Abstract
WPK4, a gene encoding a putative protein kinase, was initially identified in wheat (Triticum aestivum) and shown to be up-regulated by light, nutrient deprivation, and cytokinins. To confirm that WPK4 has protein kinase activity, the protein was produced in Escherichia coli as a fusion protein with glutathione S-transferase. The purified protein exhibited autophosphorylation activity and phosphorylated both myelin basic protein and a peptide fragment of rice 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Levels of WPK4 transcripts in wheat seedlings were increased and decreased by the removal and addition of sucrose (Suc), respectively, to the culture medium. The introduction of the N-terminal kinase region of WPK4 into the yeast snf1 mutant cells, which cannot utilize Suc as a carbon source, rescued growth in Suc-containing medium. Cytokinins up-regulated the accumulation of WPK4 transcripts, but their effects were cancelled by the addition of Suc. Our results suggest that Suc negatively regulates the signaling pathway in which transcriptional activation of WPK4 is mediated by cytokinins.
Plants have developed systems to quickly and accurately transmit stimuli from the external environment and to adjust their metabolic pathways by modulating the expression of sets of genes. Activation and/or inactivation of appropriate genes in response to particular stimuli is mediated through well-tuned signal transduction systems in which protein phosphorylation cascades play crucial roles (Crews and Erikson, 1993; Stone and Walker, 1995). Phytohormones are also thought to be indispensable in signaling pathways that involve perception and transduction of external signals (Klee and Estelle, 1991). Cytokinins have attracted particular interest because of their diverse physiological functions in response to light (Miller, 1956; Su and Howell, 1995; von Arnim and Deng, 1996), nutrients (Yu et al., 1998), growth (Chaudhury et al., 1993; Brzobohaty et al., 1994), and even wounding (Simmons et al., 1992). Recent work has revealed that cytokinins enhance the transcription of several genes involved in nutrient metabolism (Lu et al., 1990; Dominov et al., 1992; Andersen et al., 1996; Ehnes and Roitsch, 1997) and self-defense (Simmons et al., 1992; Sano et al., 1994, 1996).
During photosynthesis, when carbohydrates are synthesized, the expression of many genes is coordinately regulated by external factors including light, carbon dioxide, and temperature, and by internal factors including phytohormones such as cytokinins. It has been suggested that feedback regulation of photosynthesis is due to repression of photosynthetic gene transcription by carbohydrates (Sheen, 1990). Recent studies have suggested that hexokinase, which was postulated to play an important role in sugar sensing in yeast (Ma et al., 1989; Rose et al., 1991), could be responsible for sensing sugars to repress a number of plant genes (Graham et al., 1994; Jang and Sheen, 1994). However, there are some examples of hexose-independent changes in gene expression (Wenzler et al., 1989; Dijkwel et al., 1997) and in proton-Suc symporter activity (Chiou and Bush, 1998). Thus, mechanisms by which sugar is sensed and sugar signals are transduced are not completely clear yet.
The sugar-catabolic mechanism has been extensively investigated in yeast (Saccharomyces cerevisiae) (Carlson, 1987). A protein kinase, SNF1, functions as a regulator to derepress many Glc-repressed genes such as SUC2, which encodes a secreted invertase that hydrolyzes Suc to Glc and Fru. In higher organisms, a similar mechanism has been predicted to function in the sugar assimilation process, and genes with a significant sequence similarity to the yeast SNF1 gene have been identified in organisms ranging from mammals (Hardie et al., 1998) to plants (Halford and Hardie, 1998). To date, more than 20 of these genes have been identified from higher plants, and are called SNF1-related protein kinases (SnRKs). Based on putative amino acid sequence similarities, they are classified into three distinct subgroups (SnRK1 through SnRK3) (Halford and Hardie, 1998). Those from rye (RKIN1) (Alderson et al., 1991), tobacco (NPK5) (Muranaka et al., 1994), and Arabidopsis (AKIN10 and AKIN11) (Bhalerao et al., 1999), all of which belong to the SnRK1 group, have been shown to complement the yeast snf1 mutant cells. Antisense expression of the potato SnRK1 homolog PKIN1 resulted in a loss of sugar-inducible expression of Suc synthase gene in leaves and tubers (Purcell et al., 1998). In barley aleurone layers, a SnRK2 homolog (PKABA) has been shown to mediate abscisic acid-suppressed gene expression (Gomez-Cadenas et al., 1999). However, functional analysis of SnRK3 has not been performed.
We previously isolated WPK4, a gene encoding a SNF1-related protein kinase from wheat (Sano and Youssefian, 1994). The deduced amino acid sequence of WPK4 suggested that it belongs in the SnRK3 subgroup (Halford and Hardie, 1998). The protein kinase catalytic domain is located in the N-terminal half and has 40% to 50% amino acid sequence identity with SnRK1s. However, beyond the kinase domain, it has little sequence similarity with other plant SnRKs. WPK4 also contains the Pro-rich consensus sequence of XPYPPXP (X and Y refer to any residue and hydrophobic residues, respectively), which is predicted to be recognized by the SH3 domain present in cytoskeletal elements and signal transducing proteins (Musacchio et al., 1994). The transcript level of WPK4 is increased by both light and nutrient deprivation through the action of cytokinins (Sano and Youssefian, 1994).
We report the further characterization of WPK4, showing that it possesses protein kinase activity and partially complements the yeast snf1 mutant cells. We also report WPK4 expression being differentially regulated by Suc and cytokinins.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Wheat (Triticum aestivum cv Nanbu) seeds were germinated and grown hydroponically in 160 mL of solution with the appropriate composition indicated for each experiment, at 23°C for 6 to 7 d under continuous light conditions. For further experiments, plants were grown and maintained on soil in a greenhouse.
