Abstract
About 132 thousand cases of melanoma (more severe type of skin cancer) were registered in 2014 according to the World Health Organization. This type of cancer significantly affects the quality of life of individuals. Caffeine has shown potential inhibitory effect against epithelial cancer. In this study, it was proposed to obtain new caffeine-based molecules with potential epithelial anticancer activity. For this, a training set of 21 molecules was used for pharmacophore perception procedures. Multiple linear regression analyses were used to propose mono-, bi-, tri-, and tetra-parametric models applied in the prediction of the activity. The generated pharmacophore was used to select 350 molecules available at the ZINCpharmer server, followed by reduction to 24 molecules, after selection using the Tanimoto index, yielding 10 molecules after final selection by predicted activity values > 1.5229. These ten mole-cules had better pharmacokinetic properties than the other ones used as reference and within the clinical-ly significant limits. Only two molecules show minor hits of toxicity and were submitted to molecular docking procedures, showing BFE (binding free energy) values lower than the reference values. Statisti-cal analyses indicated strong negative correlations between BFE and pharmacophoric properties (high influence on BFE lowering) and practically null correlation between BFE and BBB. The two most prom-ising molecules can be indicated as candidates for further in vitro and in vivo analyzes.
Keywords: Epithelial cancer, caffeine, Chk1, Molecular modeling, multiple linear regression, Pharmacokinetic and toxicological properties
1. INTRODUCTION
Cancer is a disease characterized by the abnormal cell growth in an organism; it is also known as malignant neoplasm or malignant tumor. It can be caused by chemical, physical and biological agents and has its origin in genetic alterations of cells. The main effects observed in neoplastic cells are loss of function resulting from the absence of differentiation, uncontrolled proliferation, invasion of adjacent tissues and metastasis [1-3].
There are several types of cancer depending on which region of the body is affected. Diagnoses of cancer worldwide show that the most common are lung cancer, with 1.8 million cases, 1.7 million breast of breast cancer and uterine cervix and rectum cancer with 1.4 million. Regarding skin cancer, it is estimated that melanoma alone (the most aggressive of skin tumors) has an incidence of around 132 thousand cases per year, most of them registered in tropical areas [4].
Also known as epithelial cancer, skin cancer is primarily caused by the incidence of UVB rays, in addition to being influenced by the individual's lifestyle [5]. Skin cancer receives several nomenclatures, which depends on which layer of skin the cancer develops, and can be of two types: non-melanomas - Basal Cell Carcinoma (BCC) and Squamous Cell Carcinoma (SCC); And melanoma - cutaneous malignant melanoma that originates in melanocytes, melanin-producing cells, and has high potential to generate metastases [6].
The anticancer action of plant substances has been widely studied, among them there are studies related to caffeine acting as epithelial anticancer agent. Such as the effect of caffeine as a radiosensitizer on cancer cells [7], the effect of caffeine on rat skin irradiated by UVB rays [8] and the effect of caffeine and analogues on the growth of epidermal cells of rats of the JB6 P + lineage [9]. Caffeine (1,3,7-trimethylxanthine) is an alkaloid compound belonging to the xanthine group, presents basic character and of plant origin, it contains in its composition nitrogen, oxygen, hydrogen and carbon [10].
Regarding the biological receptor responsible for the epithelial anticancer action, when such neoplasia is caused by the UVB incidence, Sarkaria et al. (1999) [7] indicate that methylxanthines inhibit a phosphotransferase kinase protein required for checkpoint signaling in cells with damaged DNA and that the possible action of trialkylxanthines on protein kinases can be attributed to a prominent target candidate: Chk1. The reference to this possible target was also made by Lu et al. (2008) [8], where they consider that the key to the pro-apoptotic effect of caffeine in the epidermis of mice exposed to UVB is in the inhibition of the signal transduction pathway UVB → ATR → Chk1 → cdc25c → cdc2 → cdc2 / Cyclin B1. The presence of caffeine stimulates UVB-induced apoptosis, inhibits phosphorylation of Chk1 over Ser345, and restricts the decrease of mitotic cells acting on cyclin B1 (which occurs shortly after UVB irradiation).
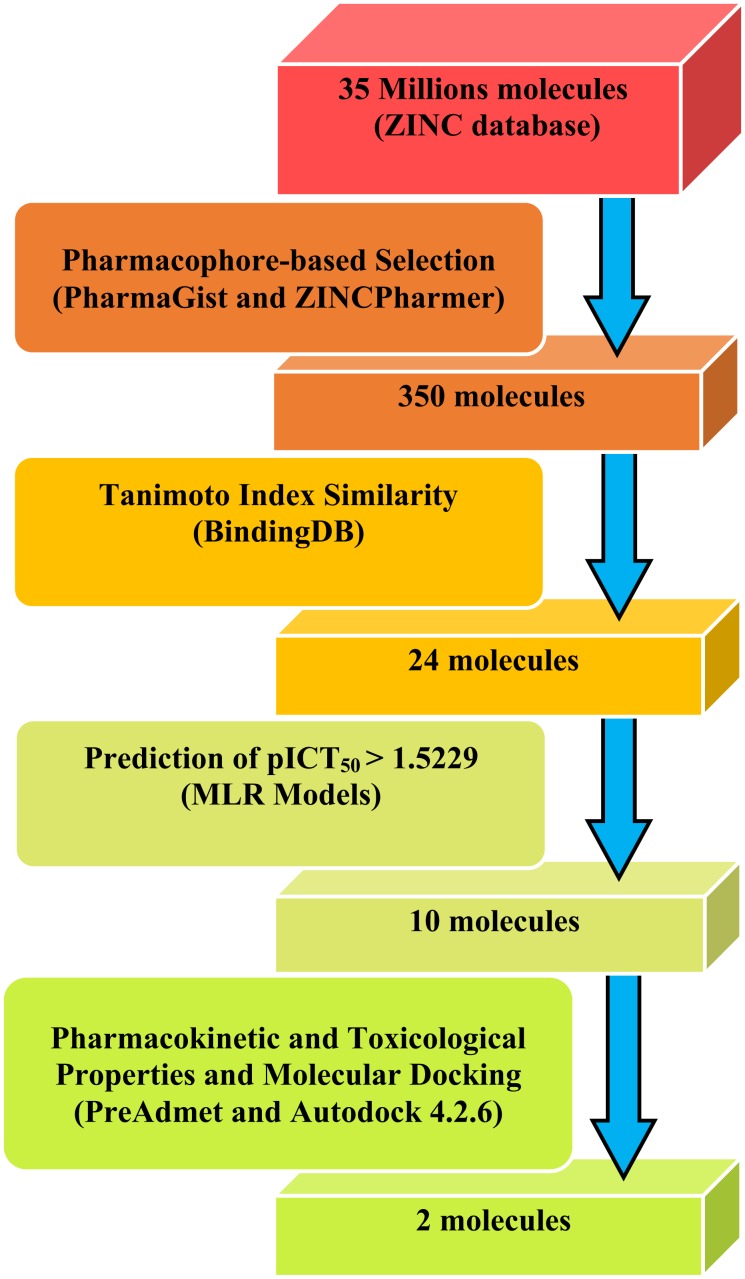
In this study, information on the biological receptor Chk1 [7, 8] and data of molecular structures and ICT50 from 30 molecules, selected from the literature [9], were used to perform virtual screening (diagram shown in Fig. 1) to obtain new caffeine analog molecules with epithelial anticancer activity.
Fig. (1).
Flowchart of the virtual screening of new caffeine analog molecules with potential epithelial anticancer activity.
2. MATERIALS AND METHODS
2.1. Data Set Selection
We selected 30 molecules derived from trialkylxanthine with experimental values of biological activity [9] related to the prevention of Epidermal Growth Factor (EGF) in the malignant transformation of epidermal cells of susceptible JB6 rats (P +) C141 (JB6 P +). Activity values were presented as ICT50 (50% inhibition of cell transformation) against epithelial cancer.
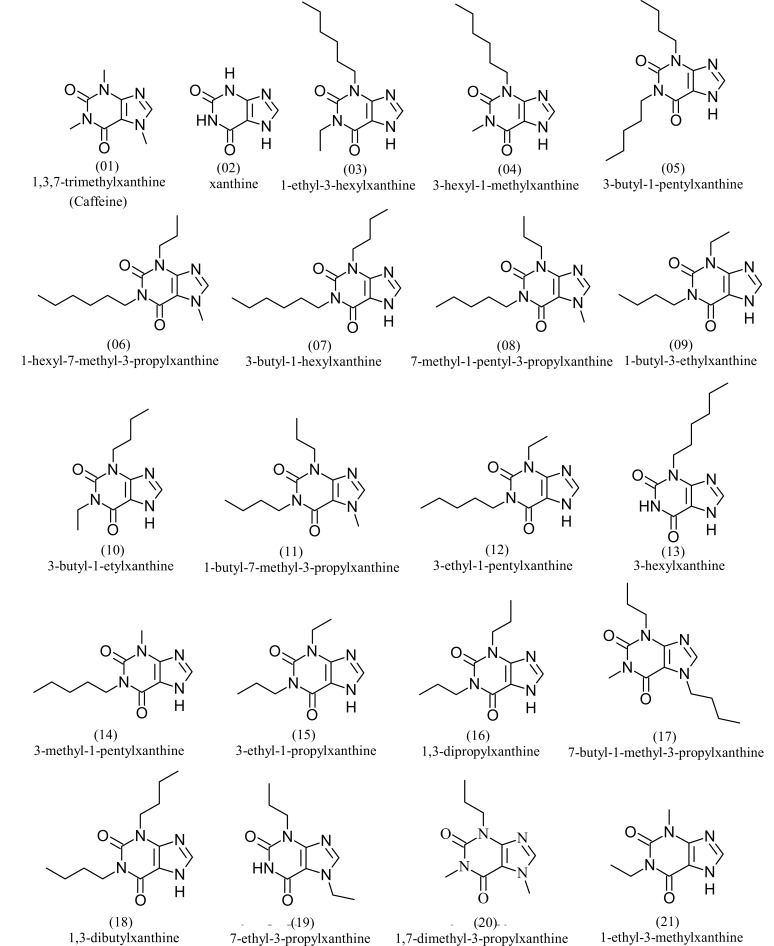
The training set molecules used for the construction of the pharmacophore model were selected in decreasing biological activity sequence (from 0.01 to 0.24 mM), containing the most active, since the activity is a critical factor for the determination of the pharmacophore characteristics and for the validation of the model Multiple Linear Regression (MLR) [11, 12]. Caffeine and xanthine molecules were introduced in the training set, according to the following considerations: (1) introduction of caffeine, here used as the reference/prototype molecule; (2) introduction of xanthine, because it has the active scaffold of the molecules here investigated. A total of 21 molecules comprise of the final training set.
The test set comprised 9 molecules randomly selected, which was here used for external validation of the MLR model. Structures were drawn using the ChemScketch software [13] and saved in the mol format, except caffeine, for which the crystallographic pose was retrieved from the Cambridge Structural Database portal at http://webcsd.ccdc.cam.ac.uk/. Both files were later.
converted to mol2 using the Open Babel tool 2.3.2 [14]. The geometries of the molecules were optimized according to the Molecular Mechanics MM+ Force Field, using the HyperChem 7 program [15].
2.2. Pharmacophore Model Perception
The mol2 files of the training set were inserted into the PharmaGist online server [16] to generate the pharmacophoric pattern of caffeine and analogues, with caffeine as pivot (keeped frozen) structure. PharmaGist generates a pharmacophore model based on the overlapping of individual pharmacohoric groups of input ligands (the training set). The method essentially aligns and overlaps the molecules with a pivot (the caffeine molecule) seeking a set of chemical and spatial characteristics that are common to the largest possible number of the input ligands. Assemblies with higher scores and with higher number of aligned ligands are considered better candidates for pharmacophoric models [16, 17].
2.3. Building and Validation of MLR Models
This step is crucial to evaluate the efficiency and predictive ability of MLR models in accurately identifying new active molecules. Pharmacophoric models indicated by PharmaGist were characterized according to their physicochemical and structural properties, such as: Number of atoms (A), general characteristics (GF), Spatial characteristics (SF), Aromatic region (Ar), Hydrophobic region (Hyd), hydrogen bond donor (Don), hydrogen bond receptor (Acc), anion (Neg) and cation (Pos). These characteristics were used to calculate the theoretical pICT50 values from multiple linear regression of the pharmacophoric characteristics of the molecules of the training set against the experimental pICT50 values.
