Abstract
Transfer RNAs (tRNAs), traditionally considered to participate in protein translation, were interspersed in the entire genome. Recent studies suggested that dysregulation was observed in not only tRNAs, but also tRNA derivatives generated by the specific cleavage of pre- and mature tRNAs in the progression of cancer. Accumulating evidence had identified that certain tRNAs and tRNA derivatives were involved in proliferation, metastasis and invasiveness of cancer cell, as well as tumor growth and angiogenesis in several malignant human tumors. This paper reviews the importance of the dysregulation of tRNAs and tRNA derivatives during the development of cancer, such as breast cancer, lung cancer, and melanoma, aiming at a better understanding of the tumorigenesis and providing new ideas for the treatment of these cancers.
Keywords: tRNA, tRNA derivatives, Breast cancer, Lung cancer, Melanoma
Background
Not participating in protein-coding, small non-coding RNA (sncRNA), including transfer ribonucleic acids (tRNA), ribosomal RNA (rRNA), small nucleolar RNA (snoRNA) and small nuclear RNA (snRNA), plays a widespread and important role both inside and outside the cell and is involved in cell proliferation, differentiation, apoptosis, and cellular metabolism [1]. With the development of high-throughput sequencing technology, new types of sncRNAs are obtained from other cellular RNA species through specific and regulated RNA processing or cleavage [2–4]. For instance, the cleaved products of mature snoRNA were mainly derived from a single gene: the C/D box type snoRNA gene methylation guide foe U6 snoRNA residue 77 (mgU6–77) [5, 6]. Also, novel sncRNAs were identified by cleavage of anti-codon loops, TψC loops, D loops, and other positions of tRNAs [4].
Previously, it was a generally accepted fact that tRNA was a housekeeping product with little regulatory function and it was rarely known by researchers the additional function of tRNAs apart from their canonical function as adapters in protein synthesis [7]. However, there are growing evidence in recent years that tRNAs and their derivatives are dysregulated in cancer and involved in the pathogenic process of cancer, for which they have recently gained significant attention [8, 9]. Furthermore, mutation of the tRNA itself and the involvement of the supplementary protein produced by the tRNA biogenesis and modification is also associated with cancer [10–12]. In this review, current understanding of the dysregulation of tRNAs and tRNA derivatives in tumor pathogenesis was summarized in terms of main cancer types such as breast cancer, lung cancer, and melanoma.
Biogenesis and structure of tRNAs and tRNA derivatives
Biogenesis and structure of tRNAs
tRNAs, with fundamental function of carrying and transporting amino acids, are a class of small noncoding ribonucleic acid folded into a “clover” secondary structure and L-shaped three-stage structure composed of 70–90 nucleotides, accounting for approximately 4–10% of all cellular RNAs [13]. As an elementary composition of the translation procedure, they convey the amino acid to the ribosome and convert the significance of the nucleotide sequence to the corresponding polypeptide chain in a manner of the interaction of codon (mRNA)-anticodon (tRNA) [14, 15]. Initial transcription product of RNA polymerase III (Pol III) is a typical precursor of tRNA that obligatorily experiences a succession of intricate biological processes to be converted to mature tRNA [16, 17], including removal of the 5′ leader by RNase P, clipping of the 3′ trailer by endonucleases and exonucleases such as RNase E, RNase PH and RNase T, addition of CCA at the 3' end via CCA-adding enzyme, shearing of introns and multiple modifications of tRNA base [18, 19]. It is important that accurate processing of pre-tRNA is essential to its successful release from the nucleus to play part in the translation of the protein. However, not all tRNAs are used for protein translation and a small percentage of them play the role of signaling molecules in response to environmental stress [20]. Under oxidative stress, tRNAs were cleaved into small molecules RNAs that repressed translation initiation [21]. Equally, oxidative stress-induced deactivation of the 3' CCA tail also shut down global translation [22].
Previously, the changes in mRNA levels in cancer cells effectively explained why cancer cells proliferate, metastasize and avoid death in their own ways [23, 24]. Researchers currently found that alteration in transcriptional level did not necessarily mean the change of protein level [25, 26], and to some extent tRNA played an irreplaceable role in the translation of proteins. The abundance, modification, and mutation of tRNA are all closely related to the protein expression. It is unexpected that the synthesis of tRNA is controlled by all kinds of oncogenes and tumor suppressors—Ras [27] and c-myc [28] promote the transcription of RNA Pol III, whereas Rb [29] and p53 [30] inhibit RNA Pol III transcription, causing the serious dysregulation of tRNA level in a wide range of cancers. In addition, Gingold et al. suggested that there were two representative cellular states in a multicellular animal, namely cell proliferation and cell differentiation corresponded to two distinctly different active tRNA pools referred as proliferative tRNA pools (pro-tRNAs) and differentiated tRNA pools (dif-tRNAs) respectively [31]. Further analysis revealed that the codons abundant in the cancer cells and inducing differentiation condition of genes were corresponded to the induced pro-tRNAs and dif-tRNAs pools, respectively. The cohort of these two genes was due to their histological modification on the chromatin [31, 32]. Moreover, several studies had shown that tRNA-modifying enzymes increased modifications to specific tRNAs in several cancers, which altered the codon preference of the tRNA that in turn led to an increase in the protein expression levels of those mRNAs found to be wealthy with a particular subset of the new “preferred” codons [33, 34]. Gerber et al. provided the evidence that the activity of adenosine deaminase (ADATs; A-to-I transformation) expanded the wobbling capacities of the tRNA base at position 34, allowing it to pair with three different codons—A, U, C [35]. Mutations in mitochondrial tRNA caused mitochondrial dysfunction also involved in tumorigenesis [36]. Finally, the binding of tRNA to cytochrome c suppressed the action of cytochrome c and apoptosis protease catalysts, thereby inhibiting apoptosis and the activity of the enzyme [37].
Biogenesis and structure of tRNA derivatives
Increasing evidence argues that tRNAs and tRNA derivatives are not only imperative ingredients of translation mechanism, but also significant signaling molecules in response to stress [38]. Furthermore, the earlier researchers reported that tRNA breakdown products existing in tumor tissue were often more frequent during stress [39], and it was noteworthy that they were observed in the sera and urine of cancerous person with expression level roughly related to cancer burden [40].
A wealth of intriguing studies discovered that tRNA derivatives, including tRNA-derived stress-induced RNAs (tiRNAs) [41, 42], tRNA-derived fragments (tRFs) [43], and tRNA-derived small RNAs (tsRNAs) [44], were generated by cleavage of the pre-tRNAs or mature tRNAs under various environmental stresses [45, 46] (Fig. 1).
Fig. 1.