Construction of Plasmids
The WPK4 cDNA was digested with EcoRI and ligated to pGEX-2T (Pharmacia Biotech, Piscataway, NJ) at the EcoRI site to fuse in-frame the coding sequence for glutathione S-transferase (GST) with that for WPK4. The resulting construct was designated pGEX-WPK4. A WPK4 mutant (WPKM4) in which the AAG codon (nucleotide positions 230–232) for a Lys was substituted for a GAT codon (Asp) was also prepared using the PCR technique. A DNA fragment encoding a peptide from rice 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGR) (corresponding to residues 471–576) was amplified by PCR using the oligonucleotides forward: 5′- GAATTCTCCCAGTGCATCAC-3′ and reverse: 5′-TACCTCGAGCATGCCACATGG-3′, and cloned into pGEX-4T-1 (Pharmacia) at the EcoRI and XhoI sites (pGEX-HMGR).
Expression and Purification of Fusion Proteins
Plasmids pGEX-WPK4, pGEX-WPKM4, and pGEX-HMGR were introduced into the Escherichia coli DH5α strain, and the transformants were grown in 2 L of 2× yeast-trypton (YT) medium with vigorous shaking at 21°C to an A600 of 0.6. Isopropyl thio-β-d-galactoside (IPTG) was then added to a final concentration of 0.1 mm, and the cultures were further incubated at 16°C for 6 h. Cells were pelleted and suspended in 20 mL of lysis buffer (20 mm Tris-HCl [pH 8.0], 1 mm EDTA [pH 8.0], 10 mm 2-mercaptoethanol, 0.1 mm phenylmethylsulfonyl fluoride [PMSF], 10% [w/v] Suc, and 0.5% [w/v] lauryl sarcosinate) and disrupted by sonication. After ultracentrifugation, the supernatants were applied to a 3-mL glutathione-Sepharose (Pharmacia) column. The columns were washed with 20 bed volumes of T buffer (20 mm Tris-HCl [pH 8.0], 1 mm EDTA [pH 8.0], 10 mm 2-mercaptoethanol), then with 5 bed volumes of T buffer containing 3 m NaCl. Absorbed proteins were eluted with 15 mL of elution buffer (20 mm Tris-HCl [pH 8.0], 1 mm EDTA [pH 8.0], 10 mm 2-mercaptoethanol, 25 mm GSH) and the fractions containing GST-WPK4, GST-WPKM4, and pGEX-HMGR were applied to a DEAE-Toyopearl (Tosoh, Tokyo) column equilibrated with T buffer. Proteins were eluted with 50 mL of T buffer with a 0 to 0.5 m NaCl linear gradient. One-milliliter fractions were collected, assayed for protein concentration, and subjected to SDS-PAGE. The gels were stained with Coomassie Brilliant Blue R250. Fractions that contained purified GST-WPK4, GST-WPKM4, and GST-HMGR fusion proteins were concentrated by ultrafiltration (Centricon, Amicon, Beverly, MA). Purified fusion proteins were used in further experiments.
Kinase Assay
To measure autophosphorylation, aliquots of 100 ng of GST-WPK4 or GST-WPKM4 fusion proteins were incubated with 0.1 mm [γ-32P]ATP (6,000 Ci/mmol) in kinase buffer containing 20 mm Tris-HCl (pH 8.0), 1 mm EDTA (pH 8.0), 0.1 mm PMSF, 20 mm MgCl2, or 20 mm MnCl2 in a final volume of 30 μL at room temperature for 30 min. A kinase assay was conducted by adding to the kinase buffer 500 ng of myelin basic proteins (MBP) or GST-HMGR as substrates. The reaction was terminated by adding one-fourth volume of 5× Laemmli's sample buffer (200 mm Tris-HCl [pH 6.8], 50 mm DTT, 5% [w/v] SDS, 50% [w/v] glycerol, and 0.1% [w/v] bromphenol blue). After heating at 95°C for 5 min, the mixture was subjected to electrophoresis in 12.5% (w/v) SDS-PAGE gel, stained with Coomassie Brilliant Blue R-250, dried, and autophotographed at −80°C for 16 h.
RNA-Blot Analysis
Total RNA was extracted from various tissues by the aurin tricarboxylic acid method (Verwoerd et al., 1989). Aliquots of 36 μg per lane were denatured, fractionated by 1.0% (w/v) formaldehyde/agarose gel electrophoresis, and transferred onto nylon membranes (Hybond-N, Amersham-Pharmacia Biotech, Uppsala). Northern hybridizations were performed according to standard protocols (Sambrook et al., 1989). As the WPK4-specific probe, a 0.8-kb fragment was amplified by PCR using primers for forward: 5′-CCTTACTAGCCTGATCATGCG-3′ and for reverse: 5′-TTGTTCCTGTCAGTTGCACC-3′. As the RBCS (RuBP carboxylase/oxygenase small subunit) probe, a 0.3-kb fragment was amplified by PCR using primers for forward: 5′-TGTCTTACTTGCCACCCCTC-3′ and for reverse: 5′-AGGGTACTCCTTCTTGACCTCC-3′. As the NR (nitrate reductase) probe, a 0.5-kb fragment was amplified by PCR using primers for forward: 5′-AAGCACATCTTCGTTTGCG-3′ and for reverse: 5′-CGATCACGTACCACACCTTG-3′. Probes were radioactively labeled using the BcaBEST labeling kit (TaKaRa Shuzo, Ltd., Kyoto). To normalize for the amount of RNA loaded, the membrane was stripped of the former probe by boiling in a 0.1% (w/v) SDS solution for 1 min before rehybridization with a wheat actin probe prepared as described earlier (Sano and Youssefian, 1994). Each mRNA level was estimated by measuring signal intensity with imaging software (NIH Image, National Institutes of Health, Bethesda, MD). The value was normalized with values for actin mRNA transcripts.
