Abstract
Background
Spindlin1 (SPIN1), a protein highly expressed in several human cancers, has been correlated with tumorigenesis and development. Alterations of drug metabolizing enzymes and drug transporters are major determinants of chemoresistance in tumor cells. However, whether the metabolizing enzymes and transporters are under the control of SPIN1 in breast cancer chemoresistance has not yet been defined.
Methods
SPIN1 expression in breast cancer cells and tissues was detected by quantitative real-time PCR (qRT-PCR) and immunohistochemistry. Chemosensitivity assays in vitro and in vivo were performed to determine the effect of SPIN1 on Adriamycin resistance. Downstream effectors of SPIN1 were screened by microarray and confirmed by qRT-PCR and Western blot. Luciferase assay and Western blot were used to identify miRNAs regulating SPIN1.
Results
We showed that SPIN1 was significantly elevated in drug-resistant breast cancer cell lines and tissues, compared with the chemosensitive ones. SPIN1 enhanced Adriamycin resistance of breast cancer cells in vitro, and downregulation of SPIN1 by miRNA could decrease Adriamycin resistance in vivo. Mechanistically, drug metabolizing enzymes and transporter CYP2C8, UGT2B4, UGT2B17 and ABCB4 were proven to be downstream effectors of SPIN1. Notably, SPIN1 was identified as a direct target of the miR-148/152 family (miR-148a-3p, miR-148b-3p and miR-152-3p). As expected, miR-148a-3p, miR-148b-3p or miR-152-3p could increase Adriamycin sensitivity in breast cancer cells in vitro. Moreover, high expression of SPIN1 or low expression of the miR-148/152 family predicted poorer survival in breast cancer patients.
Conclusions
Our results establish that SPIN1, negatively regulated by the miR-148/152 family, enhances Adriamycin resistance in breast cancer via upregulating the expression of drug metabolizing enzymes and drug transporter.
Electronic supplementary material
The online version of this article (10.1186/s13046-018-0748-9) contains supplementary material, which is available to authorized users.
Keywords: Breast cancer, Adriamycin resistance, SPIN1, miR-148/152, Drug metabolizing enzymes, Drug transporter
Background
Chemoresistance is a major obstacle for effective breast cancer chemotherapy [1]. The generation or acquisition of cancer chemoresistance is not clear understood but has been attributed to alterations in many molecular pathways, which include drug metabolizing enzymes and drug transporter genes [2]. Alterations in the efflux transporters of the ATP-binding cassette (ABC) family have been identified as major determinants of chemoresistance in tumor cells by decreasing the intracellular accumulation of drugs [3]. In addition, drug-metabolizing enzymes cytochrome P450 (CYP) and UDP-glucuronosyltransferases (UGTs) have also been reported to constitute a resistance phenotype [4–6]. So far, little is known about the regulation of efflux transporters and metabolizing enzymes, though it has been found to be governed by nuclear receptors [6].
Spindlin (SPIN), a member of the SPIN/SSTY family, was first reported as a major maternal transcript expressed in the mouse during the transition from oocyte to embryo [7]. Recent evidences showed that human Spindlin1 (SPIN1) was highly expressed in ovarian cancer and liposarcoma, and may be implicated in tumorigenesis and development [8–10]. SPIN1 has been shown to be elevated in chemoresistant and metastatic breast cancer tissues and involved in PI3K/Akt-mediated chemoresistance [11]. SPIN1 has been shown to be a histone code reader to bind histone H3 trimethylated at lysine 4 (H3K4me3) [12, 13], a chromatin mark typically located at promoters and associated with active or poised genes [14]. However, whether SPIN1 could regulate the drug metabolizing enzymes and transporters has not been defined. Moreover, we have previously found that SPIN1 was directly regulated by microRNA-489 [11]. Most microRNAs (miRNAs) only modestly affect their mRNA targets in a fine-tuning manner [15], and it has been confirmed that multiple miRNAs could target the same gene, suggesting that it is the combination of all these activities that exerts huge impacts on the expression of miRNA target genes [16]. We hypothesize that SPIN1 may also be regulated by other miRNAs.
Here we show that SPIN1 is highly expressed in drug-resistant breast cancer cells and tissues. We find that SPIN1 increases breast cancer Adriamycin resistance via enhancing the expression of drug metabolizing enzymes and transporter CYP2C8, UGT2B4, UGT2B17 and ABCB4. Mechanistically, the miR-148/152 family could directly target SPIN1 and increase Adriamycin sensitivity in breast cancer cells.
Methods
Tissue samples
We enrolled 78 cases of breast cancer patients before their first neoadjuvant Adriamycin-based chemotherapy cycle, in Qilu Hospital of Shandong University (Jinan, China) between Jul 2008 and Feb 2014. The cases included in this study were invasive breast carcinomas (invasive ductal carcinoma, n = 73; invasive lobular carcinoma, n = 5). Pre-chemotherapy needle biopsy samples and the paired surgically removed samples were collected, and the patients were divided into a drug-resistant group and a drug-sensitive group according to the pathological Miller/Payne assessment [17]. This is a five-point scale that focuses on the principal manifestation of chemotherapeutic effect on reduction in tumor cellularity of resection samples compared with pre-treatment needle biopsy tissues. The tumors were scored blindly by two pathologists and agreement by consensus was achieved if necessary. Patients were divided into a drug-resistant group (Miller/Payne grades 1–2) and a drug-sensitive group (grades 3–5) using the Miller/Payne grading system, as previously described [18, 19]. This study was approved by the Ethics Committee of School of Medicine, Shandong University (approval code: 2012028).
Immunohistochemistry (IHC)
The streptavidin-peroxidase-biotin (SP) immunohistochemical method was utilized to determine the expression of SPIN1 in breast cancer tissues and xenograft tumors. The paraffin-embedded specimens (4 μm) sections were incubated with the anti-SPIN1 antibody (1:150, Proteintech, 19531-1-AP). For negative controls, the anti-SPIN1 antibody was replaced with PBS. For each sample, 500 cells from five randomly chosen fields were counted. The samples were divided into SPIN1 low-expression (< 50% positivity) and high-expression (≥50%) groups, as previously described [11]. All slides were scanned and photographed in a Pannoramic P250 scanner (3DHistech, Hungary).
Cell culture and transfection
Human breast cancer cell lines MCF-7, MDA-MB-231, and MDA-MB-468 were obtained from the American Type Culture Collection and cultured in Dulbecco’s modified Eagle’s medium (MCF-7) or Leibovitz’s L15 medium (MDA-MB-231 and MDA-MB-468), supplemented with 10% fetal bovine serum (FBS; Gibco BRL, Grand Island, NY, U.S.). MCF-7/ADM cells were derived by treating MCF-7 cells with stepwise increasing concentrations of Adriamycin over 8 months and cultured in RPMI-1640 medium supplemented with 10% FBS. To maintain their resistance, MCF-7/ADM cells were cultured in the presence of a low concentration of Adriamycin and passaged for 1 week in drug-free medium before the experiments. The identities of the cell lines were confirmed by STR profiling in 2017 by Guangdong Hybribio Biotech Ltd. (Guangzhou, China; http://www.hybribio.cn).