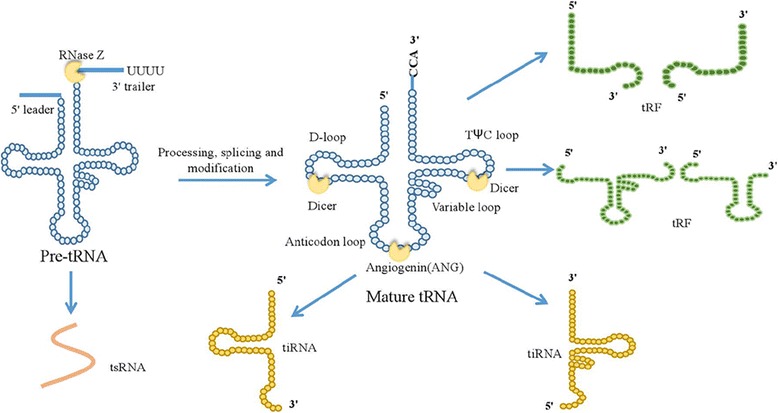
Different types of tRNA derivatives came from the cleavage of pre-tRNAs and mature tRNAs. tsRNAs were generated in the nucleus as a consequence of the pre-tRNA 3′ end cleavage. tiRNAs were generated as a consequence of angiogenin cleaving the anti-codon loop of the mature tRNA. tRFs were formed by Dicer at D-loop, T-loop and other positions of the nucleic acid ribozyme through cleavage of the mature tRNA
Not purely tRNA degradation debris, tRNA derivatives had a vital role in cancer, RNA silencing and micro-environment monitoring [47]. tiRNAs, including 5' tiRNAs and 3' tiRNAs, were generated as a consequence of angiogenin cleaving the anti-codon loop of the mature tRNA [21]. 5' tiRNAs was interacted with tRNase ZL (tRNA endonuclease) binding to the enzyme to cleave the target gene complementary to the 5′ tiRNAs sequence where 5' tiRNAs acted as a small guide RNA, thereby down-regulating the expression of the target gene [48, 49]. Several studies, furthermore, have reported that tiRNAs inhibit protein synthesis and induce the formation of stress particles [21, 50]. tRFs are relatively smaller RNAs (17-26 nt) formed by Dicer or Rnase Z at other positions of the nucleic acid ribozyme through cleavage of the mature tRNA [51]. tRFs can form a complex with Argonaute (Ago) protein, and play a similar role with miRNA by silencing a series of target gene expression [52, 53]. The high expression of the tRF in an extensive range of cancer cell lines is strongly relevant to cell proliferation [51]. tsRNAs come into being through cleaving the pre-tRNA 3'end [44]. Their conspicuous dysregulation, which may exert oncogenic or tumor-suppressor functions in cancer, have been discovered in a variety of malignant tumor onsets and progression [44, 54].
tRNAs and tRNA derivatives in breast cancer
The overexpression of tRNAs in breast cancer
Breast cancer is a fairly common malignant tumor that occurs in the glandular epithelial tissue, whose incidence has been a continuous upward trend since the late 1970s and ranks first in female malignancies [55]. Recently, the research on tRNA and breast cancer is increasing [8, 56, 57]. Mahlab et al. observed that the absolute levels of tRNA genes in breast cancer cells were significantly increased compared with healthy cells [56]. Nuclear- and mitochondrial-encoded tRNAs, also upregulated in breast tissues, were greatly enhanced in cancer cell lines based on genome-wide studies of tRNA expression [8]. tRNA profiling revealed that tRNA expression levels in breast cancer lines were different from those of non-tumorigenic cells [57].
What’s the reason for the increase in tRNA levels in cancer cells/tissues? Several pieces of evidence showed that transcription of tRNA by RNA pol III was affected by proto-oncogenes and tumor suppressor genes, which mainly impacted the subunit Brf1 of TFIII B factor in pol III, thus promoting or suppressing its function [58–60]. In estrogen receptor (ER)-positive breast cancer, the interaction of estrogen receptor alpha (ERα) with Brf1 mainly regulated the transcription of the Pol III genes—particularly tRNALeu and 5S rRNA [61]. In addition, inhibition of ERα not only reduced the expression of Brf1 and Pol III gene but also decreased the formation rate of breast cancer cell colony [61]. The researchers discovered a significantly positive correlation between the expression of telomerase reverse transcriptase (TERT) and pretranscripts of tRNA-Leu and tRNA-Tyr only in triple-negative breast cancer with highly aggressive ability by correlation analysis [62, 63]. Moreover, loss of TERT was related to reduced polyomavirus middle T oncogene-induced (PyMT-induced) mammary tumorigenesis and expression of tRNA such as pre-tRNA-Tyr [62, 64]. Recent studies suggested that all tRNALeu isoacceptors were more highly expressed only in human epidermal growth factor receptor-2 (Her2) -positive breast subtypes, but not in other subtypes, which greatly promoted the proliferation of cancer cells and their resistance to death via interaction of tRNALeuCAG with ErbB3-binding protein 1 (EBP1), subsequently enhancing the activation of ErbB2/ErbB3 pathway and the RSK1/MSK2 signaling pathway [65].
Furthermore, increased tRNAiMet in human mammary epithelial cells could enhance the capacity of cell proliferative and metabolism [66]. But there was a poor correlation between the tRNA levels induced by tRNAiMet overexpression in mammary epithelial cells and that levels in breast cancer cells, suggesting that the change of tRNA abundance and species in cancer cells had little to do with the overexpression of tRNAiMet [66]. Furthermore, Clarke et al. detected that increased tRNAiMet in carcinoma-associated fibroblasts could promote tumor growth and angiogenesis [67]. They measured tRNA levels in immortalized human breast cancer-associated fibroblasts (iCAFs) and normal fibroblasts and observed an increase in the expression of tRNAiMet and tRNAIle in iCAFs [67, 68].
It was surprising that overexpression of tRNA also affected the ability of invasion and metastasis in cancer cells [57]. Using means of a new tRNA profiling method, investigators revealed that specific tRNAs, tRNAArgCCG and tRNAGluUUC, were upregulated in highly metastatic breast cancer cells compared to poorly metastatic counterpart due to their enhanced stability and translation of the codon-rich transcripts [57]. These up-regulated tRNAs might serve as potential novel prognostic markers in addition to their involvement in the pathogenesis of breast cancer [69].
The modification of tRNAs in breast cancer
tRNA modifications such as tRNA base modifications and the enzymes catalyzing such modifications played an important role in the pathogenesis of breast cancer [70–72]. Studies have indicated that increased tRNA modifications enhanced the translational efficiency by modifying the anticodon swinging bases, increasing the decoding power of tRNA [33, 73]. In human breast cancer, the elevated expression of U34-modifying enzymes Elp3 and Ctu1/2, catalyzing the mcm5s2-U34 tRNA modification, directly promoted the translation of oncoprotein DEK in turn binding to the LEF1-IRES sequence to increase the translation of the oncogenic LEF-1 mRNA and promote the invasion and metastasis of breast cancer cells [72]. Meanwhile, RNA methyltransferase misu (NSUN2) had significantly increased expression level in squamous cell carcinoma, colorectal cancer and breast cancer, therefore it might act as a downstream target gene of myc and be involved in the proliferation of cancer cells [71]. Interestingly, another tRNA modification enzyme tRNA methyltransferase homolog 12 (TRMT12) was highly expressed in both several breast cancer cell lines and tissues [70], however, its elaborate molecular mechanisms were unclear.