Yeast Complementation Test
A fragment containing the ADH1 promoter and the CYC1 terminator was excised from pD2 and inserted into YEplac 195 (2μ,URA3) at SphI and HindIII sites to generate pF0. An EcoRI fragment containing WPK4 cDNA was ligated into the EcoRI site of pF0 (pWPK4 = sense oriented; pASWPK4 = antisense oriented). An EcoRI fragment containing the entire coding region of SUC2 was prepared by PCR using yeast genomic DNA as a template and primers tagged with EcoRI recognition sites, then ligated into pF0 (pSUC2). The WPKM4 construct was prepared with the aid of PCR, then ligated into the EcoRI site of pF0 (pWPKM4). The C-terminal-truncated WPK4 was constructed as follows: a 0.9-kb 5′-terminal fragment of WPK4 was excised from pGEX-2T by digestion with EcoRI and PvuII, and then inserted into pBluescript II KS(−) at the EcoRI and SmaI sites to generate pWB1. A 5′-terminal fragment of WPK4 was excised by BamHI and HindIII double digestion from pWB1, and then ligated to the pKF19 Km vector digested with BamHI and HindIII to generate pWK1. The 5′-terminal fragment of WPK4, generated by digestion of pWK1 with EcoRI, was ligated into pF0 (pWPK4ΔC). The orientation of the ligated fragment was confirmed by appropriate restriction endonuclease digestion. Resulting constructs were transformed into the yeast strain MCY1846 (lys 2-801, ura3-52, snf1Δ10; provided by Marian Carlson, Columbia University). For examination of the effects of various carbon sources on growth of transformed cells, the cells were plated on medium lacking uracil, supplemented with 2% (w/v) Glc or Suc, and cultured at 30°C under anaerobic conditions.
RESULTS
In Vitro Kinase Assay of WPK4
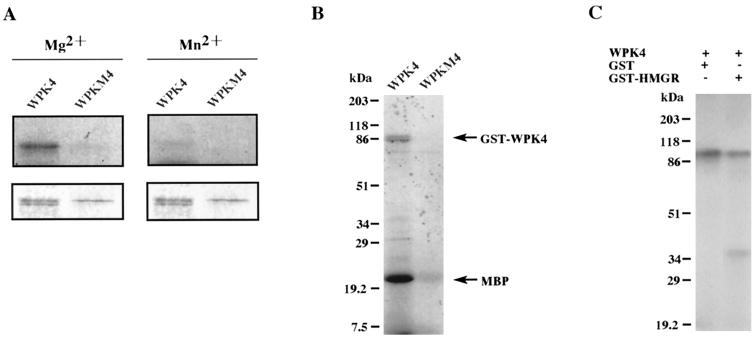
To characterize the gene product, a fusion protein of WPK4 with GST was constructed in E. coli cells, and its enzymatic activity was assayed. In the presence of Mg2+, the GST-WPK4 fused protein exhibited autophosphorylation activity. However, the reaction was not observed when Mg2+ ions were replaced with Mn2+ ions. Two protein bands were visible when the GST-WPK4 proteins were purified, possibly due to a slight difference in the size of the expressed proteins. Both proteins had protein kinase activity (Fig. 1A). WPK4 also phosphorylated MBP (Fig. 1B), with the same divalent cation requirement. When the Lys at the amino acid position 75 of WPK4, which is analogous to the Lys essential for ATP binding in most Ser/Thr protein kinases, was replaced by Asp (WPKM4), both autophosphorylation and MBP phosphorylation were completely abolished (Fig. 1). These results show that, unlike SNF1, WPK4 cannot use Mn2+ in place of Mg2+, and suggests that WPK4 differs in kinetic properties from SNF1 and other proteins of this family. To date, nitrate reductase, Suc-P synthase, and HMGR have been reported to be phosphorylated by SnRKs (Sugden et al., 1999). SAMS peptide, a synthetic peptide containing the primary phosphorylation site for AMPK of rat acetyl-CoA carboxylase (Davies et al., 1989), was also reported to be phosphorylated by plant extracts (MacKintosh et al., 1992). Taking this information into account, a peptide fragment of rice HMGR containing the putative phosphorylation site was expressed in bacteria and tested in this study. The results clearly showed that WPK4 was able to phosphorylate GST-HMGR peptide but not GST in vitro (Fig. 1C).
Figure 1.
Phosphorylation analysis of GST-WPK4 and GST-WPKM4 (mutant) fusion proteins. A, Samples were autophosphorylated in reaction mixtures containing either Mg2+ (left) or Mn2+ (right) and separated by SDS-PAGE. The autoradiogram and profiles of the gel stained with Coomassie Brilliant Blue are shown in the top and bottom panels, respectively. B and C, In vitro phosphorylation analysis of GST-WPK4 fusion proteins. Aliquots of 100 ng of GST-WPK4 or GST-WPKM4 fusion proteins were incubated in the presence of 500 ng of myelin basic protein (B), GST (C), or GST-HMGR (C) as the substrates. After the phosphorylation reaction, samples were separated by 12.5% (w/v) SDS-PAGE, dried, and exposed for autoradiography. Relative molecular masses of the standard samples are indicated on the left in kD.