Pearson's correlation was used to identify the relationship between the pharmacophoric properties of the 21 molecules in the training set associated with pICT50 values. The correlation cutoff was 0.3 according to previous studies conducted by Santos et al. (2015) [18]. After identifying the main pharmacological properties associated with biological activity, mono, bi, tri and tetra-parametric models were developed.
The experimental ICT50 values were converted to pICT50 in order to reduce inconsistencies caused by the statistical steps, with the equation (1): pICT50 = – log ICT50. The pICT50 values were predicted for the training set, test set and triaged molecules as well, by the application of the MLR models. The MLR analyses are implemented in the Statistica 7 program [19].
Data of the pharmacophoric characteristics of the test set molecules were obtained from the PharmaGist web server by the same previous way for the training set, except for the indication of a pivot molecule, where it was now selected automatically by the server itself.
2.4. Virtual Sorting in ZINCPharmer
The best pharmacophore file obtained in PharmaGist for caffeine and analogues was inserted in the ZINCPharmer web server, available at http://zincpharmer.csb.pitt.edu/ [20]. This tool performs a virtual screening from the ZINC database [20, 21], a database with approximately 35 million structures of commercially available compounds.
2.5. Tanimoto Similarity Search
The structures obtained from ZINCPharmer were selected by Tanimoto's similarity search procedure via the BindingDB web server. Molecules with similarity values greater than 0.6 were selected [22]. For this selection, 12 molecules (xanthine, caffeine and ten most active) compose reference set. Theoretical pICT50 values were calculated for the molecules obtained from the Tanimonoto test, using the MLR models. Pharmacological data for ZINC molecules were obtained in the same manner as described for both the training set and test set.
2.6. Determination of Pharmacokinetic and Toxicological Properties
This step was performed after setting a cut-off point from the pICT50 values of the five most active molecules of the training set. The resulting molecules were analyzed in the Derek software [23] (Toxicologic properties) and in the online Preadmet tool available at https://preadmet.bmdrc.kr/, This tool was used to determine selected pharmacokinetic properties (Human Intestinal Absorption - HIA, Plasma Protein Binding - PPB and Blood Brain Barrier - BBB) and toxicological (Mutagenicity and Carcinogenicity).
2.7. Molecular Docking Study
2.7.1. Selection of a Receptor-Ligand Complex Used in Molecular Docking
The selection of the Chk1 receptor was based on the propositions by Sarkaria et al. (1999) [7] and Lu et al. (2008) [8] (as previously discussed), highlighting the Chk1 receptor as a prominent target for the action of alkylxanthines against epithelial cancer.
The Protein Data Bank (PDB) provides several receptor-ligand complex alternatives for Chk1. However, the structure of this receptor complexed with caffeine is not available in the PDB, which makes it necessary to select the receptor-ligand complex (for further docking) as described below:
Selection of the receptor-ligand complex Chk1 (target protein) was performed taking into account (1) the structural similarity (visual inspection) of the ligands to the xanthine scaffold structure, regarding the presence of the heterocyclic rings; followed by (2) the overlap of the ligands with the caffeine structure, in relation to the overlap similarity values.
For PDB selection, only small ligand structures (characteristic common to the molecules here studied) from target in complex with only one ligand were selected. The structural similarity was accessed in the Discovery Studio 4.0 Client tool [24]. The ligands obtained were analyzed regarding the overlap similarity with caffeine.
2.7.2. Molecular docking of the most promising ZINC molecules
Initially, the receptor Chk1 file (ID: 2WMR, with resolution of 2.43 Å), chosen from the best caffeine overlapping ligand, was obtained from the PDB at http://www.rcsb.org/pdb/home/home.do [25], complexed with 6-morpholin-4-yl-9H-purine, PDB code ZYU [26]. Docking validation was performed by calculating the binding modes (between ZYU and Chk1) in the Autodock 4.2.6 program [27], using the standard genetic algorithm parameters, with population size 150, maximum number of Ratings 250000, maximum number of generations 27000 and crossover rate 0.8. The values for the dimensions of the grid box were: X = 25, Y = 30 and Z = 20 and the center location was x = 12.6747, y = -2.0751 and z = 7.9635. Ten solutions were calculated and the poses with lower binding energies were analyzed.
Docking validation is a process wherein a ligand (structure with crystallographic pose experimentally determined) is withdrawn from the structure of a receptor-ligand complex, and reintroduced to the receptor with the docking parameters to be validated. This process is conducted in order to verify that the coupling parameters specified in the input file for the docking method are reasonable and able to recover the structure and interactions of a known complex [27].
The best molecules obtained from pharmacokinetic and toxicological screening, caffeine and molecule 03 (the last two used as reference) were submitted to molecular docking using the same standard genetic algorithm parameters used in docking validation. The center location and grid box dimensions were also used as spatial orientation of the site of interaction between the ligand and the Chk1 receptor.
3. RESULTS and DISCUSSIONS
3.1. Pharmacophore Generation
The best model was chosen according to the pharmacophore candidate containing the highest score as well as the multiple alignment of the 21 ligands of the training set. PharmaGist generates pharmacological candidate scores based on the alignment of the ligands with the pivot molecule (here treated as reference and keeped rigid). The server algorithm uses values with standard weights for each pharmacophoric characteristic. Initially, the alignment of the pivot + ligand pair is scored by their common characteristics and then the multiple alignment between the best scored pairs is generated. Various multiple alignments are thus scored by the same way [16, 17].
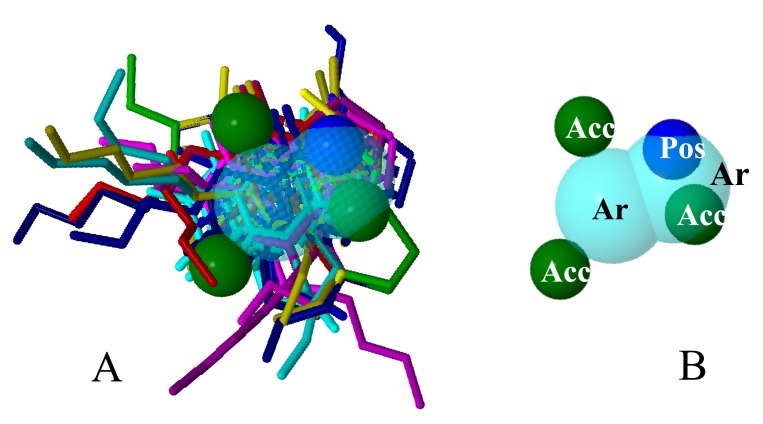
The quantitative characteristics of the best model are shown in Table 1, and the qualitative ones are shown in Figure 2.
Table 1.
Data found on the best PharmaGist pharmacophore model.
| Score | GF | SF | Ar | Hyd | Don | Acc | Neg | Pos | Molecules in multiple alignment |
|---|---|---|---|---|---|---|---|---|---|
| 60.852 | 6 | 6 | 2 | 0 | 0 | 3 | 0 | 1 | 01*, 02, 03, 04, 05, 06, 07, 08, 09, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21. |
A = number of atoms; GF = General Characteristics; SF = Spatial Characteristics; Ar = Aromatic; Hyd = Hydrophobic; Don = Hydrogen bond donor; Acc = Hydrogen bond acceptor; Neg = Anion; Pos = Cation.
*Pivot molecule (Caffeine).
Fig. (2).
Qualitative characteristics of the best model generated by PharmaGist, with aligned ligands (A) and no aligned ligands (B).
The model shows six general characteristics (GF), which represent the total pharmacophoric characteristics; six spatial characteristics (SF) related to the conformation of pharmacophoric regions; two aromatic regions (Ar); three hydrogen bond acceptor goups (Acc) and 1 cationic atom (Pos).
The training set consisted of the most active molecules, caffeine and xanthine. The structures and names of the molecules selected for the training set can be seen in Fig. 3.
Fig. (3).
Molecules of the training set used for pharmacophore generation.
The pharmacophoric data, the values of ICT50 and pICT50, and the their best correlations between with the variables of the 21 molecules of the training set are shown in Table 2. These data describe the individual pharmacophoric characteristics with significant correlations to the experimental activity values.
Table 2.
Pharmacophoric Characteristics of the Training set, pICT50 and ICT50 values, and correlation between the most Significant properties.
| Training Set Molecules | A | GF | SF | Ar | pICT50 | ICT50 |
|---|---|---|---|---|---|---|
| 1 (caffeine) | 24 | 9 | 9 | 0 | 0.3188 | 0.48 |
| 2 (xantine) | 15 | 9 | 8 | 3 | 0.3468 | 0.45 |
| 3 | 39 | 13 | 12 | 6 | 2.0000 | 0.01 |
| 4 | 36 | 13 | 12 | 6 | 1.6990 | 0.02 |
| 5 | 42 | 14 | 13 | 7 | 1.5229 | 0.03 |
| 6 | 45 | 14 | 14 | 8 | 1.5229 | 0.03 |
| 7 | 45 | 15 | 14 | 8 | 1.5229 | 0.03 |
| 8 | 42 | 13 | 13 | 7 | 1.3979 | 0.04 |
| 9 | 33 | 11 | 10 | 4 | 1.3010 | 0.05 |
| 10 | 33 | 11 | 10 | 4 | 1.3010 | 0.05 |
| 11 | 39 | 12 | 12 | 12 | 1.3010 | 0.05 |
| 12 | 36 | 12 | 11 | 11 | 1.3010 | 0.05 |
| 13 | 33 | 13 | 12 | 12 | 1.3010 | 0.05 |
| 14 | 33 | 12 | 11 | 11 | 1.0000 | 0.10 |
| 15 | 30 | 10 | 9 | 9 | 0.9208 | 0.12 |
| 16 | 33 | 11 | 10 | 10 | 0.8861 | 0.13 |
| 17 | 39 | 12 | 12 | 12 | 0.8861 | 0.13 |
| 18 | 39 | 13 | 12 | 12 | 0.8239 | 0.15 |
| 19 | 30 | 10 | 10 | 10 | 0.7959 | 0.16 |
| 20 | 30 | 10 | 10 | 10 | 0.6990 | 0.20 |
| 21 | 24 | 9 | 8 | 8 | 0.6198 | 0.24 |
| A | GF | SF | Hyd | pICT50 | ||
| A | 1.000000 | 0.904527 | 0.934078 | 0.974591 | 0.767452 | - |
| GF | 1.000000 | 0.962751 | 0.933680 | 0.800355 | - | |
| SF | 1.000000 | 0.960847 | 0.738313 | - | ||
| Hyd | 1.000000 | 0.747840 | - | |||
| pICT50 | 1.000000 | - |
Properties with absolute correlation values less than 0.3 and greater than - 0.3 were excluded. The best correlations were related to the pharmacophoric properties A, GF, SF, Hyd, with positive correlations between them greater than 0.9. The correlation values between the properties and pICT50 were also significant, all between 0.7 and 0.8. These results are considered of good statistical quality for the selection of more significant characteristics [18, 28].
3.1.1. Construction of MLR models
MLR models were built with the four best correlated pharmacophoric properties. The evaluation of the best models was carried out from the statistical descriptors of correlation coefficient (R), coefficient of determination (R2), adjusted coefficient of determination (RA2), Standard Error of Estimation (SEE), analysis of variance and T test, which can be seen in Table 3.
Table 3.
