Abstract
Gibberellin (GA) 20-oxidase (GA 20-ox) and GA 3β-hydroxylase (GA 3β-hy) are enzymes that catalyze the late steps in the formation of active GAs, and are potential control points in the regulation of GA biosynthesis by light. We have investigated the photoregulation of the GA 20-ox and GA 3β-hy transcript levels in pea (Pisum sativum L.). The GA 20-ox transcript level was higher in light-grown seedlings than in etiolated seedlings, whereas GA 3β-hy mRNA accumulation was higher in etiolated seedlings. However, transfer of etiolated seedlings to light led to a 5-fold increase in the expression of both transcripts 4 h after transfer. GA 20-ox mRNA accumulation is regulated by both phytochromes A and B. Transfer to light also resulted in a 6-fold decrease in GA1 levels within 2 h. These results suggest that the light-induced drop in GA1 level is not achieved through regulation of GA 20-ox and GA 3β-hy mRNA accumulation. The application of exogenous GA1 to apical buds of etiolated seedlings prior to light treatments inhibited the light-induced accumulation of both GA 20-ox and GA 3β-hy mRNA, suggesting that negative feedback regulation is an important mechanism in the regulation of GA 20-ox and GA 3β-hy mRNA accumulation during de-etiolation of pea seedlings.
Gibberellins (GAs) constitute a large group of natural tetracyclic diterpenoids. Biologically active GAs have a profound effect on plant growth and development (Reid and Howell, 1995). Numerous studies with GA-deficient mutants have shown that GAs are involved in processes such as stem elongation, leaf expansion, photoperiodic induction of flowering, flower development, seed development, and germination (Crozier, 1983; Swain et al., 1997).
Genes encoding GA biosynthetic enzymes have been cloned from several plant species (Hedden and Kamiya, 1997). GA 20-oxidase (GA 20-ox) is a multifunctional enzyme that catalyzes the sequential oxidation of GA53 to GA20 (Fig. 1). The product of the reaction catalyzed by GA 20-ox, GA20, is then hydroxylated by GA 3β-hydroxylase (GA 3β-hy) to produce the active GA, GA1. GA 20-ox is encoded by a small multigene family whose members are differentially regulated (Phillips et al., 1995; García-Martínez et al., 1997; Rebers et al., 1999). Two GA 20-ox cDNAs have been cloned from pea. One gene is expressed in vegetative tissues (Martin et al., 1996) and during early seed and pod development (García-Martínez et al., 1997; van Huizen et al., 1997), whereas the other gene is expressed only in immature embryos (Lester et al., 1996; Ait-Ali et al., 1997). A gene encoding a GA 3β-hy (LE) has also been cloned from pea (Lester et al., 1997; Martin et al., 1997; Fig. 1). The LE gene clearly alters stem elongation in both etiolated and de-etiolated pea (Reid, 1988). The LE transcript accumulates mainly in vegetative tissues, including the shoot and expanded internodes, although low transcript levels are also detected in leaves (Martin et al., 1997). Increasing evidence from several species suggests that GA 20 oxidation is regulated by biologically active GA through a negative feedback mechanism (Phillips et al., 1995; Martin et al., 1996). In pea, exogenous application of biologically active GA decreases not only the GA 20-ox transcript levels, but also the GA20 content (Martin et al., 1996). GA 3β-hy mRNA was demonstrated to be subject to similar negative feedback regulation in both Arabidopsis and pea (Chiang et al., 1995; Martin et al., 1996; Hedden and Kamiya, 1997; Yamaguchi et al., 1998; Ross et al., 1999).
Figure 1.
Simplified 13-hydroxylation pathway of GA biosynthesis in vegetative tissues of pea. The steps affected by mutations at the LS, LH, NA, LE, and SLN loci are indicated. GGDP, Geranylgeranyldiphosphate; CDP, copalyl diphosphate.
Several processes that are regulated by GAs (e.g. germination, stem elongation, leaf development, and flowering) are also controlled by light. This has stimulated an interest in the possibility that light responses, and in particular responses to the red- and far-red-light-absorbing phytochrome photoreceptor family, may be mediated in part through the GA signaling pathway (Hedden and Kamiya, 1997). Indeed, several aspects of GA biosynthesis and function appear to be regulated by light. For example, the daily light fluctuations of a photoperiod influence GA biosynthesis (Zeevaart and Gage, 1993; Foster and Morgan, 1995; Wu et al., 1996). In addition, Toyomasu et al. (1992) reported a decrease in GA1 content and an increase in GA20 content during de-etiolation of lettuce seedlings. These results suggest that light regulates the last steps of GA biosynthesis. In contrast, analyses of GA responsiveness in wild-type and phytochrome B (phyB)-deficient mutants of pea (Weller et al., 1994), cucumber (López-Juez et al., 1995), and Arabidopsis (Reed et al., 1996) have indicated that phyB affects responsiveness to GA1 and has little if any effect on the level of active GAs in light-grown plants, although other aspects of GA biosynthesis may be modified (Weller et al., 1994). Genetic interactions between GA-related and phytochrome-deficient mutants clearly indicate that an intact GA biosynthesis and signal transduction system is required for full expression of phytochrome-deficient phenotypes (Peng and Harberd, 1997). The relationship between GAs and light signal transduction appears to be a complex one and clearly requires further study before it is fully understood.
