 138 patients with radiographically proven IDD and 136 healthy volunteers with no history of back problems. Magnetic resonance images (MRIs) were obtained for all the patients and controls. Image evaluation for IDD was performed to evaluate the severity of IDD. All patients and controls were genotyped for rs6422747 and rs6422748. Associations between genotypes and development of IDD were analyzed.
We found that 2 SNPs in the intron region of THBS2 gene (rs6422747 and rs6422748) were associated with susceptibility of IDD. However, they were not related with severity of IDD, including the total number of degenerative disc and level of IDD. G allele in both SNPs was associated with a higher risk of IDD.
The 2 SNPs (rs6422747 and rs6422748) in the THBS2 gene were associated with susceptibility of IDD but not severity of IDD in a Chinese Han population. Our results indicated that THBS2 gene polymorphisms might be the risk factors for IDD. More studies with larger sample size need to be perfected to make sure the functions of THBS2 gene polymorphisms in IDD development.
Keywords: Chinese Han population, intervertebral disc degeneration, single nucleotide polymorphism, thrombospondin 2
1. Introduction
Lumbar disc disease (LDD) mainly caused by intervertebral disc degeneration (IDD) is a common and frequently-occurring disease of orthopedics, including lumbar disc herniation, lumbar spinal stenosis, lumbar spondylolisthesis, lumbar instability, lumbar degenerative lateral convexity. Patients often suffer from lower back pain and lower limbs pain and numbness, which leads to poor quality of life. Early onset of IDD causes not only the loss of individual labor but also the consumption of a large number of medical resources.[1,2] The etiology and pathophysiology underlying IDD remain poorly understood.[1,3] Various environmental factors increase the risk of IDD, such as smoking, age, sex, and mechanical load. However, it is hypothesized that up to 74% of the etiology of IDD is due to heritability.[4,5] With an improved understanding of the genetic variants associated with IDD, it is potential to provide personalized care and targeted therapies.
Thrombospondin proteins (THBSs) are a class of glycoproteins those bind to collagen and tissue and participate in cell-to-cell and cell-to-matrix interactions during tissue development and repair.[6,7] They have a variety of different functions in the extracellular matrix (ECM) and regulate the level of matrix metalloproteinase-2 (MMP-2) and MMP-9, which play important roles in IDD. Mice lacking THBS2 showed abnormal curvature of the spine. THBS-2 knockout mice showed increased MMP-2 levels after injury.[8] It has been reported that some functional polymorphisms in THBS2 were associated with lumbar disk herniation and lumbar spinal stenosis in the Japanese and Korean population. However, few studies on the genetic association of single nucleotide polymorphisms (SNPs) with progression of IDD in the Chinese Han population have appeared.
In this study, we collected the peripheral blood of IDD patients who have been diagnosed and controls and genotyped the 2 THBS2 SNPs to determine the contribution of them to the progression of IDD in a Chinese Han population.
2. Methods
2.1. Study design
We recruited 138 IDD patients diagnosed in Qilu Hospital of Shandong University (Qingdao) between July 2015 and July 2016. All the patients met the following criteria: diagnosis of IDD by magnetic resonance imaging (MRI) according to the evaluation standard of Schneiderman et al[9]; a history of typical IDD symptoms, such as ischialgia, osphyalgia, lower limb numbness, and so on. Exclusion criteria included: the presence of a spinal tumor, infection, tuberculosis, or other disease; body mass index (BMI) ≥28 kg/m2 to eliminate the effect of weight; heavy smoker (smoking index ≥400) to eliminate the effect of smoking; the presence of genetic related diseases, such as ankylosing spondylitis, diabetes, hypertension. The control group consisted of 136 healthy volunteers who also met the exclusion criteria. IDD was excluded in all the healthy controls by MRI. Written informed consent was obtained from each participant in this study.
2.2. Ethics approval
The study protocol was approved by the Ethics Committee of Qilu Hospital of Shandong University.