Statistical data of MLR models generated from the pharmacophoric characteristics of the training set in respect to pICT50 values.
| Mono-parametric Models | ||||||
|---|---|---|---|---|---|---|
| Eq. | Descriptor | R | R2 | R2A | SEE | F |
| 1 | GF | 0.8004 | 0.6406 | 0.6217 | 0.2703 | 33.8612 |
| 2 | A | 0.7675 | 0.5890 | 0.5674 | 0.2890 | 27.2268 |
| 3 | SF | 0.7383 | 0.5451 | 0.5212 | 0.3040 | 22.7680 |
| 4 | Ar | 0.0876 | 0.0076 | -0.0445 | 0.4491 | 0.1470 |
| Di-parametric Models | ||||||
| Eq. | Descriptor | R | R2 | R2A | SEE | F |
| 1 | GF +Ar | 0.8177 | 0.6686 | 0.6317 | 0.2666 | 18.1548 |
| 2 | GF + SF | 0.8092 | 0.6548 | 0.6164 | 0.2721 | 17.0700 |
| 3 | A + GF | 0.8068 | 0.6510 | 0.6122 | 0.2737 | 16.7865 |
| 4 | A + Ar | 0.7990 | 0.6384 | 0.5982 | 0.2786 | 15.8859 |
| 5 | A + SF | 0.7698 | 0.5926 | 0.5473 | 0.2956 | 13.0909 |
| 6 | SF + Ar | 0.7559 | 0.5714 | 0.5238 | 0.3032 | 11.9987 |
| Tri-parametric Models | ||||||
| Eq. | Descriptor | R | R2 | R2A | SEE | F |
| 1 | A + GF + SF | 0.8336 | 0.6949 | 0.6410 | 0.2632 | 12.9045 |
| 2 | A + GF + Ar | 0.8312 | 0.6909 | 0.6363 | 0.2650 | 12.6643 |
| 3 | GF + SF + Ar | 0.8238 | 0.6787 | 0.6220 | 0.2701 | 11.9706 |
| 4 | A + SF + Ar | 0.7999 | 0.6399 | 0.5764 | 0.2859 | 10.0714 |
| Tetra-parametric Model | ||||||
| Eq. | Descriptor | R | R2 | R2A | SEE | F |
| 1 | A + SF + GF + Ar | 0.8587 | 0.7374 | 0.6717 | 0.2517 | 11.2318 |
3.2. MLR Models and Validation
Data from the best MLR models resulting from combinations of one, two, three and four parameters are in bold. The values of R, R2 and RA2 increased with the increase in the number of parameters of the models, the highest values were found in tetra-parametric model and the smaller values in the mono-parametric model. The values of SEE decreased with increasing number of parameters, the tetra-parametric model presented the lowest value and the mono-parametric the highest.
The values of F decreased with increasing number of parameters. The results of the t-test for the regression coefficients (Table 4) indicate that the parameters A and GF in the tetra and tri-parametric models presented significant values, as well as the GF parameter in the bi and mono-parametric models. The results for R, R2, RA2, SEE and t-test, lead us to define, even with the low significance shown by the values of F, that the four models highlighted have a good level of statistical significance, being the tetra-parametric model the best classified.
Table 4.
Results of the t test for the regression coefficients of the best models.
| Model | Variable | t-test | p Value |
|---|---|---|---|
| Tetra-parametric A + SF + GF + Hyd |
Intercept | -0.9925 | 0.3357 |
| A | 1.1138 | 0.2818 | |
| GF | 2.2835 | 0.0364 | |
| SF | -0.9999 | 0.3323 | |
| Hyd | -0.2890 | 0.7763 | |
| Tri-parametric A+GF+SF |
Intercept | -2.1310 | 0.4880 |
| A | 1.4945 | 0.1534 | |
| GF | 2.3871 | 0.0289 | |
| SF | -1.5637 | 0.1363 |
| Model | Variable | t-test | p Value |
|---|---|---|---|
| Bi-parametric GF+Hyd |
Intercept | -1.5580 | 0.1366 |
| GF | 2.0179 | 0.0588 | |
| Hyd | 0.0112 | 0.9912 | |
| Mono-parametric GF |
Intercept | -3.0010 | 0.0073 |
| GF | 5.8190 | 0.000013 |
The regression equations for each of the best models (shown in bold, in Table 3) are shown in Table 5.
Table 5.
Regression equations for the best models.
|
Model (Eq.)
(n = 21) |
Equation |
|---|---|
| Mono-parametric (Eq1) | pICT50 = – 1.2168(±0.4055) + 0.1993(±0.0342) × GF |
| Bi-parametric (Eq2) | pICT50 = – 1.888(±0.4007) + 0.2128(±0.0355) × GF – 0.0230(±0.0187) × Hyd |
| Tri-parametric (Eq3) | pICT50 = – 0.9477(±0.4447) + 0.0334(±0.0224) × A + 0.2949(±0.1235) × GF – 0.2295(±0.1468) × SF |
| Tetra-parametric (Eq4) | pICT50 = – 0.8324(±0.4312) + 0.0415(±0.0220) × A + 0.2881(±0.1182) × GF – 0.2364(±0.1404) × SF – 0.0293(±0.0182) × Hyd |
3.2.1. Internal and external validation of MLR models
The equations proposed in Table 5 were used to predict the activity of the training set molecules (internal validation), the test set (external validation) and the selected molecules. The results for the pICT50 values calculated for each molecule of the training set from the regression equations are available in Table 6.
Table 6.
Prediction of pICT50 values of MLR models (equations 1 to 4) applied to the training set, experimental pICT50 values, mean error and maximum and minimum values
| Training set | Mono-parametric | Bi-parametric | Tri-parametric | Tetra-parametric | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Eq1 | Δ1 | Eq2 | Δ2 | Eq3 | Δ3 | Eq4 | Δ4 | pICT50 | |||||
| 1 (Caffeine) | 0.5766 | -0.2578 | 0.7262 | -0.4074 | 0.4432 | -0.1244 | 0.6297 | -0.3109 | 0.3188 | ||||
| 2 | 0.5766 | -0.2298 | 0.6572 | -0.3104 | 0.3719 | -0.0251 | 0.4043 | -0.0575 | 0.3468 | ||||
| 3 | 1.3737 | 0.6263 | 1.4393 | 0.5607 | 1.4357 | 0.5643 | 1.5200 | 0.4800 | 2.0000 | ||||
| 4 | 1.3737 | 0.3253 | 1.4393 | 0.2597 | 1.3354 | 0.3636 | 1.3954 | 0.3036 | 1.6990 | ||||
| 5 | 1.5730 | -0.0501 | 1.6290 | -0.1061 | 1.6014 | -0.0785 | 1.6670 | -0.1441 | 1.5229 | ||||
| 6 | 1.5730 | -0.0501 | 1.6060 | -0.0831 | 1.4721 | 0.0508 | 1.5259 | -0.0030 | 1.5229 | ||||
| 7 | 1.7722 | -0.2493 | 1.8188 | -0.2959 | 1.7670 | -0.2441 | 1.8140 | -0.2911 | 1.5229 | ||||
| 8 | 1.3737 | 0.0242 | 1.4163 | -0.0184 | 1.3065 | 0.0914 | 1.3790 | 0.0189 | 1.3979 | ||||
| 9 | 0.9752 | 0.3258 | 1.0597 | 0.2413 | 1.1043 | 0.1967 | 1.2260 | 0.0750 | 1.3010 | ||||
| 10 | 0.9752 | 0.3258 | 1.0597 | 0.2413 | 1.1043 | 0.1967 | 1.2260 | 0.0750 | 1.3010 | ||||
| 11 | 1.1744 | 0.1266 | 1.0885 | 0.2125 | 1.1408 | 0.1602 | 1.0561 | 0.2449 | 1.3010 | ||||
| 12 | 1.1744 | 0.1266 | 1.1115 | 0.1895 | 1.2700 | 0.0310 | 1.1971 | 0.1039 | 1.3010 | ||||
| 13 | 1.3737 | -0.0727 | 1.3013 | -0.0003 | 1.2352 | 0.0658 | 1.0949 | 0.2061 | 1.3010 | ||||
| 14 | 1.1744 | -0.1744 | 1.1115 | -0.1115 | 1.1697 | -0.1697 | 1.0725 | -0.0725 | 1.0000 | ||||
| 15 | 0.7759 | 0.1449 | 0.7320 | 0.1888 | 0.9386 | -0.0178 | 0.9032 | 0.0176 | 0.9208 | ||||
| 16 | 0.9752 | -0.0891 | 0.9217 | -0.0356 | 1.1043 | -0.2182 | 1.0501 | -0.1640 | 0.8861 | ||||
| 17 | 1.1744 | -0.2883 | 1.0885 | -0.2024 | 1.1408 | -0.2547 | 1.0561 | -0.1700 | 0.8861 | ||||
| 18 | 1.3737 | -0.5498 | 1.3013 | -0.4774 | 1.4357 | -0.6118 | 1.3441 | -0.5202 | 0.8239 | ||||
| 19 | 0.7759 | 0.0200 | 0.7090 | 0.0869 | 0.7091 | 0.0868 | 0.6375 | 0.1584 | 0.7959 | ||||
| 20 | 0.7759 | -0.0769 | 0.7090 | -0.0100 | 0.7091 | -0.0101 | 0.6375 | 0.0615 | 0.6990 | ||||
| 21 | 0.5766 | 0.0432 | 0.5422 | 0.0776 | 0.6727 | -0.0529 | 0.6315 | -0.0117 | 0.6198 | ||||
| *Mean error | 0.1989 | 0.1960 | 0.1704 | 0.1662 | |||||||||
| *Max | 0.6263 | 0.5607 | 0.6118 | 0.4800 | |||||||||
| *Min | 0.0200 | 0.0003 | 0.0101 | 0.0030 | |||||||||
* The module of the error values was considered for this determination.
The results of Table 6 allow us to observe that the models had a good predictability level for the training set molecules, since the error values (Δ - variation of the experimental pICT50 values), in general, were relatively low for all models. The tetra-parametric model should be emphasized when considering the Δ4 modules, which presented a mean error of 0.1662 and a minimum and maximum interval of 0.0030 to 0.4800, lower results for this series of data among all the models.