Cells were transfected with miRNA mimics (GenePharma, Shanghai, China), SPIN1 siRNA (si-SPIN1, RiboBio, Guangzhou, China) or the respective negative controls (NC), using X-tremeGENE transfection reagent (Roche, Indianapolis, IN, U.S.). The entire SPIN1 coding sequences were cloned into the expression plasmid pcDNA3.1(+) (pcDNA3.1(+)-SPIN1) and the empty plasmid pcDNA3.1(+) was used as a control. Plasmids were transfected with Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s instructions.
Chemosensitivity assay
For cell chemosensitivity assay, cells (5 × 103) were seeded in 96-well plates. Adriamycin (Dalian Meilun, China) was added to determine the sensitivity to chemotherapy. Cell viability was detected after 48h as previously described [11].
Microarray analysis
MCF-7/ADM cells were treated with SPIN1 siRNA or negative control for 48h, followed by RNA preparation, labeling and hybridization in NimbleGen Hybridization System. Signal intensity was calculated from digitized images captured by Axon GenePix 4000B scanner. KEGG pathway enrichment analysis was performed using the DAVID online tool. The microarray data were deposited in the Gene Expression Omnibus (GEO) database (accession number: GSE71141).
RNA isolation and quantitative real-time PCR (qRT-PCR)
For mRNA quantitative analysis, total RNAs were prepared using Trizol (Invitrogen) and were reverse transcribed with a Rever Tra Ace qPCR RT Kit (Toyobo, Osaka, Japan). Quantitative real-time PCR (qRT-PCR) was performed in a total volume of 10-μl SYBR Green Real-time PCR Master Mix (Roche, Mannheim, Germany) on a Bio-Rad CFX™ 96 C1000 Real-Time system. For quantitative detection of miRNA, reverse transcription and qPCR assay were performed using the All-in-One™ miRNA qRT-PCR detection kit (Genecopeia, Rockville, MD, USA). Primers for miR-148a-3p (HmiRQP0204), miR-148b-3p (HmiRQP0206), miR-152-3p (HmiRQP0213) and U6 (RNU6B, HmiRQP9001; reference gene) were from GeneCopoeia.
Luciferase assay
The pmirGLO vector (Promega) was used to construct the recombinant plasmid pmirGLO-SPIN1 containing the SPIN1 mRNA 3’-UTR fragments which possess binding sites of miR-148a-3p, miR-148b-3p and miR-152-3p [11]. The luciferase assay was performed as previously described [20].
Western blot
Briefly, whole-cell or tissue lysates were resolved by electrophoresis, and proteins were transferred to nitrocellulose membranes and blotted with antibodies against SPIN1 (1:1000, D152571, Sangon Biotech), ABCB4 (1:500, GTX47122, Genetex), CYP2C8 (1:200, sc-164136, Santa Cruz), UGT2B4 (1:2000, ab173580, Abcam), UGT2B17 (1:1000, abs110602, absin), or β-actin (1:1000, BA2305, Boster). The protein bands were detected using the chemiluminescent substrate with the AlphaView software (Version: 3.2.2.0) on a FluorChem Q machine (Cell Biosciences, Inc., Santa Clara, CA, USA).
In vivo mouse xenograft model
Four-week-old female nude mice (n = 20) were used for xenograft studies. MCF-7/ADM cells (2 × 106) were transplanted into the mammary fat pads of mice. We have previously shown that miRNA-489 could directly target and suppress SPIN1 expression in breast cancer [11]. Here we used miRNA-489 as a tool to downregulate SPIN1 expression in MCF-7/ADM cells. Lentiviruses miRNA-489 or miRNA control was stably transfected into cells. Chemosensitivity to Adriamycin was ascertained from mice injected with Adriamycin (5 mg/kg) through the tail vein weekly. Tumor growth was monitored once a week by Vernier caliper. At the 8th week, tumor masses were excised and further analyzed by immunohistochemistry, qRT-PCR and Western blot. Animal experiments were approved by the Laboratory Animal Center of Shandong University, and were conducted in accordance with the institutional guidelines.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 5 software. The correlation between miR-148/152 family, SPIN1, and ABCB4/CYP2C8/UGT2B4/UGT2B17 expression was determined by Spearman’s correlation. Student’s t test was used to analyze the differences between two groups. The chi-square test or Fisher’s exact test was utilized to assess the relationship between SPIN1 expression and the clinical characteristics. P values < 0.05 were considered to be statistically significant.
Results
SPIN1 was elevated in drug-resistant breast cancer cells and tissues
We have confirmed that the Adriamycin resistant breast cancer MCF-7/ADM cells displayed a more than 24-fold greater resistance to Adriamycin than the parental MCF-7 cells (Fig. 1a). SPIN1 mRNA expression level was significantly upregulated in MCF-7/ADM cells compared with MCF-7 cells (Fig. 1b). Interestingly, SPIN1 was relatively highly expressed in triple-negative metastatic cell line MDA-MB-231 and MDA-MB-468, compared with the luminal cell line MCF-7 (Fig. 1b). This observation was consistent with the previous findings that high SPIN1 expression was associated with increased aggressiveness of breast cancer [11]. Additionally, we utilized GOBO [21] to determine the expression of SPIN1 across a panel of commonly used breast cancer cell lines [22] and the analysis also revealed that SPIN1 expression was significantly highly expressed in basal-like or triple-negative breast cancer cells (Additional file 1: Figure S1).
Fig. 1.