The mutation of tRNAs in breast cancer
Additionally, mitochondrial DNA (mtDNA) depletion and mutation had been shown to be associated with increased tumorigenic and invasive phenotype [74]. The tertiary structure of mitochondrial tRNA(mt-tRNA)was affected by its genetic mutation, which brought about severely impaired mitochondrial protein synthesis [75]. Through the analysis of clinical data of breast cancer, Meng et al. concluded that mutations in mitochondrial tRNA such as mt-tRNAAsp was involved in the carcinogenesis of breast cancer [76].
tRNA derivatives in breast cancer
Many literatures reported that tRNA derivatives were dysregulated in many malignancies including breast cancer [54, 77, 78]. Performing unsupervised analysis on normal breast epithelial cells with oncogene activation mutations and cancer cells at different stages of carcinogenesis, Veronica and his colleagues found that tsRNAs expression was modulated by oncogenes, suggesting that tsRNA might be a key effector in the pathway regulated by these oncogenes [54]. Further, tsRNA expression appeared in certain obvious stages of the process of carcinogenesis: ts-3 was strongly down-regulated in aggressive late-stage breast cancer, whereas ts-67, ts-48, and ts-6 were up-regulated only in the late-stage cancer cell line [54]. Parallelly, high abundance of tRNA-derived small RNA in breast cancer extracellular vesicles (EVs) were combined with known miR signatures of tumors to differentiate EVs from those derived from other cell sources [79]. Moreover, tRFs, which are derived from tRNAGlu, tRNAAsp, tRNAGly, and tRNATyr, compete with Y-box-binding protein 1 (YB-1) for an endogenous oncogene transcript, disrupting the stability of proto-oncogene transcripts, and reducing the expression of proto-oncogenes resulting in suppression of breast cancer progression [78]. YB-1 bound to specific miRNAs, snRNAs and tRNA-derived fragments, which might cause carcinogenic effects in breast cancer [80]. Moreover, tiRNAs cooperated with YB-1 to prevent eIF4G/A from initiating translation [21].
Clinical characteristics of breast cancer were related with changes in abundance of specific tiRNA [81]. In contrast to ER-negative tumors, ER-positive tumor showed a decline in abundance of 26 specific circulating tiRNA deriving from the isoacceptors of tRNAGly, tRNAGlu and tRNALys [81]. Inflammatory breast cancer, on the other hand, was associated with increases in tiRNA-Ala in comparison to non-inflammatory breast cancer, thus suggesting that circulating tiRNA might involve in breast cancer syndromes and had potential as circulating biomarkers [81]. In ER-positive breast cancer, sex hormones and their receptors promoted the angiogenin cleavage of mature tRNA anticodon loops to produce a large number of tiRNA [77]. tiRNAs, including 5'-tiRNAAsp and 5'-tiRNAHis, had significantly higher expression in tissue or cells of breast cancer than normal epithelial counterparts [77]. Experiments manifested that specific knockdowns of 5'-tiRNA would impair cell proliferation, indicating that tiRNAs were not nonfunctionally accumulated but enhanced the cell proliferation [77]. Taken together, tRNA derivatives played different roles in different pathways.
tRNAs and tRNA derivatives in lung cancer
tRNAs in lung cancer
Lung cancer is one of the most life-threatening diseases with its morbidity and mortality increasing rapidly [82]. The dysregulation of tRNAs is closely related to the carcinogenesis of lung cancer. TERT, significantly enriched at tRNAMet, tRNAArg and tRNALys genes, regulated expression of those tRNAs and directly controlled the rate of synthesis of global cancer proteins in various cancer cell lines, mainly including HCT116, A2780 and P493 cell line, which to some extent could promote the tumorigenesis [62].
MtDNA was more readily mutated than nuclear genomic DNA owing to the lack of protective histones, introns, and efficient DNA repair systems [83]. Indeed, mutations in the mt-tRNA gene were found to be associated with various diseases including lung cancer [84]. An increasing number of literature supported that these mt-tRNA mutations, such as tRNAHis A12172G, tRNAAla T5655C, tRNALeu A12330G, tRNASer T7505C, and tRNAThr G15927A, were pathogenic and highly likely to be involved in the carcinogenesis of lung cancer [85, 86]. These mutations disrupted the secondary structure of tRNA itself, and subsequently affected tRNA post-transcriptional modifications as well as aminoacylation, which might alter the specificity or stability of the tRNA or change its affinity [87]. In all, these mutations caused a decrease in mitochondrial protein synthesis and the inability to meet the threshold of the respiratory phenotype and ATP required for normal cells [84], contributing to the tumorigenesis of lung cancer.
tRNA derivatives in lung cancer
tRNA derivatives, including tsRNAs, tRFs and tiRNAs, are associated with lung cancer development. Pekarskya et al. found that ts-3676 and ts-4521, derived from tRNA-Thr and tRNA-Ser respectively, could act as roles of not only microRNA interacted with Argonaute proteins Ago1 and Ago2, but also P-element-induced wimpy testis (Piwi)-interacting small RNAs (piRNA) interacted with Piwi-like protein 2 (Piwil2) [44, 88]. Furthermore, using Ingenuity Pathway analysis software to evaluate changes in cancer pathways in ts-4521 cells, researchers found that the cell proliferation-related pathway and apoptosis-related pathways were associated with the absence of ts-4521 [54]. Of note, these two tsRNAs were drastically down-regulated and mutated in lung cancer samples vs. matched normal lung tissues [44]. Similarly, Balatti et al. experimentally demonstrated that overexpression of ts-46 and ts-47 significantly reduced the clonal formation in lung cancer cells, which further confirmed the involvement of those tsRNAs in lung cancer pathogenesis [54]. Additionally, the high expression of tRNALeuCAG derived small molecule tiRNA had a positive correlation with non-small cell lung cancer stages by promoting cell proliferation and causing G0/G1 cell cycle progression, which would be conducive to the deterioration of the cancer [89]. The down-regulation of the proto-oncogene AURKA inhibited the expression of tiRNALeu in cancer cells, suggesting that tiRNALeu might play a part in promoting the proliferation of cancer cells by regulating the expression of AURKA [89].