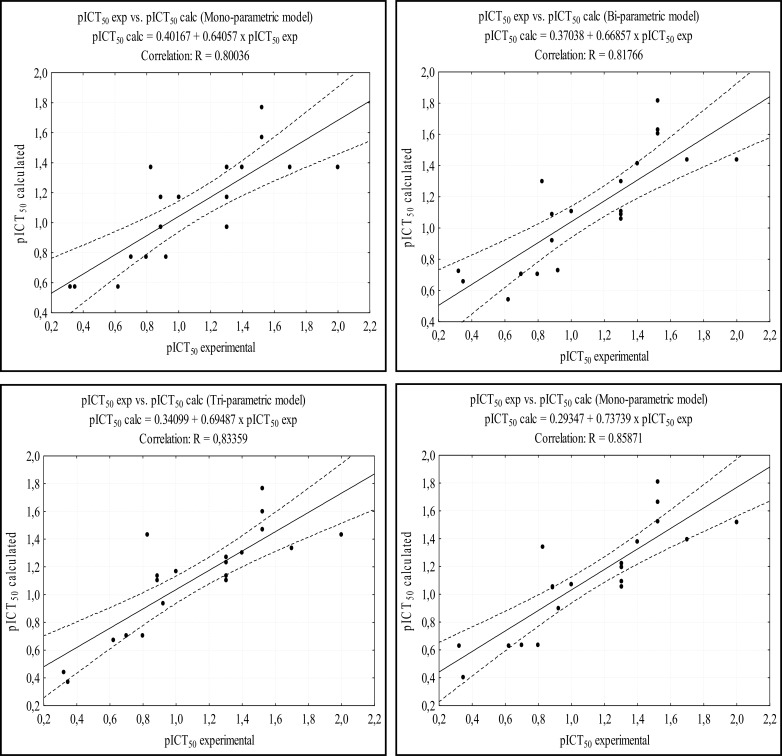
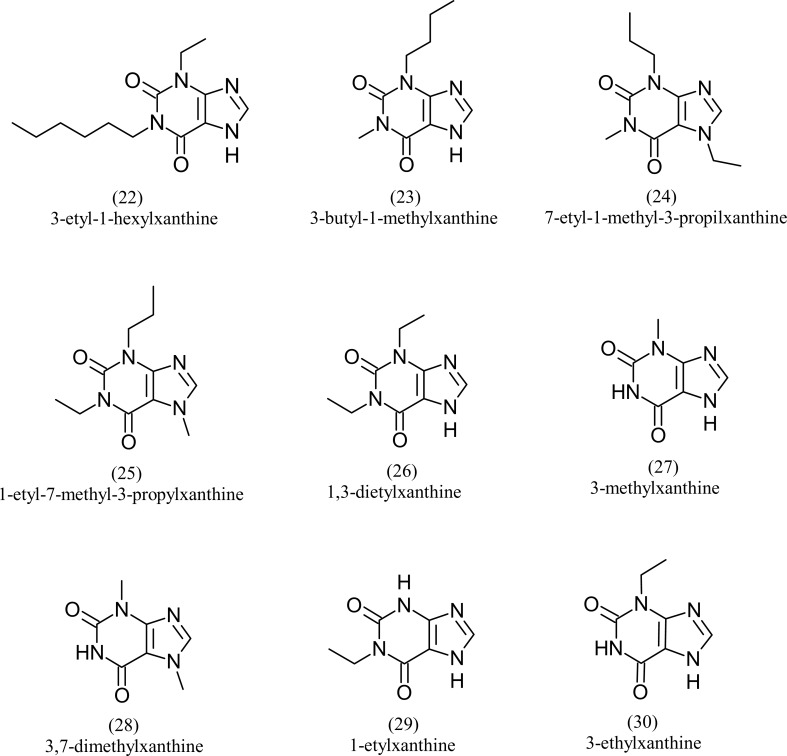
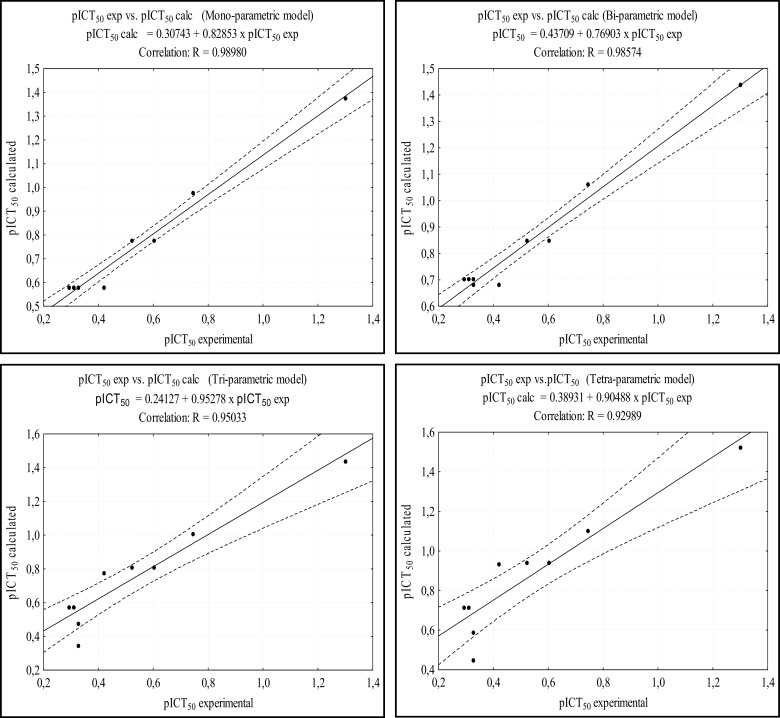
Fig. 4 shows the correlation between the calculated experimental pICT50 and calculated pICT50 for each model. The models presented good correlation indexes, with R values around 0.8. The analysis of Fig. 4 also shows good results of the tetra-parametric model, where it has the highest value of R (0.85872), with an excellent confidence level, against a value of R = 0.80037 of the mono-parametric model. Table 7 shows the pharmacophoric data, the ICT50 and pICT50 values of the 9 molecules (22 to 30) of the test set. Figure 5 shows the structures of the test set molecules. Data on the pICT50 calculations for the test set are presented in Table 8. These calculations were performed from the best MLR models, similar to those performed for the training set.
Fig. (4).
Correlation between the calculated and experimental pICT50 values of the training set are shown. The dashed lines near to the center line represent the confidence interval (95%).
Table 7.
Pharmacophoric characteristics of test set, pICT50 and ICT50 values.
| Test set | A | GF | SF | Hyd | pICT50 | ICT50 |
|---|---|---|---|---|---|---|
| 22 | 39 | 13 | 12 | 6 | 1.3010 | 0.05 |
| 23 | 30 | 11 | 10 | 4 | 0.7447 | 0.18 |
| 24 | 33 | 10 | 10 | 4 | 0.6021 | 0.25 |
| 25 | 33 | 10 | 10 | 4 | 0.5229 | 0.30 |
| 26 | 27 | 9 | 8 | 2 | 0.4202 | 0.38 |
| 27 | 18 | 9 | 8 | 1 | 0.3279 | 0.47 |
| 28 | 21 | 9 | 9 | 2 | 0.3279 | 0.47 |
| 29 | 21 | 9 | 8 | 1 | 0.3098 | 0.49 |
| 30 | 21 | 9 | 8 | 1 | 0.2924 | 0.51 |
Fig. (5).
Molecules of the test set.
Table 8.
Prediction of the pICT50 values for the MLR models (equations 1 to 4) in respect to the training set, experimental pICT50 values, mean error and maximum and minimum values.
| Mono-parametric | Bi-parametric | Tri-parametric | Tetra-parametric | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Structure | Eq1 | Δ1 | Eq2 | Δ2 | Eq3 | Δ3 | Eq4 | Δ4 | pICT50 |
| 22 | 1.3738 | -0.0727 | 1.4393 | -0.1382 | 1.4357 | -0.1347 | 1.5209 | -0.2199 | 1.3010 |
| 23 | 0.9752 | -0.2305 | 1.0597 | -0.3150 | 1.0041 | -0.2593 | 1.1020 | -0.3573 | 0.7447 |
| 24 | 0.7759 | -0.1739 | 0.8469 | -0.2449 | 0.8094 | -0.2073 | 0.9386 | -0.3365 | 0.6021 |
| 25 | 0.7759 | -0.2531 | 0.8469 | -0.3241 | 0.8094 | -0.2865 | 0.9386 | -0.4157 | 0.5229 |
| 26 | 0.5767 | -0.1565 | 0.6802 | -0.2600 | 0.7729 | -0.3527 | 0.9324 | -0.5121 | 0.4202 |
| 27 | 0.5767 | -0.2488 | 0.7032 | -0.3753 | 0.4722 | -0.1443 | 0.5877 | -0.2598 | 0.3279 |
| 28 | 0.5767 | -0.2488 | 0.6802 | -0.3523 | 0.3429 | -0.0150 | 0.4467 | -0.1188 | 0.3279 |
| 29 | 0.5767 | -0.2669 | 0.7032 | -0.3934 | 0.5724 | -0.2626 | 0.7123 | -0.4025 | 0.3098 |
| 30 | 0.5767 | -0.2842 | 0.7032 | -0.4107 | 0.5724 | -0.2800 | 0.7123 | -0.4199 | 0.2924 |
| *Mean error | 0.2150 | 0.3127 | 0.2158 | 0.3380 | |||||
| *Max | 0.2842 | 0.4107 | 0.3527 | 0.5121 | |||||
| *Min | 0.0727 | 0.1382 | 0.0150 | 0.1188 | |||||
* The module of the error values was considered for this determination.
The results of Table 8 show that the prediction using the models on the test set was satisfactory. The mono and tetra-parametric models showed mean error of 0.2150 and 0.3381, respectively. Fig. 6 shows the correlation between the calculated and experimental pICT50 values in each model. The four models presented excellent R values, all of them above 0.9,, at the 95% confidence level.
Fig. (6).
Correlation between the calculated and experimental pICT50 values of the test set. The dashed lines near to the center line represent the confidence interval (95%).
The tetra-parametric model can be considered the most reliable of the models, regarding the prediction of pICT50 values, also due to statistical consistencies observed from the regression data (R, R2, RA2, SEE e teste t) discussed above. The statistically significant values presented provide confidence to the MLR models and their predictive capacity [29, 30].
3.3. Selection from the ZINC Database
Pharmacophore-based screening performed on ZINCPharmer web server resulted in 350 ZINC molecules. Such tool selects the molecules according to the characteristics of the inserted pharmacophore. One of the main filters of this tool is the root mean square deviation (RMSD) estimation calculated between the characteristics of the input file (pharmacophile model) and the resulting molecules, as well as other filters such as molecular weight and rotational bonds [28].
3.3.1. Similarity of Tanimoto and Estimation of pICT50 Values for ZINC Molecules
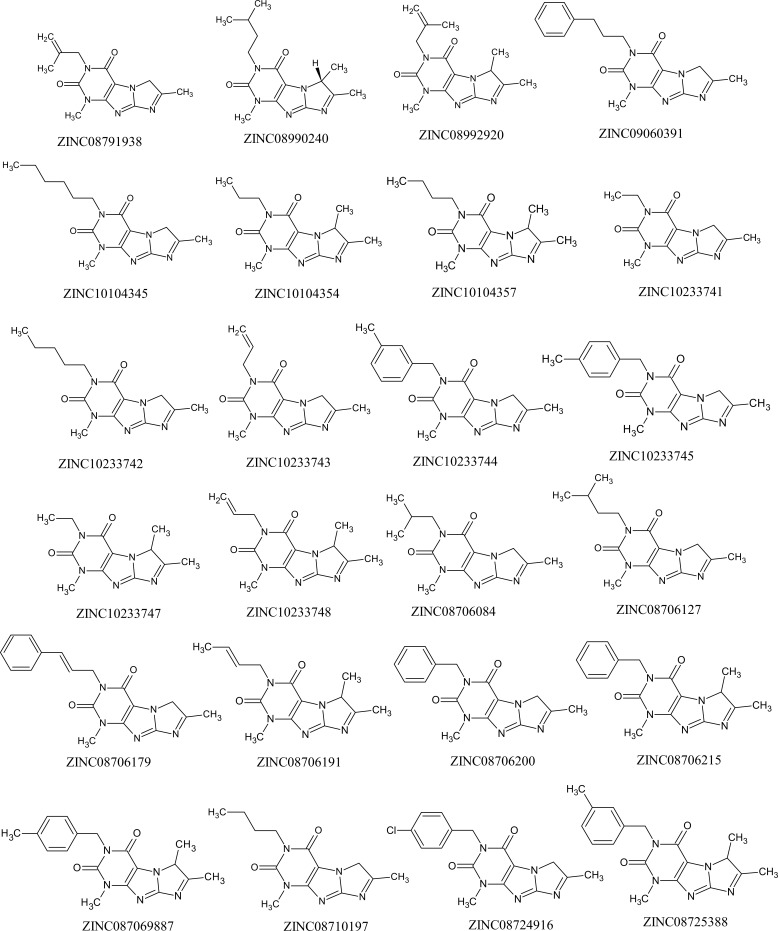
The pharmacophore data and the pICT50 values calculated for the ZINC molecules with the best Tanimoto indices can be seen in Table 9. The BindingDB server was used to select, from the 350 ZINC molecules screened in the previous step, the ones with values of maximum similarity greater than 0.6, which resulted in 24 molecules (Table 9).
Table 9.