This study examines the light regulation of GA 20-ox and GA 3β-hy transcript accumulation in vegetative tissues of pea seedlings. Since our interest was primarily in seedling de-etiolation, we restricted our analysis of GA 20-ox to transcripts of the gene expressed in vegetative tissues. We also quantified endogenous GA levels during de-etiolation. We found that the levels of both the GA 20-ox and the GA 3β-hy transcript change with seedling age, are light regulated, and exhibit strong tissue specificity.
MATERIALS AND METHODS
Plant Material
Pea (Pisum sativum L. cv Alaska) seed was purchased from Yukijirushi Subyo (Hokkaido, Japan). Seeds of the pea cv Torsdag, the phyB-deficient mutant lv (Weller et al., 1995), and the phyA-deficient mutant fun1-1 (Weller et al., 1997) were originally obtained from the mutant collection maintained at the Department of Plant Science, University of Tasmania. In this study, we used the lv-5 allele, which is stronger than previously described lv alleles (J.L. Weller, unpublished results).
cv Alaska seeds were surfaced-sterilized in 10% (v/v) commercial bleach for 10 min, rinsed with water, and imbibed for 4 h before sowing. Imbibed cv Alaska seeds and dry cv Torsdag seeds were sown at 4 pm in moist vermiculite and placed in custom-designed growth chambers (Koitotron, Koito, Tokyo). Seedlings were maintained at 25°C in constant darkness or light. Plants were watered as necessary. Light treatments (transfer from darkness to light and red-light pulses) were given at noon. Tissue for Figure 2 was harvested at noon. Stems (epicotyl between the point of cotyledon attachment and the bud) and buds were harvested from etiolated seedlings. The first two nodes of the seedlings used in this study do not produce expanded leaves. Therefore, the harvested portion of dark-grown epicotyls consisted of two nodes and three internodes. Seedlings that had been grown in constant light were divided into stem (cotyledon attachment to below the point of attachment of the second node), first leaf (second node to below the point of attachment of the fourth node), and apical bud (fourth node and above).
Figure 2.
GA 20-ox and GA 3β-hy transcript accumulation in light- and dark-grown seedlings. Seedlings were grown in constant light (L) or darkness (D) for 6 d. Buds (Bd) and stems (St) were harvested from dark-grown seedlings. Apices (Ap), first expanded nodes (Lf), and stems were harvested from light-grown seedlings. Total RNA was prepared and 5 (CAB) or 30 (GA 20-ox and GA 3-βhy) μg per lane was used for RNA-blot analysis. Blots were hybridized with probes for chlorophyll a/b-binding protein (CAB) or GA 20-oxidase (GA 20-ox) and GA 3β-hydroxylase (GA 3β-hy).
White light (70 μmol m−2 s−1) was provided by fluorescent tubes (FL20SW, National, Tokyo). Red light (30 μmol m−2 s−1) was provided by fluorescent tubes (FL20S.BRF, Toshiba, Tokyo) filtered with acrylic film (Shinkolite A102, Mitsubishi Rayon, Tokyo); far-red light (0.15 μmol m−2 s−1) was also provided by fluorescent tubes (FL20S.FR-74, Toshiba) filtered with acrylic film (Dreaglass 102, Asahikasei, Tokyo).
RNA Hybridization Analysis
Total RNA was isolated with TRIzol Reagent (GIBCO-BRL, Grand Island, NY) according to the manufacturer's protocol. RNA was glyoxylated and transferred to Hybond N+ membranes (Amersham, Little Chalfont, UK) as previously described (Frances et al., 1992). Membranes were hybridized overnight at 65°C in 50% (w/v) deionized formamide, 1× Denhardt's solution, 1% (w/v) SDS, 100 mm NaH2PO4 pH 7.0, 5× SSPE (1× SSPE is 0.18 m NaCl, 10 mm NaH2PO4, and 1 mm EDTA, pH 7.4), and 0.4 mg/mL of calf-liver RNA. Hybridization probes were prepared with an in vitro transcription system (Riboprobe, Promega, Madison, WI) from plasmids pAB96 (CAB; Coruzzi et al., 1983), pC20 (a Bluescript plasmid containing a fragment from the GA20-oxidase cDNA that was subcloned from ps27-12; García-Martínez et al., 1997), and GA 3-OH cDNA (Martin et al., 1997). Stringency washes (0.1× SSPE, 0.1% [w/v] SDS) were performed at 68°C. The hybridization signal was imaged and quantitated with a phosphor imager (BAS2000, Fuji Photo Film, Tokyo). Membranes were rehybridized with an oligo complementary to the 18S rRNA as described by Gallo-Meagher et al. (1992) to control for variations in loading and transfer efficiency.
All of the experiments were repeated at least three times. Results of all repetitions were very similar and one representative blot hybridization was chosen for the figures. Quantitated data were normalized to the maximum value of each data set and are presented as an average of data from at least three repeats. Error bars are the se of the mean.
Quantitative Analysis of Endogenous Levels of GA
Shoots (apical bud and stem) were harvested from 6-d-old seedlings. The fresh weight was recorded and the tissue immersed in methanol. Pea shoots were homogenized in 80% (v/v) aqueous methanol with a blender (Nihonseiki Kaisha, Tokyo). GA extractions and quantifications were performed as previously described (Gawronska et al., 1995; Furukawa et al., 1997). Data are presented as an average of three independent experiments. Error bars are the se of the mean.