tRNAs and tRNA derivatives in melanoma and other cancers
Melanoma, a type of malignant tumor derived from melanocytes common in the skin, mucous membranes, choroidal and other parts of the eye, is the most malignant skin tumor. Recently, researchers found that tumor growth and angiogenesis in 2 + tRNAiMet mice was significantly faster than wild-type littermate through transfection of melanoblasts into 2 + tRNAiMet transgenic mice and wild-type littermates [67]. Further study discovered that increased tRNAiMet in carcinoma-associated fibroblasts drove selective and meaningful alteration of the secretion of stromal cells, especially type II collagen which provided a convenient condition for tumor growth and metastasis, whereas there was no significant effect on the cell non-secreted protein product [67]. Similarly, Birch et al. argued that the overexpression of tRNAiMet in melanoma advanced cancer cell migration, invasiveness and elevated lung colonisation capacity leading to increased metastatic potential, but had a lesser impact on cell proliferation and primary tumor growth [90]. Increased tRNAiMet in cancer cells relied mainly on α5β1 integrin and levels of the translation initiation ternary complex to drive cell migration and invasion. Increased tRNAiMet in melanoma promoted the expression of fibronectin and α5β1integrin that were closely related to the invasion of cells [90]. Khattar et al. discovered that the increased TERT gave rise to proliferative abilities of cancer cells in melanoma, because TERT upregulated tRNA expression by its direct combination with RNA polymerase III subunit RPC32 and enhanced recruitment of chromatin resulting in an increase in the occupancy rate of RNA pol III on the tRNA gene, suggesting that TERT promoted cancer cell proliferation by augmenting tRNA expression, such as tRNAArg, tRNAAla, tRNAAsn, tRNACys, tRNALys, tRNAGlu and tRNAThr [62]. In addition, there are little researches on tRNA derivatives and melanoma, which is expected to become a hot spot for future research.
The dysregulation of tRNAs and tRNA derivatives occurs in other tumors, such as cervical cancer, prostate cancer, multiple myeloma and pancreatic cancer. In cervical cancer samples, high-risk human papillomavirus (HPV) tRNA expression was significantly increased compared to other benign lesions of HPV, such as tRNAArg and tRNASec [91]. Also, the tRFs derived from tRNAGln inhibited the translation process of the protein [92]. Dependent hormones and their receptors produced tiRNA, including 5'-tiRNAAsp and 5'-tiRNAHis as well as 5'-tiRNALys, which also promoted the proliferation of cancer cells in prostate cancer [77]. Abnormally increased tRNAs abundance promoted translation of highly active proteins in multiple myeloma [93]. tRNA modulated MEK2 function to regulate cancer cellular behavior in pancreatic cancer [94].
Conclusion & further perspectives
Indeed, a growing series of evidence had identified that the dysregulation of tRNAs and tRNA derivatives expression was of tremendous value and potential in cancer progression. As mentioned above, the increase of specific tRNAs and mutations of tRNAs promoted the proliferation, metastasis and invasiveness of cancer cell, as well as tumor growth and angiogenesis. An increase in tRNA-modifying enzymes would be better for tRNAs to adapt to the translation of oncogenes' preferred codons to promote cell proliferation in cancer cells. Beyond that, tRNA derivatives played an extremely important role in regulating the expression of cancer-related genes, RNA silencing and cell proliferation (Table 1).
Table 1.
Characteristics of representative tRNAs and their derivatives in breast cancer
| Cancer type | The change of the tRNA | Cell/tissue type | Function | References |
|---|---|---|---|---|
| ER+ breast cancer | The overexpression of tRNALeu | MCF-7 | Promoted cell proliferation and cell transformation | [61] |
| Triple-negative breast cancer | The overexpression of tRNA-Leu and tRNA-Tyr | Triple-negative breast cancer tissue | Initiated tumorigenesis | [62] |
| Her2(ErbB2)-positive breast | The overexpression of tRNALeuCAG | Her2 (ErbB2)-positive breast tissue | Increased the protein synthesis and proliferative abilities of cancer | [65] |
| Breast cancer | The overexpression of tRNAiMet and tRNAIle in immortalized human breast cancer-associated fibroblasts | MCF-7 | Promoted tumor growth and angiogenesis | [67] |
| Breast cancer | The overexpression of tRNAArgCCG and tRNAGluUUC | MCF10a, MDA-par, MDA-LM2, CN34-par, and CN-LM1a | Promoted metastasis and invasion ability | [57] |
| Breast cancer | Elevated mcm5s2-U34 tRNA modification | MDA-MB-231, MCF7, NMuMG | Promoted breast cancer cells invasion and metastasis | [72] |
| Breast cancer | The mutation of mt-tRNAAsp | Blood samples from breast cancer patients | Involved in the carcinogenesis of breast cancer | [76] |
| Breast cancer | Ts-3 was down-regulated in aggressive late-stage breast cancer, whereas ts-67, ts-48, and ts-6 were up-regulated only in the late-stage cell line | MCF7 and MDA-MB-231 cell lines | tsRNA expression appeared in certain obvious stages of the process of carcinogenesis | [54] |
| Breast cancer | High abundance of tRNA-derived miRNA such as miR-720 and miR-1274b | MCF7 EVs and MCF10A EVs | Served as biomarkers | [79] |
| Breast cancer | tRFs derived from tRNAGlu, tRNAAsp, tRNAGly, and tRNATyr | MDA-MB-231, CN34 cells, CN-LM1a and MDA-LM2 | Suppressed breast cancer progression | [78] |
| Estrogen receptor (ER)-positive breast cancer | Increased 5'-tiRNAAsp and 5'-tiRNAHis | MCF-7, BT-474 | Enhanced cell proliferation | [77] |
| Lung cancer | TERT was significantly enriched at tRNAMet,tRNAArg and tRNALys genes | A2780 | TERT regulated tRNAs expression and controlled the rate of synthesis of global cancer proteins | [62] |
| Lung cancer | Mt-tRNAHis, mt-tRNAAla, mt-tRNALeu, mt-tRNASer, and mt-tRNAThr mutation | Blood samples from lung cancer patients | Contributed to the tumorigenesis of lung cancer | [85, 86] |
| Lung cancer | tRNAThr and tRNASer derived from ts-3676 and ts-4521 down-regulated and mutated in lung cancer sample | Lung cancer sample | Acted as microRNA roles and piRNA roles | [44, 88] |
| Lung cancer | The down-regulated of ts-46 and ts-47 | A549, H1299 | Significantly reduced the clonal formation of cancer cells | [54] |
| Lung cancer | The high expression tRNALeuCAG derived small molecule tiRNA | A549, H1650, PC-9, 95-D and SPCA-1 H1299 and H23 | Promoted cell proliferation | [89] |
| Melanoma | The tRNAiMet in carcinoma-associated fibroblasts | G361, BLM, LOX-IMVI | Tumor growth and angiogenesis | [67] |
| Melanoma | Increased tRNAiMet in melanoma | Derived from the early pup skin of these Tyr: NrasQ61K/°; INK4a−/−; wild-type (wt) and Tyr: NrasQ61K/°; INK4a−/− of Melanocyte cell lines | Advanced cancer cell migration, invasiveness and lung colonisation capacity | [90] |
| Melanoma | TERT promoted the expression of tRNAArg, tRNAAla, tRNAAsn, tRNACys, tRNALys, tRNAGlu and tRNAThr | BLM, G361, LOX-IMVI | TERT promoted cancer cell proliferation by augmenting tRNA expression | [62] |
| Cervical cancer | tRNAArg and tRNASec were significantly increased in HPV | Hela and W12 cell lines | The oncoproteins E6 and E7 stimulated tRNA transcription | [91] |
| Cervical cancer | tRF derived from tRNAGln expression was decreased in conditions of slowed cell proliferation | Hela cell | Inhibited the translation process of the protein | [92] |
| Androgen receptor (AR)-positive prostate cancer | Increased expression 5'-tiRNAAsp and 5'-tiRNAHis as well as 5'-tiRNALys | LNCap-FGC | Promoted the proliferation of cancer cells | [77] |
| Multiple myeloma | High levels of tRNA abundance such as tRNAArg and tRNALeu | MM.1S, MM.1R, NCI-H929, U266 and RPMI-8266 | Increased translation of highly active proteins | [93] |
| Pancreatic cancer | tRNA interacted with MEK2 | HEK293T, Q60P, P128Q, S154F, E207K and CD18 | tRNA modulated MEK2 function to regulate cellular behavior | [94] |
tRNAs, mostly dependent on augmenting tRNA expression and acting on different pathways, regulated the progression of cancer (Fig. 2). tRNALeuCAG was involved in the progression of cancer by activating PSK1/MSK2 signaling pathway [65]. tRNAArgCCG and tRNAGluUUC accelerated the progress of cancer by enhancing the stability and translation of the transcripts [57]. tRNAiMet was related to the development of cancer by affecting the secretion of integrin and type II collagen [67, 90]. Besides, the role of other tRNAs in cancer still require further in-depth investigation.