Accumulation of WPK4 Transcripts in Green Tissues
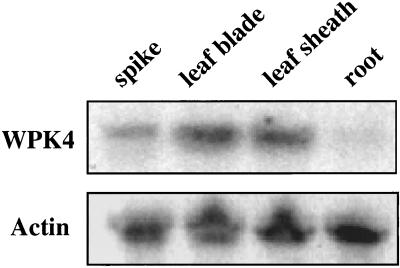
The tissue-specific accumulation of WPK4 transcripts in mature plants was examined with RNA-blot hybridization. The WPK4 transcripts accumulated predominantly in the leaf sheath and leaf blade and to a lesser extent in the spike (approximately 2 weeks after heading) but not in roots, indicating a link with photosynthetic tissues (Fig. 2). Based on this observation, the following experiments were performed mostly with light-grown green seedlings.
Figure 2.
Tissue-specific accumulation of WPK4 transcripts. Total RNA was extracted from the indicated tissue of mature wheat plants and subjected to RNA-blot hybridization. The hybridization probes were a 0.8-kb WPK4-specific sequence and a 1.2-kb wheat actin cDNA as the internal standard.
Effects of Inorganic Salts and Suc on WPK4 Transcript Levels
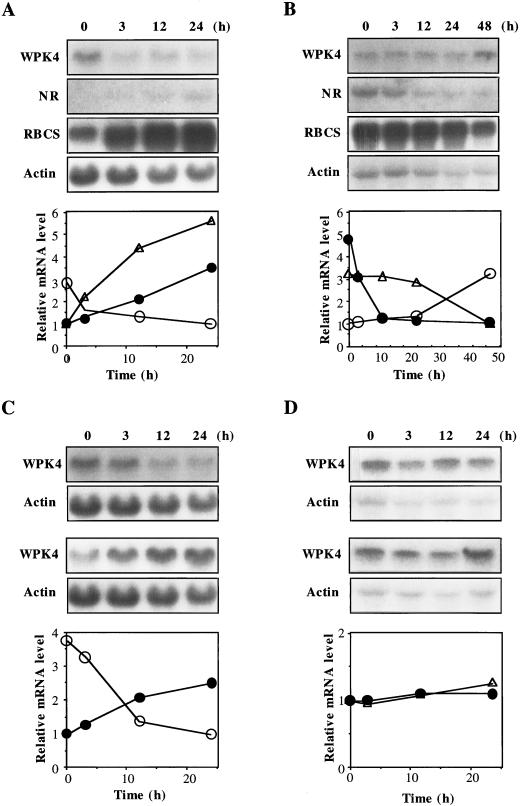
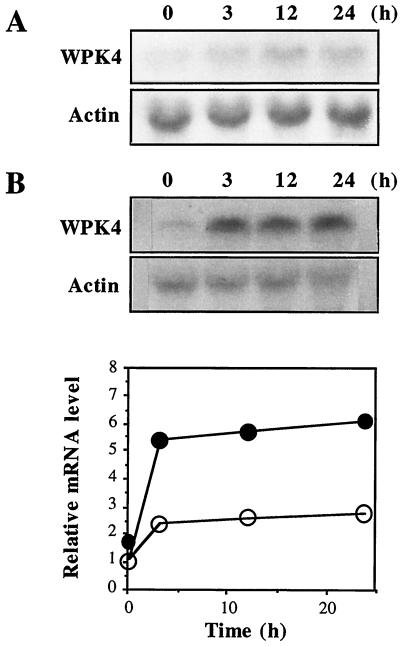
Seven-day-old seedlings were exposed to various nutrient stresses and transcript accumulation of RBCS (RuBP carboxylase/oxygenase small subunit), NR (nitrate reductase), and WPK4 was examined by northern-blot hybridization. RBCS and NR were selected because transcript accumulation of the former is typically repressed by sugars and that of the latter is rapidly induced by nitrates (Sheen, 1990; Crawford and Arst, 1993). When hydroponically grown seedlings were transferred to a one-fifth-strength Murashige and Skoog (MS) medium, WPK4 transcripts decreased within 3 h, whereas transcripts for RBCS and NR accumulated in a time-dependent manner (Fig. 3A). When seedlings initially grown in a one-fifth-strength MS medium were transferred to a nutrient-deprived medium, i.e. water, WPK4 transcripts increased by 48 h (Fig. 3B). In contrast, transcripts for RBCS decreased by 48 h, and NR transcripts rapidly declined within 10 h (Fig. 3B). When experiments were performed with Suc solution instead of MS medium, similar results were obtained. The addition of 0.7% (w/v) Suc to seedlings cultivated in water resulted in a decrease of WPK4 transcripts 3 h later (Fig. 3C, top). When seedlings were cultured initially in 0.7% (w/v) Suc and then transferred to water, WPK4 transcripts increased (Fig. 3C, bottom). Similar experiments were performed for Glc and Gal, neither of which affected WPK4 transcript levels (Fig. 3D). These results suggest that both inorganic salts and Suc are negative effectors of expression of the WPK4 gene and that the level of WPK4 transcripts is reversibly controlled by Suc.
Figure 3.