RESULTS AND DISCUSSION
Expression of Both GA 20-ox and GA 3β-hy Is Organ Specific and Differs between Dark- and Light-Grown Seedlings
Previous investigations of the relationship between GAs and light in pea have compared light- and dark-grown plants (Ross et al., 1992) or plants grown under different irradiances (Gawronska et al., 1995) or qualities (Sponsel, 1986). Results from these studies have suggested that the level of GA1 is not significantly modified by the light environment. However, the control by light of specific steps of the GA pathway in pea has not yet been examined. To determine if the expression of GA 20-ox and GA 3β-hy might be light regulated, we initially compared transcript abundance in light- and dark-grown seedlings. In addition, RNA was isolated from different organs (the buds and stems of dark-grown seedlings and the apex, first leaf, and stem of light-grown seedlings) to determine the distribution of expression. As a control to show induction and the expression pattern of light-regulated genes, the accumulation of transcripts encoding chlorophyll a/b-binding protein (CAB) was also monitored. Light regulation of the CAB gene family is well characterized (for review, see Thompson and White, 1991). As expected, CAB transcripts accumulated to much higher levels in light-grown than in dark-grown seedlings (Fig. 2). CAB transcript levels were highest in leaves and apices, although appreciable amounts of transcript also accumulated in light-grown stems.
The level of the GA 20-ox transcript was low but detectable in stems and buds of etiolated seedlings (Fig. 2). The levels in leaves and apical buds of light-grown seedlings were substantially higher than in the buds of etiolated seedlings, suggesting that GA 20-ox expression is subject to strong photoregulation in these organs. Transcript levels were affected in the stem but the change in level was not as dramatic as in the bud (Fig. 2).
The high level of GA 20-ox transcript in light-grown plants relative to dark-grown plants (Fig. 2) is consistent with the observation that the level of GA20 (the product of GA20-oxidation) is also much higher in light-grown plants (Weller et al., 1994). Moreover, the tissue distribution of the GA 20-ox transcript (i.e. high in apex and leaf and lower in stem) correlates with that of GA20 in light-grown seedlings (Smith et al., 1992). Similar conclusions have been reached for the GA 20-ox transcript levels in spinach and Arabidopsis (Wu et al., 1996; Xu et al., 1997). These correlations suggest that GA 20-ox transcript accumulation is an important regulatory point in GA biosynthesis.
In contrast to the situation for GA 20-ox, GA 3β-hy transcript levels were higher in etiolated seedlings than in light-grown seedlings (Fig. 2). This suggests that GA 3β-hy mRNA accumulation may also be regulated by light, but in an opposite and much weaker manner than GA 20-ox transcript accumulation. The organ dependence of GA 3β-hy expression differed from that observed for GA 20-ox: The GA 3β-hy transcript level in both light- and dark-grown plants was much higher in the stem than in the apex or leaf (Fig. 2; Martin et al., 1997). However, unlike GA20-ox, the GA 3β-hy transcript level cannot be directly related to the level of GA1 since it shows a marked reduction in light-grown plants (Fig. 2), whereas the GA1 level is at best only slightly reduced (Weller et al., 1994; Gawronska et al., 1995). This may be due to the rapid turnover of GA1 in etiolated seedlings than in light-grown seedlings (Sponsel, 1986). A possible explanation of these observations may be that GA 3β-hy expression is subject to post-transcriptional regulation.
Transfer from Dark to Light Induces Accumulation of Both GA 20-ox and GA 3β-hy Transcripts
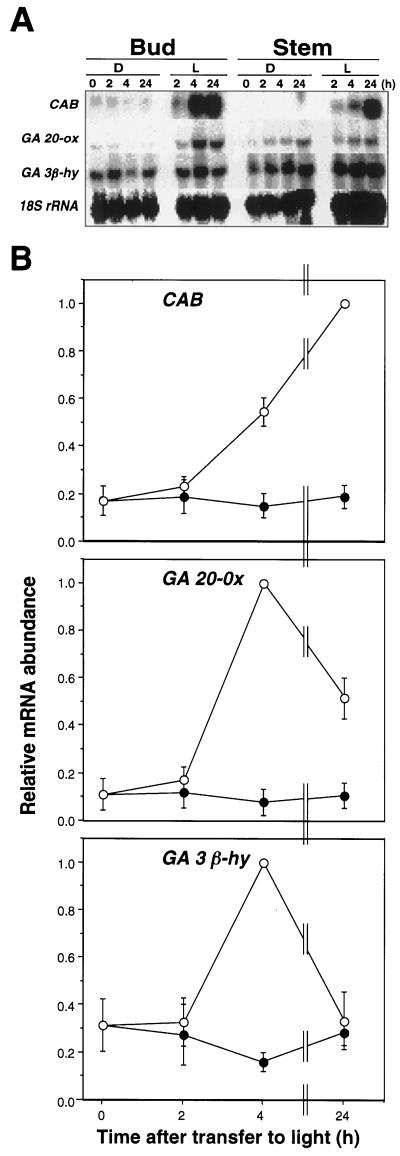
In the previous experiment, light-grown seedlings were grown from emergence in continuous light and differed substantially in morphology from etiolated seedlings. Thus, it is possible that the differences in transcript accumulation shown in Figure 2 were due to differences in the developmental states of the seedlings rather than to the light conditions. Since the change in growth rate of etiolated pea seedlings after exposure to light occurs rapidly (Behringer et al., 1990) and prior to any visible change in morphology, we focused our effort on de-etiolating seedlings. We transferred 6-d-old dark-grown seedlings to continuous light and monitored levels of the same three transcripts 0, 2, 4, and 24 h after transfer. Figure 3 shows that a 4-h exposure to light resulted in a large increase in the level of each of the three transcripts in apical buds.