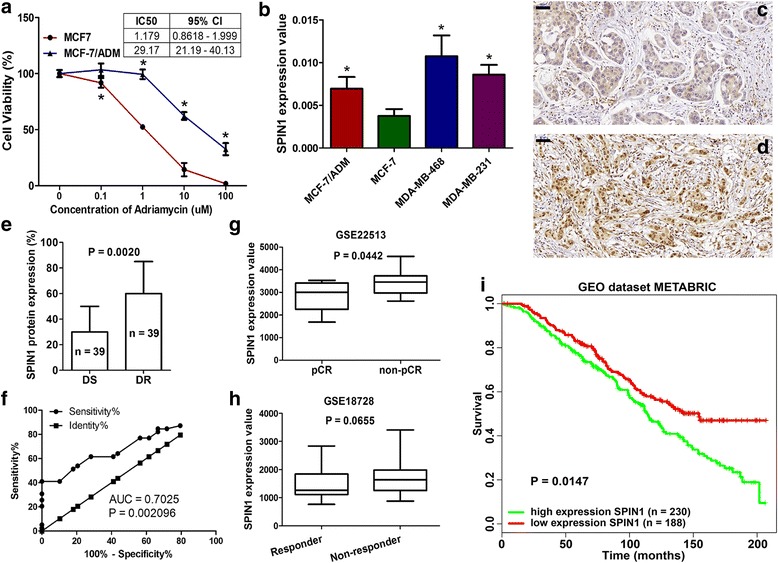
Expression of SPIN1 in breast cancer cells and tissues. a The Adriamycin resistant breast cancer MCF-7/ADM cells displayed greater resistance to Adriamycin than MCF-7 cells (IC50: 29.17 vs 1.179). b SPIN1 was highly expressed in Adriamycin resistant MCF-7/ADM cells, and metastatic MDA-MB-231 and MDA-MB-468 cells, compared with MCF-7 cells (*P < 0.05). c-e Drug-resistant (d) tissues displayed higher SPIN1 protein levels than drug-sensitive (c) tissues (e, P = 0.0020, scale bar = 50 μm). f Receiver operating characteristic (ROC) curve analysis showed that SPIN1 could significantly distinguish drug-resistant tissues from drug-sensitive tissues (area under the curve (AUC) = 0.7025, P = 0.002096). g Data from GEO datasets GSE22513 showed that SPIN1 was highly expressed in non-pCR patients compared to pCR (pathologic complete response) patients. h Data from GSE18728 showed that SPIN1 expression was higher in non-responders than responders. i Analysis of the GEO dataset METABRIC showed that high expression of SPIN1 predicted poorer survival of breast cancer patients
Next, we confirmed the correlation between SPIN1 expression and drug response in patients with breast cancer. Our results showed that drug-resistant breast cancer tissues (n = 39) exhibited higher SPIN1 protein levels compared with drug-sensitive tissues (n = 39) (Fig. 1c-f). Furthermore, the results showed positive relationship between SPIN1 expression and pN classification (Table 1, P = 0.035) and tumor stage (P = 0.020). Additionally, we determined the potential involvement of SPIN1 in breast cancer chemoresistance based on publicly available data. We obtained the normalized data of the GEO datasets GSE22513 and GSE18728 from the GENT database [23]. It was revealed that SPIN1 was upregulated in the pre-chemotherapy biopsies from patients who did not achieved a pathologic complete response (non-pCR), compared with those achieved a pathologic complete response (pCR) (GSE22513, Fig. 1g, P = 0.0442). Similarly, in the GEO dataset GSE18728, breast cancer patients who received neoadjuvant chemotherapy and classified as non-responders [24] displayed marginally higher SPIN1 expression than those classified as responders (Fig. 1h, P = 0.0655). The prognostic role of SPIN1 was determined by the PPISURV (http://www.chemoprofiling.org/cgi-bin/GEO/cancertarget/web_run_CT.V0.S1.pl) database [25, 26]. The results showed that patients with high expression of SPIN1 (n = 230) had poorer survival than those with low expression of SPIN1 (Fig. 1i, n = 188, P = 0.0147, GEO dataset METABRIC).
Table 1.
Correlation between SPIN1 expression and clinicopathological features
| Variables | SPIN1 | P value | |
|---|---|---|---|
| Low | High | ||
| Age (years) | |||
| ≤ 50 | 19 | 21 | |
| > 50 | 24 | 14 | 0.180 |
| Tumor size (cm) | |||
| < 3 | 20 | 12 | |
| ≥ 3 | 23 | 23 | 0.356 |
| Lymph node metastases | |||
| Negative | 12 | 4 | |
| Positive | 31 | 31 | 0.094 |
| Chemosensitivity | |||
| Drug-sensitive | 28 | 11 | |
| Drug-resistant | 15 | 24 | 0.006 |
| ER | |||
| Negative | 18 | 9 | |
| Positive | 25 | 26 | 0.158 |
| PR | |||
| Negative | 22 | 13 | |
| Positive | 21 | 22 | 0.257 |
| HER2 | |||
| Negative | 24 | 20 | |
| Positive | 13 | 9 | 0.796 |
| Missing | 12 | ||
| Ki-67 | |||
| Negative (< 14%) | 13 | 9 | |
| Positive (≥14%) | 30 | 26 | 0.801 |
| pT | |||
| 0 | 2 | 0 | |
| I | 13 | 11 | |
| II | 24 | 16 | |
| III | 3 | 8 | |
| IV | 1 | 0 | 0.179 |
| pN | |||
| 0 | 12 | 5 | |
| I | 13 | 4 | |
| II | 11 | 14 | |
| III | 7 | 12 | 0.035 |
| Distant metastases | |||
| M0 | 40 | 34 | |
| M1 | 3 | 1 | 0.623 |
| pTNM stage | |||
| I + II | 23 | 9 | |
| III + IV | 20 | 26 | 0.020 |
SPIN1 enhanced breast cancer resistance to Adriamycin in vitro and in vivo
To investigate the role of SPIN1 in breast cancer Adriamycin resistance, the effect of SPIN1 overexpression or inhibition on cell viability was measured by drug-sensitivity assay. As shown in Fig. 2a-c, overexpression of SPIN1 conferred greater Adriamycin resistance to MCF-7/ADM cells, with more than twofold increase in the IC50 values (43.17 μM vs 18.41 μM). However, SPIN1 knockdown in MCF-7/ADM cells significantly decreased the IC50 value by more than twofold (8.004 μM vs 22.19 μM). These findings were further confirmed in MCF-7 cells (Fig. 2d-e).
Fig. 2.
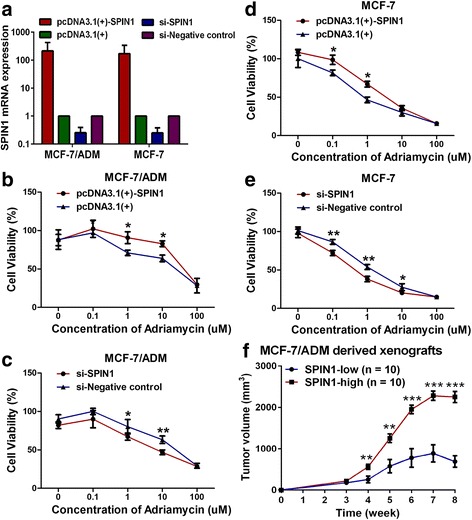
SPIN1 increased breast cancer resistance to Adriamycin in vitro and in vivo. a SPIN1 overexpression plasmid transfection increased, whereas SPIN1 siRNA transfection decreased, SPIN1 mRNA expression in breast cancer cells. b, d Overexpression of SPIN1 enhanced chemoresistance in MCF-7/ADM and MCF-7 cells. c, e Downregulation of SPIN1 increased chemosensitivity in MCF-7/ADM and MCF-7 cells. f Tumors originating from MCF-7/ADM cells expressing miRNA against SPIN1 (SPIN1-low) were significantly smaller in size than those originating from control (SPIN1-high) cells (**P < 0.01, ***P < 0.001)
To investigate whether SPIN1 enhanced resistance of breast tumor to Adriamycin in vivo, MCF-7/ADM cells stably expressing miRNA (or control) against SPIN1 were injected into mammary fat pad of female mice. Tumors were allowed to grow for 3 weeks. Subsequently, the nude mice were administered with Adriamycin, and the volumes of tumors of mice were measured. After administration with Adriamycin, the tumors originating from cells expressing miRNA against SPIN1 (SPIN1-low) [11] were significantly smaller in size than those tumors originating from control (SPIN1-high) cells (Fig. 2f). These results indicated that SPIN1 increased breast cancer cells resistance to Adriamycin in vivo.