2.3. Grading of IDD
MRI was performed using a 1.5 T scanner (Gyroscan Intera Achieva; Philips Healthcare, Amsterdam) and a Synergy Spine Coil (Philips Healthcare). Each subject was placed in the supine position with a cushion under both knees. T2-weighted fast spin-echo sagittal and axial images were obtained (TR/TE: 3500/120; slice thickness, 4 mm; slice gap, 0.4 mm). The level of IDD was assessed by evaluation standard of Schneiderman et al[9]: Level 0, normal height and signal intensity; Level 1, intermediate loss, speckled pattern, or heterogeneous decreased signal intensity; Level 2, marked loss, diffuse loss of signal; Level 3, absent signal (Fig. 1). All MR images were examined both clinically and radiologically to confirm the degree of IDD.
Figure 1.
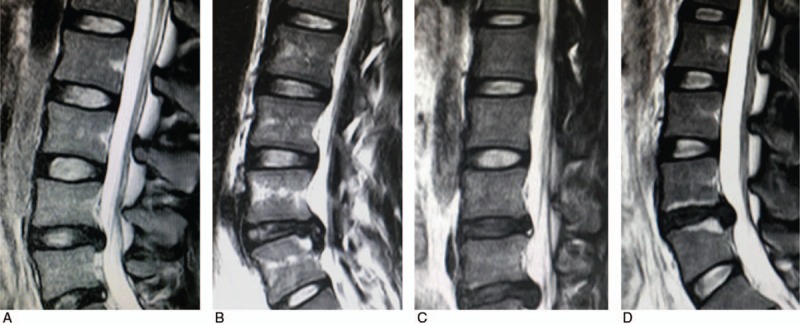
MRIs of different levels of intervertebral disc degeneration (IDD) according to evaluation standard of Schneiderman et al.[9] (A) Level 0 means normal height and signal intensity. (B) Level 1 means intermediate loss, speckled pattern or heterogeneous decreased signal intensity. (C) Level 2 means marked loss, diffuse loss of signal. (D) Level 3 means absent signal.
2.4. Genotyping
Genomic DNA for sequencing was isolated from peripheral blood cells of patients and controls using the Genomic DNA kit (Sangon Biotech, Shanghai, China) according to the manufacturer's instructions. Genotypes of SNPs (rs6422747 A>G, rs6422748 C>G) were determined by polymerase chain reaction (PCR) using sequence-specific primers to amplify fragments of 315 bp. Primer sequences were as follows: Forward: 5′-ACACCGTCATTGTCATCGTCATCAT-3′ and reverse: 5′-TCGTGAGTAGTGGTGACATAATTTC-3′. A 30 μL reaction system was used and contained 21.5 μL H2O, 1 μL dNTP, 1 μL PCR forward primer, 1 μL PCR reverse primer, 0.5 μL Taq DNA polymerase, 3 μL DNA template, and 2 μL MgCl2. The amplification protocol consisted of 1 cycle of Taq polymerase activation at 95 °C for 5 minutes, 35 cycles of denaturation at 94 °C for 60 seconds, annealing at 58 °C for 30 seconds, and extension at 72 °C for 30 seconds, and 1 cycle of a 10-minute extension at 72 °C. DNA sequencing was assessed by Sangon Biotech (Shanghai, China) to confirm the genotyping. Sequencing method was based on Sanger reaction. Sequence primer was: 5′- ACACCGTCATTGTCATCGTCATCAT-3′. BigDye Terminator v3.1 Cycle Sequencing Kit and 3730xl DNA Analyzer (Thermo Fisher Scientific, Waltham, MA) was used.
2.5. Statistical analysis
All the statistical analyses were performed using SPSS 18.0 software (SPSS, Inc., Chicago, IL). Hardy–Weinberg equilibriums (HWE) were assessed using a web tool (http://www.oege.org/software/hwe-mr-calc.shtml). Chi-square (χ2) test was used to examine the differences in allele frequencies between IDD patients and controls and the relationship between IDD grade and genotype information. The level of significance was P < .05.
3. Results
We recruited 138 IDD patients (71 [51.45%] female; mean ± SD age = 47.6 ± 12.3 years; mean ± SD body mass index [BMI] = 22.5 ± 2.7 kg/m2). The control group consisted of 136 healthy volunteers (66 [58.53%] female; mean ± SD age = 43.6 ± 15.3 years; mean ± SD BMI = 22.5 ± 3.5 kg/m2). In our study, we selected and genotyped 2 SNPs (rs6422747 and rs6422748), which showed minor allele frequencies >20% based on information in the HapMap database (International HapMap Consortium). These 2 selected SNPs covered most haplotypes and >80% of the THBS2 alleles with r2 values of ≥0.8.