Figure 3.
GA 20-ox and GA 3β-hy transcript accumulation during de-etiolation. Seedlings were grown in constant darkness for 6 d before transfer to constant light (L). Control seedlings were maintained in constant darkness (D). A, Buds and stems were harvested at 0, 2, 4, and 24 h after transfer to light and RNA-blot analysis was performed as described in Figure 2. As a loading control, blots were rehybridized with an oligo to the 18S rRNA (18S rRNA). B, RNA-blot analysis of buds were harvested 0, 2, 4, and 24 h after transfer to light, and hybridization signals were quantitated with a phosphor imager. Each data set was normalized to the maximum signal. The experiment was repeated at least three times, and bars indicate the se of the mean. ○, Constant light; ●, constant darkness.
The accumulation of CAB and GA 20-ox transcript is consistent with the results from dark- and light-grown seedlings in Figure 2. However, the accumulation of GA 3β-hy transcript after transfer was unexpected, because in the previous experiment, the expression of this gene was lower in light-grown than in dark-grown seedlings (Fig. 2). This result shows that although stimulated by a short period of exposure to light, GA 3β-hy transcript accumulation is inhibited by longer periods of exposure. This may be reflected in the fact that after exposure of dark-grown seedlings to continuous white light for 24 h (Fig. 3B), GA 3β-hy transcript levels returned to the level of the dark control, whereas GA 20-ox transcript levels were still 4-fold higher than the dark control. Relative to the etiolated control seedlings, illumination for 4 h did not substantially alter the morphology of apical buds, although 24 h after transfer, buds were visibly larger and much greener than those of etiolated control plants (data not shown). Therefore, the marked light-induced changes in transcript accumulation occured prior to visible changes in the developmental state of the seedling. This treatment regime therefore provides an appropriate and convenient system for more detailed investigations of the regulation of these transcript levels.
The effect of light on GA 20-ox and GA 3β-hy transcript levels was much stronger in apical buds than stems (Fig. 3A). The increase in GA 20-ox and GA 3β-hy transcript levels induced by light in apical buds suggests that rather than GA being transported from one location to another, it acts on cell differentiation and expansion of the cell that produces it. Transcripts for another GA biosynthesis gene, Ls (encoding the copalyl diphosphate synthase, Fig. 1), also accumulates in apical buds of etiolated seedlings (T. Ait-Ali and S. Frances, unpublished data; Ait-Ali et al., 1997).
In stems, GA 20-ox and GA 3β-hy transcript accumulation was less strongly light regulated than in buds. After transfer to light, GA 3β-hy transcript levels were higher than the levels observed in dark-grown stems. The GA 20-ox transcript abundance was similar in stems regardless of the light conditions (Fig. 3A). Interestingly, both GA 20-ox and GA 3β-hy transcript levels increased slightly in the stem of etiolated seedlings during the course of the experiments (Fig. 3A), whereas the level in buds remained relatively constant (Fig. 3B).
Phytochromes A and B Both Contribute to the Light Regulation of GA 20-ox Expression
To determine if the light induction of GA 20-ox and GA 3β-hy transcript accumulation was specifically mediated by phytochrome, we examined the effect of single, short pulses of red and/or far-red light on transcript levels in dark-grown seedlings. Initially, we examined the time course for induction by a single red light pulse to determine how closely the effects mimicked the effects of continuous light. We showed that the abundance of GA 20-ox and GA 3β-hy transcripts in 6-d-old seedlings did not differ from the dark controls over the first 2 h following a 10-min red light pulse, but increased rapidly between 2 and 3 h after the pulse and was maintained at a high level during the subsequent 3 h (data not shown). This time course of induction is similar to that observed for induction by continuous light (Fig. 3), and shows that significant induction is not detectable until more than 2 h after the commencement of the light treatment.
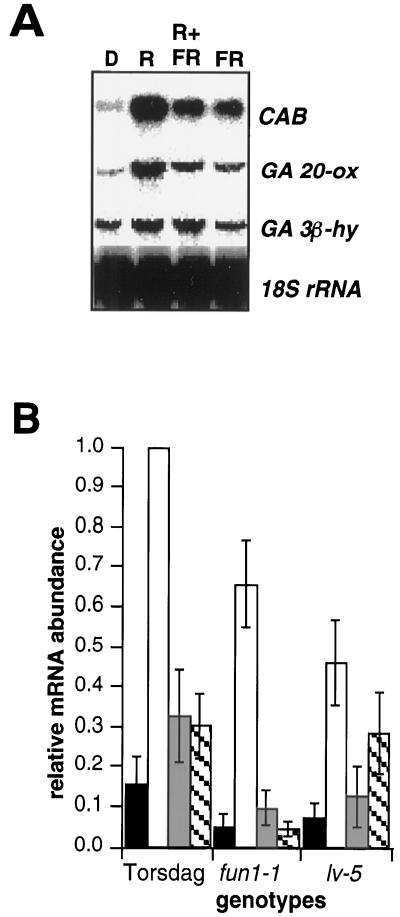
On the basis of the results of the time-course experiment, we chose 5 h as a convenient time point at which to assess the effects of red and far-red light pulses on GA 20-ox and GA 3β-hy transcript levels. As expected, CAB transcript accumulation was induced by red light, and this induction was partially reversed by far-red light (Fig. 4A). In addition, far-red light alone led to some induction of CAB gene expression. These results are consistent with previous reports that CAB expression is regulated by phytochrome in both low- and very-low-fluence-response modes (Horwitz et al., 1988). As seen for CAB, the red-light induction of GA 20-ox mRNA accumulation was also partially reversed by far-red light to the approximate level established by a single far-red light pulse (Fig. 4A). These data indicate that phytochrome mediates the red-light induction of GA 20-ox mRNA accumulation and acts in both low- and very-low-fluence-response modes.