Fig. 2.
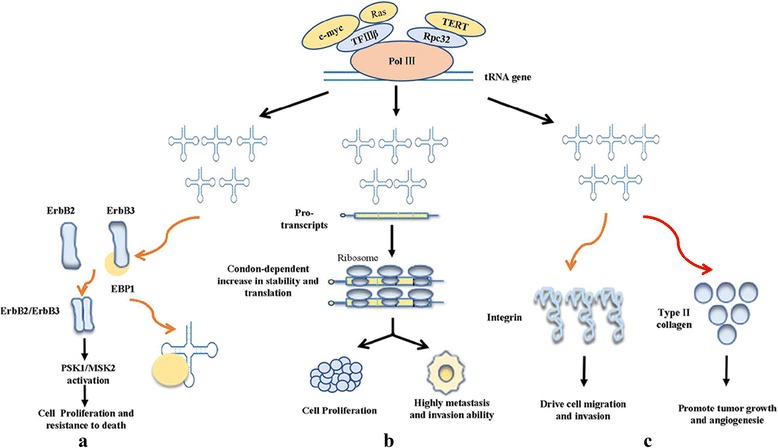
The overexpression of tRNAs regulated the progression of cancer acting on different ways: Ras, c-myc and TERT promoted the transcription of tRNA genes by binding to pol III. a The overexpression of tRNA activated the RSK1/MSK2 signaling pathway, thus influencing cell proliferation and cell apoptosis. b Overexpressed tRNA promoted the ability of invasion and metastasis by enhancing stability and translation of transcripts enriched for their cognate codons. c The overexpression of tRNA regulated the progression of cancer by increasing the secretion of integrin and type II collagen
Interestingly, Sun et al. recently reported that the biological functions of tRNA derivatives were significantly different in different type of cancers [95]. Andrea et al. found that 18 nt tRFs blocked reverse transcription, while 22 nt tRFs post-transcriptionally silenced coding-competent endogenous retroviruses in mouse stem cells [96]. Of note, another study by Kim et al. showed that a specific tsRNA, LeuCAG3'tsRNA, bound at least two ribosomal protein mRNAs (RPS28 and RPS15) to enhance their expression [97]. tRNA derivatives had double-edged sword effect on cell proliferation, whereas tRNAs mostly acted as a cell proliferation promoter (Fig. 3). Whether tRNAs may play a role in inhibiting cell proliferation needs more attention. Given that, we performed tRNAs expression profiling on normal liver tissues, adjacent tissues and liver cancer tissues, finding the different role of certain tRNAs in human hepatocellular carcinoma (HCC) tissues (data not published).
Fig. 3.
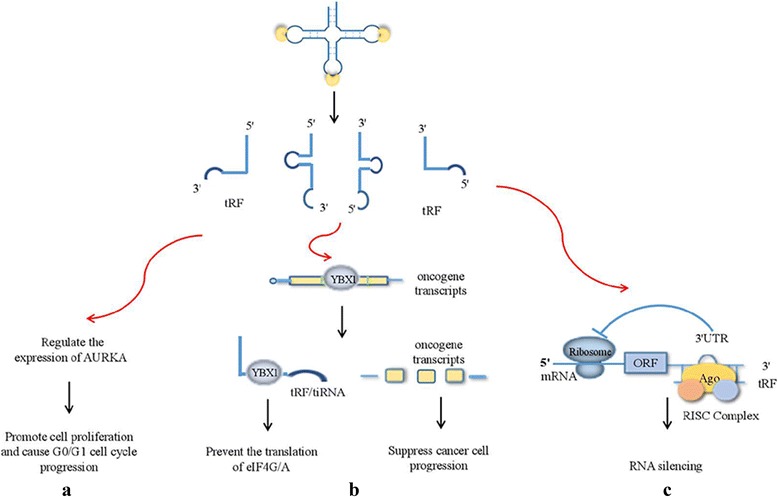
tRNA derivatives exerted their effects through different pathways. a tRNA derivatives promoted cell proliferation and the progression of G0/G1 cell cycle by regulating the expression of AURKA. b Both tRNA derivatives and endogenous oncogene transcripts competed with YBX1, suppressing the progression of cancer and preventing the translation of eIF4G/A. c tRNA derivatives promoted the translation of ribosomal protein mRNAs, subsequently enhancing cell proliferation and viability
In summary, tRNA and its derivatives may serve as an effective tool for diagnosing and treating cancer. Taking tRNAs and tRNA derivatives into account will be conducive to the treatment of malignant tumors.
Acknowledgments
Funding
This research is supported by the National Natural Science Foundation of China (81570533), Hunan province university innovation platform open fund project (16 K096).
Authors’ contributions
SQH wrote the paper; BS, ZPX,YS and WZ revised the manuscript; JX and QL revised and determined the final version. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jing Xiong, Phone: +86 731 84805380, Email: xiongjing79@csu.edu.cn.
Qing Li, Phone: +86 731 84805380, Email: liqing9251026@csu.edu.cn.