Pharmacophore characteristics and pICT50 values calculated for the sorted molecules.
|
ZINC Molecules
Selected |
Pharmacophoric Characteristics | Tanimoto Index | Calculated pICT50 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| A | GF | SF | Hyd | Eq1 | Eq2 | Eq3 | Eq4 | ||
| ZINC08791938 | 34 | 13 | 13 | 5 | 0.67 | 1.3738 | 1.4623 | 1.0391 | 1.1053 |
| ZINC08990240 | 42 | 16 | 16 | 8 | 0.71 | 1.9716 | 2.0316 | 1.5027 | 1.5047 |
| ZINC08992920 | 37 | 14 | 14 | 6 | 0.66 | 1.5730 | 1.6520 | 1.2048 | 1.2522 |
| ZINC09060391 | 43 | 13 | 13 | 4 | 0.71 | 1.3738 | 1.4853 | 1.3399 | 1.5084 |
| ZINC10104345 | 42 | 15 | 15 | 7 | 0.77 | 1.7723 | 1.8418 | 1.4373 | 1.4823 |
| ZINC10104354 | 36 | 13 | 13 | 5 | 0.70 | 1.3738 | 1.4623 | 1.1059 | 1.1883 |
| ZINC10104357 | 39 | 14 | 14 | 6 | 0.73 | 1.5730 | 1.6520 | 1.2716 | 1.3353 |
| ZINC10233741 | 30 | 11 | 11 | 3 | 0.68 | 0.9752 | 1.0827 | 0.7746 | 0.8944 |
| ZINC10233742 | 39 | 14 | 14 | 6 | 0.76 | 1.5730 | 1.6520 | 1.2716 | 1.3353 |
| ZINC10233743 | 31 | 11 | 11 | 3 | 0.65 | 0.9752 | 1.0827 | 0.8080 | 0.9359 |
| ZINC10233744 | 40 | 13 | 13 | 4 | 0.61 | 1.3738 | 1.4853 | 1.2396 | 1.3838 |
| ZINC10233745 | 40 | 13 | 13 | 4 | 0.61 | 1.3738 | 1.4853 | 1.2396 | 1.3838 |
| ZINC10233747 | 33 | 12 | 12 | 4 | 0.67 | 1.1745 | 1.2725 | 0.9403 | 1.0414 |
| ZINC10233748 | 34 | 12 | 12 | 4 | 0.64 | 1.1745 | 1.2725 | 0.9737 | 1.0829 |
| ZINC08706084 | 36 | 14 | 14 | 6 | 0.70 | 1.5730 | 1.6520 | 1.1714 | 1.2107 |
| ZINC08706127 | 39 | 15 | 15 | 7 | 0.73 | 1.7723 | 1.8418 | 1.3371 | 1.3577 |
| ZINC08706179 | 41 | 13 | 13 | 4 | 0.61 | 1.3738 | 1.4853 | 1.2730 | 1.4254 |
| ZINC08706191 | 37 | 14 | 14 | 6 | 0.62 | 1.5730 | 1.6520 | 1.2048 | 1.2522 |
| ZINC08706200 | 37 | 11 | 11 | 2 | 0.62 | 0.9752 | 1.1057 | 1.0085 | 1.2145 |
| ZINC08706215 | 40 | 12 | 12 | 3 | 0.61 | 1.1745 | 1.2955 | 1.1742 | 1.3615 |
| ZINC08709887 | 43 | 14 | 14 | 5 | 0.61 | 1.5730 | 1.6750 | 1.4053 | 1.5308 |
| ZINC08710197 | 36 | 13 | 13 | 5 | 0.75 | 1.3738 | 1.4623 | 1.1059 | 1.1883 |
| ZINC08724916 | 37 | 11 | 11 | 2 | 0.61 | 0.9752 | 1.1057 | 1.0085 | 1.2145 |
| ZINC08725388 | 43 | 14 | 14 | 5 | 0.61 | 1.5730 | 1.6750 | 1.4053 | 1.5308 |
The bindingDB web server performs similarity search as a function of chemical fingerprints (a kind of set of chemical characteristics treated as chemical fingerprints) that characterize the molecules [31, 32].
Chemical fingerprints are used in conjunction with Tanimoto's similarity by comparing each selected molecule with each molecule in the reference set and ordering the molecules in function of maximum similarity to any active molecule in the reference set [31-33]. The closer to 1 the values are, the greater the degree of similarity of the molecules to the molecules indicated as a reference.
The structures of the 24 ZINC molecules sorted out can be seen in Fig. 7.
Fig. (7).
Structure of the ZINC molecules selected by the Tanimoto index.
3.4. Prediction of Pharmacokinetic and Toxicological Properties
At this stage, the selection of the best molecules was based on the best pharmacokinetic and toxicological properties. The higher the values for the human intestinal absorption rate and Plasma Protein Binding, the more efficient the drug is in respect to each of these pharmacokinetic properties. However, for blood-brain barrier, Mutagenicity and Carcinogenicity, low values are desirable.
The predictions of pharmacokinetic and toxicological properties (Table 10) were performed for the molecules selected by Tanimoto's similarity, located within the cut-off point having as reference molecules 3-7 (only molecules with pICT50 > 1.5229 were selected). The use of the cut-off point resulted in the selection of the 10 most active molecules (Table 10). The caffeine molecule and molecule 03 (most active) from the training set were inserted into Table 10 as reference values.
Table 10.
Prediction of pharmacokinetic and toxicological properties of molecules with better pICT50 values
| Pharmacokinetic Parameters | Toxicological Parameters | |||||
|---|---|---|---|---|---|---|
| Molecules |
HIA
(%) |
PPB
(%) |
BBB
(C.brain/C.blood) |
Mutagenicity | Carcinogenicity | |
| Result | Rat | Mouse | ||||
| Caffeine | 93.82 | 14.07 | 0.33 | Mutagenic | Positive | Negative |
| Molecule 03 | 88.13 | 75.91 | 1.60 | Mutagenic | Negative | Negative |
| ZINC08706084 | 96.27 | 50.13 | 0.59 | Mutagenic | Negative | Negative |
| ZINC08706127 | 96.89 | 55.18 | 0.75 | Mutagenic | Negative | Negative |
| ZINC08706191 | 97.53 | 64.25 | 0.65 | Mutagenic | Positive | Positive |
| ZINC08709887 | 98.83 | 89.15 | 0.47 | Mutagenic | Negative | Negative |
| ZINC08725388 | 98.83 | 94.19 | 0.28 | Mutagenic | Negative | Negative |
| ZINC08990240 | 97.41 | 63.51 | 1.09 | Mutagenic | Negative | Negative |
| ZINC08992920 | 97.53 | 63.15 | 0.80 | Mutagenic | Positive | Negative |
| ZINC10104345 | 97.41 | 74.20 | 0.18 | Mutagenic | Negative | Negative |
| ZINC10104357 | 96.89 | 60.59 | 0.90 | Mutagenic | Negative | Negative |
| ZINC10233742 | 96.89 | 68.86 | 0.15 | Mutagenic | Negative | Negative |
HIA = Human Intestinal Absorption; PPB = Plasma Protein Binding; BBB = Blood-Brain Barrier Penetration
The prediction of human intestinal absorption (HIA) is measured as a function of the absorption fraction, %HIA, described as the percentage of the dose of the drug administered orally to reach the hepatic portal vein. It is also defined as the rate of total absorbed mass divided by the dose of the drug. This property is used to assess the degree of absorption of a drug, orally administered, by the intestinal epithelium [34].
Absorption levels below 25% are considered poor and greater than 80% are considered high. The results of HIA were considered excellent since, for all the sorted molecules, HIA values were higher than the two reference values (caffeine = 93.82% and molecule 03 = 88.13%) and close to 100%. These values attribute high-grade intestinal absorption to the ZINC molecules.
Plasma protein binding property (PPB) is defined as the percentage of a drug bound to plasma proteins at clinically achieved concentrations of the drug. PPB is important for evaluating the performance of a drug in the bioavailable free fraction distributed across several tissues [35]. Values of PPB above 65% are considered of high clinical significance and lower values are of low significance [36-38].
The molecules ZINC08709887 and ZINC08725388 showed higher PPB values (89.15% and 94.19%, respectively), values higher than that of the molecule 03 used as reference. All the triad molecules presented PPB values (range, 50.13% to 94.19%) higher than that of caffeine (14.07%), showing satisfactory results when compared to this reference.
Experimental PPB results for caffeine, available in the literature, show values of 35% [39], 36% [40] and 40% [41]. These values show an average error of 22.93% in respect to the PPB value of caffeine calculated by Preadmet. The margin of error of 22.93% applied to the chosen molecules theoretically allows the framing of part of this set of molecules between the clinically relevant PPB values (above 65%).
The blood-brain barrier is a specialized structure that has a protective function of the Central Nervous System (CNS). This barrier controls and regulates the homeostasis of the central nervous system through the separation of brain (Cbrain) and systemic blood (Cblood). For a drug with biological activity in the CNS, a high penetration value is required. However, for a drug without CNS activity, as herein investigated, low penetration value is required, so that side effects are minimized [35, 42].
For the ZINC molecules the values of BBB were all lower than the value presented by molecule 03 (<1.60), see Table 10. According to the literature [43], BBB values less than 1 (Cbrain / Cblood <1) give the molecule an inactive status in the CNS. This means that only the molecule ZINC08990240 (BBB = 1.09) and molecule 03 (BBB = 1.60) showed penetration into the CNS, which qualifies the majority of ZINC molecules as inactive in the CNS.
The results for the pharmacokinetic parameters found for caffeine and ZINC molecules, are consistent with the findings of Liu, Shen, Shi and Cai (2016) [44], In their study, the authors concluded that caffeine is directly related to the reduction of the risk of malignant melanoma. Through meta-analysis, the authors identified a low relative risk of malignant melanoma related to high consumption of caffeinated coffee, suggesting that caffeine present in caffeinated coffee has a chemopreventive effect against malignant melanoma. Unlike decaffeinated coffee that showed no significant relationship with the risk of malignant melanoma.
The toxicological properties, shown in Table 10, were predicted by the Ames test (mutagenicity) and carcinogenicity test. The Ames test consists of an assay that aims the mutagenic reversal of bacteria (Salmonella typhimurium) tested for histidine independence. Preadmet simulates tests for strains TA100 and TA1535, which serve to evaluate the mutagenic potential of molecules, ie the ability to generate mutation in an organism. This test simulates an environment in which a post-mitochondrial mouse liver supernatant mixture treated with a mixture of phenobarbital / β-naphthoflavone (s9) may be present or absent [45, 46]. The in silico carcinogenicity test simulates the presence of the drug in the organism of rats and mice, and it is possible to evaluate if the drug has the power to generate tumors [47].
Regarding the results for mutagenicity in Preadmet software, all screened and reference molecules were classified as mutagenic. Even showing mutagenicity, where elimination of molecules for this reason would be premature, studies with rats have shown that a mutagenic molecule when administered in combination with an antimutagenic (chemopreventive) agent may cause suppression of the mutagenic action [48, 49].
For carcinogenicity in Preadmet softtware, caffeine and the molecule ZINC08992920 showed positive results (non-carcinogenic) in rats and negative (carcinogenic) results in mice. Only the molecule ZINC08706191 presented positive results, both for mouse and rat, which allows to classify it with non-carcinogenic in Preadmet softtware.
Additional data on toxicological parameters for caffeine, molecule 03 and ZINC molecules tested in the Derek program [23] can be seen in Table 11.
Table 11.
Toxicity results obtained using the Derek software.
| Molecule |
Toxicity Prediction Alert
(Lhasa prediction) |
Toxicophoric Group | Toxicity Alert | Hits* |
|---|---|---|---|---|
| Caffeine | Chromosome damage | Xhantine | Certain | 4 |
| Teratogenicity | Xhantine | Probable | ||
| Molecule 03 | Teratogenicity | Xhantine | Plausible | 4 |
| ZINC08706084 | Skin sensitisation | Imine or alpha,beta-unsaturated imine | Plausible | 4 |
| ZINC08706127 | Skin sensitisation | Imine or alpha,beta-unsaturated imine | Plausible | 4 |
| ZINC08706191 | No Alerts | — | No Alerts | 1 |
| ZINC08709887 | Skin sensitisation | Imine or alpha,beta-unsaturated imine | Plausible | 4 |
| ZINC08725388 | Skin sensitisation | Imine or alpha,beta-unsaturated imine | Plausible | 4 |
| ZINC08990240 | Skin sensitisation | Imine or alpha,beta-unsaturated imine | Plausible | 4 |
| ZINC08992920 | Skin sensitisation | Imine or alpha,beta-unsaturated imine | Plausible | 3 |
| ZINC10104345 | Skin sensitisation | Imine or alpha,beta-unsaturated imine | Plausible | 4 |
| ZINC10104357 | Skin sensitisation | Imine or alpha,beta-unsaturated imine | Plausible | 4 |
| ZINC10233742 | Skin sensitisation | Imine or alpha,beta-unsaturated imine | Plausible | 4 |
* Sum of hits corresponding to the toxicological analyzes performed in Preadmet and Derek.
Data of Table 11 indicate that both reference molecules (caffeine and molecule 03) showed teratogenicity, representing the possibility of a substance causing abnormal formation during the gestation period. Only caffeine presented a possibility of chromosome damage, which represents the ability to generate changes in the molecular structure of the chromosome. The Derek software identified the reference scaffold/moety of xanthine present in caffeine and molecule 03 as part of the structure of the molecules responsible for the teratogenic and chromosome damage effects. None of the ZINC molecules tested showed potential for teratogenic and chromosome damage. These results allow to classify the ZINC molecules as better than the reference molecules in respect to the two toxicological parameters mentioned.
However, nine of the ten ZINC molecules tested showed toxicity to skin sensitization. This effect corresponds to an allergic response after contact of a substance with the skin [50]. This sensitization was attributed by the Derek program to the imine toxicoforic group, being able to occur nucleophilic attack of skin proteins to the carbon atom of the imine group [51].
Unlike the results presented for tests of mutagenicity and carcinogenicity carried out in Preadmet, all molecules were excluded from the mutagenicity and carcinogenicity alert in the Derek software, no toxicological groups are identified that could indicate such toxicological parameters.