3.1. The associations between rs6422747, rs6422748 polymorphisms in THBS2 gene and susceptibility of IDD
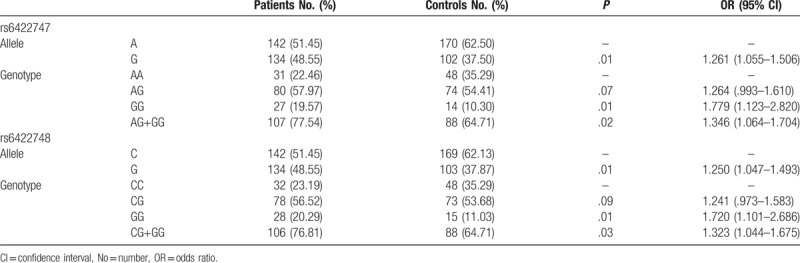
In total, 138 patients with IDD and 136 controls were involved in this study. HWE test showed P > .05 in both patient group and control group. Figure 2 showed the representative sequencing data of SNPs in THBS2 gene. Table 1 showed the distribution of rs6422747 and rs6422748 polymorphisms. For rs6422747, a higher percentage of AG, GG, and AG+GG genotypes were observed in the patients compared with the controls (P = .07, .01, and .02, respectively). G allele might be a risk factor for IDD susceptibility (P = .01). G allele was associated with a higher risk of IDD (Odds ratio [OR], 1.261; 95% confidence interval [CI], 1.055–1.506). For rs6422748, the results were similar. Based on the above, our study indicated that the G allele in rs6422747 and rs6422748 might be the risk factors for IDD.
Figure 2.
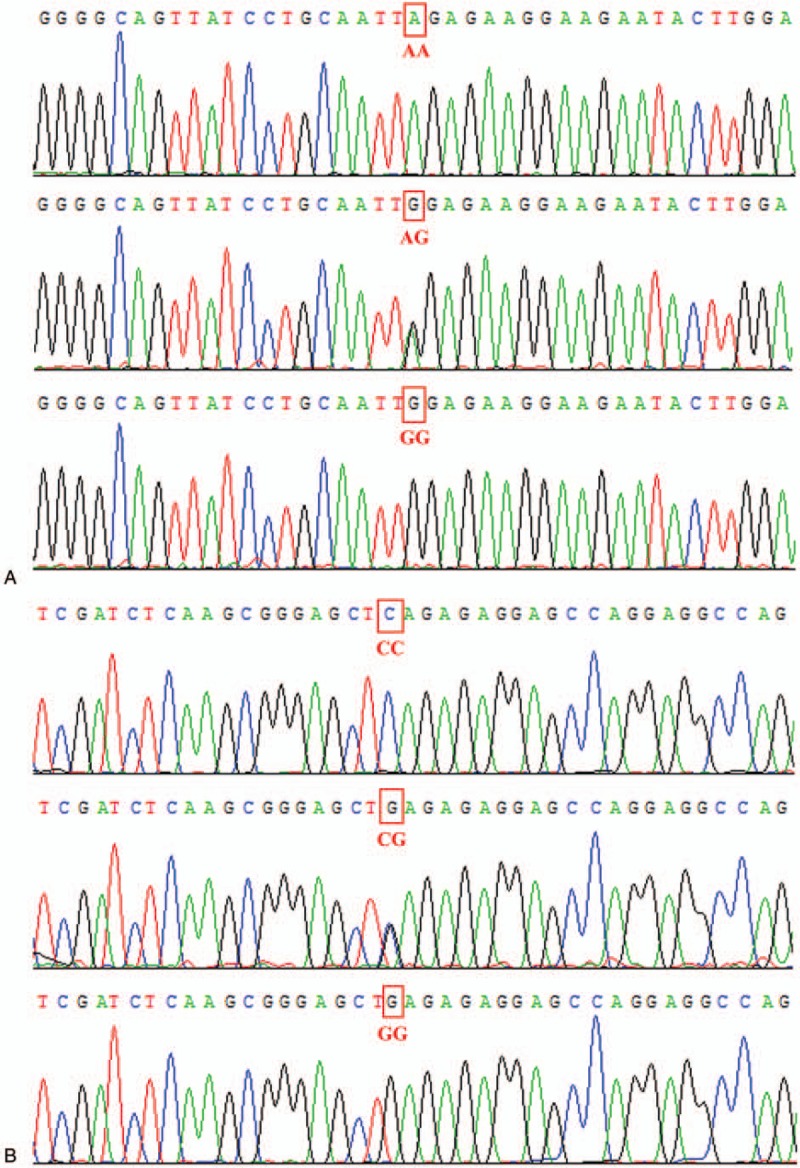
The representative sequencing data of rs6422747 and rs6422748 in THBS2 gene. (A) AA, AG, and GG genotypes of rs6422747. (B) CC, CG, and GG genotypes of rs6422748.