Figure 4.
Phytochrome mediation of GA 20-ox and GA 3β-hy transcript accumulation in wild-type and phytochrome-deficient mutants. Seedlings were grown in continuous darkness (D) for 6 d. After irradiation with 10 min of red light (R), 10 min of red light followed by 10 min of far-red light (R+FR), or 10 min of far-red light (FR), seedlings were returned to the dark. Buds were harvested at 5 h after light treatment. Control seedlings were maintained in constant darkness. RNA-blot analysis was performed as described in Figure 2. A, RNA-blot analysis of cv Alaska seedlings. B, Quantitation of hybridization signal from RNA-blot analysis for the GA 20-ox transcript. Genotypes used were cv Torsdag (WT), fun1-1 (phyA- deficient), and lv-5 (phyB-deficient). Each data set was normalized to the maximum signal (red-light induction of the wild type, cv Torsdag). Experiments were repeated five times for cv Torsdag, three times for fun1-1 and lv-5. Error bars indicate the se of the mean. Black bars, Continuous darkness; white bars, red light; shaded bars, red plus far-red light; hatched bars, far-red light.
To determine which phytochromes might be involved in the control of GA 20-ox transcript accumulation, we repeated the experiment using two phytochrome-deficient mutants. The fun1-1 and lv-5 mutants are deficient in phyA and phyB, respectively (Weller et al., 1995, 1997). Similar to cv Alaska, cv Torsdag exhibited clear red/far-red reversible induction of GA 20-ox transcript accumulation. When the data are expressed relative to the highest level of induction, red light also induced GA 20-ox expression in the phytochrome-deficient mutants fun1-1 and lv-5 (Fig. 4B). However, the red-light induction observed in both of the mutants was less than that of the wild type: about 70% of the wild-type response in the phyA-deficient mutant and about 50% in the phyB-deficient mutant. In addition, far-red light induction of GA 20-ox mRNA abundance was nearly absent in the fun1-1 mutant (Fig. 4B). These data suggest that phyA controls the very-low-fluence response. Together, these data clearly demonstrate that both phyA and phyB play roles in mediating photocontrol of GA 20-ox transcript levels during de-etiolation. However, when the data of Figure 4B are expressed as a ratio of the dark expression to treatment expression, no significant difference from the wild type was seen with either the fun1-1 or lv-5 plants (data not shown). This suggests that the mutants may be too leaky to see an effect or that other phytochromes may be involved.
In contrast, the red-light pulse induction of GA 3β-hy transcript accumulation was not reversed by a subsequent far-red light pulse, nor was it significantly induced by a pulse of far-red light alone (Fig. 4A). These data provide no clear evidence that GA 3β-hy transcript accumulation is regulated by phytochrome. This result is in contrast with the phytochrome regulation of genes encoding GA 3β-hy in germinating Arabidopsis and lettuce seeds (Toyomasu et al., 1998; Yamaguchi et al., 1998). It is difficult to clarify this discrepancy, but it is clear that the physiological events that are compared (germination versus de-etiolation) are very different. However, it is also possible that there are additional GA 3β-hy genes in pea that are subject to different regulatory control (Lester et al., 1999).
Transfer from Dark to Light Causes a Reduction in the Level of GA1
Since the ultimate purpose of regulating the expression of GA biosynthesis genes is presumably to alter the levels of bioactive GAs, we next examined whether the changes in GA 20-ox and GA 3β-hy transcript levels were reflected in changes in GA levels. We transferred 6-d-old etiolated seedlings to continuous light and quantified 13-hydroxylated GAs at 2, 4, and 24 h after transfer. Control seedlings were maintained in darkness. In contrast to the transcript accumulation experiments, we measured GA levels in whole shoots (bud and stem). Figure 5 shows the levels of GA1, GA1 precursors (GA53, GA44, GA19, and GA20), and inactive 2β-hydroxylated GAs (GA8 and GA29) in de-etiolating plants over the 24-h period following transfer to light. The most striking effect of light exposure was a reduction in the level of GA1, which dropped to less than 15% of the dark-grown control level within 2 h of transfer and was maintained at this low level over a period of 24 h. A similar decrease in the GA1 content of de-etiolating lettuce seedlings was reported by Toyomasu et al. (1992). Behringer et al. (1990) showed that substantial inhibition of elongation growth of etiolated pea seedlings develops within 2 h after a red light pulse. It is therefore possible that this growth inhibition may in part result from the drop in GA1 level. Measurement of GA1 content at more frequent time points over the first 2 h following transfer will give a better indication of whether the drop in GA1 occurs prior to the growth inhibition.
Figure 5.
Endogenous GAs in pea epicotyls during de-etiolation. 13-Hydrodroxylated GAs were determined in 6-d-old etiolated seedlings at 0, 2, 4, and 24 h after transfer to light (panels on the right side) and in etiolated seedlings maintained in darkness (panels on the left side) for the same period of time. Top panels show the levels of the inactivated GAs GA8 (▾), and GA29 (▿). Bottom panels show the levels of precursors of GA1, GA44 (▵), GA53 (○), GA19 (▴), and GA20 (●), and GA1 (▪). Three independent experiments were performed. Error bars are the se of the mean.