Nutrient effects on WPK4 transcript accumulation. Total RNA was isolated from the indicated samples and analyzed by RNA-blot hybridization. A, Seven-day-old seedlings grown in water were transferred to one-fifth-strength MS medium and harvested after the indicated time periods. Relative mRNA levels were densitometrically estimated for WPK4 (○), NR (●), and RBCS (▵). B, Seven-day-old seedlings grown in one-fifth-strength MS medium were transferred to water and harvested after the indicated time periods. Relative mRNA levels were densitometrically estimated for WPK4 (○), NR (●), and RBCS (▵). The hybridization probes were a 0.8-kb WPK4-specific sequence, a 0.5-kb wheat NR sequence, a 0.6-kb wheat RBCS sequence, and a 1.2-kb wheat actin cDNA. C, Seven-day-old seedlings grown in water were transferred to water containing 0.7% (w/v) Suc and harvested after the indicated time periods (top). Similarly, 7-d-old seedlings grown in water containing 0.7% (w/v) Suc were transferred to water and harvested after the indicated time periods (bottom). Relative mRNA levels were densitometrically estimated for Suc addition (○) and Suc removal (●). D, Seven-day-old seedlings grown in water were transferred to water containing 0.7% (w/v) Glc (top) or 0.7% (w/v) Gal (bottom) and harvested after the indicated time periods. Relative mRNA levels were densitometrically estimated for Glc addition (●) and Gal addition (▵). The hybridization probes were a 0.8-kb WPK4-specific sequence and a 1.2-kb wheat actin cDNA.
Complementation of the Yeast snf1 Mutation
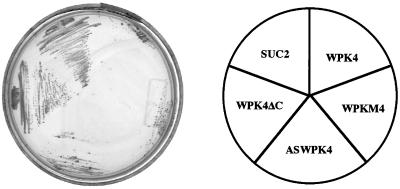
A yeast complementation test was carried out to examine whether WPK4 functions in catabolite repression in yeast cells. cDNAs encoding a full-length WPK4, WPKM4, C-terminal-truncated WPK4, or antisense-oriented WPK4 were expressed in snf1 mutant cells under the control of an ADH promoter. Mutant cells transformed with SUC2, which encodes a secreted invertase that hydrolyzes Suc to Glc and Fru, were used as a positive control. The snf1 mutant cells carrying the C-terminal truncated WPK4 containing the kinase domain but lacking the 3′-region beyond the kinase domain grew on medium supplemented with Suc, but the growth rate was low in comparison with wild-type cells (Fig. 4). Cells containing other constructs failed to grow on the medium. These results suggested that the C-terminal region functions as the regulatory domain, presumably by inhibiting the kinase activity.
Figure 4.
Complementation of snf1 mutant cells by various WPK4 constructs. Cells of S. cerevisiae MCY1846 (snf1) harboring pWPK4, pASWPK4, pWPKM4, WPK4ΔC, or pSUC2 were streaked on agar plates supplemented with 2% (w/v) Suc. Cells of MCY1846 were grown at 30°C for 6 d.
Effects of Cytokinins
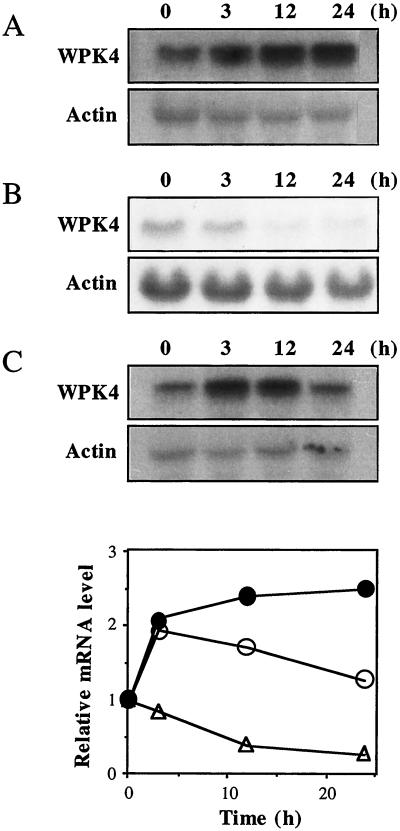
Cytokinins were previously shown to induce WPK4 (Sano and Youssefian, 1994), and that study showed that the degree of positive regulation by cytokinin was dependent upon nutritional conditions (Fig. 5). Since Suc was found to be a negative regulator of WPK4, the relationship between Suc and cytokinins was analyzed. Seedlings were cultivated in water and then treated with N6-benzylaminopurine (BA), Suc, or both. Upon treatment with BA alone, WPK4 transcripts were increased up to 3-fold (Fig. 6A). Treatment with 0.7% (w/v) Suc down-regulated WPK4 transcripts by more than 50% (Fig. 6B). When seedlings were cultivated in the presence of both BA and Suc, WPK4 transcripts were temporarily increased for 12 h, but then decreased to the initial level by 24 h (Fig. 6C). These observations indicate that Suc antagonizes the BA effect and therefore may be involved in the signal transduction pathways of WPK4.
Figure 5.
Effects of cytokinins on WPK4 transcript accumulation. Total RNAs were isolated from the indicated samples and subjected to RNA-blot hybridization. A, Seven-day-old seedlings grown in one-half-strength MS medium were treated with 100 μm BA for the indicated time periods. B, Seven-day-old seedlings grown in one-fifth-strength MS medium were treated with 100 μm BA for the indicated time periods. Relative mRNA levels were densitometrically estimated for samples extracted from seedlings grown in one-half-strength MS (○) or in one-fifth-strength MS (●) media.
Figure 6.
Antagonistic effects of cytokinins and Suc. Total RNAs were isolated from the indicated samples and subjected to RNA-blot hybridization. A, Seven-day-old seedlings grown in water were treated with 100 μm BA for the indicated time periods. B, Seven-day-old seedlings grown in water were transferred to water containing 0.7% (w/v) Suc and harvested after the indicated time periods. C, Seven-day-old seedlings grown in water were transferred to water containing 0.7% (w/v) Suc and 100 μm BA for the indicated time periods. The hybridization probes were a 0.8-kb WPK4-specific sequence and a 1.2-kb wheat actin cDNA. Relative mRNA levels were densitometrically estimated for samples extracted from seedlings grown in water treated with BA (●) or Suc (▵) or both (○).