With respect to chromosome damage and mutagenicity, a review on caffeine [52], showed that chromosome damage data from studies with caffeine were obtained at concentrations of about 6 to 100 times higher than expected for frequent coffee users and with magnitude above the lethal dose of caffeine in humans. Such as most non-mutagenic substances in in vitro tests, they are non-carcinogenic in mammals, it is unlikely that at the usual consumption level caffeine presents any mutagenic and carcinogenic risk. Such dose-effect dependency of caffeine on mutagenicity was also evidenced in a study on the mutagenic action of caffeine in higher organisms [53]. It is possible to infer from these studies that mutagenicity (existing for all ZINC molecules tested in Preadmet) has a strong dose-effect relationship when it comes to caffeine. In this context, the results of Preadmet and Derek obtained for the tested molecules can be interpreted as a possibility of toxicity depending on the level of consumption or the dose administered, which suggests that mutagenicity for the most promising molecules should be more thoroughly investigated in in vitro and in vivo analyzes, and not taken as conclusive.
Still in Table 11 it is possible to verify the hits related to the toxicity of each molecule. Such as the caffeine that has been shown to be mutagenic and carcinogenic in mice (Preadmet), besides teratogenic and promoter of chromosome damage (Derek), 4 hits of toxicity are predicted. Since the higher the hit value the greater the toxicity, the molecules ZINC08992920 (3 hits) and ZINC08706191 (1 hit and no toxicity alert on Derek) as less toxic in comparison to the other molecules (all with 4 hits). The molecule ZINC08706191 is the best one qualified in respect to toxicity, because there is no carcinogenic risk in both Preadmet and Derek softwares.
3.5. Molecular Docking
Molecular docking allows the collection of data on interactions between ligand and receptor, making possible the selection of the best ligand poses as a function of the lowest free energy which results from that interaction. For the molecular docking step only the molecules ZINC08706191 (1 hit of toxicity) and ZINC08992920 (3 hit of toxicity), classified as less toxic, were selected.
3.5.1. Receptor-ligand Complex
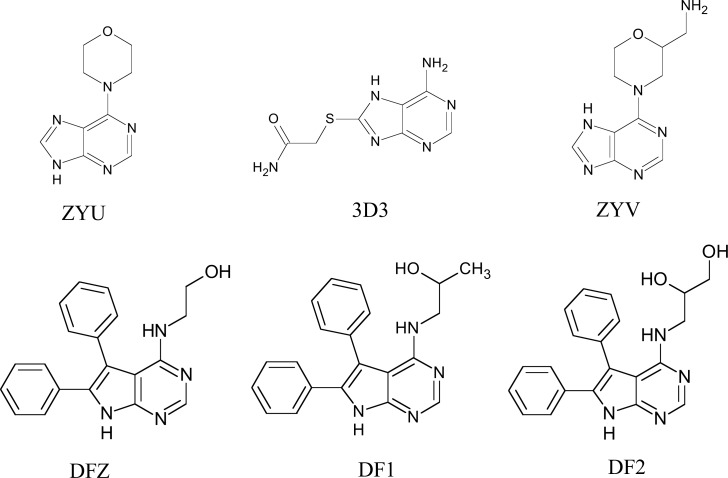
Selection of the receptor-ligand complex Chk1 was performed initially by taking into account the structure of the ligand compared to the xanthine scaffold and size of the ligands (smaller molecules), which resulted in 6 molecules (Fig. 8).
Fig. (8).
Structure of the complexed ligands with Chk1 selected from PDB.
The results for overlap similarity between the selected binders and caffeine can be seen in Table 12. Values for overlap similarity attributed to 100% estrogenic contribution (100ste), 100% electrostatic contribution (100elt), 60% steric and 40% electrostatic (60ste/40elt), 40% steric and 60% electrostatic (40ste / 60elt) and 50% of both contributions (50ste / elt), were determined.
Table 12.
Overlap similarity values of the ligands analyzed.
| Complex | Ligand | Overlap | ||||
|---|---|---|---|---|---|---|
| PDB Code | PDB Code | 100ste | 100elt | 60ste/40elt | 40ste/60elt | 50ste/elt |
| 2WMU | ZYU | 0.8967 | 0.4818 | 0.6860 | 0.6098 | 0.6476 |
| 2CGX | 3D3 | 0.8349 | 0.5361 | 0.6068 | 0.6226 | 0.5645 |
| 2WMV | ZYV | 0.8138 | 0.3378 | 0.5865 | 0.4969 | 0.5384 |
| 2BRM | DFZ | 0.7152 | 0.5368 | 0.5709 | 0.5852 | 0.5503 |
| 2BRN | DF1 | 0.6874 | 0.4597 | 0.6244 | 0.5991 | 0.6131 |
| 2BRO | DF2 | 0.6658 | 0.3912 | 0.5616 | 0.4997 | 0.5408 |
100ste = 100% of steric contribution; 100elt = 100% of electrostatic contribution; 60ste/40elt = 60% steric and 40% electrostatic; 40ste/60elt = 40% steric and 60% electrostatic; and 50ste/50elt = 50% of both contribution.
The ZYU ligand presented higher values of similarity of overlap with caffeine at the 100ste, 60ste/40elt and 50ste/elt levels (0.8967, 0.6860 and 0.6476, respectively), second higher values of 40ste/60elt (0.6098) and median value for 100elt (0.4818). The closer to 1 the values are, the greater the degree of similarity between the binder and the caffeine. While, in contrast, for smaller values, the greater the degree of structural difference [24].
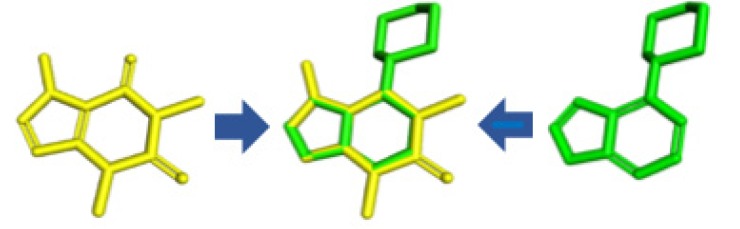
The results for overlap similarity allow us to consider the ZYU ligand with a high degree of similarity with caffeine. This ligand complexed to the Chk1 receptor, is available from the PDB code 2WMU, and its IUPAC name is 6-morpholine-4-yl-9H-purine. Fig. 9 shows the overlap of ZYU with caffeine.
Fig. (9).
Caffeine overlap (yellow) with ZYU ligand (green). (The color version of the figure is available in the electronic copy of the article).
3.5.2. Validation and Molecular Docking
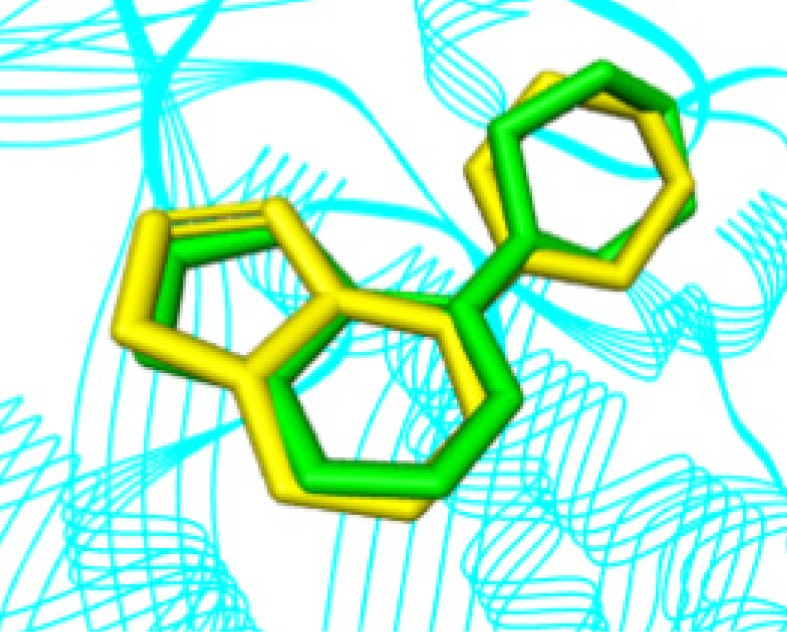
The comparison between the conformations of the crystallographic ligand (complexed with Chk1 and experimental values of crystallography) with the computational data resulting from the redocking (RMSD = 0.570), shows that the parameters used in the docking protocol were representative. RMSD values below the tolerance level of up to 2.0 Å are considered to be of good quality [54, 55]. The alignment between experimental and computational conformations, which qualitatively shows this result, can be seen in Fig. 10.
Fig. (10).
Comparison between crystallographic ligand pose (in green) and the top-ranked pose resulting from docking (yellow). (The color version of the figure is available in the electronic copy of the article).
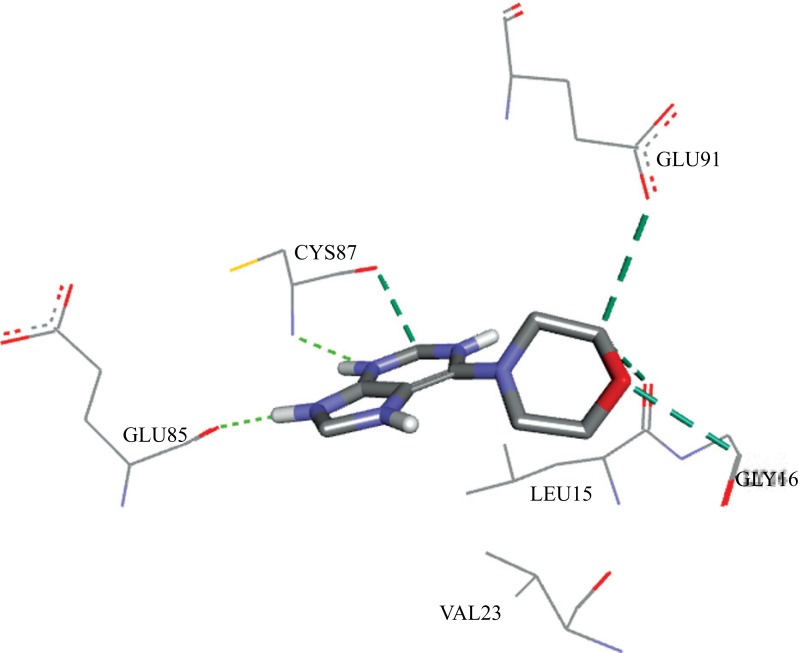
The interactions resulting from the docking pose of the ZYU with the Chk1 receptor were: two hydrogen bonds with CYS87 and GLU85, and four carbon-hydrogen bonds with CYS87, GLU91, LEU15 and GLY16, in a total of six interactions. Fig. 11 shows the interactions of ZYU with Chk1.
Fig. (11).
Interactions between the ZYU ligand and the Chk1 receptor, calculated by Autodock 4.2.6.
The parameters related to the dimensions of the grid box and coordinates of the ligand's center, used in the docking validation, were applied to the reference molecules (caffeine and molecule 03), ZINC08992920 and ZINC08706191.