The mechanism by which the drop in GA1 is achieved is also of interest. It may occur through a decrease in the level of GA1 precursors or through an increased rate of GA1 catabolism (GA 2β-hydroxylase). However, during the time period in which the drop in GA1 level was seen, no substantial change was detected in the levels of either GA1 precursors (GA53, GA44, GA20, and GA19) or of the inactive catabolite (GA8) relative to dark-grown control seedlings (Fig. 4). However, the level of GA8 in the plant is much higher than that of GA1, and it is therefore unlikely that an increased 2β-hydroxylation of GA1 sufficient to cause the observed drop would be reflected in a significant increase in the level of GA8. Feeding experiments with labeled GA1 will be necessary to resolve this issue. In addition, the recent cloning of a gene encoding a GA 2β-hydroxylase will make it possible to examine the effect of light on this step at the level of transcript accumulation (Thomas et al., 1999).
Applied GA1 Inhibits Light Induction of GA 20-ox and GA 3β-hy Expression
Transcript levels of both GA 20-ox and GA 3β-hy have been suggested to be subject to negative feedback regulation by bioactive GA (Chiang et al., 1995; Hedden and Kamiya, 1997). Since our results show that the transcript accumulation of these genes is also subject to control by light (Figs. 3 and 4), we examined the way in which GA1 and light interact to regulate the abundance of these transcripts. To do this, we tested whether exogenous application of GA1 on apical buds prior to the light treatment had an effect on the light induction of GA 20-ox and GA 3β-hy transcript accumulation.
The GA1 applications were of necessity performed under dim-green safelight and we therefore first assessed the effect of 30 min of exposure to dim-green safelight on GA 20-ox and GA 3β-hy transcript accumulation (Fig. 6A). In apical buds, dim-green safelight substantially induced GA 20-ox mRNA accumulation, but had little effect on the GA 3β-hy transcript level. Pretreatment with dim-green safelight also resulted in somewhat higher levels of accumulation of GA 20-ox transcript after transfer to light, as can be seen by comparing the results in Figures 6B and 3A. In contrast, pretreatment with dim-green safelight did not have a large effect on GA 3β-hy transcript transcript accumulation.
Figure 6.
Effect of GA1 application on light regulation of GA 20-ox and GA 3β-hy transcript accumulation. A, Effect of 30 min of exposure to dim-green safelight (G) on transcript accumulation in apical buds. B, Exogenous application of GA1 on apical buds of 6-d-old etiolated seedlings 30 min prior transfer to light. Light treatment and tissue harvestings were identical to Figure 3A. C, Two micrograms of GA1 was applied to the apical buds of 6-d-old etiolated seedlings 30 min prior to the red light pulse. Light treatments were identical to Figure 5. RNA-blot analysis was performed as described in Figure 2. D, Darkness; L, light; R, red light.
Figure 6B shows the effect of GA1 applications on GA 20-ox and GA 3β-hy transcript accumulation. Two micrograms of GA1 were applied to each apical bud under dim-green safelight 30 min before transfer to light. The GA1 application drastically reduced the GA 20-ox and GA 3β-hy mRNA levels in apical buds of etiolated and de-etiolating seedlings. However, for GA 20-ox expression, a slight induction in GA-treated apical buds was still visible 4 h after transfer to light. These results indicate that the early inductive effects of light on GA 20-ox and GA 3β-hy expression are greatly reduced in the presence of high levels of GA1.
Because the effect of GA1 application was somewhat weaker in stems than in apical buds (Fig. 6B), in the subsequent experiment we examined transcript accumulation in buds only. Figure 6C shows the effect of exogenous GA1 application on the induction of GA 20-ox and GA 3β-hy expression by a red-light pulse. As shown in Figure 6B, pretreatment with a dim-green safelight resulted in a more rapid accumulation of both transcripts following transfer to light. However, as for light-treated plants (Fig. 5B), the application of GA1 reduced GA 20-ox transcript levels in dark controls and largely prevented the inductive effect of red light on transcript accumulation.
A reduction in GA 20-ox transcript level in light-grown pea seedlings in response to the application of active GA has previously been reported by Martin et al. (1996). These authors also observed that the application of active GA resulted in a decrease in the overall metabolism rate of radiolabeled GA20, and proposed that GA 3β-hy expression might also be subject to negative feedback regulation. Our results lend further support to this suggestion, showing that applied active GA (GA1) also causes a reduction in GA 3β-hy transcript level in both dark- and light-grown pea seedlings.
CONCLUSIONS
We have shown that the transfer of etiolated pea seedlings to light results in a strong reduction in the GA1 content within 2 h. However, this reduction is not achieved through a change in the steady-state level of either GA 20-ox or GA 3β-hy mRNA, since no change in the level of either transcript was detectable after 2 h of de-etiolation. In fact, with de-etiolation periods of longer than 2 h, the levels of both transcripts increased sharply. In the case of GA 20-ox, this induction was clearly mediated by both phytochrome A and B. The levels of both GA 20-ox and GA 3β-hy transcripts showed negative feedback regulation by GA1, although it appears that they may differ in sensitivity. We suggest that the initial drop in the GA1 level may release the feedback inhibition on GA 20-ox and GA 3β-hy genes, causing an increase in the transcript level. The initial reduction in GA1 level may function as one of the signals leading to morphological changes early in de-etiolation. The subsequent effect of light GA 20-ox and GA 3β-hy transcript levels is likely to represent a homeostatic mechanism intended to restore the GA1 level to its pre-irradiation level. Further experiments will be necessary to answer the important question of how the light-induced drop in GA1 is achieved.