References
- 1.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, Fukuda S, Daub CO, Kai C, Kawai J, Yasuda J, Carninci P, Hayashizaki Y. Hidden layers of human small RNAs. BMC Genomics. 2008;9:157. doi: 10.1186/1471-2164-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, Green PJ, Barton GJ, Hutvagner G. Filtering of deep sequencing data reveals the existence of abundant dicer-dependent small RNAs derived from tRNAs. RNA (New York, NY) 2009;15:2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tycowski KT, You ZH, Graham PJ, Steitz JA. Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol Cell. 1998;2:629–638. doi: 10.1016/S1097-2765(00)80161-6. [DOI] [PubMed] [Google Scholar]
- 6.Lestrade L, Weber MJ. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 2006;34:D158–D162. doi: 10.1093/nar/gkj002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grewal SS. Why should cancer biologists care about tRNAs? tRNA synthesis, mRNA translation and the control of growth. Biochim Biophys Acta. 2015;1849:898–907. doi: 10.1016/j.bbagrm.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 8.Pavon-Eternod M, Gomes S, Geslain R, Dai Q, Rosner MR, Pan T. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 2009;37:7268–7280. doi: 10.1093/nar/gkp787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson P, Ivanov P. tRNA fragments in human health and disease. FEBS Lett. 2014;588:4297–4304. doi: 10.1016/j.febslet.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blanco S, Frye M. Role of RNA methyltransferases in tissue renewal and pathology. Curr Opin Cell Biol. 2014;31:1–7. doi: 10.1016/j.ceb.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki T, Nagao A, Suzuki T. Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu Rev Genet. 2011;45:299–329. doi: 10.1146/annurev-genet-110410-132531. [DOI] [PubMed] [Google Scholar]
- 12.Yao P, Fox PL. Aminoacyl-tRNA synthetases in medicine and disease. EMBO Mol Med. 2013;5:332–343. doi: 10.1002/emmm.201100626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirchner S, Ignatova Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat Rev Genet. 2015;16:98–112. doi: 10.1038/nrg3861. [DOI] [PubMed] [Google Scholar]
- 14.Giege R. Toward a more complete view of tRNA biology. Nat Struct Mol Biol. 2008;15:1007–1014. doi: 10.1038/nsmb.1498. [DOI] [PubMed] [Google Scholar]
- 15.Rodnina MV, Wintermeyer W. The ribosome as a molecular machine: the mechanism of tRNA-mRNA movement in translocation. Biochem Soc Trans. 2011;39:658–662. doi: 10.1042/BST0390658. [DOI] [PubMed] [Google Scholar]
- 16.Vannini A, Cramer P. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol Cell. 2012;45:439–446. doi: 10.1016/j.molcel.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 17.Torres AG, Batlle E, Ribas de Pouplana L. Role of tRNA modifications in human diseases. Trends Mol Med. 2014;20:306–314. doi: 10.1016/j.molmed.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 18.Hopper AK, Phizicky EM. tRNA transfers to the limelight. Genes Dev. 2003;17:162–180. doi: 10.1101/gad.1049103. [DOI] [PubMed] [Google Scholar]
- 19.Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24:1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 21.Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43:613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Czech A, Wende S, Morl M, Pan T, Ignatova Z. Reversible and rapid transfer-RNA deactivation as a mechanism of translational repression in stress. PLoS Genet. 2013;9:e1003767. doi: 10.1371/journal.pgen.1003767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tinker AV, Boussioutas A, Bowtell DD. The challenges of gene expression microarrays for the study of human cancer. Cancer Cell. 2006;9:333–339. doi: 10.1016/j.ccr.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 24.Prat A, Ellis MJ, Perou CM. Practical implications of gene-expression-based assays for breast oncologists. Nat Rev Clin Oncol. 2011;9:48–57. doi: 10.1038/nrclinonc.2011.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 27.Wang HD, Trivedi A, Johnson DL. Hepatitis B virus X protein induces RNA polymerase III-dependent gene transcription and increases cellular TATA-binding protein by activating the Ras signaling pathway. Mol Cell Biol. 1997;17:6838–6846. doi: 10.1128/MCB.17.12.6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gomez-Roman N, Grandori C, Eisenman RN, White RJ. Direct activation of RNA polymerase III transcription by c-Myc. Nature. 2003;421:290–294. doi: 10.1038/nature01327. [DOI] [PubMed] [Google Scholar]
- 29.White RJ, Trouche D, Martin K, Jackson SP, Kouzarides T. Repression of RNA polymerase III transcription by the retinoblastoma protein. Nature. 1996;382:88–90. doi: 10.1038/382088a0. [DOI] [PubMed] [Google Scholar]
- 30.Crighton D, Woiwode A, Zhang C, Mandavia N, Morton JP, Warnock LJ, Milner J, White RJ, Johnson DL. p53 represses RNA polymerase III transcription by targeting TBP and inhibiting promoter occupancy by TFIIIB. EMBO J. 2003;22:2810–2820. doi: 10.1093/emboj/cdg265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gingold H, Tehler D, Christoffersen NR, Nielsen MM, Asmar F, Kooistra SM, Christophersen NS, Christensen LL, Borre M, Sorensen KD, Andersen LD, Andersen CL, Hulleman E, Wurdinger T, Ralfkiaer E, Helin K, Gronbaek K, Orntoft T, Waszak SM, Dahan O, Pedersen JS, Lund AH, Pilpel Y. A dual program for translation regulation in cellular proliferation and differentiation. Cell. 2014;158:1281–1292. doi: 10.1016/j.cell.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 32.Gustafsson C, Govindarajan S, Minshull J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22:346–353. doi: 10.1016/j.tibtech.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 33.Novoa EM, Pavon-Eternod M, Pan T, Ribas de Pouplana L. A role for tRNA modifications in genome structure and codon usage. Cell. 2012;149:202–213. doi: 10.1016/j.cell.2012.01.050. [DOI] [PubMed] [Google Scholar]
- 34.Novoa EM, Ribas de Pouplana L. Speeding with control: codon usage, tRNAs, and ribosomes. Trends Genet. 2012;28:574–581. doi: 10.1016/j.tig.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 35.Gerber AP, Keller W. An adenosine deaminase that generates inosine at the wobble position of tRNAs. Sci (New York, NY) 1999;286:1146–1149. doi: 10.1126/science.286.5442.1146. [DOI] [PubMed] [Google Scholar]
- 36.Florentz C, Sohm B, Tryoen-Toth P, Putz J, Sissler M. Human mitochondrial tRNAs in health and disease. Cell Mol Life Sci. 2003;60:1356–1375. doi: 10.1007/s00018-003-2343-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mei Y, Yong J, Liu H, Shi Y, Meinkoth J, Dreyfuss G, Yang X. tRNA binds to cytochrome c and inhibits caspase activation. Mol Cell. 2010;37:668–678. doi: 10.1016/j.molcel.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA (New York, NY) 2008;14:2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borek E, Baliga BS, Gehrke CW, Kuo CW, Belman S, Troll W, Waalkes TP. High turnover rate of transfer RNA in tumor tissue. Cancer Res. 1977;37:3362–3366. [PubMed] [Google Scholar]