Drug metabolizing enzymes CYP2C8, UGT2B4 and UGT2B17, and drug transporter ABCB4 were activated by SPIN1 in breast cancer
In order to identify the downstream molecules regulated by SPIN1, MCF-7/ADM cells transfected with SPIN1 siRNA (or negative control) were subjected to gene expression microarray. Here we focused on differentially-expressed genes (fold change > 2, P < 0.05) that would affect drug metabolism or transport (Fig. 3a). Thus we selected members of ATP-binding cassette (ABC) transporters (ABCA13, ABCB4, ABCC13 and ABCC8) and genes that enriched in the “drug metabolism - cytochrome P450” pathway (CYP2B6, CYP2C8, GSTA1, GSTA4, UGT2B11, UGT2B17 and UGT2B4, P = 0.01) for qRT-PCR validation. Results showed that the mRNA expression levels of ABCB4, CYP2C8, UGT2B4, and UGT2B17 were significantly decreased after SPIN1 siRNA transfection (Fig. 3b-c). Reciprocally, overexpression of SPIN1 enhanced the ABCB4, CYP2C8, UGT2B4 and UGT2B17 mRNA expression when compared with the negative control (Fig. 3b-c). Western blot was further used to detect the protein levels of these drug metabolizing enzymes and transporters. We found that after transfection with SPIN1 siRNA, levels of ABCB4, CYP2C8, UGT2B4, and UGT2B17 were dramatically reduced as compared to the negative controls (Fig. 3d-e). Consistently, the protein levels in the SPIN1-overexpressing cells showed significant increases as compared with that of the control cells (Fig. 3d-e). Altogether these data indicate that the expression of the drug metabolizing enzymes CYP2C8, UGT2B4 and UGT2B17, and transporter ABCB4 was positively regulated by SPIN1 in breast cancer.
Fig. 3.
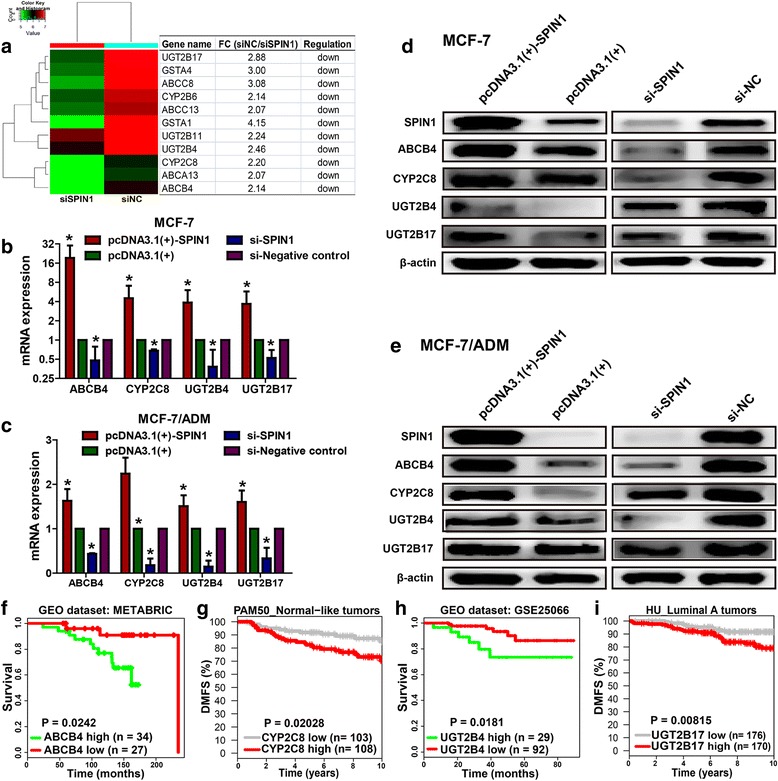
The expression of ABCB4, CYP2C8, UGT2B4 and UGT2B17 was upregulated by SPIN1 in breast cancer cells. a Candidate SPIN1 targets from microarray were presented as a heat map. b-c The mRNA expression levels of ABCB4, CYP2C8, UGT2B4, and UGT2B17 were decreased after SPIN1 siRNA transfection. Accordingly, overexpression of SPIN1 led to upregulation of ABCB4, CYP2C8, UGT2B4, and UGT2B17 mRNA expression levels. d-e Western blot analysis confirmed that overexpression of SPIN1 upregulated ABCB4, CYP2C8, UGT2B4, and UGT2B17 protein levels, however, SPIN1 downregulation decreased the expression levels of these proteins. f-i Data from PPISURV (ABCB4 and UGT2B4) and GOBO (CYP2C8 and UGT2B17) revealed that patients with high expression of ABCB4, CYP2C8, UGT2B4 or UGT2B17 had poorer survival than those with low expression
Data from PPISURV [25, 26] and GOBO [21] revealed that breast cancer patients with high expression of ABCB4 (P = 0.0242), CYP2C8 (P = 0.02028), UGT2B4 (P = 0.0181) or UGT2B17 (P = 0.00815) showed poorer survival than those with low expression (Fig. 3f-i).
SPIN1 was directly targeted and suppressed by the miR-148/152 family
MicroRNAs (miRNAs) are critical gene regulators and chemotherapy modifiers in different tumor types [27]. We suspected that SPIN1 may be potentially regulated by miRNAs and we utilized the TargetScan algorithm to screen chemotherapy-associated miRNAs that target the 3’-UTR of SPIN1 mRNA. Of the candidates, the miR-148/152 family (miR-148a-3p, miR-148b-3p and miR-152-3p), was of particular interest in light of its reported roles in regulating drug sensitivity of cancer cells [28–30]. Moreover, a microarray analysis by Kovalchuk et al. found that two members (miR-148a and miR-152) of the family displayed more than 370-fold decreases in MCF-7/ADM cells versus MCF-7 cells [31], further indicating their possible involvement in breast cancer chemoresistance.