We are grateful to Masayo Sekimoto for the GA analysis and to Yujiki Tachiyama for sequencing. We would like to thank Drs. S.M. Swain and M.J. Terry for reading and commenting on this manuscript, and Dr. William Probesting for GA 3β-hydroxylase cDNA.
Acknowledgments
AKNOWLEDGMENTS
Footnotes
This work was supported by the Frontier Research Program (RIKEN).
LITERATURE CITED
- Ait-Ali T, Swain SM, Reid JB, Sun T-P, Kamiya Y. The LS locus of pea encodes the gibberellin biosynthesis enzyme ent-kaurene synthase A. Plant J. 1997;11:443–454. doi: 10.1046/j.1365-313x.1997.11030443.x. [DOI] [PubMed] [Google Scholar]
- Behringer JF, Davies PJ, Reid JB. Genetic analysis of the role of gibberellin in the red light inhibition of stem elongation in etiolated seedlings. Plant Physiol. 1990;94:432–439. doi: 10.1104/pp.94.2.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang HH, Hwang I, Goodman HM. Isolation of the Arabidopsis GA4locus. Plant Cell. 1995;7:195–201. doi: 10.1105/tpc.7.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coruzzi G, Broghi R, Cashmore A, Chua N-H. Nucleotide sequences of two pea cDNA clones encoding the small subunit of ribulose 1,5-bisphosphate carboxylase and the major chlorophyll a/bbinding thylakoid polypeptide. J Biol Chem. 1983;258:1399–1402. [PubMed] [Google Scholar]
- Crozier A. The Biochemistry and Physiology of Gibberellins. New York: Praeger; 1983. [Google Scholar]
- Foster KR, Morgan PW. Genetic regulation of development in Sorghum bicolor: IX. The ma3R allele disrupts diurnal control of gibberellin biosynthesis. Plant Physiol. 1995;108:337–343. doi: 10.1104/pp.108.1.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frances S, White MJ, Edgerton MD, Jones AM, Elliot RC, Thompson WF. Initial characterization of a pea mutant with light-independent photomorphogenesis. Plant Cell. 1992;4:1519–1530. doi: 10.1105/tpc.4.12.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K, Yang Y-Y, Honda I, Yanagisawa T, Sakurai A, Takahashi N, Kamiya Y. Effects of ethylene and gibberellins on the elongation of rice seedlings (Oriza sativaL.) Biosci Biotech Biochem. 1997;61:864–869. doi: 10.1271/bbb.61.864. [DOI] [PubMed] [Google Scholar]
- Gallo-Meagher M, Sowinski DA, Elliott RC, Thompson WF. Both internal and external regulatory elements control expression of the pea Fed-1gene in transgenic tobacco seedlings. Plant Cell. 1992;4:389–395. doi: 10.1105/tpc.4.4.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Martínez JL, López-Díaz I, Sánchez-Beltrán MJ, Philips AL, Ward DA, Gaskin P, Hedden P. Isolation and transcript analysis of gibberellin 20-oxidase genes in pea and bean in relation to fruit development. Plant Mol Biol. 1997;33:1073–1084. doi: 10.1023/a:1005715722193. [DOI] [PubMed] [Google Scholar]
- Gawronska H, Yang YY, Furukawa K, Kendrick RE, Takahashi N, Kamiya Y. Effects of low irradiance stress on gibberellin levels in pea seedlings. Plant Cell Physiol. 1995;36:1361–1367. [Google Scholar]
- Hedden P, Kamiya Y. Gibberelin biosynthesis: enzymes, genes and their regulation. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:431–460. doi: 10.1146/annurev.arplant.48.1.431. [DOI] [PubMed] [Google Scholar]
- Horwitz BA, Thompson WF, Briggs WR. Phytochrome regulation of greening in Pisum. Plant Physiol. 1988;86:299–305. doi: 10.1104/pp.86.1.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester DR, MacKenzie-Hose AK, Davies PJ, Ross JJ, Reid JB (1999) The influence of the null le-2 mutation on gibberellin levels in developing seeds. Plant Growth Reg (in press)
- Lester DR, Ross JJ, Ait-Ali T, Martin DN, Reid JB. A gibberelin 20-oxidase cDNA (accession no. U58830) from pea (Pisum sativum L.) seed (PGR 96-050) Plant Physiol. 1996;111:1353. [Google Scholar]
- Lester DR, Ross JJ, Davies PJ, Reid JB. Mendel's stem length gene (Le) encodes a gibberellin 3 β-hydroxylase. Plant Cell. 1997;9:1435–1443. doi: 10.1105/tpc.9.8.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Juez E, Kobayashi M, Sakurai A, Kamiya Y, Kendrick RE. Phytochrome, gibberellins, and hypocotyl growth. Plant Physiol. 1995;103:15–19. doi: 10.1104/pp.107.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DN, Proebsting WM, Hedden P. Mendel's dwarfing gene: cDNAs from the Lealleles and function of the expressed proteins. Proc Natl Acad Sci USA. 1997;94:8907–8911. doi: 10.1073/pnas.94.16.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DN, Proebsting WM, Parks TD, Dougherty WG, Lange T, Lewis MJ, Gaskin P, Hedden P. Feed-back regulation of gibberellin biosynthesis and gene expression in Pisum sativumL. Planta. 1996;200:159–166. doi: 10.1007/BF00208304. [DOI] [PubMed] [Google Scholar]