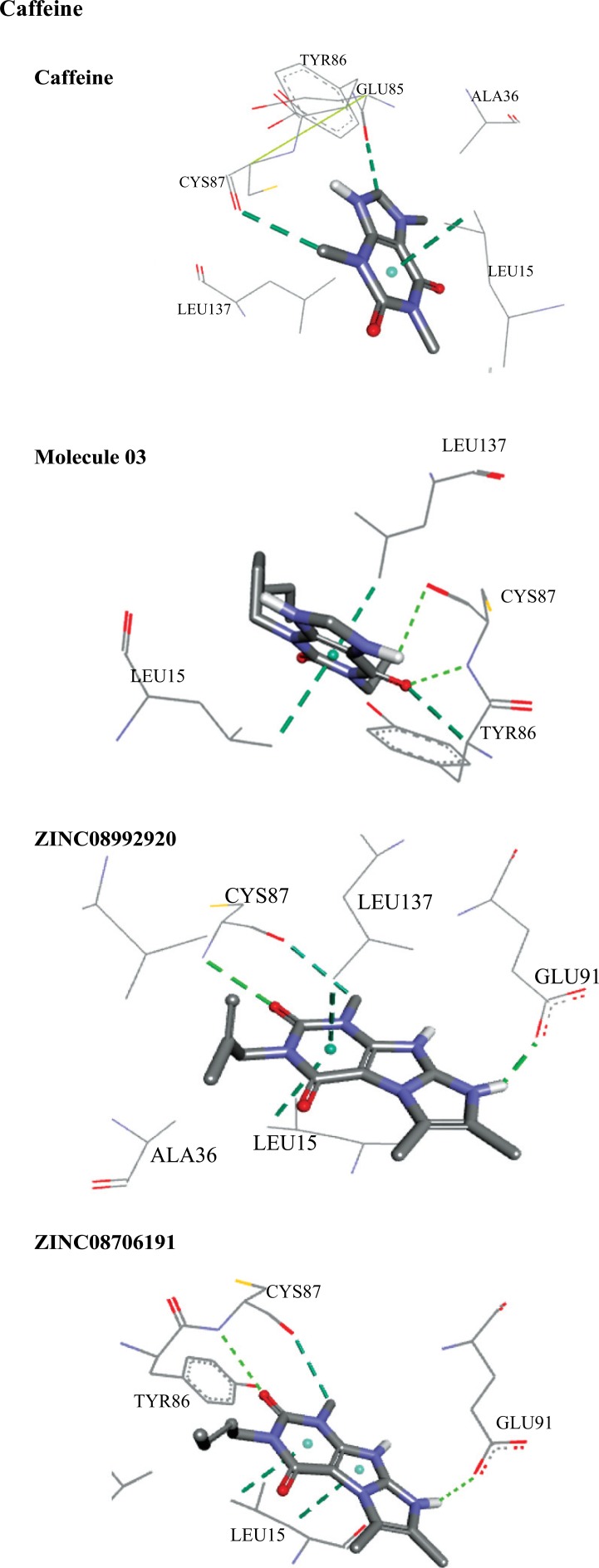
Fig. 12 shows the interactions for individually docked molecules. Caffeine showed two carbon-hydrogen bonds, with GLU85 and CYS87, and a pi-sigma bond with LEU15, with three interactions in total. The molecule 03 showed two hydrogen bonds with CYS87, one carbon-hydrogen bond with TYR86 and two pi-sigma bonds, with LEU137 and LEU15, in a total of five interactions. The molecule ZINC08992920 interacted with the receptor through two hydrogen bonds, with CYS87 and GLU91, one carbon-hydrogen bond with CYS87 and two pi-sigma bonds with LEU137 and LEU15, in a total of five interactions. The molecule ZINC08706191 interacted with two hydrogen bonds with CYS87 and GLU91, one carbon-hydrogen bond with CYS87 and two pi-sigma bonds with LEU15, in a total of five interactions.
Fig. (12).
Interactions between the best screened molecules and the Chk1 receptor calculated by Autodock 4.2.6.
Quantitative data of distances and binding free energies (BFE) between the ligands and the Chk1 receptor can be seen in Table 13. It is possible to verify that, among the reference molecules (caffeine and molecule 03), the increase in the number of interactions resulted in the lowering of free binding energy, which indicates a higher degree of spontaneity of the interactions. This effect is noticeable in molecules ZINC08992920 and ZINC08706191 with five interactions each and smaller binding energies than the reference molecules. Common interactions with the amino acids CYS87 and LEU15 occurred in all molecules analyzed, indicating that they have key importance in the epithelial anticancer activity.
Table 13.
Distances and BFE between ligands and Chk1.
| Molecule | Aminoacid | Distance (Å) | Type | BFE (kcal mol-1) |
|---|---|---|---|---|
| ZYU | CYS87 | 2.865 | Hydrogen bond | -5.37 |
| CYS87 | 3.302 | Carbon-hydrogen bond | ||
| GLU85 | 1.730 | Hydrogen bond | ||
| GLU91 | 3.402 | Carbon-hydrogen bond | ||
| LEU15 | 3.515 | Carbon-hydrogen bond | ||
| GLY16 | 3.236 | Carbon-hydrogen bond | ||
| Caffeine | GLU85 | 2.916 | Carbon-hydrogen bond | -5.14 |
| CYS87 | 2.940 | Carbon-hydrogen bond | ||
| LEU15 | 3.528 | Pi-sigma | ||
| Molecule 03 | CYS87 | 2.879 | Hydrogen bond | -5.90 |
| CYS87 | 2.401 | Hydrogen bond | ||
| TYR86 | 2.985 | Carbon-hydrogen bond | ||
| LEU137 | 3.560 | Pi-sigma | ||
| LEU15 | 3.581 | Pi-sigma | ||
| ZINC08992920 | CYS87 | 2.911 | Hydrogen bond | -7.13 |
| CYS87 | 3.337 | Carbon-hydrogen bond | ||
| GLU91 | 2.007 | Hydrogen bond | ||
| LEU137 | 3.753 | Pi-sigma | ||
| LEU15 | 3.867 | Pi-sigma | ||
| ZINC08706191 | CYS87 | 2.947 | Hydrogen bond | -7.41 |
| CYS87 | 3.165 | Carbon-hydrogen bond | ||
| GLU91 | 2.428 | Hydrogen bond | ||
| LEU15 | 3.836 | Pi-sigma | ||
| LEU15 | 3.930 | Pi-sigma |
It is noted that the pi-sigma interaction with LEU15 and LEU137 on molecules 03, ZINC08992920 and ZINC08706191, attributed lower BFE to these three molecules when compared to the interactions and BFE of ZYU and caffeine that have no Pi-sigma interactions with none of these amino acids.
Comparing the molecules ZINC08992920 and ZINC08706191, we can observe the lowering of the free energy of binding (-7.41) in ZINC08706191 that has two interactions with LEU15 in respect to the BFE (-7.13) in ZINC08992920, that has a Pi-sigma interaction with LEU15 and LEU137, indicating that the interaction with LEU15 is a favorable factor for lowering BFE between these two molecules. The pi-sigma interactions have strong hydrophobic characteristic, and when related to favorable entropic factors can promote the lowering of the free energy of ligand-receptor binding [56].
It is interesting to note that, due to structural similarity, the numerical values of the pharmacophoric characteristics, pharmacokinetics properties and the activity values calculated by the MLR models were similar for the molecules ZINC08992920 and ZINC08706191 (see Table 14), which resulted in approximate values of BFE, due to the interactions similarity.
Table 14.
Correlation between the variables analyzed and BFE.
| Pharmacophoric Characteristics | Pharmacokinetic Properties | Calculated pICT50 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecule | A | GF | SF | Hyd | HIA% | PPB% | BBB | Eq1 | Eq2 | Eq3 | Eq4 | BFE |
| Caffeine | 24 | 9 | 9 | 0 | 93.82 | 14.07 | 0.33 | 0.58 | 0.73 | 0.44 | 0.63 | -5.14 |
| Molecule 03 | 39 | 13 | 12 | 6 | 88.13 | 75.91 | 1.60 | 1.37 | 1.44 | 1.44 | 1.52 | -5.90 |
| ZINC08992920 | 37 | 14 | 14 | 6 | 97.53 | 63.15 | 0.80 | 1.57 | 1.65 | 1.20 | 1.25 | -7.13 |
| ZINC08706191 | 37 | 14 | 14 | 6 | 97.53 | 64.25 | 0.65 | 1.57 | 1.65 | 1.20 | 1.25 | -7.41 |
| CoBFE | -0.70 | -0.89 | -0.96 | -0.79 | -0.66 | -0.64 | -0.02 | -0.89 | -0.90 | -0.61 | -0.54 | |
A = number of atoms; GF = General Characteristics; SF = Spatial Characteristics; Ar = Aromatic; Hyd = Hydrophobic; HIA% = Human Intestinal Absorption; PPB = Plasma Protein Binding; BBB = Blood Brain Barrier.
3.5.3. Correlation Between Pharmacophoric Properties, Pharmacokinetics and pICT50 with BFE
Correlations between pharmacophoric, pharmacokinetic properties as well as the calculated pICT50 with BFE (Table 14) showed negative values for all analyzed variables, which indicate an inverse proportionality relationship between them and BFE. These results are consistent with the significance of each analyzed variable, which allows us to consider that an increase in the values of the pharmacophoric properties, HIA and PPB promotes increase of the activity and decrease of BFE.
The low correlation with BBB (-0.02) indicates that the value of BFE has no respect to this property, as expected, since this variable is related to CNS side effects, signalizing that molecules ZINC08992920 and ZINC08706191 do not show such effect.
The calculated pICT50 values and their correlations with BFE for the molecules ZINC08992920 and ZINC08706191, the mono (Eq1) and bi (Eq2) parametric models, with correlations -0.89 and -0.90, respectively, have a higher correlation with the BFE values than the tri (Eq3) and tetra (Eq4) parametric models, which is consistent with the activity observed for these molecules predicted by the mono and bi parametric models.
CONCLUSION
The virtual screening performed here showed reliable results in all its stages. The MLR models built from the pharmacophoric data were classified with excellent statistical quality, due to the high values of R, R2, RA2 and low SEE values, and good statistical significance confirmed by the t-test, with emphasis on the tetra-parametric model, considered the best model. The validation of the MLR models indicated excellent predictive power of the models, indicated by the low values of errors in both the predictions for the training set and the test set. These results were also qualified by the good correlations between experimental and calculated pICT50 values.
The pharmacophore model inserted in the ZINCPharmer webserver aided us to select 350 molecules, and subsequently 24 were selected using the BindingDB by Tanimoto similarity. The low amount of molecules found from the virtual screening in this webserver evidences the quality of the analytical filters used in the screening.
The pharmacokinetic properties indicated in general better results than the reference values and limits available in the literature, with HIA values higher than 90% and PPB (considered prediction errors) within the limits of clinical significance. BBB also indicated good results, where only molecules 03 (reference) and ZINC08990240 showed possible effects on the CNS.
The molecules ZINC08992920 (3 hits of toxicity, with mutagenicity and carcinogenicity alerts) and ZINC08706191 (1 hit of toxicity, only with one mutagenicity alert) presented lower toxicities among all the tested molecules. The attested mutagenicity needs to be better evaluated (in vitro e in vivo tests), because mutagenicity may have an effect depending on the level of consumption or dose of administered substance.
BFE values correlated to the properties analyzed show that BBB has no respect to BFE values, as expected, due to the low BBB values observed and this indicates side effect. High negative correlations between BFE and pharmacophoric properties, HIA, PPB, pICT50 calculated by Eq1 and by Eq2, indicate that these properties are fundamentally related to the low BFE values.
Results here obtained and discussed, which sought to evaluate the potential epithelial anticancer activity of the screened molecules, allowed us to classify the molecules ZINC08992920 and ZINC08706191 as the best ones along the observed series, and to point to ZINC08706191 (presented minor hit in toxicity) as the best among all. However, these two molecules can subsequently be subjected to further analysis to a better evaluation of their pharmacological potential against epithelial cancer.
Ethics Approval and Consent to Participate
Not applicable.
Human and Animal Rights
No Animals/Humans were used for studies that are base of this research.
Consent for Publication
Not applicable.
ACKNOWLEDGEMENTS
We acknowledge the support provided by the PROPESP/UFPA. To the Postgraduate Program in Biodiversity and Biotecnoloy of Amazon (PPG-Bionorte), Paraense Museum Emilio Goeldi (MPEG). Laboratory of Molecular Modeling and Simulation System of Federal Rural University of Amazônia (UFRA-Brazil) and Computational Laboratory of Pharmaceutical Chemistry, Faculty of Pharmaceutical Sciences of Ribeirão Preto, University of São Paulo, Ribeirão Preto (USP/RP) for computational support. To Elias Carvalho Padilha for his assistance in the revision of the English language.