Table 1.
Distribution of rs6422747 and rs6422748 polymorphisms in THBS2 gene in intervertebral disc degeneration patients and controls.
3.2. The associations between rs6422747, rs6422748 polymorphisms in THBS2 gene and severity of IDD
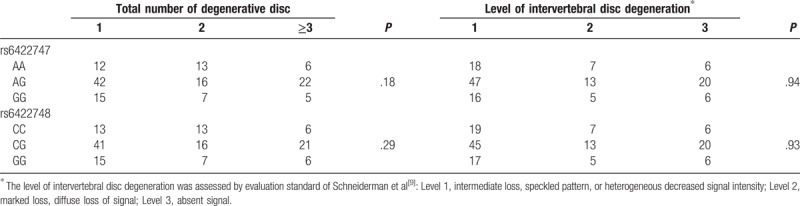
The level of IDD was assessed by evaluation standard of Schneiderman et al.[9] The associations between rs6422747, rs6422748 polymorphisms, and total number of degenerative disc, level of IDD were shown in Table 2. The results showed that the 2 SNPs were not associated with total number of degenerative disc and level of IDD (P > .05).
Table 2.
Association between rs6422747, rs6422748 polymorphisms and level of intervertebral disc degeneration.
4. Discussion
Over 80% of all people will experience lower back pain in their lifetime.[2–4] About 5% of people will suffer from IDD with sciatica as the main manifestation.[10] Genetic factors play an important role in the progression of IDD. Some research studies have investigated genes with SNPs associated with IDD and their proteins. The genes with SNPs and their proteins cover a variety of protein system, including structural protein (collagens, aggrecan, etc.),[11–13] structural support protein (carbohydrate sulfotransferase, vitamin D receptor),[14,15] cytokines (Interleukin-1α, Interleukin-6),[16] extracellular matrix-degrading enzymes (matrix metalloproteinases),[17,18] apoptotic factors (TNF [tumor necrosis factor-related apoptosis-inducing ligand, death receptor 4, caspase-9],[19,20] growth factors [growth differentiation factor 5],[19] and pain mediators [cyclooxygenase 2, catechol-O-methyltransferase]).[12,21] Understanding of genetic polymorphisms associated with IDD can provide patients with personalized and targeted therapeutics.
THBS2 belongs to the thrombospondin family. It is a disulfide-linked homotrimeric glycoprotein that mediates cell-to-cell and cell-to-matrix interactions.[6,7] Protein encoded by THBS2 gene has been shown to function as a potent inhibitor of angiogenesis. It can suppress the proliferation and migration of endothelial cell and inhibit the activity of many pro-angiogenic factors.[22] THBS2 regulated the expression of MMP-2 and MMP-9, which played important roles in IDD.[23] It has been reported that 2 polymorphisms of THBS2 were associated with degrees of Kellgren–Lawrence classification. The polymorphism of CD36 which was thrombospondin receptor was related to osteophyte and height of intervertebral disc.[24] All these studies indicated that THBS2 might play important roles in the progression of IDD.