DISCUSSION
The present study documents that WPK4, a protein kinase belonging to the SNF1-realated protein kinase family (SnPK3), phosphorylates HMGR and partially complements the yeast snf1 mutant, and that its transcription is coordinately regulated by cytokinins and Suc.
The SNF1 protein kinase, which was originally identified in yeast cells, plays an essential role in Glc utilization (Celenza and Carlson, 1986). Since its discovery, many genes encoding SNF1-related protein kinases have been reported to exist in various organisms including higher plants (Stone and Walker, 1995; Halford and Hardie, 1998). Analyses of tissue-specific expression of these genes revealed two groups: those encoding NPK5 (tobacco), AKIN10 (Arabidopsis), BKIN2 (barley), and PKIN1 (pea) appear to be ubiquitously expressed in all tissues, whereas those encoding BKIN12 (barley) and RKIN1 (rye) are specifically expressed in cereal seeds (Alderson et al., 1991). Transcripts of WPK4 in mature wheat plants were found in the present study to mostly accumulate in leaf sheaths and leaf blades, indicating that WPK4 mainly functions in photosynthetic tissues. These different expression patterns suggest different functions among structurally related protein kinases, although the physiological roles of the majority of them have yet to be determined. In this context, it should be noted that WPK4 is capable of phosphorylation of HMGR in vitro. This is consistent with biochemical experiments showing that HMGR is inactivated by phosphorylation catalyzed in vitro by calcium-dependent protein kinase and by SNF1-related protein kinases (Douglas et al., 1997; Sugden et al., 1999). However, in vivo experiments are necessary to determine whether WPK4 phosphorylates HMGR as the native substrate.
The level of WPK4 transcripts was previously shown to increase upon exposure to inorganic salt deficiency (Sano and Youssefian, 1994). Since the present kinetic analyses showed WPK4 transcripts to be increased by Suc depletion and decreased by Suc application, the question arises as to whether inorganic salts or Suc are primarily responsible for regulation of WPK4 expression. To address this question, the transcript levels of NR and RBCS were simultaneously examined with those for WPK4 upon salt or Suc treatments. The transcript level of RBCS fluctuated inversely but consistently with that of WPK4, whereas NR transcripts responded differently. These observations suggest that the change in the WPK4 transcript level upon inorganic salt deprivation possibly results from the decline in endogenous Suc caused by a decrease in photoassimilating activity. Although more experimental work is necessary to determine the exact relationship between salts and Suc, it is clear that Suc is one of the most important factors in the regulation of WPK4 gene expression.
Expression of the C-terminal-truncated form of WPK4 in the snf1 mutant of yeast complemented the mutant phenotype, but full-length WPK4 was not effective. The full-length SNFL1 from sorghum also exhibited no complementation of this yeast mutant (Annen and Stockhaus, 1998). These observations are consistent with the finding that SnRK1s (including RKIN1, AKIN10, and NPK5) are much more closely related to SNF1 than are WPK4 and SNFL1. In yeast cells, SNF1 was shown to act in a SNF1 complex with SNF4, as well as SIP1, SIP2, or GAL83 (Hardie et al., 1998). In plants, the presence of a similar complex was reported. Using the yeast two-hybrid system, AKIN10 was shown to interact with PRL1 protein, which contains repetitive sequences characteristic of a family of regulatory proteins known as WD-40 repeat proteins (Nemeth et al., 1998; Bhalerao et al., 1999). StubSNF1 from potato interacted with the yeast GAL83/SIP1/SIP2 ortholog (StubGAL83) and with SNF4 in the same system (Lakatos et al., 1999). Both PRL1 and StubGAL83 proteins bind at the C-terminal region of SnRK1 proteins.
These observations suggest that SnRK1 proteins are orthologs of SNF1 and are required for the activation of gene expression. Whether SnRK2 and SnRK3 functionally interact with those proteins has not been determined. Among three subgroups, however, the C-terminal region is structurally distinct, and it is proposed to confer specific functions to each. Contrary to SnRK1 proteins that complemented the snf1 mutation by whole protein, WPK4 only complemented the mutant in the C-terminal truncated form. It is therefore conceivable that the C-terminal region of WPK4 acts as a negative regulator of the kinase activity.
To our knowledge, WPK4 is the only protein kinase gene whose transcription is up-regulated by cytokinins and down-regulated by Suc. The fact that WPK4 transcripts first increased and then gradually decreased when nutrient-deprived seedlings were simultaneously exposed to both cytokinins and Suc indicates that cytokinins predominate over Suc. Thus, it is likely that cytokinins initially induce production of WPK4, which triggers and/or activates sugar synthesis, and that accumulated sugars block a certain step in signal transduction pathways of WPK4 to shut down cytokinin signals. This kind of feedback control system might best explain our present data. Further work is necessary to substantiate this hypothesis.
ACKNOWLEDGMENTS
The authors thank Dr. Marian Carlson for the generous gift of the yeast strain MCY1846 and Dr. Malcolm Moore for critical reading of the manuscript.
Footnotes
This work was supported by a Grant-in-Aid (no. 09274102) from the Ministry of Education, Science, Sports and Culture of Japan.