By luciferase assay, we observed a significant suppression of luciferase activity in cells transfected with miR-148a-3p, miR-148b-3p or miR-152-3p (Fig. 4a-b). On the contrary, when the binding sites of these three miRNAs in the SPIN1 pmirGLO-3’UTR were mutated, its responsiveness to miR-148a-3p, miR-148b-3p or miR-152-3p regulation was abrogated (Fig. 4b). We further investigated the miR-148/152 family-mediated SPIN1 suppression and found that expression of the SPIN1 protein was significantly downregulated in miR-148/152 overexpressing MCF-7 and MDA-MB-231 cells (Fig. 4c-d). Interestingly, downregulation of SPIN1 protein is significative after overexpression of miR-148a-3p in particular (Fig. 4c-d).
Fig. 4.
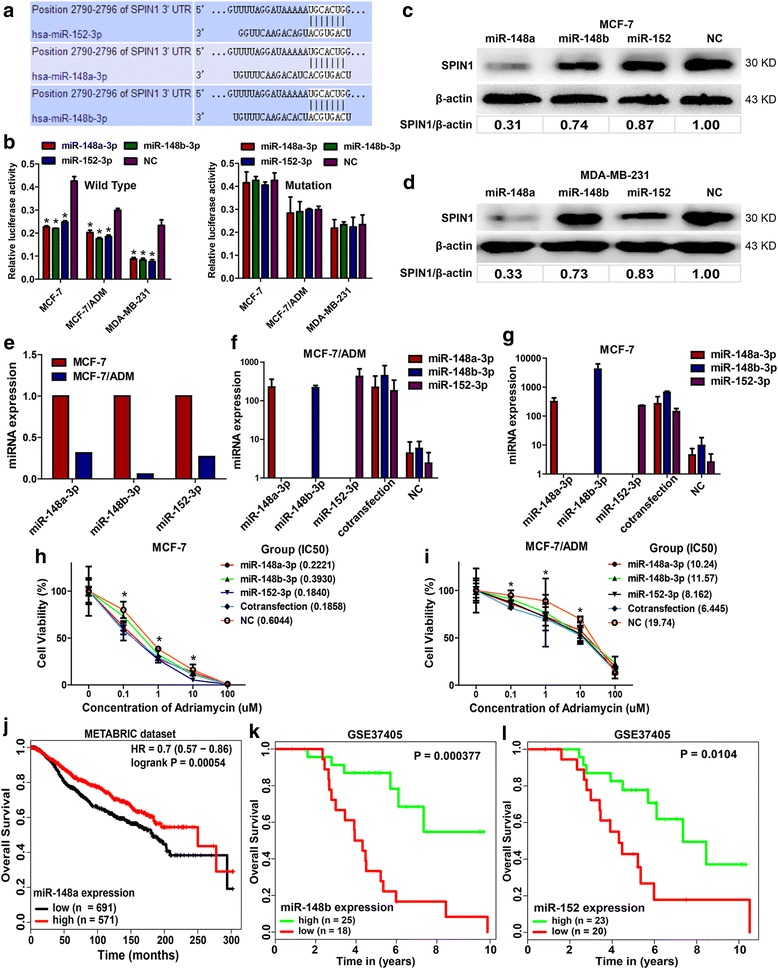
miR-148a-3p, miR-148b-3p and miR-152-3p directly targeted SPIN1 and increased chemosensitivity in breast cancer cells. a The 3’-UTR element of SPIN1 mRNA is partially complementary to miR-148a-3p, miR-148b-3p and miR-152-3p. b The relative luciferase activity was significantly reduced in the miR-148a/148b/152-3p overexpressing cells (*P < 0.05) and these effects could be abolished by mutation of SPIN1 3’-UTR. c-d The SPIN1 protein expression was clearly reduced after the transfection of miR-148a-3p, miR-148b-3p or miR-152-3p. e The expression levels of miR-148a-3p, miR-148b-3p and miR-152-3p were lower in MCF-7/ADM cells than that in MCF-7 cells. f-g Transfection of miR-148a-3p, miR-148b-3p, miR-152-3p, or cotransfection of these three miRNAs significantly increased the miRNAs levels. h-i miR-148a-3p, miR-148b-3p and miR-152-3p decreased Adriamycin resistance in MCF-7/ADM and MCF-7 cells. In MCF-7 cells, miR-148a-3p or miR-152-3p (Adriamycin = 0.1 μM and 1 μM), and miR-152-3p (Adriamycin = 10 μM) could significantly reduce cell viability, compared with the negative control (NC) group (*P < 0.05, t-test). In MCF-7/ADM cells, miR-148a-3p or miR-152-3p (Adriamycin = 0.1 μM), and miR-148a-3p, miR-148b-3p or miR-152-3p (Adriamycin = 1 μM and 10 μM) could significantly decrease cell viability compared with NC (*P < 0.05, t-test). j-l Data from MIRUMIR and miRpower showed that breast cancer patients with low expression of miR-148a, miR-148b or miR-152 had shorter survival time than those with high expression
The miR-148/152 family decreased Adriamycin resistance in breast cancer cells and was associated with patients’ survival
Our results showed the chemoresistant MCF-7/ADM cells exhibited lower miR-148a-3p, miR-148b-3p and miR-152-3p expression levels compared with chemosensitive MCF-7 cells (Fig. 4e). We further characterized the roles of miR-148a-3p, miR-148b-3p and miR-152-3p in regulating Adriamycin resistance. Transfection of miR-148a-3p, miR-148b-3p or miR-152-3p resulted in a significant decrease in survival of MCF-7/ADM and MCF-7 cells in Adriamycin-added medium (Fig. 4f-i).
Furthermore, we determined the prognostic value of the miR-148/152 family using publicly available data. As shown in MIRUMIR [26, 32], low expression of miR-148b (P = 0.000377) or miR-152 (P = 0.0104) predicted poor survival of breast cancer patients (GEO database: GSE37405, n = 43). Results from the web-tool miRpower [33] showed that breast cancer patients with low expression of miR-148a had shorter survival time (METABRIC dataset, n = 1262, P = 0.00054).
Validation of the miR-148/152–SPIN1–ABCB4/CYP2C8/UGT2B4/UGT2B17 signaling axis in xenograft tumors
In order to validate the miR-148/152–SPIN1–downstream effectors axis in vivo, we established an MCF-7/ADM derived xenograft tumor model, by suppressing SPIN1 expression [11]. Analysis on the xenograft tumors showed that expression of miR-148a-3p, miR-148b-3p or miR-152-3p was positively intercorrelated (Additional file 1: Figure S2). Moreover, expression levels of the miR-148/152 family were inversely related with SPIN1 mRNA (Fig. 5a-c) and protein (Fig. 5d-f) expression.
Fig. 5.