Conflict of Interest
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Rosenberg S.A. Progress in human tumour immunology and immunotherapy. Nature. 2001;411(6835):380–384. doi: 10.1038/35077246. [DOI] [PubMed] [Google Scholar]
- 2.Cairns J. The origin of human cancers. Nature. 1981;289(5796):353–357. doi: 10.1038/289353a0. [DOI] [PubMed] [Google Scholar]
- 3.Brentani R.R., Chammas R., Coelho F.R.G. Mecanismos de invasão e metástases. In: Brentani M.N., Coelho F.R.G., Iyeyasu H., Kowalski L.P., editors. Bases da Oncologia. 1st ed. São Paulo: Livraria e Editora Marina; 1998. pp. 1–21. [Google Scholar]
- 4.WHO - World Health Organization World Cancer Report. 2014 www.who.int/cancer/
- 5.Popim R.C., Corrente J.E., Marino J.A.G., Souza C.A. Skin cancer: use of preventive measures and demographic profile of a risk group in the city of Botucatu. Cien. Saude Colet. 2008;13(4):1331–1336. doi: 10.1590/s1413-81232008000400030. [DOI] [PubMed] [Google Scholar]
- 6.Almeida J.R.C. Farmacêuticos em oncologia: uma nova realidade. São Paulo: Atheneu; 2004. [Google Scholar]
- 7.Sarkaria J.N., Busby E.C., Tibbetts R.S., et al. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 1999;59(17):4375–4382. [PubMed] [Google Scholar]
- 8.Lu Y.P., Lou Y.R., Peng Q.Y., et al. Effect of Caffeine on the ATR/Chk1 Pathway in the Epidermis of UVB-Irradiated Mice. Cancer Res. 2008;68(7):2523–2529. doi: 10.1158/0008-5472.CAN-07-5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rogozin E.A., Lee K.W., Kang N.J., et al. Inhibitory effects of caffeine analogues on neoplastic transformation: structure–activity relationship. Carcinogenesis. 2008;29(6):1228–1234. doi: 10.1093/carcin/bgn016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alves A.B. Simultaneous determination of theobromine, theophylline and caffeine in teas by high performance liquid chromatography. Braz. J. Pharm. Sci. 2002;38(2):237–243. [Google Scholar]
- 11.Kandakatla N., Ramakrishnan G. Ligand Based Pharmacophore Modeling and Virtual Screening Studies to Design Novel HDAC2 Inhibitors. Adv. Bioinforma. 2014;2014:812148. doi: 10.1155/2014/812148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Starosyla S.A., Volynets G.P., Bdzhola V.G., Golub A.G., Protopopov M.V., Yarmoluk S.M. ASK1 pharmacophore model derived from diverse classes of inhibitors. Bioorg. Med. Chem. Lett. 2014;24(18):4418–4423. doi: 10.1016/j.bmcl.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 13. ACD/Chemsketch Freeware, version 12.00: Advanced Chemistry Development, Inc, Toronto, ON, Canada, 2010.
- 14.BABEL, Open. The open source chemistry toolbox. 2011. 2011.
- 15.ChemPlus . Modular Extensions to HyperChem, Release 6.02. Gainesville: Molecular Modeling for Windows Hyper Inc.; 2000. [Google Scholar]
- 16.Schneidman-Duhovny D., Dror O., Inbar Y., Nussinov R., Wolfson H.J. PharmaGist: a webserver for ligand-based pharmacophore detection. Nucleic Acids Res. 2008;36:223–228. doi: 10.1093/nar/gkn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneidman-Duhovny D., Dror O., Inbar Y., Nussinov R., Wolfson H.J. Deterministic Pharmacophore Detection by Multiple Flexible Alignment of Drug-Like Molecules. J. Comput. Biol. 2008;15(7):737–754. doi: 10.1089/cmb.2007.0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santos C.B.R., Lobato C.C., Braga F.S., et al. Rational Design of Antimalarial Drugs Using Molecular Modeling and Statistical. Curr. Pharm. Des. 2015;21(28):4112–4127. doi: 10.2174/1381612821666150528121423. [DOI] [PubMed] [Google Scholar]
- 19.STATISTICA (Data Analysis Software System) Version 6.1. StatSoft, Inc.; 2004. [Google Scholar]
- 20.Koes D.R., Camacho C.J. ZINCPharmer: pharmacophore search of the ZINC database. Nucleic Acids Res. 2012;40:409–414. doi: 10.1093/nar/gks378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.John J. Irwin and Brian K. Shoichet. ZINC – A Free Database of Commercially Available Compounds for Virtual Screening. J. Chem. Inf. Model. 2005;45(1):177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu T., Lin Y., Wen X., Jorissen R.N., Gilson M.K., Binding D.B. a web-accessible database of experimentally determined protein–ligand binding affinities. Nucleic Acids Res. 2007;35:198–201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanderson D.M., Earnshaw C.G. Computer prediction of possible toxic action from chemical structure; the DEREK system. Hum. Exp. Toxicol. 1991;10(4):261–273. doi: 10.1177/096032719101000405. [DOI] [PubMed] [Google Scholar]
- 24.Dassault Systèmes B.I.O.V.I.A. Discovery studio modeling environment. San Diego, CA: Dassault Systèmes; 2015. [Google Scholar]
- 25.PDB – Protein Data Bank http://www.ncbi.nlm.nih.gov/protein/
- 26.Matthews T.P., Klair S., Burns S., et al. Identification of Inhibitors of Checkpoint Kinase 1 Through Template Screening. J. Med. Chem. 2009;52(15):4810–4819. doi: 10.1021/jm900314j. [DOI] [PubMed] [Google Scholar]
- 27.Morris G.M., Goodsell D.S., Halliday R.S., et al. Automated Docking Using a Lamarckian Genetic Algorithm and and Empirical Binding Free Energy Function. J. Comput. Chem. 1998;19(14):1639–1662. [Google Scholar]
- 28.Agrawal R., Jain P., Dikshit S.N., Bahare R.S., Ganguly S. Ligand-based pharmacophore detection, screening of potential pharmacophore and docking studies, to get effective glycogen synthase kinase inhibitors. Med. Chem. Res. 2013;22(11):5504–5535. [Google Scholar]
- 29.Gupta S., Mohan C.G. Dual binding site and selective acetylcholinesterase inhibitors derived from integrated pharmacophore models and sequential virtual screening. BioMed Res. Int. 2014;2014:1–21. doi: 10.1155/2014/291214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakkiah S., Lee K.W. Pharmacophore-based virtual screening and density functional theory approach to identifying novel butyrylcholinesterase inhibitors. Acta Pharmacol. Sin. 2012;33(7):964–978. doi: 10.1038/aps.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu T., Lin Y., Wen X., Jorissen R.N., Gilson M.K. BindingDB: a web-accessible database of experimentally determined protein–ligand binding affinities. Nucleic Acids Res. 2007;35:198–201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.CSIZMADIA F. JChem: Java applets and modules supporting chemical database handling from web browsers. J. Chem. Inf. Comput. Sci. 2000;40(2):323–324. doi: 10.1021/ci9902696. [DOI] [PubMed] [Google Scholar]
- 33.Willett P., Barnard J.M., Downs G.M. Chemical similarity searching. J. Chem. Inf. Comput. Sci. 1998;38(6):983–996. [Google Scholar]
- 34.Souza J., Feitas Z.M.F., Storpirtis S. In vitro models for the determination of drug absorption and a prediction of dissolution/absorption relationships. Braz. J. Pharm. Sci. 2007;43(4):515–527. [Google Scholar]
- 35.Wang J., Hou T. Recent Advances on in silico ADME Modeling. Annu. Rep. Comput. Chem. 2009;5:101–127. [Google Scholar]
- 36.Richard TS. Protein binding: What does it mean? DICP . 1989;23(7-8 Suppl):27–31. doi: 10.1177/106002808902300706. [DOI] [PubMed] [Google Scholar]
- 37.Wilkinson G.R. Plasma and tissue binding considerations in drug disposition. Drug Metab. Rev. 1983;14(3):427–465. doi: 10.3109/03602538308991396. [DOI] [PubMed] [Google Scholar]
- 38.Rowland M. Protein binding and drug clearance. Clin. Pharmacokinet. 1984;9(Suppl.):10–17. doi: 10.2165/00003088-198400091-00002. [DOI] [PubMed] [Google Scholar]
- 39.Blanchard J. Protein binding of caffeine in young and elderly males. J. Pharm. Sci. 1982;71(12):1415–1418. doi: 10.1002/jps.2600711229. [DOI] [PubMed] [Google Scholar]
- 40.Fausch K., Uehlinger D.E., Jakob S., Pasch A. Haemodialysis in massive caffeine intoxication. Clin. Kidney J. 2012;5(2):150–152. doi: 10.1093/ckj/sfs020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zylber-Katz E., Granit L., Levy M. Relationship between caffeine concentrations in plasma and saliva. Clin. Pharmacol. Ther. 1984;36(1):133–137. doi: 10.1038/clpt.1984.151. [DOI] [PubMed] [Google Scholar]
- 42.Rojas H., Ritter C., Pizzol F.D. Mechanisms of dysfunction of the blood-brain barrier in critically ill patients: emphasis on the role of matrix metalloproteinases. Rev. Bras. Ter. Intensiva. 2011;23(2):222–227. [PubMed] [Google Scholar]
- 43.Ma X., Chen C., Yang J. Predictive model of blood-brain barrier penetration of organic compounds. Acta Pharmacol. Sin. 2005;26(4):500–512. doi: 10.1111/j.1745-7254.2005.00068.x. [DOI] [PubMed] [Google Scholar]
- 44.Liu J., Shen B., Shi M., Cai J. Higher caffeinated coffee Intake is associated with reduced malignant melanoma risk: A meta-nnalysis study. PLoS One. 2016;11(1):e0147056. doi: 10.1371/journal.pone.0147056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silva N.S.R., Santos C.F., Gonçalves L.K.S., et al. Molecular modeling of the major compounds of sesquiterpenes class in copaiba oil-resin. Br. J. Pharm. Res. 2015;7(4):247–263. [Google Scholar]
- 46.Vieira J.B., Braga F.S., Lobato C.C., et al. A QSAR, pharmacokinetic and toxicological study of new artemisinin compounds with anticancer activity. Molecules. 2014;19(8):10670–10697. doi: 10.3390/molecules190810670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woo Y.T. Mechanisms of action of chemical carcinogens, and their role in structure-activity relationships (SAR) analysis and risk assessment. In: Benigni R., editor. Quantitative Structure-Activity Relationship (QSAR) Models of Mutagens and Carcinogens. Boca Raton: CRC Press; 2003. [Google Scholar]
- 48.Primo M.S., Calliari C.M., Castro-Gómez R.J.H., Mauro M.O., Mantovan M.S., Oliveira R.J. Assessment of mutagenicity and antimutagenicity of a biopolymer extracted from the microorganism Agrobacterium radiobacter in mice. Braz J Pharmacog. 2010;20(3):340–347. [Google Scholar]
- 49.Mauro M.O., Pesarini J.R., Ishii P.L., Silva Ariane F., Oliveira R.J. Chemopreventive activity of phenylalanine against damage mutagenic prompted by the acute administration of cyclophosphamide in pregnant and non-pregnant mice using the micronucleus test. Braz J Pharmacog. 2010;20(3):334–339. [Google Scholar]
- 50.Basketter D.A. Skin sensitization: Strategies for the assessment and management of risk. Br. J. Dermatol. 2008;159(2):267–273. doi: 10.1111/j.1365-2133.2008.08625.x. [DOI] [PubMed] [Google Scholar]
- 51.Cronin M.T., Basketter D.A. Multivariate QSAR analysis of a skin sensitization database. SAR QSAR Environ. Res. 1994;2(3):159–179. doi: 10.1080/10629369408029901. [DOI] [PubMed] [Google Scholar]
- 52.Nehlig A., Debry G. Potential genotoxic, mutagenic and antimutagenic effects of coffee: A review. Mutat. Res. 1994;317(2):145–162. doi: 10.1016/0165-1110(94)90022-1. [DOI] [PubMed] [Google Scholar]
- 53.Kuhlmann W., Fromme H.G., Heege E.M., Ostertag W. The mutagenic action of caffeine in higher organisms. Cancer Res. 1968;28(11):2375–2389. [PubMed] [Google Scholar]
- 54.Hevener K.E., Zhao W., et al. Validation of molecular docking programs for virtual screening against dihydropteroate synthase. J. Chem. Inf. Model. 2009;49(2):444–460. doi: 10.1021/ci800293n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gowthaman U., Jayakanthan M., Sundar D. Molecular docking studies of dithionitrobenzoic acid and its related compounds to protein disulfide isomerase: computational screening of inhibitors to HIV-1 entry. BMC Bioinformatics. 2008;9(Suppl. 12):14. doi: 10.1186/1471-2105-9-S12-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berg J.M., Tymoczko J.L., Stryer L. 2002. [Google Scholar]