LITERATURE CITED
- Alderson A, Sabelli P, Dickinson J, Cole D, Richardson M, Kreis M, Shewry P, Halford N. Complementation of snf1, a mutation affecting global regulation of carbon metabolism in yeast, by a plant protein kinase cDNA. Proc Natl Acad Sci USA. 1991;88:8602–8605. doi: 10.1073/pnas.88.19.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen B, Chen G, Ertl J, Chen C. Transcriptional regulation of hydroxypyruvate reductase gene expression by cytokinin in etiolated pumpkin cotyledons. Planta. 1996;198:1–5. doi: 10.1007/BF00197579. [DOI] [PubMed] [Google Scholar]
- Annen F, Stockhaus J. Characterization of a Sorghum bicolor gene family encoding putative protein kinases with a high similarity to the yeast SNF1 protein kinase. Plant Mol Biol. 1998;36:529–539. doi: 10.1023/a:1005999921669. [DOI] [PubMed] [Google Scholar]
- Bhalerao RP, Salchert K, Bako L, Okresz L, Szabados L, Muranaka T, Machida Y, Schell J, Koncz C. Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases. Proc Natl Acad Sci USA. 1999;96:5322–5327. doi: 10.1073/pnas.96.9.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzobohaty B, Moore I, Palme K. Cytokinin metabolism: implications for regulation of plant growth and development. Plant Mol Biol. 1994;26:1483–1497. doi: 10.1007/BF00016486. [DOI] [PubMed] [Google Scholar]
- Carlson M. Regulation of sugar utilization in Saccharomyces cerevisiae species. J Bacteriol. 1987;169:4873–4877. doi: 10.1128/jb.169.11.4873-4877.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza J, Carlson M. A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science. 1986;233:1175–1180. doi: 10.1126/science.3526554. [DOI] [PubMed] [Google Scholar]
- Chaudhury A, Leetham S, Craig S, Dennis E. amp1-a mutant with high cytokinin levels and altered embryonic pattern, faster vegetative growth, constitutive photomorphogenesis and precocious flowering. Plant J. 1993;4:907–916. [Google Scholar]
- Chiou T-J, Bush D. Sucrose is a signal molecule in assimilate partitioning. Proc Natl Acad Sci USA. 1998;95:4784–4788. doi: 10.1073/pnas.95.8.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NM, Arst HN., Jr The molecular genetics of nitrate assimilation in fungi and plants. Annu Rev Genet. 1993;27:115–146. doi: 10.1146/annurev.ge.27.120193.000555. [DOI] [PubMed] [Google Scholar]
- Crews CM, Erikson RL. Extracellular signals and reversible protein phosphorylation: what to Mek of it all. Cell. 1993;74:215–217. doi: 10.1016/0092-8674(93)90411-i. [DOI] [PubMed] [Google Scholar]
- Davies SP, Carling D, Hardie DG. Tissue distribution of the AMP-activated protein kinase, and lack of activation by cyclic AMP-dependent protein kinase, studied using a specific and sensitive peptide assay. Eur J Biochem. 1989;186:123–128. doi: 10.1111/j.1432-1033.1989.tb15185.x. [DOI] [PubMed] [Google Scholar]
- Dijkwel P, Huijser C, Weisbeek P, Chua N-H, Smeekens S. Sucrose control of phytochrome A signaling in Arabidopsis. Plant Cell. 1997;9:583–595. doi: 10.1105/tpc.9.4.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominov J, Stenzler L, Lee S, Schwarz J, Leisner S, Howell S. Cytokinins and auxins control the expression of a gene in Nicotiana plumbaginifolia cells by feedback regulation. Plant Cell. 1992;4:451–461. doi: 10.1105/tpc.4.4.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas P, Pigaglio E, Ferrer A, Halford N, MacKintosh C. Three spinach leaf nitrate reductase-3-hydroxy-3-methylglutaryl-CoA reductase kinases that are regulated by reversible phosphorylation and/or Ca2+ ions. Biochem J. 1997;325:101–109. doi: 10.1042/bj3250101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehnes R, Roitsch T. Co-ordinated induction of mRNAs for extracellular invertase and a glucose transporter in Chenopodium rubrum by cytokinins. Plant J. 1997;11:539–548. doi: 10.1046/j.1365-313x.1997.11030539.x. [DOI] [PubMed] [Google Scholar]
- Gomez-Cadenas A, Verhey SD, Holappa LD, Shen Q, Ho TH, Walker-Simmons MK. An abscisic acid-induced protein kinase, PKABA1, mediates abscisic acid-suppressed gene expression in barley aleurone layers. Proc Natl Acad Sci USA. 1999;96:1767–1772. doi: 10.1073/pnas.96.4.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham I, Denby K, Leaver C. Carbon catabolite repression regulates glyoxylate cycle gene expression in cucumber. Plant Cell. 1994;6:761–772. doi: 10.1105/tpc.6.5.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford NG, Hardie DG. SNF1-related protein kinases: global regulators of carbon metabolism in plants? Plant Mol Biol. 1998;37:735–748. doi: 10.1023/a:1006024231305. [DOI] [PubMed] [Google Scholar]
- Hardie D, Carling D, Carlson M. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu Rev Biochem. 1998;67:821–855. doi: 10.1146/annurev.biochem.67.1.821. [DOI] [PubMed] [Google Scholar]
- Jang J-C, Sheen J. Sugar sensing in higher plants. Plant Cell. 1994;6:1665–1679. doi: 10.1105/tpc.6.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee H, Estelle M. Molecular genetic approaches to plant hormone biology. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:529–551. [Google Scholar]
- Lakatos L, Klein M, Hofgen R, Banfalvi Z. Potato StubSNF1 interacts with StubGAL83: a plant protein kinase complex with yeast and mammalian counterparts. Plant J. 1999;17:569–574. doi: 10.1046/j.1365-313x.1999.00406.x. [DOI] [PubMed] [Google Scholar]