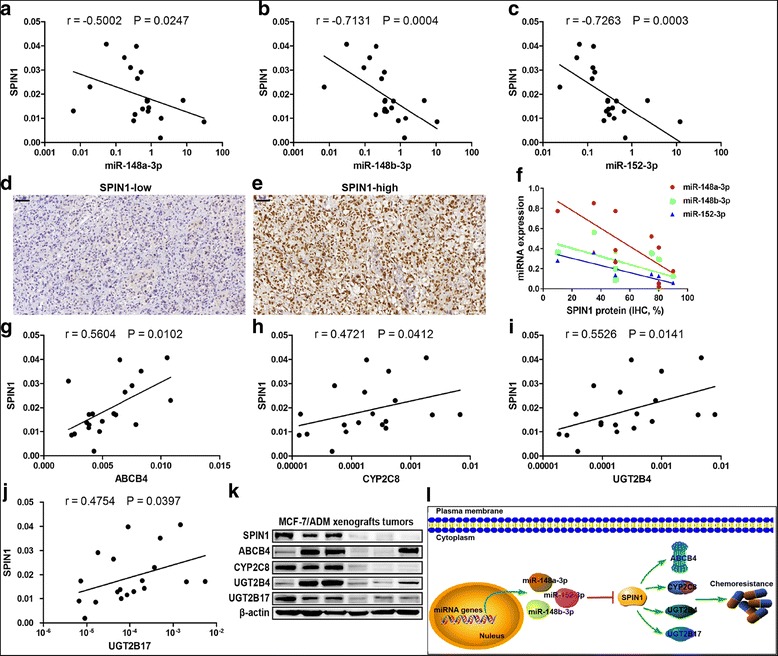
Validation of the miR-148/152–SPIN1–ABCB4/CYP2C8/UGT2B4/UGT2B17 signaling in xenograft tumors. a-c Expression levels of miR-148a-3p, miR-148b-3p or miR-152-3p were inversely related with SPIN1 mRNA expression in MCF-7/ADM xenograft tumors (n = 20). d-e SPIN1 protein expression in ten xenograft tumors was detected by immunohistochemistry. Representative images were shown. f Expression of miR-148a-3p (r = − 0.7478, P = 0.0162), miR-148b-3p (r = − 0.6524, P = 0.0473) or miR-152-3p (r = − 0.8512, P = 0.0032) were inversely related with SPIN1 protein expression. g-j A positive relationship between mRNA expression of SPIN1 and ABCB4, CYP2C8, UGT2B4 and UGT2B17 was observed (n = 20). k Three tumors with high expression of SPIN1 and the other three with low SPIN1 expression were selected for Western blot analysis. Tumors with high SPIN1 protein levels tend to show high expression of ABCB4, CYP2C8, UGT2B4 and UGT2B17. l A schematic model of miR-148/152–SPIN1–ABCB4/CYP2C8/UGT2B4/UGT2B17 regulation of breast cancer chemoresistance. SPIN1, which is direct target of miR-148a-5p/148b-5p/152-5p, promotes chemoresistance via upregulating ABCB4, CYP2C8, UGT2B4 and UGT2B17
As for the drug metabolizing enzymes and transporter, we observed a positive relationship between mRNA expression of SPIN1 and ABCB4, CYP2C8, UGT2B4 and UGT2B17 in the xenograft tumors (Fig. 5g-j). As expected, tumors with high SPIN1 protein levels displayed relatively higher protein expression of ABCB4, CYP2C8, UGT2B4 and UGT2B17, compared to tumors with low SPIN1 protein levels (Fig. 5k).
Discussion
Here, we show that SPIN1 is upregulated in drug-resistant breast cancer cells and tissues. SPIN1 is identified as a novel target of the miR-148/152 family and enhances Adriamycin resistance by regulating drug metabolizing enzymes and transporter CYP2C8, UGT2B4, UGT2B17 and ABCB4 in breast cancer (Fig. 5i). In addition, high expression of SPIN1 or low expression of the miR-148/152 family predicts poorer survival in patients with breast cancer.
In this study, by ectopic expression and loss-of-function experiments of SPIN1, we indicated that SPIN1 enhanced Adriamycin resistance of breast cancer cells. We understand that in vivo experiments with a specific depletion of SPIN1 may better reveal the involvement of SPIN1 in chemoresistance, which needs more attention in our future study. Here SPIN1 was found to lead to marked upregulation of drug metabolizing enzymes and transporter. Recently, Franz et al. have found that SPIN1, in cooperation with the transcription factor MAZ, directly enhances expression of GDNF to activate the RET signaling to increase proliferation and decrease apoptosis of liposarcoma cells [10]. These findings, together with our results, highlighted the critical roles of SPIN1 in regulating genes expression and prompted us to further investigate the underlying mechanisms of SPIN1-mediated activation of the drug metabolizing enzymes and transporter.
The drug metabolizing enzymes and transporter that identified as SPIN1 downstream effectors in this study have been shown to be highly expressed in drug-resistant tumors and associated with chemoresistance. The ATP-binding cassette (ABC) transporter gene ABCB4 is found to be amplified in Adriamycin-resistant breast cancer cells relative to drug-sensitive cells [34]. Sprouse et al. suggest that upregulation of CYP2C8 may account for paclitaxel resistance in the drug-resistant MDA-MB-231 breast cancer cells [35]. The UDP-glucuronosyltransferases UGT2B4 and UGT2B17 have been linked to breast cancer risk [36, 37]. However the upstream regulators of these drug metabolizing enzymes and transporter remain largely unknown. Here we showed that these drug metabolizing enzymes and transporter were controlled by SPIN1 and involved in breast cancer chemoresistance, which may be potential targets to reverse drug resistance in breast cancer.
We have previously found that miR-489 could directly target SPIN1 in breast cancer [11]. And here we further showed that SPIN1 was also targeted by three members (miR-148a/148b/152-3p) of the miR-148/152 family [38]. All these three miRNAs have been reported to be downregulated in breast cancer tissues and cell lines [39–41]. In addition, they are all crucial modulators for many biological processes in breast cancer. MiR-148a inhibits breast cancer migration, invasion and angiogenesis by suppressing WNT-1 [39], MMP-13 [42], ERBB3 [43]. MiR-148b is identified as a relapse-associated miRNA and suppresses breast cancer progression by targeting a series of cancer-related oncogenes [40]. MiR-152 inhibits tumor angiogenesis via targeting IGF-IR and IRS1 in breast cancer [41]. However, the function and mechanism of the miR-148/152 family in breast cancer chemoresistance have not yet been reported. Here we found that miR-148-3p, miR-148b-3p and miR-152-3p were downregulated in Adriamycin-resistant MCF-7/ADM cells compared with the parental MCF-7 cells. The three miRNAs suppressed Adriamycin resistance of breast cancer cells by directly targeting SPIN1. Moreover, analysis of publicly available data revealed that the miR-148/152 family was associated with patients’ survival in breast cancer. Our study demonstrated a newly-identified involvement of the miR-148/152 family in breast cancer Adriamycin resistance and further study is underway to confirm this in clinical samples.
In conclusion, we have presented evidence that SPIN1, a novel target of the miR-148/152 family, is upregulated in drug-resistant breast cancer cells and tissues and confers Adriamycin resistance by upregulating drug metabolizing enzymes and transporter in breast cancer. Our study may provide useful information for the development of alternative approaches to drug-resistant breast cancer.