In our study we found that 2 SNPs, rs6422747 and rs6422748, in the intron region of THBS2 gene were associated with susceptibility of IDD but not severity of IDD, including the total number of degenerative disc and level of IDD. It is worth mentioning that in our study we chose evaluation standard of Schneiderman et al[9] rather than Pfirrmann classification[25] or modified one.[26] Pfirrmann classification could apply well to young IDD patients. However, for >80% elder IDD patients it could not make a distinction between level C and level D. Modified Pfirrmann classification divided IDD to 8 levels. It was too complex in the clinical application. So, we chose standard of Schneiderman to evaluate the severity of IDD. The functions of rs6422747 and rs6422748 in progression of lumbar spinal stenosis (LSS) have been studied in the Korean population.[27] However, it was reported that the 2 SNPs might play a protective role against LSS development which was inconsistent with our results. This might be related to some reasons. Firstly, the study objects of the 2 studies were different. In our study, we chose a Chinese Han population with IDD while the published paper researched the Korean population with LSS. Different nationalities and not exactly the same disease could lead to different results. In addition the numbers of patients in our study and Korean study were a little small (138 vs 148). More studies with larger sample size or meta-analysis need to be perfected to make sure the functions of the 2 SNPs.
rs6422747 and rs6422748 both located in the intron region of THBS2 gene. The introns used to be regarded as nonsense segments and did not participate in encoding the protein. However, Mattick[28] indicated that introns could function as transposable elements and nuclear introns derived from self-splicing group II introns which then evolved in partnership with the spliceosome. The introns maybe were the exons for other specified genes. As a result, the mutations in the introns might have an effect on the mRNA splicing and processing and contribute to the expression of the genes as well as the function of the protein. Our study showed that the 2 SNPs in the intron region of THBS2 gene might be associated with IDD, which might provide some clues for researches of intron functions in THBS2 gene.
In summary we found that 2 SNPs (rs6422747 and rs6422748) in the THBS2 gene were associated with susceptibility of IDD but not severity of IDD in a Chinese Han population. Our results indicated that THBS2 gene polymorphisms might be the risk factors for IDD. More studies with larger sample size need to be perfected to make sure the functions of THBS2 gene polymorphisms in IDD development.
Footnotes
Abbreviations: BMI = body mass index, HWE = Hardy–Weinberg equilibriums, IDD = intervertebral disc degeneration, LDD = lumbar disc disease, LSS = lumbar spinal stenosis, MRI = magnetic resonance images, SNP = single nucleotide polymorphisms.
Fundings: This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of Interest: None.
References
- [1].Kepler CK, Ponnappan RK, Tannoury CA, et al. The molecular basis of intervertebral disc degeneration. Spine J 2013;13:318–30. [DOI] [PubMed] [Google Scholar]
- [2].Guo TM, Liu M, Zhang YG, et al. Association between Caspase-9 promoter region polymorphisms and discogenic low back pain. Connect Tissue Res 2011;52:133–8. [DOI] [PubMed] [Google Scholar]
- [3].Kadow T, Sowa G, Vo N, et al. Molecular basis of intervertebral disc degeneration and herniations: what are the important translational questions? Clin Orthop Relat Res 2015;473:1903–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Seki S, Kawaguchi Y, Chiba K, et al. A functional SNP in CILP, encoding cartilage intermediate layer protein, is associated with susceptibility to lumbar disc disease. Nat Genet 2005;37:607–12. [DOI] [PubMed] [Google Scholar]
- [5].Janeczko L, Janeczko M, Chrzanowski R, et al. The role of polymorphisms of genes encoding collagen IX and XI in lumbar disc disease. Neurol Neurochir Pol 2014;48:60–2. [DOI] [PubMed] [Google Scholar]
- [6].Bornstein P. Thrombospondins: structure and regulation of expression. FASEB J 1992;6:3290–9. [DOI] [PubMed] [Google Scholar]
- [7].Bornstein P, Sage EH. The thrombospondins. Methods Enzymol 1994;245:62–85. [DOI] [PubMed] [Google Scholar]