- Lu J-L, Ertl J, Chen C-M. Cytokinin enhancement of the light induction of nitrate reductase transcript levels in etiolated barley leaves. Plant Mol Biol. 1990;14:585–594. doi: 10.1007/BF00027504. [DOI] [PubMed] [Google Scholar]
- Ma H, Bloom LM, Walsh CT, Botstein D. The residual enzymatic phosphorylation activity of hexokinase II mutants is correlated with glucose repression in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:5643–5649. doi: 10.1128/mcb.9.12.5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKintosh RW, Davies SP, Clarke PR, Weekes J, Gillespie JG, Gibb BJ, Hardie DG. Evidence for a protein kinase cascade in higher plants: 3-hydroxy-3- methylglutaryl-CoA reductase kinase. Eur J Biochem. 1992;209:923–931. doi: 10.1111/j.1432-1033.1992.tb17364.x. [DOI] [PubMed] [Google Scholar]
- Miller C. Similarity of kinetin and red light effects. Plant Physiol. 1956;31:318–319. doi: 10.1104/pp.31.4.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muranaka T, Banno H, Machida Y. Characterization of tobacco protein kinase NPK5, a homolog of Saccharomyces cerevisiae SNF1 that constitutively activates expression of the glucose-repressible SUC2 gene for a secreted invertase of S. cerevisiae. Mol Cell Biol. 1994;14:2958–2965. doi: 10.1128/mcb.14.5.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Wilmanns M, Saraste M. Structure and function of the SH3 domain. Prog Biophys Mol Biol. 1994;61:283–297. doi: 10.1016/0079-6107(94)90003-5. [DOI] [PubMed] [Google Scholar]
- Nemeth K, Salchert K, Putnoky P, Bhalerao R, Koncz-Kalman Z, Stankovic-Stangeland B, Bako L, Mathur J, Okresz L, Stabel S, Geigenberger P, Stitt M, Redei GP, Schell J, Kancz C. Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev. 1998;12:3059–3073. doi: 10.1101/gad.12.19.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell PC, Smith AM, Halford NG. Antisense expression of a sucrose non-fermenting-1-related protein kinase sequence in potato results in decreased expression of sucrose synthase in tubers and loss of sucrose-inducibility of sucrose synthase transcripts in leaves. Plant J. 1998;14:195–202. [Google Scholar]
- Rose M, Albig W, Entian KD. Glucose repression in Saccharomyces cerevisiae is directly associated with hexose phosphorylation by hexokinases PI and PII. Eur J Biochem. 1991;199:511–8. doi: 10.1111/j.1432-1033.1991.tb16149.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sano H, Seo S, Koizumi N, Niki T, Iwamura H, Ohashi Y. Regulation by cytokinins of endogenous levels of jasmonic acid and salicylic acids in mechanically wounded tobacco plants. Plant Cell Physiol. 1996;37:762–769. [Google Scholar]
- Sano H, Seo S, Orudgev E, Youssefian S, Ishizuka K, Ohashi Y. Expression of the gene for a small GTP binding protein in transgenic tobacco elevates endogenous cytokinin levels, abnormally induces salicylic acid in response to wounding, and increases resistance to tobacco mosaic virus infection. Proc Natl Acad Sci USA. 1994;91:10556–10560. doi: 10.1073/pnas.91.22.10556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H, Youssefian S. Light and nutritional regulation of transcripts encoding a wheat protein kinase homolog is mediated by cytokinins. Proc Natl Acad Sci USA. 1994;91:2582–2586. doi: 10.1073/pnas.91.7.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. Metabolic repression of transcription in higher plants. Plant Cell. 1990;2:1027–1038. doi: 10.1105/tpc.2.10.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons C, Litts J, Huang N, Rodriguez R. Structure of a rice β-glucanase gene regulated by ethylene, cytokinin, wounding salicylic acid and fungal elicitors. Plant Mol Biol. 1992;18:33–45. doi: 10.1007/BF00018454. [DOI] [PubMed] [Google Scholar]
- Stone J, Walker J. Plant protein kinase families and signal transduction. Plant Physiol. 1995;108:451–457. doi: 10.1104/pp.108.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W, Howell S. The effects of cytokinin and light on hypocotyl elongation in Arabidopsis seedlings are independent and additive. Plant Physiol. 1995;108:1423–1430. doi: 10.1104/pp.108.4.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden C, Donaghy PG, Halford NG, Hardie DG. Two SNF1-related protein kinases from spinach leaf phosphorylate and inactivate 3-hydroxy-3-methylglutaryl-coenzyme A reductase, nitrate reductase, and sucrose phosphate synthase in vitro. Plant Physiol. 1999;120:257–274. doi: 10.1104/pp.120.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwoerd T, Dekker B, Hoekema A. A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 1989;17:2362. doi: 10.1093/nar/17.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim A, Deng X-W. Light control of seedling development. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:215–243. doi: 10.1146/annurev.arplant.47.1.215. [DOI] [PubMed] [Google Scholar]
- Wenzler H, Mignery G, Fisher L, Park W. Sucrose-regulated expression of a chimeric potato tuber gene in leaves of transgenic tobacco plants. Plant Mol Biol. 1989;13:347–354. doi: 10.1007/BF00015546. [DOI] [PubMed] [Google Scholar]
- Yu X, Sukumaran S, Marton L. Differential expression of the Arabidopsis Nia1 and Nia2 genes. Plant Physiol. 1998;116:1091–1096. doi: 10.1104/pp.116.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]