- 40.Speer J, Gehrke CW, Kuo KC, Waalkes TP, Borek E. tRNA breakdown products as markers for cancer. Cancer. 1979;44:2120–2123. doi: 10.1002/1097-0142(197912)44:6<2120::AID-CNCR2820440623>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 41.Saikia M, Hatzoglou M. The many virtues of tRNA-derived stress-induced RNAs (tiRNAs): discovering novel mechanisms of stress response and effect on human health. J Biol Chem. 2015;290:29761–29768. doi: 10.1074/jbc.R115.694661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shigematsu M, Kirino Y. 5’-Terminal nucleotide variations in human cytoplasmic tRNAHisGUG and its 5′-halves. RNA. 2017;23:161–168. doi: 10.1261/rna.058024.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keam SP, Hutvagner G. tRNA-Derived Fragments (tRFs): emerging new roles for an ancient RNA in the regulation of gene expression. Life (Basel, Switzerland) 2015;5:1638–1651. doi: 10.3390/life5041638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pekarsky Y, Balatti V, Palamarchuk A, Rizzotto L, Veneziano D, Nigita G, Rassenti LZ, Pass HI, Kipps TJ, Liu CG, Croce CM. Dysregulation of a family of short noncoding RNAs, tsRNAs, in human cancer. Proc Natl Acad Sci U S A. 2016;113:5071–5076. doi: 10.1073/pnas.1604266113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell. 2009;138:215–219. doi: 10.1016/j.cell.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 46.Levitz R, Chapman D, Amitsur M, Green R, Snyder L, Kaufmann G. The optional E. Coli prr locus encodes a latent form of phage T4-induced anticodon nuclease. EMBO J. 1990;9:1383–1389. doi: 10.1002/j.1460-2075.1990.tb08253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K, Dalla-Favera R. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci U S A. 2013;110:1404–1409. doi: 10.1073/pnas.1206761110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elbarbary RA, Takaku H, Uchiumi N, Tamiya H, Abe M, Takahashi M, Nishida H, Nashimoto M. Modulation of gene expression by human cytosolic tRNase Z(L) through 5′-half-tRNA. PLoS One 2009; 4:e5908. [DOI] [PMC free article] [PubMed]
- 49.Takaku H, Minagawa A, Takagi M, Nashimoto M. A novel 4-base-recognizing RNA cutter that can remove the single 3′ terminal nucleotides from RNA molecules. Nucleic Acids Res. 2004;32:e91. doi: 10.1093/nar/gnh092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA (New York, NY) 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar P, Anaya J, Mudunuri SB, Dutta A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014;12:78. doi: 10.1186/s12915-014-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Balatti V, Nigita G, Veneziano D, Drusco A, Stein GS, Messier TL, Farina NH, Lian JB, Tomasello L, Liu CG, Palamarchuk A, Hart JR, Bell C, Carosi M, Pescarmona E, Perracchio L, Diodoro M, Russo A, Antenucci A, Visca P, Ciardi A, Harris CC, Vogt PK, Pekarsky Y, Croce CM. tsRNA signatures in cancer. Proc Natl Acad Sci U S A. 2017;114:8071–8076. doi: 10.1073/pnas.1706908114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 56.Mahlab S, Tuller T, Linial M. Conservation of the relative tRNA composition in healthy and cancerous tissues. RNA (New York, NY) 2012;18:640–652. doi: 10.1261/rna.030775.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goodarzi H, Nguyen HCB, Zhang S, Dill BD, Molina H, Tavazoie SF. Modulated expression of specific tRNAs drives gene expression and Cancer progression. Cell. 2016;165:1416–1427. doi: 10.1016/j.cell.2016.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson SA, Dubeau L, Johnson DL, Enhanced RNA. Polymerase III-dependent transcription is required for oncogenic transformation. J Biol Chem. 2008;283:19184–19191. doi: 10.1074/jbc.M802872200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen Y, Kassavetis GA, Bryant GO, Berk AJ. Polymerase (pol) III TATA box-binding protein (TBP)-associated factor Brf binds to a surface on TBP also required for activated pol II transcription. Mol Cell Biol. 1998;18:1692–1700. doi: 10.1128/MCB.18.3.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rollins J, Veras I, Cabarcas S, Willis I, Schramm L. Human Maf1 negatively regulates RNA polymerase III transcription via the TFIIB family members Brf1 and Brf2. Int J Biol Sci. 2007;3:292–302. doi: 10.7150/ijbs.3.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fang Z, Yi Y, Shi G, Li S, Chen S, Lin Y, Li Z, He Z, Li W, Zhong S. Role of Brf1 interaction with ERalpha, and significance of its overexpression, in human breast cancer. Mol Oncol. 2017;11:1752–1767. doi: 10.1002/1878-0261.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khattar E, Kumar P, Liu CY, Akincilar SC, Raju A, Lakshmanan M, Maury JJ, Qiang Y, Li S, Tan EY, Hui KM, Shi M, Loh YH, Tergaonkar V. Telomerase reverse transcriptase promotes cancer cell proliferation by augmenting tRNA expression. J Clin Invest. 2016;126:4045–4060. doi: 10.1172/JCI86042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fornier M, Fumoleau P. The paradox of triple negative breast cancer: novel approaches to treatment. Breast J. 2012;18:41–51. doi: 10.1111/j.1524-4741.2011.01175.x. [DOI] [PubMed] [Google Scholar]
- 64.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12:954–961. doi: 10.1128/MCB.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kwon NH, Lee MR, Kong J, Park SK, Hwang BJ, Kim BG, Lee ES, Moon HG, Kim S. Transfer-RNA-mediated enhancement of ribosomal proteins S6 kinases signaling for cell proliferation. RNA Biol. 2017:1–14. [DOI] [PMC free article] [PubMed]
- 66.Pavon-Eternod M, Gomes S, Rosner MR, Pan T. Overexpression of initiator methionine tRNA leads to global reprogramming of tRNA expression and increased proliferation in human epithelial cells. RNA (New York, NY) 2013;19:461–466. doi: 10.1261/rna.037507.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clarke CJ, Berg TJ, Birch J, Ennis D, Mitchell L, Cloix C, Campbell A, Sumpton D, Nixon C, Campbell K, Bridgeman VL, Vermeulen PB, Foo S, Kostaras E, Jones JL, Haywood L, Pulleine E, Yin H, Strathdee D, Sansom O, Blyth K, McNeish I, Zanivan S, Reynolds AR, Norman JC. The initiator methionine tRNA drives secretion of type II collagen from stromal fibroblasts to promote tumor growth and angiogenesis. Curr Biol. 2016;26:755–765. doi: 10.1016/j.cub.2016.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kojima Y, Acar A, Eaton EN, Mellody KT, Scheel C, Ben-Porath I, Onder TT, Wang ZC, Richardson AL, Weinberg RA, Orimo A. Autocrine TGF-beta and stromal cell-derived factor-1 (SDF-1) signaling drives the evolution of tumor-promoting mammary stromal myofibroblasts. Proc Natl Acad Sci U S A. 2010;107:20009–20014. doi: 10.1073/pnas.1013805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krishnan P, Ghosh S, Wang B, Heyns M, Li D, Mackey JR, Kovalchuk O, Damaraju S. Genome-wide profiling of transfer RNAs and their role as novel prognostic markers for breast cancer. Sci Rep. 2016;6:32843. doi: 10.1038/srep32843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Towns WL, Begley TJ. Transfer RNA methytransferases and their corresponding modifications in budding yeast and humans: activities, predications, and potential roles in human health. DNA Cell Biol. 2012;31:434–454. doi: 10.1089/dna.2011.