Additional file
Figure S1 and Figure S2. SPIN1 expression in breast cancer cells and miR-148a-3p/148b-3p/152-3p expression in xenograft tumors. (DOC 261 kb)
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural Science Foundation of China (Grant No. 81372856 and 81672842) and the Taishan Scholars Program of Shandong Province (Grant No. ts201511096).
Availability of data and materials
The Microarray data were deposited in the Gene Expression Omnibus (GEO) database (accession number: GSE71141).
Abbreviations
- FBS
Fetal bovine serum
- GENT database
Gene Expression database of Normal and Tumor tissues database
- GEO
Gene Expression Omnibus
- GSTA4
Glutathione S-transferase alpha 4
- H3K4me3
Histone H3 trimethylated at lysine 4
- miRNAs
MicroRNAs
- PBS
Phosphate buffer saline
- pCR
Pathologic complete response
- PI3K/Akt
Phosphatidylinositol 3 kinase/protein kinase B
- qRT-PCR
Quantitative real-time PCR
- SP
Streptavidin-peroxidase-biotin
- SPIN1
Spindlin1
- UGT2B4
UDP glucuronosyltransferase family 2 member B4
Authors’ contributions
PG conceived and carried out experiments. YWW and XC conceived experiments and analysed data. YWW and XC carried out experiments. All authors were involved in writing the paper and had final approval of the submitted and published versions.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Shandong University (approval code: 2012028) and the procedures involving human subjects were in accordance with the Declaration of Helsinki. Animal experiments were approved by the Laboratory Animal Center of Shandong University, and all procedures were conducted in accordance with the institutional guidelines.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13046-018-0748-9) contains supplementary material, which is available to authorized users.
Contributor Information
Xu Chen, Email: chenxu999@163.com.
Ya-Wen Wang, Email: 1015408265@qq.com.
Peng Gao, Email: gaopeng@sdu.edu.cn, Email: gpzmq@sina.com.
References
- 1.Gao P, Zhou GY, Guo LL, Zhang QH, Zhen JH, Fang AJ, Lin XY. Reversal of drug resistance in breast carcinoma cells by anti-mdr1 ribozyme regulated by the tumor-specific MUC-1 promoter. Cancer Lett. 2007;256:81–89. doi: 10.1016/j.canlet.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 2.Starlard-Davenport A, Lyn-Cook B, Beland FA, Pogribny IP. The role of UDP-glucuronosyltransferases and drug transporters in breast cancer drug resistance. Exp Oncol. 2010;32:172–180. [PubMed] [Google Scholar]
- 3.Cascorbi I. Role of pharmacogenetics of ATP-binding cassette transporters in the pharmacokinetics of drugs. Pharmacol Ther. 2006;112:457–473. doi: 10.1016/j.pharmthera.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 4.Wang C, Liu KX. The drug-drug interaction mediated by efflux transporters and CYP450 enzymes. Yao Xue Xue Bao. 2014;49:590–595. [PubMed] [Google Scholar]
- 5.Decleves X, Jacob A, Yousif S, Shawahna R, Potin S, Scherrmann JM. Interplay of drug metabolizing CYP450 enzymes and ABC transporters in the blood-brain barrier. Curr Drug Metab. 2011;12:732–741. doi: 10.2174/138920011798357024. [DOI] [PubMed] [Google Scholar]
- 6.Vadlapatla RK, Vadlapudi AD, Pal D, Mitra AK. Mechanisms of drug resistance in cancer chemotherapy: coordinated role and regulation of efflux transporters and metabolizing enzymes. Curr Pharm Des. 2013;19:7126–7140. doi: 10.2174/13816128113199990493. [DOI] [PubMed] [Google Scholar]
- 7.Oh B, Hwang SY, Solter D, Knowles BB. Spindlin, a major maternal transcript expressed in the mouse during the transition from oocyte to embryo. Development. 1997;124:493–503. doi: 10.1242/dev.124.2.493. [DOI] [PubMed] [Google Scholar]
- 8.Yue W, Sun LY, Li CH, Zhang LX, Pei XT. Screening and identification of ovarian carcinomas related genes. Ai Zheng. 2004;23:141–145. [PubMed] [Google Scholar]
- 9.Wang JX, Zeng Q, Chen L, et al. SPINDLIN1 promotes cancer cell proliferation through activation of WNT/TCF-4 signaling. Mol Cancer Res. 2012;10:326–335. doi: 10.1158/1541-7786.MCR-11-0440. [DOI] [PubMed] [Google Scholar]
- 10.Franz H, Greschik H, Willmann D, et al. The histone code reader SPIN1 controls RET signaling in liposarcoma. Oncotarget. 2015;6:4773–4789. doi: 10.18632/oncotarget.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X, Wang YW, Xing AY, et al. Suppression of SPIN1-mediated PI3K-Akt pathway by miR-489 increases chemosensitivity in breast cancer. J Pathol. 2016;239:459–472. doi: 10.1002/path.4743. [DOI] [PubMed] [Google Scholar]
- 12.Yang N, Wang W, Wang Y, et al. Distinct mode of methylated lysine-4 of histone H3 recognition by tandem tudor-like domains of Spindlin1. Proc Natl Acad Sci U S A. 2012;109:17954–17959. doi: 10.1073/pnas.1208517109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W, Chen Z, Mao Z, et al. Nucleolar protein Spindlin1 recognizes H3K4 methylation and stimulates the expression of rRNA genes. EMBO Rep. 2011;12:1160–1166. doi: 10.1038/embor.2011.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voigt P, Tee WW, Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 16.Wu S, Huang S, Ding J, et al. Multiple microRNAs modulate p21Cip1/Waf1 expression by directly targeting its 3′ untranslated region. Oncogene. 2010;29:2302–2308. doi: 10.1038/onc.2010.34. [DOI] [PubMed] [Google Scholar]
- 17.Ogston KN, Miller ID, Payne S, et al. A new histological grading system to assess response of breast cancers to primary chemotherapy: prognostic significance and survival. Breast. 2003;12:320–327. doi: 10.1016/S0960-9776(03)00106-1. [DOI] [PubMed] [Google Scholar]
- 18.Kim YJ, Kim SH, Lee AW, Jin MS, Kang BJ, Song BJ. Histogram analysis of apparent diffusion coefficients after neoadjuvant chemotherapy in breast cancer. Jpn J Radiol. 2016;34:657–666. doi: 10.1007/s11604-016-0570-2. [DOI] [PubMed] [Google Scholar]
- 19.Kim T, Kang DK, An YS, et al. Utility of MRI and PET/CT after neoadjuvant chemotherapy in breast cancer patients: correlation with pathological response grading system based on tumor cellularity. Acta Radiol. 2014;55:399–408. doi: 10.1177/0284185113498720. [DOI] [PubMed] [Google Scholar]