- Peng J, Harberd NP. Gibberellin deficiency and response mutations suppress the stem elongation phenotype of phytochrome-deficient mutants of Arabidopsis. Plant Physiol. 1997;113:1051–1058. doi: 10.1104/pp.113.4.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AL, Ward DA, Uknes S, Appleford NEJ, Lange T, Huttly A, Gaskin P, Graebe JE, Hedden P. Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol. 1995;108:1049–1057. doi: 10.1104/pp.108.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebers M, Kaneta T, Kawaide H, Yamaguchi S, Sekimoto H, Imai R, Kamiya Y. Regulation of gibberellin biosynthesis genes during flower and early fruit development of tomato. Plant J. 1999;17:241–250. doi: 10.1046/j.1365-313x.1999.00366.x. [DOI] [PubMed] [Google Scholar]
- Reed JW, Foster KR, Morgan PW, Chory J. Phytochrome B affects responsiveness to gibberellins in Arabidopsis. Plant Physiol. 1996;112:337–342. doi: 10.1104/pp.112.1.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid J. Internode length in Pisum: comparison of genotypes in the light and dark. Physiol Plant. 1988;74:83–88. [Google Scholar]
- Reid J, Howell S. Peter J. Davies, ed, Plant Hormones: Physiology, Biochemistry and Molecular Biology. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. The functioning of hormones in plant growth and development; pp. 448–485. [Google Scholar]
- Ross JJ, MacKenzie-Hose A, Davies PJ, Lester DR, Twitchin B, Reid JB. Further evidence for feedback regulation of gibberellin biosynthesis in pea. Physiol Plant. 1999;105:532–538. [Google Scholar]
- Ross JJ, Willis CL, Gaskin P, Reid JB. Shoot elongation in Lathyrus odoratusL.: gibberellin levels in light- and dark-grown tall and dwarf seedlings. Planta. 1992;187:10–13. doi: 10.1007/BF00201618. [DOI] [PubMed] [Google Scholar]
- Smith VA, Knatt CJ, Gaskin P, Reid JB. The distribution of gibberellins in vegetative tissues of Pisum sativum L. Plant Physiol. 1992;99:368–371. doi: 10.1104/pp.99.2.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sponsel V. Gibberellins in dark- and red-light-grown shoots of dwarf and tall cultivars of Pisum sativum: the quantification, metabolism and biological activity of gibberellins in Progress No. 9 and Alaska. Planta. 1986;168:119–129. doi: 10.1007/BF00407018. [DOI] [PubMed] [Google Scholar]
- Swain SM, Reid JB, Kamiya Y. Gibberellins are required for embryo growth and seed development in pea. Plant J. 1997;12:423–466. [Google Scholar]
- Thomas SG, Phillips AL, Hedden P. Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc Natl Acad Sci USA. 1999;96:4698–4703. doi: 10.1073/pnas.96.8.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson WF, White MJ. Physiological and molecular studies of light regulated nuclear genes in higher plants. Annu Rev Plant Physiol Mol Biol. 1991;42:423–466. [Google Scholar]
- Toyomasu T, Kawaide H, Mitsuhashi W, Inoue Y, Kamiya Y. Phytochrome regulates gibberellin biosynthesis during germination of photoblastic lettuce seeds. Plant Physiol. 1998;118:1517–1523. doi: 10.1104/pp.118.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyomasu T, Yamane H, Yamaguchi I, Murofushi N, Takahashi N, Inoue Y. Control by light of hypocotyl elongation and levels of endogenous gibberellins in seedlings of Latuca sativaL. Plant Cell Physiol. 1992;35:695–701. [Google Scholar]
- van Huizen R, Ozga JA, Reinecke DM. Seed and hormonal regulation of gibberellin 20-oxidase expression in pea pericarp. Plant Physiol. 1997;115:123–128. doi: 10.1104/pp.115.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Murfet IC, Reid JB. Pea mutants with reduced sensitivity to far-red light define an important role for phytochrome A in daylength detection. Plant Physiol. 1997;114:1225–1236. doi: 10.1104/pp.114.4.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Nagatani A, Kendrick RE, Murfet IC, Reid JB. New lv mutants of Pisumare deficient in phytochrome B. Plant Physiol. 1995;108:525–532. doi: 10.1104/pp.108.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Ross J, Reid JB. Gibberellins and phytochrome regulation of stem elongation in pea. Planta. 1994;192:489–496. [Google Scholar]
- Wu K, Li L, Gage DA, Zeevaart JAD. Molecular cloning and photoperiod-regulated expression of gibberellin 20-oxidase from the long-day plant spinach. Plant Physiol. 1996;110:547–554. doi: 10.1104/pp.110.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Gage DA, Zeevaart JAD. Gibberellins and stem growth in Arabidopsis thaliana: effects of photoperiod on expression of the GA4 and GA5loci. Plant Physiol. 1997;114:1471–1476. doi: 10.1104/pp.114.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Smith MW, Brown RS, Kamiya Y, Sun TP. Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell. 1998;10:2115–2126. doi: 10.1105/tpc.10.12.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart JAD, Gage DA. ent-Kaurene biosynthesis is encoded by long photoperiods in long-day plants Spinacia oleracea L. and Agrostemma githagoL. Plant Physiol. 1993;101:25–29. doi: 10.1104/pp.101.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]