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Frye M, Watt FM. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Cur Biol. 2006;16:971–981. doi: 10.1016/j.cub.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 72.Delaunay S, Rapino F, Tharun L. Elp3 links tRNA modification to IRES-dependent translation of LEF1 to sustain metastasis in. breast cancer. 2016;213:2503–2523. doi: 10.1084/jem.20160397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Agris PF, Vendeix FA, Graham WD. tRNA’s wobble decoding of the genome: 40 years of modification. J Mol Biol. 2007;366:1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 74.Amuthan G, Biswas G, Zhang SY, Klein-Szanto A, Vijayasarathy C, Avadhani NG. Mitochondria-to-nucleus stress signaling induces phenotypic changes, tumor progression and cell invasion. EMBO J. 2001;20:1910–1920. doi: 10.1093/emboj/20.8.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grzybowska-Szatkowska L, Slaska B. Polymorphisms in genes encoding mt-tRNA in female breast cancer in Poland. Mitochondrial DNA. 2012;23:106–111. doi: 10.3109/19401736.2012.660925. [DOI] [PubMed] [Google Scholar]
- 76.Meng XL, Meng H, Zhang W, Qin YH, Zhao NM. The role of mitochondrial tRNA variants in female breast cancer. Mitochondrial DNA A DNA Mapp Seq Anal. 2016;27:3199–3201. doi: 10.3109/19401736.2015.1007332. [DOI] [PubMed] [Google Scholar]
- 77.Honda S, Loher P, Shigematsu M, Palazzo JP, Suzuki R, Imoto I, Rigoutsos I, Kirino Y. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers, Proc Natl Acad Sci U S A. 2015;112:E3816–25. [DOI] [PMC free article] [PubMed]
- 78.Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, Tavazoie SF. Endogenous tRNA-derived fragments suppress breast Cancer progression via YBX1 displacement. Cell. 2015;161:790–802. doi: 10.1016/j.cell.2015.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guzman N, Agarwal K, Asthagiri D, Yu L, Saji M, Ringel MD, Paulaitis ME. Breast Cancer-specific miR signature unique to extracellular vesicles includes “microRNA-like” tRNA fragments. Mol Cancer Res. 2015;13:891–901. doi: 10.1158/1541-7786.MCR-14-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Blenkiron C, Hurley DG, Fitzgerald S, Print CG, Lasham A. Links between the oncoprotein YB-1 and small non-coding RNAs in breast cancer. PLoS One. 2013;8:e80171. doi: 10.1371/journal.pone.0080171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dhahbi JM, Spindler SR, Atamna H, Boffelli D, Martin DI. Deep sequencing of serum small RNAs identifies patterns of 5’ tRNA half and YRNA fragment expression associated with breast Cancer. Biomarkers in cancer. 2014;6:37–47. doi: 10.4137/BIC.S20764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 83.Wallace DC. Mitochondrial DNA variation in human radiation and disease. Cell. 2015;163:33–38. doi: 10.1016/j.cell.2015.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lu J, Sharma LK, Bai Y. Implications of mitochondrial DNA mutations and mitochondrial dysfunction in tumorigenesis. Cell Res. 2009;19:802–815. doi: 10.1038/cr.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.He ZF, Zheng LC, Xie DY, Yu SS, Zhao J. Mutational analysis of mitochondrial tRNA genes in patients with lung cancer. Balkan J Med Genet 2016; 19:45–50. [DOI] [PMC free article] [PubMed]
- 86.Wang L, Chen ZJ, Zhang YK, Le HB. The role of mitochondrial tRNA mutations in lung cancer. Int J Clin Exp Med. 2015;8:13341–13346. [PMC free article] [PubMed] [Google Scholar]
- 87.Brule H, Holmes WM, Keith G, Giege R, Florentz C. Effect of a mutation in the anticodon of human mitochondrial tRNAPro on its post-transcriptional modification pattern. Nucleic Acids Res. 1998;26:537–543. doi: 10.1093/nar/26.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Balatti V, Rizzotto L, Miller C, Palamarchuk A, Fadda P, Pandolfo R, Rassenti LZ, Hertlein E, Ruppert AS, Lozanski A, Lozanski G, Kipps TJ, Byrd JC, Croce CM, Pekarsky Y. TCL1 targeting miR-3676 is codeleted with tumor protein p53 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2015;112:2169–2174. doi: 10.1073/pnas.1500010112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shao Y, Sun Q, Liu X, Wang P, Wu R, Ma Z. tRF-Leu-CAG promotes cell proliferation and cell cycle in non-small cell lung cancer. Chem Biol Drug Des. 2017;90:730–738. doi: 10.1111/cbdd.12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Birch J, Clarke CJ, Campbell AD, Campbell K, Mitchell L, Liko D, Kalna G, Strathdee D, Sansom OJ, Neilson M, Blyth K, Norman JC. The initiator methionine tRNA drives cell migration and invasion leading to increased metastatic potential in melanoma. Biol Open. 2016;5:1371–1379. doi: 10.1242/bio.019075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Daly NL, Arvanitis DA, Fairley JA, Gomez-Roman N, Morton JP, Graham SV, Spandidos DA, White RJ. Deregulation of RNA polymerase III transcription in cervical epithelium in response to high-risk human papillomavirus. Oncogene. 2005;24:880–888. doi: 10.1038/sj.onc.1208031. [DOI] [PubMed] [Google Scholar]
- 92.Sobala A, Hutvagner G. Small RNAs derived from the 5′ end of tRNA can inhibit protein translation in human cells. RNA Biol. 2013;10:553–563. doi: 10.4161/rna.24285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou Y, Goodenbour JM, Godley LA, Wickrema A, Pan T. High levels of tRNA abundance and alteration of tRNA charging by bortezomib in multiple myeloma. Biochem Biophys Res Commun. 2009;385:160–164. doi: 10.1016/j.bbrc.2009.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang X, Chow CR, Ebine K, Lee J, Rosner MR, Pan T, Munshi HG. Interaction of tRNA with MEK2 in pancreatic cancer cells. Sci Rep. 2016;6:28260. doi: 10.1038/srep28260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sun C, Fu Z, Wang S, Li J, Li Y, Zhang Y, Yang F, Chu J, Wu H, Huang X, Li W, Yin Y. Roles of tRNA-derived fragments in human cancers. Cancer Lett. 2018;414:16–25. doi: 10.1016/j.canlet.2017.10.031. [DOI] [PubMed] [Google Scholar]
- 96.Schorn AJ, Gutbrod MJ, LeBlanc C, Martienssen R. LTR-retrotransposon control by tRNA-derived small RNAs. Cell. 2017;170:61–71.e11. doi: 10.1016/j.cell.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kim HK, Fuchs G, Wang S, Wei W, Zhang Y, Park H, Roy-Chaudhuri B, Li P, Xu J, Chu K, Zhang F, Chua MS, So S, Zhang QC, Sarnow P, Kay MA. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature. 2017;552:57–62. doi: 10.1038/d41586-017-08387-y. [DOI] [PMC free article] [PubMed] [Google Scholar]