- [8].Hirose Y, Chiba K, Karasugi T, et al. A functional polymorphism in THBS2 that affects alternative splicing and MMP binding is associated with lumbar-disc herniation. Am J Hum Genet 2008;82:1122–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Schneiderman G, Flannigan B, Kingston S, et al. Magnetic resonance imaging in the diagnosis of disc degeneration: correlation with discography. Spine (Phila Pa 1976) 1987;12:276–81. [DOI] [PubMed] [Google Scholar]
- [10].Annunen S, Paassilta P, Lohiniva J, et al. An allele of COL9A2 associated with intervertebral disc disease. Science 1999;285:409–12. [DOI] [PubMed] [Google Scholar]
- [11].Toktas ZO, Eksi MS, Yilmaz B, et al. Association of collagen I, IX and vitamin D receptor gene polymorphisms with radiological severity of intervertebral disc degeneration in Southern European Ancestor. Eur Spine J 2015;24:2432–41. [DOI] [PubMed] [Google Scholar]
- [12].Rajasekaran S, Kanna RM, Senthil N, et al. Genetic susceptibility of lumbar degenerative disc disease in young Indian adults. Eur Spine J 2015;24:1969–75. [DOI] [PubMed] [Google Scholar]
- [13].Gu J, Guan F, Guan G, et al. Aggrecan variable number of tandem repeat polymorphism and lumbar disc degeneration: a meta-analysis. Spine (Phila Pa 1976) 2013;38:E1600–7. [DOI] [PubMed] [Google Scholar]
- [14].Song YQ, Karasugi T, Cheung KM, et al. Lumbar disc degeneration is linked to a carbohydrate sulfotransferase 3 variant. J Clin Invest 2013;123:4909–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Vieira LA, De Marchi PL, dos Santos AA, et al. Analysis of FokI polymorphism of vitamin D receptor gene in intervertebral disc degeneration. Genet Test Mol Biomarkers 2014;18:625–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Eskola PJ, Kjaer P, Sorensen JS, et al. Gender difference in genetic association between IL1A variant and early lumbar disc degeneration: a three-year follow-up. Int J Mol Epidemiol Genet 2012;3:195–204. [PMC free article] [PubMed] [Google Scholar]
- [17].Zhang Y, Gu Z, Qiu G. Association of the polymorphism of MMP2 with the risk and severity of lumbar disc degeneration in the Chinese Han population. Eur Rev Med Pharmacol Sci 2013;17:1830–4. [PubMed] [Google Scholar]
- [18].Zhang J, Sun X, Liu J, et al. The role of matrix metalloproteinase 14 polymorphisms in susceptibility to intervertebral disc degeneration in the Chinese Han population. Arch Med Sci 2015;11:801–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mu J, Ge W, Zuo X, et al. Analysis of association between IL-1beta, CASP-9, and GDF5 variants and low-back pain in Chinese male soldier: clinical article. J Neurosurg Spine 2013;19:243–7. [DOI] [PubMed] [Google Scholar]
- [20].Xu S, Liang T, Li S. Correlation between polymorphism of TRAIL gene and condition of intervertebral disc degeneration. Med Sci Monit 2015;21:2282–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Martirosyan NL, Patel AA, Carotenuto A, et al. Genetic alterations in intervertebral disc disease. Front Surg 2016;3:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Abu El-Asrar AM, Nawaz MI, Ola MS, et al. Expression of thrombospondin-2 as a marker in proliferative diabetic retinopathy. Acta Ophthalmol 2013;91:e169–77. [DOI] [PubMed] [Google Scholar]
- [23].Gruber HE, Bornstein P, Sage EH, et al. Disruption of the thrombospondin-2 gene alters the lamellar morphology but does not permit vascularization of the adult mouse lumbar disc. Arthritis Res Ther 2008;10:R96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mern DS, Beierfubeta A, Fontana J, et al. Imbalanced protein expression patterns of anabolic, catabolic, anti-catabolic and inflammatory cytokines in degenerative cervical disc cells: new indications for gene therapeutic treatments of cervical disc diseases. PLoS One 2014;9:e96870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pfirrmann CW, Metzdorf A, Zanetti M, et al. Magnetic resonance classification of lumbar intervertebral disc degeneration. Spine (Phila Pa 1976) 2001;26:1873–8. [DOI] [PubMed] [Google Scholar]
- [26].Griffith JF, Wang YX, Antonio GE, et al. Modified Pfirrmann grading system for lumbar intervertebral disc degeneration. Spine (Phila Pa 1976) 2007;32:E708–12. [DOI] [PubMed] [Google Scholar]
- [27].Hyun SJ, Park BG, Rhim SC, et al. Progression of lumbar spinal stenosis is influenced by polymorphism of thrombospondin 2 gene in the Korean population. Eur Spine J 2014;23:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mattick JS. Introns: evolution and function. Curr Opin Genet Dev 1994;4:823–31. [DOI] [PubMed] [Google Scholar]