- 20.Wang YW, Chen X, Gao JW, Zhang H, Ma RR, Gao ZH, Gao P. High expression of cAMP-responsive element-binding protein 1 (CREB1) is associated with metastasis, tumor stage and poor outcome in gastric cancer. Oncotarget. 2015;6:10646–10657. doi: 10.18632/oncotarget.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ringner M, Fredlund E, Hakkinen J, Borg A, Staaf J. GOBO: gene expression-based outcome for breast cancer online. PLoS One. 2011;6:e17911. doi: 10.1371/journal.pone.0017911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin G, Kang TW, Yang S, Baek SJ, Jeong YS, Kim SY. GENT: gene expression database of normal and tumor tissues. Cancer Inform. 2011;10:149–157. doi: 10.4137/CIN.S7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korde LA, Lusa L, McShane L, et al. Gene expression pathway analysis to predict response to neoadjuvant docetaxel and capecitabine for breast cancer. Breast Cancer Res Treat. 2010;119:685–699. doi: 10.1007/s10549-009-0651-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antonov AV, Krestyaninova M, Knight RA, Rodchenkov I, Melino G, Barlev NA. PPISURV: a novel bioinformatics tool for uncovering the hidden role of specific genes in cancer survival outcome. Oncogene. 2014;33:1621–1628. doi: 10.1038/onc.2013.119. [DOI] [PubMed] [Google Scholar]
- 26.Antonov AV. BioProfiling.de: analytical web portal for high-throughput cell biology. Nucleic Acids Res. 2011;39:W323–W327. doi: 10.1093/nar/gkr372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hummel R, Hussey DJ, Haier J. MicroRNAs: predictors and modifiers of chemo- and radiotherapy in different tumour types. Eur J Cancer. 2010;46:298–311. doi: 10.1016/j.ejca.2009.10.027. [DOI] [PubMed] [Google Scholar]
- 28.Hummel R, Watson DI, Smith C, Kist J, Michael MZ, Haier J, Hussey DJ. Mir-148a improves response to chemotherapy in sensitive and resistant oesophageal adenocarcinoma and squamous cell carcinoma cells. J Gastrointest Surg. 2011;15:429–438. doi: 10.1007/s11605-011-1418-9. [DOI] [PubMed] [Google Scholar]
- 29.Sui C, Meng F, Li Y, Jiang Y. miR-148b reverses cisplatin-resistance in non-small cell cancer cells via negatively regulating DNA (cytosine-5)-methyltransferase 1(DNMT1) expression. J Transl Med. 2015;13:132. doi: 10.1186/s12967-015-0488-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiang Y, Ma N, Wang D, et al. MiR-152 and miR-185 co-contribute to ovarian cancer cells cisplatin sensitivity by targeting DNMT1 directly: a novel epigenetic therapy independent of decitabine. Oncogene. 2014;33:378–386. doi: 10.1038/onc.2012.575. [DOI] [PubMed] [Google Scholar]
- 31.Kovalchuk O, Filkowski J, Meservy J, Ilnytskyy Y, Tryndyak VP, Chekhun VF, Pogribny IP. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther. 2008;7:2152–2159. doi: 10.1158/1535-7163.MCT-08-0021. [DOI] [PubMed] [Google Scholar]
- 32.Antonov AV, Knight RA, Melino G, Barlev NA, Tsvetkov PO. MIRUMIR: an online tool to test microRNAs as biomarkers to predict survival in cancer using multiple clinical data sets. Cell Death Differ. 2013;20:367. doi: 10.1038/cdd.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lanczky A, Nagy A, Bottai G, Munkacsy G, Szabo A, Santarpia L, Gyorffy B. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat. 2016;160:439–446. doi: 10.1007/s10549-016-4013-7. [DOI] [PubMed] [Google Scholar]
- 34.Turton NJ, Judah DJ, Riley J, et al. Gene expression and amplification in breast carcinoma cells with intrinsic and acquired doxorubicin resistance. Oncogene. 2001;20:1300–1306. doi: 10.1038/sj.onc.1204235. [DOI] [PubMed] [Google Scholar]
- 35.Sprouse AA, Herbert BS. Resveratrol augments paclitaxel treatment in MDA-MB-231 and paclitaxel-resistant MDA-MB-231 breast cancer cells. Anticancer Res. 2014;34:5363–5374. [PubMed] [Google Scholar]
- 36.Sun C, Huo D, Southard C, et al. A signature of balancing selection in the region upstream to the human UGT2B4 gene and implications for breast cancer risk. Hum Genet. 2011;130:767–775. doi: 10.1007/s00439-011-1025-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eskandari-Nasab E, Hashemi M, Rezaei H, et al. Evaluation of UDP-glucuronosyltransferase 2B17 (UGT2B17) and dihydrofolate reductase (DHFR) genes deletion and the expression level of NGX6 mRNA in breast cancer. Mol Biol Rep. 2012;39:10531–10539. doi: 10.1007/s11033-012-1938-8. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y, Song YX, Wang ZN. The microRNA-148/152 family: multi-faceted players. Mol Cancer. 2013;12:43. doi: 10.1186/1476-4598-12-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang Q, He M, Ma MT, et al. MicroRNA-148a inhibits breast cancer migration and invasion by directly targeting WNT-1. Oncol Rep. 2016;35:1425–1432. doi: 10.3892/or.2015.4502. [DOI] [PubMed] [Google Scholar]
- 40.Cimino D, De Pitta C, Orso F, et al. miR148b is a major coordinator of breast cancer progression in a relapse-associated microRNA signature by targeting ITGA5, ROCK1, PIK3CA, NRAS, and CSF1. FASEB J. 2013;27:1223–1235. doi: 10.1096/fj.12-214692. [DOI] [PubMed] [Google Scholar]
- 41.Xu Q, Jiang Y, Yin Y, et al. A regulatory circuit of miR-148a/152 and DNMT1 in modulating cell transformation and tumor angiogenesis through IGF-IR and IRS1. J Mol Cell Biol. 2013;5:3–13. doi: 10.1093/jmcb/mjs049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xue J, Chen Z, Gu X, Zhang Y, Zhang W. MicroRNA-148a inhibits migration of breast cancer cells by targeting MMP-13. Tumour Biol. 2016;37:1581–1590. doi: 10.1007/s13277-015-3926-9. [DOI] [PubMed] [Google Scholar]
- 43.Yu J, Li Q, Xu Q, Liu L, Jiang B. MiR-148a inhibits angiogenesis by targeting ERBB3. J Biomed Res. 2011;25:170–177. doi: 10.1016/S1674-8301(11)60022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 and Figure S2. SPIN1 expression in breast cancer cells and miR-148a-3p/148b-3p/152-3p expression in xenograft tumors. (DOC 261 kb)
Data Availability Statement
The Microarray data were deposited in the Gene Expression Omnibus (GEO) database (accession number: GSE71141).


